Abstract
Purpose of review
Glucokinase regulator (GCKR) encodes glucokinase regulatory protein (GKRP), a hepatocyte-specific inhibitor of the glucose-metabolizing enzyme glucokinase (GCK). Genome-wide association studies have identified a common coding variant within GCKR associated with multiple metabolic traits. This review focuses on recent insights into the critical role of GKRP in hepatic glucose metabolism that have stemmed from the study of human genetics. This knowledge has improved our understanding of glucose and lipid physiology and informed the development of targeted molecular therapeutics for diabetes.
Recent findings
Rare GCKR variants have effects on GKRP expression, localization, and activity. These variants are collectively associated with hypertriglyceridaemia but are not causal. Crystal structures of GKRP and the GCK–GKRP complex have been solved, providing greater insight into the molecular interactions between these proteins. Finally, small molecules have been identified that directly bind GKRP and reduce blood glucose levels in rodent models of diabetes.
Summary
GCKR variants across the allelic spectrum have effects on glucose and lipid homeostasis. Functional analysis has highlighted numerous molecular mechanisms for GKRP dysfunction. Hepatocyte-specific GCK activation via small molecule GKRP inhibition may be a new avenue for type 2 diabetes treatment, particularly considering evidence indicating GKRP loss-of-function alone does not cause hypertriglyceridaemia.
Keywords: diabetes therapy, glucokinase regulator, glucokinase regulatory protein, glucose homeostasis, hypertriglyceridaemia
INTRODUCTION
Glucokinase regulator (GCKR) and its cognate gene product glucokinase regulatory protein (GKRP) have been associated, both biologically and genetically, with several key metabolic pathways. In the liver, GKRP forms an inhibitory complex with glucokinase (GCK), the enzyme responsible for regulating the uptake and storage of dietary glucose [1,2]. Mechanistic studies have shown that disruption and reformation of this complex occurs in response to direct binding of glucose to GCK, and fructose 1-phosphate (F1P) (a by-product of dietary fructose metabolism) and fructose 6-phosphate (F6P, which accumulates during gluconeogenesis and glycogenolysis) to GKRP [3-5]. In this way, the GCK–GKRP complex acts as a metabolic ‘switch’ capable of initiating energy storage and release pathways in response to periods of feeding and fasting. Genome-wide association studies (GWAS) have identified multiple associations with the common coding variant p.P446L in GKRP, including type 2 diabetes (T2D) and an inverse modulation of fasting plasma glucose and triglyceride levels [6-8]. This variant affects GKRP function via a range of molecular mechanisms that ultimately result in GCK activation under conditions of normoglycaemia or hypoglycaemia [9,10]. As we move closer to an era of personalized medicine, our ability to deliver targeted and efficacious treatments for diabetes and other metabolic diseases will depend on combined insights from human genetics, molecular and structural biology, and whole-animal physiology. This review will focus on recent studies that have extended our understanding of the physiological impact of GKRP dysregulation on glucose and lipid homeostasis, and the ways in which we may be able to manipulate GKRP to provide new treatments for T2D.
GENETIC VARIATION IN GLUCOKINASE REGULATOR HAS BEEN IMPLICATED IN A WIDE RANGE OF METABOLIC TRAITS
Human genetics has played an important role in our understanding of lipid phenotypes. GWAS have identified more than 150 loci associated with lipid levels and other lipid-related traits, and the advent of next-generation sequencing has allowed for the identification of additional contributing rare and low-frequency variants [7,11-18]. The results of these studies have offered new insight into the molecular mechanisms that underpin lipid metabolism and have opened up new avenues towards the development of novel therapeutics.
The first human genetic evidence of a role for GCKR in lipid metabolism was the association of a more than 400 kb genomic region, encompassing GCKR, with plasma triglyceride levels in Europeans via GWAS [7]. A subsequent combination of imputation and fine-mapping highlighted the nonsynonymous single-nucleotide polymorphism (SNP) rs1260326 [c.1403 C>T, p.P446L, minor allele frequency (MAF) 0.34] as the likely causal variant [6,19]. Interestingly, this SNP was associated with an inverse modulation of fasting glucose and triglyceride levels, a finding that has since been replicated in other populations [8,13,18,20-23]. To date, SNP rs1260326 (or the intronic proxy SNP rs780094) has been associated with more than 25 metabolic traits, including T2D risk, fasting insulin, total cholesterol, and nonalcoholic fatty liver disease, as well as circulating levels of numerous metabolites such as uric acid, C-reactive protein, creatinine, and albumin, indicating that it is a highly pleiotropic gene [14,24-39].
GLUCOKINASE REGULATORY PROTEIN HAS A CLEAR BIOLOGICAL ROLE IN GLUCOSE HOMEOSTASIS
The abovementioned insights are of particular interest in light of an already substantial body of evidence indicating a biological role for GKRP in glucose homeostasis. As its name suggests, GKRP was first identified as a protein that bound GCK and inhibited its activity in rodent hepatocytes [2]. GCK is a key regulator of glucose disposal and storage in both liver and pancreatic beta-cells, and responds to increases in circulating glucose concentration by initiating a signalling cascade that results in insulin secretion from the beta-cell and subsequent hepatic glucose uptake and storage [1]. The vast majority of human GKRP is expressed in the liver, suggesting that its primary effect on GCK activity occurs in this organ [9]. It is, however, also expressed at low levels in other tissues, although it does not appear to be appreciably expressed in beta-cells where GCK is clearly in excess [9,40].
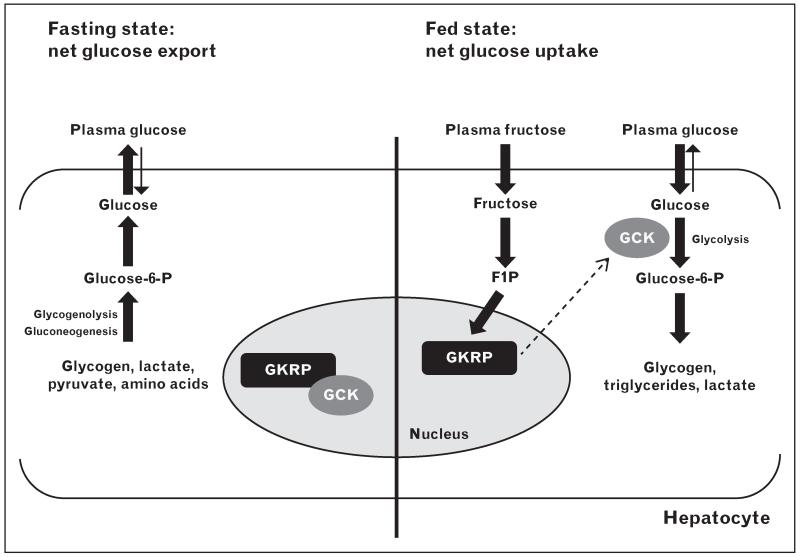
The liver is responsible for the clearance and disposal of approximately 25–35% of dietary glucose, and responds to periods of hypoglycaemia by initiating gluconeogenic and glycogenolytic pathways designed to maintain blood glucose concentrations at homeostatic levels [41-43]. Formation of the GCK–GKRP inhibitory complex is disrupted by glucose, resulting in translocation of GCK from the hepatocyte nucleus to the cytoplasm where it can assist in glucose clearance [44-48]. A similar effect is also achieved in the presence of the phosphate ester F1P, an intermediary product in the conversion of dietary fructose [2-4]. Conversely, F6P, a by-product of gluconeogenesis and glycogenolysis, enhances the interaction between GCK and GKRP, thus promoting nuclear retention and inactivation of GCK during periods of hypoglycaemia [2,3]. This shuttling mechanism effectively creates a nuclear reserve of GCK capable of mobilizing rapidly in response to fluctuations in intrahepatic glucose levels (Fig. 1). Interestingly, GKRP also acts as a post-translational stabilizer of cellular GCK, as Gckr−/− mice display WT-like Gck transcript levels but reduced GCK protein expression and activity, as well as lower hepatic glycogen concentrations and a reduced ability to respond to an acute glucose load [47,49].
FIGURE 1.
Model of GKRP regulation of hepatic glucose metabolism. In the fasting state (left), GCK is inhibited by GKRP and sequestered in the nucleus. The hepatocyte is active in producing glucose via glycogenolysis and gluconeogenesis, and exports glucose to the circulation for use by peripheral tissues. In the fed state (right), GCK is released from GKRP inhibition by glucose (binding to GCK) and F1P (binding to GKRP). Glucose phosphorylation leads to enhanced glycolytic flux and glucose disposal and storage. F1P, fructose 1-phosphate; GCK, glucokinase; GKRP, glucokinase regulatory protein.
MOLECULAR MECHANISMS AND PHENOTYPIC IMPACT OF NATURALLY OCCURRING GLUCOKINASE REGULATOR MISSENSE VARIANTS
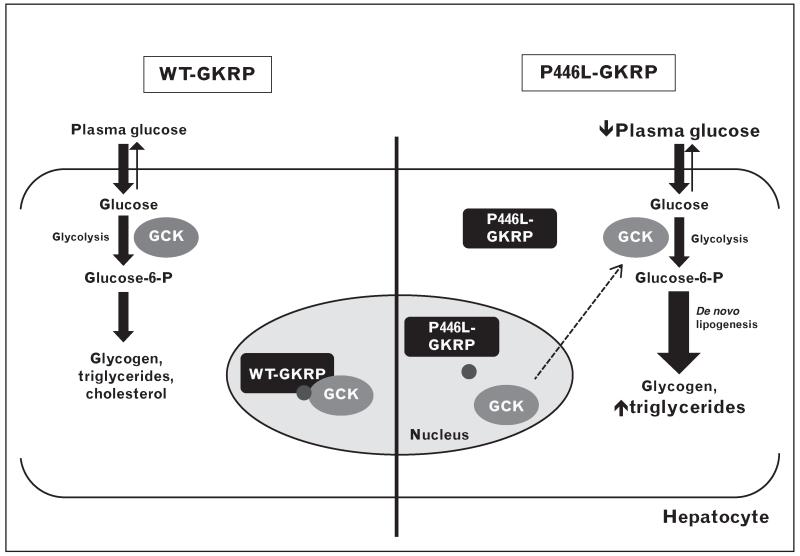
Functional analysis of the p.P446L GKRP variant provided us with the first mechanistic insights into the ways in which natural variation in GCKR may influence its cellular behaviour. A combination of in vitro and cell-based assays has shown that this variant has a reduced ability to sequester and inhibit GCK and a blunted response to F6P, both of which favour the generation of free and active cytoplasmic GCK [9,10]. Detailed biophysical experiments recently published by Zelent et al. [50■] suggest that the P > L substitution at this residue may have an overall effect on the structure of GKRP, resulting in destabilization of the GCK binding interface and altered phosphate ester binding. This explains the inverse correlation between fasting plasma glucose and triglycerides for this variant, as increased hepatic GCK activity would result in lower blood glucose levels and maintenance of energy-storing triglyceride and glycogen synthesis pathways under conditions of normoglycaemia or hypoglycaemia (Fig. 2) [51].
FIGURE 2.
Model of the effects of P446L–GKRP on hepatic glucose metabolism. Normally (left), GCK is inhibited by wild-type (WT) GKRP and sequestered in the nucleus in the fasting state. F6P (small circle) enhances complex formation. GCK is released from WT-GKRP in response to glucose and moves to the cytoplasm where it initiates glucose storage pathways. The p.P446L variant (right) creates a GKRP protein with increased cytoplasmic localization and reduced affinity for GCK and F6P, resulting in decreased plasma glucose levels and maintenance of glycolysis and de novo lipogenesis. F6P, fructose 6-phosphate; GCK, glucokinase; GKRP, glucokinase regulatory protein.
Studies by both Johansen et al. and Rees et al. have recently extended the catalogue of naturally occurring GCKR variants, and provided further insight into the mechanistic basis to GKRP dysfunction across the allelic spectrum [14,52]. In one of the first studies to identify rare (MAF <0.01) variants of (potentially) large individual effect in a gene already implicated in a particular trait via GWAS (in this case, plasma lipid levels), Johansen et al. [14] performed targeted exome sequencing of GCKR in individuals with extreme lipid phenotypes. They identified an excess of rare GCKR variants in hypertriglyceridaemic cases (defined as having fasting plasma triglyceride levels >95th percentile) relative to control individuals with normal lipid levels. Rees et al. [52] sequenced the exons of GCKR in 800 individuals from the ClinSeq cohort, who were recruited on the basis of an increased risk for coronary atherosclerosis, and identified a further 10 novel rare coding variants.
Functional studies have subsequently demonstrated a mutational mechanism for the vast majority of these variants, via a range of effects including cellular GKRP expression and localization, GCK interaction and inhibition, and phosphate ester binding [52,53■]. In one study, reclassification of variants based on empirical determination of pathogenicity increased the strength of the association with plasma triglycerides, total cholesterol, and LDL-Cholesterol, indicating that modulation of GKRP function has a demonstrable effect on clinical phenotype [52]. Interestingly, these empirical findings were repeatedly underpredicted by several widely used in silico prediction algorithms, emphasizing the need for robust analytical pipelines in the correct functional classification of novel variants [14,52,53■].
GLUCOKINASE REGULATORY PROTEIN AS A TARGET FOR DIABETES THERAPY
Although rare loss-of-function GCKR variants are associated collectively with hypertriglyceridaemia, extended family studies have demonstrated that they generally do not co-segregate with triglyceride levels [53■]. This most likely reflects the complex heritability of lipid traits, to which rare functional GCKR alleles are contributory rather than deterministic, and the influence of additional genetic and environmental factors on the penetrance of lipid phenotypes [13,14,54]. It is also encouraging news in light of a recent report describing antidiabetic effects for two small molecule GKRP inhibitors in rodents [55■■].
Lloyd et al. [55■■] used a cell-free high-throughput screening approach to identify a lead molecule (AMG-1694) that activated hepatic GCK via direct binding to GKRP, and found it to be a robust nuclear-to-cytoplasmic translocator of GCK in Wistar and Zucker diabetic fatty (ZDF) rats and primary rat hepatocytes. The drug had a specific effect on blood glucose levels – without affecting insulin or triglycerides – over a 24-h period in diabetic (ZDF) rats and did not alter blood glucose levels in normoglycaemic (Wistar) rats. Further optimization of AMG-1694 resulted in a drug that displayed greater efficacy in mice (AMG-3969), with similar translocatory effects in diet-induced obesity (DIO), ob/ob, db/db and normoglycaemic C56BL/6 mice. Once again, GCK translocation was matched by a corresponding decrease in blood glucose levels only in diabetic (DIO, ob/ob and db/db) mice [55■■,56]. The precise mechanism of action remains uncertain; however, the blood glucose-lowering effects appear to be the result of increased hepatic carbohydrate oxidation [55■■].
These results are of interest due to the potential for reduced risk of hypoglycaemia in humans, which has been a hallmark of clinical trials of glucokinase activators (GKAs) over the last 10 years. GKAs are a class of small molecules that increase the affinity of GCK for glucose by directly binding a pocket distal to its active site, thus lowering the set point for glucose-stimulated insulin secretion in the beta-cell [57,58]. In the liver, GKA binding causes GCK to dissociate from its inhibitory complex with GKRP [59]. A number of adverse side-effects to long-term GKA treatment have been reported, including hypoglycaemic episodes (due to improper insulin secretion), decreased efficacy over time, and increased hepatic triglycerides, the latter most likely due to persistent GCK activity in the liver [57,60-63].
Data suggest that increased hepatic GCK activity due to loss of GKRP function has negative long-term effects on whole-animal glucose and lipid homeostasis, particularly in the context of high fat and high sugar diets [47,61]. In humans, the p.P446L variant is associated with a modest decrease in plasma glucose but a proportionately larger increase in plasma triglycerides (8.76 mg/dl per allele in [13]), indicating that a satisfactory reduction in blood glucose levels via hepatic GCK activation may be outweighed by a correspondingly larger increase in triglyceride synthesis and storage [6,7,19,23]. The same variant is also associated with increased 2-h glucose – most likely reflecting a diminution of the nuclear GCK pool capable of being mobilized in response to a glucose challenge – making the long-term effects of chronic GCK activation on both glucose and triglycerides via GKRP inhibition difficult to predict [10,26]. As GKRP inhibitors do not increase the basal affinity of GCK for glucose, the effect on triglycerides at low glucose concentrations may be reduced relative to GKAs. Ultimately, these data emphasize that human clinical trials of AMG-1694 and AMG-3969 – or any other small molecule disruptors of the GCK–GKRP complex – will have to be closely monitored for adverse side-effects on both glucose and triglycerides, particularly in view of the fact that diabetes patients are already likely to have unfavourable lipid profiles that may be exacerbated by chronic hepatic GCK activation.
THE FUTURE FOR TARGETED DRUG DESIGN AND GLUCOKINASE REGULATORY PROTEIN
The study by Lloyd et al. [55■■] was the first to utilize crystal structure information to elucidate the precise mechanism of small molecule interference of GKRP. They crystallized the human GKRP protein bound to AMG-1694 and sorbitol 6-phosphate (an open-chain analogue of F6P), and used this information to model the effects of AMG-1694 binding on the GCK binding interface [55■■,64■■]. The very first two crystallographic studies of GKRP, however, were simultaneously published by Pautsch et al. and Beck and Miller 8 months previously. These two groups provided atomic-level information on human GKRP bound to F1P, and the human GCK–rat GKRP complex bound to F6P [65■■,66■■].
Cumulatively, these studies give a comprehensive picture of the structural changes that occur on the GKRP scaffold in response to GCK and phosphate ester binding. They demonstrate that GKRP is a trilobal protein, consisting of two sugar isomerase (SIS) homology domains capped by an alpha-helical ‘lid’. The GCK binding interface is positioned opposite the lid and is mediated by a small number of polar contacts and multiple hydrophobic interactions [64■■,65■■]. It is also distinct from the F1P–F6P binding site, which is located in a deeply buried cavity where the lid meets the SIS domains [65■■,66■■]. GCK binds to GKRP in a ‘super-open’ conformation in which its active site remains disordered and is released from GKRP in response to glucose via rearrangement into a β-hairpin structure [65■■]. Phosphate ester binding appears to modulate the strength of the interaction between the lid and the second SIS domain such that GCK binding is favoured or disfavoured depending on whether F6P or F1P is bound [65■■]. This detailed molecular information verifies several years’ worth of indirect evidence on the nature of the GKRP protein fold that had been deduced via a range of indirect methods, including homology modelling, mutagenesis, and biochemical and biophysical analyses [5,67-75].
Crystal structure information is of enormous significance in the context of current and future efforts to design targeted molecular inhibitors of GKRP, and has already been instrumental in structure-guided optimization of the initial screening hit AMG-1694 [56,76,77]. This molecule binds between the N-terminus of GKRP and the first SIS domain, revealing a novel binding pocket – distinct from the phosphate ester binding site – that clearly influences GKRP activity. This finding exemplifies the opportunities for novel, targeted therapeutics that can be inferred from structural details of previously unrecognized binding motifs, although the precise mechanism of GCK–GKRP disruption upon AMG-1694 binding remains unclear [55■■,78]. Interestingly, the p.P446 residue resides at the C-terminal end of a structural loop that interacts with GCK, and rare loss-of-function variants recently characterized by Johansen et al. and Rees et al. are distributed throughout the GKRP structure, the most severe of which are located proximal to the phosphate ester binding site [14,52,53■]. The extent to which small molecule inhibition of GKRP mimics the structural effects of these variants is yet to be elucidated, although they appear to have the same overall mechanistic effect via GCK–GKRP complex disruption.
CONCLUSION
Recent discoveries from fields as diverse as genetics, structural and cellular biology, and whole-animal physiology have underscored the importance of GKRP in hepatic glucose and triglyceride metabolism. The initial finding of multiple metabolic associations for the common coding variant p.P446L has now been extended to include collective associations for rare variants of larger individual effect with plasma lipids, and detailed functional analyses indicate that GKRP is highly sensitive to alteration in its activity via a range of molecular mechanisms [7,9,10,14,52,53■]. Rare GCKR variants, however, do not appear to be sufficient to cause hypertriglyceridaemia, giving hope for the suitability of small molecule GKRP inhibitors as an appropriate molecular therapy for hyperglycaemia and T2D [53■,55■■]. However, the long-term effects of increased hepatic GCK activity on glucose and triglycerides remain to be elucidated. In addition to recent atomic-resolution structural information about GKRP and the GCK–GKRP interaction, existing human genetic data should also prove useful in monitoring the potential side-effects of novel small molecule GKRP modulators [55■■,64■■-66■■]. Such a multifaceted approach should improve our understanding of the extent to which GKRP dysfunction is deterministic of lipid phenotypes, and the ways in which we therefore may be able to intervene and modulate its activity in a clinical setting.
KEY POINTS.
GCKR and its encoded protein GKRP are central to hepatic glucose and lipid metabolism via inhibition of GCK.
GCKR variants across the allelic spectrum have effects on glucose and lipid levels but are not deterministic for plasma triglycerides.
These variants act via a range of molecular mechanisms including protein expression, stability, localization, and GCK and phosphate ester binding.
GKRP may be an effective pharmacological target for hyperglycaemia; however, this may also negatively impact on plasma triglyceride levels.
Future efforts to design effective small molecular inhibitors of GKRP will be bolstered by resolution of the crystal structures of GKRP and the GCK–GKRP complex.
Acknowledgements
None.
Financial support and sponsorship
A.L.G. is a Wellcome Trust Fellow in Basic and Biomedical Research. The work in the Gloyn laboratory is funded by the Wellcome Trust (095101/Z/10/Z).
Footnotes
Conflicts of interest
None.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Gloyn AL, Odili S, Buettger C, et al. Glucokinase and the regulation of blood sugar. In: Matschinsky FM, Magnuson MA, editors. Glucokinase and glycaemic disease: from basics to novel therapeutics. Karger; Basel: 2004. pp. 92–109. [Google Scholar]
- 2.Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur J Biochem. 1989;179:179–184. doi: 10.1111/j.1432-1033.1989.tb14538.x. [DOI] [PubMed] [Google Scholar]
- 3.Detheux M, Vandercammen A, Van Schaftingen E. Effectors of the regulatory protein acting on liver glucokinase: a kinetic investigation. Eur J Biochem. 1991;200:553–561. doi: 10.1111/j.1432-1033.1991.tb16218.x. [DOI] [PubMed] [Google Scholar]
- 4.Vandercammen A, Detheux M, Van Schaftingen E. Binding of sorbitol 6-phosphate and of fructose 1-phosphate to the regulatory protein of liver glucokinase. Biochem J. 1992;286(Pt 1):253–256. doi: 10.1042/bj2860253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veiga-da-Cunha M, Van Schaftingen E. Identification of fructose 6-phosphate- and fructose 1-phosphate-binding residues in the regulatory protein of glucokinase. J Biol Chem. 2002;277:8466–8473. doi: 10.1074/jbc.M105984200. [DOI] [PubMed] [Google Scholar]
- 6.Orho-Melander M, Melander O, Guiducci C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 8.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beer NL, Tribble ND, McCulloch LJ, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rees MG, Wincovitch S, Schultz J, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange LA, Hu Y, Zhang H, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peloso GM, Auer PL, Bis JC, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56 000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaxillaire M, Cavalcanti-Proenca C, Dechaume A, et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57:2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam CH, Ma RC, So WY, et al. Interaction effect of genetic polymorphisms in glucokinase (GCK) and glucokinase regulatory protein (GCKR) on metabolic traits in healthy Chinese adults and adolescents. Diabetes. 2009;58:765–769. doi: 10.2337/db08-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparso T, Andersen G, Nielsen T, et al. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 22.Li N, van der Sijde MR, Bakker SJ, et al. Pleiotropic effects of lipid genes on plasma glucose, HbA1c, and HOMA-IR levels. Diabetes. 2014;63:3149–3158. doi: 10.2337/db13-1800. [DOI] [PubMed] [Google Scholar]
- 23.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamatani Y, Matsuda K, Okada Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 25.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kettunen J, Tukiainen T, Sarin AP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28 141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridker PM, Pare G, Parker A, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemaitre RN, Tanaka T, Tang W, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gieger C, Radhakrishnan A, Cvejic A, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiner AP, Beleza S, Franceschini N, et al. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am J Hum Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Seaghdha CM, Wu H, Yang Q, et al. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLoS Genet. 2013;9:e1003796. doi: 10.1371/journal.pgen.1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraja AT, Chasman DI, North KE, et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol Gen Metab. 2014;112:317–338. doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin SY, Petersen AK, Wahl S, et al. Interrogating causal pathways linking genetic variants, small molecule metabolites, and circulating lipids. Genome Med. 2014;6:25. doi: 10.1186/gm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahendran Y, Vangipurapu J, Cederberg H, et al. Association of ketone body levels with hyperglycemia and type 2 diabetes in 9398 Finnish men. Diabetes. 2013;62:3618–3626. doi: 10.2337/db12-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nica AC, Ongen H, Irminger JC, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrannini E, Bjorkman O, Reichard GA, Jr, et al. The disposal of an oral glucose load in healthy subjects. A quantitative study. Diabetes. 1985;34:580–588. doi: 10.2337/diab.34.6.580. [DOI] [PubMed] [Google Scholar]
- 42.Mari A, Wahren J, DeFronzo RA, Ferrannini E. Glucose absorption and production following oral glucose: comparison of compartmental and arteriovenous-difference methods. Metabolism. 1994;43:1419–1425. doi: 10.1016/0026-0495(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 43.Moore MC, Coate KC, Winnick JJ, et al. Regulation of hepatic glucose uptake and storage in vivo. Adv Nutr. 2012;3:286–294. doi: 10.3945/an.112.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyoda Y, Miwa I, Kamiya M, et al. Evidence for glucokinase translocation by glucose in rat hepatocytes. Biochem Biophys Res Commun. 1994;204:252–256. doi: 10.1006/bbrc.1994.2452. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda Y, Miwa I, Kamiya M, et al. Tissue and subcellular distribution of glucokinase in rat liver and their changes during fasting–refeeding. Histochem Cell Biol. 1995;103:31–38. doi: 10.1007/BF01464473. [DOI] [PubMed] [Google Scholar]
- 46.Brown KS, Kalinowski SS, Megill JR, et al. Glucokinase regulatory protein may interact with glucokinase in the hepatocyte nucleus. Diabetes. 1997;46:179–186. doi: 10.2337/diab.46.2.179. [DOI] [PubMed] [Google Scholar]
- 47.Farrelly D, Brown KS, Tieman A, et al. Mice mutant for glucokinase regulatory protein exhibit decreased liver glucokinase: a sequestration mechanism in metabolic regulation. Proc Natl Acad Sci USA. 1999;96:14511–14516. doi: 10.1073/pnas.96.25.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agius L, Peak M, Van Schaftingen E. The regulatory protein of glucokinase binds to the hepatocyte matrix, but, unlike glucokinase, does not translocate during substrate stimulation. Biochem J. 1995;309(Pt 3):711–713. doi: 10.1042/bj3090711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimsby J, Coffey JW, Dvorozniak MT, et al. Characterization of glucokinase regulatory protein-deficient mice. J Biol Chem. 2000;275:7826–7831. doi: 10.1074/jbc.275.11.7826. [DOI] [PubMed] [Google Scholar]
- 50■.Zelent B, Raimondo A, Barrett A, et al. Analysis of the co-operative interaction between the allosterically regulated proteins GK and GKRP using tryptophan fluorescence. Biochem J. 2014;459:551–564. doi: 10.1042/BJ20131363. [Detailed biophysical characterization via tryptophan fluorescence of the p.P446L GKRP variant and the way in which it alters GCK–GKRP complex formation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agius L, Peak M, Newgard CB, et al. Evidence for a role of glucose-induced translocation of glucokinase in the control of hepatic glycogen synthesis. J Biol Chem. 1996;271:30479–30486. doi: 10.1074/jbc.271.48.30479. [DOI] [PubMed] [Google Scholar]
- 52.Rees MG, Ng D, Ruppert S, et al. Correlation of rare coding variants in the gene encoding human glucokinase regulatory protein with phenotypic, cellular, and kinetic outcomes. J Clin Invest. 2012;122:205–217. doi: 10.1172/JCI46425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53■.Rees MG, Raimondo A, Wang J, et al. Inheritance of rare functional GCKR variants and their contribution to triglyceride levels in families. Hum Mol Genet. 2014;23:5570–5578. doi: 10.1093/hmg/ddu269. [This article demonstrates that rare loss-of-function GCKR variants do not co-segregate with elevated plasma triglyceride levels in families.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansen CT, Wang J, Lanktree MB, et al. An increased burden of common and rare lipid-associated risk alleles contributes to the phenotypic spectrum of hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2011;31:1916–1926. doi: 10.1161/ATVBAHA.111.226365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55■■.Lloyd DJ, St Jean DJ, Jr, Kurzeja RJ, et al. Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors. Nature. 2013;504:437–440. doi: 10.1038/nature12724. [The first published report of small molecule GKRP inhibitors that reduce blood glucose levels in rodent models of diabetes, without short-term side effects on insulin or lipids.] [DOI] [PubMed] [Google Scholar]
- 56.Ashton KS, Andrews KL, Bryan MC, et al. Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction. 1. Discovery of a novel tool compound for in vivo proof-of-concept. J Med Chem. 2014;57:309–324. doi: 10.1021/jm4016735. [DOI] [PubMed] [Google Scholar]
- 57.Matschinsky FM. Assessing the potential of glucokinase activators in diabetes therapy. Nat Rev Drug Discov. 2009;8:399–416. doi: 10.1038/nrd2850. [DOI] [PubMed] [Google Scholar]
- 58.Johnson D, Shepherd RM, Gill D, et al. Glucose-dependent modulation of insulin secretion and intracellular calcium ions by GKA50, a glucokinase activator. Diabetes. 2007;56:1694–1702. doi: 10.2337/db07-0026. [DOI] [PubMed] [Google Scholar]
- 59.Grimsby J, Sarabu R, Corbett WL, et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 60.Meininger GE, Scott R, Alba M, et al. Effects of MK-0941, a novel glucokinase activator, on glycemic control in insulin-treated patients with type 2 diabetes. Diabetes Care. 2011;34:2560–2566. doi: 10.2337/dc11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Ceuninck F, Kargar C, Ilic C, et al. Small molecule glucokinase activators disturb lipid homeostasis and induce fatty liver in rodents: a warning for therapeutic applications in humans. Br J Pharmacol. 2012;168:339–353. doi: 10.1111/j.1476-5381.2012.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nissim I, Horyn O, Daikhin Y, et al. Effects of a glucokinase activator on hepatic intermediary metabolism: study with (13)C-isotopomer-based metabolomics. Biochem J. 2012;444:537–551. doi: 10.1042/BJ20120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rees MG, Gloyn AL. Small molecular glucokinase activators: has another new antidiabetic therapeutic lost favour? Br J Pharmacol. 2013;168:335–338. doi: 10.1111/j.1476-5381.2012.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64■■.Choi JM, Seo MH, Kyeong HH, et al. Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proc Natl Acad Sci USA. 2013;110:10171–10176. doi: 10.1073/pnas.1300457110. [This article provides atomic-level resolution of the Xenopus GCK–GKRP complex bound to fructose 6-phosphate.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65■■.Beck T, Miller BG. Structural basis for regulation of human glucokinase by glucokinase regulatory protein. Biochemistry. 2013;52:6232–6239. doi: 10.1021/bi400838t. [This article provides atomic-level resolution of the mammalian GCK–GKRP complex bound to fructose 6-phosphate.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66■■.Pautsch A, Stadler N, Lohle A, et al. Crystal structure of glucokinase regulatory protein. Biochemistry. 2013;52:3523–3531. doi: 10.1021/bi4000782. [This article provides atomic-level resolution of the human GKRP protein bound to fructose 1-phosphate.] [DOI] [PubMed] [Google Scholar]
- 67.Veiga-da-Cunha M, Courtois S, Michel A, et al. Amino acid conservation in animal glucokinases. Identification of residues implicated in the interaction with the regulatory protein. J Biol Chem. 1996;271:6292–6297. doi: 10.1074/jbc.271.11.6292. [DOI] [PubMed] [Google Scholar]
- 68.Brocklehurst KJ, Davies RA, Agius L. Differences in regulatory properties between human and rat glucokinase regulatory protein. Biochem J. 2004;378:693–697. doi: 10.1042/BJ20031414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veiga-da-Cunha M, Sokolova T, Opperdoes F, Van Schaftingen E. Evolution of vertebrate glucokinase regulatory protein from a bacterial N-acetylmuramate 6-phosphate etherase. Biochem J. 2009;423:323–332. doi: 10.1042/BJ20090986. [DOI] [PubMed] [Google Scholar]
- 70.Baltrusch S, Francini F, Lenzen S, Tiedge M. Interaction of glucokinase with the liver regulatory protein is conferred by leucine-asparagine motifs of the enzyme. Diabetes. 2005;54:2829–2837. doi: 10.2337/diabetes.54.10.2829. [DOI] [PubMed] [Google Scholar]
- 71.Baltrusch S, Lenzen S, Okar DA, et al. Characterization of glucokinase-binding protein epitopes by a phage-displayed peptide library. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel interaction partner. J Biol Chem. 2001;276:43915–43923. doi: 10.1074/jbc.M105470200. [DOI] [PubMed] [Google Scholar]
- 72.Anderka O, Boyken J, Aschenbach U, et al. Biophysical characterization of the interaction between hepatic glucokinase and its regulatory protein: impact of physiological and pharmacological effectors. J Biol Chem. 2008;283:31333–31340. doi: 10.1074/jbc.M805434200. [DOI] [PubMed] [Google Scholar]
- 73.Vandercammen A, Van Schaftingen E. Competitive inhibition of liver glucokinase by its regulatory protein. Eur J Biochem. 1991;200:545–551. doi: 10.1111/j.1432-1033.1991.tb16217.x. [DOI] [PubMed] [Google Scholar]
- 74.Bourbonais FJ, Chen J, Huang C, et al. Modulation of glucokinase by glucose, small-molecule activator and glucokinase regulatory protein: steady-state kinetic and cell-based analysis. Biochem J. 2012;441:881–887. doi: 10.1042/BJ20110721. [DOI] [PubMed] [Google Scholar]
- 75.Veiga-da-Cunha M, Xu LZ, Lee YH, et al. Effect of mutations on the sensitivity of human beta-cell glucokinase to liver regulatory protein. Diabetologia. 1996;39:1173–1179. doi: 10.1007/BF02658503. [DOI] [PubMed] [Google Scholar]
- 76.Nishimura N, Norman MH, Liu L, et al. Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction. 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N′-arylpiperazine series. J Med Chem. 2014;57:3094–3116. doi: 10.1021/jm5000497. [DOI] [PubMed] [Google Scholar]
- 77.St Jean DJ, Jr, Ashton KS, Bartberger MD, et al. Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction. 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles. J Med Chem. 2014;57:325–338. doi: 10.1021/jm4016747. [DOI] [PubMed] [Google Scholar]
- 78.Hong FT, Norman MH, Ashton KS, et al. Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction. 4. Exploration of a novel binding pocket. J Med Chem. 2014;57:5949–5964. doi: 10.1021/jm5001979. [DOI] [PubMed] [Google Scholar]