Abstract
Background. Single nucleotide polymorphisms (SNPs) in genes encoding microRNAs may play important role in the development of gastric cancer. It has been reported that common SNPs rs2910164 in miR-146a and rs11614913 in miR-196a2 are associated with susceptibility to gastric cancer. The published results remain inconclusive or even controversial. A meta-analysis was conducted to quantitatively assess potential association between the two common SNPs and gastric cancer risk. Methods. A comprehensive literature search was performed in multiple internet-based electronic databases. Data from 12 eligible studies were extracted to estimate pooled odds ratios (ORs) and 95% confidence intervals (95% CI). Results. C allele of rs2910164 is associated with reduced gastric cancer risk in heterozygote model and dominant model whereas rs11614913 indicates no significant association. Subgroup analysis demonstrates that C allele of rs2910164 and rs11614913 may decrease susceptibility to diffuse type gastric cancer in dominant model and recessive model, respectively, while rs11614913 increased intestinal type gastric cancer in dominant model. Conclusion. SNPs rs2910164 and rs11614913 might have effect on gastric cancer risk in certain genetic models and specific types of cancer. Further well-designed studies should be considered to validate the potential effect.
1. Introduction
Gastric cancer is among the leading causes of cancer-related death worldwide. It is estimated that 989,600 new gastric cancer cases were diagnosed in 2008 and caused 738,000 deaths in a single year. Gastric cancer accounts for 8% of total cancer cases and 10% cancer-related death [1]. Despite decreasing incidence of gastric cancer in developed countries, gastric cancer remains a major health problem globally, especially in Eastern Asia, Eastern Europe, and South America, which may be attributed to particular dietary pattern, high prevalence of Helicobacter pylori infection, and limited availability to proper food storage [2–4]. The mechanism of gastric carcinogenesis remains elusive. Epidemiological studies have shed light on risk factors of gastric cancer including lifestyle factors, environmental carcinogens, and, importantly, Helicobacter pylori infection [5, 6]. However, these risk factors cannot fully explain the development of gastric cancer since only a minority of exposed population finally developed gastric cancer, indicating possible interplay between risk factors and personal background including genetic susceptibility [7].
In recent years, potential association between single nucleotide polymorphisms (SNPs) and risk of gastric cancer were reported [8]. Among the reported SNPs, correlation between SNPs located in genes encoding microRNAs (miRNAs) or their binding sites is of great interest [9, 10]. miRNAs are small noncoding, single-stranded RNA molecules composed of around 22 nucleotides. miRNAs bind to complementary sequences in 3′-untranslated regions of messenger RNAs and negatively regulate their stability or translational efficiency, therefore regulating posttranscriptional activity of genes [11–13]. Aberrant function or expression of miRNAs was reported to play important roles in gastric cancer. Since a single miRNA may have numerous targets, even a slight variation of a miRNA may lead to aberrance of a wide spectrum of gene expression, including many oncogenes and tumor-suppressor genes [7, 14]. SNPs in miRNA may also be involved in gastric cancer susceptibility through altering the expression or function of miRNAs, subsequently leading to aberrant expression of a set of genes [7, 15].
SNPs rs2910164 in miR-146a and rs11614913 in miR-196a2 have been reported as biomarkers of gastric cancer risk [16–27]. However, the results of these studies are controversial and inconclusive. Since the effects of SNPs in miRNAs on gastric cancer susceptibility may be slight, sample size of individual association study could be insufficient to detect minor modifications of gastric cancer risk. In this study, we performed a meta-analysis to systematically estimate the potential association between rs2910164/rs11614913 and susceptibility to gastric cancer with all available evidence.
2. Methods
2.1. Search Strategy
A systematic literature search was carried out using the combination of the following terms: “miR-146a,” “miR-196a2,” “miR-196a-2,” “rs2910164,” “rs11614913,” “gastric cancer,” “gastric carcinoma,” “gastric adenocarcinoma,” “stomach cancer,” “stomach carcinoma,” and “stomach adenocarcinoma” in multiple databases including PubMed, EMBASE, ISI Web of Knowledge, the Cochrane Library, ScienceDirect, Springer Link, Wiley Online Library, China National Knowledge Infrastructure (CNKI), Wanfang Database, and VIP Info database. Two investigators (Qing Ni and Anlai Ji) independently performed the database search. Publication language, date, and publication form (full-length article or abstract/correspondence) were not restricted. All of the search results were imported into Endnote X6 reference managing software and duplicate records were removed. The reference lists of potentially eligible studies were searched manually. The two investigators crosschecked the search results and reached consensus.
2.2. Literature Selection
We selected eligible studies based on the following criteria: (1) case-control study; (2) investigated associations between rs2910164 and/or rs11614913 and gastric cancer susceptibility; (3) provided sufficient data of allele and genotype frequencies of SNPs or required information could be calculated; (4) if serial studies on the same population were published, only the most recent study was included; (5) proper methodology design. Quality of methodology was evaluated by (1) comparable demographic characteristics between patients and control population; (2) proper diagnosis of gastric cancer; (3) appropriate methods and quality control for genotype determination; (4) Hardy-Weinberg equilibrium that was reached in control group; (5) proper statistical methods that were used. Two independent investigators (Qing Ni and Anlai Ji) performed study selection and reached final consensus. The details of literature search and selection were shown in Figure 1 in standard PRISMA flow diagram style.
Figure 1.
Flow diagram of study search and selection.
2.3. Data Extraction
Data for meta-analysis were extracted from eligible studies by two independent investigators (Qing Ni and Anlai Ji). Authors of study, publish year, origin country, ethnicity of studied population, study design (hospital based, HB, or population based, PB), genotyping method, and allele/genotype frequencies were collected. Two investigators crosschecked the results of data abstraction and discussed them to reach mutual agreement by discussion.
2.4. Statistical Analysis
Quantitative data synthesis was performed by Review Manager 5.2.11 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration). Statistical heterogeneity among studies was estimated by χ 2-based Q test. A P value less than 0.1 for Q test indicated the existence of significant statistical heterogeneity [28]. If no significant heterogeneity was detected, the pooled odds ratios (ORs) with corresponding 95% confidence interval (95% CI) were estimated by the Mantel-Haenszel fixed-effects model [29]. Otherwise, the DerSimonian-Liard random-effects model was used to calculate pooled ORs [30, 31]. The amount of heterogeneity was measured by the I 2 statistic [32]. I 2 value less than 25%, between 25% and 50%, greater than 50% indicated low, moderate, and high heterogeneity, respectively. The statistical significance of pooled ORs was determined by Z test. A P value for Z test less than 0.05 was considered statistically significant. Forest plots were provided generated to summarize the results of meta-analysis. The strength of associations between SNPs and the risk of gastric cancer were determined under the following genetic models: (1) allele frequency (C versus G for rs2910164 and C versus T for rs11614913); (2) heterozygous model (GC versus GG for rs2910164 and TC versus TT for rs11614913); (3) homozygous model (CC versus GG for rs2910164 and CC versus TT for rs11614913); (4) dominant model (GC + CC versus GG for rs2910164 and TC + CC versus TT for rs11614913); (5) recessive model (CC versus GG + GC for rs2910164 and CC versus TT + TC for rs11614913).
Sensitivity analysis was conducted by excluding one individual study in turn to observe whether the significance of heterogeneity test and pooled ORs changed. Subgroup analyses were performed by stratified analysis according to Lauren's histology classification of gastric cancer (intestinal or diffuse), cardiac or noncardiac gastric cancer, and lymph node status (N0 or N1) when sufficient data were available.
2.5. Publication Bias
Publication bias of the included studies was assessed by funnel plots generated by Review Manager. Begg's test and Egger's test were performed using STATA 11 software. A symmetrical plot suggested no obvious publication bias.
3. Results
3.1. Characteristics of Included Studies
A total of 582 papers were retrieved after electronic search and duplicate removal. As shown in Figure 1, after initial screening and review of full-text, 12 studies were included in this meta-analysis [16–27]. Characteristics of included studies were presented in Tables 1 and 2. For rs2910164 in miR-146a, 9 studies consisting of 4468 cases and 6844 controls were analyzed [16, 17, 19–22, 24, 25, 27]. For rs11614913, 9 studies involving 3992 cases and 5418 controls were included [16–20, 23, 25–27]. The genotyping methods in these studies include polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), polymerase chain reaction with confronting two-pair primers (PCR-CTPP), and TaqMan probe-based genotyping.
Table 1.
Characteristics of included studies on rs2910164.
| Author | Year | Country | Ethnicity | Study design | SNP | Genotyping methods | HWE | Case genotype | Control genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | GG | GC | CC | ||||||||
| Ahn et al. [20] | 2013 | South Korea | Asian | HB | rs2910164 | PCR-RFLP | 0.362 | 71 | 231 | 159 | 62 | 221 | 164 |
| Dikeakos et al. [19] | 2014 | Greece | Caucasian | HB | rs2910164 | PCR-RFLP | 0.289 | 13 | 45 | 105 | 24 | 149 | 307 |
| Hishida et al. [21] | 2011 | Japan | Asian | HB | rs2910164 | PCR-CTPP | 0.738 | 82 | 271 | 230 | 229 | 775 | 633 |
| Kupcinskas et al. [25] | 2014 | Germany/ Lithuania/ Latvia |
Caucasian | HB | rs2910164 | TaqMan | 0.531 | 252 | 94 | 16 | 223 | 108 | 16 |
| Okubo et al. [17] | 2010 | Japan | Asian | HB | rs2910164 | PCR-RFLP | 0.278 | 73 | 243 | 236 | 121 | 322 | 254 |
| Parlayan et al. [16] | 2014 | Japan | Asian | HB | rs2910164 | TaqMan | 0.640 | 20 | 79 | 61 | 71 | 237 | 216 |
| Pu et al. [27] | 2014 | China | Asian | HB | rs2910164 | PCR-RFLP | 0.080 | 36 | 96 | 65 | 96 | 274 | 143 |
| Zeng et al. [22] | 2010 | China | Asian | HB | rs2910164 | PCR-RFLP | 0.122 | 62 | 153 | 89 | 53 | 132 | 119 |
| Zhou et al. [24] | 2012 | China | Asian | HB | rs2910164 | TaqMan | 0.544 | 248 | 380 | 122 | 236 | 424 | 175 |
| 0.929 | 330 | 442 | 164 | 315 | 527 | 218 | |||||||
SNP, single nucleotide polymorphism; HB, hospital based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-CTPP, polymerase chain reaction with confronting two-pair primers; HWE, P value for Hardy-Weinberg equilibrium test.
Table 2.
Characteristics of included studies on rs11614913.
| Author | Year | Country | Ethnicity | Study design | SNP | Genotyping methods | HWE | Case genotype | Control genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | ||||||||
| Ahn et al. [20] | 2013 | South Korea | Asian | HB | rs11614913 | PCR-RFLP | 0.322 | 119 | 242 | 100 | 128 | 232 | 87 |
| Dikeakos et al. [19] | 2014 | Greece | Caucasian | HB | rs11614913 | PCR-RFLP | 0.850 | 15 | 46 | 102 | 172 | 229 | 79 |
| Kupcinskas et al. [25] | 2014 | Germany/ Lithuania/ Latvia |
Caucasian | HB | rs11614913 | TaqMan | 0.161 | 35 | 184 | 144 | 46 | 145 | 159 |
| Okubo et al. [17] | 2010 | Japan | Asian | HB | rs11614913 | PCR-RFLP | 0.510 | 166 | 281 | 105 | 223 | 350 | 124 |
| Parlayan et al. [16] | 2014 | Japan | Asian | HB | rs11614913 | TaqMan | 0.410 | 44 | 72 | 44 | 146 | 270 | 108 |
| Peng et al. [18] | 2010 | China | Asian | HB | rs11614913 | PCR-RFLP | 0.936 | 43 | 94 | 76 | 50 | 107 | 56 |
| Pu et al. [27] | 2014 | China | Asian | HB | rs11614913 | PCR-RFLP | <0.01 | 25 | 95 | 39 | 86 | 324 | 101 |
| Wang et al. [66] | 2013 | China | Asian | HB | rs11614913 | TaqMan | 0.898 | 226 | 371 | 152 | 232 | 448 | 220 |
| 0.058 | 293 | 480 | 167 | 292 | 492 | 262 | |||||||
| Yang et al. [26] | 2013 | China | Asian | PB | rs11614913 | TaqMan | 0.100 | 21 | 109 | 102 | 42 | 136 | 72 |
SNP, single nucleotide polymorphism; HB, hospital based; PB, population based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; HWE, P value for Hardy-Weinberg equilibrium test.
3.2. Association between rs2910164 in miR-146a and Gastric Cancer Susceptibility
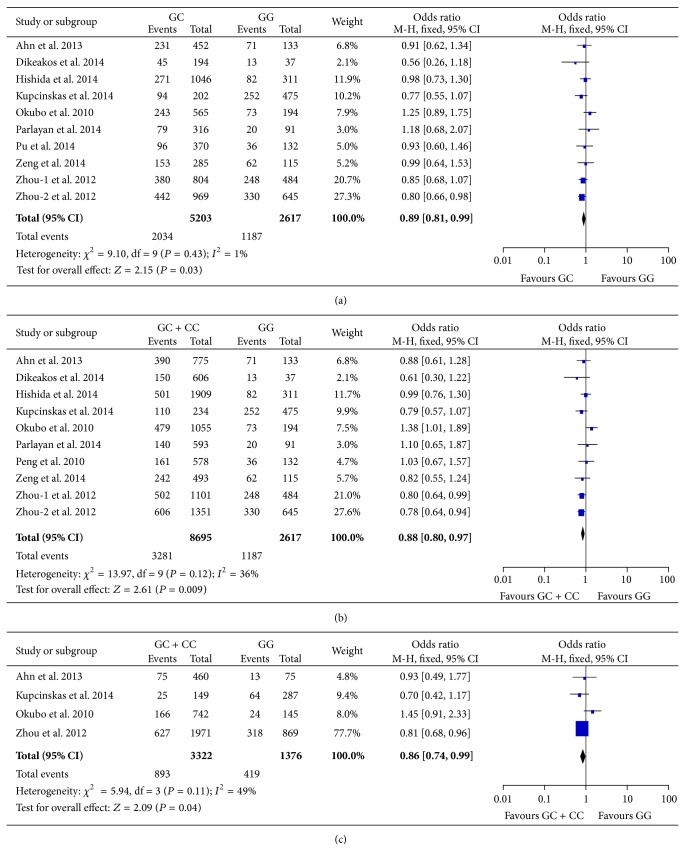
The association between rs2910164 and the risk of gastric cancer were analyzed based on data from 9 studies [16, 17, 19–22, 24, 25, 27]. The report from Zhou et al. [24] is comprised of two independent populations. In this meta-analysis, the two population groups were included separately. Significant heterogeneity was detected in allele frequency model, homozygote model, and recessive model and random-effects model was employed to calculate pooled ORs and 95% CIs in these comparisons. The results of the meta-analyses on rs2910164 were summarized in Table 3. Heterozygous C allele carrier showed decreased risk of gastric cancer compared with GG genotype (OR = 0.89, 95% CI 0.81–0.99, P = 0.03, Figure 2(a)). Similarly, in dominant model, GC and CC genotypes were associated with less susceptibility to gastric cancer compared with GG carriers (OR = 0.88, 95% CI 0.80–0.97, P = 0.009, Figure 2(b)). No significant association was demonstrated in allele frequency model, homozygote model, and recessive model. Interestingly, in sensitivity analysis, after removal of Okubo et al.'s study, statistical heterogeneity in allele frequency model, homozygote model, and recessive model all became nonsignificant and the pooled ORs showed reduced risk of gastric cancer with statistical significance. Therefore, the study from Okubo et al. may represent an outlier among the included studies. We next performed subgroup analyses stratified by Lauren's histology classification in dominant model and recessive model (Table 5). The results indicated that, in dominant model, GC and CC carriers had reduced risk of diffuse type gastric cancer (OR = 0.86, 95% CI = 0.74–0.99, P = 0.04, Figure 2(c)). No significant association was suggested in other models and intestinal type gastric cancer.
Table 3.
Summary of pooled ORs in meta-analyses of rs2910164.
| Genetic model | Pooled OR [95% CI] | P | P hetero |
|---|---|---|---|
| C versus G | 0.94 [0.85–1.04] | 0.21 | 0.003 |
| GC versus GG | 0.89 [0.81–0.99] | 0.03 | 0.43 |
| CC versus GG | 0.89 [0.72–1.08] | 0.23 | 0.009 |
| GC + CC versus GG | 0.88 [0.80–0.97] | 0.009 | 0.12 |
| CC versus GC + GG | 0.94 [0.81–1.08] | 0.38 | 0.008 |
OR, odds ratio; 95% CI, 95% confidence interval; P hetero, P value for heterogeneity test.
Figure 2.
Forest plots of meta-analyses on rs2910164 in miR-146a. (a) Meta-analysis comparing heterozygous GC with GG. (b) Meta-analysis under dominant model (GC + CC versus GG). (c) Subgroup analysis in diffuse type gastric cancer using dominant model (GC + CC versus GG). The blue squares and corresponding horizontal lines indicate odds ratio of individual study. The area of the squares reflects weight of indicated study. The black filled diamond represents pooled odds ratio and 95% confidence interval.
Table 5.
Summary of pooled ORs in subgroup analyses.
| SNP | Number of studies | Subgroup | Genetic model | Pooled OR [95% CI] | P | P hetero |
|---|---|---|---|---|---|---|
| rs2910164 | 4 | Intestinal type | GC + CC versus GG | 0.95 [0.72–1.25] | 0.7 | 0.04 |
| rs2910165 | 4 | Diffuse type | GC + CC versus GG | 0.86 [0.74–0.99] | 0.04 | 0.11 |
| rs2910164 | 3 | Intestinal type | CC versus GG + GC | 0.91 [0.76–1.11] | 0.36 | 0.33 |
| rs2910165 | 3 | Diffuse type | CC versus GG + GC | 0.88 [0.68–1.14] | 0.33 | 0.26 |
| rs11614913 | 3 | Cardiac lesion | CC versus TT + TC | 0.91 [0.51–1.64] | 0.76 | 0.04 |
| rs11614913 | 3 | Noncardiac lesion | CC versus TT + TC | 1.10 [0.63–1.89] | 0.74 | 0.0002 |
| rs11614913 | 3 | Lymph node negative | CC versus TT + TC | 0.89 [0.74–1.07] | 0.22 | 0.15 |
| rs11614913 | 3 | Lymph node positive | CC versus TT + TC | 1.54 [0.54–4.35] | 0.42 | <0.00001 |
| rs11614913 | 3 | Intestinal type | TC + CC versus TT | 1.27 [1.03–1.55] | 0.02 | 0.23 |
| rs11614913 | 3 | Diffuse type | TC + CC versus TT | 1.01 [0.78–1.32] | 0.92 | 0.55 |
| rs11614913 | 4 | Intestinal type | CC versus TT + TC | 0.91 [0.64–1.28] | 0.58 | 0.003 |
| rs11614913 | 4 | Diffuse type | CC versus TT + TC | 0.83 [0.71–0.97] | 0.02 | 0.4 |
OR, odds ratio; 95% CI, 95% confidence interval; P hetero, P value for heterogeneity test.
3.3. Association between rs11614913 in miR-196a2 and Gastric Cancer Susceptibility
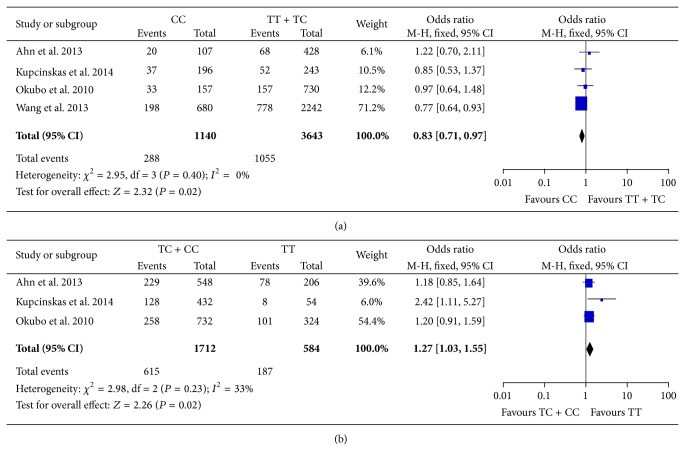
Potential association between rs11614913 and susceptibility to gastric cancer was evaluated using the data reported in 9 studies [16–20, 23, 25–27]. Wang et al.'s study [23] included two sets of independent cases and controls which were analyzed as separate populations in this meta-analysis. Heterogeneity test in all of the genetic models showed statistical significance and random-effects model was used. The results of the comparisons were listed in Table 4. To our surprise, rs11614913 in miR-196a2 demonstrated no significant association with gastric cancer risk in any genetic model tested. Although exclusion of the studies from Dikeakos et al., Kupcinskas et al., and Wang et al., respectively, diminished statistical heterogeneity in heterozygote model (TC versus CC), pooled ORs remained nonsignificant. In subgroup analyses (Table 5), rs11614913 was not associated with either cardiac or noncardiac lesions. In recessive model, this SNP also presented no association with lymph node metastasis. Interestingly, CC genotype may correlate with a decreased risk of diffuse gastric cancer in recessive model, as suggested by a pooled OR = 0.83 (95% CI 0.71–0.97, P = 0.02, Figure 3(a)). TC and CC genotypes may predispose carrier to intestinal type cancer in dominant model (OR = 1.27, 95% CI 1.03–1.55, P = 0.02, Figure 3(b)).
Table 4.
Summary of pooled ORs in meta-analyses of rs11614913.
| Genetic model | Pooled OR [95% CI] | P | P hetero |
|---|---|---|---|
| C versus T | 1.25 [0.97–1.60] | 0.09 | <0.00001 |
| TC versus TT | 1.09 [0.94–1.28] | 0.25 | 0.06 |
| CC versus TT | 1.52 [0.96–2.39] | 0.07 | <0.00001 |
| TC + CC versus TT | 1.26 [0.98–1.63] | 0.07 | <0.00001 |
| CC versus TC + TT | 1.36 [0.90–2.05] | 0.14 | <0.00001 |
OR, odds ratio; 95% CI, 95% confidence interval; P hetero, P value for heterogeneity test.
Figure 3.
Forest plots of meta-analyses on rs11614913 in miR-196a2. (a) Subgroup analysis in diffuse type of gastric cancer using recessive model (CC versus TT + TC). (b) Subgroup analysis in intestinal type of gastric cancer under dominant model (TC + CC versus TT). The blue squares and corresponding horizontal lines indicate odds ratio of individual study. The area of the squares reflects weight of indicated study. The black filled diamond represents pooled odds ratio and 95% confidence interval.
3.4. Publication Bias
The distribution of studies in funnel plots for analyses of rs2910164 was symmetrical, indicating no evidence for significant publication bias. Begg's test and Egger's test in meta-analyses demonstrating significant outcome also suggested no statistically significant publication bias in these comparisons (GC versus GG: Begg's test P = 0.474 and Egger's test P = 0.544; GC + CC versus GG: Begg's test P = 0.474 and Egger's test P = 0.481). However, the funnel plots for rs11614913 showed asymmetrical distribution. Publication bias may exist in studies on potential association between rs11614913 and gastric cancer susceptibility.
4. Discussion
Recent research on miRNAs has led to new insight into molecular mechanisms of gastric cancer development [10, 33, 34]. Variations in miRNAs may have profound impact on individual's susceptibility to gastric cancer through regulating a wide spectrum of oncogenes and tumor-suppressor genes. SNPs in miRNA-coding genes and their influence on gastric cancer risk have drawn much attention and related results may help broaden our horizon of gastric cancer. Better understanding of SNPs in miRNAs could improve current management of this detrimental disease by early detection of gastric cancer in high risk populations [35]. Functional SNPs rs2910164 in miR-146a and rs11614913 in miR-196a2 are reported to have association with gastric cancer susceptibility though the results are inconclusive or even controversial [16–27]. In this present study, we conducted a meta-analysis by quantitatively synthesizing available data from 12 published papers to demonstrate potential effects of these two common SNPs on gastric cancer susceptibility.
Located in the stem region opposite to mature miR-146a sequence, rs2910164 G > C polymorphism changed G : U pair to C : U mispair in the stem region of the precursor of miR-146a. C allele of rs2910164 resulted in decreased production of mature miR-146a and subsequently reduced the inhibition of multiple target genes in thyroid cells and hepatocellular carcinoma [36, 37]. In contrast, another two studies reported that C allele of rs2910164 elevated the expression level of miR-146a in breast cancer cells and cervical cancer tissues [38, 39]. The different regulation of this SNP on mature mir-146a may reflect complex gene background between different tissues. The influenced genes by miR-146a include IL-1 receptor-associated kinase 1 (IRAK1), TNF receptor-associated factor 6 (TRAF6), and papillary thyroid carcinoma 1 (PTC1) [36]. Interestingly, IRAK1 and TRAF6 are involved in the regulation of Toll-like receptor (TLR-4) pathway, which has important role in innate immunity against Helicobacter pylori [40, 41]. Hishida et al.'s indeed elucidated interaction between miR-146a rs2910164 and TLR4 + 3725 polymorphisms. Their study found that GG genotype of rs2910164 and TLR4 + 3725 C allele increased the risk of severe gastric atrophy in Helicobacter pylori-infected Japanese population [21]. miR-146a itself also has important role in cancer cell proliferation [37]. Association between rs2910164 and gastric cancer susceptibility has been reported [17, 20, 22, 24]; however other studies demonstrated no correlation of this SNP with gastric cancer risk [16, 19, 21, 25, 27].
In this meta-analysis, a total of 9 case-control studies were systematically summarized to generate a comprehensive evaluation of the association between rs2910164 in miR-146a and gastric cancer susceptibility. Our result indicated that rs2910164 GC genotype displayed reduced risk of gastric cancer compared with GG carriers. In dominant model, GC and CC genotype also showed decreased susceptibility to gastric cancer with statistical significance. This association was not found in other genetic models. However, the study from Okubo et al. [17] had a significant influence on pooled ORs. In sensitivity analyses, removal of this study not only diminished statistical heterogeneity among included studies but also changed pooled ORs towards significant reduced risk of gastric cancer in allele frequency model (C versus G), homozygote model (CC versus GG), and recessive model (CC versus GC + GG). This study may be the source of heterogeneity with potential bias and could cause a major distortion on the analysis of association between rs2910164 and gastric cancer risk. The possible effect of rs2910164 on gastric cancer susceptibility in allele frequency, homozygote model, and recessive model should not be ruled out. In subgroup analysis, our results demonstrated a significant reduction of diffuse type cancer in dominant model. This result is of considerable importance since diffused type gastric cancer is correlated with poorer prognosis [42]. C allele of rs2910164 may represent a protective factor against diffused type cancer and could serve as a reference in the screening among high risk population.
The other SNP investigated in this study is rs11614913 in miR-196a2. It was initially reported as a prognostic factor of non-small cell lung cancer [43]. The role of rs11614913 in esophageal cancer [44], hepatocellular carcinoma [45], and head and neck cancer [46] was also demonstrated. C allele of rs11614913 increased the expression of mature miR-196a2 in HCC tissues [47] and may cause aberrant expression of downstream genes, including several carcinogenesis-related genes such as homeobox (HOX) family, annexin A1 (ANXA1), and high mobility group AT-hook1 (HMGA1) [48]. Aberrance in HOX family transcription factors plays a significant role in gastric carcinogenesis and cancer stemness [49]. Acting as a mediator of apoptosis and an inhibitor of proliferation, ANXA1 participates in many pathological processes of human disease [50–52]. Deregulation of ANXA1 was found in both precancerous gastric lesions and gastric cancer [53, 54]. Similarly, HMGA1 was also reported to maintain cell proliferation in gastric cancer [55]. Therefore, SNP rs11614913 in miR-196a2 could cause multiple expression change of gastric cancer-related genes and contribute to susceptibility of gastric cancer.
Our meta-analysis systematically summarized data from 9 studies involving 10 study populations and to our surprise, rs11614913 in miR196a2 did not associate with gastric cancer risk in any genetic model tested. Nevertheless, in subgroup analyses, CC genotype of rs11614913 was found to reduce the risk of diffuse type gastric cancer in recessive model compared with TT and TC carriers. Interestingly, TC and CC carriers showed higher risk of intestinal type cancer in dominant model. These findings were not suggested in comparisons in tumor location (cardiac or noncardiac lesion) and lymph node status.
4.1. Comparison with Other Meta-Analyses
Before this meta-analysis, several papers from other authors have been published on the effects of rs2910164 and rs11614913 on cancer risk [56–68]. However, most of these studies did not distinguish type of cancers and investigated overall effect of the SNPs on all types of cancer [56–59, 62]. Some papers narrowed the aim of study to digestive cancers or gastrointestinal cancers but still included several cancers from different tissues [60, 61, 64–66]. A major concern is that different cancers from different tissue origins have distinct mechanism of pathogenesis. The clinical heterogeneity brought by this inherent difference could distort the result of meta-analysis. Only one study from Hua et al. summarized potential effect of these two common SNPs on gastric cancer by meta-analysis [7]. Their study found no association between rs2910164 and rs11614913 and gastric cancer susceptibility. Several additional studies have been reported after they published their paper, which is added to this updated meta-analysis. Therefore, this present study included all available evidence up to date and provided most comprehensive analysis regarding the effect of these two common SNPs on gastric cancer risk. Additionally, we also performed subgroup analyses to explore potential association between SNPs and cancer histological types, tumor locations, and lymph node status. Our results may expand our knowledge on rs2910164 and rs11614913 and their role in altering the risk of gastric cancer.
4.2. Limitations
Of note, this meta-analysis has its limitations and the results should be interpreted with caution. First, although we carried out the comparisons in similar backgrounds, significant heterogeneity still exists, especially in the analyses of rs11614913. Okubo et al.'s study [17] brought statistical heterogeneity with significance in the comparisons of rs2910164. The heterogeneity may distort the results of this meta-analysis and potential association between rs2910164 and susceptibility to gastric cancer should not be ruled out in genetic models that did not derive statistical significance. Second, due to limited number of studies, the subgroup analyses should be interpreted cautiously even if they indicated positive results. The role of these SNPs in different histological types should be further explored. Third, publication bias existed in studies on rs11614913, which implies the true effect of rs11614913 may not be fully discovered or reported.
In summary, despite the limitations, this meta-analysis suggests that rs2910164 in miR-146a and rs11614913 in miR-196a2 might be associated with reduced gastric cancer risk in certain genetic models and cancer histological types. More future studies with good methodology design are warranted.
Acknowledgment
This study was supported by Yangzhou Key Discipline/Laboratory of Medicine and Key Provincial Talents Program.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Qing Ni and Xinnong Liu conceived this study. Qing Ni, Anlai Ji, Junfeng Yin, and Xiangjun Wang collected data. Anlai Ji, Junfeng Yin, and Xiangjun Wang analyzed data. Qing Ni, Anlai Ji, and Xinnong Liu composed the paper. All of the authors have read the paper and gave final approval for publication. Qing Ni and Anlai Ji contributed equally to this study.
References
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin D. M. The global health burden of infection-associated cancers in the year 2002. International Journal of Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 3.Bertuccio P., Chatenoud L., Levi F., et al. Recent patterns in gastric cancer: a global overview. International Journal of Cancer. 2009;125(3):666–673. doi: 10.1002/ijc.24290. [DOI] [PubMed] [Google Scholar]
- 4.Yang L. Incidence and mortality of gastric cancer in China. World Journal of Gastroenterology. 2006;12(1):17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crew K. D., Neugut A. I. Epidemiology of upper gastrointestinal malignancies. Seminars in Oncology. 2004;31(4):450–464. doi: 10.1053/j.seminoncol.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Uemura N., Okamoto S., Yamamoto S., et al. Helicobacter pylori infection and the development of gastric cancer. The New England Journal of Medicine. 2001;345(11):784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 7.Hua H.-B., Yan T.-T., Sun Q.-M. miRNA polymorphisms and risk of gastric cancer in Asian population. World Journal of Gastroenterology. 2014;20(19):5700–5707. doi: 10.3748/wjg.v20.i19.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeki N., Ono H., Sakamoto H., Yoshida T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Science. 2013;104(1):1–8. doi: 10.1111/cas.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Link A., Kupcinskas J., Wex T., Malfertheiner P. Macro-role of microRNA in gastric cancer. Digestive Diseases. 2012;30(3):255–267. doi: 10.1159/000336919. [DOI] [PubMed] [Google Scholar]
- 10.Pan H.-W., Li S.-C., Tsai K.-W. MicroRNA dysregulation in gastric cancer. Current Pharmaceutical Design. 2013;19(7):1273–1284. doi: 10.2174/138161213804805621. [DOI] [PubMed] [Google Scholar]
- 11.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla G. C., Singh J., Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Molecular and Cellular Pharmacology. 2011;3(3):83–92. doi: 10.4255/mcpharmacol.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovat F., Valeri N., Croce C. M. MicroRNAs in the pathogenesis of cancer. Seminars in Oncology. 2011;38(6):724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Saunders M. A., Liang H., Li W.-H. Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parlayan C., Ikeda S., Sato N., Sawabe M., Muramatsu M., Arai T. Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pacific Journal of Cancer Prevention. 2014;15(5):2101–2107. doi: 10.7314/APJCP.2014.15.5.2101. [DOI] [PubMed] [Google Scholar]
- 17.Okubo M., Tahara T., Shibata T., et al. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15(6):524–531. doi: 10.1111/j.1523-5378.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 18.Peng S., Kuang Z., Sheng C., Zhang Y., Xu H., Cheng Q. Association of MicroRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Digestive Diseases and Sciences. 2010;55(8):2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 19.Dikeakos P., Theodoropoulos G., Rizos S., Tzanakis N., Zografos G., Gazouli M. Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Molecular Biology Reports. 2014;41(2):1075–1080. doi: 10.1007/s11033-013-2953-0. [DOI] [PubMed] [Google Scholar]
- 20.Ahn D. H., Rah H., Choi Y.-K., et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the korean population. Molecular Carcinogenesis. 2013;52(supplement 1):39–51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 21.Hishida A., Matsuo K., Goto Y., et al. Combined effect of miR-146a rs2910164 G/C polymorphism and toll-like receptor 4 +3725 G/C polymorphism on the risk of severe gastric atrophy in Japanese. Digestive Diseases and Sciences. 2011;56(4):1131–1137. doi: 10.1007/s10620-010-1376-1. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Y., Sun Q. M., Liu N. N., et al. Correlation between pre-miR-146a C/G polymorphism and gastric cancer risk in Chinese population. World Journal of Gastroenterology. 2010;16(28):3578–3583. doi: 10.3748/wjg.v16.i28.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Tao G., Wu D., et al. A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Molecular Carcinogenesis. 2013;52(supplement 1):87–95. doi: 10.1002/mc.22017. [DOI] [PubMed] [Google Scholar]
- 24.Zhou F., Zhu H., Luo D., et al. A functional polymorphism in pre-miR-146a is associated with susceptibility to gastric cancer in a chinese population. DNA and Cell Biology. 2012;31(7):1290–1295. doi: 10.1089/dna.2011.1596. [DOI] [PubMed] [Google Scholar]
- 25.Kupcinskas J., Wex T., Link A., et al. Gene polymorphisms of micrornas in helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0087467.e87467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Q., Jie Z., Wang J., et al. Association between miR-196a rs11614913 C/T polymorphisms and gastric cancer susceptibility. Academic Journal of Guangzhou Medical College. 2013;41 [Google Scholar]
- 27.Pu J.-Y., Dong W., Zhang L., Liang W.-B., Yang Y., Lv M.-L. No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iranian Journal of Basic Medical Sciences. 2014;17(2):128–133. [PMC free article] [PubMed] [Google Scholar]
- 28.Lau J., Ioannidis J. P. A., Schmid C. H. Quantitative synthesis in systematic reviews. Annals of Internal Medicine. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 29.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22:719–748. [PubMed] [Google Scholar]
- 30.DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 31.DerSimonian R., Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemporary Clinical Trials. 2007;28(2):105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 33.Ma J., Hong L., Chen Z., Nie Y., Fan D. Epigenetic regulation of microRNAs in gastric cancer. Digestive Diseases and Sciences. 2014;59(4):716–723. doi: 10.1007/s10620-013-2939-8. [DOI] [PubMed] [Google Scholar]
- 34.Wang F., Sun G. P., Zou Y. F., Hao J. Q., Zhong F., Ren W. J. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomarkers. 2012;11(6):259–267. doi: 10.3233/CBM-2012-00284. [DOI] [PubMed] [Google Scholar]
- 35.Tong F., Cao P., Yin Y., Xia S., Lai R., Liu S. MicroRNAs in gastric cancer: from benchtop to bedside. Digestive Diseases and Sciences. 2014;59(1):24–30. doi: 10.1007/s10620-013-2887-3. [DOI] [PubMed] [Google Scholar]
- 36.Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R., De La Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu T., Zhu Y., Wei Q.-K., et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29(11):2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 38.Shen J., Ambrosone C. B., DiCioccio R. A., Odunsi K., Lele S. B., Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29(10):1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 39.Yue C., Wang M., Ding B., et al. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecologic Oncology. 2011;122(1):33–37. doi: 10.1016/j.ygyno.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Ishihara S., Rumi M. A. K., Kadowaki Y., et al. Essential role of MD-2 in TLR4-dependent signaling during Helicobacter pylori-associated gastritis. The Journal of Immunology. 2004;173(2):1406–1416. doi: 10.4049/jimmunol.173.2.1406. [DOI] [PubMed] [Google Scholar]
- 41.Akira S., Takeda K. Toll-like receptor signalling. Nature Reviews Immunology. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 42.Qiu M.-Z., Cai M.-Y., Zhang D.-S., et al. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. Journal of Translational Medicine. 2013;11(1, article 58) doi: 10.1186/1479-5876-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Z., Chen J., Tian T., et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. The Journal of Clinical Investigation. 2008;118(7):2600–2608. doi: 10.1172/JC134934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Y., Wang K. K., Gu J., et al. Genetic variations in MicroRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prevention Research. 2008;1(6):460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi P., Dou T.-H., Geng L., et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Human Immunology. 2010;71(6):621–626. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Christensen C. L., Gjetting T., Poulsen T. T., Cramer F., Roth J. A., Poulsen H. S. Targeted cytosine deaminase-uracil phosphoribosyl transferase suicide gene therapy induces small cell lung cancer-specific cytotoxicity and tumor growth delay. Clinical Cancer Research. 2010;16(8):2308–2319. doi: 10.1158/1078-0432.CCR-09-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X. D., Li Z. G., Song X. X., Liu C. F. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology. 2010;42(7):669–673. doi: 10.3109/00313025.2010.522175. [DOI] [PubMed] [Google Scholar]
- 48.Chen C. J., Zhang Y., Zhang L., Weakley S. M., Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. Journal of Cellular and Molecular Medicine. 2011;15(1):14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhavan-Niaki H., Samadani A. A. Molecular insight in gastric cancer induction: an overview of cancer stemness genes. Cell Biochemistry and Biophysics. 2014;68:463–473. doi: 10.1007/s12013-013-9749-7. [DOI] [PubMed] [Google Scholar]
- 50.Perretti M., Dalli J. Exploiting the Annexin A1 pathway for the development of novel anti-inflammatory therapeutics. British Journal of Pharmacology. 2009;158(4):936–946. doi: 10.1111/j.1476-5381.2009.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damazo A. S., Flower R. J., Solito E., Oliani S. M. Annexin-A1 gene expression during liver development and post-translation modification after experimental endotoxemia. Inflammation Research. 2008;57(3):97–103. doi: 10.1007/s00011-007-7114-2. [DOI] [PubMed] [Google Scholar]
- 52.Maschler S., Gebeshuber C. A., Wiedemann E.-M., et al. Annexin A1 attenuates EMT and metastatic potential in breast cancer. EMBO Molecular Medicine. 2010;2(10):401–414. doi: 10.1002/emmm.201000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z.-Q., Li X.-J., Liu G.-T., Ping Y., Xia Y., Wen H. Identification of Annexin A1 protein expression in human gastric adenocarcinoma using proteomics and tissue microarray. World Journal of Gastroenterology. 2013;19(43):7795–7803. doi: 10.3748/wjg.v19.i43.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi A. F. T., Duarte M. C., Poltronieri A. B., et al. Deregulation of annexin-A1 and galectin-1 expression in precancerous gastric lesions: intestinal metaplasia and gastric ulcer. Mediators of Inflammation. 2014;2014:11. doi: 10.1155/2014/478138.478138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akaboshi S.-I., Watanabe S., Hino Y., et al. HMGA1 is induced by Wnt/β-catenin pathway and maintains cell proliferation in gastric cancer. The American Journal of Pathology. 2009;175(4):1675–1685. doi: 10.2353/ajpath.2009.090069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He B., Pan Y., Cho W. C., et al. The association between four genetic variants in MicroRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049032.e49032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma X. P., Zhang T., Peng B., Yu L., Jiang D. K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079584.e79584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y. Q., Gu L., Pan Y. Q., et al. Different effects of three polymorphisms in microRNAs on cancer risk in Asian population: evidence from published literatures. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0065123.e65123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin Z., Yan L., Cui Z., Li X., Ren Y., Zhou B. Effects of common polymorphisms rs2910164 in miR-146a and rs3746444 in miR-499 on cancer susceptibility: a meta-analysis. Molecular Biology Reports. 2013;40(4):3003–3013. doi: 10.1007/s11033-012-2372-7. [DOI] [PubMed] [Google Scholar]
- 60.Li Y.-J., Zhang Z.-Y., Mao Y.-Y., et al. A genetic variant in MIR-146a modifies digestive system cancer risk: a meta-analysis. Asian Pacific Journal of Cancer Prevention. 2014;15(1):145–150. doi: 10.7314/APJCP.2014.15.1.145. [DOI] [PubMed] [Google Scholar]
- 61.Guo J., Jin M., Zhang M., Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0030585.e30585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Bi J., Liu X., Li K., Di J., Wang B. Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Molecular Biology Reports. 2012;39(4):4571–4579. doi: 10.1007/s11033-011-1247-7. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H., Su Y.-L., Yu H., Qian B.-Y. Meta-analysis of the association between miR-196a-2 polymorphism and cancer susceptibility. Cancer Biology & Medicine. 2012;9(1):63–72. doi: 10.3969/j.issn.2095-3941.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X. H., Yang X. D., Ru G., et al. miR-146a gene polymorphism rs2910164 and the risk of digestive tumors: a meta-analysis of 21 case-control studies. Oncology Reports. 2014;31(1):472–479. doi: 10.3892/or.2013.2854. [DOI] [PubMed] [Google Scholar]
- 65.Wu D., Wang F., Dai W.-Q., et al. The miR-146a rs2910164 G > C polymorphism and susceptibility to digestive cancer in chinese. Asian Pacific Journal of Cancer Prevention. 2013;14(1):399–403. doi: 10.7314/APJCP.2013.14.1.399. [DOI] [PubMed] [Google Scholar]
- 66.Wang F., Sun G. P., Zou Y. F., Fan L. L., Song B. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and gastrointestinal cancer risk. Molecular Biology Reports. 2013;40(1):109–116. doi: 10.1007/s11033-012-2039-4. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Wu J., Zhang G., Cao Y., Jiang C., Ding Y. Associations of miR-499 and miR-34b/c polymorphisms with susceptibility to hepatocellular carcinoma: an evidence-based evaluation. Gastroenterology Research and Practice. 2013;2013:8. doi: 10.1155/2013/719202.719202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z., Cao Y., Jiang C., Yang G., Wu J., Ding Y. Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to hepatocellular carcinoma: a meta-analysis. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0040039.e40039 [DOI] [PMC free article] [PubMed] [Google Scholar]