Abstract
Cell fates change dynamically in response to various extracellular signals, including growth factors that stimulate differentiation and proliferation. The processes underlying cell-fate decisions are complex and often include large cell-to-cell variations, even within a clonal population in the same environment. To understand the origins of these cell-to-cell variations, we must detect the internal dynamics of single cells that reflect their changing chemical milieu. In this study, we used the Raman spectra of single cells to trace their internal dynamics during the early stages of growth factor stimulation. This method allows nondestructive and inclusive time-series analyses of chemical compositions of the same single cells. Applying a Gaussian mixture model to the major principal components of the single-cell Raman spectra, we detected the dynamics of the chemical states in MCF-7 cancer-derived cells in the absence and presence of differentiation and proliferation factors. The dynamics displayed characteristic variations according to the functions of the growth factors. In the differentiation pathway, the chemical composition changed directionally between multiple states, including both reversible and irreversible state transitions. In contrast, in the proliferation pathway, the chemical composition was homogenized into a single state. The differentiation factor also stimulated fluctuations in the chemical composition, whereas the proliferation factor did not.
Introduction
The morphologies and functions of cells change dramatically through proliferation and differentiation during the developmental process. These changes are supported by intracellular reactions among many species of biological molecules, which generate complicatedly diversified developmental pathways within populations of cells (1). In addition, there are large cell-to-cell variations in these developmental processes, even under the influence of similar extracellular cues. These variations can even be observed in model systems of clonal cells under the same culture conditions (2). Some of these variations are attributable to the intrinsically stochastic nature of chemical reactions, and others are determined by differences in the initial and boundary conditions of individual cells before they are affected by extracellular cues. Although the detection of intracellular dynamics is essential if we are to understand and control cellular behaviors including these variations, we have yet to perfect a technology to detect the complex and individual intracellular dynamics within the whole chemical milieu inside cells along the pathways of cellular events. Current genomic, proteomic, and metabolomic technologies can detect cellular components with very fine and multicomponent resolution (3,4). However, these technologies are destructive and cannot trace the dynamics in single cells over time. Most fluorescence imaging technologies, which are currently very popular, are insufficient to make multidimensional measurements and require prior knowledge to determine the target molecules (5).
Raman microspectroscopy is a technology that complements the omic technologies and conventional fluorescence microscopy (6). From the Raman spectra obtained from single cells, we can detect the cell-to-cell distributions and/or time-series changes in the chemical compositions of the cells. The Raman signals are derived from the inelastic light scattering caused by interactions between molecular vibrations and light. The spectrum of Raman signals carries information about the compositions of chemical species, including proteins, nucleic acids, carbohydrates, and lipids, in a biological specimen (7,8). Of particular importance, Raman spectra provide highly multidimensional information noninvasively and without labeling. These features allow Raman spectroscopy to be applied to various medical and biological research fields. At the tissue level, Raman spectroscopy has recently been used for melanoma diagnosis (9), to detect differences in the chemical components of bonelike cells (10), and to discriminate between cancerous and normal cells in the skin (11). In single cells, Raman imaging has been used to observe the differentiation of mouse (12,13) and human embryonic stem cells (14,15), to determine the differences between human skin fibroblast cells and the induced pluripotent cells derived from them (16), and to investigate the apoptosis of human epithelial cells (17,18). Although only a few studies have used Raman spectroscopy for single-cell time-series analyses, a multivariate Raman spectral analysis of the yeast cell cycle (19) and coherent anti-Stokes Raman scattering imaging of hormone-stimulated adipocyte lipolysis (20) have demonstrated that these techniques are useful for detecting the dynamics of the chemical compositions of single living cells.
We have used Raman microspectroscopy to study the differentiation process of the MCF-7 human breast cancer cell line (21), a model of cell fate changes, because MCF-7 cells can be stimulated by heregulin (HRG) to differentiate or by epidermal growth factor (EGF) to proliferate. The differentiation induced by HRG is morphologically characterized by the appearance of oil droplets in the cytoplasm. The cell-surface receptors for HRG (ErbB3 and B4) and EGF (ErbB1) all belong to the same ErbB family and activate the intracellular RAS-MAPK pathway. However, because HRG and EGF induce cell-fate changes in different directions (22,23), MCF-7 cells provide a good experimental system in which to compare the cell differentiation and proliferation pathways. Genomic (23,24) and biochemical analyses (25) of the growth-factor-induced fate decisions in MCF-7 cells have already been reported. Using Raman microspectroscopy, we observed the cyclic and oscillating dynamics of the cellular chemical compositions of a population of cells during the HRG-induced differentiation process (21).
In this study, we used Raman microspectroscopy to follow the time course of the chemical composition of the cytoplasm in the same single MCF-7 cells in the early stages of HRG or EGF stimulation. Our results indicate that the chemical compositions of single cells fluctuate dynamically in tens of minutes, even under resting conditions, and that the dynamics of the major chemical components after stimulation with HRG are more diverse than those in resting cells or EGF-stimulated cells. Although these two stimulants use the same RAS-MAPK signal transduction pathway, the cellular responses to the differentiation and proliferation pathways are significantly different.
Materials and Methods
Cell culture and sample preparation
The human breast cancer cell line MCF-7 (obtained from the American Type Culture Collection) was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37°C under 5% CO2. The cells were subcultured on 35 mm cell culture dishes with a quartz coverslip on the bottom (SF-S-D27, Fine Plus International, Kyoto, Japan), for Raman microspectroscopy. The cells were rinsed twice with Hank’s balanced salt solution containing 10 mM PIPES-Na (pH 7.2; PIPES-HBSS) and incubated at 25°C for 30 min in PIPES-HBSS before the Raman measurements were made. To stimulate cell differentiation or proliferation, 30 nM recombinant human HRG (NRG1-β1/HRG1-β1 EGF domain, R&D Systems, Minneapolis, MN) or 15 nM murine recombinant EGF (PeproTech, Rocky Hill, CT), respectively, was added to the culture medium. For measurements made during the first 2 h (0–2 h) of stimulation, the growth factors were added to the medium under the microscope immediately before the second measurement was made (time 0), and successive measurements were made until the cells had been stimulated for 2 h. For measurements made from 24 to 26 h of the stimulation period, the cells were cultured in the presence of growth factor for 23.5 h, then washed and measured in the presence of growth factor from 24 to 26 h. The cells in the resting condition were measured for 2 h in PIPES-HBSS in the absence of growth factor.
Raman microspectroscopy
The cells were measured with a confocal Raman microspectrometer (inVia, Renishaw, United Kingdom). Measurements were made seven to eight times in the cytoplasm of 48 resting cells and 60 growth-factor-treated cells at 20 min intervals. Raman scattering was excited by a 532 nm laser focused on the sampling point through a 63× dip-type water-immersion objective (HCX APO LU-V-1, NA 0.9, Leica, Wetzlar, Germany) with 6.5 mW irradiation after the objective. The spatial resolutions of the lateral and focal directions were 1 μm and 3–4 μm, respectively. The laser focus was set at 3 μm above the coverslip surface. Scattered light from the sampling point was collected through the same objective, dispersed with a grating (1800 lines/mm), and accumulated on a Peltier-cooled charge-coupled device detector (1024 × 256 pixels) for 1.5 s. At every measurement time, the laser spot was moved along a line at ∼2 μm intervals to obtain spectra from 15 different points in a single cell. No significant phototoxic damage to the cells by laser irradiation was observed, even after the last measurements (Fig. 1 A). The measurement error of the wavenumber was corrected using the peak of the ring-breathing mode of phenylalanine (1002 cm–1) as the standard.
Figure 1.
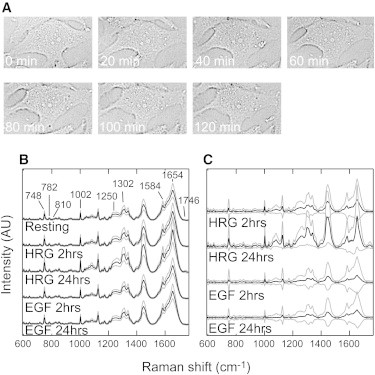
Raman measurements of single living cells. (A) Transmission-illumination micrographs of a single MCF-7 cell just after the Raman measurements made at the indicated times. No significant changes were observed in the cell morphology. (B) Averaged Raman spectra from the cytoplasm of cells under the indicated conditions. Positions of the major Raman bands derived from proteins (1002, 1250, and 1654 cm–1), cyt c (748 and 1584 cm–1), lipids (1302, 1654, and 1746 cm–1), and nucleic acids (782 and 810 cm–1) are indicated. (C) The difference spectra against the averaged spectrum in the resting condition. The black and gray lines in (B) and (C) indicate the average and width of the standard deviations, respectively. AU, arbitrary units.
Data processing and spectral analysis
To reduce the point-to-point deviations in single cells, the spectra obtained in the cytoplasm of a single cell at each measurement were selected and averaged as the single-cell spectrum at that single time point. The Raman spectrum of the quartz coverslip was subtracted from the averaged spectrum. The background signal (predominantly from the autofluorescence of the cells) was then subtracted. The background was determined by linearly connecting the signals of 20 points (592, 610, 630, 652, 706, 736, 770, 796, 866, 910, 992, 1012, 1022, 1138, 1184, 1378, 1500, 1522, 1722, and 1768 cm–1) in the individual spectra, where the averages and standard deviations (SDs) of the signal intensities were small (21). The spectra at 592–1768 cm–1 (after background subtraction) were used for further analyses.
Principal component analysis (PCA) was used to obtain an overview of the cell-state changes by reducing the dimensions of the Raman spectra. The data matrix of the measured Raman spectra was composed of m rows (Raman shifts) and n columns (samples). In our measurements, m = 589 (592–1768 cm–1, 2 cm–1/channel) and n = 2136 (48 control cells × 7 times + 60 stimulated cells × 7 or 8 times × 2 growth factors). The average of all 2136 spectra was subtracted from each single spectrum to obtain the mean-centered data matrices, X. The PCA then decomposed X as the linear combination of vectors t and p, using a singular value decomposition algorithm:
so the p vectors were orthogonal to each other. The t and p pairs were ordered from PC1 to PCm according to the lengths of the t vectors (t1 is the longest). In PCA, t and p are called scores and loadings, respectively.
The spectral points in the PC1-PC2 plane were grouped using two-dimensional Gaussian model separation. The distributions of the spectral points were fitted with the function
Here, μ and Σ correspond to average and variance-covariance metrics, respectively. The best-fit values for the parameters were estimated with the expectation-maximization algorithm (26). To avoid trapping in a local minimum, the estimation cycle was repeated >500 times for each data set using different initial conditions. The number of components, k, was changed from 1 to 4, and the most probable number was determined based on the Bayesian information criterion (BIC) to avoid over fitting. The BIC is given by , where L, k, and n correspond to the likelihood function in the expectation-maximization algorithm, the number of groups, and the number of spectra in each condition, respectively. The model (i.e., the value of k) that yielded the smallest BIC was adopted (Table S2 in the Supporting Material).
The transition probability between the cell states was estimated using a nonparametric bootstrap method (27). The values of the transition probabilities were resampled 10,000 times to obtain their medians and confidence intervals.
Spectral data processing, PCA, dynamics analysis, Gaussian mixture model separation, and the statistical test were performed with MATLAB (MathWorks, Natick, MA).
Results
Changes in Raman spectra in the early stages of growth factor stimulation
Raman spectra were measured in the cytoplasm of single MCF-7 cells with or without stimulation by the growth factors HRG (a differentiation factor) or EGF (a proliferation factor). The signals came from the cytosol, membrane organelles, and cytoskeleton. Measurements were made for 2 h at 20 min intervals in unstimulated (resting) cells and for 0–2 h and 24–26 h of the stimulation period in cells undergoing stimulation. The averages and SDs of the Raman spectra under each condition were compared (Fig. 1 B). The spectra from different cells and at different times during each 2 h period of successive measurements were mixed and averaged. The characteristic Raman bands from the major biological molecules (proteins, nucleic acids, lipids, and carbohydrates) were observed in the spectra. Resonant Raman signals for cytochrome c (cyt c) were also observed at the 532 nm excitation used in this study (see Table S1 for the assignment of Raman peaks). These spectra were not vector-normalized, so the intensity changes at each Raman shift indicate concentration changes in the corresponding compounds. The spectra obtained during 24–26 h in the presence of HRG (hereafter this condition is called HRG 24 h; other conditions are referred to in a similar way) showed a larger SD than the other spectra. Because the SD reflects the variation in the cytoplasmic chemical composition among single measurements, this result indicates that the cell-to-cell and/or time-to-time deviations in the chemical state increased after HRG stimulation for 24 h.
Difference spectra against the average spectrum in the resting condition were calculated as the average and SD (Fig. 1 C). The intensities of the Raman signals derived from proteins, including cyt c, increased in all spectra after growth factor stimulation. The signals from nucleic acids increased slightly after HRG treatment. It is highly likely that these increases correspond to the genetic and metabolic responses to differentiation and proliferation. With HRG stimulation, especially after 24 h, large increases in the intensities at 1250, 1302, 1448, 1654, 1746, and 1100–1150 cm–1 were observed. These Raman bands correspond to the chemical bonds in lipid molecules (Table S1). After stimulation with HRG for 24 h, a small proportion of cells accumulated oil droplets in the cytoplasm, as confirmed by fluorescence staining with BODIPY (Fig. S1). The increased lipid signals reflect both the formation of oil droplets and the increase in the amount of lipid in the cytoplasm before the production of oil droplets.
PCA of cellular chemical compositions
We used PCA based on the Raman spectra from all the single cells under different conditions and at different times of growth factor stimulation to examine the population and single-cell dynamics in the phase space of possible compositions of the major chemicals in single cells under various conditions (Fig. 2). The contribution ratio of the first principal component (PC1) was markedly larger than the contribution ratios of the other components, and PC2–PC4 were the next major components. However, because there were many minor components with similar low contribution ratios, the total contribution ratio of PC1–PC8 was only 26.8% (Fig. 2 A). We used PC1 and PC2 to analyze the cellular dynamics. The reconstructed single-cell spectra composed of the mean, PC1, and PC2 (or mean, PC3, and PC4) are shown in Fig. S2, and suggest that most of the dynamics characteristic of growth factor stimulation were visualized with PC1 and PC2. The spectral variations in PC3 and PC4 were much smaller than those in PC1 and PC2, which suggests that it is difficult to detect significant changes in higher PCs.
Figure 2.
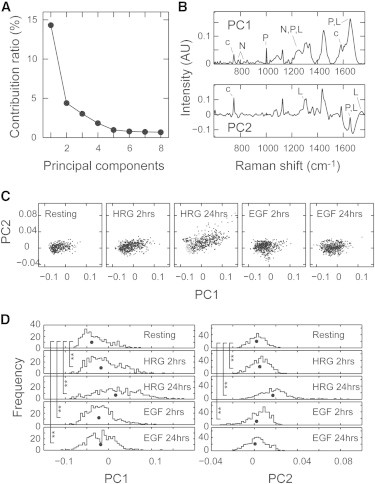
PCA of the Raman spectra. (A) Contribution ratios of PC1–PC8. The sum of the contribution ratios of PC1 and PC2 was 18.7%. PCA was performed based on all 2136 spectra. (B) Loadings for PC1 and PC2. PC1 mainly corresponded to proteins (P), lipids (L), nucleic acids (N), and cytochrome c (c). PC2 mainly corresponded to lipids, cytochrome c, and water (broad band around 1640 cm−1) (29). (C) Score plots in the PC1-PC2 plane. Black dots indicate the data set of cells under the indicated conditions, and gray dots indicate the data set of resting cells. (D) Histograms of the PC1 (left) and PC2 (right) scores. Black dots indicate the medians. ∗∗p < 0.01 on the Wilcoxon test. AU, arbitrary units.
The loadings for the PCs do not directly indicate the compositions of individual chemical components, but suggest the chemicals that predominantly contribute to the variations in each PC. PC1 was mainly comprised of signals from proteins (including cyt c), lipids, carbohydrates, and nucleic acids. PC2 was mainly comprised of signals from lipids, cyt c, and water (Fig. 2 B). In the two-dimensional scatter plots in the PC1-PC2 phase space, the populations of Raman spectra show overlapping but characteristic distributions according to the culture conditions (Fig. 2 C). The distribution at HRG 24 h was broadest, as expected from the size of the SD shown in Fig. 1 B. The distributions of the PC scores were compared among the culture conditions (Fig. 2 D). Significant changes in the distributions of the PCs, i.e., the metabolic pathways in cells, had already begun in the first 2 h of growth factor stimulation. Both the PC1 and PC2 scores increased with time during HRG stimulation, indicating increases in the metabolite concentrations in the cytoplasm. (The signal for water is negative in PC2; Fig. 2 B, lower.) Although the PC scores also increased in the cells stimulated with EGF for 2 h, the increases were smaller than those in the HRG-stimulated cells. After EGF stimulation for 24 h, the distribution of PC2 became statistically indistinguishable from that of the resting cells. The spectral distributions of different conditions of cells were almost overlapped in the PC3-PC4 plane (Fig. S3).
Dynamics of the chemical compositions of single cells in response to growth factors
The trajectories of the same single cells during 2 h of measurements were observed in the PC1 and PC2 scores (Figs. 3 A and S4). The scores fluctuated but showed no clear tendency to increase or decrease with time under all conditions. The fluctuations in single cells, similar to the width of the cell-to-cell distributions (Fig. 2), increased in PC1 after stimulation with growth factors. In the PC2 scores, HRG 24 h showed especially large fluctuations. The mean-square displacements (MSDs) in the two-dimensional PC1-PC2 plane were plotted with time (Fig. 3 B). The time evolution of MSDs for all conditions suggested random diffusion with a restriction in the motion area, but no directional movement was suggested, as expected from the single trajectories (Fig. 3 A). The diffusion coefficient and the size of the diffusion area (initial slope and asymptote of the MSD plot, respectively) were different for each condition. The histograms of the average step sizes in the trajectories of single cells along the PC1-PC2 plane, which reflect the single-cell distributions of the diffusion coefficients, were compared among the different conditions of the cells (Fig. 3 C). The step sizes were large and diverse in the cells after growth factor stimulation, especially for HRG 2 h and 24 h. The step size indicates the rate of the random changes in the chemical compositions of the cytoplasm.
Figure 3.
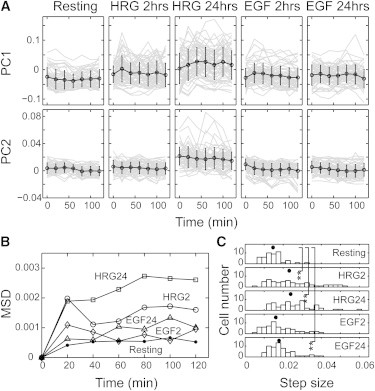
Single-cell trajectories of the major chemical components. (A) Trajectories of PC1 (upper) and PC2 (lower) in single cells. Black lines indicate averages with SDs. Gray lines show single-cell trajectories with time. (B) Time evolution of the two-dimensional MSDs of the trajectories under each condition in the PC1-PC2 plane. (C) Histograms of the average step sizes of single cells for 2 h under the indicated conditions. Black dots indicate the median. ∗∗p < 0.01 on the Wilcoxon test.
As shown here, the growth factors, especially HRG, induced not only changes in the chemical composition in average of cells and measurement times (Fig. 2, C and D), but also vigorous fluctuations on a 10 min timescale (Fig. 3). Although the chemical composition averaged over the cells drifted directionally across time periods of hours and days (Fig. 2 D), its fluctuation in single cells was large and the trajectories of single cells were almost diffusive within 2 h (Fig. 3).
Grouping the chemical compositions of single cells in the PC1-PC2 plane
The cell-to-cell distributions of the major chemical compositions (PC1 and PC2) were wide and under some conditions had shoulders and long tails, suggesting the inclusion of multiple components (Fig. 2, C and D). To understand the dynamics of the chemical compositions of cells, we examined whether it is possible to group the chemical compositions based on the PC scores. The Gaussian mixture model in the two-dimensional plane was used for grouping, because this model is simple and the distributions of the PC scores look like normal distributions in the simplest cases (for example, the PC2 distribution of resting cells; Fig. 2 D). As a result, the presence of one to three components was suggested for each condition (Fig. 4 A).
Figure 4.
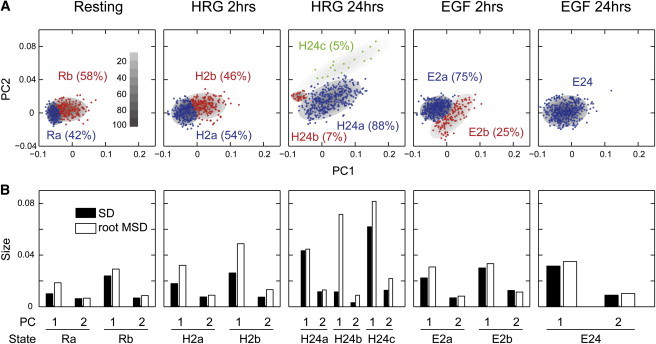
Grouping the cellular states based on the Gaussian mixture model. (A) Results of grouping the single-cell chemical compositions in the PC1-PC2 phase space. Elliptical contours show the positions and distributions of the chemical states of the cells. Fractions of each state are indicated. Gray scale indicates the relative depth of the distributions. (B) Comparison of the size of the chemical state distribution (SD) and the root MSDs of single-cell trajectories at the plateau. To see this figure in color, go online.
The single-cell dynamics in the MSD plots (Fig. 3 B) can be explained as diffusion in restricted areas of the PC space. If the areas of restriction corresponded to the distributions of single spectra belonging to each of the chemical states suggested by the Gaussian mixture model (Fig. 4 A), the sizes of the restriction in the MSD plots should be similar to the widths of the Gaussian distributions. This requirement comes from the assumption of the quasisteady state of the chemical compositions fluctuating in the Gaussian distributions. We compared the SD of the Gaussian distribution and the square root of the MSD at the plateau (Fig. S5) in each PC axis (root MSD (Fig. 4 B)). In most cases, the SD and root MSD were similar, indicating that during 2 h of measurements, individual cells diffused within the area of the distribution of the chemical composition to which the cells belonged, but that the transition to other chemical distributions was somehow restricted.
These cell dynamics are not inconsistent with the assumption that multiple chemical states were present. Therefore, we provisionally called each component estimated in the Gaussian mixture model a state, although further examination is required to define true cellular states. We named each state as indicated in Fig. 4 A. For the Ra state of the resting cells, and the H2a and H2b states of HRG 2 h cells, the root MSD of PC1 was significantly larger than the corresponding SD (Fig. 4 B). For the Ra and H2a states, this could be caused by the misassignment of some trajectories because of large overlaps with the distributions of the other state with larger root MSDs (Rb and H2b, respectively). However, for the H2b state, it could be caused by the transition between the H2a and H2b states, because the root MSD was larger than the SD for both the H2a and H2b distributions. Actually, some trajectories at HRG 2 h covered the areas of H2a and H2b (Fig. S4). For PC1 of H24b of HRG 24 h, the difference between the SD and root MSD was very large, but because the area of H24b was much smaller than those of the other states, it is unclear whether the H24b state was significant.
Time evolution of the cellular states during stimulation with growth factors
A comparison among positions of the estimated chemical states of the cells in the PC1-PC2 plane (Fig. 5 A) indicated that HRG and EGF induce cellular responses with completely different time evolutions. The chemical states of the HRG-stimulated cells moved in the direction of increased scores for both PC1 and PC2 in the HRG 24 h condition. As a result of this process, a new state with a high PC2 score (H24c) appeared. In the area of this state, no cells were observed before or after HRG stimulation for 2 h. At the same time, the states with the lowest scores for PC1 and PC2 (Ra and H2a) disappeared. These dynamics induced by HRG reflect increases in the chemical concentrations in the cytoplasm, especially the concentration of newly synthesized lipid molecules (PC2), in the early stages of cell differentiation (Fig. S1). In contrast, with EGF stimulation, no directional change in the chemical state was observed over the 24 h period. Although a new state (E2b) was observed once in the first 2 h of stimulation, it disappeared after 24 h, and the two states observed before stimulation merged into a single state (E24). Thus, the chemical state was homogenized in the process of cell proliferation.
Figure 5.
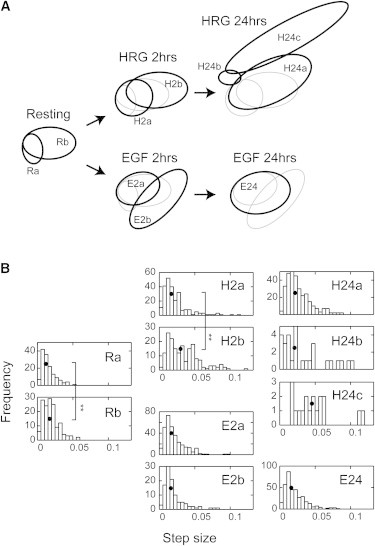
Time evolution and fluctuations in the cellular chemical states. (A) Distributions of the chemical states of cells under different conditions are compared to show their time evolutions during growth factor stimulation. Black ellipses show the state at the indicated stage. Gray ellipses show the state before the indicated stage. (B) Histograms of the step sizes of single-cell trajectories in each chemical state. The step sizes indicate the rate of fluctuation in the chemical compositions of single cells. Black dots indicate the median values. ∗∗p < 0.01 in the Wilcoxon test.
We compared the step sizes of the single-cell PC dynamics in each chemical state (Fig. 5 B). Because the dynamics were diffusive under every condition, the step size indicates the rate of the chemical fluctuations in single cells over time. The fluctuations were again characteristic of the chemical states and growth factors. In the resting cells, the state with larger PC scores (Rb) showed slightly but significantly larger fluctuations (on average) than those of the other state (Ra), i.e., the state with higher concentrations of chemicals showed larger chemical fluctuations. This relationship between the concentrations and fluctuations in the chemical compositions was preserved and enhanced during HRG stimulation, and the new state at 24 h (H24c) showed further large PC scores and fluctuations. Taking these data together, the fluctuations in the chemical compositions of the cells undergoing differentiation increased with time, at least up to 24 h. However, the fluctuations in the EGF-stimulated cells were completely different. Although changes in the positions and numbers of the chemical states in the PC1-PC2 plane were observed during stimulation with EGF (Fig. 5 A), the rate of fluctuations in the chemical composition did not change at all (Fig. 5 B). Thus, the fluctuation and the time evolutions of the chemical states shown in the PC1-PC2 plane were specific for the differentiation and proliferation of cells.
Dynamics of single cells immediately after stimulation with growth factors
The state transitions of single cells were examined from the resting state to the initial 2 h of growth factor stimulation (Fig. 6). In cells stimulated with HRG, the cells grouped in the Ra state before stimulation tended to move into the H2a state, which was distributed in a similar region to the Ra state in the PC1-PC2 plane, and to stay within this state for 2 h (Fig. 6 A). In contrast, cells that initially grouped in the Rb state distributed into both the H2a and H2b states (Fig. 6 A), and some of them moved around the combined H2a and H2b areas during the 2 h period (Fig. S4). Because the state assignment used only a single time point (0 or 120 min) and the state distributions overlapped, the probability values in the diagrams shown in Fig. 6 B may contain considerable errors. However, it is useful to understand the tendencies in the state transitions. As illustrated in Fig. 6 D, 78% of the cells that started from the Ra state moved into the H2a state, and the cells that started from the Rb state were distributed to the H2a (61%) and H2b (39%) states (Fig. 6 D).
Figure 6.
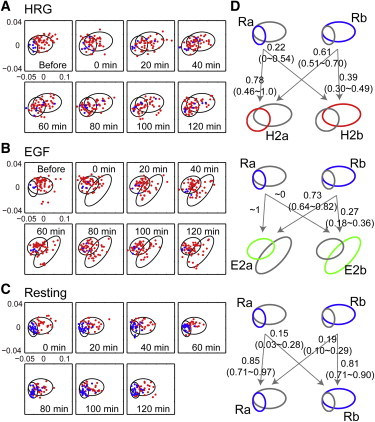
Initial chemical states and responses to the growth factors. (A–C) Distributions of the chemical compositions of single cells before and during 2 h of stimulation with HRG (A) or EGF (B). Distributions of cells in the resting condition for 2 h (C) are shown for comparison. Blue and red cells were grouped into Ra (blue) or Rb (red) before stimulation (A and B) or at the first measurement (C). Ellipses show the distributions of the cell states in the corresponding conditions. (D) State transition diagrams between the first and last measurements. The transition rates were estimated from the weighted distributions of single Raman spectra to the two states using the bootstrap method. Numbers indicate the medians of the transition probabilities, with the 90% confidence intervals in parentheses. Because the fraction of cells was small, it was difficult to accurately estimate the confidence intervals for the transition rates from the Ra state in (B). To see this figure in color, go online.
These dynamics, including closely distributed transitions from a single state to two different states, were specific for the HRG-stimulated cells. The direction of the state transitions was much more deterministic for cells stimulated with EGF (Fig. 6 B). When the state transitions of the resting cells after EGF stimulation were examined, we noticed that most of the cells starting from Ra moved to E2a, and transitions into E2b were only observed in cells starting from Rb before stimulation. In the resting condition, most cells stayed within a single state during 2 h (Fig. 6 C).
Discussion
In this study, we detected changes in the chemical compositions of the cytoplasm of single MCF-7 cells using Raman microspectroscopy. Although the cell-to-cell and time-to-time deviations in the Raman spectra were large, both among the resting cells and among the cells stimulated with growth factors, populations of cells showed characteristic distributions of the spectra under each condition, responding differently to the differentiation factor (HRG) and the proliferation factor (EGF) with time (Fig. 1). Despite the complicated chemical composition of the cytoplasm, the responses of the cell populations to the growth factors could be detected using the first two PCs (Fig. 2). In the initial 2 h of stimulation with the growth factors, the distributions of the PCs shifted significantly from those of resting cells, and the shifts increased (HRG) or decreased (EGF) in the subsequent 24 h (Fig. 2 D). These results indicate that we can discuss the dynamics of the chemical composition inside cells in response to growth factors based on Raman spectra.
We focused on the cell dynamics for 2 h in the resting condition, and those immediately after and 24 h after the application of growth factors to the cell culture medium. The dynamics of the Raman spectra of single cells during the 2 h in which the measurements were made were not directional but diffusive under every condition (Fig. 3). However, changes in the average and distributions of the spectra indicate that on long timescales (>2 h), the chemical compositions responded systematically to the growth factors. Previously, we have observed changes in the populations of Raman spectra for the cytoplasm of MCF-7 cells in response to HRG treatment over a much longer timescale (up to 12 days) (21). In this study, we detected the dynamics in the same single cells on a much shorter timescale (20 min to 2 h).
The distributions of the PCs showed long tails or shoulders, even for the resting cells, and after the growth factor treatments, these distributions changed in shape and width, especially in response to HRG (Fig. 2 D). To characterize the shapes of the PC distributions, i.e., the variation in the chemical compositions, we used a Gaussian mixture model to subdivide the distributions into several components, and identified one to three components for each condition. From the outset, this analysis was performed with the convenience of visualization in mind. However, the trajectories of the single-cell dynamics with time suggest that there are substantial meanings in this grouping. The trajectories of the single cells in the PC1-PC2 phase space were diffusive in restricted areas (Figs. 3 and S4), and the confinement size of the diffusion was generally similar to the width of the PC distribution to which the cells belonged (Fig. 4 B). These results are consistent with the notion that individual cells belonged to one of the chemical states at a certain position in the PC space with a limited distribution width. From this perspective, the chemical composition of a single cell is randomly drifting within the range of its chemical state, and transitions across the boundary between chemical states are restricted. Although such chemical states of cells must be carefully validated in the future, the Gaussian mixture model is useful for discussing the dynamics of the chemical compositions of cells.
The Raman spectra of the resting cells without growth factor stimulation consist of two Gaussian components (Fig. 4 A, Ra and Rb). The cellular states represented by these two components mainly differ in the concentrations of the chemicals that were responsible for PC1. The fluctuations in the chemical compositions also differed between these two states, i.e., the cells in Rb showed larger step sizes within 20 min (Fig. 5 B). These differences could reflect differences in the conditions of the cells, e.g., in their cell cycles and/or cell-to-cell communications. However, we could not distinguish these two cellular states based on morphological information. A wide distribution of the single-cell Raman spectra under undifferentiated conditions was observed in mouse embryonic stem cells (13), which may indicate the presence of multiple states. After differentiation, the distribution narrowed. In contrast, in MCF-7 cells, the width of the spectral distribution did not change markedly, although the overall shape of the spectrum changed after differentiation (21). Various types of cells must be examined to find the general principles underlying the changes in the Raman spectra that occur during cell differentiation.
The population and single-cell dynamics of the cellular chemical compositions were diversified according to the type of growth factor used to stimulate the cells (Figs. 2 and 5). During the first 2 h of HRG treatment, two chemical components of the cells remained at locations similar to those before the treatment, with a slight shift toward higher contents of PC1, but with increased fluctuations within individual cells, especially for the higher PC1 state (H2b). After HRG treatment for 24 h, the chemical distribution of the lower PC1 state (H24a) was enlarged to cover the distributions of both Rb and H2b, with fluctuations as large as that of H2b. The content of PC1 in H24a was higher than the content in Rb and H2b, on average. No separable states with PC1 contents as small as those of Ra and H2a were found at this stage. The other chemical states found after 24 h of HRG treatment (H24b and H24c) were distributed in completely different regions from H24a and any other states identified with our measurements. H24c was characteristic of a high PC2 content, indicating high concentrations of lipids in the cytoplasm. In response to HRG, MCF-7 forms lipid droplets in the cytoplasm as an indication of cell differentiation (28). Although MCF-7 cells usually require 10 days or more for the complete differentiation of the population (21), the cells classified into H24c had already formed lipid droplets in the cytoplasm (Fig. S1). The fluctuations in the chemical compositions of single cells became larger as PC1 and PC2 increased (H2b, H24a, and H24c), indicating that in the early stage of differentiation, the chemical composition of the cytoplasm fluctuates vigorously, probably attributable to the rearrangement of the metabolic pathways producing the various chemicals included in PC1 and PC2. In particular, the increases in the lipid components, which are suggested from the dynamics of PC2, clearly characterize populations of cells moving toward differentiation.
In the PC1-PC2 phase plane, the arrangement of the two states of the resting cells, Ra and Rb, was similar to that of the two states, H2a and H2b, observed for cells during the first 2 h of HRG stimulation (Fig. 5 A). However, the state transition dynamics of the single cells differed between the cells in the resting condition and those in the HRG-stimulated conditions, i.e., in the resting condition, cells tended to stay in one of the single states for 2 h (Fig. 6 C), whereas HRG stimulation induced active state transitions (Fig. 6 A). This means that the shift in the cell states observed in the first 2 h of differentiation included highly fluctuating dynamics, covering different states of the cells. On the contrary, no transition was observed between the two states, H24a and H24c, detected after 24 h of cell differentiation (Fig. S4). These findings suggest that the initial differentiation process is composed of successive reversible and irreversible state transitions.
Treatment with EGF, a proliferation factor, induced completely different dynamics in the cells from those observed during HRG stimulation. For the first 2 h, a new transient state (E2b) was observed in the higher PC1 region. Its distribution was different from those of the states observed after HRG treatment. This component disappeared after 24 h, leaving one population covering the entire region of the two states observed for resting cells. The remaining state (E24) largely overlapped with the state (E2a) observed after 2 h of EGF treatment. Interestingly, the fluctuations in the chemical composition (Fig. 5 B) did not increase after EGF stimulation for any of the states we observed. Therefore, during EGF stimulation, the cellular chemical states converged into a single state, with slow fluctuations. Although the cells in the two states in the resting condition (Ra and Rb) were mixed into the E2a state during the 2 h of EGF stimulation, only the cells in the cell population that started from the Rb state moved into the E2b state (Fig. 6 D). This suggests that the Rb state contained at least two substates, although these states could not be distinguished based on the composition of the major chemicals in the cytoplasm.
Using Raman microspectroscopy, we characterized the dynamics of the chemical compositions in single cells and populations of cells in the resting condition and in the early stages of growth factor stimulation. Single-cell Raman spectroscopy revealed that the chemical composition and its fluctuations were not identical between the cells, even in the resting condition, but that the cell populations could be subdivided into at least two groups, which displayed different responses to the differentiation factor, HRG, and the proliferation factor, EGF. Surprisingly, the chemical compositions of the cells fluctuated vigorously on a 10 min timescale. HRG increased these fluctuations, whereas EGF did not. Directional drift and homogenization of the cell states were induced by HRG and EGF, respectively, in populations of cells on a timescale of hours. We have demonstrated that Raman microspectroscopy is a powerful tool for detecting dynamics of the global chemical states in single cells.
Author Contributions
S.T. performed the experiments. S.T. and S.M. performed data analyses. S.T., Y.O., and Y.S. planned the study. S.T. and Y.S. wrote the manuscript. All the authors commented on the manuscript.
Acknowledgments
The authors thank Michio Hiroshima at RIKEN for experimental advice and helpful discussion and Renishaw for allowing us to use the Raman microscope.
S.T. was supported by a Junior Research Associate Fellowship from RIKEN.
Supporting Material
References
- 1.Avraham R., Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 2.Mouri K., Sako Y. Optimality conditions for cell-fate heterogeneity that maximize the effects of growth factors in PC12 cells. PLOS Comput. Biol. 2013;9:e1003320. doi: 10.1371/journal.pcbi.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffin J.L., Shockcor J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 4.Spratlin J.L., Serkova N.J., Eckhardt S.G. Clinical applications of metabolomics in oncology: a review. Clin. Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang B., Wu H., Zare R.N. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- 6.Spiller D.G., Wood C.D., White M.R. Measurement of single-cell dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 7.Notingher I., Hench L.L. Raman microspectroscopy: a noninvasive tool for studies of individual living cells in vitro. Expert Rev. Med. Devices. 2006;3:215–234. doi: 10.1586/17434440.3.2.215. [DOI] [PubMed] [Google Scholar]
- 8.Puppels G.J., de Mul F.F., Jovin T.M. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature. 1990;347:301–303. doi: 10.1038/347301a0. [DOI] [PubMed] [Google Scholar]
- 9.Gniadecka M., Philipsen P.A., Wulf H.C. Melanoma diagnosis by Raman spectroscopy and neural networks: structure alterations in proteins and lipids in intact cancer tissue. J. Invest. Dermatol. 2004;122:443–449. doi: 10.1046/j.0022-202X.2004.22208.x. [DOI] [PubMed] [Google Scholar]
- 10.Gentleman E., Swain R.J., Stevens M.M. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat. Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- 11.Kong K., Rowlands C.J., Notingher I. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc. Natl. Acad. Sci. USA. 2013;110:15189–15194. doi: 10.1073/pnas.1311289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notingher I., Bisson I., Hench L.L. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal. Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura T., Chiu L.D., Fujita H. Visualizing cell state transition using Raman spectroscopy. PLoS ONE. 2014;9:e84478. doi: 10.1371/journal.pone.0084478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze H.G., Konorov S.O., Turner R.F. Assessing differentiation status of human embryonic stem cells noninvasively using Raman microspectroscopy. Anal. Chem. 2010;82:5020–5027. doi: 10.1021/ac902697q. [DOI] [PubMed] [Google Scholar]
- 15.Pascut F.C., Goh H.T., Notingher I. Noninvasive detection and imaging of molecular markers in live cardiomyocytes derived from human embryonic stem cells. Biophys. J. 2011;100:251–259. doi: 10.1016/j.bpj.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pliss A., Kuzmin A.N., Prasad P.N. Nucleolar molecular signature of pluripotent stem cells. Anal. Chem. 2013;85:3545–3552. doi: 10.1021/ac303806j. [DOI] [PubMed] [Google Scholar]
- 17.Zoladek A., Pascut F.C., Notingher I. Non-invasive time-course imaging of apoptotic cells by confocal Raman micro-spectroscopy. J. Raman Spectrosc. 2011;42:251–258. [Google Scholar]
- 18.Okada M., Smith N.I., Fujita K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA. 2012;109:28–32. doi: 10.1073/pnas.1107524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C.K., Ando M., Shigeto S. Disentangling dynamic changes of multiple cellular components during the yeast cell cycle by in vivo multivariate Raman imaging. Anal. Chem. 2012;84:5661–5668. doi: 10.1021/ac300834f. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto T., Segawa H., Osumi T. Active involvement of micro-lipid droplets and lipid-droplet-associated proteins in hormone-stimulated lipolysis in adipocytes. J. Cell Sci. 2012;125:6127–6136. doi: 10.1242/jcs.113084. [DOI] [PubMed] [Google Scholar]
- 21.Morita S., Takanezawa S., Sako Y. Raman and autofluorescence spectrum dynamics along the HRG-induced differentiation pathway of MCF-7 cells. Biophys. J. 2014;107:2221–2229. doi: 10.1016/j.bpj.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citri A., Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 23.Nagashima T., Shimodaira H., Hatakeyama M. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2007;282:4045–4056. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 24.Saeki Y., Endo T., Okada-Hatakeyama M. Ligand-specific sequential regulation of transcription factors for differentiation of MCF-7 cells. BMC Genomics. 2009;10:545. doi: 10.1186/1471-2164-10-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakakuki T., Birtwistle M.R., Kholodenko B.N. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell. 2010;141:884–896. doi: 10.1016/j.cell.2010.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLachlan G., Peel D. John Wiley and Sons; New York: 2000. Finite Mixture Models. [Google Scholar]
- 27.Efron B. Bootstrap methods: another look at the jackknife. Ann. Stat. 1979;7:1–26. [Google Scholar]
- 28.Bacus S.S., Kiguchi K., Huberman E. Differentiation of cultured human breast cancer cells (AU-565 and MCF-7) associated with loss of cell surface HER-2/neu antigen. Mol. Carcinog. 1990;3:350–362. doi: 10.1002/mc.2940030607. [DOI] [PubMed] [Google Scholar]
- 29.Walrafen G.E., Blatz L.A. Weak Raman bands from water. J. Chem. Phys. 1973;59:2646–2650. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


