Abstract
Bacterial cell division is driven by an FtsZ ring in which the FtsZ protein localizes at mid-cell and recruits other proteins, forming a divisome. In Escherichia coli, the first molecular assembly of the divisome, the proto-ring, is formed by the association of FtsZ polymers to the cytoplasmic membrane through the membrane-tethering FtsA and ZipA proteins. The MinCDE system plays a major role in the site selection of the division ring because these proteins oscillate from pole to pole in such a way that the concentration of the FtsZ-ring inhibitor, MinC, is minimal at the cell center, thus favoring FtsZ assembly in this region. We show that MinCDE drives the formation of waves of FtsZ polymers associated to bilayers by ZipA, which propagate as antiphase patterns with respect to those of Min as revealed by confocal fluorescence microscopy. The emergence of these FtsZ waves results from the displacement of FtsZ polymers from the vicinity of the membrane by MinCD, which efficiently competes with ZipA for the C-terminal region of FtsZ, a central hub for multiple interactions that are essential for division. The coupling between FtsZ polymers and Min is enhanced at higher surface densities of ZipA or in the presence of crowding agents that favor the accumulation of FtsZ polymers near the membrane. The association of FtsZ polymers to the membrane modifies the response of FtsZ to Min, and comigrating Min-FtsZ waves are observed when FtsZ is free in solution and not attached to the membrane by ZipA. Taken together, our findings show that the dynamic Min patterns modulate the spatial distribution of FtsZ polymers in controlled minimal membranes. We propose that ZipA plays an important role in mid-cell recruitment of FtsZ orchestrated by MinCDE.
Introduction
Bacterial division is a fundamental biological process that is strictly regulated in time and space, and is carried out by macromolecular assemblies whose components transiently interact to form, toward the end of the cell cycle, a contractile ring at mid-cell. At subsequent stages, the recruitment of other proteins results in the assembly of the functionally active molecular machinery that drives cytokinesis, the divisome. The most abundant and conserved element of the division ring is the protein FtsZ, which in Escherichia coli is anchored to the cytoplasmic membrane by the action of the bitopic membrane protein ZipA and the amphitropic protein FtsA. These three proteins assemble together to form the first essential complex of the division machinery, the proto-ring, which is ultimately joined by the rest of the division proteins (1). Positioning of FtsZ, forming the Z ring at mid-cell, is directed by the Min proteins, which oscillate from pole to pole, blocking polymerization of FtsZ at both poles (2), and by nucleoid occlusion (NO), which prevents polymerization in the vicinity of the bacterial chromosome (3–5). Recently, it was demonstrated that MinCDE oscillations are sufficient to establish distinct concentration gradients of FtsZ assembly to the membrane of cell-shaped compartments (6).
FtsZ is a GTPase that is related to eukaryotic tubulin and, in the case of E. coli, self-assembles, forming single-stranded protofilaments. These protofilaments are structurally flexible and tend to further associate into higher-order polymers that are sufficiently plastic to adopt multiple geometries depending on the environment in which they are located. These assembly reactions, linked to GTP hydrolysis, are considered to be crucial for Z-ring function (7,8). The detailed arrangement of FtsZ polymers in the proto-ring is not known, but current models stress the importance of the presence of lateral interactions between FtsZ protofilaments that loosely arrange to form a structure that is active in division (9). FtsZ has no intrinsic affinity for phospholipid bilayers and is anchored to the cytoplasmic membrane through the interaction of its carboxy-terminal region with the other proto-ring proteins, ZipA and FtsA (10,11). Apparently, both of these proteins can support the attachment of FtsZ to the membrane, but no localization of FtsZ at the membrane occurs if both are absent (12,13). FtsA is an actin-like amphitropic protein with a short amphipathic helix that is thought to mediate its association with the membrane (14). ZipA contains a short amino-terminal region that is integrated in the membrane and connected to the carboxy-terminal FtsZ-interacting domain by a flexible, unstructured linker region (15). It is known that ZipA protects FtsZ from degradation by the protease system ClpXP, suggesting a specific function of ZipA in proto-ring stability, as this role cannot be fulfilled by FtsA or the gain-of-function FtsA∗ mutant (16,17).
The Min system comprises three proteins (MinD, MinE, and MinC) that oscillate between the cell poles. Together, MinD (a reversibly membrane-bound ATPase) and MinE (MinD’s ATPase activator) account for the dynamic behavior of the system, which arises from self-organization and exchange of protein subunits between the membrane and the cytosol (18,19). As a result of the oscillations, the concentration of membrane-bound MinC (an inhibitor of FtsZ assembly), which is passively transported with the wave, exhibits a time-averaged local minimum at mid-cell, where it targets the formation of the Z ring (20,21). Although there is evidence that inhibition of Z-ring assembly by MinC occurs through the disassembly of FtsZ filaments and the decrease of lateral contacts between the filaments in the presence of MinC (22–24), the impairment of membrane attachment, specifically the FtsZ-ZipA interaction, has also been suggested to play a role in this process (25).
Because the assembly of the proto-ring complex in vivo takes place at the inner bacterial membrane, investigators have made a considerable effort to study the assembly properties of FtsZ and its interaction with companion proteins in reconstructions of the proto-ring on biomimetic membrane systems (26–28). Among these systems, supported lipid bilayers (SLBs) are well suited for investigating the reactivity (i.e., dynamic interactions) and the membrane structural organization of division proteins by surface-sensitive imaging techniques under biochemically controlled and well-defined experimental conditions (29–31).
Studies employing this bottom-up strategy showed that FtsZ polymers bound to SLBs through a soluble variant of ZipA, lacking the transmembrane region, formed two-dimensional networks that retained their capacity to dynamically restructure, as revealed by atomic force microscopy (AFM) (32–34). Reconstituted copolymers of wild-type FtsZ and a variant form containing a membrane-targeting sequence from MinD (FtsZ-YFP-mts) were found to preferentially align along the surface of microstructured SLBs, with curvatures similar to the those of the inner face of E. coli cells, as visualized by AFM and total internal reflection fluorescence microscopy (TIRFM) (35). Similarly, the Min proteins were found to assemble in wave-like patterns, powered by ATP hydrolysis, that travel across the membrane surface of SLBs (36). This wave-like behavior exhibits many features of the Min oscillations observed in vivo (37–39).
In this work, we co-reconstituted the essential division FtsZ protein and the site-selection MinCDE complex from E. coli on SLBs containing full-length ZipA, which confers membrane tethering to FtsZ. We followed events occurring on the membrane surface in real time by confocal fluorescence microscopy. In particular, we addressed the question of how the propagating Min waves can modulate the dynamic reorganization of FtsZ polymers when tethered to the membrane by ZipA. This work complements previous studies in which artificial membrane anchors were employed. Specifically, we explored the influence of ZipA surface density and crowding-induced formation of high-order FtsZ polymers on the coupling of the two systems.
Materials and Methods
Materials
Reagents, salts, and buffer components were purchased from Sigma, Merck, and Invitrogen. E. coli polar lipid extract was obtained from Avanti Polar Lipids (Alabaster, AL). All reagents were of analytical or spectroscopic grade.
Protein purification and labeling
The proteins used in this work were overproduced, purified, and fluorescently labeled essentially as previously described (FtsZ (40), ZipA (41), His-tagged MinC, eGFP-MinC (eGFP-MinC fusion protein), MinD, and MinE (36,37)). The degree of labeling of the proteins was typically 0.5–0.7 mol fluorophore/mol protein. The dynamic behavior of the Min proteins, as characterized by velocity, wave period, and wavelength, was essentially the same regardless of the ratio of unlabeled and labeled proteins used.
Small unilamellar vesicles
E. coli polar extract in chloroform was dried by nitrogen flow for 30 min and kept under vacuum for another 2 h to remove organic traces. Multilamellar vesicles (MLVs) were obtained upon hydration of the dried film in SLB buffer (50 mM Tris-HCl, 150 mM KCl, pH 7.5). Small unilamellar vesicles (SUVs) were obtained after a 10 min sonication of the MLVs.
ZipA proteoliposomes
Full-length purified ZipA was reconstituted in E. coli polar extract liposomes according to the standard method described by Rigaud and Lévy (42). Briefly, a fraction of purified ZipA was solubilized in 0.2% Triton-X100. The mixture was added together with E. coli polar extract SUVs described above already solubilized with Triton-X100 (1.5 detergent/lipid molar ratio) and then incubated for 1 h at 4°C under mild stirring. The detergent was removed gradually in four successive dialysis treatments (2 h at 4°C each). In each dialysis step, 10 mg BioBeads (Bio-Rad) per milligram of Triton-X100 was added. After complete detergent removal, the reconstituted proteoliposomes were fractionated and stored at −80°C. To quantify the ZipA/lipid ratio of the proteoliposomes prepared, we used in parallel the methods of quantification of inorganic phosphorus (43) and BCA assay (Pierce) to quantify the amount of lipid and protein, respectively.
SLBs and ZipA-containing bilayers
SLBs and ZipA-containing bilayers (ZipA-SLBs) were obtained by vesicle fusion from SUVs or proteoliposomes, respectively, on a glass surface (44). Thus, a 1 mg/mL suspension of SUVs or proteoliposomes in SLB buffer was added to a glass-bottom chamber together with 2 mM CaCl2 to promote vesicle fusion and deposition of the bilayer. The preparations were incubated at 38°C for 20 min and then washed with the same buffer to remove nonfused vesicles. Exchange of the SLB buffer for the Z buffer (50 mM Tris-HCl, 500 mM KCl, 5 mM MgCl2, pH 7.5) was carried out in a final washing step. The final volume of the samples was 200 μL. The surface density of ZipA (ρZipA) in these bilayers could be controlled by varying the ZipA/lipid ratio on the proteoliposomes. The ρZipA values corresponding to the ZipA-SLBs used in this work are summarized in Table 1. Fluorescence recovery after photobleaching (FRAP) experiments on ZipA-SLBs containing ZipA-Alexa488 revealed that the reconstituted ZipA remained immobile and evenly distributed in the SLBs. The fluorescence intensity of ZipA in the bilayers did not recover completely after photobleaching (data not shown), most probably due to interactions of the ZipA molecules with the glass support underneath hindering diffusion. The dynamics of the Min system did not affect the spatial distribution of ZipA, which, as mentioned above, remained evenly distributed in these bilayers. This was also the case when the Min system and FtsZ were co-reconstructed on the same bilayer.
Table 1.
Five different surface densities of ZipA (ρZipA) reconstructed in our ZipA-SLBs
| ZipA/lipid (molar ratio) | 1:1200 | 1:1600 | 1:2500 | 1:4500 | 1:5500 |
| Distance between two ZipA molecules (nm) | 30 | 35 | 43 | 58 | 64 |
| ZipA content (molar %) | 0.083 | 0.062 | 0.040 | 0.022 | 0.018 |
For each condition assayed, the number of lipids per ZipA molecule in the bilayer, the theoretical average distance between two ZipA molecules, and the ZipA content in the bilayer (molar %) are displayed.
Self-organization assays
Self-organization of Min proteins on ZipA-SLBs was conducted in Z buffer. The experimental concentrations in the assays described in this work were 0.75 μM MinD and 0.75 μM MinE (with or without 0.06 μM MinC) containing ∼40% of labeled proteins and supplemented with 2.5 mM ATP. The experiments were carried out at 23°C. For the coupling assays, Min proteins were incubated on ZipA-SLBs, and then 2 μM FtsZ (15% labeled FtsZ) was added along with 3 mM GTP, 1 U/mL acetate kinase, and 15 mM acetyl phosphate (Sigma) (enzymatic GTP-regenerating system as previously described (45)). In the crowding assays, samples containing preassembled FtsZ networks on ZipA-SLBs were supplemented with Ficoll70 at the corresponding concentrations. Control experiments precluded the addition of an oxygen-scavenging system, which is commonly used to improve dye stability, as unspecific bundling of FtsZ on ZipA-SLBs occurred in the presence of 0.7 μM of glucose oxidase (Fig. S1 in the Supporting Material).
Imaging
Image acquisition was performed on a Zeiss (Jena, Germany) LSM780 confocal laser scanning microscope equipped with a Zeiss Plan-Apochromat 40×/NA 1.2 W corr and Plan-Apochromat 20×/NA 0.8 objectives. Image processing and analyses were carried out with ImageJ (http://rsb.info.nih.gov/ij/). Images were colored in red and green, respectively, to facilitate identification of the labeled protein. The FtsZ waves were quantified by a modulation parameter, m, defined as the relative difference between the maximum and minimum fluorescence intensity measured within the wave: m = (Fmax − Fmin)/Fmax (see Fig. S2). Measurements were taken using the 633 nm laser line at 0.8% laser power in the photon-counting mode. The m values presented correspond to the mean values of at least four independent experiments.
FRAP
The half-time turnover (τ1/2) of FtsZ subunits attached to ZipA-SLBs were obtained from FRAP measurements on the surface of the bilayer for the different samples. Since ZipA remained immobile in these bilayers (ZipA-SLBs), fluorescence recovery after bleaching in these samples was just due to exchange of FtsZ monomers with the solution. For the bleaching experiments, a Zeiss Plan-Apochromat 40×/NA 1.2 W corr objective was used. After a time series of prebleaching images was recorded, a circular area of 5 μm diameter (region of interest) on the bilayer was photobleached by 30 iterations of the 633 nm laser under 100% laser power. Recovery of the bleached area was monitored at 0.8% laser power for 2 min. The fluorescence recovery curves were corrected and normalized for the bleaching caused by imaging and then fitted by a single exponential curve: I(t) = A(1 − e − kt), where I(t) is the fluorescence of the bleached region at time t, A is the mobile fraction, and τ1/2 = ln2/k. The τ1/2 values presented correspond to the mean values of at least three independent experiments (see Fig. S3 A).
Image correlation
A sequence of images of ZipA-SLB with FtsZ under variable conditions (described in the text) was acquired with the Zeiss LSM 780 microscope. Typically, 400 images with pixel size 0.208 μm, 256 × 256 pixels, and 0.243 s per image were recorded. The correlation g(τ) between the fluorescence intensities in the same pixel with a time difference of τ was calculated as (46)
| (1) |
where Fij(t) is the intensity in the pixel (i, j), nij is the number of elements in the sum, and the time t indexes the images in the sequence. Before correlation, an average image, calculated over the whole sequence, was subtracted from all images Fij(t) to eliminate the static features in the images. The correlations were found to decay exponentially and thus were fitted to a single exponential function, with the decay of correlation characterized by the half-time τ1/2 = τc ln2 (see also Fig. S3 B).
Direct stochastical optical reconstruction microscopy imaging
The FtsZ filaments on the ZipA-SLB were imaged by direct stochastical optical reconstruction microscopy (dSTORM), a high-resolution technique based on emitter localization (47). To induce switching of the fluorescent label, mercaptoethyl-amine (MEA) at a concentration of 15 mM was added to the Z buffer. The sample was illuminated with a 640 nm laser in total internal reflection mode using the Zeiss Plan-Apochromat 100×/NA 1.46 oil TIRF objective. Images were recorded with an iXon Ultra DU-897U emCCD camera. In total 20,000 frames with an exposure time of 0.05 s were recorded, and the resulting high-resolution image was reconstructed using rapidSTORM software (48). To create movies, 30 original images with localized emitters were merged to create one frame, corresponding to one frame per 1.5 s in real time.
Estimation of ZipA concentration in vivo
In vivo, ZipA is found evenly distributed in the inner membrane but is concentrated in the septum during division (49). Considering a rod-shaped bacteria with dimensions (2 × 0.8) μm (50) and taking into account that the number of ZipA copied per cell is ∼1000–1500 (49,51), the estimated distance between two ZipA molecules, assuming they were distributed uniformly, would be ∼50 nm. This situation is compatible with an early stage of the division process in which the Min system starts its pole-to-pole oscillation but the Z ring is not targeted at mid-cell yet. This value is equivalent to an ∼1:3200 ZipA/lipid molar ratio or 0.03 molar % of ZipA.
Results
FtsZ polymers anchored to lipid bilayers through ZipA form dynamic networks
The full-length ZipA protein, which provides membrane tethering to FtsZ, was incorporated in E. coli SLBs to reproduce the lipid composition found in the bacterial inner membrane. The surface density of ZipA (ρZipA) in these bilayers (ZipA-SLBs) could be controlled because they were formed by vesicle fusion of previously prepared proteoliposomes containing a known amount of ZipA (see Materials and Methods).
ZipA in the bilayer was found to recruit GDP-FtsZ, as revealed by fluorescence confocal microscopy. Then, upon addition of GTP, an immediate recruitment (within seconds) of FtsZ polymers to ZipA-SLBs was observed, resulting in a 10-fold increase of the relative fluorescence intensity in the vicinity of the membrane when compared with the signal of GDP-FtsZ (Fig. 1, A and B). Furthermore, it was observed that the concentration of GTP-FtsZ at the membrane increased less than linearly with the ZipA concentration (Fig. 1 C). The turnover time of FtsZ subunits within the polymer network assembled (∼3.6 s), as measured by FRAP, was found to be similar to the value determined for FtsZ protofilaments in solution (45) and much faster than that measured for pronounced polymer bundles (24). Thus, it can be assumed that the FtsZ network on the membrane in these reconstitution assays is essentially assembled by protofilaments tethered to the membrane via ZipA. Together with the fast dynamics of the polymer network (Movie S1) and the flexibility of the unstructured domain of ZipA, this might explain why the structural organization of the FtsZ polymers integrating the network could not be resolved by confocal microscopy or by subdiffraction microscopy (dSTORM; Figs. 1 B and S4).
Figure 1.
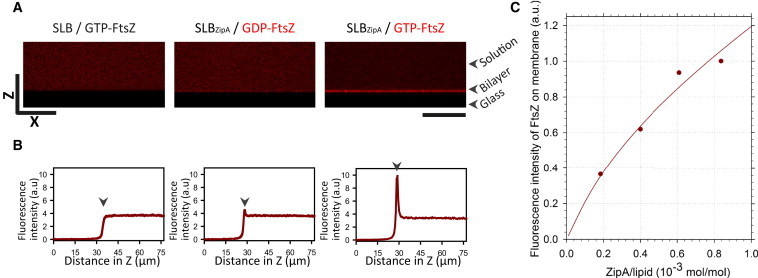
Recruitment of FtsZ to lipid bilayers by the action of ZipA. (A) Vertical confocal image (XZ) showing the distribution of GDP- and GTP-FtsZ on SLBs with and without ZipA. No FtsZ is recruited in the absence of ZipA. Scale bars are 50 μm. (B) FtsZ fluorescence intensity profiles corresponding to the cross sections shown in A. The arrows indicate the position of the bilayer. (C) Fluorescence intensity of FtsZ on the membrane depending on the surface densities of ZipA. To see this figure in color, go online.
Dynamic and plastic multistranded polymer networks compatible with the ones reported here were previously observed in experimental conditions similar to those used in this work (32). Thicker and less dynamic FtsZ polymer bundles were found at greater ZipA surface densities (1–8 molar % of a soluble variant of ZipA (lacking the transmembrane region) attached to the bilayers) and under milder salt conditions (52), which are known to favor interactions between FtsZ polymers. This is consistent with the well-known plasticity and polymorphic nature of FtsZ polymers. These previous studies used high concentrations of membrane-associated FtsZ, lower salt concentrations, and an oxygen-scavenging system. These experimental conditions, which are known to favor the formation of higher-order polymer structures, may explain the discrepancies between those studies and ours. Indeed, we confirmed the formation of FtsZ polymer bundles in low-salt buffers using the soluble form of ZipA attached to the bilayer at high surface density (results not shown). On the other hand, dense filament bundles of FtsZ were observed in supported bilayers when an FtsZ chimera was used at a 1:1 molar ratio together with wild-type FtsZ (24). FtsZ-YFP-mts bypasses the requirement of an external anchor by binding to the membrane through its membrane-targeting sequence. Experiments with FtsZ-YFP-mts recreate conditions of high surface densities of the anchor in which one of every two FtsZ molecules is membrane bound. Once in the membrane, these hybrid protofilaments collide and anneal in thick bundles.
FtsZ polymer networks anchored to the membrane via ZipA reorganize into propagating waves by the action of the Min system
The dynamic behavior of the Min proteins was successfully reconstructed on ZipA-SLBs containing 0.083 molar % of ZipA (equivalent to an average distance of 30 nm between two ZipA molecules). The dynamic behavior of the Min system observed in these experiments, as characterized by velocity, wave period, and wavelength, was similar in the absence or presence of ZipA (data not shown). At the same time, neither the FtsZ binding to ZipA nor the dynamics of the Min system affected the spatial distribution of ZipA in our assays, as it remained evenly distributed and immobile in the bilayers (see Materials and Methods).
Upon reconstitution of the MinCDE complex on the ZipA-SLBs, FtsZ and GTP were added to allow the formation of ZipA-anchored FtsZ polymer networks as described in the previous section. Time-lapse fluorescence confocal microscopy revealed the formation of FtsZ dynamic wave-like patterns (Fig. 2). Those propagating FtsZ waves resulted from the exclusion of FtsZ polymers on the membrane by the Min traveling waves. In particular, it is expected that MinC disrupts interactions between FtsZ subunits, as well as ZipA-FtsZ interactions to a lesser extent.
Figure 2.
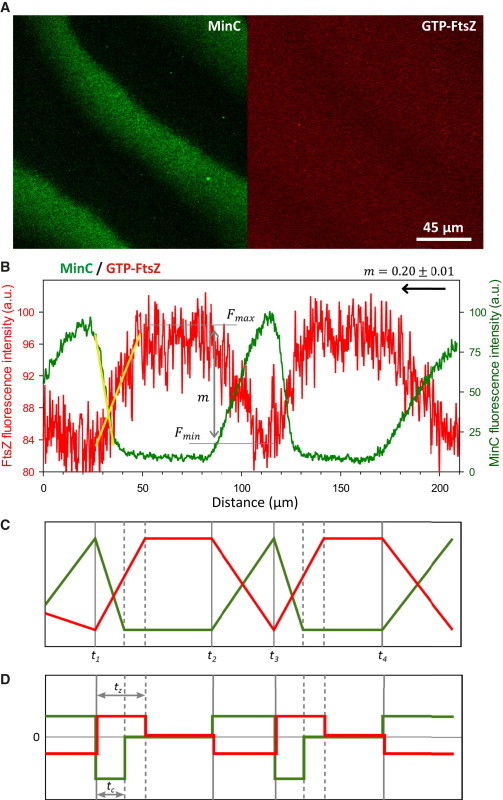
Dynamic coupling of MinCDE and FtsZ propagating waves on ZipA-SLBs. (A) Confocal fluorescence micrographs showing MinCDE (labeled protein: eGFP-MinC) and FtsZ (labeled protein: FtsZ-Alexa647) waves observed upon joint reconstruction on ZipA-SLBs. (B) Fluorescence intensity profiles of MinC and FtsZ acquired from the image shown in A. MinC displaces FtsZ in such a way that the maximal intensity of FtsZ coincides with the minimal signal of MinC. Whereas MinC has a sharp concentration maximum toward the rear of the wave, the maximum of FtsZ is relatively broad. The FtsZ wave observed in these conditions was characterized by a modulation value, m, of 0.20 ± 0.01. The arrow indicates the direction of the propagating wave. The slopes of the FtsZ and MinC profiles are highlighted with yellow lines. The wavelength of this wave is 95 μm, the period is 130 s, and the velocity is 0.73 μm/s. (C) Scheme of the MinC and FtsZ profiles. FtsZ reacts almost immediately to the decrease (times t1 and t3) and increase (times t2 and t4) of MinC concentration on the membrane. (D) Scheme of the derivatives of the MinC and FtsZ profiles. The response of FtsZ to MinC during the membrane attachment phase is somewhat slower than that observed during detachment: MinC reaches its steady state after time tc after the time point t1, whereas FtsZ requires a longer time tz. To see this figure in color, go online.
The fluorescence intensity of the propagating wave-like pattern of FtsZ, which is proportional to the protein concentration at the membrane, reaches its maximium when the fluorescence signal of MinC is minimal, showing that oscillations of FtsZ and Min are in antiphase (Fig. 2 B). Although MinC, like MinE (36,37), has a sharp concentration maximum toward the rear of the wave, the maximum of FtsZ is relatively broad, coinciding with the broad minimum of the MinC wave (Fig. 2 B). The temporal sequence of protein detachment and the shapes of the concentration profiles of Min proteins along the wave in these experiments did not significantly differ from the distributions previously obtained in the absence of FtsZ in vitro (36).
The ability of the MinCDE system to dynamically modulate the dislodgement (and subsequent recruitment) of FtsZ from (to) the membrane, i.e., the degree of coupling between the MinC and FtsZ waves in vitro, can be described by a modulation parameter (m). It is defined as the relative difference between the maximum and minimum fluorescence intensity of the FtsZ wave: m = (Fmax − Fmin)/Fmax (Figs. 2 and S2; see Materials and Methods). If the two systems were not coupled, FtsZ waves would not be observed in the presence of MinC, resulting in a value of m equal to zero. On the other hand, complete depletion of FtsZ in the wave minima would correspond to an m value equal to one. Under the conditions used in our assays (0.083 molar % of ZipA in the bilayer), the modulation value of the FtsZ waves was found to be 0.20 ± 0.01, suggesting a moderate regulation of FtsZ distribution by the Min system. The MinC and FtsZ waves are tightly coupled in time by the inhibitory interaction. Whenever the concentration of MinC starts to change, either by decreasing (times t1 and t3 in Fig. 2 C) or increasing (times t2 and t4), FtsZ reacts almost immediately with a change in the opposite direction. However, the increase of FtsZ is somewhat slower than the change in MinC concentration: after the time t1 (Fig. 2, C and D) MinC reaches a steady state within tc ∼10 s, whereas FtsZ requires a longer time, tz ∼20 s. This slower response of FtsZ is consistent with the response of the FtsZ network to MinC occurring on a timescale on the order of 10 s (24). In contrast, the opposite process—the decline of FtsZ with an increase of MinC—is tightly coupled in duration.
The presence of MinC, the element of the Min system that impairs FtsZ assembly, was essential in these experiments, as FtsZ waves were not observed on ZipA-SLBs with only MinD and MinE present (data not shown). The concentration of MinC in solution did not significantly affect the modulation value m within the range of 0.06–0.3 μM (data not shown). Since the recruitment of MinC to the membrane by MinD results in a great increase in the concentration at the membrane (53), small changes in the bulk concentration of MinC are not expected to have a big impact on the dynamics of the system. We consistently obtained these results using different combinations of fluorescently labeled proteins as tracers (data not shown).
The modulation of FtsZ waves by MinC depends on the ZipA concentration
We tested the recruitment of FtsZ to the bilayer in our system by reconstituting ZipA at different surface densities, from 0.018 to 0.083 molar % of ZipA in the bilayer, covering the estimated range of ZipA concentrations in the bacterial membrane (see Supporting Material). Preparations containing higher ZipA concentrations resulted in the formation of nonhomogeneous bilayers due to their high protein content, thus precluding their use. We observed that ZipA density modulates the above-mentioned coupling, as evidenced by the direct correlation observed between the modulation of the FtsZ waves and the amount of ZipA incorporated in the bilayer (Figs. 1 C and 3, A and B). The wave modulation is stronger at higher ZipA densities, suggesting that the interaction of FtsZ with the membrane is modified in such a way that makes it more susceptible to the action of MinC.
Figure 3.
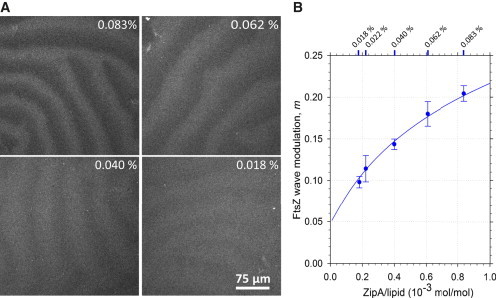
Effect of ZipA surface density on modulation of the FtsZ waves mediated by the Min system. (A) Confocal fluorescence micrographs showing FtsZ waves (traced by FtsZ-Alexa647) on bilayers with different surface densities of ZipA (ZipA content as molar %). (B) Modulation of the FtsZ waves, m, depending on the surface density of ZipA on the membrane (●). The data shown correspond to the mean values of at least four independent experiments plotted with the corresponding standard error. The line shows the fitting of the proposed model to the experimental data. The fit parameters were k2/k−2 = 10.1 × 103 lipid, k−1/k1 = 1.89. Fig. S6B shows the concentrations bi of FtsZ filaments anchored with i ZipA molecules for this fit. To see this figure in color, go online.
The increase in wave modulation with the ZipA concentration observed in our experiments suggests that the structural organization of membrane-tethered FtsZ and/or its interaction with ZipA is modified at higher concentrations of ZipA. This leads to an increased susceptibility of FtsZ to MinC at the membrane. In addition, the experiments show that the FtsZ concentration at the membrane increases less than linearly with the ZipA concentration (Fig. 1 C). One plausible explanation for this is that at higher ZipA densities, single FtsZ filaments are anchored to the membrane by several ZipA molecules, bringing them closer to the membrane and therefore increasing their rate of encounters with membrane-bound MinC (Fig. S6 A). This idea leads to a model based on the following reactions:
| (2) |
| (3) |
Free ZipA binds FtsZ from the solution (with a rate constant k1) to form FtsZ complexes anchored to the membrane with one ZipA molecule (species FtsZ1, concentration b1). FtsZ can also dissociate from ZipA (with a rate constant k − 1) resulting in its dislodgment from the membrane. Additionally, FtsZ anchored to the membrane with i ZipA molecules (species FtsZi, concentration bi) can either bind one more ZipA (with a rate constant k2) to form FtsZ complexes anchored with i + 1 molecules, or dissociate one ZipA (with a rate constant k − 2) to form FtsZ anchored with i − 1 molecules. The total concentration of ZipA on the membrane is z0 and the concentration of free (not bound to FtsZ) ZipA is z, with the rest of ZipA being bound to FtsZ. The following rate equations then describe the system:
| (4) |
| (5) |
| (6) |
| (7) |
where N is the maximum number of ZipA molecules bound to one FtsZ filament (limited by the filament length). We assume steady state, with all time derivatives on the left side set to zero. The total concentration of membrane-bound FtsZ b is then
| (8) |
and the FtsZ fluorescence on the membrane is proportional to b: F = αb (Fig. 1 C). The FtsZ filaments with more ZipA anchors will be more susceptible to MinC (we can assume that the susceptibility of a filament will be directly proportional to the number of its ZipA anchors). The susceptibility of all of the bound FtsZ will then be proportional to , and the wave modulation m will be proportional to the fraction of susceptible FtsZ relative to the total FtsZ:
| (9) |
Fitting this model to our data, the dependencies of FtsZ intensity on the membrane (Fig. 1 C) and FtsZ wave modulation (Fig. 3 B) show that this model can describe our main experimental observations, i.e., that the FtsZ concentration increases less than linearly with ZipA (Fig. 1 C), and the FtsZ wave modulation increases with ZipA (Fig. 3 B). Fig. S6 B shows the concentrations bi of FtsZ filaments anchored with i ZipA molecules for this fit.
Notably, we found that alternative schemes to explain the Min-FtsZ coupling did not describe our experimental observations. One of them assumes that MinC could have a maximum capacity of FtsZ that it can remove from the membrane during the pass of the wave. This would mean that always the same amount of FtsZ would be removed, regardless of the total amount of FtsZ on the membrane. This would lead to decreasing modulation m with higher ZipA, which is the opposite of what was observed in this work. Another possibility is that MinC is able to remove FtsZ from the membrane with an effective rate proportional to the concentrations of both MinC and FtsZ. In this case, always the same fraction of FtsZ would be removed independently of the ZipA concentration, resulting in a constant wave modulation m, which again does not describe the experimental observations.
The anchoring of FtsZ to the membrane through ZipA was required for the formation of anticorrelated FtsZ and MinCDE patterns. In the absence of ZipA, a small fraction of FtsZ was found to be transported within the MinCDE wave, giving rise to distinct correlated MinCDE and FtsZ patterns (referred to as MinCDE-FtsZ waves; Fig. 4). In contrast to the FtsZ waves in the presence of ZipA (Fig. 2), in its absence the FtsZ profile is correlated with the MinC profile (Fig. 4). Although the MinC and MinD profiles are similar, when compared with the FtsZ profile without ZipA, FtsZ is closer to the MinC wave than to the MinD wave (Fig. S5). The maxima of the FtsZ profile on these MinCDE-FtsZ waves are ∼4-fold less intense than the maxima reached by FtsZ waves on ZipA-SLBs. In this situation, where ZipA is not present in the bilayer to tether FtsZ, the colocalization of MinC and FtsZ might indicate a weak transient interaction between MinC and GTP-FtsZ (no patterns were observed in the presence of GDP).
Figure 4.
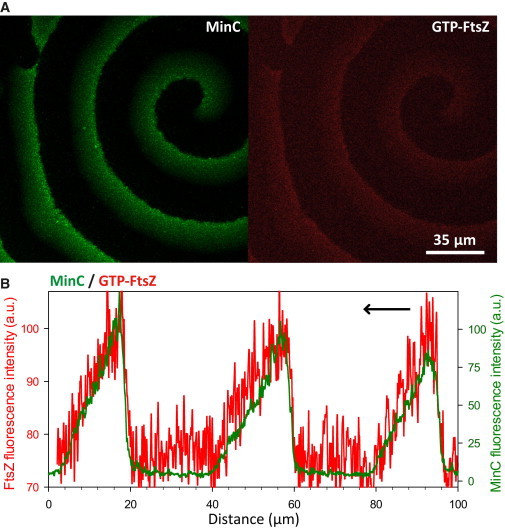
ZipA is required for the anticorrelated coupling of MinCDE and FtsZ. (A) Confocal fluorescence micrographs showing MinCDE-FtsZ waves (labeled proteins: eGFP-MinC and FtsZ-Alexa647) observed upon joint reconstruction on bilayers without ZipA. (B) Fluorescence intensity profiles of MinC and FtsZ acquired from the image shown in (A). In the absence of ZipA, a small fraction of FtsZ was found to be transported with the MinCDE wave by binding to MinC. In contrast to the situation described for ZipA-SLBs, where anticorrelated FtsZ and MinCDE patterns were observed, here FtsZ colocalizes with MinC, matching its profile. To see this figure in color, go online.
Crowding enhances the coupling of membrane-anchored FtsZ polymers to Min waves
High concentrations of biochemically inert macromolecules, such as Ficoll, have commonly been used to simulate the natural crowding conditions found on the cytoplasm of E. coli. In the case of FtsZ, Ficoll70 is known to promote the formation of dynamic, multistranded FtsZ polymer networks resulting from lateral interactions between protofibrils, which are thermodynamically favored in crowded environments because they occupy less of the total volume (45,54). As expected, bundles of FtsZ fibrils on ZipA-SLBs were found in the presence of high concentrations of Ficoll70 (Fig. 5). The addition of this crowding agent retarded the exchange of FtsZ subunits within the membrane-tethered polymer by up to 3-fold when compared with the exchange measured in the absence of Ficoll (see above), as revealed by FRAP and correlation measurements (Figs. 5 C and S3). The slowdown of the subunit turnover within the network by the increase of Ficoll70 illustrates the formation of higher-order structures in a Ficoll-dependent manner, being compatible with the values previously measured in solution (45,55). At the same time, the effect of Ficoll70 on the Min wave propagation was addressed. In the presence of increasing amounts of this crowder, both the wavelength (λ) and velocity (ν) of the Min waves decreased, whereas the period (T, T= λ/ν) remained practically independent of Ficoll70 (∼100 s; Fig. S7). This effect can be tentatively linked to the lower diffusion coefficient of the Min proteins at higher Ficoll70 concentrations.
Figure 5.
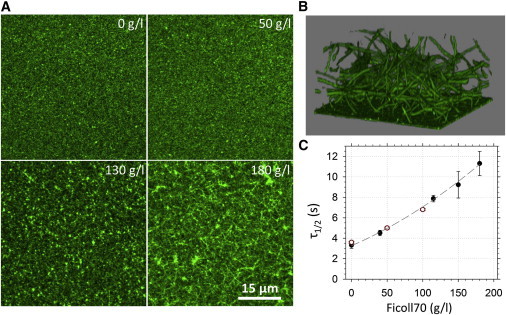
Confocal microscopy analysis of crowding-induced FtsZ assembly in ZipA-SLBs. (A) Confocal fluorescence images showing FtsZ networks formed on ZipA-SLBs in the presence of 0, 50, 130, and 180 g/L of Ficoll70. (B) 3D reconstruction of confocal fluorescence images showing Ficoll-induced FtsZ bundles on ZipA-SLBs. The image was taken at 190 g/L Ficoll70. These dense and dynamic 3D networks localize in solution and are connected to the bilayer by some fibers. Frame size: 45 μm × 45 μm (4.4 nm per pixel). (C) Half-time turnover (τ1/2) of FtsZ subunits on the membrane depending on the Ficoll concentration, as determined by FRAP (●) and image correlation (○). Ficoll70 significantly retards the subunit exchange within the polymer. The dashed line is an arbitrary curve drawn to guide the eye. The data shown correspond to the mean values of at least three independent measurements plotted with the corresponding standard error. To see this figure in color, go online.
The turnover rate of FtsZ at ∼135 g/L Ficoll70 was found to be ∼9 s, which is similar to the half-time of recovery of the bleached FtsZ region at the Z ring measured in vivo (55). Above this threshold concentration of Ficoll70, high-order FtsZ dynamic polymer filaments and networks were thick enough to be resolved by fluorescence confocal microscopy (Fig. 5, A and B). Under these conditions, which maximize the lateral interactions between FtsZ fibers, the modulation of the membrane-tethered FtsZ waves by MinC was substantially enhanced (m = 0.55 ± 0.01) when compared with the coupling between the two systems measured in the absence of crowders (m = 0.20 ± 0.01) (Fig. 6; Movie S2). As noted above, the presence of crowders enhances the lateral interactions between FtsZ protofilaments, thus increasing the effective concentration of FtsZ on the membrane. This FtsZ network would contain more stretches of FtsZ not directly anchored to the membrane by ZipA. Thus, the substantial increase in wave modulation could be explained as follows: if MinC broke the network at these nonanchored parts, larger fragments of FtsZ could be released upon breakage, effectively enhancing the action of MinC, meaning that more FtsZ would be released after the same number of breakage events.
Figure 6.
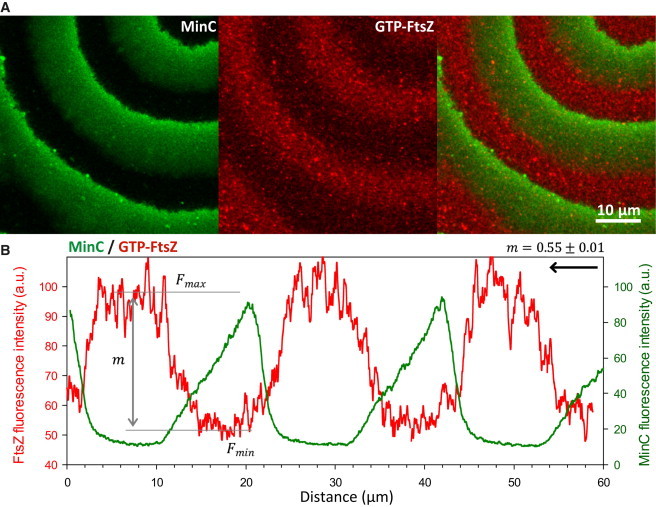
Dynamic coupling of crowding-induced FtsZ polymers in ZipA-SLBs to Min propagating waves. (A) Confocal fluorescence micrographs showing MinCDE waves (traced by eGFP-MinC) and the FtsZ network (traced by FtsZ-Alexa647) to ZipA-SLBs in the presence of 130 g/L Ficoll. (B) Fluorescence intensity profiles of MinC and FtsZ acquired from the image shown in (A). The modulation value, m, of the FtsZ observed in these conditions was 0.55 ± 0.01. The arrow indicates the direction of the propagating wave. See also Movie S2. To see this figure in color, go online.
Discussion
In this work, we investigated the interaction between the site-selection MinCDE system and two proteins that assemble in the E. coli proto-ring (the soluble FtsZ and the membrane-bound ZipA) in lipid bilayers as minimal membrane systems. We found that the Min system drives the dynamic distribution of FtsZ polymers when they are tethered to the membrane by ZipA, as revealed by confocal microscopy analysis, resulting in the dislodgment of FtsZ from the membrane and the subsequent generation of FtsZ wave-like patterns (Fig. 7 A). These rearrangements are mediated by MinC, the element of the Min system that acts as an inhibitor of FtsZ assembly. The dynamic coupling between the Min proteins and membrane-associated FtsZ polymers is facilitated by experimental conditions that promote the accumulation of more FtsZ molecules in the vicinity of the membrane, as an increase in the ZipA surface concentration or crowding induces the formation of dynamic FtsZ polymer networks, leading to a relatively stronger exclusion of FtsZ from regions in which MinC waves reaches a maximum (Figs. 4 and 6).
Figure 7.
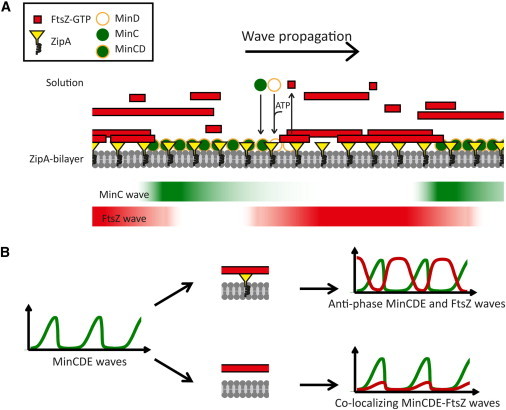
(A) Model showing the dynamic distribution of FtsZ polymers coupled to Min oscillations in membranes containing ZipA. In the Min system, MinD and MinE govern this oscillation behavior. MinC is a weak inhibitor of FtsZ polymerization that together with the membrane-bound MinD forms an inhibitory complex, preventing FtsZ polymers from accumulating in its vicinity. MinE (omitted in this figure for simplicity) acts on the membrane-bound MinCD complex and induces its dissociation. In this work, the MinCDE waves were observed by time-resolved confocal imaging as they displaced the fluorescent signal of FtsZ from the membrane. Thus, a maximal concentration of MinC correlates with a minimal concentration of FtsZ on the membrane. The Min-FtsZ coupling is facilitated at high surface densities of ZipA and enhanced by crowding-induced lateral interactions of FtsZ polymers (for simplicity, crowders are not represented here). (B) Effect of membrane recruitment of FtsZ polymers by ZipA: FtsZ waves in antiphase with the MinCDE waves are formed when FtsZ is anchored to the membrane through ZipA, resulting in an efficient dislodgment of FtsZ from the membrane regions where MinC is present. Correlated MinCDE and FtsZ waves are found when FtsZ is not anchored to the membrane by ZipA. To see this figure in color, go online.
The reconstruction strategy we used allowed us to investigate the influence of membrane tethering on the response of FtsZ polymers to the action of the oscillating Min system (Fig. 7). In this way, we found that although Min-mediated FtsZ waves are formed when FtsZ is associated to the membrane through ZipA, they are not observed in the absence of ZipA, when FtsZ is free in solution. Interestingly, in this case, correlated MinCDE and FtsZ waves emerged in the presence of GTP to trigger FtsZ polymerization, as further elaborated below (Fig. 7 B).
From our results, we can conclude that the association of FtsZ to the membrane by ZipA plays an important role in the counter-oscillations of FtsZ polymers promoted by the Min system. This is consistent with the Min-driven FtsZ waves observed using a variant of FtsZ (FtsZ-YFP-mts) containing the membrane-targeting sequence of MinD to bypass the need for the natural proto-ring tethering proteins (6,24,56). The interaction of FtsZ with ZipA takes place through the C-terminal end of FtsZ (57), which also interacts with the C-terminal domain of MinC (22) and other division-related proteins (10,58), acting as a central hub to integrate multiple interactions that modulate the assembly of the division machinery. Our findings suggest that the affinity of MinC (in complex with MinD at the membrane) for FtsZ is higher than the association of FtsZ with ZipA, thus resulting in the displacement of FtsZ polymers from the membrane. The latter is compatible with the well-established role of the interaction between the C-terminal regions of MinC and FtsZ in disrupting the lateral associations of FtsZ molecules in the polymer (22). A significant fraction of the FtsZ subunits in the membrane-bound polymer network will have C-terminal regions accessible for interactions with MinC, allowing them to respond also to MinC and leading to a strong FtsZ coupling to the Min waves, as occurs in the case of multistranded polymers formed in macromolecularly crowded solutions.
The experiments showing that a fraction of soluble FtsZ polymers comigrated with Min waves in the absence of ZipA provide additional clues as to the mechanisms of interaction between MinC and FtsZ. These correlated MinCDE and FtsZ waves were not found in the presence of GDP, suggesting that GTP-containing FtsZ polymers, but not GDP forms, bind to MinCD at the membrane. This behavior may indicate that the interaction between MinCD and GTP-FtsZ polymers at the membrane is stronger than the interaction between MinCD and GDP-FtsZ species. Structural changes in MinC as it interacts with MinD at the membrane may explain why MinC alone, in contrast, preferentially binds to GDP-bound FtsZ (24,59).
The observations reported here strongly suggest that ZipA may play an important regulatory role in Z-ring placement guided by the Min system in E. coli cells. It is known that the recruitment of ZipA and FtsA to the division ring at mid-cell depends on FtsZ (60,61). Consistent with our findings, one of the functions of the Min proteins may be to couple their dynamic oscillating behavior to guide the attachment of FtsZ to the membrane by the action of ZipA and, to a lesser extent by FtsA, given the rapidly reorganizing FtsZ polymers driven by FtsA (52).
Our results may also help to clarify some of the puzzling features of the proposed stages in the assembly of the E. coli proto-ring. FtsA may provide a link between FtsZ and the membrane weaker than that conferred by ZipA, as both the interactions between FtsA and the membrane, and between FtsZ and FtsA are thought to be weaker than the equivalent interactions provided by ZipA. Additionally, FtsA is displaced from the Z ring in vivo more readily than ZipA (22). However, the initial stage of proto-ring assembly may still be orchestrated by the association of FtsZ to the membrane through FtsA if, for example, soluble FtsZ-FtsA hetero-complexes are the ones that preferentially bind to the membrane. In the absence of ZipA, these FtsZ polymers, once associated to the membrane, would be very dynamic, in agreement with the coordinated streams of FtsZ polymers and chirally rotating rings recently observed upon the co-reconstitution of FtsZ and FtsA in lipid bilayers (52). This observation can be reconciled with the role of ZipA in proto-ring stabilization if after the first stage of assembly, driven by FtsA, a second stage involving a competition between ZipA and FtsA for the C-terminus of FtsZ (given the relatively stronger binding affinity for FtsZ of ZipA) occurs, as a result of which FtsA is replaced by ZipA in the attachment of FtsZ to the membrane. In this way, FtsA is then free to recruit late division proteins, such as FtsN and FtsI (62), to assemble the molecular machinery for cytokinesis.
In line with the results described here, dynamic FtsZ oscillations have also been observed in E. coli cells, showing a period of oscillation similar to that of the Min proteins (56,63,64). Interestingly, in these in vivo studies it was found that MinCDE drives the counter-oscillation of early division proteins, such as ZipA, along with FtsZ (56), suggesting that a fraction of ZipA moves along the membrane with FtsZ. Our observations of anticorrelated waves of FtsZ driven by the Min system in minimal membranes, obtained under defined conditions, complement the in vivo counter-oscillation of FtsZ along with ZipA reported by Bisicchia et al. (56). The high complexity of the cellular environment precludes us from proposing a simple molecular description of the observed co-oscillation of FtsZ and ZipA. Therefore, it remains to be determined whether the observed mobility is conferred by, for example, the interaction between ZipA and FtsZ-GDP or even the interaction of ZipA with other division proteins.
Conclusions
The data presented here show that the bacterial division protein FtsZ, when associated to the membrane through ZipA, assembles into antiphase dynamic patterns by the action of the site-selection Min system. The presence of MinC is required for the formation of the FtsZ propagating waves, suggesting that this FtsZ-ring inhibitor, when complexed to MinD, has a higher affinity for the central hub of FtsZ than ZipA, resulting in the displacement of FtsZ polymers from the membrane. The reconstruction approach adopted here, using ZipA as a natural membrane tether of FtsZ, is uniquely suited for exploring the impact of FtsZ membrane attachment on Min-FtsZ coupling. In this way, we have shown that when FtsZ is free in solution, a fraction of FtsZ polymers comigrate with the Min waves instead of forming the Min-mediated antiphase FtsZ waves observed in ZipA-containing membranes or membrane-targeted FtsZ-mts mutants. This evidence suggests that ZipA, in addition to its role in proto-ring stability, is of central importance for the dynamic coupling between the self-oscillating Min system and the remodeling of FtsZ polymers at the early stages of division in E. coli, contributing to the efficient selection of the division site. This study may help to elucidate the conditions needed to generate Min-FtsZ dynamic patterns inside cell-like compartments, eventually leading to a controlled division of these containers.
Acknowledgments
We are grateful to Silvia Zorrilla and Begoña Monterroso for useful discussions and comments, to Armin Lambacher and Daniela-Azucena García for assistance with the experiments, and to Katja Zieske and Beatrix Scheffer for MinC and MinDeGFP purification, respectively. We also thank the anonymous reviewers for useful remarks.
This work was supported in part by the Human Frontiers Science Program through grant RGP0050/2010-C102 (to P.S. and G.R.), the DFG Leibniz Prize (to P.S.), the European Commission through contract HEALTH-F3-2009-223431 (to G.R.), and the Spanish Government through grant BIO2011-28941-C03-03 (to G.R.).
Contributor Information
Germán Rivas, Email: grivas@cib.csic.es.
Petra Schwille, Email: schwille@biochem.mpg.de.
Supporting Material
Thirty original images with localized emitters were merged to create one movie frame, resulting in one frame per 1.5 s in real time. Frame size: 10 μm × 10 μm. See also Fig. S4.
Sequence of confocal images showing MinCDE waves (traced by MinC-eGFP) and the FtsZ network (traced by FtsZ-Alexa647) on a ZipA-SLB in the presence of 130 g/L Ficoll70. The modulation value, m, of the FtsZ observed in these conditions was 0.55 ± 0.01. Images were taken every 20 s. Frame size: 53 μm × 53 μm. See also Fig. 6.
References
- 1.Rico A.I., Krupka M., Vicente M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J. Biol. Chem. 2013;288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowlett V.W., Margolin W. The bacterial Min system. Curr. Biol. 2013;23:R553–R556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 3.Adams D.W., Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 4.Lutkenhaus J., Pichoff S., Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu L.J., Errington J. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 2012;10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 6.Zieske K., Schwille P. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife. 2014;3:3. doi: 10.7554/eLife.03949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson H.P., Anderson D.E., Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mingorance J., Rivas G., Vicente M. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 2010;18:348–356. doi: 10.1016/j.tim.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Meier E.L., Goley E.D. Form and function of the bacterial cytokinetic ring. Curr. Opin. Cell Biol. 2014;26:19–27. doi: 10.1016/j.ceb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Haney S.A., Glasfeld E., de Boer P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 2001;276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 11.Pichoff S., Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geissler B., Shiomi D., Margolin W. The ftsA∗ gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology. 2007;153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichoff S., Shen B., Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoff S., Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi T., Hale C.A., Erickson H.P. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 2002;184:4313–4315. doi: 10.1128/JB.184.15.4313-4315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazos M., Natale P., Vicente M. A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J. Biol. Chem. 2013;288:3219–3226. doi: 10.1074/jbc.M112.434944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geissler B., Elraheb D., Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Boer P.A., Crossley R.E., Rothfield L.I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer P.A., Crossley R.E., Rothfield L.I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z., Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 21.Meinhardt H., de Boer P.A. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA. 2001;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen B., Lutkenhaus J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol. Microbiol. 2009;72:410–424. doi: 10.1111/j.1365-2958.2009.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen B., Lutkenhaus J. Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol. Microbiol. 2010;75:1285–1298. doi: 10.1111/j.1365-2958.2010.07055.x. [DOI] [PubMed] [Google Scholar]
- 24.Arumugam S., Petrašek Z., Schwille P. MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl. Acad. Sci. USA. 2014;111:E1192–E1200. doi: 10.1073/pnas.1317764111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dajkovic A., Lan G., Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 26.Martos A., Jiménez M., Schwille P. Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 2012;22:634–643. doi: 10.1016/j.tcb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Natale P., Pazos M., Vicente M. The Escherichia coli divisome: born to divide. Environ. Microbiol. 2013;15:3169–3182. doi: 10.1111/1462-2920.12227. [DOI] [PubMed] [Google Scholar]
- 28.Jiménez M., Martos A., Rivas G. Giant vesicles: a powerful tool to reconstruct bacterial division assemblies in cell-like compartments. Environ. Microbiol. 2013;15:3158–3168. doi: 10.1111/1462-2920.12214. [DOI] [PubMed] [Google Scholar]
- 29.Schwille P., Diez S. Synthetic biology of minimal systems. Crit. Rev. Biochem. Mol. Biol. 2009;44:223–242. doi: 10.1080/10409230903074549. [DOI] [PubMed] [Google Scholar]
- 30.Loose M., Schwille P. Biomimetic membrane systems to study cellular organization. J. Struct. Biol. 2009;168:143–151. doi: 10.1016/j.jsb.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Vogel S.K., Schwille P. Minimal systems to study membrane-cytoskeleton interactions. Curr. Opin. Biotechnol. 2012;23:758–765. doi: 10.1016/j.copbio.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Mateos-Gil P., Márquez I., Vélez M. FtsZ polymers bound to lipid bilayers through ZipA form dynamic two dimensional networks. Biochim. Biophys. Acta. 2012;1818:806–813. doi: 10.1016/j.bbamem.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 33.López-Montero I., Mateos-Gil P., Monroy F. Active membrane viscoelasticity by the bacterial FtsZ-division protein. Langmuir. 2012;28:4744–4753. doi: 10.1021/la204742b. [DOI] [PubMed] [Google Scholar]
- 34.Mateos-Gil P., Paez A., Vélez M. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. USA. 2012;109:8133–8138. doi: 10.1073/pnas.1204844109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arumugam S., Chwastek G., Schwille P. Surface topology engineering of membranes for the mechanical investigation of the tubulin homologue FtsZ. Angew. Chem. Int. Ed. Engl. 2012;51:11858–11862. doi: 10.1002/anie.201204332. [DOI] [PubMed] [Google Scholar]
- 36.Loose M., Fischer-Friedrich E., Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 37.Loose M., Fischer-Friedrich E., Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat. Struct. Mol. Biol. 2011;18:577–583. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- 38.Schweizer J., Loose M., Schwille P. Geometry sensing by self-organized protein patterns. Proc. Natl. Acad. Sci. USA. 2012;109:15283–15288. doi: 10.1073/pnas.1206953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zieske K., Schwille P. Reconstitution of pole-to-pole oscillations of min proteins in microengineered polydimethylsiloxane compartments. Angew. Chem. Int. Ed. Engl. 2013;52:459–462. doi: 10.1002/anie.201207078. [DOI] [PubMed] [Google Scholar]
- 40.Rivas G., López A., Andreu J.M. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. J. Biol. Chem. 2000;275:11740–11749. doi: 10.1074/jbc.275.16.11740. [DOI] [PubMed] [Google Scholar]
- 41.RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigaud J.L., Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 43.Rouser G., Siakotos A.N., Fleischer S. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1966;1:85–86. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- 44.Brian A.A., McConnell H.M. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. USA. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González J.M., Jiménez M., Rivas G. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 2003;278:37664–37671. doi: 10.1074/jbc.M305230200. [DOI] [PubMed] [Google Scholar]
- 46.Wiseman P.W., Petersen N.O. Image correlation spectroscopy. II. Optimization for ultrasensitive detection of preexisting platelet-derived growth factor-beta receptor oligomers on intact cells. Biophys. J. 1999;76:963–977. doi: 10.1016/S0006-3495(99)77260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Linde S., Löschberger A., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 48.Wolter S., Löschberger A., Sauer M. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods. 2012;9:1040–1041. doi: 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]
- 49.Rueda S., Vicente M., Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundararaj S., Guo A., Wishart D.S. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 2004;32:D293–D295. doi: 10.1093/nar/gkh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hale C.A., de Boer P.A. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 2002;184:2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loose M., Mitchison T.J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Z., Mukherjee A., Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivas G., Alfonso C., Zorrilla S. Macromolecular interactions of bacterial cell division FtsZ protein: from quantitative biochemistry and crowding to reconstructing minimal divisomes in the test tube. Biophys. Rev. 2013;5:63–77. doi: 10.1007/s12551-013-0115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson D.E., Gueiros-Filho F.J., Erickson H.P. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bisicchia P., Arumugam S., Sherratt D. MinC, MinD, and MinE drive counter-oscillation of early-cell-division proteins prior to Escherichia coli septum formation. MBio. 2013;4 doi: 10.1128/mBio.00856-13. e00856–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosyak L., Zhang Y., Somers W.S. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma X., Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernández-Rocamora V.M., García-Montañés C., Rivas G. MinC protein shortens FtsZ protofilaments by preferentially interacting with GDP-bound subunits. J. Biol. Chem. 2013;288:24625–24635. doi: 10.1074/jbc.M113.483222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Addinall S.G., Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hale C.A., de Boer P.A. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rico A.I., García-Ovalle M., Vicente M. Role of Escherichia coli FtsN protein in the assembly and stability of the cell division ring. Mol. Microbiol. 2010;76:760–771. doi: 10.1111/j.1365-2958.2010.07134.x. [DOI] [PubMed] [Google Scholar]
- 63.Thanedar S., Margolin W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tonthat N.K., Milam S.L., Schumacher M.A. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc. Natl. Acad. Sci. USA. 2013;110:10586–10591. doi: 10.1073/pnas.1221036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Thirty original images with localized emitters were merged to create one movie frame, resulting in one frame per 1.5 s in real time. Frame size: 10 μm × 10 μm. See also Fig. S4.
Sequence of confocal images showing MinCDE waves (traced by MinC-eGFP) and the FtsZ network (traced by FtsZ-Alexa647) on a ZipA-SLB in the presence of 130 g/L Ficoll70. The modulation value, m, of the FtsZ observed in these conditions was 0.55 ± 0.01. Images were taken every 20 s. Frame size: 53 μm × 53 μm. See also Fig. 6.


