Abstract
Nanopore sensing involves an electrophoretic transport of analytes through a nanoscale pore, permitting label-free sensing at the single-molecule level. However, to date, the detection of individual small proteins has been challenging, primarily due to the poor signal/noise ratio that these molecules produce during passage through the pore. Here, we show that fine adjustment of the buffer pH, close to the isoelectric point, can be used to slow down the translocation speed of the analytes, hence permitting sensing and characterization of small globular proteins. Ubiquitin (Ub) is a small protein of 8.5 kDa, which is well conserved in all eukaryotes. Ub conjugates to proteins as a posttranslational modification called ubiquitination. The immense diversity of Ub substrates, as well as the complexity of Ub modification types and the numerous physiological consequences of these modifications, make Ub and Ub chains an interesting and challenging subject of study. The ability to detect Ub and to identify Ub linkage type at the single-molecule level may provide a novel tool for investigation in the Ub field. This is especially adequate because, for most ubiquitinated substrates, Ub modifies only a few molecules in the cell at a given time. Applying our method to the detection of mono- and poly-Ub molecules, we show that we can analyze their characteristics using nanopores. Of particular importance is that two Ub dimers that are equal in molecular weight but differ in 3D structure due to their different linkage types can be readily discriminated. Thus, to our knowledge, our method offers a novel approach for analyzing proteins in unprecedented detail using solid-state nanopores. Specifically, it provides the basis for development of single-molecule sensing of differently ubiquitinated substrates with different biological significance. Finally, our study serves as a proof of concept for approaching nanopore detection of sub-10-kDa proteins and demonstrates the ability of this method to differentiate among native and untethered proteins of the same mass.
Introduction
Ubiquitin (Ub) is an evolutionarily conserved, highly stable, 8.5 kDa protein that adopts a compact fold with a flexible six-residue C-terminal tail (1). Its well-conserved structure throughout all eukaryotes, from yeast to human, suggests high evolutionary pressure to maintain its crucial function for the survival of any eukaryotic cell (2). Ubiquitination is a posttranslational-modification cellular process, in which the C-terminal glycine residue of Ub is covalently attached to the ε-NH2 of a lysine side chain in a substrate protein (3). The Ub conjugation system is complex due to the versatility of Ub modifications (4). Ub can attach to the substrate as one moiety on one or multiple lysines, modifying the target by mono- or multiubiquitination, respectively. Furthermore, Ub forms polyUb chains, where one Ub moiety is conjugated to another Ub via one of the seven lysine residues on its amino acid sequence (5). It is important to point out that the specific lysine linkage type has unique biological significance; thus, it is often referred to as the Ub code (5). Thus far, all seven possible linkage types have been found to be relevant in vivo, as have mixed linkages, branched chains, and linear chains (5). Protein ubiquitination serves as a signal to target proteins for degradation by the proteasome or in lysosomes, modulate protein function or localization (4). Often, just a few substrate molecules conjugated to Ub are sufficient for initiating the processes signaled by the Ub modification. Hence, there is interest in developing single-molecule approaches for discrimination of the Ub chain properties.
One of the main challenges of such a tool is to identify the poly-Ub chain type on a particular substrate and thereby decipher the physiological importance of this modification. Often regarded as a model ubiquitinated substrate, the proliferating nuclear antigen, the DNA polymerase clamp essential for replication, is differentially modified on the same residue, activating different DNA damage tolerance pathways (6,7). The common approach to study the poly-Ub chain type is to use various Ub mutants in which six of seven lysines are replaced by arginines, thus allowing only one type of chain to form in each case. As an alternative, researchers often employ Western blot analysis using linkage-specific antibodies, the recognition epitopes of which lie in the Ub linkage site (8,9). However, producing antibodies with high reactivity and specificity is a highly laborious and complicated process. Mass spectrometry studies conducted in recent years have contributed enormously to our understanding of the Ub field, revealing thousands of ubiquitinated targets (10). Nonetheless, identification of the ubiquitinated site or the polyUb linkage type is still often precluded by technical limitations (11). Interestingly, two Ub dimers that are identical in molecular weight but differ in linkage type may attain different conformations, thus providing a structural basis for their biological difference (12,13). This was revealed by structural studies using tools such as nuclear magnetic resonance and x-ray crystallography (13,14). The disadvantage of the latter is that it can only capture a snapshot of the structure in the crystal, whereas NMR experiments are performed in solution and can dissect a range of conformations (15). However, both techniques require a large amount of highly purified material, and to date, the ability to distinguish among similar protein complexes at the single-molecule level remains limited.
Nanopore (NP) sensing is a relatively new method for physical characterization of single biomolecules. It typically consists of an ultrathin, electrically insulating membrane in which a single nanometer-scale pore is fabricated. The membrane is immersed in an electrolyte aqueous solution, creating two small chambers connected only by the NP. Two electrodes dipped in either of the chambers are used to apply an electrical potential across the pore, resulting in an electrophoretic force to attract charged biomolecules to the pore, where the molecules are sensed. As a biomolecule translocates through the pore it partially blocks the ionic current flowing through the NP, giving rise to an electrical signal that changes with the biomolecule length, cross section, and charge, as well as other properties (16). Solid-state NPs offer a large flexibility, as their size and surface properties can be tailored to match a broad range of analytes. Specifically, studies to date have focused on the characterization of nucleic acids, such as DNAs or RNAs and related complexes. Some examples of such studies are the use of NPs to characterize double-stranded DNA lengths (17) and RNA forms, and to discriminate among viral genomes using sequence-specific probes (18,19,20). More recently, biological NPs, as well as solid-state NPs, have been used to analyze proteins and protein complexes, in particular to resolve polypeptide structure and kinetics, folding/unfolding of proteins (21–27), protein charge (28,29), protein size (30), prion structure (31), protein aptamer interactions (32), and protein phosphorylation state (33). Furthermore, NPs have been used to control protein unfolding and to discriminate among large protein variants (34). Taken together, these studies show that NPs are a valuable tool for protein characterization studies at the single-molecule level.
Recent protein detection studies using solid-state NPs (27–29,35–37) highlight an exceptional challenge: in many cases, proteins translocate through the pore at a rate that exceeds the measurement bandwidth. Specifically, Plesa and co-workers reported that most of the protein translocation events in their study remained undetected, since the mean protein translocation times (∼1 μs) were beyond the resolution of the electrical sensing (37). To address this issue, Di Fiori and co-workers employed a laser-induced electroosmotic flow in a direction opposite to the analyte electrophoretic motion, which slowed down the translocation speed of the analytes, permitting the characterization of small proteins such as Ub (38). More recently, Larkin et al. utilized an improved electrical amplifier that permitted them to detect smaller proteins (>10 kDa) in thinned NP sensors (39).
Protein translocation direction and rate are governed by a combined action of electrophoretic and electroosmotic forces, as well as by their interactions with the NP walls. One way to significantly reduce the electrophoretic component is to use a buffer consisting of a pH close to the isoelectric point (pI) of the proteins, where their net charge vanishes (40). It is thus expected that at a pH near the pI, the electrophoretic mobility (μe) of the proteins will be decreased, resulting in longer residence time in the NP and allowing better temporal resolution. Electroosmotic flow, on the other hand, can either slow down or accelerate the motion of the analyte, depending on its respective charge. At the pH values used in our experiments, SiNx solid-state NPs typically possess a weak negative surface charge, which does not dominate pore conductivity at salt concentrations >∼100 mM (41–43). Therefore, if the pH is adjusted such that the protein remains also slightly negatively charged, the cation-based electroosmotic flow will tend to further prolong the protein’s dwell time in the pore, facilitating its detection.
In this work, we utilize the above-mentioned strategy and combine it with small NPs ∼3 nm in diameter drilled in ultrathin SiNx membrane (<10 nm thick) to characterize Ub and Ub chains at the single-molecule level. We show that upon optimizing the pH level, single Ub, as well as short Ub polymers of two and five moieties, can be readily detected and discriminated. Strikingly, under these conditions, and by combining information from the dwell times and blockade amplitude, we were able to discriminate between two nearly identical Ub dimers (linked by either Lys48 or Lys63). Finally, we show that deconjugation of Ub dimer, performed by the deubiquitination enzyme Usp8, can be analyzed using the NP system, allowing us to quantify and monitor the product accumulation in time. These results show good correspondence with gel-based quantification, and illustrate the ability of NPs, as compared with the bulk methods, to characterize very small quantities of Ub chains in a faster and less laborious way.
Materials and Methods
Chip and NP fabrication
NP chips were fabricated on a 4-inch silicon wafer coated with SiO2 (500 nm) and low-stress amorphous silicon nitride (SiNx; 60 nm). The SiNx was locally thinned to ∼8 nm in 1.5- to 2-μm circular wells, using a lithographic-mask whole wafer protection followed by a controlled reactive-ion etch process (44). Freestanding membranes of SiNx (20 × 20 μm2) were created by through-etching the wafer with KOH, with the reactive-ion-etched wells aligned to the centers of the freestanding SiNx membranes in a mask aligner. NPs were fabricated in the thinned SiNx regions using a high-resolution aberration-corrected transmission electron microscope (Titan 80-300 FEG-S/TEM, FEI, Hillsboro, OR), as previously reported (45). Pore formation proceeded with visual feedback by iterating through a uniformly expanded beam for imaging the NP diameter during formation and converging the beam to locally sputter to melt the membrane. Pores of 3 ± 0.2 nm could be consistently formed. See the Supporting Material for a detailed description.
Data acquisition and analysis
Electrical NP measurements were performed in a dark, double-insulated Faraday cage using a custom cell (17). Before their assembly in the Teflon cell, the nanochips were thoroughly washed with solvents (acetone, ethanol, and isopropanol) and were made hydrophilic by boiling in hot piranha solution (3:1 H2SO4/H2O2 at 300°C) followed by extensive washing in MilliQ ddH2O (Millipore, Billerica, MA). The nanochips were immersed in Tris-HCl buffer containing 1.5 M KCl at different pHs, as indicated. All buffers were filtered using a 0.02-μm syringe filter before use, and the pH of the measurement buffer was verified immediately before the experiment. All experiments were performed at a constant temperature (22.0 ± 0.5°C) and controlled relative humidity (40%) environment. Two Ag/AgCl pellet electrodes (A-M Systems, Sequim, WA) encapsulated in heat-shrinkable Teflon tubing were used to apply voltage and measure the ion current through the NP. To maximize consistency among different molecules or pH conditions, we performed each set of measurements using the same NP, washing the cis chamber extensively with fresh buffer before the addition of a new molecule. Measurements were repeated using different NPs, as indicated. The VC100 (Chimera Instruments, New York, NY) was used as the current amplifier in all experiments. It was operated at a sampling rate of 4.17 MHz and digitally low-pass filtered using a Butterworth filter before analysis (IRMS < 0.6 nA at 1 MHz, IRMS < 0.1 nA at 200 KHz, consistent with Balan et al. (46)). The raw data were read by a custom LabView code (National Instruments, Austin, TX) for real-time event identification and data storage. Data were then analyzed off-line using a dedicated LabView code to extract the translocation events. Heat maps were generated using a MatLab (MathWorks, Natick, MA) script (see Supporting Material), and all histograms and curve fittings were performed using Igor Pro 6 (Wavemetrics, Lake Oswego, OR).
Ub samples and catalysis
Ub monomers, Lys48 dimers, Lys63 dimers, Lys48 pentamers, and USP8 samples (U-100H, UC-200, UC-300, UC-216, and E-520, respectively) were purchased from BostonBiochem (Cambridge, MA). The lyophilized Ub proteins were reconstituted using 20 mM HEPES, pH 7, 10% glycerol, 100 mM NaCl, and 1 mM dithiothreitol buffer, divided into aliquots and frozen with liquid nitrogen. USP8 was used in its appropriate buffer provided by the manufacturer. Protein stocks were prepared at 1 mg/mL by suspending a known number of grams in a known amount of buffer, according to the data sheets provided by Boston Biochem. The stock concentrations for the di-Ubs and the penta-Ub were further validated in comparison with mono-Ub using a Coomassie-stained gel. Typical final concentrations of the proteins loaded in the cis chamber were 0.0025–0.01 μg/μL.
The catalysis assay was performed in 50 mM Hepes, pH 8, so upon adding 4 μL of the sample to the cis chamber, another 1 μL of 0.64% HCl was added to lower the pH to 7 (the ratio of 204:1 was checked empirically with the pH meter using the buffers at larger volumes).
Results and Discussion
Our system is schematically described in Fig. 1. An applied voltage of 125 mV typically resulted in a steady ion current of ∼2.7 nA, which is consistent with the expected conductance of a NP of diameter d ≈ 3.6 nm (see inset for a typical TEM image) fabricated in a membrane with an effective thickness of h = 8 nm using , where is the buffer conductance at 1.5 M KCl. Upon addition of Ub, we observe stochastic downward blockades of the ion current. The apparent event rate (average number of blockade events per unit time) of Ub and Ub chain molecules measured at similar molar concentrations was significantly smaller than that for DNA molecules, which typically exhibit a relatively high event rate. To obtain sufficient statistics (typically 500–1000 events), we allowed each experiment to run over 4–6 h.
Figure 1.
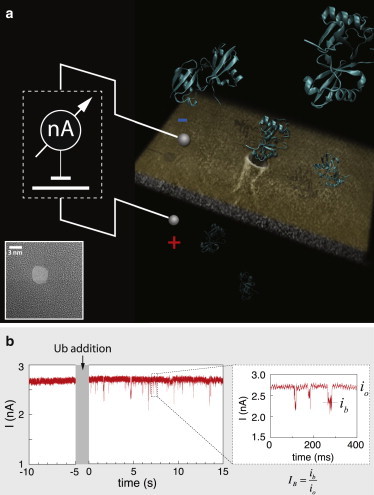
Schematic diagram of the experimental system and sample ion-current trace before and after addition of Ub molecules. (a) Solid-state NPs of ∼3.5 nm are used as single-molecule sensors for Ub and Ub chains (see inset for a typical TEM image). The passage of di-Ub molecules through the NP is illustrated. (b) NP current before (t < −5 s) and after (t > 0 s) addition of mono-Ub molecules to the cis chamber (V = 125 mV). (Inset) Enlargement of three translocation events, the open-pore (io) and the blocked-pore levels (ib). Current traces were digitally low-pass filtered (50 kHz) for display purpose only. To see this figure in color, go online.
Translocation dynamics of mono-Ub proteins as a function of pH
The characteristic translocation dwell time of mono-Ub through solid-state NPs was recently estimated to be <12 μs (38). However, the actual dwell times of Ub under the conditions used in this measurement (pH 8.5) were likely much shorter than its bandwidth-limited value. Given that the isoelectric point of Ub is 6.7 (47,48), we hypothesized that performing experiments at a reduced pH value that fulfills pI < pH < 8.5, would result in smaller negatively charged Ub molecules and hence a much longer translocation dwell time. This effect is expected to be greatly amplified in small NPs with an effective diameter only slightly larger than the size of the analyte (17). Thus, we hypothesized that employing NPs with diameters close to that of Ub (4.2 nm by 3.1 nm) and a pH value close to 6.7 could increase the apparent dwell time sufficiently to allow full characterization of the translocation events.
To check this hypothesis, we performed translocation experiments at two different pH values, 7.2 and 7.0, and used the same SiNx NP (d = 3.5 nm) to detect the result. In each case >500 Ub translocation events were acquired and analyzed. Fig. 2 summarizes these results. In Fig. 2 a, we present semilog heat maps of the fractional blocked ion current (IB) (49) versus the event dwell time (tD) for mono-Ub measured either at pH 7.2 (upper) or pH 7.0 (lower) using the same NP (io = 2.8 nA). In these plots, the temperature color reflects the local density of events. Also displayed in Fig. 2 a are the corresponding histograms of IB and tD fitted by Gaussian and exponential functions, respectively. Typical events for mono-Ub measured at these two pH values, using the same NP are shown in Fig. 2 b (note that the time between events was truncated to allow us to fit multiple events).
Figure 2.
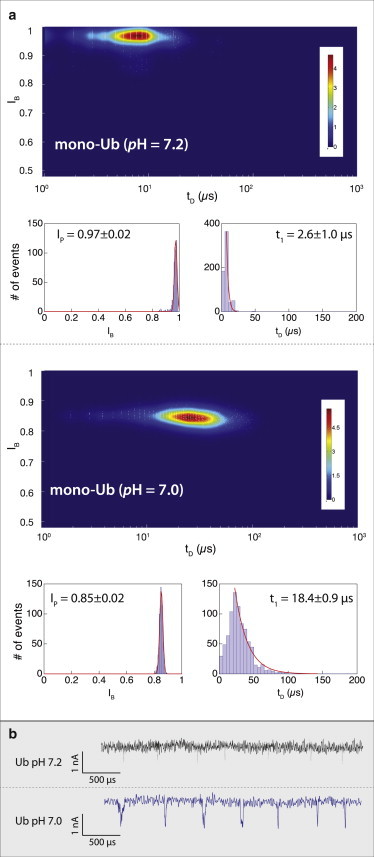
Dependence of the translocation dynamics of mono-Ub through a solid-state NP at pH close to its isoelectric point. (a) Heat maps (IB versus tD) at pH 7.2 (upper) and pH 7.0 (lower), showing a substantial shift toward longer translocation times and deeper blockade levels for Ub at pH 7.0 (N > 500 for each case). Histograms of the IB and tD, shown beneath the heat maps, are fitted to Gaussian and exponential functions, respectively (red curves). (b) Representative sets of translocation events of mono-Ub measured using the same ∼3.5 nm pore at pH 7.2 (black) and pH 7.0 (blue). At pH 7.2, the event amplitudes and dwell times are bandwidth-limited, but at pH 7.0, both are well defined (n = 6 NPs). To see this figure in color, go online.
Focusing first on the pH 7.2 data, we note that the measured event dwell times and amplitudes are essentially bandwidth-limited (estimated temporal resolution is ∼2.5 μs at 200 KHz) and therefore were not used to characterize the molecules. Consistent with previous studies, we cannot rule out that a large fraction of the events were not detected by our system due to their ultra short dwell times. It is interesting that lowering the pH to 7.0 results in a substantial increase in the typical translocation dwell time, bringing these events well within the reliable measurement bandwidth. We characterize the dwell time using a monoexponential tail fit (Fig. 2 a, red curves) with a time constant of 18.4 ± 0.9 μs. We also note that at this pH, the event fractional blockades are well-resolved and fit well to a Gaussian distribution with a mean ± SD of 0.85 ± 0.02, corresponding to mean event amplitudes of 0.42 ± 0.06 nA. These results show that tuning the net electrical charge of Ub by fine modulation of the buffer pH, combined with small NPs, is an effective way to achieve substantial slowing down of its translocation. Specifically, we obtained roughly an order-of-magnitude increase in tD by lowering the pH by only 0.2 units. At the same time, we observed a decrease in the event capture rate, which although smaller, did not prohibit us from performing extensive characterization of Ub and Ub chains at physiological pH (pH 7.0). Our attempts to perform measurements at even lower pH values resulted in an even smaller capture rate, rendering the measurement nearly impractical.
Discriminating Ub chain lengths using NPs
Taking advantage of our ability to reliably sense Ub in the NP system, we moved on to compare the translocation signals of multiunit Ub chains. To this end, we repeated the measurement of mono-Ub (using a different NP) and measured the translocation dynamics of two K48-linked Ub chains, di-Ub and penta-Ub. As before, we used an ∼3-nm-diameter pore and obtained >500 events in each case. Fig. 3 displays the resulting heat maps as well as detailed dwell times and fractional-blockade current distributions. Two salient features are readily observed. 1) The characteristic dwell time of di-Ub is roughly threefold larger than the typical dwell time of mono-Ub (68.2 ± 2.4 μs and 19.6 ± 1.0, respectively). We observe a much larger increase of the penta-Ub dwell time (780 ± 27 μs), suggesting a nonlinear dependence of the dwell time on Ub chain length. 2) The peak of the fractional current blockage histogram is consistently shifted toward deeper blockades for the longer Ub chains: 0.84 ± 0.03, 0.70 ± 0.057, and 0.45 ± 0.11 for the mono-, di-, and penta-Ub, respectively. Furthermore, the penta-Ub chain displays a nearly twofold broader distribution of blockade currents compared with the shorter Ub chains.
Figure 3.
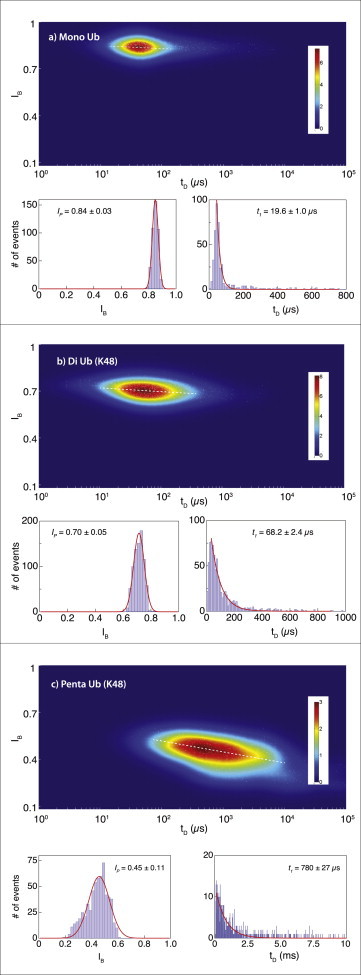
Translocation dynamics of mono- and multiunit Ub chains. Two-dimensional heat maps, blockade current, and dwell-time histograms for mono-Ub (a), di-Ub (K48-linked) (b), and penta-Ub (K48-linked) (c). Measurements were performed using the same NP to maximize consistency among the different molecules (n = 8 NPs). To see this figure in color, go online.
The shift in the peak of the fractional blocked current can be rationalized if we note that the effective NP thickness, h, is larger than the mean size of a mono-Ub chain (∼8 nm vs. ∼4 nm). In these cases, we expect that the fractional blocked current, IB, will be proportional to the fraction of unoccupied volume in the NP. Previous reports using single-stranded DNA translocations through the α-HL NP indicated that in this regime of short polymers, the dependence of IB on polymer length is nonlinear (50). Furthermore, unlike linear polymers such as single-stranded DNA, which are well characterized by a fairly uniform charge distribution along their length, multiunit protein chains, such as Ub polymers, can adopt a complex and nonuniform three-dimensional (3D) superstructure. This feature could complicate the relationship between the overall size of the proteins (i.e., their total molecular weight), or their chain length, and the observed blockade current and dwell times. Although it is plausible to assume that IB is in fact determined by complex, and yet to be discovered, contributions of multiple properties, such as its 3D shape, charge distribution, local hydrophobicity, and perhaps other features, our results clearly show that it is possible to discriminate among Ub chain lengths based on their current/dwell-time patterns.
Discriminating between Ub chain linkage type using NPs
We further challenged our NP characterization method by examining its ability to discriminate between two nearly identical di-Ub chains of the same molecular weight, differing only in their chain linkage type. Ubiquitination of proteins by a polymeric chain of Ub moieties linked through the K48 side chain lead to their degradation via the proteasome (3), whereas a polymeric chain of the same length linked through the K63 side chain participates in DNA repair processes as well as signaling processes leading to activation of the transcription factor NF-kB (51). These Ub chains adopt either closed conformations, where adjacent moieties interact with each other, or open conformations, where no interfaces are present except for the linkage site. The canonical K48-linked chains adopt closed conformations, whereas K63-linked chains mostly display open conformations, as shown by NMR analysis (13,14) and crystal structures of these chain types (12,52–54). Gel-based analysis of the two di-Ubs showed a nearly identical migration pattern (see Supporting Material). We therefore hypothesized that K63-linked di-Ub, which has higher conformational freedom than K48-linked di-Ub, will occupy a larger volume within the NP, thus leading to lower IB values.
The experiment consisted of translocating either one of the two di-Ubs (K63- or K48-linked di-Ub), or a mixed population (molar ratio 1:1) of the two proteins using the same NP. As seen in Fig. 4 a, each of the individual species gives rise to a single population of events characterized by single-peak IB histograms (IP = 0.67 ± 0.06 and IP = 0.86 ± 0.06 for K63- and K48-linked di-Ub, respectively), and the mixture produces two distinct current blockade populations that can be readily distinguished. The IB histogram of the mixture displays two statistically distinguishable peaks (0.68 ± 0.06 and 0.87 ± 0.04, mean ± SD), which we fit by a double Gaussian function (Fig. 4 a, red line). By examining the heat maps, one can also see that the dwell times associated with K63-linked di-Ub are shifted to longer timescales compared to those associated with K48-linked di-Ub. This effect is quantified by the detailed dwell-time histograms in Fig. S3, which give characteristic time constants of 95.9 ± 9.3 μs and 34.2 ± 1.2 μs for the K63- and K48-linked di-Ubs, respectively. This effect is more pronounced when we analyze the two species separately. Nonetheless, the resemblances of the two-dimensional patterns of the two populations reflected in the heat maps can be used to identify the higher IB population as that associated with K48-linked di-Ub and the lower IB (showing also longer dwell times) as that associated with the K63-linked di-Ub molecules.
Figure 4.
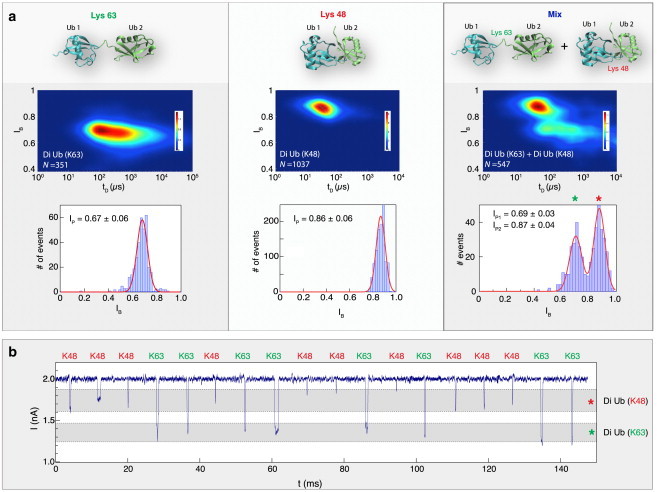
Single-molecule discrimination among the two forms of di-Ub molecules. (a) The expected equilibrium conformation of di-Ub linked by K63 (left), K48 (middle), or a mixture of the two (right). The structure cartoons were created using Visual Molecular Dynamics (53) based on Protein Data Bank structures 1AAR (12) and 2JF5 (54). Heat maps of the two different di-Ub conformers (measured using the same NP) show a distinct pattern for each dimer. The two molecules can be distinguished by their corresponding fractional current blockades shown below the heat maps. An experiment consisting of an equal molar ratio of both dimers yielded two populations in the heat maps, corresponding to two distinct Ip values. (b) Instantaneous discrimination among K63- and K48-linked di-Ubs. A collection of single-molecule events is shown (time between events was truncated to allow fitting of multiple events on the graph). Gray bands correspond to the two Ip (mean ± SD) values of the two dimers shown in (a) (n = 10 NPs). To see this figure in color, go online.
Previous studies indicated that the equilibrium conformation of K48-linked di-Ub is pH-dependent (13). This explained the various different crystal structures that were reported for this molecule. Specifically, at pH 6.8 (close to our working conditions), the predominant conformation of K48-linked chain is the closed conformation, with a pI of 5.8 (13). In contrast, at this pH, the K63-linked chain conformation is open or extended, as the two Ub moieties are not in contact with each other, and has a pI of 6.7, similar to that of mono-Ub. These observations are in line with our results: Working at a pH value closer to its pI value, the K63-linked dimer may be less charged compared to the K48-linked dimer, thus explaining the much longer translocation dwell time of the former. At the same time, the extended form of the K63-linked di-Ub allows it to exclude a larger volume from the pore during translocation, hence resulting in a smaller average IB.
Furthermore, a smaller negative charge of the K63-linked dimer is expected to diminish its event rate compared to that of the K48-linked dimer. This prediction is borne out by our data: integration of the blockade-level histogram fit in Fig. 4 a, right, from IB = 0 to 0.8 (the minima separating the double Gaussian fit) and from IB = 0.8 to 1, can be used to obtain the ratio of 1:1.5 detected K63- versus K48-linked Ub dimer. Thus, the higher capture rate of the K48-linked di-Ub (we used a 1:1 molar ratio in this experiment) can be explained by its stronger charge.
To examine the possibility of discriminating among individual di-Ub molecules, we mixed the K63- and K48-linked dimers at an equal molar ratio. In Fig. 4 b, we present a series of translocation events obtained from a mixture of K63- and K48-linked di-Ub proteins. Using the double-Gaussian fit of the IB histogram of the mixture, with gray shading around the mean values and widths equal to twice their standard deviations obtained from the fit, we can tentatively classify each event as having originated from either K63- or K48-linked dimer, as indicated at the top of the chart. Although this method of NP-based classification of protein complexes may require further validation, perhaps by introduction of a simultaneous single-molecule fluorescence sensing (38,44), it could pave the way for powerful and nondestructive protein quantification of biological relevance.
Quantification of Ub chain catalysis
Protein ubiquitination can be enzymatically reversed by deubiquitinating enzymes (DUBs). To specifically control Ub-dependent signaling, human cells contain ∼85 different kinds of enzymes to deal with chains of distinct linkage, topology, and length (55). For example, proteasome-bound DUBs, such as USP14, UCH37/UCHL5, and RPN11/POH1, protect ubiquitin from degradation (56). This process is vital for keeping sufficient levels of free ubiquitin that can be used for chain assembly.
To analyze and quantify the Ub chain catalysis process, an assay was established using USP8 and K48-linked di-Ub. We chose to work with USP8 (and not with IsoT), since its high pI (8.7) ensures that it would not translocate through the pore in the same direction as Ub at pH 7. USP8 (0.024 μg/μL) was preincubated with di-Ub (K48) (0.2 μg/μL) on ice for 5 min in a buffer containing 50 mM Hepes-NaOH, pH 8, 100 mM NaCl, and 1 mM dithiothreitol. Deubiquitination reactions were carried at 37°C for 30, 60, 120, and 240 min. A sample was taken out at each time point and the reaction was terminated by immediately freezing the samples using liquid nitrogen. Each sample was divided into two parts; the first part was loaded onto a polyacrylamide gel to be analyzed by gel electrophoresis (see Fig. 6 a), and the second was loaded into the NP (Fig. 5). The data from counting single Ub molecules, using the same ∼3-nm NP, are presented in Fig. 5. For each time point, we collected ∼500 translocation events and, as previously, produced heat maps and fractional-blockade histograms. Consistent with our previous results, at t = 0, we obtain a single population with Ip = 0.71 ± 0.06, attributed to the uncleaved di-Ub. After 4 h of incubation, we obtain a primary population of events with Ip = 0.86 ± 0.01, attributed to mono-Ub, and a much smaller population at Ip = 0.71 ± 0.01, attributed to the residual uncleaved di-Ub molecules. Consistent with these two end points, the other three time points in the figure give intermediate population fractions of these two blocking currents, thus providing a mean to quantify the enzymatic deconjugation process.
Figure 6.
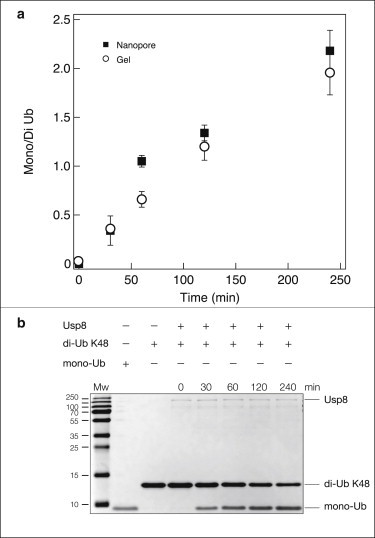
Comparison between deubiquitination reaction results quantified by NP analysis and by Coomassie-stained band intensity. (a) Product/substrate ratios of each deubiquitination time point are plotted against time. The fraction of mono- to di-Ub was calculated by integration under the fractional blocked current histogram (squares). Open circles represent gel quantification. (b) Mono-Ub (1.5 μg), di-Ub (K48) (1.5 μg), and samples from the deubiquitination reaction (described in Fig. 5 legend) were separated on 15% SDS-PAGE and stained with Coomassie.
Figure 5.
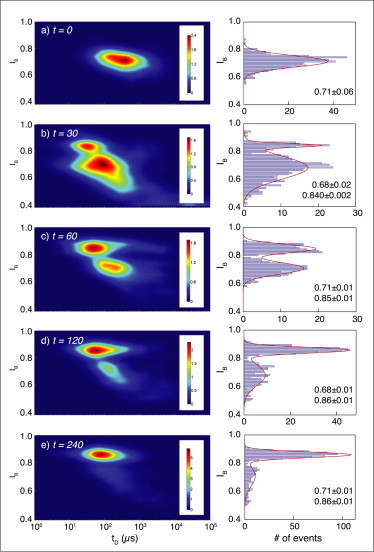
Monitoring a deubiquitination reaction using NPs. (a) USP8 was preincubated with di-Ub (K48) on ice before analysis by the NP. At t = 0, a single-event population can be seen, consistent with di-Ub (K48). Deubiquitination reactions were carried out at 37°C for 30 (b), 60 (c), 120 (d), and 240 min (e). The reaction was terminated by immediately freezing the samples using liquid nitrogen. Defrosted samples were analyzed using NPs and the distributions of IB values for the different time points are presented along their heat maps. All measurements were performed using a single NP to maximize consistency (n = 6 NPs). To see this figure in color, go online.
The most common approach for following the progression of an enzymatic reaction using gel electrophoresis involves the integration of intensity in a spot or a rectangle drawn on the gel image on top of the Coomassie-stained bands of the substrate or product. Fig. 6 b displays our analysis for the Ub products shown in Fig. 5 against a protein molecular-weight marker. Three repeats of this gel-based analysis were performed to establish error bars (see details in Supporting Material). Comparing the NP analysis and the gel-based analysis (Fig. 6 a, solid squares), we can see a good correspondence between the two methods.
We note that although the gel-based technique has been successfully applied in numerous experiments (57–59), it can only be used to determine individual band intensity in cases for which bands are extremely well-resolved. As a result, this approach is often limited to the integration of a group of closely spaced bands (60). This method is also error prone due to imaging equipment and image compression methods for ensemble in pixels. Moreover, to see a significant band, one must load the gel with tens of nanomoles (∼1016) of the analyzed protein. In contrast, NP-based analysis is a single-molecule approach requiring only a few thousand copies of each of the products, thus potentially offering possibilities for biomolecular quantification in cases where sample concentrations are very low.
Conclusions
In this article, we have demonstrated that by combining a protein-specific conditioned current measurement platform with small and ultrathin solid-state NPs, Ub and Ub chains can be efficiently detected and analyzed. By utilizing the Ub intrinsic isoelectric point, we were able to fit buffer conditions to slow down translocations and thus detect thousands of events. Under these conditions, we were able to differentiate among Ub chain lengths and Ub chain linkage types, and to quantify the products of di-Ub deconjugation reaction. As shown, varying the pH can be used to assess more quantitatively protein translocations, yet this method also affects the charge properties of the protein and of the pore (28,40), which needs to be independently investigated.
The challenge of differentiating proteins with small structural differences is not new to the NP platform. Conformational differences of the same protein in solid-state NPs were detected (21–27), as were single amino acid mutations in biological NPs (31). However, to our knowledge, discrimination of proteins of the same molecular weight at their native folded state using solid-state NPs has not been reported. To our knowledge, our work opens a novel approach for analyzing proteins in unprecedented detail using solid-state NPs. It serves as a proof of concept for approaching NP detection of sub-10-kDa proteins and presents, to our knowledge, the first ever differentiation of native proteins of the same mass. Future studies may include completing a full catalysis assay using the NPs platform in real-time, identification of single amino acid mutations in native proteins and detection of Ub-like proteins that share a similar 3D structure to Ub but regulate a strikingly diverse set of cellular processes (61). Furthermore, one of the biggest future challenges would be to use NPs to directly detect ubiquitinated substrates isolated from cells.
Author Contributions
I.N. prepared samples, performed nanopore measurements, analyzed data, and helped write the article. D.H. prepared samples, performed gel analysis, designed the experiments, and helped write the article. A.M. conceived the project goals and methods, developed the measurement system, analyzed data, and helped write the article.
Acknowledgments
We acknowledge generous assistance with nanopore fabrication from Ossama Assad and Daniel Bar.
Financial support for this article was provided by a Marie Curie People award (GA-2010-277060), the Israeli Centers of Research Excellence (I-CORE) program (Center no. 1902/12), and the Israeli Science Foundation (award no. 845/11). We also thank the staff at the Technion Electron Microscopy Center for their dedicated support.
Supporting Material
References
- 1.Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. The ubiquitin system. Trends Biochem. Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Komander D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 5.Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 6.Hoege C., Pfander B., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 7.Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 8.Newton K., Matsumoto M.L., Dixit V.M. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 9.Phu L., Izrael-Tomasevic A., Matsumoto M.L., Bustos D., Dynek J.N., Fedorova A.V., Bakalarski C.E., Arnott D., Deshayes K., Dixit V.M., Kelley R.F., Vucic D., Kirkpatrick D.S. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003756. M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heride C., Urbé S., Clague M.J. Ubiquitin code assembly and disassembly. Curr. Biol. 2014;24:R215–R220. doi: 10.1016/j.cub.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Polge C., Uttenweiler-Joseph S., Taillandier D. Deciphering the ubiquitin proteome: Limits and advantages of high throughput global affinity purification-mass spectrometry approaches. Int. J. Biochem. Cell Biol. 2013;45:2136–2146. doi: 10.1016/j.biocel.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Cook W.J., Jeffrey L.C., Pickart C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J. Biol. Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 13.Varadan R., Walker O., Fushman D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 14.Varadan R., Assfalg M., Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 15.Eddins M.J., Varadan R., Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 16.Meller A. Nanopores: single-molecule sensors of nucleic acid-based complexes. In: Rice S.A., Dinner A.R., editors. Vol. 149. John Wiley & Sons; Hoboken, NJ: 2012. pp. 251–268. (Advances in Chemical Physics). [Google Scholar]
- 17.Wanunu M., Sutin J., Meller A. DNA translocation governed by interactions with solid-state nanopores. Biophys. J. 2008;95:4716–4725. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer A., Rapireddy S., Meller A. Electronic barcoding of a viral gene at the single-molecule level. Nano Lett. 2012;12:1722–1728. doi: 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wanunu M., Dadosh T., Drndić M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 20.Milchev A. Single-polymer dynamics under constraints: scaling theory and computer experiment. J. Phys. Condens. Matter. 2011;23:103101. doi: 10.1088/0953-8984/23/10/103101. [DOI] [PubMed] [Google Scholar]
- 21.Freedman K.J., Jürgens M., Kim M.J. Chemical, thermal, and electric field induced unfolding of single protein molecules studied using nanopores. Anal. Chem. 2011;83:5137–5144. doi: 10.1021/ac2001725. [DOI] [PubMed] [Google Scholar]
- 22.Goodrich C.P., Kirmizialtin S., Movileanu L. Single-molecule electrophoresis of β-hairpin peptides by electrical recordings and Langevin dynamics simulations. J. Phys. Chem. B. 2007;111:3332–3335. doi: 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- 23.Merstorf C., Cressiot B., Pelta J. Wild type, mutant protein unfolding and phase transition detected by single-nanopore recording. ACS Chem. Biol. 2012;7:652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 24.Oukhaled G., Mathé J., Auvray L. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 2007;98:158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Larrea D., Bayley H. Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefureac R.I., Lee J.S. Nanopore analysis of the folding of zinc fingers. Small. 2008;4:1646–1650. doi: 10.1002/smll.200800585. [DOI] [PubMed] [Google Scholar]
- 27.Talaga D.S., Li J. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firnkes M., Pedone D., Rant U. Electrically facilitated translocations of proteins through silicon nitride nanopores: conjoint and competitive action of diffusion, electrophoresis, and electroosmosis. Nano Lett. 2010;10:2162–2167. doi: 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- 29.Fologea D., Ledden B., Li J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007;91:539011–539013. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusko E.C., Prangkio P., Mayer M. Single-particle characterization of Aβ oligomers in solution. ACS Nano. 2012;6:5909–5919. doi: 10.1021/nn300542q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jetha N.N., Semenchenko V., Marziali A. Nanopore analysis of wild-type and mutant prion protein (PrPC): single molecule discrimination and PrPC kinetics. PLoS ONE. 2013;8:e54982. doi: 10.1371/journal.pone.0054982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedzwiecki D.J., Iyer R., Movileanu L. Sampling a biomarker of the human immunodeficiency virus across a synthetic nanopore. ACS Nano. 2013;7:3341–3350. doi: 10.1021/nn400125c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen C.B., Rodriguez-Larrea D., Bayley H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat. Biotechnol. 2014;32:179–181. doi: 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nivala J., Marks D.B., Akeson M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat. Biotechnol. 2013;31:247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowalczyk S.W., Kapinos L., Dekker C. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 2011;6:433–438. doi: 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- 36.Oukhaled A., Cressiot B., Pelta J. Dynamics of completely unfolded and native proteins through solid-state nanopores as a function of electric driving force. ACS Nano. 2011;5:3628–3638. doi: 10.1021/nn1034795. [DOI] [PubMed] [Google Scholar]
- 37.Plesa C., Kowalczyk S.W., Dekker C. Fast translocation of proteins through solid state nanopores. Nano Lett. 2013;13:658–663. doi: 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Fiori N., Squires A., Meller A. Optoelectronic control of surface charge and translocation dynamics in solid-state nanopores. Nat. Nanotechnol. 2013;8:946–951. doi: 10.1038/nnano.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin J., Henley R.Y., Wanunu M. High-bandwidth protein analysis using solid-state nanopores. Biophys. J. 2014;106:696–704. doi: 10.1016/j.bpj.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han A., Creus M., Staufer U. Label-free detection of single protein molecules and protein-protein interactions using synthetic nanopores. Anal. Chem. 2008;80:4651–4658. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- 41.Ho C., Qiao R., Timp G. Electrolytic transport through a synthetic nanometer-diameter pore. Proc. Natl. Acad. Sci. USA. 2005;102:10445–10450. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoogerheide D.P., Garaj S., Golovchenko J.A. Probing surface charge fluctuations with solid-state nanopores. Phys. Rev. Lett. 2009;102:256804. doi: 10.1103/PhysRevLett.102.256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeets R.M., Keyser U.F., Dekker C. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 44.Assad O.N., Di Fiori N., Meller A. Two color DNA barcode detection in photoluminescence suppressed silicon nitride nanopores. Nano Lett. 2015;15:745–752. doi: 10.1021/nl504459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M.J., Wanunu M., Meller A. Rapid fabrication of uniformly αsized nanopores and nanopore arrays for parallel DNA analysis. Adv. Mater. 2006;18:3149–3153. [Google Scholar]
- 46.Balan A., Machielse B., Drndić M. Improving signal-to-noise performance for DNA translocation in solid-state nanopores at MHz bandwidths. Nano Lett. 2014;14:7215–7220. doi: 10.1021/nl504345y. [DOI] [PubMed] [Google Scholar]
- 47.Low T.L., Goldstein A.L. The chemistry and biology of thymosin. II. Amino acid sequence analysis of thymosin α1 and polypeptide β1. J. Biol. Chem. 1979;254:987–995. [PubMed] [Google Scholar]
- 48.Wilkinson K.D., Urban M.K., Haas A.L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 1980;255:7529–7532. [PubMed] [Google Scholar]
- 49.Eid J., Fehr A., Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 50.Meller A., Nivon L., Branton D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z.J., Sun L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Weeks S.D., Grasty K.C., Loll P.J. Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture. Proteins. 2009;77:753–759. doi: 10.1002/prot.22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 54.Komander D., Reyes-Turcu F., Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 56.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenowitz M., Senear D.F., Ackers G.K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 58.Morrison T.B., Parkinson S. Quantifying radiolabeled macromolecules and small molecules on a single gel. Biotechniques. 1994;17:922–926. [PubMed] [Google Scholar]
- 59.Stankus A., Goodisman J., Dabrowiak J.C. Quantitative footprinting analysis of the chromomycin A3-DNA interaction. Biochemistry. 1992;31:9310–9318. doi: 10.1021/bi00153a026. [DOI] [PubMed] [Google Scholar]
- 60.Shadle S.E., Allen D.F., Tullius T.D. Quantitative analysis of electrophoresis data: novel curve fitting methodology and its application to the determination of a protein-DNA binding constant. Nucleic Acids Res. 1997;25:850–860. doi: 10.1093/nar/25.4.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


