Abstract
Capillary electrophoresis has been used to characterize the interaction of monovalent cations with 26-basepair DNA oligomers containing A-tracts embedded in flanking sequences with different basepair compositions. A 26-basepair random-sequence oligomer was used as the reference; lithium and tetrabutylammonium (TBA+) ions were used as the probe ions. The free solution mobilities of the A-tract and random-sequence oligomers were identical in solutions containing <∼100 mM cation. At higher cation concentrations, the A-tract oligomers migrated faster than the reference oligomer in TBA+ and slower than the reference in Li+. Hence, cations of different sizes can interact very differently with DNA A-tracts. The increased mobilities observed in TBA+ suggest that the large hydrophobic TBA+ ions are preferentially excluded from the vicinity of the A-tract minor groove, increasing the effective net charge of the A-tract oligomers and increasing the mobility. By contrast, Li+ ions decrease the mobility of A-tract oligomers because of the preferential localization of Li+ ions in the narrow A-tract minor groove. Embedding the A-tracts in AT-rich flanking sequences markedly alters preferential interactions of monovalent cations with the B∗ conformation. Hence, A-tracts embedded in genomic DNA may or may not interact preferentially with monovalent cations, depending on the relative number of A·T basepairs in the flanking sequences.
Introduction
DNA A-tracts, runs of four or more A·T basepairs not interrupted by a TpA basepair step, exhibit a unique conformation, often called the B∗ conformation, that differs from that of normal B-DNA by having a narrow minor groove, propeller twisted basepairs, and bifurcated hydrogen bonds between the two strands (1–5). NMR experiments have shown that DNA A-tracts are intrinsically curved, although the curvature is delocalized and extends into the nucleotides flanking the A-tracts (6–8). Bending at the junctions between B-form and B∗-DNA can lead to macroscopic curvature of the helix backbone when the A-tracts are repeated in phase with the helix screw (3–5,9).
DNA A-tracts have been the focus of much attention in recent years because they are overrepresented in genomic DNA (10) and are found near many origins of replication and transcription factor binding sites (5,10–12). However, the biological role of DNA A-tracts in the cell is not well understood (5), most likely because the B∗ conformation is in rapid equilibrium with normal B-DNA under physiological conditions. The midpoint of the B∗ ↔ B transition occurs at temperatures between 30° and 40°C, depending on the A-tract sequence and the ionic strength of the solution (13–18). Furthermore, the macroscopic curvature of the DNA backbone caused by A-tract phasing is nearly eliminated in solutions containing ∼200 mM monovalent cations (19).
To better understand the role of DNA A-tracts in the cell, it is important to characterize the interaction of various monovalent cations with A-tract and non-A-tract DNAs in various sequence contexts. Recent experimental studies (20,21) and molecular dynamics simulations (22–24) have indicated that monovalent cations in the counterion cloud (25) fill the major and minor grooves of the DNA, as well as forming a shell of condensed ions around the helix. Various x-ray and NMR experiments have shown that monovalent cations can be preferentially localized in the A-tract minor groove, displacing some of the water molecules in the spine of hydration at the base of the groove (6,26–30). Whether the localization of monovalent cations in the A-tract minor groove leads to narrowing of the groove and the formation of the B∗ conformation, or whether cation localization is due to the presence of the intrinsically narrow A-tract minor groove, is still a matter of debate (6,24,31–37).
Most studies of the interaction of monovalent cations with DNA A-tracts have focused on A-tracts flanked by G·C basepairs, even though A-tracts are surrounded by a variety of flanking sequences in genomic DNA. Here, we use free solution capillary electrophoresis (CE) to analyze the effect of the flanking sequences on the interaction of monovalent cations with DNA A-tracts. CE is a useful technique for such studies because the electrophoretic mobility of a small DNA oligomer is directly proportional to its effective charge after counterion condensation (38–42) and inversely proportional to its translational friction coefficient (19,43,44). Because the translational diffusion coefficients of small DNA oligomers containing the same number of basepairs are essentially independent of the presence or absence of A-tracts (42,43,45), the free solution mobilities of small DNA oligomers reflect differences in effective charge due to sequence-dependent cation interactions.
We have previously used CE to demonstrate that A-tract oligomers migrate more slowly than non-A-tract oligomers containing the same number of basepairs (45–48). We showed that the decreased mobility depends on the length and sequence of the A-tract(s), as well as the identity of the monovalent cation in the solution (46–48). The decreased mobility is due to the preferential localization of monovalent cations in the A-tract minor groove, because no mobility decrease is observed if the minor groove is blocked by the binding of netropsin (46). We have also characterized the dependence of DNA electrophoretic mobilities on the ionic strength of the solution and the charge density of the polyion (49–51). Finally, we showed that the curvature of A-tract-containing DNA molecules is markedly reduced in solutions containing ∼200 mM cation, even though monovalent cations are still preferentially localized in the A-tract minor groove (19). Hence, curvature of the DNA helix backbone and the preferential localization of monovalent cations in the A-tract minor groove are not necessarily linked. That being the case, we define the B∗ conformation as one in which the A-tract has a narrow minor groove and is capable of preferentially interacting with monovalent cations.
In this study, we use CE to analyze the effect of different flanking sequences on the interaction of DNA A-tracts with monovalent cations. The oligomers were 26 bp in size, had A+T contents ranging from 38% to 81%, and contained one or two A-tracts of different lengths. The reference was a 26-bp random-sequence oligomer containing 46% A+T. Two different monovalent cations were investigated, the large hydrophobic tetrabutylammonium ion (TBA+) and the small hydrophilic ion, Li+. The A-tract oligomers and the reference have the same mobility in solutions containing <∼100 mM cation, suggesting that A-tract oligomers exhibit the normal B conformation in low ionic strength solutions. At higher cation concentrations, TBA+ ions are preferentially excluded from A-tracts in the B∗ conformation, increasing the effective net charge of the A-tract oligomers and increasing the mobility. By contrast, Li+ ions are preferentially localized in the A-tract minor groove, decreasing the effective net charge and decreasing the mobility of the A-tract oligomers. Embedding the A-tracts in AT-rich flanking sequences alters the preferential interactions of TBA+ and Li+ ions with DNA A-tracts.
Materials and Methods
DNA samples
The 26 bp DNA oligomers used in this work were synthesized by IDT (Coralville, IA) and purified by polyacrylamide gel electrophoresis. Duplexes were prepared by mixing equimolar quantities of two complementary strands (1 μg/μL) in 10 mM Tris-chloride buffer, pH 8.0, heating to 94°C for 5 min and slowly cooling to room temperature. The concentrated stock solutions were stored at −20°C until needed. The oligomers are identified in the following text by acronyms denoting the A-tract sequence and phasing, followed by a number in parentheses indicating the number of A·T basepairs in the flanking sequences. The acronyms and sequences of the various oligomers are given in Tables 1 and 2. The reference, Ra(12), was a random-sequence 26-bp oligomer containing 12 A·T basepairs and 14 G·C basepairs; its sequence is given as the first entry in Table 2.
Table 1.
Acronyms and sequences of oligomers containing two phased A4T1-tracts and variable numbers of flanking A·T basepairs
| Acronym | Sequence |
|---|---|
| A4T1in(0) | CGCAAAATCGGGCAAAATCGGCGGCG |
| A4T1in(1) | CGCAAAATCGGGCAAAATCGCTGGCG |
| A4T1in(2) | CGCAAAATCGGTCAAAATCGGCTGCG |
| A4T1in(3) | CGCAAAATCTGTCAAAATCGCTGGCG |
| A4T1in(4) | CGCAAAATCTGTCAAAATCTGCGTCG |
| A4T1in(5) | CGCAAAATCTGTCAAAATCTGTCTGC |
| A4T1in(6) | CGTAAAATCTGTCAAAATCTGTCTCG |
| A4T1in(7) | CGTAAAATCTGTCAAAATCTATCTCG |
| A4T1in(8) | CGTAAAATCTGTCAAAATCTATTACG |
| A4T1in(9) | CGTAAAATCTATCAAAATCTATTACG |
| A4T1in(10) | CGTAAAATATATCAAAATCTATTACG |
| A4T1in(11) | CGTAAAATATATCAAAATATATTACG |
The A-tracts are underlined for clarity.
Table 2.
Acronyms and sequences of the reference, Ra(12), and oligomers with variable A-tracts and flanking sequences
| Acronym | Sequence |
|---|---|
| Ra(12) | CGCAGTGTACGACTAGACTACAGACG |
| A3(9) | CGCAAAGCGATCGACACTAGTACTCG |
| A3(13) | CGCAAAGTGTCTATACATATGTATCG |
| A2T2(8) | CGCAATTCTATATGCTCCGCAGACCG |
| A3T3(6) | CGCAAATTTCTATGCTCCGCAGACCG |
| A4T4(4) | CGCAAAATTTTCTGCTCCGCAGACCG |
| A5T5(2) | CGCAAAAATTTTTACTCGCCGCGCCG |
| A4in(4) | CGCAAAAGCGTCGAAAACCTCTCTCG |
| A4in(8) | CGCAAAAGTGTCTAAAATCTGTTCTG |
| A5in(2) | CGCAAAAAGCCGCAAAAACCTCGTCG |
| A5out(2) | CGCAAAAAGCCGCCTGAAAAACCTCG |
| A5=T5in(2) | GCCAAAAAGCTCGTTTTTCCGCACCG |
| A5=T5out(2) | CGCAAAAAGCTCGCCTTTTTGCACCG |
| A2T2in(4) | CGCAATTCGTCACAATTCTGCGACCG |
| A3T3in(0) | CGCAAATTTCGCCAAATTTCGCGCCG |
| A3T3in(4) | CGCAAATTTCGACAAATTTCTAGTCG |
| A3T3out(4) | CGCAAATTTCGAGTCAAATTTCTACG |
| A3T4in(2) | CGCAAATTTTCGCAAATTTTCAGACG |
| A4T2in(4) | CGCAAAATTCGTCAAAATTCTGTCTG |
| A4T3in(2) | CGCAAAATTTGTCAAAATTTCGCTCG |
| A4T4in(0) | CGCAAAATTTTCGAAAATTTTCGCCG |
| A4T4out(0) | CGCAAAATTTTCGCGCAAAATTTTCG |
| A4T4′in(0) | CGCCGCAAAATTTTCGAAAATTTTCG |
| ApT3in(0) | CAAATTCGGCAAATTTCGCAAATTCG |
All oligomers contained 46% or 62% A+T. The A-tracts are underlined for clarity.
Buffers
Most buffers used as the background electrolyte (BGE) contained 200 mM diethylmalonic acid ((CH3CH2)2C(COOH)2, Sigma-Aldrich, St. Louis, MO), titrated to pH 7.3, the pKa of the second carboxyl group at 25°C, with a concentrated solution of the hydroxide of Li + or TBA+ (Sigma-Aldrich). Because the second carboxyl group of diethylmalonic acid is half ionized at pH 7.3, the total cation concentration in the diethylmalonate buffers was 300 mM; the ionic strength was 400 mM. Mobilities measured as a function of BGE concentration used Li+ or TBA+ as the cation and acetate as the anion.
Capillary electrophoresis
Capillary zone electrophoresis measurements were carried out using a Beckman Coulter P/ACE System MDQ Capillary Electrophoresis System (Fullerton, CA), run in the reverse polarity mode (anode on the detector side) with ultraviolet detection at 254 nm, as described previously (52). Migration times and peak profiles were analyzed using the 32 Karat software. The capillaries were internally coated LPA (linear poly-acrylamide) capillaries from Bio-Rad (Hercules, CA). The LPA coating minimizes the electroosmotic flow (EOF) of the solvent without affecting the mobility of the analyte (38). The capillaries were 40.0 cm in total length, with external diameters of 375 μm and internal diameters of 75 μm, mounted in a liquid-cooled cartridge. The capillary was conditioned at the beginning of each day, and between experiments, by rinsing with the BGE for 5 min at high pressure (25 psi, 0.17 Mpa). The capillary was rinsed with deionized water at 25 psi for 5–10 min at the end of each day and stored in deionized water overnight. All samples contained an A-tract oligomer and the reference, Ra(12), mixed in different molar ratios to enable easy identification. The samples were hydrodynamically injected into the capillary by applying low pressure (0.5 psi, 0.0035 Mpa) for 3 s. The sample volume was 22 nL; the length of the sample plug was 0.51 cm, ∼1.3% of capillary length. The DNA concentration ranged from 10 to 50 ng/μL; mobilities measured within this range are independent of DNA concentration (38).
Unless otherwise indicated, the temperature of all experiments was 20.0° ± 0.1°C. The applied electric field was typically 90–110 V/cm (3.5 to 4.5 kV applied voltage); the mobilities were independent of the applied field within this range. The current ranged between 40 and 70 μA, depending on the ionic strength and cation identity. The residual EOF in the capillary, measured every day by the fast method of Williams and Vigh (53), was typically ∼5 × 10−6 cm2/Vs for a new capillary. When the EOF increased to ∼2 × 10−5 cm2/Vs, indicating deterioration of the capillary coating, the capillary was replaced.
Mobility calculations
Because the EOF was negligibly small, DNA electrophoretic mobilities, μ, were calculated from Eq. 1:
| (1) |
where Ld is the distance to the detector (in cm), E is the electric field strength (in V/cm), and t is time required for the sample to migrate from the capillary inlet to the detector (in seconds). The mobilities were analyzed either by calculating mobility differences [μ(A-tract) – μ(random)] or mobility ratios [μ(A-tract) /μ(random)]. Mobility ratios were used when comparing mobilities measured at different temperatures or in BGEs containing different cation concentrations; the more intuitive mobility differences were used when comparing mobilities measured in the same BGE. Negative mobility differences, or mobility ratios <1.000, indicate that the A-tract oligomers migrated more slowly than the reference; positive mobility differences, or mobility ratios >1.000, indicate the opposite. For convenience, the mobility differences are reported in mobility units (m.u., 1 m.u. = 1 ×10−4 cm2V−1s−1). Transitions were analyzed either by 4-parameter sigmoidal fits or 3-parameter exponential decays using the equations given in SigmaPlot; the transition midpoints and estimated uncertainties were taken from the fits.
All mobility measurements were repeated at least twice; duplicate measurements made on the same day usually differed by <±0.2%. Mobility measurements made on different days could differ by up to ±1.0% because of transient changes in the capillary coating, which affected the EOF. These EOF variations had no effect on the mobility differences and only a small effect on the mobility ratios, as shown below in Fig. 1 A.
Figure 1.
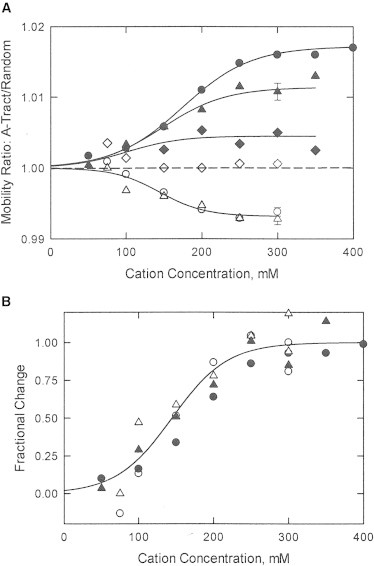
(A) Dependence of the mobility ratios, μ(A-tract)/μ(random), observed for: (•,○), A4T1in(0); (▲,Δ), A4T1in(6); and (♦,⋄), A4T1in(10) as a function of cation concentration in BGEs containing: TBA+ (solid symbols) or Li+ (open symbols). The error bars correspond to the standard deviation of replicate measurements in each BGE. (B) Fractional change of the mobility ratios observed for A4T1in(0) and A4T1in(6) as a function of cation concentration; the transition midpoint occurs at 150 ± 10 mM cation.
Results and Discussion
TBA+ and Li+ ions have opposite effects on the mobility of A-tract oligomers
The free solution mobilities of oligomers A4T1in(0), A4T1in(6), and A4T1in(10) were determined as a function of cation concentration in BGEs containing various concentrations of TBA+ or Li+ ions, with the results shown in Fig. 1 A. Mobility ratios are used to eliminate differences in the viscosities and dielectric constants of BGEs containing different concentrations of Li+ or TBA+. The mobility ratios observed in TBA+ (solid symbols) were close to 1.000 at low [TBA+] and increased sigmoidally to values >1.000 before leveling off at constant plateau values at TBA+ concentrations of ∼300 mM and greater. The amplitudes of the plateau mobility ratios decreased with the increasing number of flanking A·T basepairs in the oligomer.
In BGEs containing Li+, the mobility ratios observed for A4T1in(0) and A4T1in(6) were equal to 1.000 at low [Li+], and then decreased sigmoidally at higher [Li+], as shown by the open circles and triangles in Fig. 1 A. The mobility ratios leveled off at a plateau value of 0.993 at Li+ ion concentrations of ∼250 mM and greater. Mobility ratios <1.000 indicate that oligomers A4T1in(0) and A4T1in(6) migrated more slowly than the reference at high [Li+]. By contrast, oligomer A4T1in(10) migrated with the same mobility as the reference, as shown by the open diamonds in Fig. 1 A, so that the mobility ratio was equal to 1.000 at all [Li+]. Mobility ratios equal to 1.000 are also observed for oligomers without A-tracts (46).
Fig. 1 B shows that the mobility ratios observed for oligomers A4T1in(0) and A4T1in(6) in Li+ and in TBA+, normalized by dividing each data set by the plateau mobility ratio observed at high cation concentrations, exhibited a common dependence on cation concentration with a common midpoint, 150 ± 10 mM. Hence, even though the mobility ratios differed in sign and amplitude in TBA+ and Li+, the mobility ratios exhibited the same dependence on ionic strength. The results suggest that the A-tracts have the normal B conformation at low ionic strengths and the B∗ conformation becomes more highly populated with increasing cation concentration. Alternatively, it is possible that the mobilities of B∗ and B-DNAs are equal at low ionic strengths and that preferential interactions with monovalent cations depend on cation concentration. The preferential interactions of TBA+ and Li+ ions with the A-tract oligomers reached their limiting plateau values in solutions containing 250 to 300 mM cation. Therefore, all further experiments were carried out in BGEs containing 300 mM TBA+ or Li+.
The mobility ratios depend on temperature
The mobility ratios observed for oligomer A4T1in(6) as a function of temperature are illustrated in Fig. 2. In 300 mM Li+ (open circles), the mobility ratio gradually approached a value of 1.000 with increasing temperature, indicating a gradual loss of the B∗ conformation with increasing temperature. This transition, with a midpoint of 39° ± 1°C, cannot be attributed to duplex melting; strand separation gives an unmistakable CE signature with a decrease in the amplitude of the duplex peak and the simultaneous appearance of a peak with a much longer migration time (54,55). Because these effects did not occur, the transition observed in Li+ with increasing temperature is a premelting transition characterizing the loss of the B∗ conformation with increasing temperature. Similar premelting transitions with similar melting temperatures have been observed for other DNA A-tracts, using a variety of experimental methods (14,16,56,57).
Figure 2.
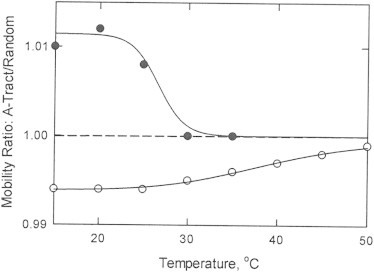
Dependence of the mobility ratios observed for oligomer A4T1in(6) on temperature in: (○), 300 mM Li+; and (•), 300 mM TBA+. The midpoints of the thermal transitions are 39 ± 1°C in Li+ and 27 ± 1°C in TBA+.
A premelting transition was also observed for oligomer A4T1in(6) in 300 mM TBA+, as shown by the solid circles in Fig. 2. Hence, A-tract oligomers have the B∗ conformation in 300 mM TBA+ as well as in 300 mM Li+. The premelting transition observed in TBA+ was highly cooperative, with a midpoint of 27 ± 1°C. The sharper transition and the relatively low premelting temperature compared with Li+ suggest that TBA+ ions preferentially interact with B-DNA, pulling the B∗ ↔ B conformational equilibrium toward B-DNA with increasing temperature. By contrast, Li+ ions apparently stabilize the B∗ conformation, increasing the premelting temperature. The differing results observed in Li+ and TBA+ suggest that the intrinsic premelting temperature of the B∗ conformation of oligomer A4T1in(6) lies somewhere between the premelting midpoints observed in Li+ and TBA+.
The observed mobilities are independent of DNA shape
The mobility ratios and mobility differences observed for the A-tract oligomers in TBA+ and Li+ could have been due either to differences in shape or to differences in effective charge. Three observations suggest that the mobility differences are not due to differences in shape. First, differences in shape are not observed in BGEs containing 300 mM cation (19). Second, previous studies have shown that the translational diffusion constants of small DNA oligomers containing the same number of basepairs are independent of the presence or absence of A-tracts, within experimental error (43,45). Third, mobility differences due to differences in shape should depend on A-tract phasing. Table 3 compares the mobility differences observed for four pairs of oligomers with A-tracts in- and out-of-phase. The ΔΔμ values [ΔΔμ = Δμ(in) – Δμ(out)] ranged from 0.0001 to 0.0037 m.u. for the four phasing pairs in TBA+ and in Li+, with mean values of 0.0017 ± 0.0015 m.u. and 0.0016 ± 0.0006 m.u., respectively. The ΔΔμ values exhibited no particular trend with A-tract length or the number of A·T basepairs in the flanking sequences. Hence, differences in shape did not contribute significantly to the mobility ratios and mobility differences observed for the A-tract oligomers in Li+ or TBA+.
Table 3.
Effect of A-tract phasing on the mobility differences observed in TBA+ and Li+
| TBA+ | Li+ | |||||
|---|---|---|---|---|---|---|
| A-tract | Δμ(in) | Δμ(out) | ΔΔμ | −Δμ(in) | −Δμ(out) | ΔΔμ |
| A5=T5(2) | 0.0151 | 0.0154 | 0.0003 | 0. 0124 | 0.0107 | 0.0017 |
| A5(2) | 0.0183 | 0.0220 | 0.0037 | 0.0109 | 0.0098 | 0.0011 |
| A3T3(4) | 0.0236 | 0.0262 | 0.0028 | 0.0088 | 0.0078 | 0.0010 |
| A4T4(0) | 0.0402 | 0.0401 | 0.0001 | 0.0138 | 0.0114 | 0.0024 |
| Mean | 0.0017 | 0.0016 | ||||
| SD | 0.0015 | 0.0006 | ||||
The mobility differences, in m.u., observed for each phasing pair, Δμ(in) and Δμ(out), and the absolute value of the difference in the mobility differences, ΔΔμ = , are indicated. For brevity, the oligomers are identified only by their A-tracts and the number of flanking A·T basepairs; the phasing is given in the column headings.
The observed mobility differences depend on effective charge
If differences in shape were not responsible for the mobility differences observed in Li+ and TBA+, the observed mobility ratios and mobility differences must have been due to differences in the effective charge of the A-tract and reference oligomers. The decreased mobility ratios observed in Li+ (Fig. 1 A) indicate that the A-tract oligomers migrated more slowly than the reference in this BGE, most likely because of the preferential localization of Li+ ions in the narrow A-tract minor groove (46), decreasing the net negative charge of the A-tract oligomers and decreasing the observed mobility. Similar mobility decreases have been observed for other small A-tract-containing oligomers in free solution (46–48).
A different type of preferential interaction must have been occurring in solutions containing TBA+ ions, because the increased mobility ratios indicate that the A-tract oligomers were migrating faster than the reference. Any effect of TBA+ size on the proximity of the counterion cloud to the DNA helix would have been the same for both A-tract and reference oligomers. Similarly, any electrostatic effects due to the reduced surface charge density of TBA+ ions would have been the same for both A-tract and reference oligomers. However, if the narrow A-tract minor groove of the B∗ conformation prevented the localization of some of the TBA+ ions in or near that groove, whereas the TBA+ ions could localize in the normal manner in or near the grooves of the reference (20–23), the effective negative charge of the A-tract oligomers would have been greater than the effective negative charge of the reference, leading to the observed increase in the mobility ratios and mobility differences.
Alternatively, it is possible that TBA+ ions preferentially interact with isolated A·T basepairs in DNA, similar to the preferential interaction of tetramethylammonium ions with A·T basepairs (58,59). In this case, the reference oligomer contained 12 isolated A·T basepairs, whereas oligomer A4T1in(6), for example, contained 6 flanking A·T basepairs. The preferential interaction of TBA+ ions with the 12 A·T basepairs in the reference oligomer would have decreased the effective charge of the reference oligomer more than the preferential interaction of TBA+ ions with the 6 flanking A·T basepairs in oligomer A4T1in(6), explaining the faster mobility of oligomer A4T1in(6) in TBA+. This topic will be discussed further below.
The mobility differences depend on A-tract length
Figs. 1 and 2 indicate that monovalent cations can either increase or decrease the mobility of an A-tract oligomer in the B∗ conformation, depending on the size and hydrophobicity of the cation, as well as the temperature and ionic strength of the solution. Fig. 3 summarizes the dependence of the average mobility differences observed per A-tract on A-tract length. Mobility differences are used here instead of mobility ratios because the viscosities of BGEs containing 300 mM of a given cation are constant at constant temperature. The oligomers contained one, two, or three A-tracts of the indicated length, variable numbers of flanking A·T basepairs, and either 10 or 14 flanking G·C basepairs (see Table 2). The average mobility differences observed per A-tract in TBA+ (solid circles) and in Li+ (open circles) are plotted as a function of the number of A·T basepairs in each A-tract. The error bars correspond to the standard deviation of the mobility differences from the average, due to variations in the A-tract sequences and the number of flanking A·T basepairs. The mobility differences observed in TBA+ and Li+ were equal to zero for A-tracts containing three basepairs, consistent with the observation that an A-tract must contain at least four contiguous An or AnTm basepairs to exhibit the narrow minor groove characteristic of the B∗ conformation (3–5).
Figure 3.
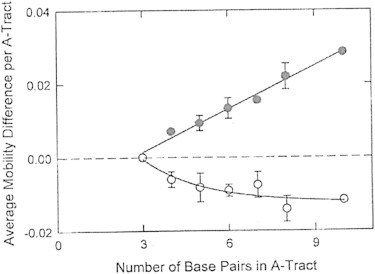
Dependence of the average mobility differences observed per A-tract, in m.u., on A-tract length. The BGEs contained 300 mM TBA+ (solid symbols); or 300 mM Li+ (open symbols). The error bars represent the standard deviation of the mobility differences from the average; the standard deviations of the individual mobility differences were smaller than the sizes of the symbols. The straight line through the TBA+ data was drawn by linear regression (r2 = 0.985); the Li+ data were fitted to a three-parameter exponential decay (r2 = 0.839).
For A-tracts containing four or more basepairs, the average mobility differences observed in TBA+ increased linearly with increasing A-tract length, most likely because the number of TBA+ ions excluded from the vicinity of the A-tract minor groove increased linearly with increasing A-tract length. In Li+, the average mobility differences increased in absolute value with increasing A-tract length, but leveled off at an approximately constant plateau value for A-tracts containing six or more A·T basepairs. The results are consistent with gel electrophoresis experiments showing that the anomalous mobilities of A-tract oligomers reach a maximum for A-tracts containing six basepairs (9).
The results in Fig. 3 suggest that A-tract length is the primary determinant of the mobility differences, modulated by A-tract sequence (An or AnTm) and the number of flanking A·T basepairs. The flanking G·C basepairs contributed relatively little to the observed mobility differences, because the mobility differences were essentially independent of whether the oligomer contained 10 or 14 flanking G·C basepairs.
The mobility differences observed for a given A-tract are modulated by the number of A·T basepairs in the flanking sequences
The contribution of the flanking A·T basepairs to the observed mobility differences was investigated by keeping the A-tract sequence constant and varying the number of flanking A·T basepairs, using the oligomers given in Table 1. The mobility differences observed for oligomers A4T1in(0) to A4T1in(11) are plotted in Fig. 4 as a function of the number of flanking A·T basepairs. In 300 mM TBA+ (solid circles), the mobility differences decreased gradually with the increasing number of flanking A·T basepairs, as though the effective length of the A-tract was gradually decreasing (see Fig. 3). It is well known that the terminal A·T basepairs in an A-tract are bridging residues with conformations that are intermediate between B∗ and B-DNA (8,12). The lifetimes of the terminal A·T basepairs in DNA A-tracts are intermediate between the lifetimes of the interior A·T basepairs and the lifetimes of isolated A·T basepairs that are not part of an A-tract (60–62). The chemical reactivities of the terminal basepairs in DNA A-tracts are also greater than the reactivities of the interior A·T basepairs (63). These results, together with Fig. 4, suggest that the terminal residues in DNA A-tracts may be somewhat flexible in TBA+ and/or may have a conformation intermediate between B∗ and B-form DNA. As a result, the TBA+ ions are not excluded from interacting with the terminal phosphate residues in the A-tracts, decreasing the effective length of the A-tract and decreasing the mobility differences between the A-tract and reference oligomers.
Figure 4.
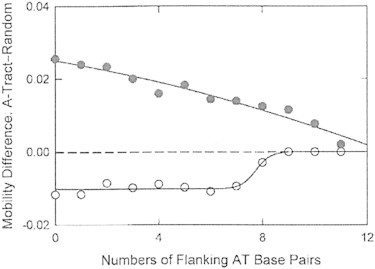
Mobility differences, in m.u., observed for oligomers A4T1in(0) to A4T1in(11) as a function of the number of flanking A·T basepairs, in BGEs containing: (•), 300 mM TBA+; or (○), 300 mM Li+. Error bars corresponding to the reproducibility of the measurements are smaller than the sizes of the symbols.
Alternatively, or in addition, TBA+ ions may interact preferentially with isolated A·T basepairs in the reference oligomer and in the A-tract flanking sequences, as discussed previously. Because the reference oligomer contained 12 isolated A·T basepairs, whereas the A-tract oligomers contained up to 11 flanking A·T basepairs (Table 1), the mobility differences would be expected to decrease approximately linearly to zero with the increasing number of flanking A·T basepairs, as observed. Further studies are needed to distinguish between these two possibilities.
In 300 mM Li+ (open circles in Fig. 4), the mobility differences were negative in sign and constant in amplitude for oligomers containing 0 to 7 flanking A·T basepairs, most likely because the localization of Li+ ions in the A-tract minor groove (46) stabilized the B∗ conformation even when the flanking sequences contained a significant number of A·T basepairs. Surprisingly, the mobility differences observed in Li+ decreased to zero for oligomers containing 9 or more flanking A·T basepairs, suggesting that Li+ ions cannot stabilize the B∗ conformation when the flanking sequences contain more A·T basepairs (9, in this case) than G·C basepairs (7). Under such conditions, the B∗ ↔ B conformational equilibrium shifts to B-DNA and the mobilities of the A-tract and random oligomers become equal.
The results in Fig. 4 indicate not only that Li+ and TBA+ ions interact differently with DNA A-tracts, but that significant numbers of A·T basepairs in the flanking sequences can alter these interactions. Previous studies have not detected the effect of flanking A·T basepairs on the B∗ conformation because the A-tracts were always flanked by G·C basepairs (1–6,9,12,26–31). The studies presented here have shown that cation interactions with DNA A-tracts depend primarily on A-tract length (Fig. 3), modulated by the number of A·T basepairs in the flanking sequences (Fig. 4). Hence, DNA A-tracts may not exhibit the B∗ conformation and may not interact preferentially with monovalent cations when the flanking sequences contain a significant number of A·T basepairs.
Conclusions
The results in this study indicate that preferential interactions of monovalent cations with DNA A- tracts depend on the identity of the cation, the length and sequence of the A-tract, and the relative proportions of A·T and G·C basepairs in the flanking sequences. Hence, DNA A-tracts embedded in AT-rich sequences may not have a narrow minor groove and may not preferentially interact with monovalent cations in the surrounding medium. Such sequence-dependent variations in minor groove width are likely to contribute to indirect readout mechanisms (5,64–68) and nucleosome positioning (69).
DNA A-tracts appear to exist in a conformational equilibrium between B∗ and B-DNA. The B∗ conformation is stabilized in solutions containing physiological concentrations of monovalent cations, but is destabilized by the physiological temperature of 37°C. The B∗ ↔ B conformational equilibrium is also affected by A-tract length, the relative number of flanking A·T and G·C basepairs, and preferential interactions of the A-tract oligomers with various types of cations in the solution. Small hydrophilic cations such as Li+ stabilize the B∗ conformation by preferential localization in the A-tract minor groove. Large hydrophobic cations such as TBA+ appear to destabilize the B∗ conformation, possibly by exclusion from the vicinity of the A-tract minor groove or by preferential interactions with isolated A·T basepairs in the flanking sequences, pulling the B∗ ↔ B equilibrium toward B-DNA. Although TBA+ ions do not exist in the cell, other large hydrophobic cations such as histones and polyamines are present and may interact with DNA A-tracts in a similar manner. A detailed study of the competing interactions of hydrophobic and hydrophilic monovalent cations with DNA A-tracts embedded in a variety of flanking sequences will be presented separately.
Author Contributions
E.S., Q.D., and N.C.S. designed research; Q.D. and E.S. performed research; E.S., Q.D., and N.C.S. analyzed data; E.S. and N.C.S. wrote the article.
Acknowledgments
This work was supported in part by Grants GM61009 from the National Institute of General Medical Sciences and CHE748271 from the Analytical and Surface Chemistry Program of the National Science Foundation (to N.C.S.).
References
- 1.Coll M., Frederick C.A., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc. Natl. Acad. Sci. USA. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson H.C.M., Finch J.T., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 3.Hagerman P.J. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 4.Olson W.K., Zhurkin V.B. Twenty years of DNA bending. In: Sarma R.H., Sarma M.H., editors. Biological Structure and Dynamics. Adenine Press; Schenectady, NY: 1996. pp. 341–370. [Google Scholar]
- 5.Haran T.E., Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 6.Stefl R., Wu H., Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl. Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald D., Herbert K., Lu P. Solution structure of an A-tract DNA bend. J. Mol. Biol. 2001;306:1081–1098. doi: 10.1006/jmbi.2001.4447. [DOI] [PubMed] [Google Scholar]
- 8.Barbič A., Zimmer D.P., Crothers D.M. Structural origins of adenine-tract bending. Proc. Natl. Acad. Sci. USA. 2003;100:2369–2373. doi: 10.1073/pnas.0437877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koo H.-S., Wu H.-M., Crothers D.M. DNA bending at adenine.thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 10.Segal E., Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal M., Kumar A., Yella V.R. Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr. Opin. Struct. Biol. 2014;25:77–85. doi: 10.1016/j.sbi.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Hizver J., Rozenberg H., Shakked Z. DNA bending by an adenine—thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. USA. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera J.E., Chaires J.B. A premelting conformational transition in poly(dA)-poly(dT) coupled to daunomycin binding. Biochemistry. 1989;28:1993–2000. doi: 10.1021/bi00431a006. [DOI] [PubMed] [Google Scholar]
- 14.Park Y.-W., Breslauer K.J. A spectroscopic and calorimetric study of the melting behaviors of a “bent” and a “normal” DNA duplex: [d(GA4T4C)]2 versus [d(GT4A4C)]2. Proc. Natl. Acad. Sci. USA. 1991;88:1551–1555. doi: 10.1073/pnas.88.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan S.S., Breslauer K.J., Hogan M.E. Thermodynamics and premelting conformational changes of phased (dA)5 tracts. Biochemistry. 1993;32:11776–11784. doi: 10.1021/bi00095a005. [DOI] [PubMed] [Google Scholar]
- 16.Jerkovic B., Bolton P.H. The curvature of dA tracts is temperature dependent. Biochemistry. 2000;39:12121–12127. doi: 10.1021/bi0014507. [DOI] [PubMed] [Google Scholar]
- 17.Augustyn K.E., Wojtuszewski K., Mukerji I. Examination of the premelting transition of DNA A-tracts using a fluorescent adenosine analogue. Biochemistry. 2006;45:5039–5047. doi: 10.1021/bi0518343. [DOI] [PubMed] [Google Scholar]
- 18.Chan S.S., Austin R.H., Spiro T.G. Temperature-dependent ultraviolet resonance Raman spectroscopy of the premelting state of dA.dT DNA. Biophys. J. 1997;72:1512–1520. doi: 10.1016/S0006-3495(97)78799-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stellwagen E., Peters J.P., Stellwagen N.C. DNA A-tracts are not curved in solutions containing high concentrations of monovalent cations. Biochemistry. 2013;52:4138–4148. doi: 10.1021/bi400118m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Y., Greenfeld M., Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husale S., Grange W., Hegner M. Interaction of cationic surfactants with DNA: a single-molecule study. Nucleic Acids Res. 2008;36:1443–1449. doi: 10.1093/nar/gkm1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo J., Aksimentiev A. Competitive binding of cations to duplex DNA revealed through molecular dynamics simulations. J. Phys. Chem. B. 2012;116:12946–12954. doi: 10.1021/jp306598y. [DOI] [PubMed] [Google Scholar]
- 23.Giambaşu G.M., Luchko T., Case D.A. Ion counting from explicit-solvent simulations and 3D-RISM. Biophys. J. 2014;106:883–894. doi: 10.1016/j.bpj.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mocci F., Saba G. Molecular dynamics simulations of A. T-rich oligomers: sequence-specific binding of Na+ in the minor groove of B-DNA. Biopolymers. 2003;68:471–485. doi: 10.1002/bip.10334. [DOI] [PubMed] [Google Scholar]
- 25.Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 26.Howerton S.B., Sines C.C., Williams L.D. Locating monovalent cations in the grooves of B-DNA. Biochemistry. 2001;40:10023–10031. doi: 10.1021/bi010391+. [DOI] [PubMed] [Google Scholar]
- 27.Tereshko V., Wilds C.J., Egli M. Detection of alkali metal ions in DNA crystals using state-of-the-art x-ray diffraction experiments. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods K.K., Maehigashi T., Williams L.D. High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2. J. Am. Chem. Soc. 2004;126:15330–15331. doi: 10.1021/ja045207x. [DOI] [PubMed] [Google Scholar]
- 29.Hud N.V., Sklenár V., Feigon J. Localization of ammonium ions in the minor groove of DNA duplexes in solution and the origin of DNA A-tract bending. J. Mol. Biol. 1999;286:651–660. doi: 10.1006/jmbi.1998.2513. [DOI] [PubMed] [Google Scholar]
- 30.Cesare Marincola F., Denisov V.P., Halle B. Competitive Na+ and Rb+ binding in the minor groove of DNA. J. Am. Chem. Soc. 2004;126:6739–6750. doi: 10.1021/ja049930z. [DOI] [PubMed] [Google Scholar]
- 31.Hamelberg D., McFail-Isom L., Wilson W.D. Flexible structure of DNA: ion dependence of minor-groove structure and dynamics. J. Am. Chem. Soc. 2000;122:10513–10520. [Google Scholar]
- 32.Hud N.V., Plavec J. A unified model for the origin of DNA sequence-directed curvature. Biopolymers. 2003;69:144–158. doi: 10.1002/bip.10364. [DOI] [PubMed] [Google Scholar]
- 33.Pérez A., Luque F.J., Orozco M. Dynamics of B-DNA on the microsecond time scale. J. Am. Chem. Soc. 2007;129:14739–14745. doi: 10.1021/ja0753546. [DOI] [PubMed] [Google Scholar]
- 34.Egli M. DNA-cation interactions: quo vadis? Chem. Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 35.Subirana J.A., Soler-Lopez M. Cations as hydrogen bond donors: a view of electrostatic interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003;32:27–45. doi: 10.1146/annurev.biophys.32.110601.141726. [DOI] [PubMed] [Google Scholar]
- 36.McConnell K.J., Beveridge D.L. DNA structure: what’s in charge? J. Mol. Biol. 2000;304:803–820. doi: 10.1006/jmbi.2000.4167. [DOI] [PubMed] [Google Scholar]
- 37.Lavery R., Maddocks J.H., Zakrzewska K. Analyzing ion distributions around DNA. Nucleic Acids Res. 2014;42:8138–8149. doi: 10.1093/nar/gku504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stellwagen N.C., Gelfi C., Righetti P.G. The free solution mobility of DNA. Biopolymers. 1997;42:687–703. doi: 10.1002/(SICI)1097-0282(199711)42:6<687::AID-BIP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Ma C., Bloomfield V.A. Gel electrophoresis measurement of counterion condensation on DNA. Biopolymers. 1995;35:211–216. doi: 10.1002/bip.360350209. [DOI] [PubMed] [Google Scholar]
- 40.Li A.Z., Qi L.J., Marx K.A. Trivalent counterion condensation on DNA measured by pulse gel electrophoresis. Biopolymers. 1996;38:367–376. doi: 10.1002/(sici)1097-0282(199603)38:3<367::aid-bip9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 41.Li A.Z., Huang H., Marx K.A. A gel electrophoresis study of the competitive effects of monovalent counterion on the extent of divalent counterions binding to DNA. Biophys. J. 1998;74:964–973. doi: 10.1016/S0006-3495(98)74019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank S., Winkler R.G. Mesoscale hydrodynamic simulation of short polyelectrolytes in electric fields. J. Chem. Phys. 2009;131:234905. doi: 10.1063/1.3274681. [DOI] [PubMed] [Google Scholar]
- 43.Stellwagen E., Lu Y., Stellwagen N.C. Unified description of electrophoresis and diffusion for DNA and other polyions. Biochemistry. 2003;42:11745–11750. doi: 10.1021/bi035203p. [DOI] [PubMed] [Google Scholar]
- 44.Viovy J.-L. Electrophoresis of DNA and other polyelectrolytes: physical mechanisms. Rev. Mod. Phys. 2000;72:813–872. [Google Scholar]
- 45.Stellwagen N.C., Magnusdóttir S., Righetti P.G. Measuring the translational diffusion coefficients of small DNA molecules by capillary electrophoresis. Biopolymers. 2001;58:390–397. doi: 10.1002/1097-0282(20010405)58:4<390::AID-BIP1015>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 46.Dong Q., Stellwagen E., Stellwagen N.C. Monovalent cation binding in the minor groove of DNA A-tracts. Biochemistry. 2009;48:1047–1055. doi: 10.1021/bi8020718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stellwagen N.C., Magnusdóttir S., Righetti P.G. Preferential counterion binding to A-tract DNA oligomers. J. Mol. Biol. 2001;305:1025–1033. doi: 10.1006/jmbi.2000.4362. [DOI] [PubMed] [Google Scholar]
- 48.Stellwagen E., Lu Y., Stellwagen N.C. Curved DNA molecules migrate anomalously slowly in free solution. Nucleic Acids Res. 2005;33:4425–4432. doi: 10.1093/nar/gki748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stellwagen E., Stellwagen N.C. The free solution mobility of DNA in Tris-acetate-EDTA buffers of different concentrations, with and without added NaCl. Electrophoresis. 2002;23:1935–1941. doi: 10.1002/1522-2683(200206)23:12<1935::AID-ELPS1935>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 50.Stellwagen E., Stellwagen N.C. Probing the electrostatic shielding of DNA with capillary electrophoresis. Biophys. J. 2003;84:1855–1866. doi: 10.1016/S0006-3495(03)74993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stellwagen N.C., Peters J.P., Stellwagen E. The free solution mobility of DNA and other analytes varies as the logarithm of the fractional negative charge. Electrophoresis. 2014;35:1855–1863. doi: 10.1002/elps.201400040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stellwagen E., Dong Q., Stellwagen N.C. Quantitative analysis of monovalent counterion binding to random-sequence, double-stranded DNA using the replacement ion method. Biochemistry. 2007;46:2050–2058. doi: 10.1021/bi062132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams B.A., Vigh G. Fast, accurate mobility determination method for capillary electrophoresis. Anal. Chem. 1996;68:1174–1180. doi: 10.1021/ac950968r. [DOI] [PubMed] [Google Scholar]
- 54.Stellwagen E., Renze A., Stellwagen N.C. Capillary electrophoresis is a sensitive monitor of the hairpin-random coil transition in DNA oligomers. Anal. Biochem. 2007;365:103–110. doi: 10.1016/j.ab.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stellwagen E., Abdulla A., Stellwagen N.C. Electrophoretic mobility is a reporter of hairpin structure in single-stranded DNA oligomers. Biochemistry. 2007;46:10931–10941. doi: 10.1021/bi701058f. [DOI] [PubMed] [Google Scholar]
- 56.Chan S.S., Breslauer K.J., Wiles N.C. Physical studies of DNA premelting equilibria in duplexes with and without homo dA.dT tracts: correlations with DNA bending. Biochemistry. 1990;29:6161–6171. doi: 10.1021/bi00478a008. [DOI] [PubMed] [Google Scholar]
- 57.Mukerji I., Williams A.P. UV resonance Raman and circular dichroism studies of a DNA duplex containing an A3T3 tract: evidence for a premelting transition and three-centered H-bonds. Biochemistry. 2002;41:69–77. doi: 10.1021/bi010918i. [DOI] [PubMed] [Google Scholar]
- 58.Portella G., Germann M.W., Orozco M. MD and NMR analyses of choline and TMA binding to duplex DNA: on the origins of aberrant sequence-dependent stability by alkyl cations in aqueous and water-free solvents. J. Am. Chem. Soc. 2014;136:3075–3086. doi: 10.1021/ja410698u. [DOI] [PubMed] [Google Scholar]
- 59.Nakano M., Tateishi-Karimata H., Sugimoto N. Choline ion interactions with DNA atoms explain unique stabilization of A-T base pairs in DNA duplexes: a microscopic view. J. Phys. Chem. B. 2014;118:379–389. doi: 10.1021/jp406647b. [DOI] [PubMed] [Google Scholar]
- 60.Leroy J.-L., Charretier E., Guéron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988;27:8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- 61.Moe J.G., Russu I.M. Proton exchange and base-pair opening kinetics in 5′-d(CGCGAATTCGCG)-3′ and related dodecamers. Nucleic Acids Res. 1990;18:821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leroy J.L., Kochoyan M., Guéron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J. Mol. Biol. 1988;200:223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 63.McCarthy J.G., Williams L.D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990;29:6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- 64.Parker S.C.J., Hansen L., Margulies E.H. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohs R., West S.M., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oguey C., Foloppe N., Hartmann B. Understanding the sequence-dependence of DNA groove dimensions: implications for DNA interactions. PLoS ONE. 2010;5:e15931. doi: 10.1371/journal.pone.0015931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris L.-A., Watkins D., Koudelka G.B. Indirect readout of DNA sequence by p22 repressor: roles of DNA and protein functional groups in modulating DNA conformation. J. Mol. Biol. 2013;425:133–143. doi: 10.1016/j.jmb.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Hancock S.P., Ghane T., Johnson R.C. Control of DNA minor groove width and Fis protein binding by the purine 2-amino group. Nucleic Acids Res. 2013;41:6750–6760. doi: 10.1093/nar/gkt357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X., Ben Imeddourene A., Hartmann B. NMR studies of DNA support the role of pre-existing minor groove variations in nucleosome indirect readout. Biochemistry. 2014;53:5601–5612. doi: 10.1021/bi500504y. [DOI] [PubMed] [Google Scholar]


