Abstract
The notorious unresponsiveness of metastatic cutaneous melanoma to current treatment strategies coupled with its increasing incidence constitutes a serious worldwide clinical problem. Moreover, despite recent advances in targeted therapies for patients with BRAFV600E mutant melanomas, acquired resistance remains a limiting factor and hence emphasises the acute need for comprehensive pre-clinical studies to increase the biological understanding of such tumours in order to develop novel effective and longlasting therapeutic strategies. Autophagy and ER stress both have a role in melanoma development/progression and chemoresistance although their real impact is still unclear. Here, we show that BRAFV600E induces a chronic ER stress status directly increasing basal cell autophagy. BRAFV600E-mediated p38 activation stimulates both the IRE1/ASK1/JNK and TRB3 pathways. Bcl-XL/Bcl-2 phosphorylation by active JNK releases Beclin1 whereas TRB3 inhibits the Akt/mTor axes, together resulting in an increase in basal autophagy. Furthermore, we demonstrate chemical chaperones relieve the BRAFV600E-mediated chronic ER stress status, consequently reducing basal autophagic activity and increasing the sensitivity of melanoma cells to apoptosis. Taken together, these results suggest enhanced basal autophagy, typically observed in BRAFV600E melanomas, is a consequence of a chronic ER stress status, which ultimately results in the chemoresistance of such tumours. Targeted therapies that attenuate ER stress may therefore represent a novel and more effective therapeutic strategy for BRAF mutant melanoma.
Cutaneous melanoma represents one of the most aggressive and difficult to treat forms of human cancer, with a worldwide incidence that has steadily increased over the past 40 years.1, 2
Notoriously unresponsive to conventional chemotherapy, metastatic disease is highly invasive and evolves with an extensive repertoire of molecular defences against immunological and cytotoxic attack.3
Although linked to exposure to ultraviolet light, it is widely accepted that both genotypic and phenotypic changes in melanocytes predispose to melanocyte transformation and the onset of melanoma.4, 5
Surprisingly, p53 mutations are very rare in melanoma, but activity is, however, impaired through direct or indirect inactivation of key elements of this pathway, including through the suppression of APAF-1 expression,6 loss of PTEN function,7 dysregulation of Bcl-2 expression,8 upregulation of the anti-apoptotic protein Mcl-1 together with its altered slice variant expression 9, 10 and the ER chaperone GRP78.11, 12, 13 Oncogenic mutations, however, in the Ras/Raf pathway are the most well-described genetic mutations associated with melanoma development and progression.14 Indeed, up to 90% of all melanomas harbour activating NRAS or BRAF mutations, with BRAFV600E representing more than 90% of BRAF mutations,15, 16 the consequence of which is the constitutive activation of RAF-extracellular signal-regulated kinase/ERK signalling promoting melanoma proliferation and resistance to apoptosis.17 Nevertheless, mutation of NRAS/BRAF alone is not sufficient to initiate melanomagenesis, because these common mutations are also present in benign nevi, thereby highlighting the requirement of other factors to drive melanocyte transformation and melanoma development.15, 16 Dysregulation of autophagy has accordingly been postulated as a secondary event contributing to melanoma progression and, importantly, also has a key role in chemoresistance.18, 19, 20
Autophagy is the principal catabolic process for the bulk degradation and recycling of aged/damaged cellular components, organelles and proteins through the formation of a double-membraned cytosolic vesicle able to wrap targeted material. The subsequent fusion with lysosomes and degradation of cargo provide nutrients in times of environmental stress, such as nutrient deprivation or hypoxia.21 Though essential for the maintenance of cellular homeostasis under conditions of nutrient deprivation, paradoxically, autophagy promotes both tumour suppression and tumour development.22 Although the accumulation of damaged organelles/cytosolic proteins may lead to cellular transformation, autophagy may also sustain tumour growth in a microenvironment which is commonly poor of oxygen and nutrients.22 Thus, not surprisingly, autophagy activation is frequently observed in late-stage malignancy although the molecular mechanisms mediating its activation/regain of function remain unclear.
ER stress may also constitute a key secondary event in melanoma development.23 Primarily a cytoprotective pro-survival process, ER stress is activated as a result of accumulated unfolded proteins, protein overload or depletion of ER calcium stores and mediated through the activity of the master ER chaperone Grp78 and three signalling pathways; PERK/eIF2α/ATF4, IRE-1/Xbp-1 and ATF6 which collectively maintain ER homeostasis through the instigation of an unfolded protein response (UPR)24 or sustained ER stress may lead to the induction of apoptosis.25, 26 Increasing evidence indicates that nutrient deprivation and hypoxia lead to activation of the UPR in various solid tumours, frequently correlating with resistance to chemotherapy.27 The accepted hypothesis is thus that activation of the UPR in cancer cells enables their adaption to such ER stress resulting in the resistance to apoptosis through the persistent expression of pro-survival instead of pro-apoptotic proteins.28
Although under stress conditions, autophagy and ER stress seem to act in parallel, indeed they are closely related, because one can regulate the other and vice versa. In fact, ER stress is able to promptly stimulate autophagy,26 whereas autophagy selectively removes the membranes of the endoplasmic reticulum at the end of the UPR, although the molecular mechanisms are still largely unclear.29
In the present study, we investigated the link between oncogenic BRAFV600E and increased basal autophagy in melanoma cells, highlighting the pivotal role played by ER stress, possibly responsible for tumour growth and chemoresistance.
Results
BRAFV600E increases basal autophagy in melanoma cells
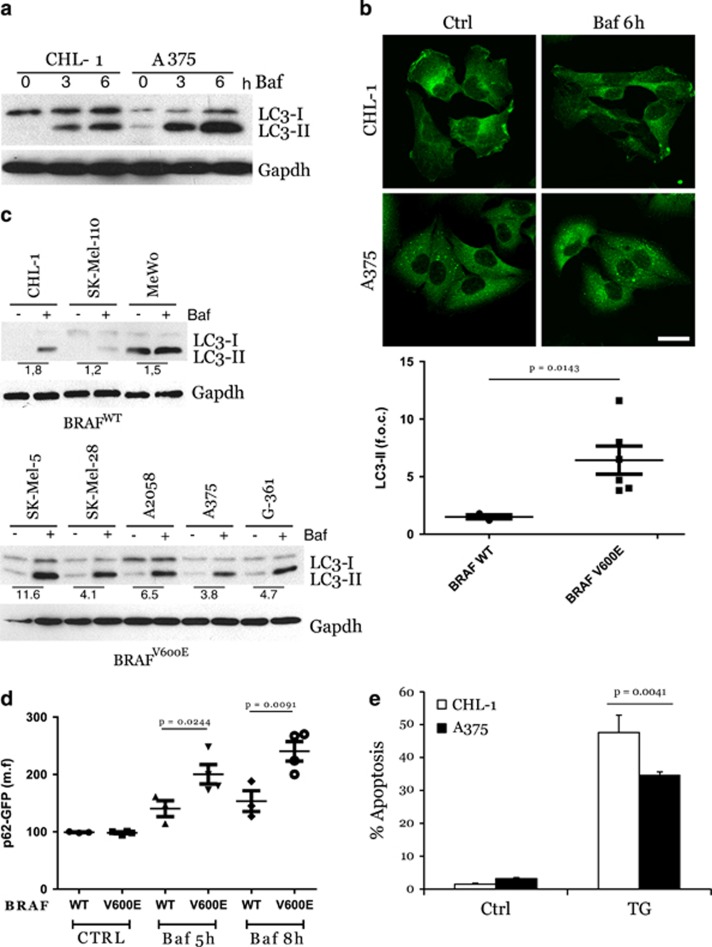
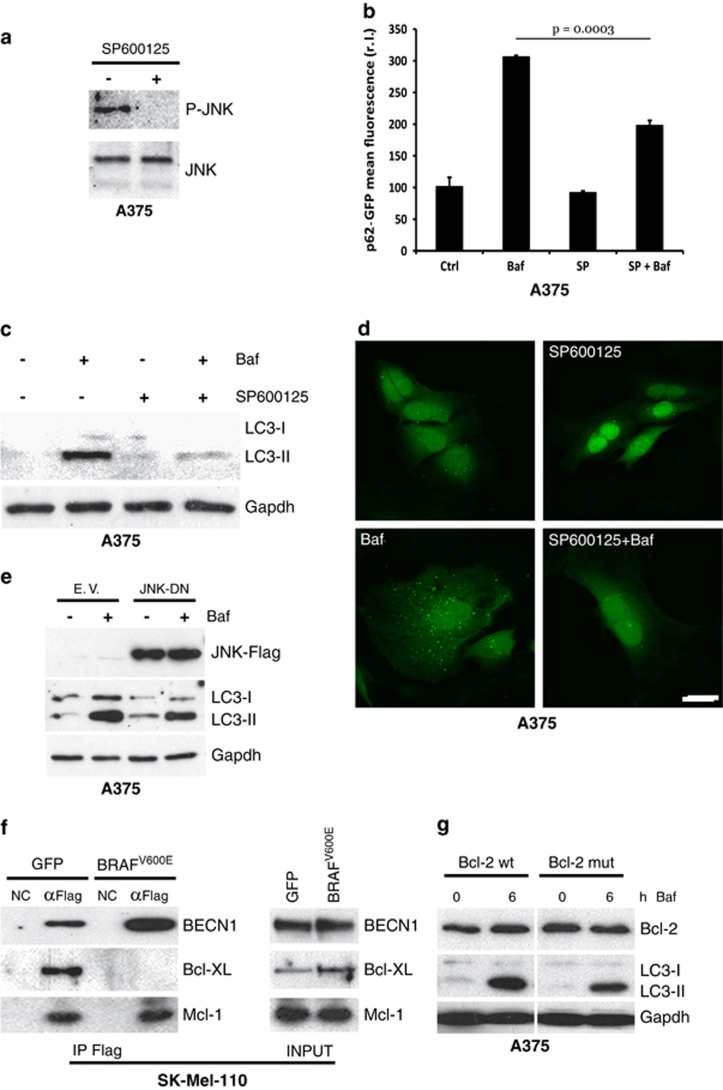
The deregulation of autophagy is frequent in malignancy, including in cutaneous melanomas. In particular, we and others have recently demonstrated that melanomas harbouring mutant BRAFV600E display an increased basal autophagic rate.3, 20, 30, 31 In order to confirm this observation, we compared basal autophagic flux in melanoma cell lines, CHL-1 and A375, with BRAF wild-type (wt) or V600E-mutated alleles, respectively. As shown in Figure 1a, A375 cells displayed increased expression of lipidated LC3 compared with CHL-1 cells, which was sustained by co-treatment with bafilomycin A (Baf), to prevent autophagic flux. To further confirm these observations, analysis of LC3 conversion was also performed in an extended panel of BRAF wt or mutated melanoma cells (Supplementary Table 1,Figure 1c). Results confirmed an overall increase in basal autophagy in BRAF mutant compared with BRAF wt cells (Figure 1c). Moreover, increased basal degradation of the autophagy cargo p62 was also observed in A375 compared with CHL-1 cells, as evidenced by the accumulation of this protein in the presence of Baf (Figure 1b), and confirmed comparing p62 accumulation in BRAF-mutated versus BRAF wt melanoma cell lines, in a time-dependent manner (Figure 1d).
Figure 1.
Enhanced basal autophagy and less apoptotic responsiveness of BRAFV600E melanoma cells. BRAF wt (CHL-1) and BRAFV600E (A375) melanoma cells were exposed 3 and 6 h (a) to bafilomycin A (Baf) and basal autophagy was evaluated by monitoring both the LC3 conversion by western blotting analysis (a; Gapdh was used as the loading control) and p62 accumulation by immunofluorescence (b; bar=10 μm). Basal autophagy was also evaluated in a panel of BRAFWT or BRAFV600E melanoma cell lines, in presence or absence of Baf (4 h), by monitoring LC3 conversion by western blotting analysis (c, left panels; Gapdh was used as the loading control). Densitometric analysis of LC3-II bands is shown in each cell lines, comparing treated with untreated cells (with Baf); mean±S.D. of LC3-II bands in BRAFWT and BRAFV600E cell lines is also reported (c, right panel). Autophagic flux was quantitated in all melanoma cell lines stably expressing a p62-GFP recombinant protein, treated with Baf in a time-dependent manner, by cytofluorimetric analysis (d; n=3). Apoptosis induction was evaluated by cytofluorimetric analysis of propidium iodide-stained CHL-1 and A375 cells exposed 24 h to thapsigargin (TG, 10 μg/ml; (e; n=3; P=0.004)
As autophagy is primarily a pro-survival pathway actively counteracting the apoptotic process, high basal autophagy levels may account for, at least in part, the notorious resistance of BRAF-mutated melanoma cells to ER stress-induced apoptosis,17 as confirmed in Figure 1e also showing that A375 cells are more resistant to thapsigargin-induced apoptosis, compared with CHL-1 cells.
Oncogenic BRAF results in increased basal autophagy through chronic ER stress induction
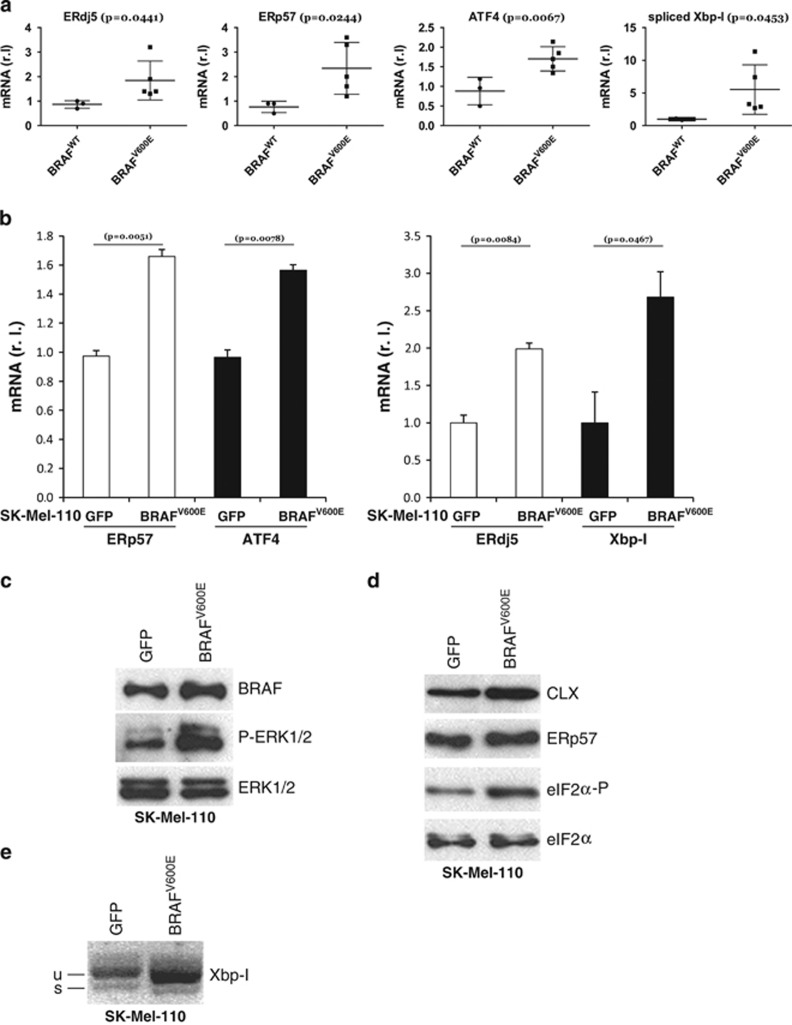
We have previously shown that BRAF-mutated melanoma cells display reduced sensitivity to ER stress activation compared with wt cells, suggesting that an abrogated ER stress response may limit drug-induced apoptosis in these cells20, 32 (Figure 1). We therefore hypothesised that the reduced ER stress response observed in BRAF mutant melanoma cells results from chronic ER stress that desensitizes them to further stress stimuli.28 To test this hypothesis, we analysed the expression levels of ER stress markers in our extended panel of BRAFV600E melanoma cells compared with BRAFWT cells. As shown in Figure 2a, BRAFV600E melanoma cell lines displayed approximately twofold greater expression of mRNA coding for ERdj5, ERp57, ATF4 and Xbp-I33 compared with BRAF wt cells, indicating that a mild but consistent induction of ER stress is conferred by an activating mutation in the BRAF protein kinase.28, 32, 33
Figure 2.
BRAF induces a chronic ER stress status. (a) BRAF-mediated ER stress was evaluated in both BRAFWT and BRAFV600E melanoma cell lines by measuring the mRNA levels of ER stress markers ERdj5, ERp57, ATF4 and Xbp-1 (spliced, mature form) by qRT-PCR (n=3). BRAFV600E and GFP were transduced in BRAF wt SK-Mell-110 melanoma cells. BRAF and P-ERK1/2 (ERK1/2 was used as the loading control) protein levels were evaluated by western blotting analysis (c). ER stress status was monitored in GFP or BRAFV600E expressing cells comparing: (b) mRNA levels of ERp57, ATF4, ERdj5 and Xbp-1 (spliced, mature form) by qRT-PCR (n=3); (d) protein level of Calnexin (CLX), ERp57 and eIF2a-P (eIF2a was used as loading control) by western blotting analysis and (e) Xbp-1 mRNA splicing by RT-PCR
To rule out the possibility that differences in ER stress induction were due to effects other than BRAF mutation, we expressed constitutively active BRAFV600E in the BRAF wt SK-Mel-110 melanoma cell line in order to compare melanoma cells with identical genetic background except for BRAF status. Cells stably expressing BRAFV600E or GFP were selected for more than 1 month in culture, in order to better represent in vivo tumour development and adaptation to BRAF activating mutation. Expression of oncogenic BRAF in SK-Mel-110 cells, as evidenced by ERK1/2 hyper-phosphorylation (Figure 2c), resulted in a clear upregulation of mRNA coding for ERp57, ATF4, ERdj5 and Xbp-I (Figure 2b), increased levels of both calnexin and ERp57 proteins and eIF2α phosphorylation (Figure 2d), as well as enhanced Xbp-I splicing (Figure 2e), confirming that BRAF mutation is solely responsible for ER compartment deregulation, and compatible with a chronic ER stress status. These data thus support recent observations of increased ER stress as a consequence of BRAF therapy-induced resistance by Ma and co-workers,34 and provide additional insight into the mechanisms mediating increased basal ER stress in BRAF-mutated melanoma cells.
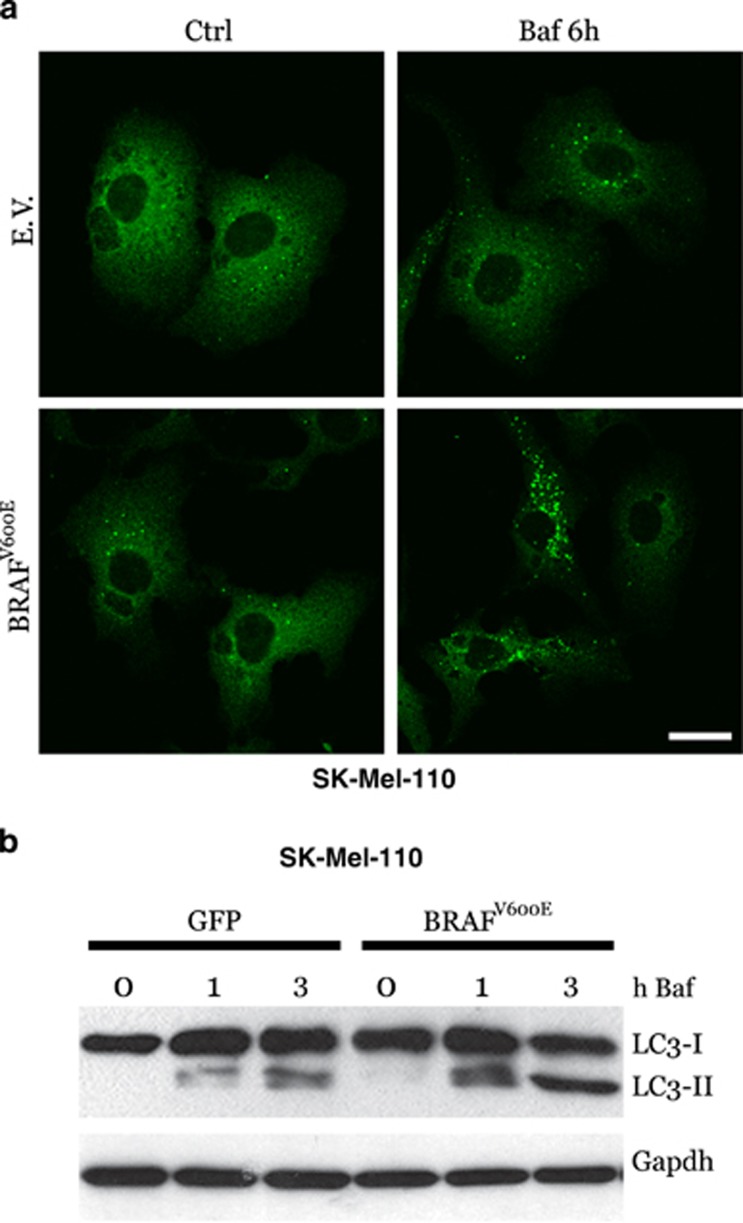
Furthermore, overexpression of BRAFV600E also resulted in increased basal autophagy in SK-Mel-110 cells, as indicated by both the cytosolic accumulation of p62 puncta (Figure 3a) and LC3 conversion (Figure 3b), in a time-dependent manner and under conditions of bafilomycin exposure, suggesting a correlation between chronic ER stress and increased basal autophagy in BRAFV600E mutant cells, as previously evidenced in Figure 1.
Figure 3.
BRAFV600E expression and basal autophagy. BRAFV600E and GFP or empty vector (EV) were transduced in BRAF wt SK-Mell-110 melanoma cells. Cells were incubated with bafilomycin A as indicated, and basal autophagy was evaluated by both confocal analysis of p62 puncta (a; bar=10 μm) or LC3 conversion by western blotting (b; Gapdh was used as the loading control)
ER stress-mediated JNK activation contributes to basal autophagy induction
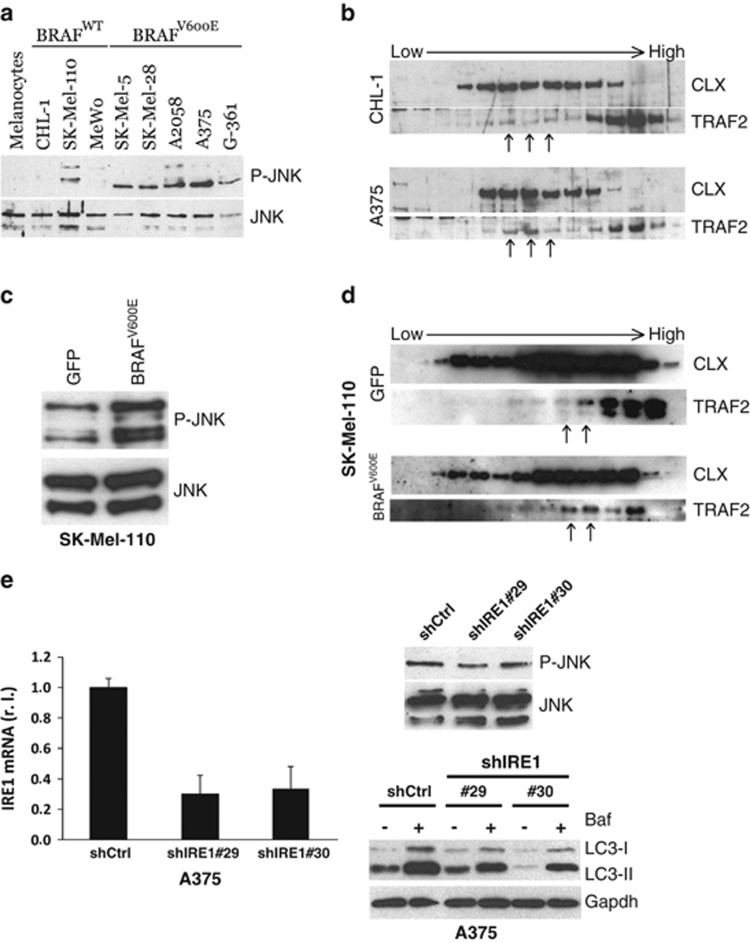
To unveil the possible link connecting oncogenic BRAF-induced chronic ER stress and the elevation of basal autophagy, we explored the activation of known pathways linking these two processes. Results demonstrated constitutive phosphorylation of JNK in BRAFV600E compared with BRAF wt cells and normal melanocytes, confirming protein kinase activation (Figure 4a). To verify whether JNK activation was specifically due to the presence of oncogenic BRAF, we analysed JNK phosphorylation in SK-Mel-110 cells stably expressing BRAFV600E or GFP. As shown in Figure 4c, phosphorylated JNK is strictly associated with the expression of mutant BRAF.
Figure 4.
Autophagy modulation by the IRE1/TRAF2/JNK axis. JNK activation (P-JNK) was evaluated in primary melanocytes and in a panel of BRAFWT or BRAFV600E melanoma cell lines (a), or in SK-Mell-110-expressing GFP or BRAFV600E (c) cells by western blotting analysis (JNK was used as loading control). The recruitment of TRAF2 by activated IRE1 on ER was monitored by western blotting analysis of subcellular fractions from CHL-1 and A375 (b) or SK-Mell-110-expressing GFP or BRAFV600E (d) cells, by using specific anti-TRAF2 and anti-CLX antibodies; arrows have been used to highlight the different distribution of TRAF2 onto ER membranes, between compared cell lines. (e) A375 cells were infected with indicated shIRE1- or shCtrl-carrying lentiviruses and IRE1 levels were evaluated by qRT-PCR (right panel; P=0.0005; n=3); P-JNK was evaluated by western blotting analysis (JNK was used as the loading control; upper panel) and LC3 conversion was monitored by western blotting analysis in the presence or absence of bafilomycin (Baf, 3 h; Gapdh was used as the loading control; bottom panel)
ER stress-mediated JNK activation has also been reported to be associated with the activation of the transmembrane ER protein IRE1 and subsequent recruitment of cytosolic ASK1 through the adapter molecule TRAF2.35 This multiprotein complex determines the activation of the kinase ASK1, thus mediating the phosphorylation of JNK.35 To verify the involvement of this pathway in oncogenic BRAF-induced ER stress-mediated JNK activation, we evaluated the translocation of TRAF2 onto ER membranes using a subcellular fractionation assay. Results reported in Figures 4b and d confirmed a mild but significant relocalization of TRAF2 onto the ER compartment, as indicated by the colocalization with the ER marker calnexin, in cells expressing the mutated BRAF (see arrows in Figures 4b and d, compare each upper panel with corresponding bottom panel), consistent with a chronic rather than an acute ER stress status. Importantly, this relocalization was specifically induced by BRAFV600E because GFP-transduced SK-Mel-110 cells exhibited TRAF2 distribution similar to that observed in CHL-1 cells (see arrows as marked in Figures 4b and d, upper panels). This evidence was further confirmed by evaluating the relocalization of TRAF2 onto ER membranes of other BRAF-mutated cell lines compared with BRAF wt once (Supplementary Figure S1).
To confirm the role of the IRE-1/TRAF2/ASK1 axes in the JNK activation and autophagy induction in BRAFV600E cells, we downregulated the expression of IRE1 using two specific shRNAs in A375 cells (Figure 4e, left panel) and evaluated the level of both phosphorylated JNK and LC3 conversion. As shown in Figure 4e, the impairment of the IRE1 signalling pathway resulted in decreased JNK activation (upper right panel) and concomitant reduction of A375 basal autophagy (bottom right panel). To further confirm these results, A375 cells were treated with SP600125 (10 μM) to inhibit JNK activation36 prior to analysing of basal autophagic activity, in the presence or absence of bafilomycin. As shown in Figures 5a–d, inhibiting the activation of JNK resulted in a clear decrease in basal autophagy, indicating that this pathway significantly contributes to sustained basal autophagic activity in BRAF-mutated melanoma cells. These data were also confirmed in A2058 and SK-Mel-5 cell lines (Supplementary Figure S2A). To further support these observations, we also inhibited the activity of JNK by transient overexpression of a JNK dominant negative (Flag-JNK-DN) into A375 and A2058 cells. As shown in Figure 5e and Supplementary Figure S2B, expression of JNK-DN clearly resulted in the inhibition of basal autophagy, as evidenced by reduced LC3 conversion compared with LC3-II expression in control vector transduced cells (EV).
Figure 5.
JNK and basal autophagy. A375 cells were exposed to SP600125, and JNK activation (P-JNK) was evaluated by western blotting analysis (a; JNK was used as the loading control). A375 cells expressing a p62-GFP recombinant protein were treated or untreated with bafilomycin A (Baf, 4 h) and SP600125 (6 h) alone or in combination, and the occurrence of autophagy was analysed by measuring p62-GFP levels by citofluorimetric analysis (b; n=3), LC3 conversion by western blotting analysis (c; Gapdh was used as the loading control), and by evaluating the presence of p62-GFP cytosolic puncta by confocal analysis (d; bar=10 μm). A Flag-tagged JNK dominant negative (JNK-DN) was ectopically expressed in A375 cells by transient transfection and expression levels of JNK-DN protein and LC3 conversion, and accumulation was evaluated in presence or absence of Baf by western blotting (e). GFP or BRAFV600E expressing SK-Mel-110 cells were transiently transfected with expression plasmids encoding Flag-tagged Beclin 1 and protein extracts were subjected to IP using an anti-Flag antibody. Purified complexes were analysed together with the corresponding total extracts by western blotting using anti-Flag (f, top), anti-Bcl-XL (f, middle) and anti-Mcl-1 (bottom) antibodies. A375 cells were transiently transfected with expression plasmids encoding wild-type or a T69A/S70A/S87A mutant Bcl-2. Cells were treated or untreated with bafilomycin, as indicated, and total Bcl-2 protein expression together with LC3 conversion were evaluated by western blotting analysis (g; Gapdh was used as the loading control)
The known link between JNK and autophagy resides in the ability of active JNK to phosphorylate the inhibitory partners of Beclin 1, Bcl-XL and Bcl-2. Phosphorylated Bcl-XL and Bcl-2 are thus released by Beclin 1 enabling its subsequent ability to stimulate the autophagic process.37, 38 To verify the involvement of Bcl-XL in our model, we immunoprecipitated a Flag-Beclin 1 recombinant protein ectopically expressed in both GFP and BRAFV600E-expressing SK-Mel-110 cells, and analysed the presence of Bcl-XL by western blotting. As shown in Figure 5f, the interaction between Beclin 1 and Bcl-XL was abrogated in BRAFV600E cells. On the other hand, to verify the activity of activated JNK on Bcl-2, we overexpressed wt or a mutated (T69A S70A S87A) Bcl-2 protein39 in A375 cells and analysed LC3 conversion in the presence or absence of bafilomycin in a time-course experiment. As reported in Figure 5g, the presence of mutant Bcl-2 in which the phosphorylation sites have been mutated inhibited basal autophagy in A375 cells, compared with Bcl-2 wt expression.
Taken together, these data indicate BRAFV600E-induced ER stress-mediated constitutive activation of JNK results in the abrogation of Beclin 1/Bcl-XL and Beclin 1/Bcl-2 inhibitory interactions leading to constitutive induction of autophagy, whereas it does not affect the Beclin 1/Mcl-1 inhibitory bond, which is not regulated by phosphorylation of Mcl-1 (Figure 5f).
ER stress-mediated TRB3 induction contributes to basal autophagy induction
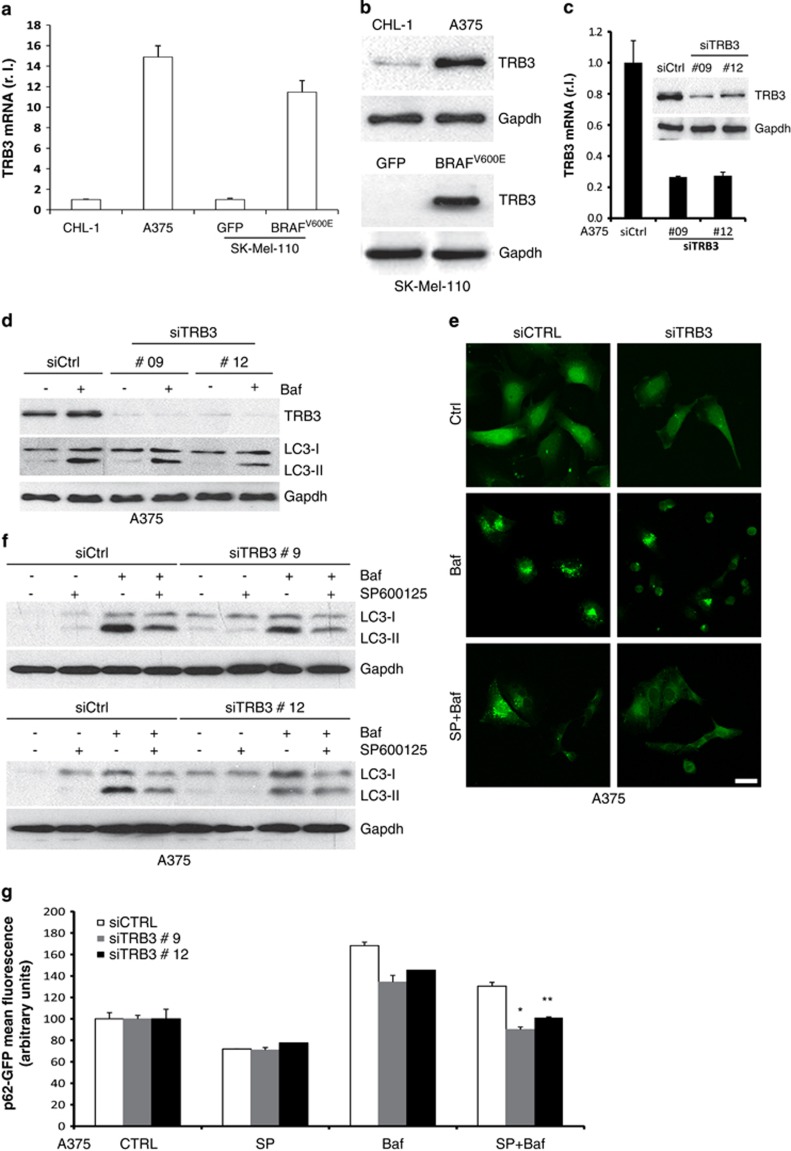
Our comparative analysis between wt and BRAF-mutated melanoma cells also evidenced an altered expression of an additional factor linking ER stress and autophagy, the mammalian homolog of Drosophila tribbles TRB3.40 qRT-PCR and western blotting revealed consistent upregulation of TRB3 at both the mRNA and protein level in A375 compared with CHL-1 cells (Figure 6a left and 6b upper panel, respectively). A similar result was obtained comparing TRB3 expression in GFP- and BRAFV600E-expressing SK-Mel-110 cells (Figure 6a right and 6b bottom panel), indicating that TRB3 upregulation directly correlates with the presence of mutant BRAF. Moreover, we also confirmed the increased expression of TRB3 in BRAFV600E compared with BRAF wt cells in an extended cohort of melanoma cell lines (Supplementary Figure S3).
Figure 6.
Modulation of TRB3 expression by BRAF. TRB3 expression was evaluated in CHL-1, A375 and SK-Mel-110 cells expressing GFP or BRAFV600E cells by qRT-PCR (a) and western blotting (b). A375 cells were transiently transfected with specific siRNA oligos and TRB3 downregulation was evaluated by both qRT-PCR and western blotting (c). Cells were then treated or untreated with bafilomycin and LC3 conversion or p62 puncta accumulation were evaluated by western blotting (d; Gapdh was used as the loading control) or confocal (e) analysis, respectively. Cells were also treated in the presence or absence of both bafilomycin and SP600125 as indicated, and autophagic rate was evaluated by measuring the LC3 conversion by western blotting (f; Gapdh was used as the loading control) or by p62 puncta accumulation by confocal microscopy (e; bar=10 μm). A375 cells expressing a p62-GFP recombinant protein were transiently transfected with specific siRNA oligos (siCtrl or siTRB3), treated as in (b) and autophagic flux was evaluated by measuring the levels of p62-GFP by flow cytometry (g; *P=0.0002; **P=0.003; n=3)
As ER stress-induced upregulation of TRB3 induces autophagy and the inhibition of the Akt/mTORC1 axis,41 we investigated the contribution of TRB3 to BRAFV600E-induced basal autophagy. RNAi-mediated knockdown of TRB3 was carried out in A375 cells (Figure 6c) and the effect on basal autophagy rate analysed in the presence or absence of bafilomycin. Similarly to the effects of JNK activity inhibition previously shown in Figures 4 and 5, our results also demonstrated that knockdown of TRB3 inhibited BRAFV600E-induced enhanced basal autophagy (Figures 6d and e, upper and middle panels).
Finally, to determine the effect of dual inhibition of JNK and TRB3 on oncogenic BRAF-enhanced basal autophagy, we analysed LC3 conversion (by both western blotting and immunofluorescence analysis) and p62 degradation in A375 cells in which the JNK or the TRB3 pathways were individually or collectively abrogated. The concomitant inactivation of both TRB3 and JNK pathways resulted in significant reduction of LC3 conversion (Figure 6f), LC3 puncta accumulation (Figure 6e) and p62 degradation (Figure 6g), indicating an additive contribution of both signalling pathways to oncogenic BRAF-induced ER stress-mediated basal autophagy.
p38 activation contributes to BRAF-induced ER stress
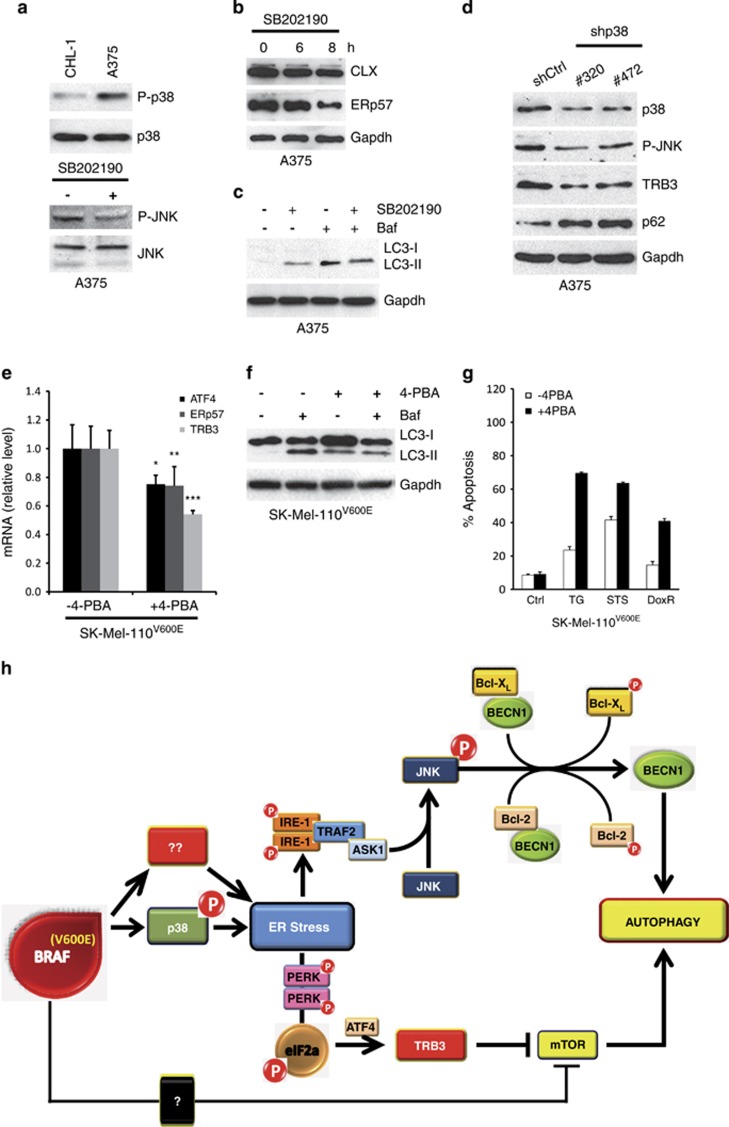
Published data demonstrate that melanoma cells harbouring hyperactivating mutations in BRAF or NRAS display increased activation of the stress-activated protein kinase (SAPK)-p38.42 Moreover, it has also been reported that active p38 in cancer cells is able to induce an ER stress response to coordinate cell survival through the activation of PERK and IRE-1.43
To evaluate the potential role of p38-mediated induction of chronic ER stress in BRAF mutant melanoma cells, we first compared the expression of activated p38 (P-p38) in CHL-1 and A375 cells, demonstrating increased P-p38 levels in BRAF mutated compared with BRAF wt cells (Figure 7a, upper panel), also confirmed in our cohort of melanoma cell lines (Supplementary Figure S4 A and B). To unveil the impact of constitutively activated p38 on melanoma basal autophagy, we inhibited the activity of p38 by ectopic expression of a p38 dominant negative (p38-DN) into A375 cells prior to evaluating the effect on LC3 conversion by western blotting. As shown in Supplementary Figure S4C, the expression of p38-DN resulted in clear inhibition of basal autophagy in A375 cells.
Figure 7.
p38 activation and ER stress induction in BRAF-mutated cells. p38 activation (P-p38) was evaluated by western blotting analysis in both CHL-1 and A375 (a, upper panel; p38 was used as the loading control). A375 cells were treated or untreated with SB202190 inhibitor as indicated and JNK activation (P-JNK), Calnexin (CLX) and ERp57 expression were evaluated by western blotting analysis (a, bottom panel, and b; JNK or Gapdh were used as the loading control). LC3 conversion was evaluated in A375 exposed to SB202190 (8 h) in presence or absence of bafilomycin A by western blotting analysis (c; Gapdh was used as the loading control). A375 cells were transiently transfected with specific siRNA oligos and p38, P-JNK, TRB3 and p62 protein levels were evaluated by western blotting analysis (d; Gapdh was used as the loading control). The expression of TRB3 and ER stress markers (ATF4, ERp57), and LC3 conversion were evaluated in SK-Mel-110 BRAFV600E cells continuously exposed to 4-PBA (3 mM), by qRT-PCR or western blotting (e and f; Gapdh was used as the loading control; *P=0.012; **P=0.025; ***P=0.002; n=3), in the presence or absence of bafilomycin A, as indicated. Apoptotic rates were compared in SK-Mel-110 BRAFV600E cells in the presence or absence of 4-PBA, treated or untreated 24 h with thapsigargin (TG), staurosporine (STS) or doxurubicin (DoxR), by cytofluorimetric analysis of PI-stained cells (g). Schematic representation of BRAF-induced ER stress and basal autophagy modulation (h)
Secondly, we inhibited the kinase activity of p38 by treatment of BRAFV600E-mutated A375 cells with 10 μM SB202190 for 6 or 8 h. As shown in Figure 7, impairment of p38 activity resulted in: (i) the inhibition of JNK phosphorylation (Figure 7a, bottom panel), (ii) ER stress response attenuation (Figure 7b) and (iii) a decrease in basal levels of autophagy (Figure 7c and Supplementary Figure S4D). Furthermore, we also inhibited the expression of p38 in A375 cells by using two specific shRNA oligos (shp38#320 and #472), which, as shown in Figure 7d and Supplementary Figure S4 (E & F), resulted in the inhibition of p38 expression as well as decreased expression of ERdj5, ERp57 and TRB3, JNK phosphorylation and the accumulation of p62. The inhibition of p38 expression was also carried out in SK-Mel-5 cells by shRNA, and resulted in both TRB3 expression attenuation and decreased JNK activation (Supplementary Figure S4G), confirming results obtained in A375 cells.
Collectively, these data suggest p38 signalling contributes to the induction of a chronic ER stress status in BRAF-mutated melanoma cells, resulting in an increase in basal autophagic activity.
Chemical chaperones sensitizes BRAFV600E melanoma cells to apoptosis induction
Autophagy is primarily a pro-survival process that is upregulated in advanced stages of melanoma,3 particularly in those tumours harbouring hyperactivating mutations in BRAF, to sustain cell survival and counteract apoptotic signalling induced by chemotherapeutic agents.3, 20, 30
The contribution of chronic ER stress to increased basal autophagy in BRAF-mutated melanoma cells prompted us to test whether the inhibition of incurred ER stress by chemical chaperones would lead to a decrease in basal autophagy and the increased susceptibility to cell death induction. As 4-phenylbutyric acid (4-PBA) has been shown to reduce ER stress both in vivo and in vitro,44 we exposed SK-Mel-110 cells expressing BRAFV600E to 4-PBA (3 mM) for 3 weeks and compared the expression of the ER stress markers ATF4, ERp57 and TRB3 in either treated or untreated cells. 4-PBA treatment significantly decreased the expression of all markers, indicating the relief of ER stress imposed by BRAFV600E (Figure 7e). In addition, 4-PBA consistently and significantly reduced basal autophagy in SK-Mel-110 BRAFV600E cells, as evidenced by the reduced conversion and accumulation of LC3, in presence of bafilomycin (Figure 7f). These findings therefore indicate that 4-PBA alleviates chronic ER stress in BRAFV600E mutant melanoma cells, with consequent reduction in basal autophagic activity.
To determine the effect of 4-PBA-induced reduction of ER stress and basal autophagy on susceptibility to apoptosis induction, SK-Mel-110 BRAFV600E cells were subsequently treated in the presence of absence of a panel of pro-apoptotic drugs, able to activate different apoptotic pathways; thapsigargin (TG, 10 μg/ml), staurosporine (STS, 0,2 μM) and doxorubicin (DoxR, 10 μM) (Figure 7g). Results revealed that 4-PBA significantly increased the sensitivity of SK-Mel-110 BRAFV600E cells to drug-induced apoptosis by all agents tested, thus suggesting chronic ER stress imposed by oncogenic BRAF expression and the subsequent increase in basal autophagy represents a skilled pro-survival mechanism used by these cells to escape cell death induction.
Discussion
It is now quite clear that both autophagy and ER stress have pivotal roles in cancer development and progression, as well as response to pharmacological therapeutic intervention.45 Although autophagy may represent a primary barrier to cellular transformation and the prevention of tumour development, paradoxically, both pathways promote pro-survival cell responses, allowing tumour growth and progression, and possibly conferring chemoresistance.22, 46 Thus, targeting ER stress and/or autophagy may present considerable benefit for cancer therapy.47 However, such potential is complicated by the inter-connection of both processes, and the fact that both ER stress and autophagy are able to modulate each other and that their specific function is strictly dependent on both tumour type and stage progression.45 In cutaneous metastatic melanoma, we have recently demonstrated the clinical potential of ER stress-induced apoptosis, with BRAF wt melanomas showing greater sensitivity, compared with tumours harbouring hyperactivating mutations in the BRAF protein kinase.32, 33 Moreover, we have also shown (and confirmed in the present study) that BRAF mutation is associated with increased levels of basal autophagy.20 Interestingly, ER stress-induced apoptosis is reduced in melanoma cells harbouring oncogenic BRAF compared with the observed induction in BRAF wt melanoma cells, and in this context and in contrast to observations in BRAF mutant melanoma cells, we have also shown that autophagy inhibition significantly sensitizes BRAF wt melanoma cells to ER stress-mediated apoptosis.20 On the basis of established evidence that: (i) melanoma development and progression is accompanied by the induction of ER stress,30 (ii) BRAFV600E melanomas are characterized by increased rates in basal autophagy compared with BRAF wt malignancies 20 and that (iii) ER stress and autophagy are strictly linked,26 we now show that mutation of BRAF confers a chronic ER stress status responsible for increased basal autophagy in melanoma cells. Specifically, the presence of BRAFV600E increases the expression of typical ER stress markers, compatible with a chronic ER stress status, possibly accounting for reduced sensitivity to further ER stress stimulation. Importantly, the ectopic expression of BRAFV600E mutation not only induced a chronic ER stress status but was also responsible for increased basal autophagy. Furthermore, hyperactivation of BRAF, due to mutation, is directly responsible for p38 activation and subsequent ER stress induction. Moreover, the IRE1/TRAF2/ASK1/JNK branch of the UPR together with the TRB3 axis are key to ER-stress-mediated autophagy induction observed in BRAF-mutated melanoma cells. While JNK mediates the phosphorylation of both Bcl-XL and Bcl-2, responsible for Beclin 1 release and autophagy induction, TRB3, in turn, mediates Akt/mTORC1 inhibition and, thus, autophagy induction.
Collectively these data demonstrate that oncogenic mutations in BRAF impose a chronic ER stress status within melanoma cells, resulting in enhanced basal levels of autophagy and that the increased activity of these two pro-survival signalling mechanisms contributes to the notorious chemoresistance of such tumours (schematically represented in Figure 7h). Inhibiting ER stress in BRAF mutant melanomas may thus represent a novel and valuable therapeutic strategy through which both pro-survival ER stress and autophagy can be inhibited, and these tumours can be re-sensitized to chemotherapeutic intervention. Perhaps this approach may also overcome the potential for tumour recurrence in patients treated with autophagy inhibitors given autophagy inhibition per se, may drive secondary tumourigenesis. To our knowledge chemical chaperones are, at present, the only available molecules able to effectively reduce ER stress both in vitro and in vivo44 and because in the present study, prolonged 4-PBA treatment of BRAF-mutated melanoma cells resulted in the reduced expression of both markers of ER stress and basal autophagy as well as increased sensitivity to pro-apoptotic drugs, the clinical use of chemical chaperones may thus harness ER stress and autophagy modulation for the therapeutic benefit of patients bearing BRAF V600E tumours.
Recent studies by Ma et al.34 identified ER stress and autophagy induction as a mechanism mediating resistance to BRAF inhibitor therapy in patients bearing BRAFV600E mutant melanomas suggesting a direct link between these two pathways. Our data thus complement the studies of Ma et al.34 and provide additional novel molecular insight into how oncogenic BRAF leads to a chronic ER stress state and the promotion of basal autophagy. Moreover, given observations of ER chemical chaperone-mediated resensitization of BRAFV600E melanoma cells to a panel of pro-apoptotic drugs, able to activate different apoptotic pathways, the use of drugs able to harness ER stress or, alternatively, to target the molecular pathways linking BRAFV600E-induced ER stress and basal autophagy (such as the IRE1/TRAF2/ASK1/JNK and the TRB3 axes) for the clinical benefit of such tumours is warranted. Observations demonstrating chloroquine/hydroxychloroquine can reduce tumour immunogenicity,48 and accelerate tumour formation,49 have questioned the benefit of autophagy inhibition in cancer therapy, and hence our novel proposed therapeutic approaches (ER stress buffering or JNK and TRB3 axes targeting) may also increase the sensitivity of BRAF mutant melanomas to clinical BRAF inhibition.
Materials and Methods
Cell culture and treatments
All cell lines were cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) except for A2058 cells that were cultured in RPMI (Sigma-Aldrich). Cell culture medium was supplemented with 10% foetal bovine serum (Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), 1% penicillin/streptomycin solution (Sigma-Aldrich) at 37 °C under 5% CO2, except medium. Primary melanocytes have been previously described.10 Cells were treated with thapsigargin (Sigma-Aldrich) at a final concentration of 10 μg/ml, staurosporine (Sigma-Aldrich), 2 μM, doxorubicin (Sigma-Aldrich), 10 μM, Baf (Sigma-Aldrich), 5 nM, 4-PBA (Sigma-Aldrich), 3 mM, SP600125 (Sigma-Aldrich), or 10 μM, SB202190 (Sigma-Aldrich). All agents were added in DMSO or methanol with an equal volume of vehicle used to treat control cells (0.1–0.5% DMSO or methanol).
Retroviral generation and Infection
Fifteen micrograms of retroviral vectors were co-transfected with 5 μg of expression plasmid for the vesicular stomatitis virus G protein into the 293 cell line gp/bsr using calcium phosphate.50 Forty-eight hours later, the supernatant containing retroviral particles was recovered and supplemented with polybrene (4 mg/ml). Cells were infected by incubation with retroviral containing supernatant for 6–8 h as previously described.50
Lentiviral generation and infection
Ten micrograms of lentiviral vectors (shRNA-pLKO) were co-transfected with 2.5 μg of an expression plasmid for the vesicular stomatitis virus G protein and psPAX2 plasmid, containing gag, pol and rev genes, into the 293T packaging cell line using calcium phosphate as outlined above. Forty-eight hours later, the supernatant containing the lentiviral particles was recovered and supplemented with polybrene (4 μg/ml). Cells were infected by incubation with lentiviral containing supernatant for 6–8 h.
Cell transfection
Expression vectors for BRAFV600E mutant,12 GFP or pcDNA4 (Invitrogen, Carlsbad, CA, USA), wt/mutant Bcl-2,39 JNK-DN 51 and p38-DN 52 (Addgene plasmids 8768, 13340, 13846, 20356, respectively; Cambridge, MA, USA) vectors were transiently transfected using lipofectamine LTX, according to the manufacturer's recommendations.
PCR for XBP-1 splicing
The human XBP-1 sequence was amplified by PCR using the primer pair AAACAGAGTAGCAGCTCAGACTGC and CCTTCTGGGTAGACCTCTGGGAG as previously described.33
Quantitative RT-polymerase chain reaction
RNA was extracted by using Trizol reagent (Invitrogen) as indicated by the supplier. cDNA synthesis was generated using a reverse transcription kit (Promega, Madison, WI, USA) according to the manufacturer's recommendations. Quantitative PCR reactions were performed with the Rotor-Gene 6000 (Corbett Research Ltd, Cambridgeshire, UK) thermocycler. Primer sets for all amplicons were designed using the Primer-Express 1.0 software system (Roche, Basel, Switzerland):
L34 forward: 5′-GTCCCGAACCCCTGGTAATAGA-3′
L34 reverse: 5′-GGCCCTGCTGACATGTTTCTT-3′
ERp57 forward: 5′-TGTGGCATCACGCAGTTTCA-3′
ERp57 reverse: 5′-GGGAAGTTAAAGGGCCACACC-3′
ATF4 forward: 5′-GTGGCCAAGCACTTCAAACC-3′
ATF4 reverse: 5′-CCCGGAGAAGGCATCCTC-3′
ERdj5 forward: 5'-TCATGTTACCACGCTTGGACC-3'
ERdj5 reverse: 5′-GTAAAGCTCGACATGGTGGACAC-3′
Xbp-I (spliced) forward: 5′-GAATGAAGTGAGGCCAGTG-3′
Xbp-I (spliced) reverse: 5′-GAGTCAATACCGCCAGAATC-3′
IRE1 forward: 5′-GAGACCCTGCGCTATCTGACC-3′
IRE1 reverse: 5′-CAGAGTGGCGTCAGCTTG-3′
TRB3 forward: 5′-TCAAGCTGTGTCGCTTTGTCTTCG-3′
TRB3 reverse: 5′-TGCTTGTCCCACAGGGAATCATCT-3′
L34 mRNA level was used as an internal control and results were expressed as previously described.50
Western blotting
Total proteins were extracted from cells using the Cell Lytic buffer (Sigma-Aldrich) following addition of protease inhibitors and resolved by electrophoresis through NuPAGE Bis-Tris gel (Invitrogen) and electroblotted onto nitrocellulose (Protran, Sigma-Aldrich) membrane. Blots were incubated with indicated primary antibodies in 5% non-fat dry milk in PBS plus 0.1% Tween20 overnight at 4 °C. Primary antibodies were: anti-Beclin1 (1 : 500; Becton-Dickinson, Franklin Lakes, NJ, USA); anti-Gapdh (1 : 106; Calbiochem, Merck Millipore, Darmstadt, Germany); anti-Flag (1 : 3000; Sigma-Aldrich), BRAF (1 : 500), CLX (1 : 500), TRAF2 (1 : 300) (Santa Cruz Biotechnology, Dallas, TX, USA); LC3 (1 : 2000), ERK (1 : 2000), P-ERK (1 : 500), eIF2 (1 : 1000), P-eIF2 (1 : 500), JNK (1 : 1000), P-JNK (1 : 500), p38 (1 : 500), P-p38 (1 : 500) (Cell Signaling Technology, Beverly, MA, USA); ERp57 (1 : 5000, Stressgen, San Diego, CA, USA), Bcl-XL (1 : 300; Enzo Life Sciences, Farmingdale, NY, USA), Bcl-2 (1 : 500; DAKO, Glostrup, Denmark). Detection was achieved using horseradish peroxidase-conjugate secondary antibody (1 : 5000; Jackson ImmunoResearch, Suffolk, UK) and visualized with ECL plus (Amersham Biosciences, GE Healthcare, Buckinghamshire, UK).
Velocity sedimentation by sucrose gradient
Cells were suspended in buffer containing 0.25 M sucrose, 10 mM Hepes, and 1 mM EDTA and protease cocktail inhibitors and homogenized (Sigma-Aldrich) by 100 strokes in a dounce potter homogenizer and centrifuged for 10 min at 600 × g to obtain a post-nuclear supernatant. The post-nuclear supernatant was re-centrifuged for 15 min at 11 000 × g to obtain a post-mitochondrial supernatant. The post-mitochondrial supernatant was layered onto a discontinuous four-step gradient consisting of 2 ml each of 2.0 M, 1.3 M, 1.0 M and 0.6 M sucrose in 10 mM Hepes. Centrifugation was performed using a rotor (SW41 Ti; Beckman Coulter, High Wycombe, UK) at 27 000 × g for 18 h, and 0.4-ml fractions were manually collected and checked for density.
IP assay
Cells were suspended in lyses buffer (10 mM Tris-HCl, pH 8.0, 0.5% NP40,) plus protease and phosphatase inhibitors (protease inhibitor cocktail plus 1 mM sodium fluoride, 1 mM sodium orthovanadate and 1 mM sodium molibdate; Sigma-Aldrich). One to three milligrams of lysates was incubated at 4 °C for 30 min. After a centrifugation at 4 °C for 10 min at 13 000 × g to remove insoluble debris, equal amounts of protein were incubated with 20 μl monoclonal anti-Flag antibody conjugated with protein A agarose beads (Takara, Otsu, Japan and Sigma-Aldrich, respectively) with rotation at 4 °C for 4 h followed by 60-min incubation with 30 μl protein A–Sepharose beads (Roche). The beads were collected by centrifugation and washed four times with lyses buffer. Proteins bound to the beads were eluted with 30 μl FLAG-peptide (200 ng/l, Sigma-Aldrich) with rotation at 4 °C for 1 h followed by a centrifugation at 4 °C for 10 min at 13 000 × g to remove beads. A 2 × SDS-PAGE sample buffer was added and samples were boiled at 95 °C for 10 min. Western blotting analyses were performed as described above.
Apoptosis analysis
Flow cytometry of propidium iodide-stained cells was used to estimate the level of cell death or apoptosis by measuring the percentage of cells in the sub-G1 fraction.33
Confocal microscopy
Cells were grown on coverslips and fixed with 4% PFA in PBS followed by permeabilization with 0.1% Triton X-100 in PBS. p62 primary antibody (1 : 500; MBL, Woburn, MA, USA) was incubated for 1 h at RT and visualized by means of AlexaFluor-488 (1 : 300, Invitrogen). Coverslips were mounted in antifade (SlowFade; Invitrogen) and examined under a confocal microscope (TCS SP2; Leica, Solms, Germany) equipped with a 63 × 1.40–0.60 NA HCX Plan Apo oil BL objective at RT.
Autophagy assay
Cells infected with a retroviral vector encoding p62-GFP were seeded at a concentration of 25 × 104 cells/well in 6-well plates and treated as indicated. Cells were fixed in 4% PFA and fluorescence analysed by FACS analysis (FacsScan, Becton-Dickinson).
For immunodetection of p62 puncta, cells were grown on coverslips and fixed with 4% PFA in PBS, washed three times and incubated with anti-p62 antibody, as described above, prior to analysis confocal microscopy. Results were recorded as percentage of p62-positive cells with p62 punctate as previously described.53 A minimum of 50–100 cells per sample was counted for triplicate samples per condition per experiment.
Densitometric analysis
In order to measure protein expression levels, intensities of specific bands, corresponding to the proteins of interest are measured using Gel Doc 2000 and QuantyOne software (Bio-Rad, Hercules, CA, USA). Briefly, blots were acquired using the Gel Doc 2000 apparatus; images were imported into the QuantyOne software; contrast was adjusted such that the bands were clearly visible on the blot image; area around each band was selected; background intensity was subtracted from the blot image; bands were then selected by drawing a tight boundary around them; intensities of the selected bands was exported in excel format which was used to perform further analysis; statistical analysis (t-test) was performed using Prism 5 software (GraphPad, La Jolla, CA, USA).
Statistical analysis
Unless indicated otherwise, all data are presented as the means±S.D. of the mean, and statistical differences were evaluated by two-tailed Student's t-tests. For all analyses, we considered P<0.05 to be statistically significant.
Acknowledgments
We thank Mr V. Santolini for helpful technical assistance, Dr. Derijard for JNK-DN vector, Dr. Enslen for p38-DN vector, and Dr. Yamamoto for Bcl-2wt and Bcl-2mut plasmids. Financial support: This work was supported by grants from AIRC (MFAG-11743 to MC, IG2012 to GMF and IG2010 to MP), the Italian Ministry of University and Research (PRIN 2012 and FIRB Accordi di Programma 2011), the Italian Ministry of Health (Ricerca Finalizzata and Ricerca Corrente) and the British Skin Foundation, The JGW Patterson Foundation and The Newcastle healthcare Charity, UK.
Author Contributions
MC conceived the project, designed and performed experiments and analysed the data. FR, FC, PG, MA and BC performed experiments. GMF, PEL and MP contributed to the study and experimental design as well as data analysis. This manuscript was written by MC, PEL, GMF and MP. All authors reviewed the manuscript.
Glossary
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- Baf
bafilomycin A
- Wt
wild-type
- DN
dominant negative
- 4-PBA
4-phenylbutyric acid
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- Cockerell CJ. The pathology of melanoma. Dermatol Clin. 2012;30:445–468. doi: 10.1016/j.det.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Corazzari M, Fimia GM, Lovat P, Piacentini M. Why is autophagy important for melanoma? molecular mechanisms and therapeutic implications. Semin Cancer Biol. 2013;23:337–343. doi: 10.1016/j.semcancer.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- Arkenau HT, Kefford R, Long GV. Targeting BRAF for patients with melanoma. Br J Cancer. 2011;104:392–398. doi: 10.1038/sj.bjc.6606030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, et al. Inactivation of the apoptosis effector apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen S, Hernberg M, Pyrhonen S, Tarkkanen J, Asko-Seljavaara S. Ki-67, bcl-2 and p53 expression in primary and metastatic melanoma. Melanoma Res. 2005;15:375–381. doi: 10.1097/00008390-200510000-00005. [DOI] [PubMed] [Google Scholar]
- Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the bcl-2 family members PUMA, bim, and mcl-1. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- McKee CS, Hill DS, Redfern CP, Armstrong JL, Lovat PE. Oncogenic BRAF signalling increases mcl-1 expression in cutaneous metastatic melanoma. Exp Dermatol. 2013;22:767–769. doi: 10.1111/exd.12254. [DOI] [PubMed] [Google Scholar]
- Jiang CC, Chen LH, Gillespie S, Wang YF, Kiejda KA, Zhang XD, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67:9750–9761. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- Martin S, Lamb HK, Brady C, Lefkove B, Bonner MY, Thompson P, et al. Inducing apoptosis of cancer cells using small-molecule plant compounds that bind to GRP78. Br J Cancer. 2013;109:433–443. doi: 10.1038/bjc.2013.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell-Dorris ER, O'Leary JJ, Sheils OM. BRAFV600E: Implications for carcinogenesis and molecular therapy. Mol Cancer Ther. 2011;10:385–394. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- Aguissa-Toure AH, Li G. Genetic alterations of PTEN in human melanoma. Cell Mol Life Sci. 2012;69:1475–1491. doi: 10.1007/s00018-011-0878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Marais R.BRAF signaling and targeted therapies in melanoma Hematol Oncol Clin North Am 200923529–545.ix. [DOI] [PubMed] [Google Scholar]
- Lazova R, Klump V, Pawelek J. Autophagy in cutaneous malignant melanoma. J Cutan Pathol. 2010;37:256–268. doi: 10.1111/j.1600-0560.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–3489. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JL, Corazzari M, Martin S, Pagliarini V, Falasca L, Hill DS, et al. Oncogenic B-RAF signaling in melanoma impairs the therapeutic advantage of autophagy inhibition. Clin Cancer Res. 2011;17:2216–2226. doi: 10.1158/1078-0432.CCR-10-3003. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci. 2010;67:1581–1588. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersey P, Zhang XD. Adaptation to ER stress as a driver of malignancy and resistance to therapy in human melanoma. Pigment Cell Melanoma Res. 2008;21:358–367. doi: 10.1111/j.1755-148X.2008.00467.x. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. A trip to the ER: Coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Gorman AM, Healy SJ, Jager R, Samali A. Stress management at the ER: Regulators of ER stress-induced apoptosis. Pharmacol Ther. 2012;134:306–316. doi: 10.1016/j.pharmthera.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Verfaillie T, Salazar M, Velasco G, Agostinis P. Linking ER stress to autophagy: Potential implications for cancer therapy. Int J Cell Biol. 2010;2010:930509. doi: 10.1155/2010/930509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: Friend or foe. Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazzari M, Lovat PE. Harnessing autophagy for melanoma benefit. Cell Biol: Res Ther. 2013;2 [Google Scholar]
- Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V. Induction of autophagy and inhibition of melanoma growth in vitro and in vivo by hyperactivation of oncogenic BRAF. J Invest Dermatol. 2010;130:1657–1667. doi: 10.1038/jid.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovat PE, Corazzari M, Armstrong JL, Martin S, Pagliarini V, Hill D, et al. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 2008;68:5363–5369. doi: 10.1158/0008-5472.CAN-08-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, et al. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: The role of stress proteins ERdj5 and ERp57. Br J Cancer. 2007;96:1062–1071. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O'Leary EC, Sakata ST, Xu W, et al. SP600125, an anthrapyrazolone inhibitor of jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Wang Q, Srinivasan B, Xu J, Padilla MT, Li Z, et al. A JNK-mediated autophagy pathway that triggers c-IAP degradation and necroptosis for anticancer chemotherapy. Oncogene. 2013;33:3004–3013. doi: 10.1038/onc.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernández-Tiedra S, Egia A, Lorente M, et al. TRB3 links ER stress to autophagy in cannabinoid anti-tumoral action. Autophagy. 2009;5:1048–1049. doi: 10.4161/auto.5.7.9508. [DOI] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernández-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada Y, Dong J, Ossowski L. Positive crosstalk between ERK and p38 in melanoma stimulates migration and in vivo proliferation. Pigment Cell Melanoma Res. 2009;22:66–76. doi: 10.1111/j.1755-148X.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–1711. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazzari M. ER stress & autophagy in cancer: Contenders or partners in crime. Int J Mol Biol Biochem. 2013;1:23. [Google Scholar]
- Lee AS, Hendershot LM. ER stress and cancer. Cancer Biol Ther. 2006;5:721–722. doi: 10.4161/cbt.5.7.3120. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. P53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, et al. PMC3422474; proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012;19:1495–1504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, et al. JNK1: A protein kinase stimulated by UV light and ha-ras that binds and phosphorylates the c-jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Enslen H, Raingeaud J, Davis RJ. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.