Abstract
Gap genes are involved in segment determination during early development in dipteran insects (flies, midges, and mosquitoes). We carried out a systematic quantitative comparative analysis of the gap gene network across different dipteran species. Our work provides mechanistic insights into the evolution of this pattern-forming network. As a central component of our project, we created a high-resolution quantitative spatio-temporal data set of gap and maternal co-ordinate gene expression in the blastoderm embryo of the non-drosophilid scuttle fly, Megaselia abdita. Our data include expression patterns in both wild-type and RNAi-treated embryos. The data—covering 10 genes, 10 time points, and over 1,000 individual embryos—consist of original embryo images, quantified expression profiles, extracted positions of expression boundaries, and integrated expression patterns, plus metadata and intermediate processing steps. These data provide a valuable resource for researchers interested in the comparative study of gene regulatory networks and pattern formation, an essential step towards a more quantitative and mechanistic understanding of developmental evolution.
Subject terms: Evolutionary developmental biology, Developmental biology, Systems biology, Gene expression analysis
Background & Summary
Understanding the function and evolution of complex regulatory networks is a challenge that lies at the heart of modern biology1–5. In the context of developmental evolution (evo-devo), tackling this challenge requires systematic and quantitative comparative studies of pattern-forming gene regulatory networks. We use a combination of genetics, dynamical systems modelling and reverse engineering to achieve this aim6,7. As a case study, we focus on the gap gene network involved in pattern formation and segment determination during early development of the vinegar fly Drosophila melanogaster and other dipteran insects (flies, midges, and mosquitoes)8. Our approach relies on quantitative gap gene expression data with high spatial and temporal resolution, not only in standard laboratory models, but also in non-model organisms9–19. Such data must be based on quantitative microscopy and image bioinformatics20–22 to preserve the spatial aspect of gene expression, which is crucial for the study of pattern formation.
Here, we present a quantitative data set of segmentation gene expression from a non-drosophilid species, the scuttle fly Megaselia abdita 23. It comprises a high-resolution spatio-temporal atlas for the expression of the maternal-coordinate genes bicoid (bcd), hunchback (hb), and caudal (cad), as well as the gap genes hb, Krüppel (Kr), knirps (kni), giant (gt), tailless (tll), and huckebein (hkb) along the major embryonic axis—the antero-posterior or A–P axis—during the cleavage and blastoderm stages of early development24–26. We are using these data, in combination with our previously published expression data set from D. melanogaster 14, to study the evolution of maternal gradients and gap gene regulation across dipteran species27,28.
Quantified spatio-temporal expression data form the foundation of our investigation in two complementary ways. First, high-resolution measurements of gene expression dynamics—in combination with gene knock-down by RNA interference (RNAi)—allows us to study maternal regulatory inputs28 and gap-gap cross-regulatory interactions27 at an unprecedented level of rigour and detail. Our work reveals how small changes in regulatory interactions lead to different mechanisms establishing embryo polarity and the dynamic positioning of shifting gap domain boundaries in M. abdita compared to D. melanogaster. Careful characterisation of expression timing, in combination with the dosage-dependent nature of RNAi, allows us to assess the effects of subtle changes in regulatory interaction strength on expression dynamics. This would not be possible without a high-resolution quantitative data set such as the one presented in this paper. Second, in addition to direct analysis, we also used our data to fit quantitative dynamic models of the gap gene network (publication currently under review). These models, which have been verified by RNAi knock-down, go beyond genetic analysis in that they predict specific and precise regulatory mechanisms for gap gene expression dynamics and its evolution by quantitative system drift.
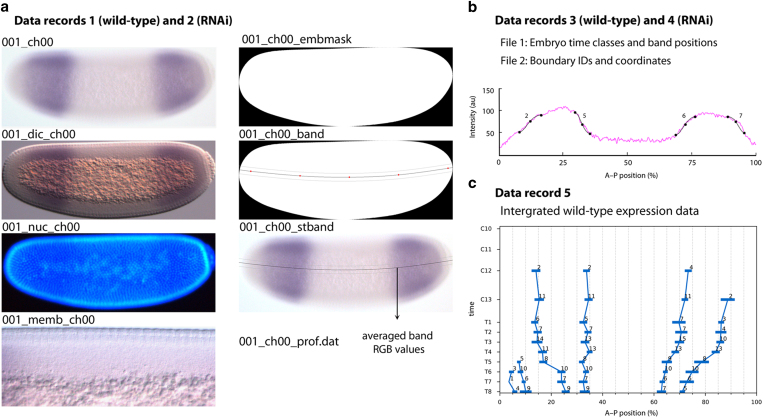
The expression data presented here consist of (1) images of carefully curated21 and staged26 laterally aligned M. abdita blastoderm embryos stained against one or two maternal co-ordinate or gap mRNA gene products by whole-mount in situ hybridisation14 in a wild-type (Fig. 1a; Data Record 1, Data Citation 1) and RNAi-treated background (Fig. 1a; Data Record 2, Data Citation 1); (2) the associated quantified spatial gene expression profiles21 classified by gene and developmental time point (cleavage cycles 1–14A, C1–C14A; cleavage cycle C14A is subdivided into eight time classes, C14-T1–T8)26 for wild-type (Fig. 1b; Data Record 3, Data Citation 1) and RNAi-treated embryos (Fig. 1b; Data Record 4, Data Citation 1); and (3) integrated spatio-temporal expression data, averaged across all embryos stained for a given gene at a given time point in wild-type embryos (Fig. 1c; Data Record 5, Data Citation 1)27,28.
Figure 1. Overview of data records presented in this paper.
(a) Data Records 1 (wild-type) and 2 (RNAi). These data records include cropped and rotated images of gap gene expression taken using bright field and differential interference contrast optics (ch00 and dic_ch00 suffixes respectively), an image of the nuclear counterstain (nuc_ch00) and an unprocessed image of the dorsal membrane morphology (memb_ch00). These data records also include embryo masks (embmask) and images of the band location superimposed onto the embryo mask (band) and brightfield (stband) images. The file 001_ch00_prof.dat contains averaged band RGB values seen plotted in magenta in (b). (b) Data Records 3 (wild-type) and 4 (RNAi): these records are composed of two CSV files that document the time class and position of the 10% strip used for profile extraction for each embryo (File 1), plus the associated boundary IDs and coordinates (File 2). The graph shows an example, where splines fit to boundaries 2, 5, 6 and 7 of gt are plotted against the extracted profile (magenta; see Fig. 2 for boundary numbers). Images in (a) and (b) correspond to embryo 001 that can be found in Data Record 1 in ish/megaselia/ma_gt_260911/proc. (c) Data Record 5. This data record is composed of a CSV file containing averaged coordinates and standard deviations of expression boundaries for each gene. Embryos are oriented with anterior to the left, dorsal up.
Methods
Blastoderm-stage embryos of M. abdita—corresponding to embryonic stages 4 and 5 (ref. 26)—were collected 4 h (hrs) after egg laying (see refs 29,30, for fly culture and embryo collection/fixation). Gap gene mRNA expression patterns were visualised using an enzymatic (colorimetric) in situ hybridisation protocol as previously described14. Visualisation was performed using NBT/BCIP (purple) and/or FastRed (red) staining reagents. We used single RNA probes for the majority of stains (mostly NBT/BCIP). Embryos double-stained for gap genes, or a gap gene plus the pair-rule gene even-skipped (eve), were used to confirm and register the position of expression domains relative to each other.
RNAi treatment was carried out as described in refs 23,31,32. Two distinct dsRNA constructs were injected per gene, one being the knock-down agent, the other one—with conserved domains excluded—serving as a control. RNAi-treated embryos were then stained for each of the trunk gap genes as described above.
Stained wild-type and RNAi-treated embryos were imaged, staged and processed as previously described21,27 (Data Records 1–4, Data Citation 1). In brief, four images were taken of each stained embryo (Fig. 1a)21: a bright-field image from which the expression profile was extracted, a differential interference contrast (DIC) image used to create the whole-embryo mask, and two images used to determine the time class of each embryo26: one of fluorescently counterstained nuclei, and the other showing details of membrane morphology with DIC optics. Images of wild-type embryos were further processed through a quantification pipeline as previously described21. In summary, we automatically create a whole-embryo mask used for image cropping, rotation, and alignment; we then locate a 10%-strip along the dorso-ventral midline of the embryo with a cubic spline (see ref. 33 for mathematical details) that is defined by five equidistant knots—that is, five control points; knot position is automatically pre-set using a skeleton algorithm and can be manually adjusted if necessary (Fig. 1a); we extract expression profiles along the antero-posterior axis along this strip as follows (Fig. 1b): for each pixel on the spline curve we extract average RGB values, calculated across a perpendicular column spanning the entire width of the 10%-strip; intensity profiles of NBT/BCIP (purple) stains were extracted using the inverse red RGB channel (that is: intensity=255—red; the blue RGB channel was ignored as its signal is very similar to the red channel with a weaker signal-to-background ratio); the profile of FastRed (red) stains was determined by subtracting the green from the red RGB channel and inverting the outcome. In this manner both the blue component of NBT/BCIP and the yellow-like background of the unstained embryo regions are removed; once profiles are extracted, we visually identify expression domain boundaries, which are manually fitted by clamped splines (first derivative set to 0.0 at the ends of the spline) to measure their width and position (Fig. 1b). Subsequently, we calculate median boundary positions and expression variability for each boundary at each time class, which results in an integrated high-resolution data set of spatio-temporal gene expression (Fig. 1c)27,28. RNAi-treated embryos were processed through the same pipeline as wild-type embryos to measure the position of gene expression boundaries. Due to the variability in gene knock-down effect, we did not evaluate median boundary positions in this data set27.
Data Records
Data record 1—image data and extracted profiles from wild-type M. abdita embryos stained for maternal co-ordinate and gap gene expression
This data record contains series of images stored in Portable Network Graphics (PNG) format displaying wild-type M. abdita embryos that have been stained for gap gene expression at various intermediate stages of data processing. Each series of images comes with a data file containing an extracted expression profile. Data are archived as a ZIP-compressed file, which is organised into a hierarchy of directories (Data Record 1, Data Citation 1). The top directory ‘ish’ (in situ hybridisation) contains the sub-folder ‘megaselia’, which in turn contains a number of directories corresponding to individual batches of stained embryos, which are each given a specific batch ID. Each embryo within these batches is given an individual embryo ID and each processing step receives an identifying suffix in our file-naming scheme. Details are as follows.
Batch ID
Each batch directory is named in the following format ‘species_stain_date’. For example, ma_gt_260911 contains data from M. abdita (ma) stained for the gap gene giant (gt) on the 26th September 2011 (260911). Each of these directories contains a folder called ‘proc’ (short for stored PROCessing steps), which contains the processed images.
Embryo ID
Images from specific embryos within the ‘proc’ directories receive a numerical ID: 001, 002, 003... followed by a suffix that identifies each of the processing steps. Suffixes are:
Suffix: _ch00
Cropped and rotated bright-field images (taken with a 10X objective) used to extract gene expression boundary positions.
Suffix: _dic_ch00
Cropped and rotated differential interference contrast (DIC) images (taken with a 10X objective) used to create the embryo mask.
Suffix: _nuc_ch00
Fluorescent images (taken with a 10X objective) with DAPI nuclear counterstain used for staging embryo into separate cleavage cycles (C1–C14A).
Suffix: _memb_ch00
DIC images (taken with a 40X objective) of mid-dorsal membrane morphology used to classify embryos within cleavage cycle 14A (C14A) into time-classes T1–8.
Suffix: _embmsk
Binary whole-embryo masks generated from DIC images used to automatically crop/rotate embryo images.
Suffix: _band
These images show the 10%-strip (band) used to extract gene expression profiles in overlay with the binary embryo mask.
Suffix: _stband
These images show the 10%-strip (band) used to extract gene expression profiles in overlay with the original bright-field image from which expression profiles are extracted.
Suffix: _prof.dat
Text file containing extracted expression profiles along the 10%-strip. Channel extraction is performed as described above.
Data record 2—image data and extracted profiles from RNAi-treated M. abdita embryos stained for maternal co-ordinate and gap gene expression
This data record contains series of images stored in Portable Network Graphics (PNG) format displaying RNAi-treated M. abdita embryos that have been stained for gap gene expression at various intermediate stages of data processing. Each series of images comes with a data file containing an extracted expression profile. Data are archived as a ZIP-compressed file, which is organised into a hierarchy of directories (Data Record 2, Data Citation 1). The top directory ‘RNAi’ (RNA interference) contains the sub-folder ‘megaselia’, which in turn contains a number of directories corresponding to separate RNAi experiments, which are each given a specific batch ID. Each embryo within these batches is given an individual embryo ID and each processing step receives an identifying suffix in our file naming scheme as described for data record 1.
Data record 3—data tables for gap gene expression in wild-type embryos
These tables contain positions of extracted boundaries and associated meta-data on time classification and embryo regions used for boundary extraction. The data have been deposited with figshare as two tables in comma-separated value (CSV) format (Data Record 3, Data Citation 1).
File 1: wt_timeclasses_bands.csv
This file contains information on time classification and positions of 10%-strips from which expression data have been extracted from wild-type embryos. Columns are structured as follows:
1. embryo
Shows the embryo ID and its directory path.
2. timeclass
This column contains the cleavage cycle (C1 to C13), or time class if in C14A (C14_T1–T8). Our staging scheme is based on ref. 26: all cleavage cycles up to C9 are of a very similar duration (approximately 10 min). Blastoderm stages C10 to C14A last for 13, 11, 14, 23, and 58 min respectively. C14A is further subdivided into eight equal time classes (T1–8) of around 7 min each.
3. spline_height
Indicates the absolute height of the 10%-strip in pixels calculated as 10% of the height of each particular embryo mask.
4. – 13. spline_x1 to x5 and spline_y1 to y5
These columns indicate the x and y co-ordinates of the five equidistant points placed on the midline spline. Positions are given in pixels with an origin at the upper left corner of the image.
File 2: wt_boundary_data.csv
This file contains the co-ordinates of expression boundaries extracted by fitting clamped splines to profiles from wild-type embryos. Columns are structured as follows:
1. embryo
Shows the embryo ID and its directory path.
2. boundary_id
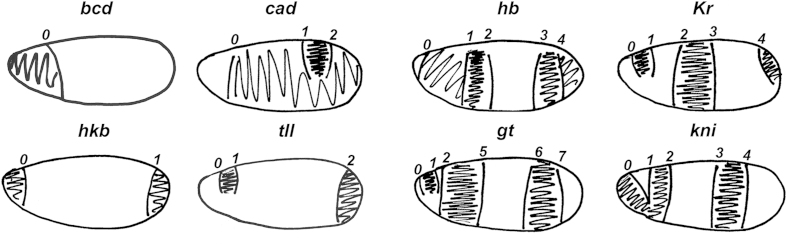
This number represents a specific expression boundary as shown in Fig. 2.
Figure 2. M. abdita boundary naming scheme.
This figure displays schematic drawings of embryos showing expression boundary numbers and relative positions for each M. abdita gene listed in File 2 of Data Records 3 and 4. gt boundaries 3 and 4 are omitted to provide consistent numbering with homologous D. melanogaster boundaries14. Maternal inputs (bcd, cad) and terminal gap genes (tll, hkb) are shown on the left. Trunk gap genes (hb, Kr, gt, kni) are shown on the right. Embryos drawn anterior to the left, dorsal up. Figure reproduced from (ref. 27, Supplementary File 2).
3. facing
This column indicates the position of a boundary with regard to its expression domain (i.e. which direction it is facing). There are two possible values: ‘anterior’ and ‘posterior’.
4. gene
This column indicates the gene name (given as standard gene abbreviation; for example, ‘gt’ for giant).
5. channel
This column records the colour of the stain (purple: NBT/BCIP, or red: Fast Red) from which the boundaries were extracted.
6. – 11. boundary_x1 to x3 and boundary_y1 to y3
These columns contain the start-, mid-, and end-points of the clamped splines used to measure boundary positions and width. Positions are indicated in pixels with an origin in the upper left corner of the graph.
Data record 4—data tables for gap gene expression in RNAi-treated embryos
These tables contain positions of extracted boundaries and associated meta-data on time classification and embryo regions used for boundary extraction. The data have been deposited with figshare as two tables in CSV format (Data Record 4, Data Citation 1).
File 1: RNAi_timeclasses_bands.csv
This file contains information on time classification and positions of 10%-strips from which expression data have been extracted from RNAi-treated embryos. Columns are structured as described for Data Record 3, File 1.
File 2: RNAi_boundary_data.csv
This file contains the co-ordinates of expression boundaries extracted by fitting clamped splines to profiles from RNAi-treated embryos. Columns are structured as described for Data Record 3, File 2.
Data record 5—integrated wild-type expression data
These data have been deposited with figshare as a table in CSV format (Data Record 5, Data Citation 1).
File 1: wt_integrated_boundaries.csv
This file details averaged coordinates of expression boundaries. Averages and deviations are computed as both arithmetic means and medians, and standard deviations and median absolute deviations (MAD), respectively. The file consists of the following columns:
1. gene
This column indicates the gene name (given as standard gene abbreviation; for example, ‘gt’ for giant).
2. timeclass
This column contains the cleavage cycle (C1 to C13), or time class if in C14A (C14_T1–T8). Our staging scheme is based on ref. 26. See Data Record 3 for details.
3. boundary id
This number represents a specific expression boundary as shown in Fig. 2 (ref. 27).
4. mean
Contains the (arithmetic) mean A–P position (in % embryo length) of the boundary at a given time point.
5. median
Contains the median A–P position (in % embryo length) of the boundary at a given time point.
6. standard deviation
Shows the standard deviation (in % embryo length) for each boundary and time class.
7. MAD
Shows the median absolute deviation (MAD, in % embryo length) for each boundary and time class.
Technical Validation
Our data processing pipeline21 produces high-quality spatio-temporal mRNA expression data, which are quality-checked and validated in several ways. Quality control is implemented as follows: embryo morphology, orientation, and quality of the staining (for example, signal-to-background ratio) are carefully assessed and recorded during data processing, and only high-quality data are selected for further use. Moreover, processing steps that are sensitive to observer bias—such as fitting of splines to extract boundaries (Fig. 1b) or time classification of embryos26—are always checked by two independent researchers.
In terms of data validation, a D. melanogaster expression data set—quantified in a manner equivalent to our M. abdita data—is in close agreement with previously published, independent qualitative and quantitative evidence. This includes the vast literature with qualitative observations on maternal co-ordinate and gap gene expression patterns in D. melanogaster that has accumulated over the past 30 years (reviewed in ref. 8) and, more importantly, a number of more recent quantitative studies documenting gap gene expression patterns in D. melanogaster and related drosophilid species both at the mRNA and protein level11,12,14,17,34–36. Another line of evidence also confirms the accuracy and validity of our data: we have used our D. melanogaster mRNA data set to fit gene network models of the gap gene system. The regulatory network structure predicted by these models is in strong agreement, not only with previous models fitted to protein expression data9,10,13,14, but also with the evidence on regulatory interactions gained from a vast amount of genetic experiments8. Especially the latter case, where gap gene network structures derived from completely independent and complementary evidence—gained by reverse-engineering and genetics, respectively—agree, is a strong indication that our data accurately represent the actual expression dynamics in the embryo.
Gene knock-down by RNAi is an established protocol in M. abdita 23,31,32. We control for possible off-target effect by the injection of two dsRNA fragments, one of which lacks conserved regions (that is, zinc finger or homeobox domains). In each case this resulted in identical knock-down phenotypes. Using this RNAi protocol, we have been able to reproduce previously published gene expression patterns and cuticle phenotypes in RNAi-treated M. abdita embryos27,31,32. In addition, knock-down of gap genes that had not been examined previously, closely resemble the equivalent null mutant phenotypes in D. melanogaster 27. Finally, RNAi using an identical protocol in D. melanogaster produced exact phenocopies of the respective null mutants28.
Usage Notes
Our data set is available online through the SuperFly database: http://superfly.crg.eu (ref. 37). SuperFly provides a user-friendly web interface for browsing, searching, and downloading batches of data or individual embryos. It includes an online help system with instructions on how to use it.
Additional information
How to cite this article: Wotton, K. R. et al. High-resolution gene expression data from blastoderm embryos of the scuttle fly Megaselia abdita. Sci. Data 2:150005 doi: 10.1038/sdata.2015.5 (2015).
Supplementary Material
Acknowledgments
Urs Schmidt-Ott and Steffen Lemke provided essential start-up help and support for our work with M. abdita. We thank Brenda Gavilán (Universitat Autònoma de Barcelona) and Núria Bosch Guiteras (Universitat Pompeu Fabra), who assisted with the maintenance of our M. abdita culture. We thank the CRG Bioinformatics Core, and Antonio Hermoso Pulido in particular, for setting up and maintaining the SuperFly database. This work was funded by the MEC-EMBL agreement for the EMBL/CRG Research Unit in Systems Biology, by European Commission grant FP7-KBBE-2011-5/289434 (BioPreDyn), by Grant 153 (MOPDEV) of the ERANet: ComplexityNET programme, by AGAUR SGR Grant 406, as well as grants BFU2009-10184 and BFU2012-33775 from the Spanish Ministry of the Economy and Competitiveness (MINECO, formerly MICINN). The Centre for Genomic Regulation (CRG) acknowledges support from MINECO, 'Centro de Excelencia Severo Ochoa 2013-2017', SEV-2012-0208.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Wotton K. R, Jiménez-Guri E., Crombach A., Cicin-Sain D., Jaeger J. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1252195 [DOI] [PMC free article] [PubMed]
References
- Davidson E. H. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution (Academic Press, 2006). [Google Scholar]
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits (Chapman and Hall/CRC, 2007). [Google Scholar]
- Stern D. L. & Orgogozo V. Is genetic evolution predictable? Science 323, 746–751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. & Sharpe J. in Towards a Theory of Development (eds Minelli A., Pradeu T.) 56–78 (Oxford University Press, 2014). [Google Scholar]
- Jaeger J. & Monk N. Bioattractors: dynamical systems theory and the evolution of regulatory processes. J. Physiol. 592, 2267–2281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. & Crombach A. in Evolutionary Systems Biology (ed. Soyer O.) 93–119 (Springer, 2012). [Google Scholar]
- Jaeger J. & Monk N. in Learning and Inference in Computational Systems Biology (eds. Lawrence N. D., Girolami M., Rattray M., Sanguinetti G.) 9–34 (MIT Press, 2010). [Google Scholar]
- Jaeger J. The gap gene network. Cell. Mol. Life Sci. 68, 243–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. et al. Dynamic control of positional information in the early Drosophila embryo. Nature 430, 368–371 (2004). [DOI] [PubMed] [Google Scholar]
- Jaeger J. et al. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics 167, 1721–1737 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J., Sharp D. H. & Reinitz J. Known maternal gradients are not sufficient for the establishment of gap domains in Drosophila melanogaster. Mech. Dev. 124, 108–128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S. et al. Characterization of the Drosophila segment determination morphome. Dev. Biol. 313, 844–862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashyraliyev M. et al. Gene circuit analysis of the terminal gap gene huckebein. PLoS Comput. Biol. 5, e1000548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombach A., Wotton K. R., Cicin-Sain D., Ashyraliyev M. & Jaeger J. Efficient reverse-engineering of a developmental gene regulatory network. PLoS Comput. Biol. 8, e1002589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H. et al. Lack of tailless leads to an increase in expression variability in Drosophila embryos. Dev. Biol. 377, 305–317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S. et al. Quantitative dynamics and increased variability of segmentation gene expression in the Drosophila Krüppel and knirps mutants. Dev. Biol. 376, 99–112 (2013). [DOI] [PubMed] [Google Scholar]
- Becker K. et al. Reverse-engineering post-transcriptional regulation of gap genes in Drosophila melanogaster. PLoS Comput. Biol. 9, e1003281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H. et al. A quantitative atlas of Even-skipped and Hunchback expression in Clogmia albipunctata (Diptera: Psychodidae) blastoderm embryos. Evodevo 5, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombach A., García-Solache M. A. & Jaeger J. Evolution of early development in dipterans: Reverse-engineering the gap gene network in the moth midge Clogmia albipunctata (Psychodidae). Biosystems 123, 75–85 (2014). [DOI] [PubMed] [Google Scholar]
- Surkova S. et al. Pipeline for acquisition of quantitative data on segmentation gene expression from confocal images. Fly 2, 58–66 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombach A., Wotton K. R., Cicin-Sain D. & Jaeger J. Medium-throughput processing of whole mount in situ hybridisation experiments into gene expression domains. PLoS ONE 7, e46658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S. et al. in Imaging in Developmental Biology (eds Sharpe J., Wong R.) 683–698 (Cold Spring Harbor Press, 2011). [Google Scholar]
- Rafiqi A., Lemke S. & Schmidt-Ott U. The scuttle fly Megaselia abdita (Phoridae): A link between Drosophila and mosquito development. Cold Spring Harb. Protoc. 4, 349–353 (2011). [DOI] [PubMed] [Google Scholar]
- Foe V. E. & Alberts B. M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31–70 (1983). [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. & Hartenstein V. The Embryonic Development of Drosophila melanogaster, 2nd edn, (Springer, 1997). [Google Scholar]
- Wotton K. R., Jiménez-Guri E., García Matheu B. & Jaeger J. A staging scheme for the development of the scuttle fly Megaselia abdita. PLoS ONE 9, e84421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton K. R. et al. Quantitative System drift compensates for altered maternal inputs to the gap gene network of the scuttle fly Megaselia abdita. eLIFE 4, e04785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton K., Jiménez-Guri E. & Jaeger J. Axis polarity in the scuttle fly Megaselia abdita. PLoS Genet. 10.1371/journal.pgen.1005042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi A. M., Lemke S. & Schmidt-Ott U. Megaselia abdita: culturing and egg collection. Cold Spring Harb. Protoc. 2011, 423–425 10.1101/pdb.prot5600 (2011). [DOI] [PubMed] [Google Scholar]
- Rafiqi A. M., Lemke S. & Schmidt-Ott U. Megaselia abdita: fixing and devitellinizing embryos. Cold Spring Harb. Protoc. 2011, 429–431 10.1101/pdb.prot5602 (2011). [DOI] [PubMed] [Google Scholar]
- Stauber M., Taubert H. & Schmidt-Ott U. Function of bicoid and hunchback homologs in the basal cyclorrhaphan fly Megaselia (Phoridae). Proc. Natl Acad. Sci. USA 97, 10844–10849 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke S. et al. Bicoid occurrence and Bicoid-dependent hunchback regulation in lower cyclorrhaphan flies. Evol. Dev. 10, 413–420 (2008). [DOI] [PubMed] [Google Scholar]
- Weisstein E. W. “Cubic Spline.” From MathWorld—A Wolfram Web Resource http://mathworld.wolfram.com/CubicSpline.html.
- Fowlkes C. C. et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell 133, 364–374 (2008). [DOI] [PubMed] [Google Scholar]
- Fowlkes C. C. et al. A conserved developmental patterning network produces quantitatively different output in multiple species of Drosophila. PLoS Genet. 7, e1002346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z. et al. Dissecting sources of quantitative gene expression pattern divergence between Drosophila species. Mol. Syst. Biol. 8, 604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicin-Sain D. et al. SuperFly: a comparative database for quantified spatio-temporal gene expression patterns in early dipteran embryos. Nucleic Acids Res 43(Database Issue): D752–D756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wotton K. R, Jiménez-Guri E., Crombach A., Cicin-Sain D., Jaeger J. 2015. Figshare. http://dx.doi.org/10.6084/m9.figshare.1252195 [DOI] [PMC free article] [PubMed]