Abstract
Rapid and high wing-beat frequencies achieved during insect flight are powered by the indirect flight muscles, the largest group of muscles present in the thorax. Any anomaly during the assembly and/or structural impairment of the indirect flight muscles gives rise to a flightless phenotype. Multiple mutagenesis screens in Drosophila melanogaster for defective flight behavior have led to the isolation and characterization of mutations that have been instrumental in the identification of many proteins and residues that are important for muscle assembly, function, and disease. In this article, we present a molecular-genetic characterization of a flightless mutation, flightless-H (fliH), originally designated as heldup-a (hdp-a). We show that fliH is a cis-regulatory mutation of the wings up A (wupA) gene, which codes for the troponin-I protein, one of the troponin complex proteins, involved in regulation of muscle contraction. The mutation leads to reduced levels of troponin-I transcript and protein. In addition to this, there is also coordinated reduction in transcript and protein levels of other structural protein isoforms that are part of the troponin complex. The altered transcript and protein stoichiometry ultimately culminates in unregulated acto-myosin interactions and a hypercontraction muscle phenotype. Our results shed new insights into the importance of maintaining the stoichiometry of structural proteins during muscle assembly for proper function with implications for the identification of mutations and disease phenotypes in other species, including humans.
Keywords: Drosophila, flight muscles, troponin, muscle hypercontraction, protein stoichiometry
THE indirect flight muscles (IFMs) of Drosophila serve as a good genetic model system to study muscle development, assembly of structural proteins, and regulation of muscle contraction (Vigoreaux 2006; Nongthomba et al. 2007). Like the vertebrate skeletal muscles, IFM contraction is regulated by the influx of neurally stimulated intracellular Ca2+. Stretch activation allows IFM to contract at a much higher frequency and is similar to the asynchronous muscle contraction in the human heart (Peckham et al. 1990; Josephson et al. 2000; Agianian et al. 2004; Moore et al. 2006). In addition, major structural proteins like myosin, actin, tropomyosin (Tm), troponin (Tn), α-actinin, etc., that are involved in the assembly of sarcomeres, are conserved in vertebrates and invertebrates and dispense similar functions. Mutations in these sarcomeric proteins disrupt muscle structure and function (summarized in Vigoreaux 2006). Since IFMs are the only fibrillar muscles present in a fly’s body, many of the myofibrillar proteins have IFM-specific isoforms (Cripps 2006; Nongthomba et al. 2007) and are dispensable under laboratory conditions, enabling the isolation of mutations without affecting other physiological activities. The majority of mutations affecting the IFMs were isolated during mutagenesis screens for flightless behavior (Deak 1977; Homyk and Sheppard 1977; Mogami and Hotta 1981; Deak et al. 1982; Cripps et al. 1994; summarized in Cripps 2006). However, molecular lesions for many of these mutations are yet to be identified; as a result, plausible mechanisms that give rise to muscle phenotype of these mutations remain elusive.
Many of these flightless mutants are known to show a muscle phenotype that has been categorized as “hypercontraction.” Hypercontraction is a phenomenon that leads to muscle defects like thinning and tearing, following uncontrolled acto-myosin interactions of otherwise normally assembled sarcomeric structures (Nongthomba et al. 2003). Mutations leading to hypercontraction have been localized in structural genes such as the upheld gene (up101) (Fyrberg et al. 1990; Nongthomba et al. 2003), flightin (fln0) (Reedy et al. 2000), Actin88F (An and Mogami 1996; Nongthomba et al. 2003), Myosin heavy chain (Mhc6, Mhc13, and Mhc19) (Kronert et al. 1995), wings up A (wupAhdp-2) (Beall and Fyrberg 1991; Nongthomba et al. 2003), the protein phosphatase genes flapwing (flw1, flw6, and flw7) (Raghavan et al. 2000; Pronovost et al. 2013), and calcineurin B2 (canB2EP(2)0774) (Gajewski et al. 2006). Mutations producing hypercontraction fall in a large repertoire of proteins, suggesting that there could be multiple grounds for reaching the phenotype. Suppressor studies of the hypercontraction phenotype have led to the identification of mutations in structural genes like Tm2 (Naimi et al. 2001), Mhc (Kronert et al. 1999; Nongthomba et al. 2003), and the integrin adhesive complex protein PINCH (Pronovost et al. 2013), providing very fruitful insights into the structure–function relationship of these proteins. Studies of these hypercontracting alleles have led to the conclusion that defects in Ca2+ regulation, structural defects, troponin–tropomyosin (Tn-Tm) regulation defects, and mechanical stress can lead to IFM hypercontraction (Nongthomba et al. 2003; Cammarato et al. 2004; Pronovost et al. 2013). However, further work is required to identify genes/proteins and pathways involved in the pathogenesis of hypercontraction to unravel new players involved in the regulation of muscle contraction and possible mechanisms leading to muscle dysfunction, as is the case with most human myopathies.
Detailed characterization of the flightless mutations, which were isolated decades ago, has led to the identification of many proteins and residues that are important for muscle assembly, function and diseases (reviewed in Cripps 2006; Vigoreaux 2006). Previously, we have shown detailed characterization of the mutation up1, which was isolated in 1958 (Fahmy and Fahmy 1958) and which yielded new insights into troponin-T (TnT) isoform switching, muscle assembly, and function (Nongthomba et al. 2007). In the present study, we have used a similar approach to characterize flightless-H (fliH), a flightless mutation generated by Homyk and Sheppard (1977) using a chemical mutagen ethyl methanesulfonate (EMS). fliH was considered to be an allele of the heldup-a (hdp-a) mutation at the Beadex (Bx) locus. We found that fliH is a regulatory mutation of troponin-I (TnI), rather than Bx, with a hypercontraction muscle phenotype. All the isoforms of TnI in Drosophila are encoded by a single wupA gene. Many mutations already exist for the wupA gene, which showed developmental defects during sarcomere assembly leading to muscle degeneration (Beall and Fyrberg 1991; Nongthomba et al. 2004). wupAhdp-2, as mentioned above, is a hypercontracting allele of TnI and involves a point mutation changing alanine to valine at position 116 (Beall and Fyrberg 1991). Our study reveals that fliH is a unique allele that confers a temperature-sensitive muscle phenotype. This is the first mutation found in the regulatory region of any structural gene that leads to muscle hypercontraction. This study also emphasizes the importance of maintaining proper stoichiometry of structural proteins for proper functioning of the muscle.
Materials and Methods
Fly strains and crosses
All flies were maintained on corn flour–glucose–yeast–agar–medium. Canton-S served as the wild-type control, unless otherwise mentioned. The fliH stock was obtained from the Bloomington Drosophila Stock Center (BS #6028), and flies were raised at 25° and 18° for various experiments. UH3-Gal4, line specific for adult IFM, was generated in our lab (Singh et al. 2014), and UAS-L9 was procured from Alberto Ferrus (Cajal Institute, Madrid). wee-P26 (Mhc-GFP) (Clyne et al. 2003), Tm2-GFP (Morin et al. 2001), and sls-GFP (Burkart et al. 2007) are fusion constructs wherein GFP-coding sequence has been inserted at the C-terminal end of the native protein. Act88F-GFP transgenic line leads to GFP expression in IFM under the Act88F promoter (Barthmaier and Fyrberg 1995). Mhc null allele and Y97 (transgenic fly line carrying Headless myosin) have been described previously (Cripps et al. 1999). up101 (Fyrberg et al. 1990), wupAhdp-3 (Barbas et al. 1993; Nongthomba et al. 2003), and wupAhdp-2 (Beall and Fyrberg 1991) are mutations of troponin-T and troponin-I, respectively. All chromosomes and gene symbols are as in FlyBase (http://www.flybase.org). Pupae were aged according to a method previously described (Fernandes et al. 1991).
Behavioral test
Flight test was performed as described previously (Drummond et al. 1991). A walking and jumping test was performed as described previously (Naimi et al. 2001).
Polarized light microscopy
Fly hemithoraxes were prepared for polarized microscopy as described (Nongthomba et al. 1999). Briefly, fly thoraxes were frozen in liquid nitrogen, bisected longitudinally using a razor blade, dehydrated in an alcohol series, cleared in methyl salicylate, and mounted using Di-n-butyl phthalate in Xylene mounting medium. The hemithoraxes were observed under an Olympus SZX12 microscope and photographed using an Olympus C-5060 camera under polarized light optics.
Confocal microscopy
Flies were bisected as mentioned above, fixed in 4% paraformaldehyde, washed four times with PBT× (0.3% Triton-X100 in phosphate buffer saline) for 15 min each, and stained with 1:200 diluted phalloidin–tetramethylrhodamine B isothiocyanate (phalloidin–TRITC) (50 μg/ml stock; Sigma) for 20 min. Anti-Mlp60A antibody raised in our lab was used as a marker for Z-disc staining. Finally, the sections were washed four times with PBT× and mounted in Vectashield mounting medium (Vector Labs). Imaging was done using a Carl Zeiss LSM 510 META confocal microscope.
Genomic DNA isolation and PCR
Genomic DNA isolation was done by following Berkeley Drosophila Genome Project (http://www.flybase.org) protocol. DNA was diluted 1:100 times in milli-Q water and quantified using a Bio-Rad spectrophotometer. Approximately 200 ng of DNA was used as template for amplifying the specified genomic region. The following primers were used: TnI-URE1039F—5′-GGGATTCCCCAATTTTATCT-3′; TnI-URE1526R—5′-CCGCTTGGAATTCAATGC-3′; TnI-URE1818R—5′-AACTGACATGGCAGAGCACA-3′; TnI-URE57F—5′-AACGCTCGGAACGAGAATGA-3′; TnI-URE2078R—5′-CTGAACGGGCCGACGATCCA-3′; TnI-URE840F—5′-GCGGCCAACATGCAAGATA-3′; TnI-URE932R—5′-TTCTTAGACCGTGCCACT-3′; TnI-IRE1453F—5′-ACTATACGGATAGGCTAGCA-3′; TnI-IRE2021R—5′-ATCGCACACGCCTACGATCT-3′; TnI-IRE7F—5′-CGATCCGTATCTGTATCCGT-3′; TnI-IRE2495R—5′-GGTTGCATGTTGCGTGGTTG-3′; TnI-IRE1111F—5′-CCGAAGGTCGTCATTGTCAGAA-3′; and TnI-IRE1158R—5′-CTTAGCGAAGGTAAGGCGTG-3′.
Gel purification, cloning, and sequencing of PCR products
PCR products were purified using a gel purification kit (Qiagen), ligated to pGEM-T Easy cloning vector (Promega), and transformed into Escherichia coli DH5α cells. Plasmid preparations were done using a miniprep kit (Qiagen). DNA sequencing was done at Macrogen Inc. (Seoul, Korea) using T7 sequencing primers or primers used to generate the fragments. Both the DNA strands, for a minimum of three clones from each PCR, were sequenced; output was analyzed using Chromas Lite and ClustalW softwares.
Semiquantitative RT-PCR
Newly eclosed flies were kept in 70% ethanol and frozen at −80° freezer. IFMs were dissected from these flies, and total RNA was isolated using Trizol reagent (Sigma). The RNA amount was quantified by taking optical density (OD) at 260 nm, and purity was assessed by calculating the OD ratio at 260- and 280-nm wavelengths. Total RNA (2 μg) in a volume of 20 µl was used for complementary DNA (cDNA) preparation using a first-strand cDNA synthesis kit (Fermentas). cDNA (1 μl) was used for carrying out the PCR. All the PCR reactions were performed at a predetermined nonsaturating cycle number for each gene. All the primers used have been described previously (Nongthomba et al. 2007). rp-49 (ribosomal protein-encoding RNA) was used as internal control. The PCR products were resolved on 1.2% agarose gels, and images were captured using the JHBIO (JH BIO Innovations) gel documentation system, and gel quantification was done using the SpotDenso tool of AlphaEaseFC software (Alpha Innotech). The data were processed using MS Excel.
Quantitative real-time RT-PCR
Real-time quantitation was performed using cDNA equivalent to 20 ng of total RNA isolated from fly IFM. All the PCR reactions were carried out using Dynamo SYBRgreen mix (Finnzymes, Finland) in the ABI Prism 7900HT sequence detection system (Applied Biosystems) and analyzed with SDS 2.1 software (Applied Biosystems). rp-49 primers were used for normalization of RT-PCR data, and fold change over control was calculated. Primers used in the present study were tested beforehand for the presence of a single peak in the dissociation curve, which suggests that there is a single amplicon. The following primers were used: ribosomal gene rp-49F—5′-AAGCTGTCGCACAAATGG-3′; rp-49R—5′-ATCCGTAACCGATGTTGG-3′; TnI-Ex4F—5′-GGCTGATGATGAGGCTAAGA-3′; TnI-Ex4R—5′-TACGCAGCAGCAACCTGAGT-3′; TnI-Ex6F—5′-CCAGCGAAGGCGAATTG-3′; and TnI-Ex6R—5′-GATCGTTGATCTCCCAGTCT-3′.
Protein extraction and Western blot
IFMs were removed from bisected flies preserved in 70% alcohol and homogenized in 1× buffer (0.1 M NaCl, 10 mM potassium phosphate, pH 7.0, 2 mM EGTA, 2 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 0.5% Triton-X). The IFM lysate was spun down to obtain protein pellet, which was further washed with the same 1× buffer without Triton-X and then boiled in SDS–sample buffer (0.0625 M Tris–Cl, pH 6.8, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 5 μg bromophenol blue) for 4 min at 95°. Samples were then resolved in 12% PAGE gel in a mini electrophoresis unit (Amersham) at 100 V. The protein was then transferred from gel to PVDF membrane (Immobilon-P, Millipore) in transfer buffer (20% methanol, 25 mM Tris–base, and 150 mM glycine). The membrane was blocked with 8% milk solution in Tris buffer saline (TBS, pH 7.4) for 1 hr and then probed with primary antibody at prescribed dilution overnight at 4°. The following antisera were used: anti-Drosophila troponin-I (1:1000, raised in rabbit, gift from Alberto Ferrus); antiacetylated α-tubulin [1:1000, raised in mouse (Sigma)]. After washing three times with TBS, the membrane was incubated with HRP-conjugated secondary antibody (1:1000, Bangalore Genei, Bangalore, India) for 3 hr at room temperature. The membrane was then washed three times (15 min each) with TBST (TBS with 0.05% Tween 20) and 5 min with 0.5 M NaCl. Bands were detected by adding 3,3′-diaminobenzidine substrate along with 0.5% H2O2 on the membrane or developed using the enhanced chemiluminescence method (Supersignal WestPico Chemiluminescent substrate, Pierce).
Protein expression
Mef-2 forward primer (5′-ATCTGTTCATATGGGCCGCA-3′) and Mef-2 reverse primer (5′-CTGCTGCTCGAGATGGTGTT-3′) were used to amplify the Mef-2 sequence from the pBluescript vector containing Mef-2 cDNA (provided by Richard Cripps, University of New Mexico). The cDNA was cloned in pET15b expression vector (Novagen) under T7 promoter using NdeI and XhoI restriction enzymes. The protein was expressed in TNT Quick Coupled Transcription/Translation systems (Promega) by adding 1 µg of purified Mef-2-pET plasmid vector, 1 mM methionine, and 40 µl of TNT Quick Master Mix and incubating the cocktail at 30° for 30 min. The lysate from the reaction was used for further experiments.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assay (EMSA) was performed essentially as described (Sambrook et al. 1989). Probe DNA and unlabeled competitors were generated by annealing complementary oligonucleotides to generate a double-stranded DNA molecule with GG overhangs, thus creating recessed 3′ ends. The following oligonuleotides were used for EMSA: Dmef2(+)—5′-GGTGTCTATATTTAGCCC-3′; Dmef2(−)—5′-GGGGGCTAAATATAGACA-3′ (Cripps et al. 2004); CS1(+) (control sequence1)—5′-GGCATTTATCCTCATAAACATAACTAATATTTCGATGA-3′; CS1(−)—5′-GGTCATCGAAATATTAGTTATGTTTATGAGGATAAATG-3′; CS2(+)—5′-GGAGTTGACTGAATACAAATTACTGTTTTCA-3′; CS2(−)—5′-GGTGAAAACAGTAATTTGTATTCAGTCAACT-3′; FM1(+) (fliH mutant sequence1) —5′-GGCATTTATTTATTCTCATAAACATAACTAATATTTCTATGA-3′; FM1(−)—5′-GGTCATAGAAATATTAGTTATGTTTATGAGGATAAATAAATG-3′; FM2(+)—5′-GGAGTTGACTGAATTACAAATACTGTTTCA-3′; and FM2(−)—5′-GGTGAAACAGTATTTGTAATTCAGTCAACT-3′. Annealed sequences were radioactively labeled with 32P-dCTP (BRIT, Hyderabad, India) using Klenow enzyme (Fermentas Life Sciences) and purified using G-25 columns. Mef-2 lysate was pre-incubated for 15 min on ice with 50 mg/ml of poly (dI-dC) and binding buffer (20 mM Hepes–KOH, 60 mM KCl, 200 µM EDTA, 10% (w/v) glycerol). Competitors were then added and incubated for 15 min on ice. Finally, labeled probes were added and reaction was incubated for 30 min. The entire reactions mixes were loaded onto an 8% nondenaturing polyacrylamide gel that was run at 100 V at 4°. The gel was then dried for 1 hr and exposed overnight. The image was captured using a Typhoon image scanner (Amersham Bioscience).
Luciferase assay
To determine the promoter activities of wild-type and fliH mutant, luciferase assay was performed. The 979-bp-long DNA fragment from the regulatory region of wupA was PCR-amplified from wild-type and fliH mutant genomic DNA using the primer set TnI-URE840F—5′-GCGGCCAACATGCAAGATA-3′ and TnI-URE1818R—5′-AACTGACATGGCAGAGCACA-3′. The resultant fragments were inserted into the pTZ57R/T vector (Fermentas) and sequenced. These fragments were further subcloned into the KpnI/XhoI sites of the pGL3-luciferase vector (Promega). The C2C12 myoblast cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco). The C2C12 cells (6 × 104 cells/well) were grown in a 24-well plate (Nunc,Thermo Scientific) for 24 hr until 70% confluence was reached. The cells were then transfected with pGL3-Basic (Control) wild-type 979 bp (Control-URE), and mutant 979 bp (fliH-URE) along with that of pRL-TK Renilla luciferase (Promega) using lipofectamine 2000 (Invitrogen) as per the manufacturer’s instruction. Luciferase reporter activity was estimated after 36 hr using the dual luciferase assay kit (Promega) and a TD-20/20 luminometer (Turner Design, Sunnyvale, CA) following the manufacturer’s protocol, and the Renilla luciferase signal was normalized to the firefly luciferase signal. Human foreskin fibroblast (HFF) cells were used to reconfirm the promoter activation of wild type and the fliH mutant. The HFF cells (devoid of the transcription factor Mef2) were cultured in DMEM (Sigma Aldrich) supplemented with 10% FBS and 100 units/ml penicillin and 100 µg/ml streptomycin (Invitrogen Life Sciences). The HFF cells (6 × 104 cells/well) were grown in a 12-well plate (Nunc, Thermo Scientific) for 24 hr until 70% confluence was reached. The cells were then transfected with the pGL3-Basic, wild-type 979-bp (Control-URE) and mutant 979-bp (fliH-URE) plasmids along with the pRL-TK (Promega) plasmid as transfection control. The cells were also transfected with pcDNA3.1(−) and pcDNA3.1-Mef2 along with pGL3-Basic, wild-type 979 bp (Control-URE), and mutant 979 bp (fliH-URE) along with that of the pRL-TK Renilla luciferase (Promega) using lipofectamine 2000 (Invitrogen) per manufacturer’s instructions. The luciferase reporter activity was measured after 24 hr using the dual luciferase assay kit (Promega) and a TD-20/20 luminometer (Turner Design) following the manufacturer’s protocol. The Renilla luciferase signal was normalized to the firefly luciferase signal. The DLR assay system kit (Promega) was used to perform dual luciferase assays. Briefly, HFF cells were grown in 12-well plates and transfected with 0.5 µg of pGL3-Basic, wild-type 979 bp (Control-URE), and mutant 979 bp (fliH-URE) along with 0.5 µg of either the pCDNA3.1 empty vector or pCDNA3.1-Mef2 and 50 ng of pRL-TK (Renilla luciferase under the constitutively active thymidine kinase promoter) construct for 6 hr in plain DMEM using lipofectamine 2000 (Invitrogen) reagent. After 6 hr cells were washed with Dulbecco's Phosphate-Buffered Saline, and fresh reconstituted media was added for a period of 24 hr. Cells were lysed in 200 µl of 1× passive lysis buffer. The lysates were transferred into microfuge tubes and centrifuged at 10,000 × g for 10 min at 4° to pellet-down the debris. The supernatant (10 μl) was taken in a fresh microfuge tube to which 10 µl of the luciferase assay reagent II was added, and the activity of the firefly luciferase was recorded using a luminometer (TD-20/20, Turner Designs). The activity of the Renilla luciferase was also measured after adding the Renilla substrate, Stop and Glo. The ratio of firefly and Renilla (relative luciferase units) was calculated and plotted.
The cDNA was prepared from HFF cells alone and HFF cells transfected with pCDNA3.1 + pGCL3-Basic empty vector and pCDNA3.1-MEF2 + pGCL3-Basic empty vector. The following primers were used to see the expression of Drosophila Mef2 in HFF cells: RPL-35A-F—5′-GGGTACAGCATCACTCGGA; RPL-35A-R—5′-ACGCCCGAGATGAAACAG; dMef2-F—5′-ACGAGTCCCTCACCAACAAG; and dMef2-R—5′-CGTGTAGCTGCTGTTTGGAA.
Results
fliH mutation leads to IFM hypercontraction phenotype
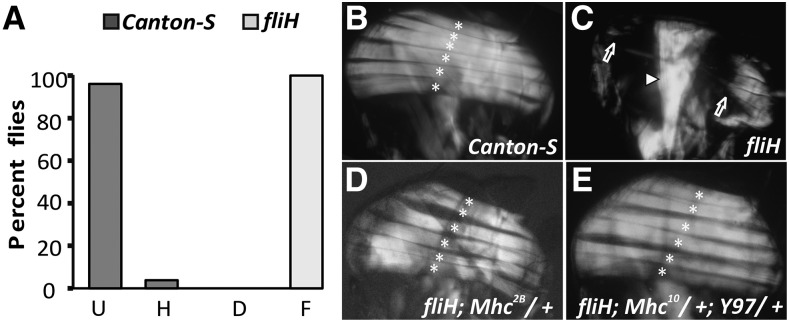
The fliH mutant was isolated based on a recessive flightless phenotype in an EMS screen for flightless behavior (Homyk and Sheppard 1977). fliH flies indeed showed complete flightlessness compared to their wild-type Canton-S counterpart (Figure 1A). Mutations affecting flight muscles or neurons innervating these muscles are the major cause of decreased flight ability (reviewed in Lloyd and Taylor 2010). Two opposing groups of IFMs, the dorsal longitudinal muscles (DLMs) and the dorso-ventral muscles, assist the direct flight muscles linked to wing hinge to produce higher wing-beat frequencies required for flight by distorting the thorax (reviewed in Josephson 2006; Lehmann 2006). Upon examining the bisected thorax that houses these major muscles, we noted that DLMs showed a hypercontraction-like muscle phenotype as reported earlier (Nongthomba et al. 2003), with muscles torn from the middle with few remnants attached at both ends (Figure 1C), compared to the six DLMs in wild type (Figure 1B).
Figure 1.
IFM abnormalities in 3- to 5-day-old fliH flies. (A) Flight data of Canton-S and fliH flies grown at 25°, where all the mutant flies show flightless phenotype (n = 50). Flight ability was measured using Sparrow Box, with gradation on their ability to fly upward (U), horizontally (H), and downward (D) and those that were could not fly (F). (B) Polarized light micrograph of wild-type DLMs. (C) fliH hemithorax showing hypercontraction muscle phenotype (arrows). Arrowhead points to normal jump muscles in the mutant. (D) Rescue of fliH phenotype. Mhc2B harbors mutation in actin-binding loop and completely rescues fliH hypercontraction phenotype. (E) Significant rescue of the muscle phenotype is also achieved with headless myosin construct (Y97) and a copy of myosin null (to reduce functional myosin). All six DLMs are marked with asterisks. Anterior is to the left and dorsal is on the top.
All hypercontraction muscle phenotypes can be rescued by removing myosin or reducing acto-myosin interaction through headless myosin construct (Y97) or myosin mutations (Nongthomba et al. 2003). Mhc2B (MhcP401S) is a mutation in the actin-binding loop (Nongthomba et al. 2003) of myosin that inhibits its interaction with actin, and this mutation completely rescued the fliH (Figure 1D) muscle phenotype. The headless myosin construct (Y97) completely rescued the fliH phenotype (Figure 1E). Both these experiments confirmed that the fliH muscle phenotype is due to hypercontraction, which may result from increased or unregulated acto-myosin interaction or mechanical stress.
Early IFM myofibrillogenesis proceeds normally in fliH, and hypercontraction of muscles is seen at eclosion
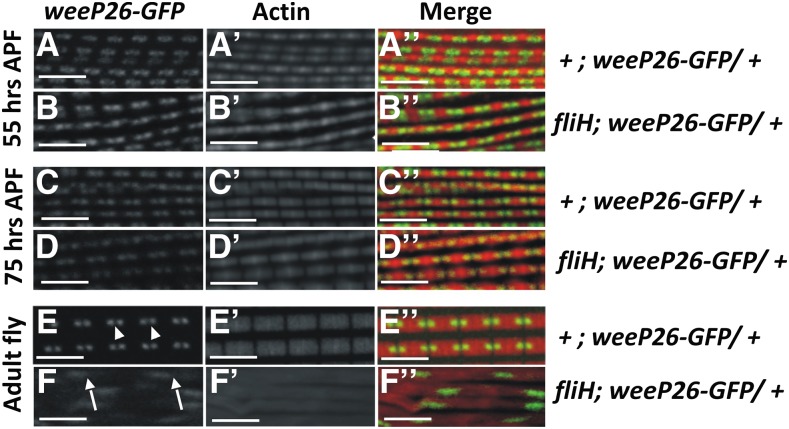
During IFM development, well-demarcated sarcomere structures are seen by 42 hr after puparium formation (APF), and increase in the length and breadth of the sarcomere takes place throughout the pupal development stages (Reedy and Beall 1993; Nongthomba et al. 2004). Development and assembly of sarcomeric structure in fliH was tracked using confocal microscopy utilizing wee-P26, a “protein trap construct” expressing GFP-tagged myosin heavy chain (Mhc-GFP) fusion protein, which enables tracking of thick filaments in the sarcomere (Clyne et al. 2003). The muscles from this genetic background were counterstained with phalloidin-TRITC to visualize F-actin. Assembly of sarcomeres in fliH IFM during early developmental stages, i.e., 55 hr APF (Figure 2, B–B′′) and 75 hr APF (Figure 2, D–D′′), were normal with properly organized thick and thin filaments that are comparable to controls (Figure 2, A–A′′ and C–C′′). However, sarcomere organization in just-eclosed adult mutant flies showed completely disrupted structures (Figure 2, F–F′′ and Supporting Information, Figure S1, B–B′′), suggesting that hypercontraction of the muscles takes place in at the late pupal stage, as with other hypercontracting alleles (Nongthomba et al. 2003). The normal F-actin banding pattern was lost in mutant IFMs (Figure 2F′; compare with Figure 2E′). Mutant IFMs also showed disarrayed myofibrils moving in and out of the confocal plane (Figure S1B′′) as opposed to their parallel arrangement in wild type (Figure S1A′′).
Figure 2.
Developmental profile of the fliH DLMs. The sarcomeric structure of muscles was observed using a confocal microscope. Interdigitating thick filaments were followed with wee-P26 (green; see Materials and Methods for details) and thin filaments (red) with phalloidin-TRITC. (A–D′′) Myofibril develops normally in fliH pupa until 75 hr APF. (E–E′′) Wild-type sarcomeres show uniform distribution of thick filaments (E, arrowhead). (F–F′′) Myofibrillar disorganization is seen in the IFMs of freshly eclosed fliH flies. (F) Thick filaments are diffused with increased spacing between them (arrows). (F′′) Regular arrangement of thin filaments is also disturbed (compare with E′′). Bar, 5 µm.
The mutant muscle phenotypes in adult IFM were also studied using tropomyosin-2-GFP (TM2-GFP, a protein trap expressing GFP-tagged Tropomyosin-2), a thin filament protein (Figure S1, C and D), and sallimus-GFP (SLS-GFP, a protein trap expressing GFP-tagged sallimus), a Z-disc protein (Figure S1, E–G). Thin filament proteins are seen in small clumps in the fliH hypercontracted muscles (Figure S1D′′). The regular banding patterns of Z-discs and size and spacing of the thin filaments were lost in freshly eclosed fliH IFMs (Figure S1, F–F′) when compared to control (Figure S1E′′). More pronounced dissolution of Z-discs and myofilaments is observed in 2-day-old flies (Figure S1, G–G′′).
fliH shows temperature-dependent IFM and other locomotor behavior defects
Most of the mutations isolated during Homyk and Sheppard’s (1977) screen showed temperature-dependent flight behavior. Since fliH was also isolated from the screen, we wanted to know if fliH is a temperature-sensitive mutant. Indeed, the muscle hypercontraction phenotype of fliH was a temperature-dependent phenotype (Figure S3). As expected, polarized light microscopy showed that DLM morphology is completely rescued in the mutant flies raised at lower temperature (data not shown). A confocal micrograph of the DLM fibers of these mutants showed proper assembly of the thick filaments (Figure S2, A–A′′), thin filaments (Figure S2, B–B′′), and Z-discs (Figure S2, C–C′′) reflecting normal sarcomere development. Since flight behavior directly reflects the muscle organization, it can be concluded that hypercontraction of the IFMs in fliH is temperature-dependent.
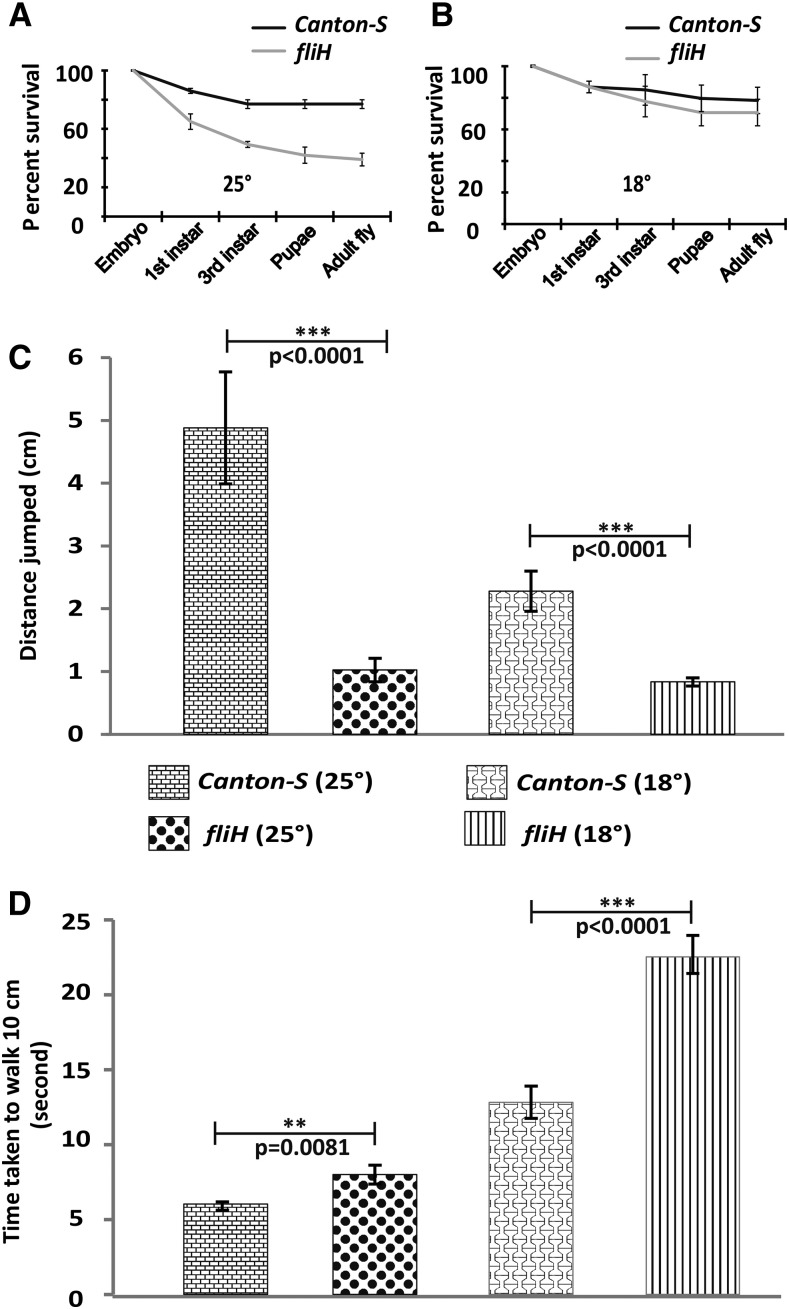
Many muscle mutants show a phenotype restricted to IFMs and thus affecting only flight. Such mutations in question have a genetic lesion in the protein isoforms that are IFM-specific (Nongthomba et al. 2004, 2007). We wanted to check if the phenotype associated with fliH is restricted to IFMs or not. A survival test was performed to assay for any associated lethality during development. We found that the fliH mutation leads to partial lethality at various stages of development at 25° (Figure 3A) compared to much reduced lethality at 18° (Figure 3B). Jumping (Figure 3C) and walking (Figure 3D) abilities were also found to be significantly compromised in fliH as compared to Canton-S, suggesting that the tergal depressor of trochanter (TDT) and leg muscles are also affected. As with the flight ability, we also found reduced jumping and walking abilities in wild-type and fliH flies when raised at 18°. However, unlike flight ability, walking and jumping abilities were not rescued in fliH raised at 18°; rather showed a more pronounced phenotype (Figure 3, C and D), suggesting that TDT and leg muscles may have abnormalities that are independent of temperature. Jumping and walking abilities when monitored for several days post-eclosion did not show any deterioration, ruling out any enhanced age-dependent progressive TDT damage or leg muscle defects (data not shown), which have been shown for other hypercontracting alleles (Nongthomba et al. 2003). Overall, we found that the fliH mutation also affects other muscles, but the most pronounced phenotype is seen in the IFMs.
Figure 3.
Behavioral experiments. (A) Survival curve of fliH flies raised at 25° shows partial lethality dispersed over all the developmental stages although the flies do not have any associated lethality at permissive temperature (18°) (B). (C) Jumping ability is severely compromised in mutants at 25° and 18°. (D) Mutant flies take longer to walk a distance of 10 cm as compared to the wild-type counterpart at 18° and 25°. Error bar denotes SEM. Statistical analysis was performed using unpaired two-tailed t-test.
fliH mutation is present in the wupA regulatory region
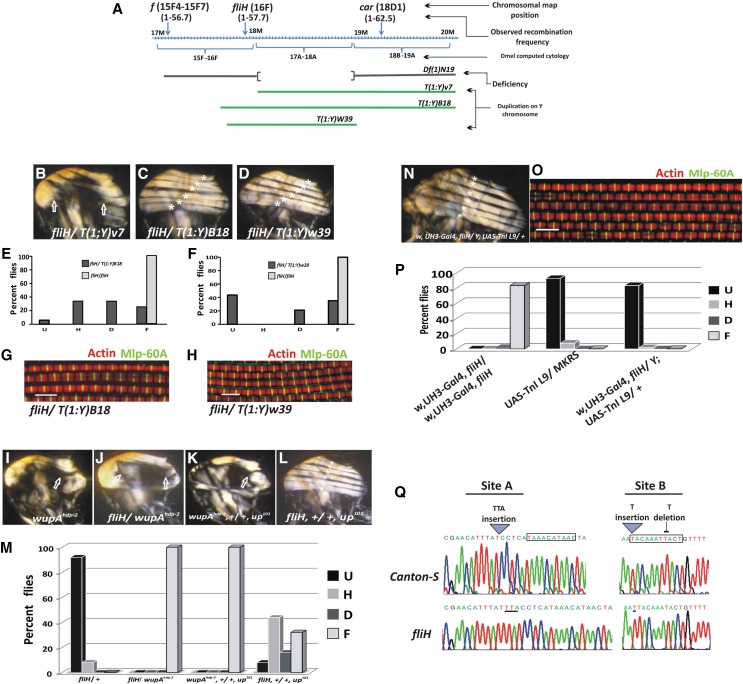
fliH is considered as a hdp-a mutation for the Bx locus with cytogenetic map position at 17A1-18A2. However, we did not find any communication that has reported the cytogenetic mapping or locus associated with fliH or hdp102 (which is synonymous with the fliH reported in Homyk and Emerson 1988). Hypomorphs of Bx do show a held-up wing phenotype (Lifschytz and Green 1979; Shoresh et al. 1998), but IFMs do not show a hypercontraction phenotype (our unpublished data). The fliH mutation was reported to be proximal to forked (f) and close to wupAhdp-2 and wupAhdp-3 (Homyk and Sheppard 1977). Using f and carnation (car) recessive markers, we found the recombination map location of fliH to be at 1–57.69 ± 0.87 (16F chromosomal locus) (Figure 4A). Unlike the FlyBase report, fliH is not uncovered by Df(1)N19, which deletes the chromosomal segment 17A1-18A2, which includes the Bx gene (Figure 4A). Recombination mapping suggested that fliH could be localized near the chromosome locus 16F. We have used multiple translocation and duplication lines to fine-map the mutation. We could not rescue the mutant phenotype with Y-chromosome translocation T(1;Y)V7 [translocated chromosome regions of 16F5-16F7; h18-h25 (Ferrus et al. 1990)] (Figure 4B). However, fliH was rescued by the lines T(1;Y)B18 (16F-17A; h1-h17) (Stewart and Merriam 1973) and Dp(1;Y)W39 (duplicated region spanning 16F1-18A7 (Prado et al. 1999) (Figure 4A). Rescued flies showed six DLM fascicles like the wild-type counterpart in polarized light imaging of hemithoraxes (Figure 4, B–D). These flies also showed slight rescue of the flight ability (Figure 4, E and F). Confocal micrographs of the rescued flies showed marked regularity in Z-disc spacing as well as regular banding pattern of thin filaments (Figure 4, G and H). Thus, genetic analyses using duplication and deletion mapping place the fliH mutation at the cytogenetic map location 16F1-17A. The only structural gene reported in this region that gives muscle hypercontraction similar to fliH is the wupA gene, which codes for TnI, an inhibitory component of the troponin complex.
Figure 4.
Genetic mapping and rescue of fliH. (A) X-chromosome map showing cytological location 15F-19A. Green lines denote X-chromosome segments duplicated on Y chromosome for various transposition lines used in the study. Meiotic recombination mapping with forked (f) and carnation (car) places fliH mutation at 1–57.69 ± 0.87 recombination map unit. (B) fliH was not rescued with T(1:Y)v7. Complete rescue of the DLM fiber morphology with T(1:Y)B18 and T(1:Y)W39 transposition line (C and D, respectively). (E and F) Partial rescue of the flight ability of fliH mutants with above-mentioned transposition lines. (G and H) Confocal micrograph showing normal sarcomeric organization of DLMs in fly rescued with duplicated segments on Y chromosome. (I–M) fliH interaction with other hypercontracting alleles. (I) wupAhdp-2 is a hypercontracting allele of TnI that shows muscle tearing. (H) fliH genetically interacts with wupAhdp-2 at 25°, and all the flies show muscle hypercontraction. (K) wupAhdp-2 genetically interacts with TnT hypercontracting allele up101 in trans, leading to muscle rupturing. (L) fliH and up101 trans-heterozygotes do not show muscle tearing. Arrows I–K point to muscle tearing due to hypercontraction. (M) Flight data reveal that all the flies are flightless in fliH/wupAhdp-2and wupAhdp-2/up101 trans-heterozygotes, whereas fliH/up101 females show gradation in flight ability. Flies heterozygous for the fliH mutation serve as control for the flight test. (N–P) fliH mutants rescued by targeted overexpression of TnI transgenic line in IFM. (N) Polarized light image showing the rescue of DLM fibers with overexpression of a copy of TnI (w, UH3, fliH/Y; UAS-TnI-L9). (O) Myofiber showing complete rescue of the sarcomere organization as visualized by confocal imaging of w, UH3, fliH/Y; UAS-TnI-L9 flies. (P) Rescued fliH flies with TnI transgene show normal flight. (Q) Chromatogram showing mutations in fliH as analyzed by DNA sequencing. Mutations lie in upstream regulatory region of the wupA gene coding for TnI. Box indicates transcription factor Mef-2-binding site as predicted by MatInspector. Phallodin-TRITC for F-actin (red) and anti-Mlp60A antibody localizing on Z-discs (green). Flight data and muscle analysis: n = 25, and all the flies were grown at 25°.
The wupA gene function is impeded in the case of mutations such as in the alleles wupAhdp-2 (Beall and Fyrberg 1991) and wupAhdp-3 (Barbas et al. 1993; Nongthomba et al. 2004). wupAhdp-2 is a recessive mutation involving the single-amino-acid change A116V in constitutive exon 5 in the region that interacts with TnC. This mutation is recessive in nature, and homozygous flies show muscle hypercontraction (Figure 4I). However, wupAhdp-3 is a splice mutation in the intron preceding the alternative exon 6b1, which is IFM- and TDT-specific. wupAhdp-3 leads to a TnI-null condition in IFM and TDT. wupAhdp-3 heterozygotes are flightless and show hypercontraction muscle defects in 30% of the flies (Nongthomba et al. 2004). fliH fails to complement wupAhdp-3 and all the individuals show muscle tearing (data not shown). Similarly, fliH/wupAhdp-2 heterozygotes show muscle hypercontraction, suggesting that the interaction between the two mutations could be intragenic in nature (Figure 4J). wupAhdp-2 mutant is known to show muscle tearing in a trans-heterozygous condition with another hypercontracting mutation, up101, in the upheld gene (Figure 4K). The upheld gene codes for TnT, and up101 is a recessive single-point mutation (Fyrberg et al. 1990). Such interaction studies have revealed that mutation in one component of muscle can either enhance or relieve the effect of mutation at any other locus that takes part in normal muscle assembly and function. These studies have been very fruitful in deciphering gene functions and the pathways involved (Mogami and Hotta 1981; Homyk and Emerson 1988). up101 and wupAhdp-2 mutants show genetic interaction possibly due to the fact that they are mutations in the TnT and TnI proteins, respectively, which are part of the same complex that regulates muscle contraction. Since fliH fails to complement the wupAhdp-2 allele, and wupAhdp-2 in turn interacts with up101, the fliH interaction with up101 was studied. fliH/up101 trans-heretozygotes showed gradation of phenotypes from flightlessness to individuals with normal flight (Figure 4M). In addition, none of the flies showed any muscle defect (Figure 4L). This suggests that the nature of mutation involved in fliH is completely different from wupAhdp-2, if fliH and wupAhdp-2 are intragenic. Targeted overexpression of TnI using UAS TnI L9 [embryonic isoform of TnI (Sahota et al. 2009)] under the control of UH3-GAL4, which expresses in developing IFM (Singh et al. 2014), brings about rescue of the fliH muscle phenotype (Figure 4, N and O). Six DLM fascicles were seen (Figure 4N) with normal banding pattern sarcomeres (Figure 4O). Rescued flies also showed functional recovery of the muscles as indicated by their flight ability (Figure 4P). Overall, genetic mapping, interaction, and rescue experiments showed that fliH is a wupA allele. Keeping the FlyBase genetic nomenclature of the wupA gene, fliH is renamed as wupAfliH. However, for clarity we will still address the mutation with the original name fliH in this article.
The wupA gene consists of 13 exons, and alternative splicing gives rise to isoforms containing exon 6b1 in the IFMs. There are two major isoforms in IFMs, one with exon 3 and another lacking it (Nongthomba et al. 2004). Sequencing of the TnI-coding region as well as UTRs of messenger RNA (mRNA) did not reveal any mutation (data not shown). Quantitative expression of TnI is controlled by two regulatory sequences, the upstream regulatory element (URE) present upstream to the 5′ UTR, and the intronic regulatory element (IRE), lying within first intron of the wupA gene. Both these elements work synergistically to control the correct expression of TnI in most of the Drosophila muscles (Marin-Cruz et al. 2004). Both the regulatory regions were amplified and sequenced using genomic DNA isolated from fliH and Canton-S. Sequencing results revealed no mutation in IRE and few changes in the URE region. A schematic showing mutations as well as changes in the chromatogram is shown in Figure 4Q. Changes include a TTA insertion at one site (site A) and changes in another site (site B) that include insertion of a T at one location and deletion of a T from a nearby sequence. However, for clarity these mutations are addressed as site A and site B mutations. MatInspector (Genomatix Software, Munich, Germany), a program to find transcription-factor-binding sites, predicted Myocyte enhancer factor-2 (Mef-2)-binding sites present near site A and another falling at site B. The predicted Mef2 sites are not similar to a previously published consensus sequence (YTAWWWWTAR) Haberland et al. 2007). However, Mef2 has also been known to bind to different TA-rich sequences (Andrés et al. 1995).
Mutation abrogates Mef-2 binding to URE, leading to reduced TnI transcript and protein
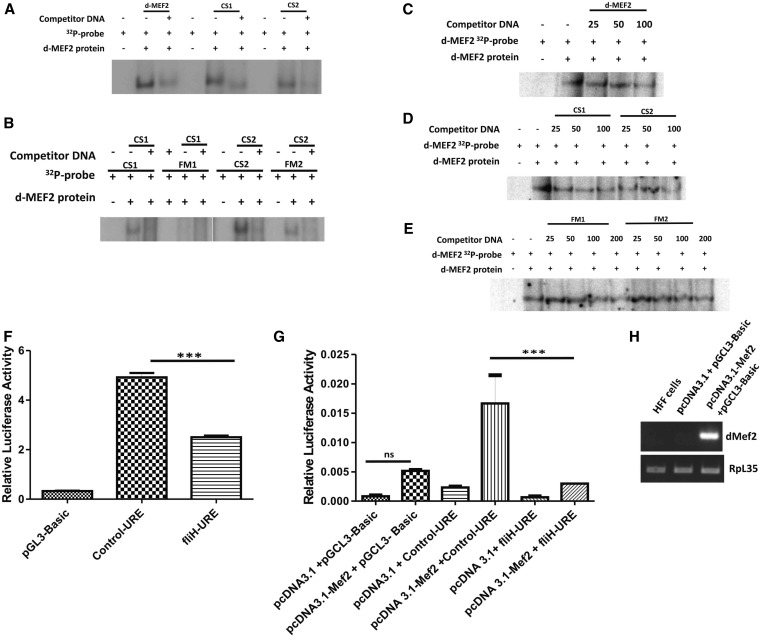
Gene regulation is an elaborate process involving multiple players at various levels. The Mef-2 transcription factor is known to bring about the transcription of the muscle genes and muscle differentiation (Lin et al. 1996; Black and Olson 1998; Kelly et al. 2002; Sandmann et al. 2006; Elgar et al. 2008; Tanaka et al. 2008). Mef-2 is also involved in spatio-temporal expression of downstream targets by altering its own activity levels (Elgar et al. 2008). Previous studies done on wupA gene regulation in Drosophila by Marin-Cruz et al. (2004) have shown the significance of Mef-2 (dMef-2) transcription factor binding in TnI IRE for quantitative wupA expression pattern in different muscle sets. They also suggested multiple Mef-2-binding sites in the URE region that could work synergistically with IRE to bring about optimal expression levels of TnI in the IFMs. To assay for the Mef-2-binding affinity, CS1 and CS2 oligos (for the DNA sequence of the wupA gene regulatory region containing site A and site B Mef-2-binding sites predicted near fliH mutation; Figure 4Q; see Materials and Methods) were used as control. Their corresponding mutant fragments (carrying the in vivo mutation present in fliH) are labeled as FM1 and FM2, respectively. dMef2 oligos that had already been shown to bind dMef2 protein were used as positive controls (Cripps et al. 2004). dMef2, CS1, and CS2 oligos bind to the dMef-2 lysate, and the binding is out-competed by their respective cold oligos (Figure 5A). Mutant FM1 oligos showed no binding whereas FM2 showed lesser complex formation as compared to CS2 when an equal amount of radioactivity was taken (Figure 5B). CS1 and CS2 oligos could out-compete when its own specific cold was used. An excess of wild-type CS2 cold was able to out-compete the FM2 complex formation. To verify the quantitative difference in dMef2 binding between the control and mutant sequences, we performed cross-competition EMSAs. Our results in Figure 5C indicate that the cold dMef2 probe gradually out-competes the labeled dMef2 at 50- and 100-fold excess, thus confirming the specificity of the protein binding to the control dMef2 sequence. The cold CS1 and CS2 probe was able to out-compete the labeled dMEf2 at 50- and 100-fold excess (Figure 5D), but the cold FM1 and FM2 can only out-compete at 200-fold excess but remain same at 100-fold (Figure 5E). A greater amount of mutant cold is required to out-compete the dMef2–protein complex. These results verify that CS1 and CS2 binds to dMef2 protein in a specific and stronger fashion than the mutant FM1 and FM2 sequence. The cross-competition experiments show that the binding is not an all-or-none phenomenon.
Figure 5.
Mef-2 binding is affected in fliH, leading to reduced TnI transcript and protein. (A) The dMef2, CS1, and CS2 oligos bind to the Mef2 protein lysate to form a complex, and the binding is out-competed by the respective cold oligos. (B) Mutation in fliH abrogates binding of dMef-2 to FM1 oligo whereas the level of binding is much reduced in FM2 as compared to CS2. The binding of FM2 is out-competed by CS2 cold. Cross-competition EMSAs. (C) Published dMef2 probe (Cripps et al. 2004) was used as a control for the competition assay. Cross-competition using cold dMef2 oligos (25-, 50-, and 100-fold excess) shows sequential abrogation of the complex. (D) Cross-competition with cold CS1 and CS2 (25-, 50-, and 100-fold excess) shows better competition than the (E) cold FM1 and FM2 (25-, 50-, 100-, and 200-fold excess) (compare the lanes with 100-fold excess). But at 200-fold excess even FM1 cold out-competes the complex but not FM2. These results were estimated quantitatively as the results do not show all or no competition. (F) The 979-bp upstream region of the TnI gene was used for promoter activity studies by subcloning the region into luciferase vector pGL3 basic. The assay was carried out using the C2C12 myoblast cell line. The fliH-URE shows a 50% reduction compared to the control (Control-URE) in luciferase activity and was significant at P < 0.0001. (G) HFF cells were cotransfected with pcDNA3.1 and pcDNA3.1–Mef2 constructs with mutant and control promoters independently. There is a significant increase in the luciferase activity in the Control-URE where the Mef2 protein was overexpressed compared to that of fliH-URE at P < 0.0001. (H) RT-PCR shows that dMEF2 transcripts are present only in the HFF cells where the MEf2 protein was overexpressed. HFF cells have vertebrate MEf2 protein present at very basal levels (Neelam et al. 2005)
To confirm the above results further, the 979-bp stretch of genomic DNA harboring the fliH mutation (fliH-URE) was checked for promoter activity in C2C12 myoblast cell culture by subcloning it in luciferase vector pGL3 Basic. C2C12 is a mouse myoblast cell line that expresses the Mef-2 ortholog of Drosophila dMef2 (Tomczak et al. 2003). fliH-URE showed a 50% reduction in luciferase activity compared to the control (Control-URE) (significant at P < 0.0001 (Figure 5F). To verify whether Drosophila Mef2 protein alone can activate the promoter, we used HFF cells, where it is known that the vertebrate MEF2 protein is very minimally expressed (Neelam et al. 2005), thus serving as a good model to study promoter activity in a vertebrate Mef2-null background. There was no significant difference in the luciferase activity when Control-URE and fliH-URE constructs were transfected in HFF cells (data not shown). When these same constructs were cotransfected in the Drosophila Mef2 overexpression background, a significant level of luciferase activity was seen in the Control-URE, suggesting that the activation of promoter is due to Drosophila Mef2 protein binding to the promoter region (Figure 5G). The confirmation of the overexpression of dMef2 was seen in the transcript level of the plasmid transfected cells as compared to the naive HFF cells (Figure 5H). Thus, electrophoretic mobility shift assay, supported by promoter activity assay, suggests that the fliH mutation abrogates the proper binding of dMef-2 transcription factor to TnI URE, which may cause decreased expression of TnI and also confirms that Drosophila Mef2 binding activates the promoter of TnI.
To assess the relative levels of TnI transcript accumulation in mutant vs. wild-type control, quantitative RT-PCR was performed using mRNA isolated from 2-day-old adult fly IFMs (Figure 6A). Transcripts were detected by primers that amplified exon 6b1 (IFM- and TDT-specific exon) and exon 4 (constitutive exon). Transcript level was found to be significantly reduced in fliH mutants raised at 25° as compared to Canton-S wild type. There were no differences in the TnI expression pattern between the constitutive exon and IFM-TDT-specific primers, supporting that the URE mutations could be the reason. The level of TnI transcript in the fliH mutant raised at 18° was higher, reflecting the normal muscle phenotype. Flies heterozygous for the wupAhdp-3 mutation were used as the internal control, and results showed that transcript accumulation was much reduced in wupAhdp-3/+ as compared to fliH at 25°. Consistent with the RNA levels, Western blot from the mutant also showed reduced accumulation of TnI protein in adult IFMs raised at 25° as compared to wild-type Canton-S flies (Figure 6B). Protein accumulation was checked in 70- to 80-hr pupae IFMs that had not undergone hypercontraction. Protein accumulation in the mutant pupae grown at 25° and 18° is comparable (Figure 6C), suggesting that accumulation of TnI is similar at both temperatures before the initiation of hypercontraction. However, the fliH genotype where there is no hypercontraction as a result of removal of thick filaments still showed a slight decrease in expression of TnI when raised at 25° (Figure 6D) as compared to 18° flies, suggesting that temperature has a bearing on the expression pattern of TnI as a result of the URE mutations.
Figure 6.
Reduction in TnI transcripts and protein levels. (A) Quantitative RT-PCR from 1- to 2-day-old adult IFM shows significant reduction in the level of TnI transcript in fliH background at 25° with slight elevation in transcript levels at 18°. Error bar denotes SD. Statistical analysis using one-way ANOVA reveals that the means of fold expression of the TnI transcript of different genotypes when analyzed in groups of two show significant difference in mRNA accumulation in fliH (P < 0.001). (B) TnI protein accumulation in mutant adult IFMs. Western blot analysis using Drosophila TnI antibody reveals that final accumulation of protein in 1- to 2-day-old adult IFMs is much reduced in fliH raised at 25° (P < 0.05). However, protein levels are comparable to wild type in fliH flies grown at 18°. wupAhdp-3/+ flies also show significantly reduced TnI accumulation (P < 0.05). Anti-tubulin antibody was used as control. Error bar denotes SEM. (C) TnI accumulation in 70- to 80-hr-old pupae. The protein accumulation is not significantly different in all the genotypes. Error bar denotes SEM. (D) TnI protein accumulation in fliH flies in Mhc7 background. Mhc mutations do not affect the thin filament protein accumulation (Nongthomba et al. 2004). Mhc7 was used to suppress the hypercontraction of 25°-grown fliH flies to obtain enough muscle materials for protein and RNA assays.
Coordinated down-regulation of structural gene transcripts working together in a complex
Mogami and Hotta (1981) showed for the first time that mutations in a single myofibrillar protein gene affect accumulation of other structural proteins. This has been specially studied elaborately in the case of the null alleles, Act88F and Mhc alleles, which lead to their corresponding protein null in the IFMs (Beall et al. 1989). Similar results have been reported in the cases of the TnI null allele wupAhdp-3 (Nongthomba et al. 2004) and the TnT major isoform TnT10a null in IFM (Nongthomba et al. 2007). However, hypercontracting alleles present in the coding region of TnI (wupAhdp-2) or TnT (up101) do not show such down-regulation (Nongthomba et al. 2003). Since fliH is a hypercontracting allele that does not fall in the above categories of mutations, transcript levels of other structural proteins were checked through semiquantitative RT-PCR.
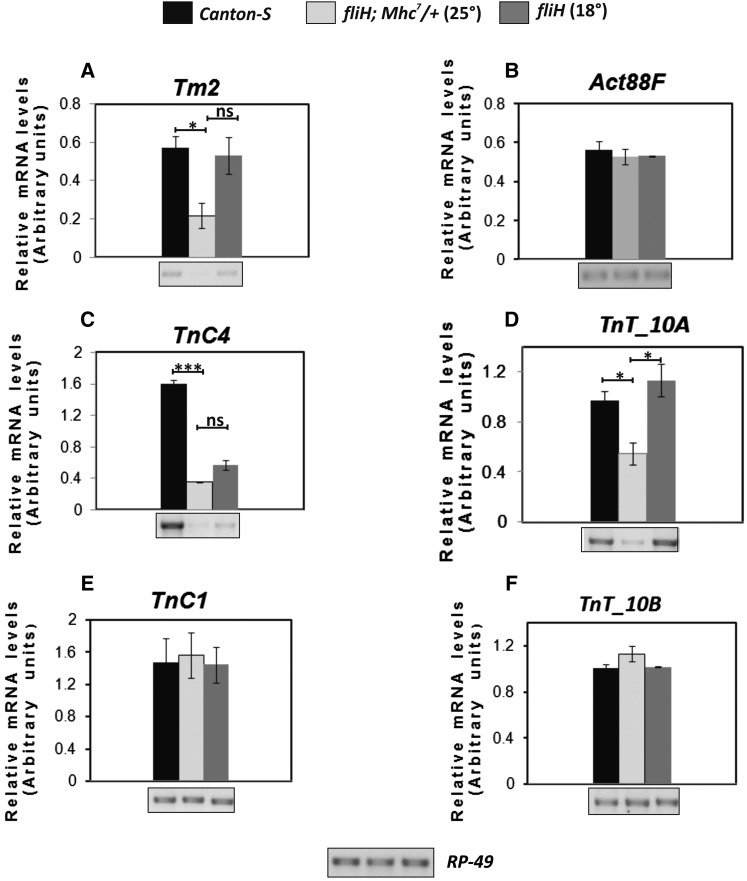
Coordinated down-regulation of RNA levels of the genes, which function together in muscle to generate regulated force, was observed in the case of fliH mutation (Figure 7). The Tm2 gene that encodes for tropomyosin showed down-regulation of transcript in fliH raised at 25° (Figure 7A). However, no significant difference was seen in Act88F RNA levels (Figure 7B). Semiquantitaive RT-PCR also showed reduced levels of troponin-C (TnC4) transcript in fliH (Figure 7C). TnC4 is the major isoform of TnC expressed in IFMs (Qiu et al. 2003). Like TnC4, TnT10a is the major TnT isoform specific for IFM and is expressed at much higher levels than other ubiquitous TnT10b isoforms (Nongthomba et al. 2007). TnT10a transcript was found to be significantly reduced in fliH (Figure 7D). No reduction in the transcript levels for minor isoforms of TnC (TnC1) and TnT (TnT10b) was observed (Figure 7, E and F).
Figure 7.
Coordinated downregulation of transcripts of other thin filament proteins in fliH. Semiquantitative RT-PCRs of (A) Tm2, (C) TnC4, and (D) TnT10a show reduced levels in fliH flies raised at 25°. Significant rescue in RNA levels is observed in 18°-raised fliH flies. (E) TnC1, (F) TnT10b, and (B) Act88F RNA levels remain unchanged at both 25° and 18°. Semiquantitative RT-PCR plot showing relative RNA levels in arbitrary units. Error bar represents mean from standard deviation for minimum of three RT-PCR runs.
Discussion
Maintaining the right stoichiometry of the structural proteins is important for the proper assembly of myofibrillar structure and function. This is evident from the myofibrillar defects shown by overexpression of TnT (Marco-Ferreres et al. 2005), Mhc (Cripps et al. 1994), and heterozygotes for most of the structural proteins (Prado et al. 1999). Beall et al. (1989) showed in an elegant experiment that concomitant reduction of both actin and myosin (hetrozygote condition for null alleles of both) restores normal myofibrillar structure and flight ability than the haploinsufficiency of either proteins alone (heterozygote condition of each alone) (Beall et al. 1989). Many factors are responsible for regulating the right levels of expression of the structural proteins in different muscles, including the transcription factors Mef-2 and Chorion factor-2 (CF-2) (Elgar et al. 2008; Garcia-Zaragoza et al. 2008; Tanaka et al. 2008). Mef-2 has been shown to dictate the differential expression of its downstream target genes through its spatio-temporal activities (Elgar et al. 2008), whereas CF-2 has been shown to be involved in regulation of the actin and myosin filament stoichiometry (Gajewski and Schulz 2010). For TnI and TnT, correct levels of expression in different muscles are brought about by two enhancer elements, namely the URE and the IRE that work synergistically (Marin-Cruz et al. 2004; Mas et al. 2004). Both the elements harbor multiple Mef-2-binding sites, suggesting that integration of Mef-2 in these regions could be important for bringing the correct levels of expression of associated genes. In fliH, two Mef-2-binding sites in the URE region are compromised, leading to 50% reduction in promoter activity and overall reduction in TnI transcripts and protein levels (Figure 6, A–D). Overexpression of larval isoform of TnI (L9) rescued not only the muscle phenotype but also the flight abilities, suggesting that correct stoichiometry of TnI is restored. It is likely that reduction in the TnI levels impedes proper troponin complex formation, thus leading to uncontrolled acto-myosin interaction, leading to IFM hypercontraction (Figure 8). Even though function of other muscles is also affected, a more pronounced phenotype is seen in the IFMs. Other hypercontracting alleles like wupAhdp-2 and up101, with mutations in constitutive exons, also show a more pronounced phenotype in the IFMs (Nongthomba et al. 2003), which may result from the fact that IFMs are the only muscles that require stretch activation and have to sustain the mechanical stress and strain produced through cuticular movement. Such heterogeneity in the myopathic phenotypes in different muscles in a single individual is also observed in other species, including humans (reviewed in Emery 2002). Reduction of TnI level is also seen in the case of flies heterozygous for the wupAhdp-3 mutation that is null for TnI in the IFMs. wupAhdp-3/+ flies also show muscle hypercontraction (Nongthomba et al. 2003), suggesting that the mechanistic aspects of muscle dysfunction and tearing are similar between wupAhdp-3/+ and fliH. Unlike wupAhdp-3 (Nongthomba et al. 2004), fliH does not show any defect in sarcomeric assembly until 75 hr APF, suggesting that TnI protein levels during pupal development is sufficient to drive proper assembly. This implies that a certain threshold of TnI is required for proper assembly; however, a proportion of TnI-containing troponin complexes may not be adequate for proper regulation of muscle contraction. Thus, mutants will not be able to sustain prolonged unregulated force leading to muscle damage once activated at ∼72–75 hr APF.
Figure 8.
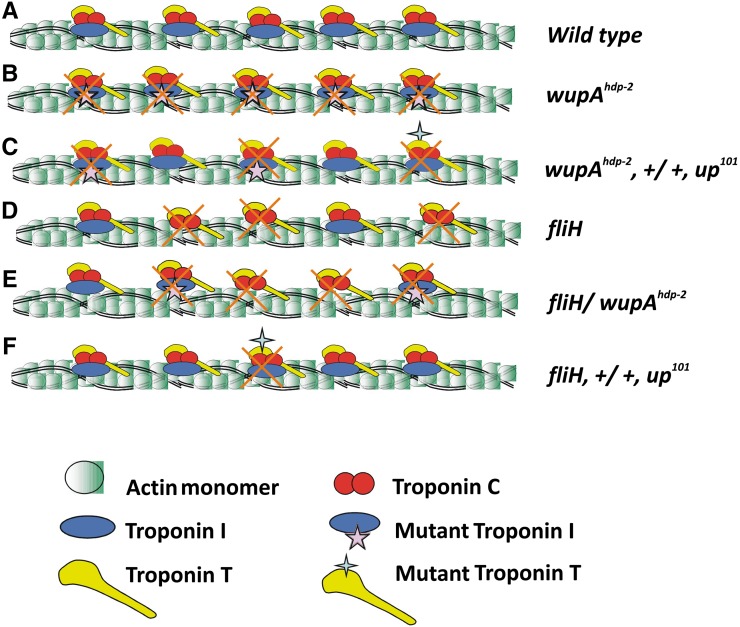
Schematics for genetic interactions between hypercontracting alleles. (A) Thin filament protein organization in the wild-type flies. (B) Troponin complex is functionally abnormal in wupAhdp-2 (troponin I mutant) flies, leading to aberrant regulation of muscle contraction and relaxation and hence resulting in tearing of muscles. (C) The recessive mutations up101 (troponin T mutant) and wupAhdp-2 interact in trans, rendering most of the troponin complex abnormal as they must harbor either mutation. (D) fliH shows downregulation of TnI, and hence troponin complex formation is impeded. (E) fliH shows cis interaction with wupAhdp-2. (F) Because fliH is a recessive regulatory mutation, it does not interact with the TnT mutation up101 to bring about significant IFM tearing.
Another interesting observation was the difference in the penetrance of the hypercontraction phenotype in fliH and wupAhdp-3/+. As opposed to 100% in fliH, only 30% of flies showed the hypercontraction phenotype in wupAhdp-3/+ (Nongthomba et al. 2003). Molecular data show that fliH has elevated TnI levels greater than those of wupAhdp-3/+, which will explain normal myofibril assembly during pupal development stages, whereas wupAhdp-3/+ has been shown to have defective assembly during early development (Nongthomba et al. 2004), which does not allow proper generation of force to produce hypercontraction. Based on heterogeneity in development, some of the flies that generate enough uncontrolled acto-myosin force may produce the hypercontraction phenotype in wupAhdp-3/+. Other factors like the difference in the rate of accumulation of the proteins cannot be ruled out. The rate of actin accumulation with respect to myosin is slower in Act88FKM88/+ flies; as a result, the flies are rendered flightless even though actin amounts are comparable to those of wild type. This can be rescued, to some extent, by balancing the stoichiometry with myosin (Mhc10/+; Myosin null) heterozygotes (J. Sparrow and U. Nongthomba, unpublished data.). It is likely that in wupAhdp-3/+ the rate of accumulation of TnI could be slower with respect to other thin filament proteins with which it forms a complex. One can speculate, based on the difference in TnI levels, that there could be a difference in the sarcomeric assembly and the troponin complex formed between the two genotypes. We propose that this difference leads to more compromised sarcomeric assembly in wupAhdp-3/+ flies, thus producing less acto-myosin interaction owing to sarcomeric defects and manifestation of muscle hypercontraction in fewer flies.
Hypercontracting alleles such as wupAhdp-2 and up101 harbor point mutations in their coding sequence that lead to defects in regulation of muscle contraction, owing to which IFMs degenerate once they start functioning (Nongthomba et al. 2003; Cammarato et al. 2004). Both these mutations are recessive in nature, but, when brought into a trans-heterozygous condition, show genetic interaction leading to muscle hypercontraction in all the flies. It is likely that more than half the troponin complexes formed in this genetic background will carry one or the other mutation (Figure 8). Once activated by Ca2+, these complexes will move away from steric blocking sites on actin, leading to uncontrolled acto-myosin interactions. Since the troponin complex plays a central role in Ca2+ regulation of muscle contraction, absence of it will lead to muscle hypercontraction. fliH and wupAhdp-3/+ show reduced accumulation of TnI, thus impeding formation of the functional troponin complex, which leads to unregulated acto-myosin interaction and hence IFM hypercontraction. Similarly, one can explain the intragenic interaction between fliH and wupAhdp-2 based on formation of the functional troponin complex. In this case, too, the troponin complex formed either will lack TnI or will carry a mutated version leading to hypercontraction. fliH and up101 trans-heterozygotes show reduced flight ability with no obvious muscle tearing or hypercontraction. In this genetic background, up101 contributes a normal copy of the TnI locus, and one copy of the regulatory mutations is enough to express sufficient protein (fliH/+ flies are normal); very few troponin complexes that carry mutant TnT will be nonfunctional (such as up101/+), so flies show partial complementation.
Coordinated down-regulation of the expression of thin filament genes in response to mutation in a single thin filament protein has been very well documented in the case of the null alleles of myosin (Mhc7), actin (Act88FKM88), TnI (wupAhdp-3), and TnT (up1). Such a phenomenon has not been reported for known hypercontracting alleles such as wupAhdp-2 and up101. Our result emphasizes that there is coordinated down-regulation of major isoforms of TnC and TnT (TnC4 and TnT10a, respectively) as compared to the minor isoforms TnC1 and TnT10b, which do not show any change in RNA levels in the IFMs. This could be explained based on the formation of the functional troponin complex and the interdependence of the expression level of each member of the complex by an unknown mechanism. Thus, in the absence of a single player of the complex, the message for the other major interacting partners is also down-regulated. Studies involving coordinated down-regulation of proteins that functions together in the same complex have also been reported in other model organisms such as zebrafish (Sehnert et al. 2002).
Genetic variations lead to changes at the cellular and molecular level affecting performance of the flight muscles. Apart from the intrinsic variables, an extrinsic factor such as temperature also plays an important role in flight ability. Certain species of moth cannot fly until they have prewarmed their flight muscles (Esch 1988). Flight performance of the Canton-S flies, used as control in the present study, showed reduced flight at 18° (Figure 3). They could fly better at an elevated temperature, suggesting a physiological difference at the two extreme temperatures. The fliH mutation also shows a temperature-dependent effect in its viability, walking, and jumping. However, a more profound phenotype is seen in the IFMs that are raised at 25°, which correlates with a reduced level of TnI transcript and protein as compared to those at 18° and wild type. Other hypercontracting alleles, such as flapwing, are known to show increased viability and reduced hypercontraction when flies are cultured in reduced temperature (Pronovost et al. 2013). Initiation of hypercontraction correlates with the movement of the thorax of the pupae within the pupal case, which is more pronounced in 25°-raised flies than in those raised at 18°. Moreover, one needs to keep the 18°-raised flies for at least 2–3 hr at 25° to see flight, suggesting a limited movement and lethargic nature of pupae and muscles at a lower temperature. It is likely that the reduced activity of the muscles and less mechanical stress will allow the muscles to assemble completely, so that the muscle will show suppression of the hypercontraction after eclosion. Our work has shown that binding of Mef-2, a transcripton factor, which brings about correct expression of the structural genes, is affected in the fliH mutant. However, defective binding of Mef-2 or any other transcription factors or changes in local structure of the DNA that brings about the temperature-sensitive effect need to be worked out. Little is known about the mechanisms that confer different temperature-dependent phenotypes. Research done on lower organisms like bacteria (Tamai et al. 1998) and yeast (Nouraini et al. 1996) and in plants (Gilmour et al. 1998; Zhu et al. 2007) has shown that mutations in the cis-regulatory region show temperature-dependent phenotypes. Work done on the W3133 operon in bacteria showed that an insertional mutation in the promoter region could confer a temperature-sensitive phenotype and affect transcription efficiency by stabilizing the DNA stem loop structure (Tamai et al. 1998). Such results have not been reported in higher organisms, which may be due to higher-order organization of the genome. However, temperature-sensitive mutations in protein-coding sequence have been isolated in eukaryotes wherein temperature difference might affect the folding, stability, and function of the proteins (Reese and Katzenellenbogen 1991; Mondal et al. 2007; Hoeberichts et al. 2008).
In this study, we have shown that mutation in the regulatory region of TnI can lead to IFM hypercontraction. Mutations in the coding region of myofibrillar protein causing myopathies in humans are very well documented. However, there is no record of a mutation or single nucleotide polymorphism in the noncoding region and regulatory region being associated with disease. This may be one reason for the large number of myopathic cases where the causative nature of mutation remains unknown. We propose from our study that regulatory mutations as well as mutations leading to stoichiometric changes (splice site mutants or nonsense mutations resulting in null protein) that may cause myopathic conditions can easily be identified by simple quantitation of transcripts and proteins by applying the whole-genome approach. In humans, many mutations in sarcomeric proteins that lead to myopathic conditions have been identified. Cellular fiber disarray seen in hypercontracted IFM is also observed in the case of human hypertrophic cardiomyopathies (Seidman and Seidman 2001) and dystrophic muscles (Amato et al. 1998), suggesting that there may be parallel genetic pathways for hypercontraction-induced cellular phenotypes. Conservation of expression of many remodeling proteins has been already shown for hypercontracting Mhc alleles (Montana and Littleton 2006). Mutations have also been uncovered in human TnI that lead to various cardiomyopathies and skeletal myopathies (Kimura et al. 1997; Murphy et al. 2004; Gomes et al. 2005). The molecular mechanism by which these mutations lead to pathogenesis of myopathies remains unclear. One needs to study the pathogenesis of these mutations in a model organism and follow the effects of other factors like environmental stress as well as different genetic backgrounds. Overall, our results shed new insights into the importance of the maintenance of structural protein stoichiometry during muscle assembly for proper function with implications for identification of mutations and disease phenotypes in other species, including humans.
Supplementary Material
Acknowledgments
We thank Sneha Raghuram and Meenakshi Sen at the Indian Institute for Science-Confocal Facility for their technical assistance; anonymous reviewers, John Sparrow (University of York), S. Mahadevan, and our lab members for their critical comments and suggestions; Richard Cripps (University of New Mexico) for the Mef-2 construct; Alberto Ferrus (Cajal Institute, Madrid) for flies and antibodies; Sathees C. Raghavan and M. Nishana (Indian Institute of Science) for help with EMSA experiments; Sunita Chopra for help in the cotransfection luciferase assays; and the Bloomington Drosophila Stock Center and the National Center for Biological Sciences-Stock Centre (Bangalore, India) for providing flies. We thank the Indian Institute of Science and the Department of Science and Technology and Department of Biotechnology, Government of India, for financial assistance.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175604/-/DC1.
Communicating editor: L. Cooley
Literature Cited
- Agianian B., Krzic U., Qiu F., Linke W. A., Leonard K., et al. , 2004. A troponin switch that regulates muscle contraction by stretch instead of calcium. EMBO J. 23: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A. A., Kagan-Hallet K., Jackson C. E., Lampkin S., Wolfe G. I., et al. , 1998. The wide spectrum of myofibrillar myopathy suggests a multifactorial etiology and pathogenesis. Neurology 51: 1646–1655. [DOI] [PubMed] [Google Scholar]
- An H., Mogami K., 1996. Isolation of 88F actin mutants of Drosophila melanogaster and possible alterations in the mutant actin structures. J. Mol. Biol. 260: 492–505. [DOI] [PubMed] [Google Scholar]
- Andrés , V., M. Cervera, and V. Mahdavi, 1995. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J. Biol. Chem. 270: 23246–23249. [DOI] [PubMed] [Google Scholar]
- Barbas J. A., Galceran J., Torroja L., Prado A., Ferrús A., 1993. Abnormal muscle development in the heldup3 mutant of Drosophila melanogaster is caused by a splicing defect affecting selected troponin I isoforms. Mol. Cell. Biol. 13: 1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthmaier P., Fyrberg E., 1995. Monitoring development and pathology of Drosophila indirect flight muscles using green fluorescent protein. Dev. Biol. 169: 770–774. [DOI] [PubMed] [Google Scholar]
- Beall C. J., Fyrberg E., 1991. Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J. Biol. Chem. 114: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. J., Sepanski M. A., Fyrberg E. A., 1989. Genetic dissection of Drosophila myofibril formation: effects of actin and myosin heavy chain null alleles. Genes Dev. 3: 131–140. [DOI] [PubMed] [Google Scholar]
- Black B. L., Olson E. N., 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14: 167–196. [DOI] [PubMed] [Google Scholar]
- Burkart C., Qiu F., Brendel S., Benes V., Haag P., et al. , 2007. Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J. Mol. Biol. 367: 953–969. [DOI] [PubMed] [Google Scholar]
- Cammarato A., Hatch V., Saide J., Craig R., Sparrow J. C., et al. , 2004. Drosophila muscle regulation characterized by electron microscopy and three-dimensional reconstruction of thin filament mutants. Biophys. J. 86: 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P. J., Brotman J. S., Sweeney S. T., Davis G., 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P-elements. Genetics 165: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps, R. M., 2006 The contributions of genetics to the study of insect flight muscle function, pp. 2–15 in Nature’s Versatile Engine: Insect Flight Muscle Inside and Out, edited by J. Vigoreaux. Springer/Landes Bioscience, New York. [Google Scholar]
- Cripps R. M., Becker K. D., Mardahl M., Kronert W. A., Hodges D., et al. , 1994. Transformation of Drosophila melanogaster with the wild-type myosin heavy-chain gene: rescue of mutant phenotypes and analysis of defects caused by overexpression. J. Cell Biol. 126: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps R. M., Suggs J. A., Bernstein S. I., 1999. Assembly of thick filaments and myofibrils occurs in the absence of the myosin head. EMBO J. 18: 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps R. M., Lovato T. L., Olson E. N., 2004. Positive autoregulation of the Myocyte enhancer factor-2 myogenic control gene during somatic muscle development in Drosophila. Dev. Biol. 267: 536–547. [DOI] [PubMed] [Google Scholar]
- Deak I. I., 1977. Mutations of Drosophila melanogaster that affect muscles. J. Embryol. Exp. Morphol. 40: 35–63. [PubMed] [Google Scholar]
- Deak I. I., Bellamy P. R., Bienz M., Dubuis Y., Fenner E., et al. , 1982. Mutations affecting the indirect flight muscles of Drosophila melanogaster. J. Embryol. Exp. Morphol. 69: 61–81. [PubMed] [Google Scholar]
- Drummond D. R., Hennessey E. S., Sparrow J. C., 1991. Characterization of missense mutations in the Act88F gene of Drosophila melanogaster. Mol. Gen. Genet. 226: 70–80. [DOI] [PubMed] [Google Scholar]
- Elgar S. J., Han J., Taylor M. V., 2008. Mef2 activity levels differentially affect gene expression during Drosophila muscle development. Proc. Natl. Acad. Sci. USA 105: 918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery A. E., 2002. The muscular dystrophies. Lancet 359: 687–695. [DOI] [PubMed] [Google Scholar]
- Esch H., 1988. The effect of temperature on flight muscle potentials in honeybees and cuculiinid winter moths. J. Exp. Biol. 135: 109–117. [Google Scholar]
- Fahmy O. G., Fahmy M., 1958. New mutants report. Drosoph. Inf. Serv. 32: 67–78. [Google Scholar]
- Fernandes J., Bate M., and K. VijayRaghavan, 1991. Development of the indirect flight muscle of Drosophila. Development 113: 67–77. [DOI] [PubMed] [Google Scholar]
- Ferrús A., Llamazares S., de la Pompa J. L., Tanouye M. A., Pongs O., 1990. Genetic analysis of the Shaker gene complex of Drosophila melanogaster. Genetics 125: 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg E. A., Fyrberg C. C., Beall C., Saville D. L., 1990. Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J. Mol. Biol. 216: 657–675. [DOI] [PubMed] [Google Scholar]
- Gajewski K., Schulz R. A., 2010. CF2 represses actin 88F gene expression and maintains filament balance during indirect flight muscle development in Drosophila. PLoS ONE 5: e10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski K. M., Wang J., Schulz R. A., 2006. Calcineurin function is required for myofilament formation and troponin I isoform transition in Drosophila indirect flight muscle. Dev. Biol. 289: 17–29. [DOI] [PubMed] [Google Scholar]
- García-Zaragoza E., Mas J. A., Vivar J., Arredondo J. J., Cervera M., 2008. CF2 activity and enhancer integration are required for proper muscle gene expression in Drosophila. Mech. Dev. 125: 617–630. [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold induced COR gene expression. Plant J. 16: 433–442. [DOI] [PubMed] [Google Scholar]
- Gomes A. V., Liang J., Potter J. D., 2005. Mutations in human cardiac troponin I that are associated with restrictive cardiomyopathy affect basal ATPase activity and the calcium sensitivity of force development. J. Biol. Chem. 280: 30909–30915. [DOI] [PubMed] [Google Scholar]
- Haberland M., Arnold M. A., McAnally J., Phan D., Kim Y., et al. , 2007. Regulation of HDAC9 gene expression by Mef2 establishes a negative feedback loop in the transcriptional circuitry of muscle differentiation. Mol. Cell Biol. 27: 518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts F. A., Vaeck E., Kiddle G., Coppens E., van de Cotte B., et al. , 2008. A temperature-sensitive mutation in the Arabidopsis thaliana phosphomannomutase gene disrupts protein glycosylation and triggers cell death. J. Biol. Chem. 283: 5708–5718. [DOI] [PubMed] [Google Scholar]
- Homyk T., Emerson Jr C. P., 1988. Functional interactions between unlinked muscle genes within haploinsufficient regions of the Drosophila genome. Genetics 119: 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homyk T., Sheppard D. E., 1977. Behavioral mutants of Drosophila melaonogaster I. Isolation and mapping of mutations which decrease flight ability. Genetics 87: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson, R. K., 2006 Comparative physiology of insect flight muscle, pp. 34–43 in Nature’s Versatile Engine: Insect Flight Muscle Inside and Out, edited by J. Vigoreaux. Springer/Landes Bioscience, New York. [Google Scholar]
- Josephson R. K., Malamud J. G., Stokes D. R., 2000. Asynchronous muscle: a primer. J. Exp. Biol. 203: 2713–2722. [DOI] [PubMed] [Google Scholar]
- Kelly K. K., Stryder M. M., Cripps R. M., 2002. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech. Dev. 110: 39–50. [DOI] [PubMed] [Google Scholar]
- Kimura A., Harada H., Park J., Nishi H., Satoh M., et al. , 1997. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat. Genet. 16: 379–382. [DOI] [PubMed] [Google Scholar]
- Kronert W. A., O’Donnell P. T., Fieck A., Lawn A., Vigoreaux J. O., et al. , 1995. Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J. Mol. Biol. 249: 111–125. [DOI] [PubMed] [Google Scholar]
- Kronert W. A., Acebes A., Ferrus A., Bernstein S. I., 1999. Specific myosin heavy chain mutations suppress troponin I defects in Drosophila muscles. J. Cell Biol. 144: 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, F. O., 2006 Muscle system design and integration, pp. 230–241 in Nature’s Versatile Engine: Insect Flight Muscle Inside and Out, edited by J. Vigoreaux. Springer/Landes Bioscience, New York. [Google Scholar]
- Lifschytz E., Green M. M., 1979. Genetic identification of dominant overproducing mutations: the Beadex gene. Mol. Gen. Genet. 171: 153–159. [DOI] [PubMed] [Google Scholar]
- Lin M. H., Nguyen H. T., Dybala C., Storti R. V., 1996. Myocyte-specific enhancer factor-2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc. Natl. Acad. Sci. USA 93: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd T. E., Taylor J. P., 2010. Flightless flies: Drosophila models of neuromuscular disease. Ann. N. Y. Acad. Sci. 1184: e1–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Ferreres R., Arredondo J. J., Fraille B., Cervera M., 2005. Overexpression of troponin T in Drosophila muscles causes a decrease in the levels of thin-filament proteins. Biochem. J. 386: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Cruz M., Jose-Rodrigo R., Ferrus A., 2004. Transcription of Drosophila troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol. Biol. Cell 15: 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas J. A., Garcia-Zaragoza E., Cervera M., 2004. Two functionally identical modular enhancers in Drosophila troponin T gene establish the correct protein levels in different muscle types. Mol. Biol. Cell 15: 1931–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami K., Hotta Y., 1981. Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscle. Mol. Gen. Genet. 183: 409–417. [DOI] [PubMed] [Google Scholar]
- Mondal K., K. VijayRaghavan, and R. Varadarajan, 2007. Design and utility of temperature-sensitive Gal4 mutants for conditional gene expression in Drosophila. Fly (Austin) 1: 282–286. [DOI] [PubMed] [Google Scholar]
- Montana E. S., Littleton J. T., 2006. Expression profiling of a hypercontraction-induced myopathy in Drosophila suggests a compensatory cytoskeletal remodeling response. J. Biol. Chem. 281: 8100–8109. [DOI] [PubMed] [Google Scholar]
- Moore S. A., Shilling C. J., Westra S., Wall C., Wicklund M. P., et al. , 2006. Limb-girdle muscular dystrophy in the United States. J. Neuropathol. Exp. Neurol. 65: 995–1003. [DOI] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W., 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. T., Mogensen J., Shaw A., Kubo T., Hughes S., et al. , 2004. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet 363: 371–372. [DOI] [PubMed] [Google Scholar]
- Naimi B., Harrison A., Cummins M., Nongthomba U., Clark S., et al. , 2001. A tropomyosin mutation suppresses troponin I myopathy in Drosophila. Mol. Biol. Cell 12: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Harinivas H. K., Pramod P. N., Ling Z., Marilyn S. S., et al. , 2005. ERK1/2 and MEK1/2 induced by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79: 10308–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Ramachandra N. B., 1999. A direct screen identifies new flight muscle mutants on the Drosophila second chromosome. Genetics 153: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Cummins M., Vigoreaux J., Sparrow J. C., 2003. Suppression of muscle hypercontraction by mutations in the myosin heavy chain gene of Drosophila melanogaster. Genetics 164: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongthomba U., Clark S., Cummins M. C., Ansari M., Stark M., et al. , 2004. Troponin I is required for myofibrillogenesis and sarcomere formation in Drosophila flight muscle. J. Cell Sci. 117: 1795–1805. [DOI] [PubMed] [Google Scholar]
- Nongthomba U., Ansari M., Thimmaiya D., Stark M., Sparrow J. C., 2007. Aberrant splicing of an alternative exon in the Drosophila troponin-T gene affects flight muscle development. Genetics 177: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouraini S, J. Hu, L. D. McBroom, and J. D. Friesen, 1996. Mutations in an Abflp binding site in the promoter of yeast rp026 shift the transcription start sites and reduce the level of RP026 mRNA. Yeast 12: 1339–1350. [DOI] [PubMed] [Google Scholar]
- Peckham M., Molloy J. E., Sparrow J. C., White D. C., 1990. Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J. Muscle Res. Cell Motil. 11: 203–215. [DOI] [PubMed] [Google Scholar]
- Prado A., Canal I., Ferrus A., 1999. The haplolethal region at the 16F gene cluster of Drosophila melanogaster: structure and function. Genetics 151: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronovost S. M., Beckerle M. C., Kadrmas J. L., 2013. Elevated expression of the integrin-associated protein PINCH suppresses the defects of Drosophila melanogaster muscle hypercontraction mutants. PLoS Genet. 9: e1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F., Lakey A., Agianian B., Hutchings A., Butcher G. W., et al. , 2003. Troponin C in different insect muscle types: identification of two isoforms in Lethocerus, Drosophila and Anopheles that are specific to asynchronous flight muscle in the adult insect. Biochem. J. 371: 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S., Williams I., Aslam H., Thomas D., Szoor B., et al. , 2000. Protein phosphatase 1 is required for the maintenance of muscle attachments. Curr. Biol. 10: 269–272. [DOI] [PubMed] [Google Scholar]
- Reedy M. C., Beall C., 1993. Ultrastructure of developing flight muscle in Drosophila. Dev. Biol. 160: 443–465. [DOI] [PubMed] [Google Scholar]
- Reedy M. C., Bullard B., Vigoreaux J. O., 2000. Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J. Cell Biol. 151: 1483–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J. C., Katzenellenbogen B. S., 1991. Mutagenesis of cysteines in the hormone binding domain of the human estrogen receptor. Alterations in binding and transcriptional activation by covalently and reversibly attaching ligands. J. Biol. Chem. 266: 10880–10887. [PubMed] [Google Scholar]
- Sahota V. K., Grau B. F., Mansilla A., Ferrus A., 2009. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J. Cell Sci. 122: 2623–2631. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sandmann T., Jensen L. J., Jakobsen J. S., Karzynski M. M., Eichenlaub M. P., 2006. A temporal map of transcription factor activity: Mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 7: 797–807. [DOI] [PubMed] [Google Scholar]
- Sehnert A. J., Huq A., Weinstein B. M., Walker C., Fishman M., 2002. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 31: 106–110. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Seidman C., 2001. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104: 557–567. [DOI] [PubMed] [Google Scholar]
- Shoresh M., Orgad S., Shmueli O., Werczberger R., Gelbaum D., et al. , 1998. Overexpression Beadex mutations and loss-of function heldup-a mutations in Drosophila affect the 3′ regulatory and coding components, respectively, of the Dlmo gene. Genetics 206: 283–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. H., Kumar P., Ramachandra N. B., Nongthomba U., 2014. Roles of the troponin isoforms during indirect flight muscle development in Drosophila. J. Genet. 93: 379–388. [DOI] [PubMed] [Google Scholar]
- Stewart B., Merriam J. R., 1973. Segmental aneuploidy of the X-chromosome. Drosoph. Inf. Serv. 50: 167–170. [Google Scholar]
- Tamai E., Shimamoto T., Tsuda M., Mizushima T., Tsuchiya T., 1998. Conversion of temperature-sensitive to -resistant gene expression due to mutations in the promoter region of the melibiose operon in Escherichia coli. J. Biol. Chem. 273: 16860–16864. [DOI] [PubMed] [Google Scholar]
- Tanaka K. K. K., Bryantsev A. L., Cripps R. M., 2008. Myocyte enhancer factor 2 and Chorion factor 2 collaborate in activation of the myogenic program in Drosophila. Mol. Cell. Biol. 28: 1616–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K. K., Marinescu V. D., Ramoni M. F., Sanoudou D., Montanaro F., et al. , 2003. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 18: 403–405. [DOI] [PubMed] [Google Scholar]
- Vigoreaux, J., 2006 Molecular basis of muscle structure, pp. 143–156 in Muscle Development in Drosophila, edited by Helen Sink. Springer/Landes Bioscience, New York. [Google Scholar]
- Zhu J., Dong C., Zhu J., 2007. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 10: 290–295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.