Abstract
Pattern formation during epithelial development requires the coordination of multiple signaling pathways. Here, we investigate the functions of an ovary-enriched miRNA, miR-318, in epithelial development during Drosophila oogenesis. mir-318 maternal loss-of-function mutants were female-sterile and laid eggs with abnormal morphology. Removal of mir-318 disrupted the dorsal–anterior follicle cell patterning, resulting in abnormal dorsal appendages. mir-318 mutant females also produced thin and fragile eggshells due to impaired chorion gene amplification. We provide evidence that the ecdysone signaling pathway activates expression of miR-318 and that miR-318 cooperates with Tramtrack69 to control the switch from endocycling to chorion gene amplification during differentiation of the follicular epithelium. The multiple functions of miR-318 in oogenesis illustrate the importance of miRNAs in maintaining cell fate and in promoting the developmental transition in the female follicular epithelium.
Keywords: miR-318, pattern formation, gene amplification
THE small noncoding RNAs known as microRNAs (miRNAs) act to silence target messenger RNAs (mRNAs) by promoting mRNA degradation and/or protein translation inhibition. The importance of miRNAs in animal development and human disease has been well established through genetic and genomic studies (Kloosterman and Plasterk 2006; Bushati and Cohen 2007; Flynt and Lai 2008; Ebert and Sharp 2012; Mendell and Olson 2012; van Rooij and Olson 2012; Sun and Lai 2013). However, understanding the cellular functions of individual miRNAs in different biological settings remains a major challenge as many miRNAs have been identified in different species.
miRNAs have been implicated in a variety of important roles during Drosophila oogenesis (Dai et al. 2012). Oogenesis takes place within the ovarioles, each of which contains an assembly line of developing egg chambers. Each egg chamber consists of 16 interconnected germline cells, including 15 nurse cells and one oocyte, surrounded by a monolayer of ∼1000 somatically derived follicle cells (Sprading 1993). A complex exchange of signals between the germline cells and the surrounding follicle cells is required for oocyte development and eggshell patterning (Dobens and Raftery 2000; Berg 2005).
Previous studies reported that mutations for components of miRNA biogenesis pathway, including Dicer-1, loquacious, pasha, drosha, and AGO1, resulted in defects in germline stem cell division and in follicle cell development (Hatfield et al. 2005; Jin and Xie 2007; Park et al. 2007; Azzam et al. 2012). Ninety-three miRNAs are expressed in the Drosophila ovary (Czech et al. 2008), but functions have been assigned to only a few of them. miR-184 controls germline stem cell differentiation and dorsoventral patterning by regulating Saxophone and K10 (Iovino et al. 2009). miR-7 and miR-278 target Dacapo (Dap) to regulate cell cycle progression in germline stem cells (Yu et al. 2009). miR-7 also regulates Tramtrack69 (Ttk69) to control a developmental switch in the follicle cells (Huang et al. 2013). miR-279 represses signal transducer and activator of transcription (STAT) in both the follicle cells and migratory border cells to control cell fate (Yoon et al. 2011). miR-989 regulates border cell migration through multiple target genes (Kugler et al. 2013). Thus, it appears that individual miRNAs act in a variety of ways to control different aspects of oogenesis.
Here, we report on the role of an ovary-enriched miRNA, miR-318. Drosophila miR-318 shares the same “seed” sequence with two other miRNAs, miR-3 and miR-309, composing the miR-3 seed family (Supporting Information, Figure S1). Although all three miRNAs in principle can target the same mRNAs, their spatial and temporal expression differs. miR-3 and miR-309 are part of a polycistronic miRNA complex expressed strongly in the early embryo, with roles in regulation of the maternal-zygotic transition (Bushati et al. 2008). miR-3 and miR-309 are expressed at very low levels, if at all, in the ovary (Ruby et al. 2007; Czech et al. 2008). In contrast, miR-318 is the seventh most abundant miRNA in the ovary, comprising >6% of total miRNA sequence reads (Czech et al. 2008). We present evidence that miR-318 acts in the somatic follicle cells to regulate eggshell patterning and biogenesis during Drosophila oogenesis.
Materials and Methods
Drosophila stocks and genetics
Fly strains used were the following: w1118, Df(3R)BSC479 (Bloomington Stock Center BL24983 removes mir-318), ttk1e11 (BL4164), Df(3R)ED6361 (BL24143), P{neoFRT}82B (BL2035), P{neoFRT}82Bttk1e11(W. M. Deng), P{GSV6}GS15934 (Drosophila Genetic Resource Center, DGRC206424), P{GawB}c323 (c323-GAL4, B. Calvi), mir-318 control sensor (lab stock), P{UAS-LUC-mir-318.T} (UAS-mir-318, BL41161), P{UAS-EcR.B2.W650A} (UAS-EcRDN, BL9449), P{TRiP.JF01368} (UAS-EgfrRNAi, BL25781), P{TRiP.JF01371} (UAS-dppRNAi, BL25782), P{UAS-Egfr.DN.B} (UAS-EgfrDN, J. C. Pastor-Pareja), HsFLP; Act5C > y+>Gal4 UAS-GFP.S56T/(CyO) (J. C. Pastor-Pareja), HsFlp; Act5C > y+>GAL4 UAS-myr.RFP/CyO (J. C. Pastor-Pareja) and hsFlp; P{neoFRT}82B arm-LacZ/TM3 Ser (P. Rørth).
The mir-318 genomic rescue construct was created by PCR amplification of genomic fragments containing the mir-318 region. The DNA was amplified in two fragments so that the mir-994 sequences could be left out. The fragments were then cloned into the site-specific integration vector pAttB. Primers for fragment 1 were 5′-CGTCTAGAAAAAATCTATGTTGGTTCGATAC-3′ with 5′-CGGCGGCCGCTAAATTCAGGACGCGATCGAAG-3′ and for fragment 2 (with mir-318) were 5′-CGGCGGCCGCAAACAGCCTAGATCCGAATGCG-3′ and 5′-CGGCGGCCGCTTGGGTGTACTTTGTTTTTCTG-3′.
To generate the mir-318 sensor construct, oligonucleotides containing two copies of the sequence complementary to mir-318 were annealed and cloned downstream of the enhanced green fluorescence protein (EGFP)-coding region of Tub-EGFP in pCaSpeR4 (Brennecke et al. 2003).
Mutant generation
A modified targeting vector was used to make a GFP knock-in allele by homologous recombination. An EGFP fragment cut from pEGFP-N1 was subcloned into pW25 to generate the targeting vector pW25-EGFP. Approximately 4-kb fragments of genomic DNA flanking mir-318 were amplified and cloned into pW25-EGFP using the following primers: 5′-GCGGCCGCGAGAACAGATTCCAATTGACAT-3′ and 5′-GCGGCCGCCACGCAAGGCACTCGGATACTC-3′ for upstream flanking sequence and 5′-GGCGCGCCGGAAACCTTAAATCATACCAAT-3′ and 5′-GGCGCGCCGTCAGGCAATGTCAAGTAGAAG-3′ for downstream flanking sequence. Targeting was performed as described (Weng et al. 2009).
The mir-318Δ2 mutant was made by mobilization of P{GSV6}GS15934, using standard procedures. The deletion removed a 1.8-kb genomic DNA (from Ch3R:10407286 to Ch3R:10409088), determined by sequencing the DNA fragment spanning the breakpoints.
Clonal analysis
We used FLP/FRT recombination technique to generate loss-of-function mutant clones in the follicle cells. mir-318Δ1 was first recombined onto a P{neoFRT}82B chromosome. To induce mir-318 mutant clones, adult female flies with genotype HsFLP/+;P{neoFRT}82Bmir-318Δ1/P{neoFRT}82B arm-lacZ were heat-shocked at 37° for 1 hr and then incubated at 25° for 3–4 days before ovary dissection. As mir-318Δ1 is a GFP knock-in allele, null mutant clones were marked by the presence of two copies of GFP, and wild-type twin clones were labeled by the loss of a GFP marker. We used the flip-out system to generate overepression clones in the follicle cells. To induce ectopic expression of miR-318 or EcRDN, adult female flies with genotype HsFLP/+; Act5C > y+>Gal4 UAS-GFP.S56T/+; UAS-mir-318/+ or HsFLP/+; Act5C > y+>Gal4 UAS-myr.RFP /UAS-EcRDN were heat-shocked at 37° for 1 hr and then incubated at 25° for 3–4 days before ovary dissection. Clones were marked by the presence of GFP or red fluorescent protein (RFP).
Immunocytochemistry, 5-ethynyl-2′-deoxyuridine labeling, and microscopy
For immunostaining, ovaries of 3- to 5-day-old adult female flies were dissected in ice-cold PBS and fixed for 15 min in PBS containing 4% paraformaldehyde. Fixed ovaries were washed three times with PBS and 0.1% Triton X100 (PBT) and blocked in PBT with 5% normal goat serum (NGS) for 1 hr at room temperature. Samples were incubated with primary antibodies overnight at 4°, washed three times, and incubated with secondary antibodies for 2 hr and with DAPI for 20 min at room temperature. Samples were washed three times and mounted in 1:1 PBS–glycerol or Vectashield (Vector Labs). Primary antibodies used were the following: chicken anti-GFP (1:2000, Abcam), rabbit anti-RFP (1:500, Abcam,ab62341), mouse anti-β-galactosidase (1:2000, Promega), mouse anti-Broad-core [25E9.D7; 1:100, Devlopmental Studies Hybridoma Bank (DSHB) Iowa City, IA], mouse anti-Cut (2B10;1:15, DSHB), mouse anti-hnt (1G9; 1:15, DSHB), mouse anti-FasIII (7G10; 1:100, DSHB). Anti-Ttk69 was raised in rabbit against a GST-Ttk69 fusion protein containing the C-terminal amino acids 475–641 of Ttk69. Secondary antibodies were Alexa Fluor 488-, 555-, or 633-conjugated (1:250 or 1:500, Molecular Probes). DAPI (1 μg/ml, Sigma, St. Louis) was used to stain for nuclei. 5-Ethynyl-2′-deoxyuridine (EdU) staining was performed using the Click-iT EdU Alexa Fluor 555 Imaging Kit (C10338, Life Technologies). Dissected ovaries were incubated for 1 hr at 25° in Grace’s medium (Life Technologies) containing 10 μm EdU and then fixed and immunostained as described above. Samples were washed for 20 min in PBT and then incubated with a EdU reaction cocktail for 30 min. Ovaries were then washed for 2 hr before adding the mounting medium. Optical sections were acquired using a Zeiss LSM510 confocal microscope at 1-μm intervals and converted into a single image using maximum intensity projection. For scanning electron microscope analysis, eggs were mounted directly and imaged with a JEOL JSM-6360LV scanning electron microscope.
Cell culture, transfection, and luciferase assays
Transfections were performed with X-tremeGENE reagent according to the manufacturer’s protocol (Roche). S2 cells were cultured in Schneider’s medium supplemented with 10% FBS. 20-Hydroxyecdysone (20E) (Enzo Life Sciences, ALX-370-012-M005) was added to the medium at a final concentration of 10 × 10−6 M at 6 hr after transfection. The luciferase reporter vector containing a wild-type mir-318 regulatory sequence was prepared as follows. A 852-bp sequence upstream of the mir-994/318 stem loops was amplified and cloned into pGL3 using the primers CGGGTACCAAAAATCTATGTTGGTTCGATAC and CGCTCGAGTAAATTCAGGACGCGATCGAAG. A mutated version that lacked the putative EcRE was cloned into the same vector by a two-step PCR method. Two additional primers used were TGACCCACAAATACGCCCTGAATAGTCACGTTGTGGGATTC and CAGGGCGTATTTGTGGGTCA. Transfections were done in 24-well plates with 500 ng of firefly luciferase reporter plasmid and 50 ng of Renilla luciferase plasmid as a control. Transfections were done with three technical replicates in four independent experiments. Firefly and Renilla luciferase were measured using the dual luciferase assays kit protocol 48 hr after transfection.
Quantitative real-time PCR for mature miRNAs
Total RNA was isolated from 10 pairs of ovaries with TRIzol reagent (Invitrogen). Reverse transcription reactions were done on 10 ng of total RNA with miRNA-specific stem-loop reverse transcription primers. Real-time PCR was performed using the TaqMan miRNA assay kit protocol on an ABI7500 fast real-time PCR machine. miRNA levels were normalized to miR-184.
Results
mir-318 mutant females produce defective eggs
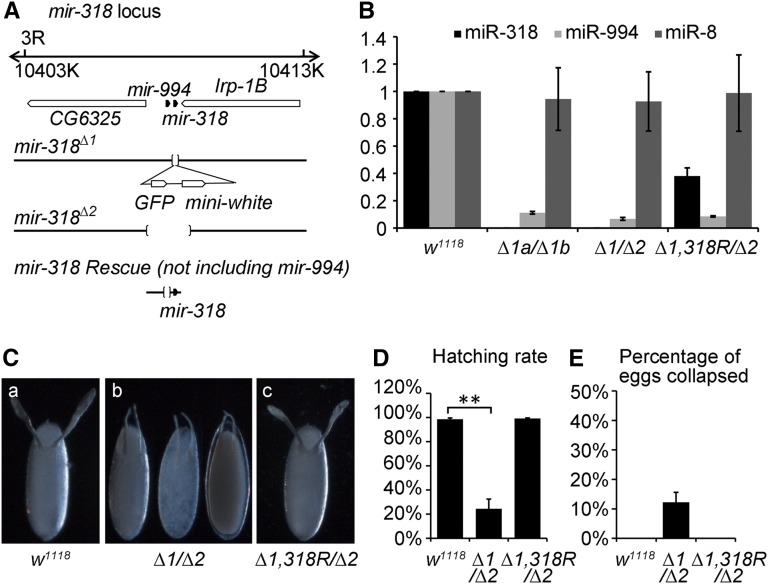
mir-318 is located in close proximity to mir-994 in the interval between two protein-coding genes (Figure 1A). To study mir-318 function, we made use of a targeted knock-out allele in which a 126-bp genomic fragment including the mir-318 hairpin was replaced with GFP and mini-white reporter genes (Figure 1A) (Chen et al. 2014). A second genetically independent allele, mir-318Δ2, was produced by imprecise excision of the P-element insertion line P{GSV6}GS15934, resulting in a deletion of 1.8 kb. mir-318Δ2 removed mir-318 and mir-994, as well as part of the coding region of Irp-1B (Figure 1A). miRNA quantitative PCR confirmed that the mature miR-318 miRNA was absent in RNA samples prepared from mir-318Δ1a/mir-318Δ1b (Δ1a and Δ1b are equivalent alleles from two independent homologous recombination events) and mir-318Δ1/mir-318Δ2 mutants (Figure 1B). In addition, we noted that the expression of mir-994 was reduced to 11% in the mir-318Δ1a/mir-318Δ1b mutant and to 7% in the mir-318Δ1/mir-318Δ2 mutant, compared to the control, suggesting that mir-318Δ1 is a hypomorphic allele for mir-994 (Figure 1B). The trans-heterozygous combination mir-318Δ1/mir-318Δ2 was used for phenotypic assays. We also made use of a mir-318 transgene, consisting of the interval between the flanking protein genes, but lacking mir-994 sequences (Figure 1A). The transgene was recombined onto the mir-318Δ1 chromosome and restored miR-318 expression to ∼40% of wild-type control levels, without restoring mir-994 expression (Δ1, 318R/Δ2, Figure 1B).
Figure 1.
Generation and characterization of mir-318 mutant. (A) Organization of the mir-318 locus. mir-318 is located on chromosome 3R at 86B4 on the cytogenetic map. Solid pentagons indicate the positions of mir-318 and mir-994. Open pentagons indicate the positions of adjacent predicted genes. Deletions of mir-318Δ1 and mir-318Δ2 mutant alleles are indicated by the bracketed area. mir-318Δ1 was generated by ends-out gene targeting. A total of 126 bp, including the mir-318 stem loop, was replaced by the GFP and miniwhite genes. The integrated DNA is indicated below. mir-318Δ2 was produced by imprecise excision of P{GSV6}GS15934, and the deletion spans 1.8 kb. The mir-318 genomic rescue transgene is shown at the bottom. The bracketed area indicates the removed DNA, including mir-994. (B) Mature miRNA for miR-318 is absent in mir-318Δ1 and mir-318Δ2 mutants. Quantitative RT-PCR analysis of mature miR-318 and miR-994 miRNA levels in RNAs extracted from ovaries of wild-type control and two combinations of mir-318 mutant alleles and the mir-318 rescue transgenic flies. Δ1 denotes the mir-318Δ1 mutant allele. Δ1a and Δ1b are from two independent homologous recombination events. Δ2 denotes the mir-318Δ2 mutant allele. 318R denotes the mir-318 genomic rescue transgene. miR-8 served as a control miRNA that is expressed in ovaries. miRNA expression levels were normalized to miR-184. Error bars represent standard deviation from three independent experiments. (C) mir-318 maternal loss-of-function mutants produce morphologically abnormal eggs. Shown are phase-contrast images of dorsal views of eggs from wild-type control (a), mir-318 mutant (b), and mir-318 rescue (c) female flies. All the genotypes are indicated. (D) The hatching rate of eggs laid by mir-318 mutant females is reduced. Shown is the percentage of hatched eggs from female flies of indicated genotypes. Error bars represent standard deviation from three independent experiments. n > 200 for each experiment. P < 0.05. (E) Eggs laid by mir-318 mutant females are fragile. Shown is the percentage of collapsed eggs from female flies of indicated genotypes. Error bars represent standard deviation from three independent experiments. n > 200 for each experiment.
mir-318 mutant flies were fully viable and did not show obvious morphological defects. However, females lacking mir-318 laid eggs with a range of defects in eggshell formation and patterning (Figure 1C). More than 90% of eggs from mir-318 mutant mothers also showed defects in dorsal pattern: the dorsal appendages were shifted toward posterior and were closer together and shorter than in wild-type embryos (Figure 1C). The operculum was also slightly enlarged. These defects were fully rescued by introducing the mir-318 rescue transgene in the mutant background (Δ1, 318R/Δ2, Figure 1C).
In addition, 76% of eggs from mir-318 mutant mothers failed to hatch into first instar larvae (Figure 1D). Most of the eggs appeared not to be fertilized, and the micropyle was reduced or absent (visualized by scanning electron microscopy, Figure S2, A and B). In addition, eggs had a transparent and thin eggshell and were fragile (Figure 1C). Twelve percent of eggs had collapsed by 48 hr after laying (Figure 1E). All these defects were fully rescued by introducing the mir-318 rescue transgene into the mutant background. Suppression of these phenotypes by expression of the rescue transgene suggests that the defects are attributable to the loss of mir-318. mir-994 appears to be dispensable in this context. Thus miR-318 is required for egg maturation and for eggshell patterning.
miR-318 is expressed in the follicle cells
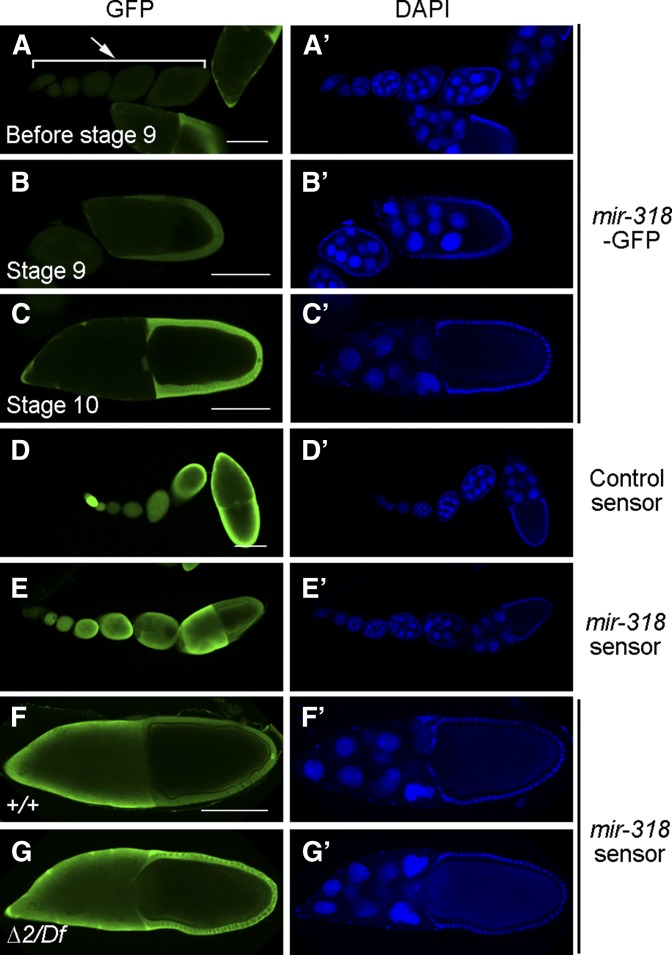
Consistent with previous deep sequencing data, quantitative miRNA PCR showed that miR-318 was undetectable in adult males, but was expressed in adult females and enriched in ovaries (Figure S3, A and B). To visualize miR-318 expression in vivo, we made use of the mir-318Δ1 allele, in which GFP was integrated into the mir-318 locus in place of the miRNA hairpin (Figure 1A). This placed GFP expression under control of the endogenous mir-318 regulatory elements. Ovaries were dissected from mir-318Δ1/+ heterozygous adult females and stained with antibody to GFP (Figure 2, A–C′). GFP was not detected in early stage egg chambers (Figure 2, A and A′). Beginning at stage 9, GFP was observed in most follicle cells surrounding the oocyte (Figure 2, B and B′). From stage 10 onward, GFP persisted in the follicle cells, including the border cells, the function of which is required to make the micropyle (Figure 2, C and C′; data not shown for later stages). We noted that miR-318 expression was undetectable in the polar cells within the border cell cluster and the posterior polar cells (labeled with a polar cell-specific marker FasIII, Figure S4, A–C′′′).
Figure 2.
Expression of miR-318 during oogenesis. (A–C′′) miR-318 is expressed in the follicle cells in stage-9 (B and B′) and stage-10 (C and C′) egg chambers, but not in early stage egg chambers (A and A′). Shown are confocal images of egg chambers from mir-318Δ1/+ heterozygous females, stained with anti-GFP (green) and DAPI (blue). As GFP is integrated into the mir-318 locus in the mir-318Δ1 allele, the pattern of GFP expression reflects miR-318 expression. Arrow in A indicates egg chambers at early stages. (D–E′) Mature miR-318 is expressed in the follicle cells in a stage-10 egg chamber. Shown are confocal images of egg chambers from wild-type females expressing control sensor or mir-318 sensor transgenes, stained with anti-GFP (green) and DAPI (blue). Note that mir-318 sensor GFP expression is repressed in the follicle cells in a stage-10 egg chamber. (F–G′) Repression of the mir-318 sensor GFP in the follicle cells is due to the presence of miR-318. Shown are confocal images of stage-10 egg chambers from wild-type and mir-318Δ2/Df mutant females expressing the mir-318 sensor transgene, stained with anti-GFP (green) and DAPI (blue). Df denotes the deficiency chromosome Df(3R)BSC479. Note the difference of GFP expression level in the follicle cells. Bars, 100 um.
As an independent means to visualize miR-318 activity, we used a sensor transgene that expresses GFP under the control of a ubiquitously expressed tubulin promoter and has two perfect mir-318 target sites in its 3′ UTR. miRNA activity represses GFP expression from this type of sensor transgene. GFP expression from a control sensor transgene (lacking the miRNA target sites) was uniform in egg chambers of various stages (Figure 2, D and D′). However, GFP expression was suppressed in the follicle cells in stage-10 egg chambers of flies carrying the mir-318 sensor (Figure 2, E–F′). The reduced GFP expression in the follicle cells was not observed when the mir-318 sensor was introduced into the mir-318 mutant (Figure 1, G and G′), confirming that the difference in expression in the follicle cells vs. the germline was due to the activity of the miRNA in the follicle cells. Expression of mir-318 in the follicle cells beginning in stage 9 is consistent with the phenotypic evidence for a role in eggshell formation and patterning.
Eggshell patterning
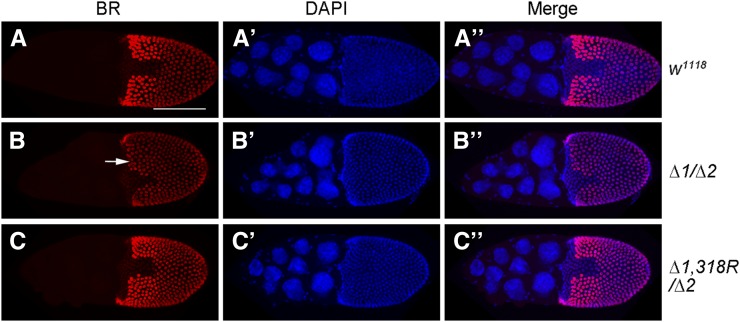
Patterning the eggshell along the dorsal–ventral and anterior–posterior axis requires the EGFR- and Dpp-signaling pathways (Dobens and Raftery 2000; Berg 2005). Broad (Br), a zinc-finger transcription factor, functions downstream of these two signaling pathways to specify the dorsal appendages (Deng and Bownes 1997). Beginning at stage 6, Broad is expressed in all oocyte-associated follicle cells and subsequently repressed in the anterior midline region in early stage-10B egg chambers. Later, Broad expression increases in the two groups of dorsal–anterior follicle cells that play an essential function in the specification of the dorsal appendages (Deng and Bownes 1997). In mir-318 mutant ovaries, Broad expression was not repatterned to clear the dorsal–anterior region in 8 of the 15 stage-10B egg chambers examined (Figure 3, A–B′′). Fewer cells showed reduced Broad expression in the anterior midline region, and the domain was irregular in shape. In addition, only very few of the laterally positioned follicle cells adjacent to this domain displayed the normal increase in Broad expression in the dorsal appendage field (14/15 mutant samples, arrow in Figure 3B). Introducing the mir-318 transgene rescued these defects (Figure 3, C–C′′). Although miR-318 activity is required for the normal pattern of changes in Br expression that depend on EGFR and DPP activity, miR-318 expression itself does not appear to be regulated by either of these pathways (Figure S5, A–D′′). This suggests that miR-318 activity may be required for the correct interpretation of these patterning cues or for the ability of cells to respond normally to them.
Figure 3.
Loss of mir-318 causes defects in follicle cell patterning. (A–C′′) The expression pattern of BR is abnormal in stage-10B egg chambers from mir-318 mutants. Shown are maximum intensity projections of Z-stack confocal images of stage-10B egg chambers from female flies of indicated genotype. Samples were stained with anti-BR (red) and DAPI (blue) to visualize the nuclei. Arrow indicates the dorsal–anterior region. (Right) Merged images. Bar, 100 um.
Chorion gene amplification
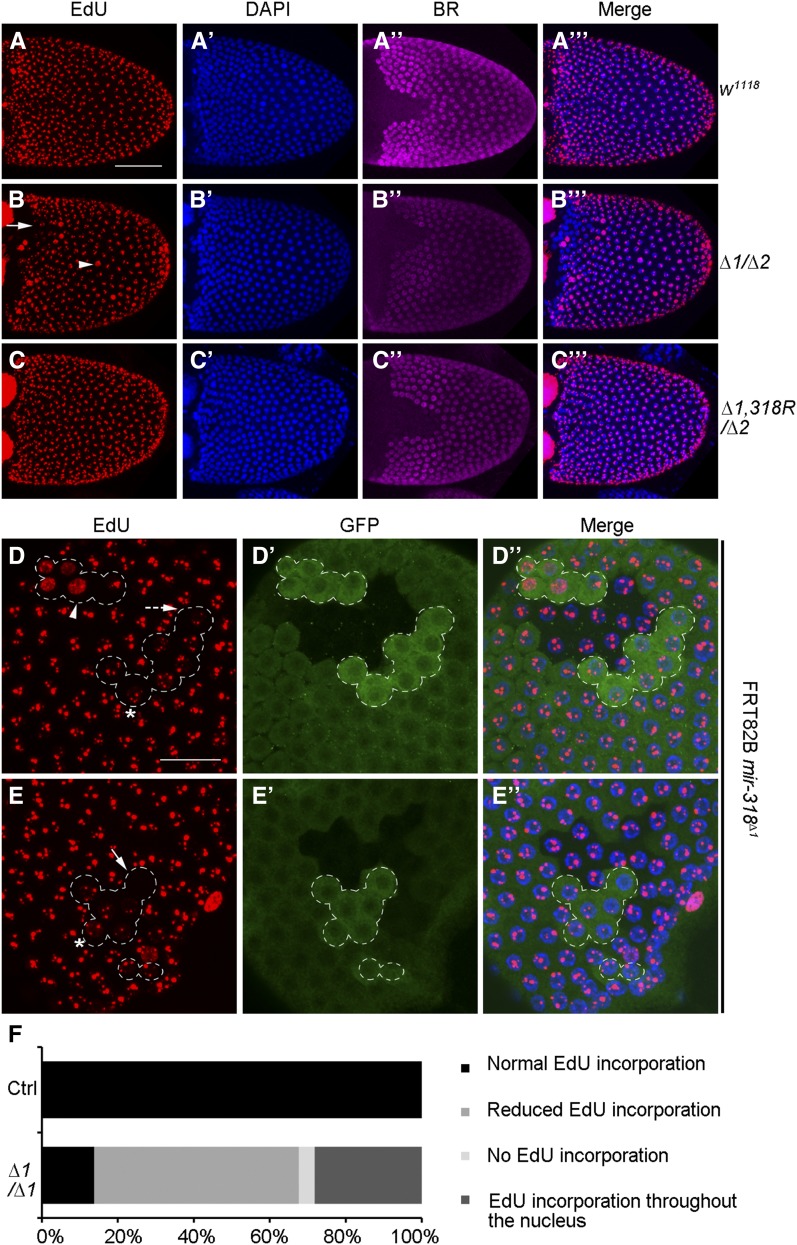
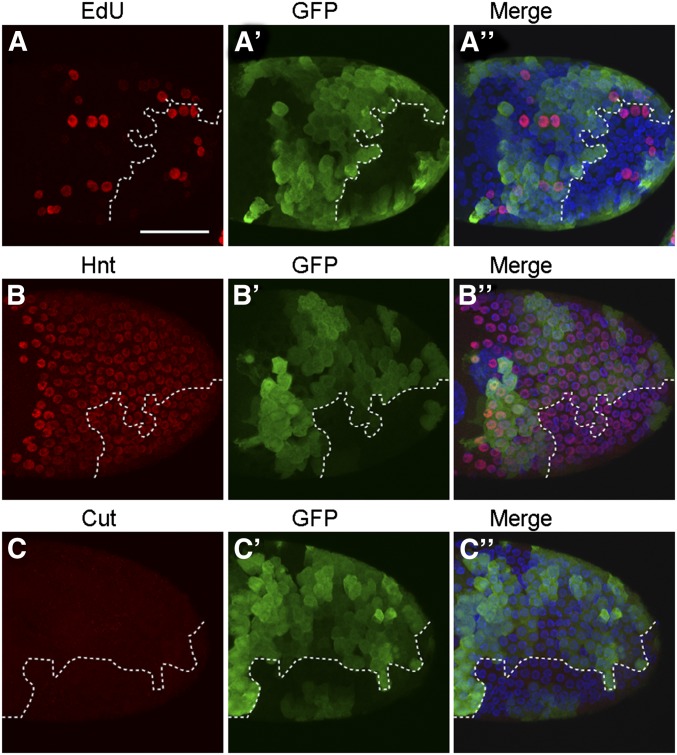
During late oogenesis, follicle cells produce the chorion proteins that make the eggshell. Molecular and genetic studies have shown that developmentally regulated amplification of the chorion genes is required to support eggshell biogenesis (Calvi et al. 1998; Calvi and Spradling 1999; Tower 2004). The fragile eggshell phenotype led us to explore the possibility that miR-318 might be involved in chorion gene amplification. To test this, we used incorporation of a thymidine analog, EdU, to monitor DNA replication. In wild-type controls, the sites of chorion gene amplification are seen as four to six subnuclear foci of EdU incorporation in each of the follicle cell nuclei (Figure 4, A–A′′′). Higher magnification views are shown in Figure S6, A–A′′. mir-318 mutant egg chambers showed a range of abnormalities in EdU incorporation (Figure 4, B–B′′′ and Figure S6, B–B′′). In each egg chamber, a few cells showed EdU incorporation uniformly throughout the nucleus (e.g., arrowhead in Figure 4B and Figure S6B), suggesting that all of the DNA was replicating. Cells in the dorsal–anterior region (where the Broad expression domain was affected) showed little or no incorporation of EdU (arrow in Figure 4B; arrow and dotted arrow in Figure S6B). This suggested a reduction in chorion gene amplification in this region. In addition, a generally reduced EdU incorporation was also observed in 15/20 mutant samples examined. These defects were rescued by restoring miRNA expression with the rescue transgene (Figure 4, C–C′′′ and Figure S6, C–C′′).
Figure 4.
Removal of mir-318 impairs chorion gene amplification. (A–C′′′) The EdU incorporation pattern is abnormal in intact stage-10B egg chambers from mir-318 mutants. Shown are maximum intensity projections of confocal Z-stacks of stage-10B egg chambers from females of indicated genotypes. Samples were labeled with EdU (red), anti-BR (purple) and DAPI (blue) to visualize the nuclei. Gene amplification was assayed by incorporation of EdU, which labels sites of active DNA replication as subnuclear foci. In wild-type stage-10B egg chambers, four to six EdU foci with different intensity and size were observed in each follicle cell nucleus. Synchronous EdU incorporation was seen. In contrast to this, mir-318 mutant egg chambers showed apparent defects in EdU incorporation, e.g., EdU incorporation throughout the nucleus (arrowhead in B) and absent EdU incorporation (arrow in B). (Right) Merged images. See Figure S6 for higher magnification. Bar, 50 um. (D–E′′) The EdU incorporation pattern is abnormal in mir-318 mutant clones of stage-10B egg chambers. Shown are maximum intensity projections of Z-stacks of confocal images of stage-10B egg chambers with mir-318 mutant clones in the follicle cells. mir-318 mutant clones were marked by two copies of GFP, and the lack of GFP indicated wild-type clones. Dashed line marks the mutant clone areas. Samples were labeled with EdU (red), anti-GFP (green) and DAPI (blue). Four mutant clones from two egg chambers are shown. Different classes of DNA replication patterns were seen according to the intensity and size of EdU foci: EdU incorporation throughout the nucleus (arrowhead in D), reduced EdU incorporation (dashed arrow in D) and absent EdU incorporation (arrow in E). Star indicates a weaker diffuse Edu incorporation in the rest of nucleus. Bar, 10 μm. (F) Quantitative analysis of EdU incorporation patterns. The level of EdU fluorescence intensity in each nucleus was measured with ImageJ. Comparison was made for the follicle cells within the same egg chamber. Reduced EdU incorporation was defined as the EdU intensity in the mutant nucleus is <90% of the average level of wild-type nuclei. n = 167 nuclei from 10 separate egg chambers.
To more precisely examine the effects of removing mir-318, we used the FLP-FRT system (Xu and Rubin 1993) to create clones of the mir-318 mutant in the follicle cells. Use of genetic mosaics allows direct comparison of mutant cells with their age-matched neighbors in the same egg chamber. In this experiment the mir-318 mutant cells were marked by two copies of GFP (brighter green), compared to the heterozygous GFP/+ cells, which carried one mutant copy of mir-318 and the +/+ cells with two wild-type copies of mir-318. As in the intact egg chamber, we observed a proportion of cells in the clones with EdU incorporation throughout the nucleus (Figure 4, D–D′′, arrowhead). Twenty-eight percent of nuclei in the mutant clones showed this phenotype (Figure 4F). Fifty-four percent of the nuclei in the mutant clones showed reduced EdU incorporation (Figure 4, D–D′′, dashed arrow; Figure 4F), while 4% showed no Edu incorporation (Figure 4, E–E′′, arrow; Figure 4F). We also noted that mutant nuclei with reduced Edu signals in chorion amplification foci appeared to show a weaker diffuse Edu incorporation in the rest of nucleus (Figure 4, D–D′′ and E–E′′, star).
Delayed exit from follicle cell endocyling
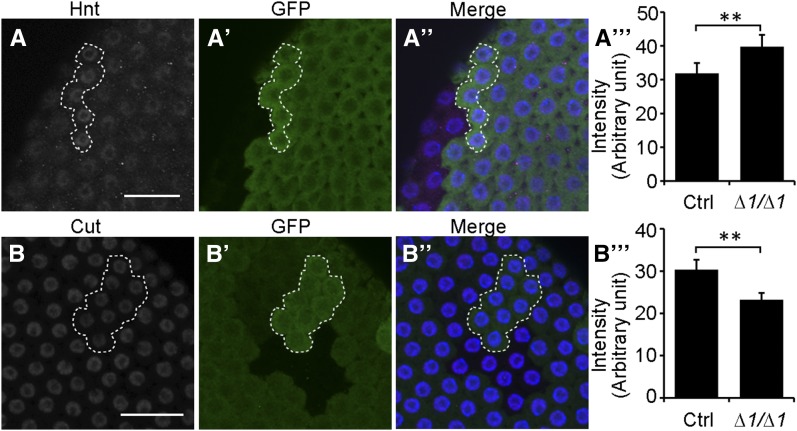
The observation of uniform incorporation of EdU in some mir-318 mutant cells at stage 10B suggested that miR-318 might be required for the transition of follicle cells from whole-genome replication to selective chorion gene amplification. Follicle cell ploidy increases by endoreplication of the DNA without subsequent cell division. Endoreplication stops during stage 10A/B. Cut expression is normally very low during endocycles but increases to higher levels in most follicle cells by stage 10B (Sun and Deng 2005). Conversely, expression of another transcription factor, Hindsight (Hnt), shows the opposite pattern, being higher during the endocycles and decreasing at stage 10B (Sun and Deng 2007). Increased Cut expression and decreased Hnt expression provide markers for the transition from endocycles to gene amplification (Huang et al. 2013). We compared the expression patterns of Cut and Hnt in mutant and wild-type clones in stage 10B egg chambers. As shown in Figure 5, A–A′′′, Hnt expression was higher in mir-318 mutant clones compared to control clones. Consistently, Cut expression appeared to be lower in mir-318 mutant clones compared to control clones (Figure 5, B–B′′′). These results suggest that loss of mir-318 compromises the transition from endocycles to chorion gene amplification in the follicular epithelium.
Figure 5.
mir-318 mutation impairs the developmental switch from endocycles to amplification. (A–B′′′) Hnt is up-regulated and Cut is down-regulated in mir-318 mutant follicle cell clones in stage-10B egg chambers. Shown are maximum intensity projections of Z-stacks of confocal images of stage-10B egg chambers with mir-318 mutant clones in the follicle cells. Samples were stained with anti-Hnt (gray in A; purple in A′′), anti-Cut (gray in B; purple in B′′), anti-GFP (green) and DAPI (blue). (A′′′ and B′′′) Quantitative analysis. n = 5 (A′′′) and 8 (B′′′). P < 0.05 for each comparison. Bars, 10 um.
Given that loss of mir-318 delayed exit from follicle cell endocyling, we next asked whether miR-318 overexpression would be able to advance this process and lead to premature chorion gene amplification. To this end, we induced ectopic expression of miR-318 using the flip-out GAL4 system in the follicle cells. In stage-9 egg chambers, miR-318 overexpression cells showed an oscillating genomic EdU incorporation pattern similar to that observed in adjacent wild-type cells (Figure 6, A–A′′). Consistent with this, the expression of Hnt and Cut were unchanged in miR-318-overexpressing cells (Figure 6, B–C′′). Thus, overexpression of miR-318 was not sufficient to activate premature gene amplification.
Figure 6.
Overexpression of miR-318 does not cause premature gene amplification. (A–C′′) Overexpression of miR-318 does not affect EdU incorporation or Hnt and Cut expression in the follicle cells in stage-9 egg chambers. Shown are maximum intensity projections of Z-stacks of confocal images of stage-9 egg chambers with miR-318 overexpression clones in the follicle cells. miR-318 overexpression clones were generated using a Flip-out system and marked by the presence of GFP. Samples were labeled with EdU (red in A), anti-Hnt (red in B), anti-Cut (red in C), anti-GFP (green) and DAPI (blue). Bar, 50 um.
Ecdysone receptor signaling is required for miR-318 expression
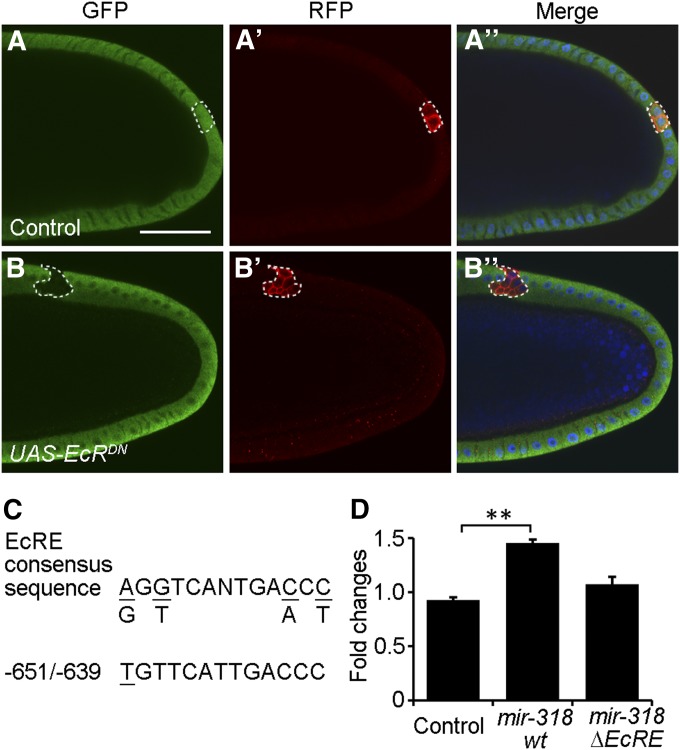
Activation of the ecdysone-signaling pathway promotes the transition from endocyling to gene amplification in the follicle cells (Sun et al. 2008). We therefore asked whether the steroid hormone 20E and its nuclear receptor were involved in regulating miR-318 expression. Expression of a dominant-negative form of the ecdysone receptor (EcR) in the clones of follicle cells led to loss of a mir-318-GFP reporter transgene (Figure 7, A–B′′). To further test whether this regulation is directly through the regulatory elements of mir-318, we prepared a reporter construct containing a 852-bp fragment of genomic DNA upstream of mir-318. This fragment contains a putative ecdysone response element (EcRE) (Figure 7C), similar to the consensus EcRE sequence through which the EcR/Usp heterodimeric complex binds to DNA to mediate ecdysone-regulated gene expression (Cherbas et al. 1991; Yao et al. 1992). 20E treatment increased miR-318 luciferase reporter activity by 1.5-fold in S2 cells (Figure 7D). A mutated luciferase reporter that lacked the EcRE showed a blunted response to 20E treatment (Figure 7D), indicating that the EcRE was functionally important in mediating the response to ecdysone in S2 cells.
Figure 7.
EcR signaling is required for miR-318 expression. (A–B′′) miR-318 expression is absent in the follicle cells overexpressing EcRDN. Shown are confocal images of stage-10B egg chambers from mir-318Δ1/+ heterozygous female flies with control or EcRDN overexpression clones in the follicle cells. GFP expression reflects miR-318 expression in the mir-318Δ1 allele as described previously. Clones were marked by the presence of RFP. Samples were stained with anti-GFP (green), anti-RFP (red) and DAPI (blue). Bar, 50 um. (C) Predicted EcRE sequence in the mir-318 regulatory region is shown relative to the EcRE consensus sequence. Mismatches are underlined. The position is shown relative to the mir-318 stem loop region. (D) Luciferase activity assays of wild-type and mutated mir-318 regulatory region luciferase reporters upon 20E treatment in S2 cells. Fold changes were measured as luciferase activity of 20E-treated vs. untreated samples. Control samples were cotransfected with the empty vector. Error bars represent standard deviation from four independent experiments. P < 0.05.
mir-318 interacts with ttk69
Previous studies have shown that regulation of Ttk69, a zinc-finger protein, plays an important role during late oogenesis (French et al. 2003). At stage 10B, Ttk69 functions to promote dorsal appendage tube elongation and chorion gene amplification (Sun et al. 2008; Boyle and Berg 2009; Peters et al. 2013). A recent report has found that miR-7 miRNA regulates Ttk69 during the transition from endocycles to gene amplification. miR-7 overexpression was able to repress Ttk69 expression and delay the switch from endocycles to amplification, but loss of miR-7 was not sufficient to advance this process (Huang et al. 2013).
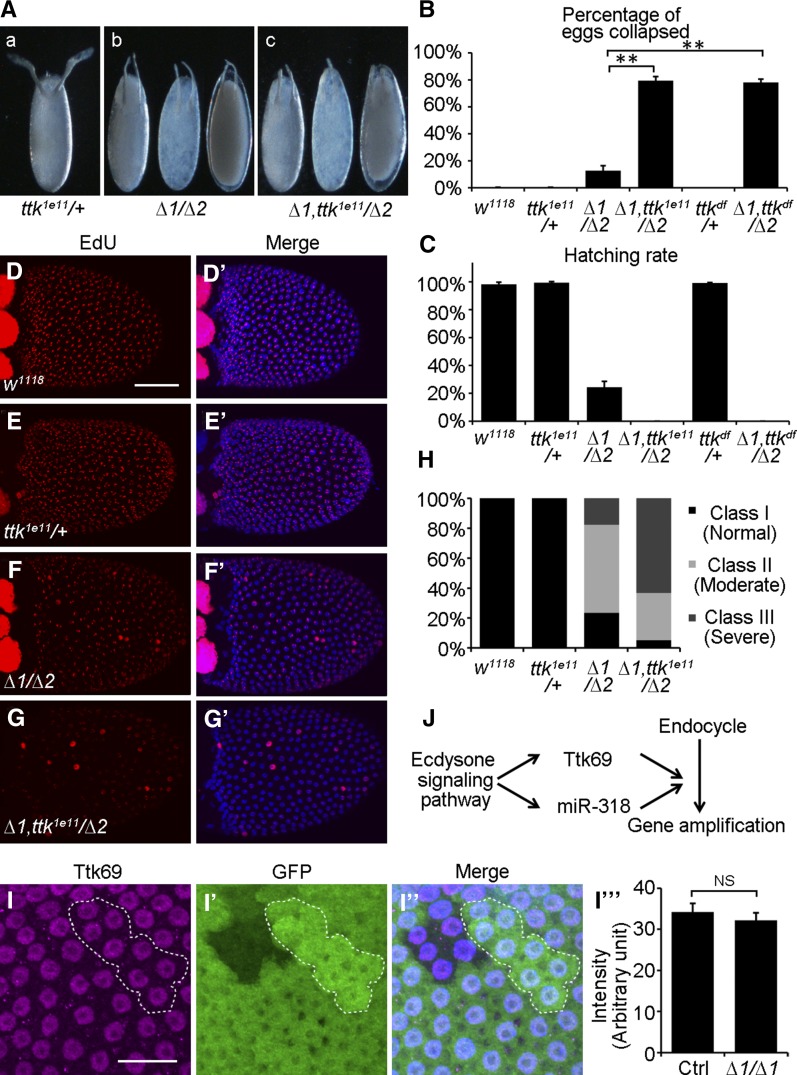
In light of the gene amplification defect that we observed in mir-318 mutants, we asked whether miR-318 interacts with Ttk69. To test this, we examined the effects of introducing one copy of the ttk69 mutant allele ttk1e11 into the mir-318 mutant background. Heterozygous ttk1e11/+females laid normal eggs. However, removing one copy of ttk69 increased the severity of the mir-318 mutant egg defect, including short dorsal appendages (Figure 8A). Seventy-nine percent of eggs from mir-318Δ1 ttk1e11/mir-318Δ2 mothers collapsed, compared to 13% in the mir-318Δ1/mir-318Δ2 mutants (Figure 8B). Whereas 24% of eggs from mir-318 mutant females were able to develop into first instar larvae, none of eggs from mir-318Δ1 ttk1e11/mir-318Δ2 females hatched into first instar larvae (Figure 8C). Similar results were obtained when using a deficiency chromosome that covered the ttk69 locus (Figure 8, B and C).
Figure 8.
mir-318 interacts with ttk69 to promote gene amplification. (A) Removal of one copy of ttk69 enhances the morphologically abnormal egg phenotype in the mir-318 mutant background. Shown are phase-contrast images of dorsal views of eggs from ttk1e11 heterozygous control (a), mir-318 mutant (b), and mir-318 mutant with one copy of ttk1e11 (c) female flies. (B) Removal of one copy of ttk69 increases the percentage of collapsed eggs in the mir-318 mutant background. Shown is the percentage of collapsed eggs from female flies of indicated genotypes. Error bars represent standard deviation from three independent experiments. n > 200 for each experiment. P < 0.05. (C) Removal of one copy of ttk69 causes severely reduced hatching rate of eggs in the mir-318 mutant background. Shown is the percentage of hatched eggs from female flies of indicated genotypes. Error bars represent standard deviation from three independent experiments. n > 200 for each experiment. (D–G′) Removal of one copy of ttk69 enhances gene amplification defects in the mir-318 mutant background. Shown are maximum intensity projections of confocal Z-stacks of stage-10B egg chambers from females of indicated genotypes. Samples were labeled with EdU (red) and DAPI (blue). Bar, 50 um. (H) Quantitative analysis of EdU intensity from D to G′. Three classes of egg chambers were observed according to the intensity of EdU foci: class I (normal), class II (moderate) and class III (severe). n = 50 for w1118, n = 53 for ttk1e11/+, n = 34 for Δ1/Δ2, and n = 38 for Δ1 ttk1e11/Δ2. (I–I′′′) Ttk69 expression is not affected in mir-318 mutant follicle cells. Shown are maximum intensity projections of Z-stacks of confocal images of stage-10B egg chambers with mir-318 mutant clones in the follicle cells. Samples were stained with anti-Ttk69 (purple), anti-GFP (green), and DAPI (blue). (I′′′) Quantitative analysis. n = 6. Bar, 20 um. (J) Diagram of Ttk69 and miR-318 regulation by the ecdysone-signaling pathway during the endocyle/gene amplification switch.
To determine whether these more severe defects in eggshell morphology were associated with the impairment of chorion gene amplification, we analyzed the pattern of gene amplification using EdU incorporation. Egg chambers from mir-318Δ1 ttk1e11/mir-318Δ2 females showed a more severe reduction of EdU incorporation in the chorion gene amplification pattern and a higher frequency of nuclei showing overall EdU labeling, compared to the mir-318Δ1/mir-318Δ2 mutants (Figure 8, D–G′; quantified in Figure 8H). This analysis revealed that 63% of stage-10B egg chambers from mir-318Δ1 ttk1e11/mir-318Δ2 females had severe defective EdU incorporation and 31% had moderate defective EdU incorporation, compared to 18% of stage-10B egg chambers from mir-318Δ1/mir-318Δ2 females that had severe defects and 59% had moderate defects (Figure 8H). This suggests that removal of one copy of ttk69 enhanced the gene amplification defects in mir-318 mutant females.
The genetic interaction between mir-318 and ttk69 prompted us to test whether miR-318 was able to activate Ttk69 expression in the follicle cells. To do this, we performed immunofluorescence staining with an antibody against Ttk69. However, Ttk69 expression was not affected in mir-318 mutant clones compared to control clones in the follicle cells in stage-10B egg chambers (Figure 8, I–I′′′). These results suggest that miR-318 acts cooperatively with Ttk69 to control gene amplification.
Discussion
In this study, we have shown that miR-318, one member of the miR-3 seed family, was highly expressed in ovaries and acted as a novel regulator in pattern formation and chorion gene amplification during Drosophila oogenesis. In contrast to this, miR-3 and miR-309, two other members of this family, function exclusively during embyrogenesis (Bushati et al. 2008). Because miRNAs within the same miRNA family share the same “seed” sequence, multiple members of the same miRNA family could function redundantly by acting on the same set of target genes. For example, miR-11 and miR-6, two members of the miR-2 seed family miRNAs, function to control the level of apoptosis by repressing expression of the proapoptotic genes including rpr, hid, and skl (Ge et al. 2012). Different roles between miR-318 and miR-3/309 provided one example that members of the same miRNA family could have distinct functions during development, potentially by acting on different sets of target genes. As a large number of miRNA knock-out mutants have been generated in Drosophila, the phenotypical analysis of individual miRNA mutants will help us to understand the function redundancy and diversity of miRNAs within the same miRNA family in different developmental processes (Chen et al. 2014).
Our analysis of miR-318 in Drosophila oogenesis revealed several features of its expression and function: (1) The spatial and temporal expression pattern of miR-318 is tightly controlled during oogenesis. miR-318 is uniformly expressed in somatically derived follicle cells during late oogenesis. It has been shown that spatial gene expression patterns in the follicle cells follow a simple combinatorial code based on six basic shapes, including the uniform pattern (Yakoby et al. 2008; Cheung et al. 2011). The uniform expression pattern of miR-318 in the follicle cells may reflect its function as a uniform repressor. Although EGFR and Dpp pathways control the expression of multiple genes in the follicle cells, they appear not to be involved in regulating miR-318 expression. In contrast, miR-318 expression is activated by the ecdysone-signaling pathway through the EcRE site at the mir-318 locus. The direct regulation of miR-318 by EcR highlights the importance of miRNAs in mediating downstream effects of EcR signaling. (2) There are multiple functions of miR-318 during late oogenesis. The morphological defects of mir-318 mutant eggs suggest connections between miR-318 and EGFR- and Dpp-signaling pathways. The enlarged operculum in the mir-318 mutant resembles the phenotype observed when Dpp is overexpressed (Twombly et al. 1996). Given that reduced Dpp activity did not compromise miR-318 expression, it is possible that miR-318 might function to limit Dpp activity. In the main body of follicle cells, mutation of mir-318 resulted in decreased Br expression in two dorsal–anterior domains and reduced size of the midline region. These defects would be consistent with reduced EGFR activity (Schupbach 1987; Roth 1998). Except for its roles in patterning follicular epithelium, miR-318 also functions in eggshell biogenesis. Reduced EdU incorporation at chorion gene loci in mir-318 mutant follicle cells suggests that miR-318 is important for efficient chorion gene amplification in the follicle cells. The chorion gene amplification programs are also coupled with the endocycles-to-amplification transition in the development of follicle cells (Mcconnell et al. 2012). Delayed exit from follicle cell endocycling in mir-318 mutants indicates that miR-318 is essential for this developmental switch. (3) There is a cooperation between miR-318 and the transcriptional repressor Ttk69 in controlling the endocycles-to-amplification switch. It has been reported that the cell cycle transition from endocyling to gene amplification requires downregulation of the Notch pathway, activation of the EcR pathway, and upregulation of Ttk69 (Sun et al. 2008). The enhancement of gene amplification defects in mir-318 mutants with reduced Ttk69 activities demonstrates that these two negative regulators of gene expression function together to control the switch from endocycling to gene amplification. The EcR pathway promotes the activity of Ttk69 and in turn triggers gene amplification (Sun et al. 2008). The fact that EcR signaling controls miR-318 expression further supports the notion that miR-318 acts cooperatively with Ttk69 (Figure 8J). (4) There are spatial differences in miR-318 effects on gene amplification. In stage 10B, gene amplification occurs synchronously in all follicle cells over the oocyte (Calvi et al. 1998). We have shown that some cells had more reduced EdU incorporation than others in the mir-318 mutant follicular epithelium. Especially, EdU incorporation was severely affected in dorsal–anterior follicle cells. These results raise the possibility that the roles of miR-318 vary in different domains of the follicular epithelium. How does the uniformly expressed miR-318 affect gene amplification with a spatial difference? It is conceivable that miR-318 might interact with its potential targets in specific regions of the follicular epithelium.
As miRNAs normally downregulate target gene expression, the mRNA levels of miR-318 targets whose expression leads to pattern formation and chorion gene amplification defects in mir-318 mutants would be expected to be increased in the mutant ovaries. We performed RNA deep sequencing analysis from RNA samples prepared from both wild-type and mir-318 mutant ovaries and identified a number of genes that were upregulated in the mutant ovaries. However, the overlapping hits between our identified genes and predicted miR-318 targets from TargetScanFly and http://miRNA.org were not apparently involved in regulating pattern formation and chorion gene amplification. Thus, we did not verify this further. As miR-318 is expressed only in a subpopulation of cells, assaying target RNA or protein level through RNA in situ hybridization or antibody staining in egg chambers with mir-318 mutant clones would be a better way to verify the predicted targets. Although we have been unable to identify the specific targets for miR-318, our results indeed demonstrate that miR-318 is one important regulator in oogenesis, and future investigations of its targets could potentially provide more mechanistic insights into pattern formation and developmental gene amplification.
Supplementary Material
Acknowledgments
We thank B. R. Calvi, P. Rørth, J. C. Pastor-Pareja, W. M. Deng, Z. H. Li, the Developmental Studies Hybridoma Bank, the Bloomington Drosophila Stock Center, and the Drosophila Genetic Resource Center for fly stocks and antibodies; Z. Nan for generating Ttk69 antibody; N. Bushati for generating mir-318 mutant; K. Tan, S. Lim, and S. Song for technical support; and P. Verma for discussion. This work was supported by the Institute of Molecular and Cell Biology, Singapore; the National Natural Science Foundation of China (grant 31371319); the National Key Basic Research Program of the Ministry of Science and Technology of China (grant 2013CB945600); and the fundamental research funds for the central universities, China. The authors declare no conflict of interest.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.174748/-/DC1.
Communicating editor: L. Cooley
Literature Cited
- Azzam G., Smibert P., Lai E. C., Liu J. L., 2012. Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division. Dev. Biol. 365: 384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. A., 2005. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 21: 346–355. [DOI] [PubMed] [Google Scholar]
- Boyle M. J., Berg C. A., 2009. Control in time and space: Tramtrack69 cooperates with Notch and Ecdysone to repress ectopic fate and shape changes during Drosophila egg chamber maturation. Development 136: 4187–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M., 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S. M., 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23: 175–205. [DOI] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M., 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18: 501–506. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Spradling A. C., 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18: 407–417. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., Spradling A. C., 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. W., Song S., Weng R., Verma P., Kugler J. M., et al. , 2014. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell 31: 784–800. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Lee K., Cherbas P., 1991. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 5: 120–131. [DOI] [PubMed] [Google Scholar]
- Cheung L. S., Schupbach T., Shvartsman S. Y., 2011. Pattern formation by receptor tyrosine kinases: analysis of the Gurken gradient in Drosophila oogenesis. Curr. Opin. Genet. Dev. 21: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., et al. , 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Smibert P., Lai E. C., 2012. Exploiting Drosophila genetics to understand microRNA function and regulation. Curr. Top. Dev. Biol. 99: 201–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. M., Bownes M., 1997. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development 124: 4639–4647. [DOI] [PubMed] [Google Scholar]
- Dobens L. L., Raftery L. A., 2000. Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev. Dyn. 218: 80–93. [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Sharp P. A., 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A. S., Lai E. C., 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R. L., Cosand K. A., Berg C. A., 2003. The Drosophila female sterile mutation twin peaks is a novel allele of tramtrack and reveals a requirement for Ttk69 in epithelial morphogenesis. Dev. Biol. 253: 18–35. [DOI] [PubMed] [Google Scholar]
- Ge W., Chen Y. W., Weng R., Lim S. F., Buescher M., et al. , 2012. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 19: 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield S. D., Shcherbata H. R., Fischer K. A., Nakahara K., Carthew R. W., et al. , 2005. Stem cell division is regulated by the microRNA pathway. Nature 435: 974–978. [DOI] [PubMed] [Google Scholar]
- Huang Y. C., Smith L., Poulton J., Deng W. M., 2013. The microRNA miR-7 regulates Tramtrack69 in a developmental switch in Drosophila follicle cells. Development 140: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino N., Pane A., Gaul U., 2009. miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 17: 123–133. [DOI] [PubMed] [Google Scholar]
- Jin Z., Xie T., 2007. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 17: 539–544. [DOI] [PubMed] [Google Scholar]
- Kloosterman W. P., Plasterk R. H., 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell 11: 441–450. [DOI] [PubMed] [Google Scholar]
- Kugler J. M., Verma P., Chen Y. W., Weng R., Cohen S. M., 2013. miR-989 is required for border cell migration in the Drosophila ovary. PLoS ONE 8: e67075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell K. H., Dixon M., Calvi B. R., 2012. The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development 139: 3880–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T., Olson E. N., 2012. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. K., Liu X., Strauss T. J., McKearin D. M., Liu Q., 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17: 533–538. [DOI] [PubMed] [Google Scholar]
- Peters N. C., Thayer N. H., Kerr S. A., Tompa M., Berg C. A., 2013. Following the ‘tracks’: Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror. Dev. Biol. 378: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., 1998. Drosophila development: the secrets of delayed induction. Curr. Biol. 8: R906–R910. [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., Lai E. C., 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome research. 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T., 1987. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell 49: 699–707. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., 1993. Developmental genetics of oogenesis, pp. 1–70 in The Development of Drosophila melanogaster, edited by M. Bate, A. A. Martinez. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sun J., Deng W. M., 2005. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng W. M., 2007. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Smith L., Armento A., Deng W. M., 2008. Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Lai E. C., 2013. Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 14: 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., 2004. Developmental gene amplification and origin regulation. Annu. Rev. Genet. 38: 273–304. [DOI] [PubMed] [Google Scholar]
- Twombly V., Blackman R. K., Jin H., Graff J. M., Padgett R. W., et al. , 1996. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development 122: 1555–1565. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Olson E. N., 2012. microRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 11: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R., Chen Y. W., Bushati N., Cliffe A., Cohen S. M., 2009. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics 183: 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yakoby N., Bristow C. A., Gong D., Schafer X., Lembong J., et al. , 2008. A combinatorial code for pattern formation in Drosophila oogenesis. Dev. Cell 15: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. P., Segraves W. A., Oro A. E., McKeown M., Evans R. M., 1992. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71: 63–72. [DOI] [PubMed] [Google Scholar]
- Yoon W. H., Meinhardt H., Montell D. J., 2011. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol. 13: 1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Y., Reynolds S. H., Hatfield S. D., Shcherbata H. R., Fischer K. A., et al. , 2009. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development 136: 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.