Abstract
Genetic analysis requires the ability to identify the genotypes of individuals in a segregating population. This task is straightforward if each genotype has a distinctive phenotype, but is difficult if these genotypes are phenotypically similar or identical. We show that Arabidopsis seeds homozygous or heterozygous for a mutation of interest can be identified in a segregating family by placing the mutation in trans to a chromosome carrying a pair of seed-expressed green and red fluorescent transgenes (a “traffic line”) that flank the mutation. Nonfluorescent seeds in the self-pollinated progeny of such a heterozygous plant are usually homozygous for the mutation, whereas seeds with intermediate green and red fluorescence are typically heterozygous for the mutation. This makes it possible to identify seedlings homozygous for mutations that lack an obvious seedling phenotype, and also facilitates the analysis of lethal or sterile mutations, which must be propagated in heterozygous condition. Traffic lines can also be used to identify progeny that have undergone recombination within a defined region of the genome, facilitating genetic mapping and the production of near-isogenic lines. We produced 488 transgenic lines containing single genome-mapped insertions of NAP:dsRED and NAP:eGFP in Columbia (330 lines) and Landsberg erecta (158 lines) and generated sets of traffic lines that span most regions of the Arabidopsis genome. We demonstrated the utility of these lines for identifying seeds of a specific genotype and for generating near-isogenic lines using mutations of WUSCHEL and SHOOTMERISTEMLESS. This new resource significantly decreases the effort and cost of genotyping segregating families and increases the efficiency of experiments that rely on the ability to detect recombination in a defined chromosomal segment.
Keywords: recombination, wus, stm, T-DNA, mapping
ARABIDOPSIS thaliana has had a major impact on plant biology over the past 20 years because of the advantages it offers for genetic analysis (Somerville and Koornneef 2002; Koornneef and Meinke 2010). The ease of performing large-scale phenotype-based genetic screens, the availability of sequence-indexed mutations in most genes (Sundaresan et al. 1995; Budziszewski et al. 2001; McElver et al. 2001; Alonso et al. 2003), and techniques for site-specific mutagenesis and gene silencing (Chuang and Meyerowitz 2000; Alvarez et al. 2006; Schwab et al. 2006; Franco-Zorrilla et al. 2007; Todesco et al. 2010; Cermak et al. 2011) have all contributed to the popularity of Arabidopsis as a model system in plant biology. But, although there are many ways to obtain mutations in genes of interest in Arabidopsis, there are surprisingly few ways to efficiently manipulate these mutations. Unless a mutation has an obvious, fully penetrant and viable phenotype, or is associated with a selectable marker, the only way it can be followed in genetic crosses is by molecular genotyping or progeny testing. Mutations that have invisible, weak, or variable phenotypes, as well as mutations that cannot be maintained under homozygous conditions (e.g., lethal or sterile mutations), are difficult to work with. A tool that makes it possible to unambiguously identify plants homozygous or heterozygous for these types of mutations—without the need for molecular genotyping—would significantly enhance the efficiency of genetic analysis in this important experimental system.
In Drosophila melanogaster, this problem is solved with balancer chromosomes (Ashburner et al. 2005). Balancer chromosomes contain multiple inversions and are marked with a dominant, viable, mutation/transgene and a recessive lethal mutation. These features ensure that chromosomes in trans to the balancer are transmitted intact and provide a convenient way to maintain stocks of lethal or sterile mutations. Although balancer chromosomes exist in other species (Zheng et al. 1999; Wilson et al. 2005; Edgley et al. 2006), they have not been widely used outside of Drosophila because of the technical difficulty of generating such chromosomes.
Here we describe an approach that has some of the features of a balancer chromosome, but does not require extensive chromosome engineering. This approach was originally developed to study recombination in Arabidopsis and involves using a pair of linked, differently colored fluorescent transgenes to mark a chromosome interval (Melamed-Bessudo et al. 2005; Berchowitz et al. 2007; Francis et al. 2007; Berchowitz and Copenhaver 2008; Berchowitz and Copenhaver 2009; Pecinka et al. 2011; Melamed-Bessudo and Levy 2012). The first use of this approach (Melamed-Bessudo et al. 2005) employed fluorescent reporters (eGFP and dsRED) fused to the seed-specific napA storage protein promoter (Stuitje et al. 2003). We recognized that in addition to revealing the products of recombination, these doubly marked chromosomes provide a convenient way to follow nonrecombinant chromosome segments in genetic crosses. When NAP:eGFP and NAP:dsRED are linked on a single chromosome, most of the individuals that inherit a doubly marked chromosome from a parent hemizygous for these transgenes (NAP:eGFP NAP:dsRED/−) will possess a nonrecombinant version of the chromosome segment flanked by these two transgenes, and most of the individuals that lack NAP:eGFP and NAP:dsRED will contain a nonrecombinant version of the segment in trans to these markers; the exceptions are cases in which double recombination has occurred within the marked interval, and these will be rare for small intervals. This means that mutations present on the chromosome in trans to a doubly marked chromosome can be followed in genetic crosses by screening for nonfluorescent seeds in the progeny of heterozygous plants. Furthermore, because the intensity of eGFP and dsRED fluorescence is sensitive to gene dose, seeds heterozygous for a mutation within the marked interval can be distinguished from seeds homozygous for the NAP:eGFP–NAP:dsRED chromosome by their intermediate level of fluorescence. These properties are particularly useful for mutations with weak or invisible phenotypes, which are typically identified in segregating families by PCR genotyping or progeny testing and for mutations that cannot be maintained in homozygous condition, such as lethal or sterile mutations.
Transgenic lines containing NAP:eGFP or NAP:dsRED at a single site in the Arabidopsis genome were screened to identify lines that had no obvious homozygous phenotype, and the position of these transgenes was then determined by DNA sequencing. Linked NAP:eGFP and NAP:dsRED insertions were then joined by recombination to create a series of traffic lines (TLs) that span most of the Arabidopsis genome. This new collection of marker lines and examples of the ways in which they can be used for genetic analysis are described in this article.
Materials and Methods
Vector construction
Plasmid pFLUAR100 harboring NAP:eGFP, and plasmid pFLUAR101 harboring NAP:dsRED were kindly provided by Dr. Antoine R. Stuitje (Department of Genetics, Institute for Molecular Cell Biology, The Netherlands). Due to the low genetic transformation frequencies associated with these two constructs and the low recovery rate of T-DNA border sequence, these constructs were modified using overlapping PCR and the high-fidelity Phusion DNA polymerase (NEB). For the pCAM–NAP:eGFP construct, the napA promoter sequence was PCR amplified using the forward primer 5′-CCCAAGCTTAGGAAGCTTGCATGCTGCAG-3′ and the reverse primer 5′-TCCTCGCCCTTGCTCACCATGGGGTATGTTTTTAATCTTG-3′, and the eGFP open reading frame was PCR amplified using the forward primer 5′-ATGGTGAGCAAGGGCGAGGA-3′ and the reverse primer 5′-TGCCAAATGTTTGAACGATCTTACTTGTACAGCTCGTCCA-3′. The nos sequence was PCR amplified using the forward primer 5′-GATCGTTCAAACATTTGGCAATAAAG-3′ and the reverse primer 5′-CCGCTCGAGGATCTAGTAACATAGATGACAC-3′ and was used as the terminator for both the pCAM–NAP:eGFP and pCAM–NAP:dsRED constructs. For the pCAM–NAP:dsRED construct, the napA promoter sequence was PCR amplified using the forward primer 5′-CCCAAGCTTAGGAAGCTTGCATGCTGCAG-3′ and the reverse primer 5′-ACATTCTTGGAAGACCGCATGGGGTATGTTTTTAATCTTG-3′, and the dsRED open reading frame was PCR amplified using the forward primer 5′-ATGCGGTCTTCCAAGAATGT-3′ and the reverse primer 5′-TGCCAAATGTTTGAACGATCTTAAAGGAACAGATGGTGGC-3′. After overlapping PCR, the NAP:eGFP–NOS expression cassette was cloned into the XhoI and EcoRI-linearized CAMBIA3300 vector, and the NAP:dsRED–NOS expression cassette was cloned into the XhoI and BamHI-linearized CAMBIA3300 vector. The resulting pCAM–NAP:eGFP and pCAM–NAP:dsRED plasmids were sequenced before they were electroporated into Agrobacterium competent cells for genetic transformation.
Selecting transgenic plants
Transformation was carried out by floral dipping (Clough and Bent 1998). Mature T1 seeds were screened using a Leica MZ FLIII stereomicroscope and long-bandpass GFP (ET480/40x/ET510 LP) and RFP (ET546/10×/ET590 nm LP) filter sets. It is also possible to identify transgenic seeds using the stereomicroscope adaptors available from Nightsea, Inc. (http://www.nightsea.com/galleries/arabidopsis/). Seeds were manually classified as low, medium, or high fluorescence and planted individually in 96 well flats. T1 plants were subsequently screened for semisterility and embryo lethal mutations by opening immature siliques and looking for low seed set or defective seeds. Siliques with ∼50% aborted seeds were classified as semisterile, whereas siliques with ∼25% albino seeds were classified as having an embryo lethal mutation. Mature T2 seeds from fully fertile plants were screened with a stereomicroscope to identify families segregating 3:1 fluorescent:nonfluorescent seeds, and several of the brightest seeds from these families were planted to obtain lines homozygous for the insertion. If all of the homozygous plants appeared phenotypically normal, the sequence of the insertion site was determined.
Mapping transgenes
Insertion sites were sequenced using an adapter ligation-mediated PCR method as described previously (O’Malley et al. 2007) with some modifications. Genomic DNA was prepared by phenol-chloroform extraction. About 100 ng of genomic DNA was digested overnight at room temperature with HindIII and EcoRI in a cocktail containing 10× NEB ligase buffer, 1 μl. HindIII adapter, EcoRI adapter, and NEB T4 DNA ligase. Digested genomic DNA was used as the template in a 20-μl system for the first-round PCR with an adapter primer, 5′-GTAATACGACTCACTATAGGGC-3′, and a NAPIN–NOS–LB1 primer, 5′-CCTGTTGCCGGTCTTGCGATGATT-3′. PCR was carried out using TAKARA Ex Taq Hot Start DNA polymerase as follows: predenaturation at 95° for 3 min, and then 29 cycles of 98° for 10 sec, 60° for 20 sec, 72° for 2 min and 30 sec, and extension at 72° for 7 min. PCR products were diluted with ddH2O to 100μl. Diluted PCR products, 1 μl, was used as the template for a second-round PCR with a AP2-C primer, 5′-TGGTCGACGGCCCGGGCTGC-3′, and a NAPIN-NOS-LB2 primer, 5′-GCGCGGTGTCATCTATGTTACTAG-3′. The second round of PCR was done as described above except that the annealing temperature was increased to 62°. PCR products were gel purified and sequenced, and the T-DNA border sequences were then mapped to the Arabidopsis genome using the SIGnAL T-DNA express mapping tool (http://signal.salk.edu/cgi-bin/tdnaexpress).
Stock distribution
The transgenic lines described in this article may be obtained from the Arabidopsis Biological Resource Center (ABRC) (https://abrc.osu.edu). ABRC stock numbers and other information about individual lines are available in the supporting information (Table S1, Table S2, Table S3, and Table S4), and online at https://sites.sas.upenn.edu/poethig-lab/pages/data. Complete sets of the lines described in this article are available from ABRC using the following set numbers: CS71568 (CG and CR lines), CS71569 (LG and LR lines), CS71570 (CTL lines), and CS71571 (LTL lines).
Results
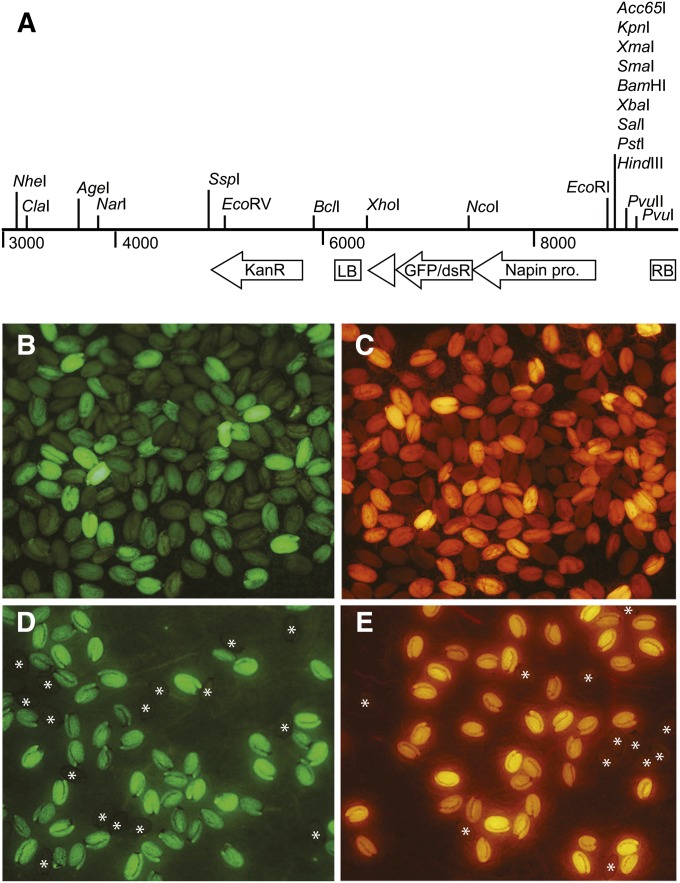
Lines transformed with NAP:eGFP and NAP:dsRED were initially produced using the pFLUAR vectors (Stuitje et al. 2003). Consistent with a previous report (Melamed-Bessudo et al. 2005), these vectors had low transformation efficiency and their insertion sites were difficult to sequence. To solve these problems, we cloned NAP:eGFP and NAP:dsRED fragments from pFLUAR100 and pFLUAR101 into pCAMBIA3300. These new vectors, pCAM–NAP:dsRED and pCAM–NAP:eGFP (Figure 1A), had significantly higher transformation efficiencies than the pFLUAR vectors, and we were able to determine the insertion site of the transgene in >80% of the lines we sequenced.
Figure 1.
Dosage-sensitive expression of NAP:eGFP and NAP:dsRED in dry seeds. (A) Structure of the T-DNA vectors (pCAM–NAP:eGFP and pCAM–NAP:dsRED) used in this study. (B) T1 seeds transformed with NAP:eGFP. (C) T1 seeds transformed with NAP:dsRED. (D) T2 seeds from a transgenic plant segregating 3:1 for NAP:eGFP. (E) T2 seeds from a transgenic plant segregating 3:1 for NAP:dsRED. Nonfluorescent seeds are marked with an asterix.
The fluorescence intensity of primary transformed seed (T1 seed) varied widely (Figure 1, B and C). To explore the basis for this phenomenon, Columbia (Col) seeds transformed with NAP:eGFP were divided into low, medium, and high fluorescence classes, and the number of insertion sites in these seeds was determined by counting the frequency of fluorescent and nonfluorescent seed in their T2 progeny. T1 plants that produced 3:1 fluorescent:nonfluorescent seeds were classified as having a single insertion site, whereas plants that produce >3:1 ratio of fluorescent:nonfluorescent seed were categorized as having more than one insertion site (Table 1). Only 18% of the T1 seeds in the high-fluorescence class contained a single insertion. In contrast, 60–70% of T1 seeds with low or medium fluorescence had a single insertion site. This result is consistent with previous studies indicating that plants transformed with Agrobacterium tumefaciens often contain T-DNA insertions at multiple sites (McElver et al. 2001; Alonso et al. 2003) and demonstrates that it is possible to enrich for plants containing single T-DNA insertions using seed fluorescence as a transformation marker.
Table 1. eGFP fluorescence intensity predicts transgene copy number.
| Fluorescence intensity of T1 seed | Frequency of single insertion lines |
|---|---|
| High | 6/33 (18%) |
| Medium | 25/41 (61%) |
| Low | 31/45 (69%) |
The seeds of plants segregating for a single insertion usually varied in fluorescence intensity (Figure 1, D and E). In a previous study of a single transgenic line containing GFP under the regulation of the seed-specific OLE1 promoter, it was reported that fluorescence intensity was correlated with homo- or hemizygosity for the transgene (Shimada et al. 2010). To determine if fluorescence intensity can indeed be used to determine the genotype of a seed, 10–15 bright fluorescent seeds from 52 independently derived T1 lines segregating 3:1 for NAP:eGFP were planted. The seeds produced by these 643 T2 plants were then examined to determine if the plant was homozygous for the transgene. In 95% of cases (610/643), the T2 plant was homozygous for the transgene. This result demonstrates that fluorescence intensity can be used to identify seeds homozygous for a T-DNA insertion.
Marker lines expressing NAP:eGFP and NAP:dsRED
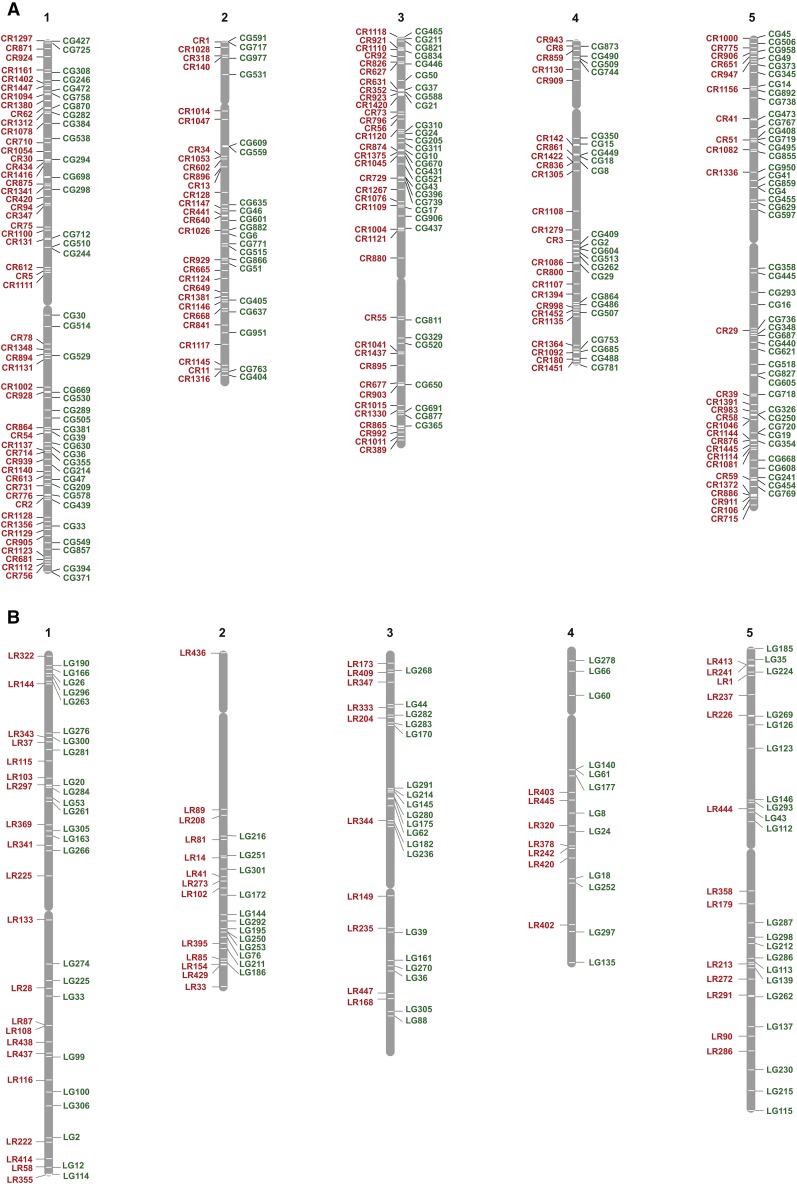
The primary goal of this study was to develop a large set of transgene insertions useful as genetic markers. For this purpose, T1 seeds expressing moderate to low levels of eGFP and dsRED were planted, and their T2 seeds were examined to identify lines segregating 3:1 fluorescent:nonfluorescent seeds. Analyses of the first 40 of these lines revealed that 4 were semisterile when the transgene was hemizygous, but completely fertile when it was homozygous; these lines also displayed reduced recombination in the vicinity of the transgene. This behavior is characteristic of reciprocal translocations or large inversions and supports a previous study indicating that Agrobacterium-mediated transformation frequently produces large chromosomal rearrangements (Clark and Krysan 2010). Because reciprocal translocations make it difficult to use these lines for genetic mapping, immature siliques of T1 plants were screened to identify those with 50% undeveloped ovules (semisterile) or 25% aborted seed (embryo lethal mutation). Of T1 plants (Col), 16% were semisterile (Table 2), which is similar to the frequency of reciprocal translocations (19%) in the SALK T-DNA collection (Clark and Krysan 2010). Of T1 plants (Col), 54% were fully fertile and produced T2 families segregating fluorescent and nonfluorescent seeds in a 3:1 ratio (Table 2). Three highly fluorescent seeds from these families were planted to obtain plants homozygous for the transgene. This also enabled us to identify insertions associated with a mutant phenotype, which was apparent when all three plants had the same aberrant phenotype. The insertion site in phenotypically normal families was amplified by adaptor-ligation PCR (O’Malley et al. 2007), gel purified, and sequenced. To date, we have determined the genomic locations of 330 NAP:eGFP and NAP:dsRED transgene insertions in Col (Figure 2A and Table S1) and 158 transgene insertions in Landsberg erecta (Ler) (Figure 2B and Table S2).
Table 2. Characterization of T2 families transformed with NAP:eGFP and NAP:dsRED.
| Fertile | ||||
|---|---|---|---|---|
| Transgene | 3:1a | >3:1b | Semisterilec | Embryo lethald |
| NAP:eGFP | 175 (57%) | 74 (24%) | 45 (15%) | 14 (4%) |
| NAP:dsRED | 140 (50%) | 77 (28%) | 48 (17%) | 13 (5%) |
| Total | 315 (54%) | 151 (26%) | 93 (16%) | 27 (5%) |
Number of completely fertile T2 families segregating 3:1 fluorescent:nonfluorescent seeds.
Number of completely fertile T2 families segregating greater than 3:1 fluorescent:nonfluorescent seeds.
Number of T2 families with 50% ovule abortion.
Number of T2 families segregating 25% aborted seed.
Figure 2.
Genomic locations of NAP:eGFP and NAP:dsRED insertions. (A) Transgene insertions in Col. (B) Transgene insertions in Ler. Maps were generated using the TAIR chromosome map tool (https://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). CG and CR, NAP:eGFP and NAP:dsRED insertions in Columbia. LG and LR, NAP:eGFP and NAP:dsRED in Ler.
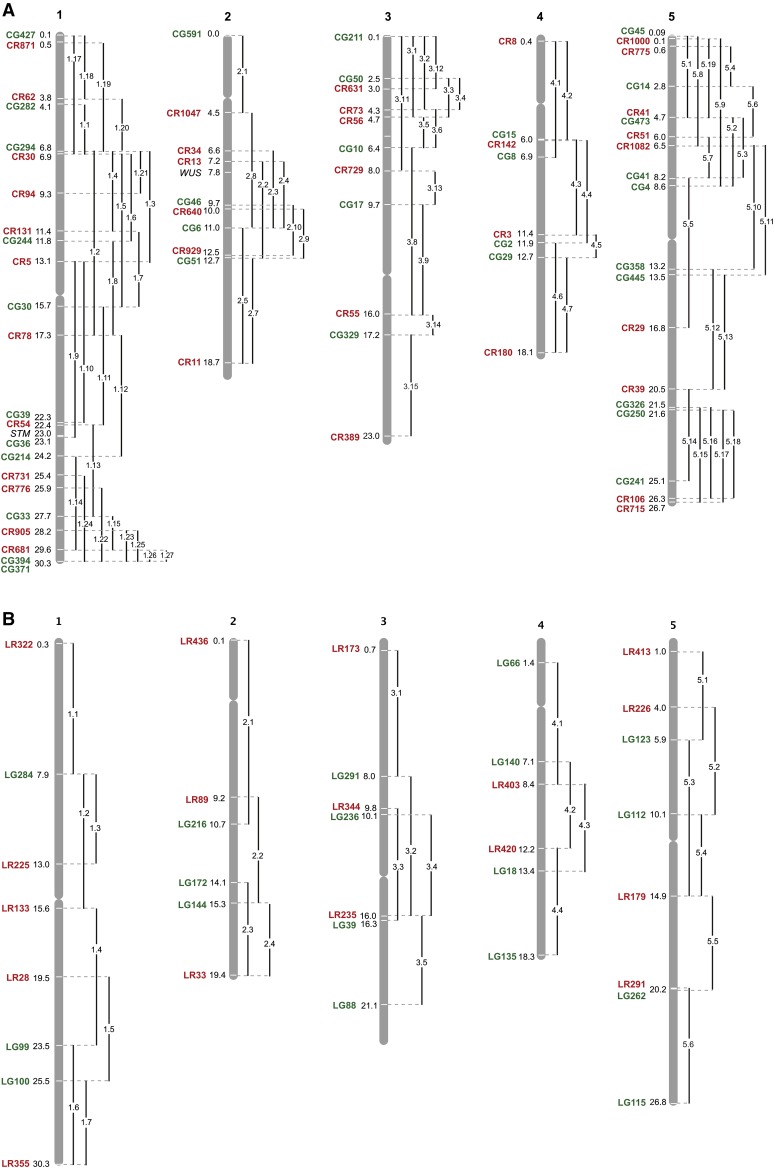
The recombination distance between linked NAP:EGP and NAP:dsRED transgenes was estimated using high-resolution mapping data from recombinant inbred lines (Singer et al. 2006). Lines containing insertions <30 cM apart were intercrossed, and the resulting hybrids were crossed as pollen parents to the appropriate parental ecotype (Col or Ler) to obtain recombinant chromosomes containing both transgenes (Figure 3). Stocks homozygous for the recombinant chromosome were produced by selecting brightly fluorescent seed from the F2 progeny of recombinant plants. We generated 74 recombinant lines in Col (Figure 4A and Table S1) and 26 recombinant lines in Ler (Figure 4B and Table S2), which cover most of the Arabidopsis genome. These stocks are named traffic lines (TLs) because their seeds fluoresce red, green, and yellow under appropriate illumination.
Figure 3.
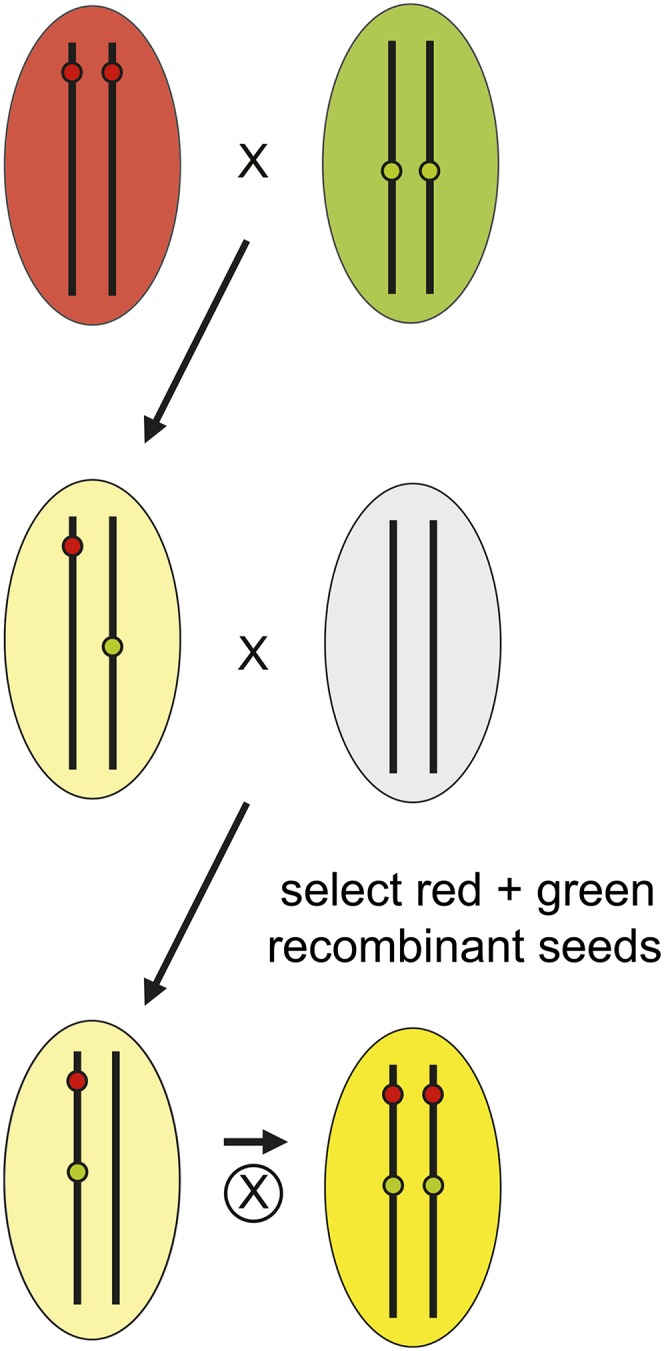
Crossing scheme used to produce TLs
Figure 4.
Location of NAP:eGFP and NAP:dsRED insertions in TLs. (A) TLs in Col; (B) TLs in Ler. Numbers indicate the nucleotide position of the insertion in megabases. Vertical lines connect the red and green transgenes in a TL. TLs are identified by the ecotype, chromosome, and the order in which the line was created (e.g., CTL1.2, Columbia, chromosome 1, second line).
Using TLs to identify genotypes in segregating populations
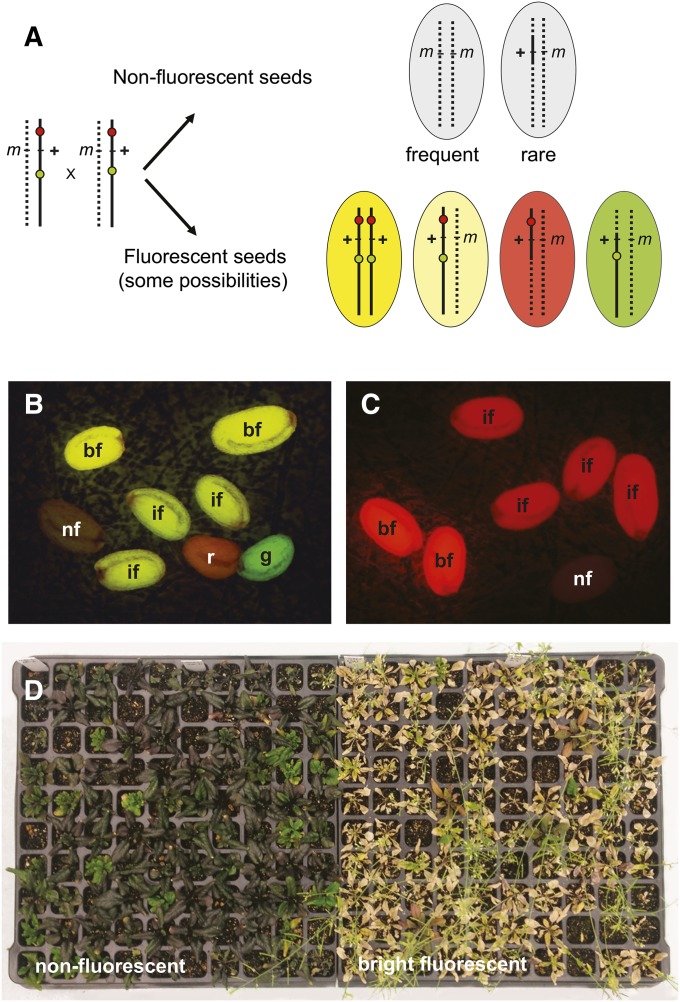
Chromosome segments bounded by NAP:eGFP and NAP:dsRED are useful because both the marked segment, and the segment in trans to the marked chromosome, can be followed in genetic crosses: self or outcross progeny that inherit the marked chromosome will have green and red fluorescent seeds, whereas progeny that inherit the nontransgenic chromosome will have nonfluorescent seeds. Because double recombination within a short chromosome segment is rare, the vast majority of these nonfluorescent seeds will contain a parental version of the region in trans to the marked interval (Figure 5A). Additionally, the dosage sensitivity of NAP:eGFP and NAP:dsRED makes it possible to distinguish F2 seeds that are heterozygous for the marked chromosome from seeds homozygous for this chromosome by their fluorescence intensity (Figure 5, A and B).
Figure 5.
Using a TL to identify seeds homozygous for a recessive mutation. (A) The most common genotypes in the F2 progeny of a plant heterozygous for a TL and a mutation (m) of interest. (B) F2 seeds from a plant of the genotype CTL2.4/wus-5 photographed using a broadband GFP filter set (see Materials and Methods), which allows GFP and dsRED fluorescence to be visualized simultaneously. (C) F2 seeds from a CR128/wus-5 plant. nf, nonfluorescent; bf, bright fluorescent; if, intermediate fluorescent, r, red; g, green. (D) Phenotype of plants derived from nonfluorescent and bright fluorescent seeds in the F2 progeny of CTL2.4/wus-5. Plants were sprayed with Basta after flowering to determine the genotype of the phenotypically wild-type plants. All nonfluorescent seeds were resistant to Basta and had a wus-5 mutant phenotype, indicating they were wus-5/wus-5, and most of the bright fluorescent seeds were homozygous for the wild-type WUS allele, as indicated by their sensitivity to Basta.
To test the value of TLs as a genotyping tool, we used them to identify seeds homozygous or heterozygous for wuschel-5 (wus-5) (Sonoda et al. 2007; Chatfield et al. 2013) and shootmeristmless-1 (stm-1) (Barton and Poethig 1993). wus-5 is caused by a T-DNA insertion (SAIL_150_G06) and is identifiable in homozygous condition by its shoot defective phenotype and in heterozygous condition by the Basta-resistance conferred by the transgene insertion. stm-1 is an EMS-induced allele of SHOOTMERISTEMLESS. Both mutations produce defective shoot meristems and therefore cannot be maintained in homozygous condition. Three TLs encompassing WUS—CTL2.1, CTL2.3, and CTL2.4—were crossed to plants heterozygous for wus-5, and nonfluorescent and bright fluorescent seeds in the F2 progeny of TL/wus-5 plants were selected (Figure 5B). One hundred percent of the nonfluorescent F2 progeny of CTL2.1/wus-5 and CTL2.4/wus-5, and 99% of the nonfluorescent F2 progeny of CTL2.3/wus-5, were homozygous for wus-5 (Table 3). All of the plants from bright fluorescent seeds were morphologically normal (Table 3) and most were homozygous for the wild-type WUS allele, as evident from their sensitivity to Basta (Figure 5D). Similar results were obtained with stm-1. Plants heterozygous for stm-1 were crossed to CTL1.4 and CTL1.9, and nonfluorescent F2 seeds of CTL/stm-1 individuals were planted. All of these nonfluorescent seeds produced seedlings with defective meristems, indicating that they were homozygous for stm-1 (Table 3). These results demonstrate that TLs can be used to identify seeds homozygous for a mutation located within the marked interval. To determine if TLs can be used to identify seeds heterozygous for a mutation, we selected seeds with intermediate red and green fluorescence in the F2 progeny of CTL2.4/wus-5 (Figure 5B). Of these seeds 100% (90/90 plants) were phenotypically wild type and resistant to Basta, indicating that they were heterozygous for wus-5.
Table 3. Phenotype of the F2 progeny of plants heterozygous for a TL or a single transgene.
| Parental genotype | Nonfluorescent F2 seeds | Bright fluorescent F2 seeds | ||||
|---|---|---|---|---|---|---|
| Mutanta | Wild-typeb | % mutant | Mutanta | Wild-typeb | % wild-type | |
| CTL2.1/wus-5 | 92 | 0 | 100 | 0 | 74 | 100 |
| CTL2.3/wus-5 | 92 | 1 | 99 | 0 | 79 | 100 |
| CTL2.4/wus-5 | 91 | 0 | 100 | 0 | 76 | 100 |
| CR128/wus-5 | 116 | 7 | 94 | 0 | 37 | 100 |
| CR13/wus-6 | 139 | 6 | 96 | 0 | 82 | 100 |
| CTL1.9/stm-1 | 83 | 0 | 100 | NDc | ND | ND |
| CTL1.14/stm-1 | 93 | 0 | 100 | ND | ND | ND |
Number of seedlings displaying a wus or stm phenotype.
Number of phenotypically wild-type seedlings.
Not determined.
Single NAP:EGFP or NAP:dsRED insertions can also be used to identify seeds of a desired genotype if the insertion is tightly linked to the gene of interest. CR13 and CR128 contain NAP:dsRED insertions located, respectively, 600 kb above and 900 kb below WUS on chromosome 2 (Figure 4A). Ninety-six percent of the nonfluorescent seeds in the F2 progeny of CR13/wus-5 and 94% of the nonfluorescent seeds in the F2 progeny of CR128/wus-5 were homozygous for wus (Figure 5C; Table 3). Of the plants from seeds with intermediate fluorescence, 98% (84/86) were heterozygous for wus-5, as indicated by their morphologically normal, Basta-resistant phenotype.
It should be emphasized that distinguishing seeds homozygous and heterozygous for NAP:eGFP or NAP:dsRED can be difficult in some lines. Some transgenes produce similar levels of fluorescence in heterozygous and homozygous condition, whereas others produce variable fluorescence even when homozygous. Nevertheless, by selecting seeds at the ends of the fluorescence spectrum it is usually possible to reliably identify homozygous and heterozygous genotypes in a segregating population.
Using traffic lines to select for recombination in a defined region
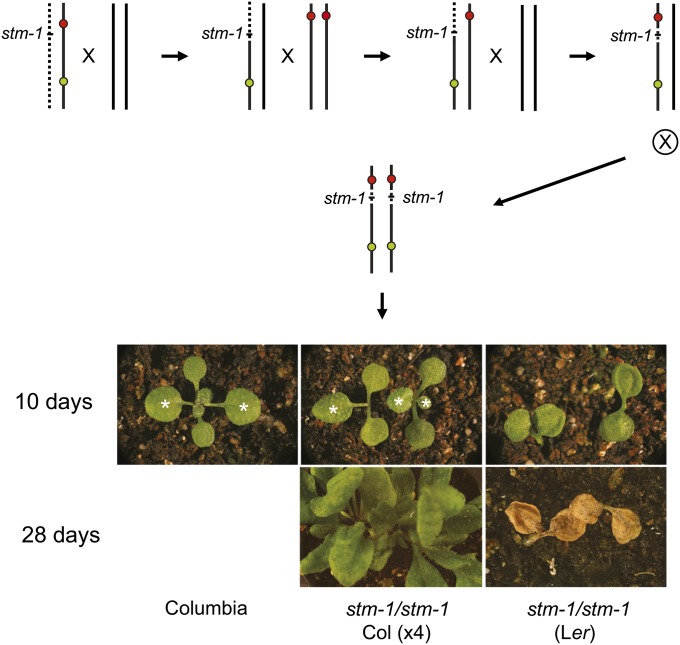
To test the value of TLs for generating near-isogenic stocks, we introgressed stm-1 (in Ler) into Col, using the strategy illustrated in Figure 6. Plants heterozygous for stm-1 were crossed to CTL1.13 (Figure 4A), and F1 progeny containing stm-1 were backcrossed to Col. Seeds from this backcross were screened to identify recombinants expressing green, but not red fluorescence. Seven of these recombinants were genotyped for markers within the CR54–CG33 interval to identify individuals with recombination sites close to stm-1. One plant with a recombination site ∼300 kb below stm-1 was crossed to CR54. The CR54 + −/− stm-1 CG33 progeny of this cross were then crossed to Col, and red–green recombinants [CR54 (stm-1) CG33/− + −] were planted and screened for stm-1 by PCR. Of 54 recombinants, 8 were CR54 stm-1 CG33/− + −. Seeds with the brightest red and green fluorescence were selected from the F2 progeny of these plants in order to obtain plants homozygous for the CR54 stm-1 CG33 chromosome. In contrast to the shootmeristemless phenotype of stm-1 in a Ler background, these seedlings produced one or two rosette leaves from the primary axis and multiple defective axillary shoots; some plants produced an inflorescence with defective flowers (Figure 6). stm-1 has a nonsense mutation upstream of the DNA binding domain of STM (Long et al. 1996) and consequently is likely to be a null allele. This result therefore implies that Col has one or more genes that can partially compensate for the loss of STM.
Figure 6.
Using a TL to select recombinants in a specific chromosome segment. Procedure used to introgress stm-1 into a Col genetic background. The transgenic chromosome is CTL1.13. Recombinants containing stm-1 were identified by progeny testing or by molecular genotyping. stm-1 has a much weaker phenotype in a Col background than in Ler. Col flowered 28 days after planting.
Discussion
Many problems in genetic analysis depend on the ability to determine the genotype of an individual independent of its phenotype. In Arabidopsis, this is usually done after the molecular nature of the relevant polymorphisms has been determined, so that they can be detected by a PCR-based technique. Although this approach is effective, it is time consuming and expensive and of course requires the destruction of at least a part of the individuals being genotyped. TLs make it possible to visually determine the genotype of a seed prior to planting, providing significant savings in time, space, and resources. They can be used either to propagate a chromosome segment intact in genetic crosses or to identify progeny resulting from recombination within a defined interval. These features enable a variety of experiments that are currently very difficult or impossible to perform in Arabidopsis.
The properties of plants transformed With NAP:eGFP and NAP:dsRED
TLs were generated using the seed-specific, semidominant fluorescent reporters, NAP:eGFP and NAP:dsRED. Aside from their value as genetic markers, these fluorescent reporters have several advantages over the herbicide or antibiotic resistance genes that are traditionally used for the identification of transgenic plants. Antibiotic and herbicide resistance are dominant traits, meaning that it is impossible to distinguish homozygous from hemizygous individuals on the basis of their resistance phenotype. In addition, these traits must be scored after germination, which requires both time and space. To identify a line homozygous for a single transgene using these dominant markers it is necessary to: (1) plant a large number of T1 seeds and treat with a selective agent to identify transgenic plants; (2) harvest T2 seeds from resistant plants and screen these families for lines segregating 3:1 for the selectable trait; (3) harvest seeds from five to six plants in families with single insertions; and (4) plant these T3 seeds to identify families homozygous for the insertion. In contrast, fluorescent reporters can be scored in dry seed (Stuitje et al. 2003; Lu and Kang 2008; Shimada et al. 2010; Untergasser et al. 2012) and are dosage sensitive (Shimada et al. 2010; this report). Using these markers, the procedure for identifying lines homozygous for single transgenic insertions consists of: (1) visually selecting transgenic T1 seeds prior to planting; (2) screening the F2 seed from these T1 plants to identify lines segregating 3:1 for the fluorescent transgene, and (3) planting a few (three to five) of the brightest seeds from these families to obtain homozygous lines. The phenotype of the plants produced by these bright seeds will also reveal whether the transgene has generated a recessive mutation; in this case, all of these seeds will produce plants with a mutant phenotype. Our observation that seed fluorescence is correlated with the number of transgenic insertions also means that it is possible to enrich for T1 plants with single insertions prior to planting. Finally, it is worth noting that fluorescent proteins expressed under the regulation of the napA promoter are only visible late in seed development and for a short time following germination and thus do not interfere with the use of these proteins as gene expression markers. Seed-specific fluorescent markers such as NAP:eGFP or NAP:dsRED therefore provide enormous advantages for the generation and analysis of transgenic lines in plant biology. They are particularly useful for the molecular breeding of crop plants (Lu and Kang 2008) because they significantly reduce the time and space required to obtain homozygous transgenic lines in these large, relatively long-lived plants.
Although the NAP:eGFP and NAP:dsRED transgenes described in this study have many advantages, they are still susceptible to the problems inherent in T-DNA transformation. Consistent with previous studies (Szabados et al. 2002; Alonso et al. 2003), the insertion sites were distributed in a nonrandom fashion. Some regions had relatively few T-DNA insertions, whereas others were T-DNA insertion hotspots. Although this property makes it difficult to obtain even coverage of the genome, it is not a serious impediment to the use of these insertions as genetic markers because, as we show in this article, intervals that lack transgenes can be marked by a flanking pair of transgenes of different colors. The fluorescence intensity of lines containing a single T-DNA insertion site also varied quite significantly. This could reflect the presence of variable numbers of T-DNAs at each site, or the position of these T-DNAs in the genome. Whatever its basis, this phenomenon demonstrates the difficulty of generating transgenic lines with predictable levels of transgene expression. Many T1 plants were semisterile. Consistent with the conclusions of a previous study (Clark and Krysan 2010), we found that in most cases the transgene in these lines was completely fertile when homozygous, suggesting that it was associated with either a reciprocal translocation or a large inversion. We try to select against such chromosomal rearrangements by prescreening T1 lines for semisterility, but this approach does not detect small inversions, which are nearly completely fertile even in heterozygous condition. Investigators interested in selecting for recombination in the vicinity of a specific transgene should be aware that some transgenes may be associated with a small inversion and guard against this possibility by using overlapping TLs. Finally, it should be emphasized that while we made an effort to select stably fluorescent lines, we have not propagated these lines for enough generations to predict their long-term stability; several TLs had to be discarded because one or both of the transgenes began to silence in advanced generations.
Some uses of traffic lines
TLs facilitate the use of segregating families for genetic analysis because seeds of a desired genotype can be accurately identified prior to planting. This is particularly important in the case of lethal or sterile mutations because these mutations must be propagated as heterozygotes; the ability to visually identify seeds heterozygous for these mutations in segregating families greatly reduces the effort required for stock maintenance. It is sometimes also important to identify plants homozygous for such mutations before the mutant phenotype becomes obvious (e.g., for gene expression studies); TLs make it possible to collect large numbers of homozygous mutant seeds from a segregating family in a few hours. TLs are also useful for analyses of mutations with weak or environmentally sensitive phenotypes, as well as for phenotypic evaluation of mutations that exist in heterogeneous genetic backgrounds. In Arabidopsis, mutations are typically maintained as homozygous lines and compared to an independently maintained homozygous line of a different genotype. This approach requires that the lines being compared are isogenic for the genes of interest and also assumes that the relevant phenotype is insensitive to maternal influence or seed age effects. However, unless the mutation has been backcrossed to the wild-type control multiple times, the mutant line will differ from wild type at numerous sites in the genome, making it difficult to obtain meaningful phenotypic information. One way to avoid this problem is to compare the phenotype of mutant and wild-type siblings in a segregating family. Siblings share the same maternal environment and, on average, have the same genetic background because second-site mutations unlinked to the mutation of interest will segregate randomly among both mutant and wild-type siblings. Although mutations closely linked to the mutation of interest will more often be homozygous in mutant than in wild-type plants, this approach is still a significant improvement over the use of independent mutant and wild-type lines for phenotypic analysis. TLs make it possible to identify seeds homozygous and heterozygous (or wild type) for a mutation of interest in a segregating family, so that the phenotypes of these genotypes can be compared.
TLs can also be used to detect and preserve the products of rare recombination events (Melamed-Bessudo et al. 2005; Shaked et al. 2006; Pecinka et al. 2011; Melamed-Bessudo and Levy 2012). This is particularly valuable for the creation of near-isogenic lines from mutagenized populations and for the analysis of quantitative trait loci (QTL). Newly identified mutations from a forward mutagenesis experiment must be backcrossed to the progenitor line several times to remove the second-site mutations present in the mutagenized line. Second-site mutations unlinked to the mutation-of-interest can be eliminated by five to six backcrosses. However, second-site mutations closely linked to the desired mutation are difficult to eliminate because crossing over in the vicinity of this mutation will be relatively rare. Furthermore, recombination in this region is difficult to detect because—with the exception of newly induced mutations—the mutagenized line is genetically identical to the progenitor line. To remove second-site mutations linked to a gene of interest, these mutations must be identified by sequencing the genome of the mutagenized line and then selected against in a backcross population using PCR primers specific for the second-site mutation. TLs make it possible to detect recombination in the regions flanking a gene of interest, so that these regions can be removed without the need for information about any mutations present in these regions. TLs can also be used to introgress small chromosome segments from different accessions or ecotypes into a common genetic background, which is important for QTL analysis or for the purpose of comparing the phenotypes of mutations isolated in different genetic backgrounds.
Unlike traditional balancer chromosomes—which require extensive engineering—a fluorescent marker-based system can be easily developed. Because all major model genetic systems are capable of being transformed, and many different colored fluorescent markers are currently available, it should be straightforward to generate doubly marked strains using promoters appropriate for the system of interest. In principle, the approach described here could be used in any system to facilitate the maintenance and manipulation of useful genotypes.
Supplementary Material
Acknowledgments
We are grateful to the members of the Poethig laboratory for helpful discussions and to Kriya Patel and Mitchelle Matesva for technical support. This project is funded by an National Science Foundation grant (MCB 1243754) to R.S.P.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173435/-/DC1
Communicating editor: J. O. Borevitz
Literature Cited
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., et al. , 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Alvarez J. P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., et al. , 2006. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K., Hawley R. S., 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Barton M. K., Poethig R. S., 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shootmeristemless mutant. Development 119: 823–831. [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2008. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 3: 41–50. [DOI] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2009. Visual markers for detecting gene conversion directly in the gametes of Arabidopsis thaliana. Methods Mol. Biol. 557: 99–114. [DOI] [PubMed] [Google Scholar]
- Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewski G. J., Lewis S. P., Glover L. W., Reineke J., Jones G., et al. , 2001. Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield S. P., Capron R., Severino A., Penttila P. A., Alfred S., et al. , 2013. Incipient stem cell niche conversion in tissue culture: using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordia into shoot meristems. Plant J. 73: 798–813. [DOI] [PubMed] [Google Scholar]
- Chuang C., Meyerowitz E., 2000. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., Krysan P. J., 2010. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 64: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Edgley, M. L., D. L. Baillie, D. L. Riddle, and A. M. Rose, 2006 Genetic balancers. WormBook /10.1895/wormbook.1.89.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Francis K. E., Lam S. Y., Harrison B. D., Bey A. L., Berchowitz L. E., et al. , 2007. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3913–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., et al. , 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Meinke D., 2010. The development of Arabidopsis as a model plant. Plant J. 61: 909–921. [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K., 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Lu C. F., Kang J. L., 2008. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 27: 273–278. [DOI] [PubMed] [Google Scholar]
- McElver J., Tzafrir I., Aux G., Rogers R., Ashby C., et al. , 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Levy A. A., 2012. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Yehuda E., Stuitje A. R., Levy A. A., 2005. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 43: 458–466. [DOI] [PubMed] [Google Scholar]
- O’Malley R. C., Alonso J. M., Kim C. J., Leisse T. J., Ecker J. R., 2007. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2: 2910–2917. [DOI] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A. A., Mittelsten Scheid O., 2011. Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol. 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D., 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Avivi-Ragolsky N., Levy A. A., 2006. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. L., Shimada T., Hara-Nishimura I., 2010. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Singer T., Fan Y., Chang H. S., Zhu T., Hazen S. P., et al. , 2006. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Koornneef M., 2002. A fortunate choice: the history of Arabidopsis as a model plant. Nat. Rev. Genet. 3: 883–889. [DOI] [PubMed] [Google Scholar]
- Sonoda Y., Yao S. G., Sako K., Sato T., Kato W., et al. , 2007. SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. Plant J. 50: 586–596. [DOI] [PubMed] [Google Scholar]
- Stuitje A. R., Verbree E. C., van der Linden K. H., Mietkiewska E. M., Nap J. P., et al. , 2003. Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol. J. 1: 301–309. [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J. D., et al. , 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Szabados L., Kovacs I., Oberschall A., Abraham E., Kerekes I., et al. , 2002. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 32: 233–242. [DOI] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D., 2010. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Bijl G. J., Liu W., Bisseling T., Schaart J. G., et al. , 2012. One-step Agrobacterium-mediated transformation of eight genes essential for Rhizobium symbiotic signaling using the novel binary vector system pHUGE. PLoS ONE 7: e47885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Ching Y. H., Farias M., Hartford S. A., Howell G., et al. , 2005. Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 15: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Sage M., Cai W. W., Thompson D. M., Tavsanli B. C., et al. , 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22: 375–378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.