Abstract
Interest in the study of marine sponges and their associated microbiome has increased both for ecological reasons and for their great biotechnological potential. In this work, heterotrophic bacteria associated with three specimens of the marine sponge Erylus deficiens, were isolated in pure culture, phylogenetically identified and screened for antimicrobial activity. The isolation of bacteria after an enrichment treatment in heterotrophic medium revealed diversity in bacterial composition with only Pseudoalteromonas being shared by two specimens. Of the 83 selected isolates, 58% belong to Proteobacteria, 23% to Actinobacteria and 19% to Firmicutes. Diffusion agar assays for bioactivity screening against four bacterial strains and one yeast, revealed that a high number of the isolated bacteria (68.7%) were active, particularly against Candida albicans and Vibrio anguillarum. Pseudoalteromonas, Microbacterium, and Proteus were the most bioactive genera. After this preliminary screening, the bioactive strains were further evaluated in liquid assays against C. albicans, Bacillus subtilis and Escherichia coli. Filtered culture medium and acetone extracts from three and 5 days-old cultures were assayed. High antifungal activity against C. albicans in both aqueous and acetone extracts as well as absence of activity against B. subtilis were confirmed. Higher levels of activity were obtained with the aqueous extracts when compared to the acetone extracts and differences were also observed between the 3 and 5 day-old extracts. Furthermore, a low number of active strains was observed against E. coli. Potential presence of type-I polyketide synthases (PKS-I) and non-ribosomal peptide synthetases (NRPSs) genes were detected in 17 and 30 isolates, respectively. The high levels of bioactivity and the likely presence of associated genes suggest that Erylus deficiens bacteria are potential sources of novel marine bioactive compounds.
Keywords: sponge associated heterotrophic bacteria, Erylus deficiens, bioactivity screening assay, PKS-I and NRPS genes, Candida albicans
Introduction
Marine sponges (Phylum Porifera) are sessile filter-feeding organisms and represent a significant component of benthic communities' worldwide (Sarà and Vacelet, 1973). They harbor a large community of diverse microorganisms, representing up to 50–60% of the sponge biomass (Thomas et al., 2010), which develop symbiotic relationships essential to their biology. Sponges have been the focus of many studies as they have proved to be a rich source of biologically active secondary metabolites. These compounds may serve as a defense strategy to escape from predators, thus playing a crucial role in sponge survival in the marine ecosystem (Thakur and Müller, 2004). Since the discovery of spongothymidine and spongouridine in the early 50's (Bergmann and Feeney, 1950, 1951), a great number of biologically active compounds were isolated from marine sponges and their associated microorganisms (Taylor et al., 2007; Thomas et al., 2010). About 300 novel compounds were reported in 2011 from the phylum Porifera (Blunt et al., 2013) and more sponge-derived compounds are subjected to clinical and preclinical trials than from any other marine phylum (Martins et al., 2014). The chemical diversity of sponge-derived products is remarkable and their biological activity ranges from anti-inflammatory, antitumour, immuno- or neurosuppressive, antiviral, antimalarial, antibiotic to antifouling (Imhoff et al., 2011). However, many of the bioactive compounds found in sponges are, in fact, produced by their associated microbial communities (Wang, 2006; Khan et al., 2014). This is the case, for instance, of the peptide thiocoraline, produced by a sponge-associated Micromonospora species (Romero et al., 1997) and the cytotoxic and antibacterial tetrabromodiphenyl ethers produced by a Vibrio spp. associated with the sponge Dysidea sp. (Elyakov et al., 1991). This microbial community represents a treasure trove of novel molecules for marine biotechnology. Due to the increasing resistance of bacteria against the common antibiotics there is still a pressing need to find new drugs (Bull and Stach, 2007; Kumarasamy et al., 2010). By the end of 2008, about 3000 natural products had been identified from marine microorganisms (Laatsch, 2008). The most relevant phyla of bacterial producers of new compounds are Actinobacteria (40%), Cyanobacteria (33%), Proteobacteria (12%), and Firmicutes and Bacteroidetes (5% each) (Williams, 2009). In addition to these, archaea, fungi, unicellular algae and other bacterial phyla namely Acidobacteria, Chloroflexi, Deinococcus-Thermus, Firmicutes, Gemmatimonadetes, Nitrospira, Planctomycetes, Poribacteria, Spirochaetes, and Verrucomicrobia, are also present in the sponge microbial community as revealed mainly by culture-independent molecular methods (Taylor et al., 2007).
In a genomic era, it is easy to assess by molecular methods the bacterial potential for the production of bioactive molecules. These natural bioactive secondary metabolites can be the result of the activity of non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) that are large multi-modular, multi-domain enzymes. The minimal set of domains that have to be present in these biosynthetic pathways are ketosynthase (KS), acyltransferase (AT) and acyl carrier proteins in PKSs (Khan et al., 2014) and adenylation (A), condensation (C) and peptidyl carrier proteins (PCPs) for peptide elongation in NRPSs (Jenke-Kodama and Dittmann, 2009). In addition to the synthesis of these defined domains, optional domains can also be produced like ketoreductase. The intra and inter variability of the enzymatic modules codified by PKS and NRPS genes results in the production of diverse bioactive compounds. Antibiotic molecules such erythromycin, tetracycline and glycopeptides of the vancomycin family were reported to have origin in PKS and NRPS productions (Fischbach and Walsh, 2006). In fact, many of the complex polyketides isolated from sponges are the most promising compounds with pharmacological application (Hochmuth and Piel, 2009). Therefore, the search for the genes responsible for the expression of these enzymes is indicative of the biotechnological potential of a specific organism.
The marine sponge genus Erylus, Gray, 1867, contains approximately 70 species and is known to inhabit the Mediterranean Sea, and the Atlantic and Pacific Oceans (Adams and Hooper, 2001). Species of this genus have been reported to possess characteristic bioactive glycosides named, according to their aglycone skeleton, erylosides or erylusamines (Kalinin et al., 2012). Some of these glycolipids have been shown to inhibit tumor cells, fungi, bacteria, viruses including the human immunodeficiency virus (HIV), neuraminidase, interleukin-6, thrombin receptor antagonist, and in vitro human platelet aggregation (Carmely et al., 1989; Fusetani et al., 1993; Gulavita et al., 1994; Sata et al., 1994; Stead et al., 2000; Kubanek and Fenical, 2001; Shin et al., 2001; Takada et al., 2002; Van Altena et al., 2003; Fouad et al., 2004; Sandler et al., 2005; Okada et al., 2006; Afiyatullov et al., 2007; Antonov et al., 2007). Furthermore, Erylus spp. are well known producers of oligoglycosides with antifeedant activity involved in chemical defenses of sponges against predatory fish (Kubanek et al., 2000; Kubanek and Fenical, 2001; Antonov et al., 2011) and with larvicidal activity (Carballeira and Negrón, 1991). Crude extracts of Erylus deficiens from the Portuguese coast were found to possess indoleamine 2,3-dioxygenase (IDO) inhibitory activity (Gaspar et al., 2012). This enzyme plays a fundamental role in the kynurenine pathway, one of the major tryptophan catabolism routes with implications in cancer and in CNS disorders like Alzheimer's disease and cerebral malaria (Chen and Guillemin, 2009). The bioactive potential of bacteria associated with Erylus discophorus was evidenced by Graça et al. (2013). The work showed that 31% of the isolated bacteria were able to produce antimicrobial metabolites being this the only study performed with bacteria associated with the Erylus sponges. This lack of results associated with the low yield of metabolites normally originated from marine sponges, together with the importance of obtaining new bioactive bacteria to avoid the undesired exploitation of natural sponges, lead us to assess in three specimens of E. deficiens collected in the Gorringe Bank, an isolated seamount in the Atlantic, the potential of heterotrophic culturable bacteria as actual producers of antimicrobial compounds by screening and molecular assays. As glycolipids from these sponges are already being studied by others, NRPS and PKS appeared as good gene targets for the search of new potential bioactivities.
Materials and methods
Sponge sampling and identification
Three specimens of E. deficiens Topsent, 1927 (Demospongiae, Astrophorida, Geodiidae) were collected by scuba diving at approximately 38 m depth on Gettysburg (specimens #66 and #91—lat. 36° 31′ 10″ N, long. 11° 34′ 10″ W) and Ormonde (specimen #118—lat. 36° 42′ 70″ N, 11° 09′ 70″ W) Peaks, in the course of LusoExpedição Olympus 2008 to the Gorringe Bank, a large seamount located 150 Km off the southwest coast of Portugal. The samples were placed individually in ziploc® plastic bags and then carried to the laboratory for identification and microbial isolation. Voucher samples were preserved in 90% ethanol for taxonomic identification and deposited in the Biology Department's zoological collection of the University of the Azores (collection DBUA.Por). Specimens were identified using general external and internal morphological characters analysis, i.e., shape, type, size and arrangement of skeletal structures (spicules) following the Systema Porifera classification system (Hooper and Van Soest, 2002). This species was previously reported in this seamount and identified as Erylus sp. (Xavier and Van Soest, 2007) and further examination of additional samples, including specimens from Madeira Island, the type-locality of E. deficiens, enabled its identification to species level.
Heterotrophic bacterial isolation
Fragments of sponge tissues were placed under aseptic conditions in sterile Erlenmeyers containing M514 medium (Bacto-peptone (Difco) 5 g/L; yeast Extract (Oxoid) 1 g/L and sea salts (Red Sea®) 36 g/L). This enrichment was performed to promote faster growing and abundant heterotrophic bacteria. When noticeable bacterial growth occurred, usually after 1 week without stirring, several dilutions (10−1, 10−6, and 10−12) were prepared and, 100 μL of each, spread in several solid marine media prepared with 0.2 μm filtered natural seawater and 1.8% agar: modified M13 (Lage and Bondoso, 2011), PYGV (0.025% peptone, 0.025% yeast extract, 0.025% glucose, 10 mL.L−1 vitamin solution n°6, 20 mL.L−1 Hutner's basal salts (Cohen-Bazire et al., 1957), Marine Agar (MA - Difco), M17 (Difco), Glycerol Asparagine (1% glycerol, 0.1% L-asparagine, 0.1% K2HPO4), M4 (Zhang et al., 2006). Cultures were incubated in the dark at 26°C. Growth was monitored daily and distinct colony morphotypes were transferred to fresh medium for isolation. Pure bacterial cultures were cryopreserved in seawater supplemented with 20% glycerol at −80°C.
Molecular identification of isolates
Isolates were identified by 16S rRNA gene sequence analysis after amplification with the primers 27f and 1492r (Lane, 1991). The PCR was carried out as described by Bondoso et al. (2011), using, as template, material from a single colony or genomic DNA extracted with the Bacterial DNA Isolation Kit (Omega), according to the manufacturer' instructions. PCR products were directly sequenced at Macrogen Europe (www.Macrogen.com).
The obtained 16S rRNA gene sequences were processed with the Sequencing Analysis 5.2 (Applied Biosystems) and assembled with Vector NTI Advance™ 10.3. The resulting 16S rRNA gene sequences were then blasted on SeqMatch (Ribossomal Database Project) (Cole et al., 2014) against the nucleotide database of GenBank (NCBI) and the closest neighbors were downloaded. The sequences were corrected manually using the alignment generated. Subsquently, sequences with 818 bp were used for molecular evolutionary analyses in MEGA version 5.2 (Tamura et al., 2011). A final maximum likelihood (ML) phylogenetic tree was generated applying General Time Reversible model and Gamma distributed with Invariant sites (G+I). Validation of reproducibility of the branching patterns was made by bootstrap based on 1000 resamplings. Pairwise sequence similarities based on Jukes-Cantor model were calculated in MEGA. Alignment gaps and missing data were not included in the calculations. Different phylotypes were considered based on a 97% 16S rRNA gene threshold (Stackebrandt and Goebel, 1994). The sequences of the isolated bacteria used in this study were deposited in GenBank with the serial accession numbers KP120772- KP120854.
Bioactivity of the bacterial isolates
To assess the antimicrobial bioactivity of the sponge-isolated bacteria, designated hereunder as test bacteria, a simple co-culture agar diffusion assay (pre-screening) was performed against a panel of target organisms. This panel was constituted by Gram negative and positive bacteria commonly used in bioactivity bioassays, a relevant environmental species and a human pathogenic yeast. Thus, the bioactivity against four bacterial strains (Bacillus subtilis ATCC 6633, B. cereus FCUP collection, Escherichia coli ATCC 25922 and the fish pathogen Vibrio anguillarum FCUP collection) and a yeast (clinical isolate of Candida albicans) was evaluated. The agar diffusion assay was performed in MA medium, solidified with 1.4% agar as this medium allowed the simultaneous growth of the test bacteria and the target microorganisms. The test bacteria were heavily inoculated on half a Petri dish containing MA medium and incubated vertically to favor the diffusion of the metabolites to the non-inoculated part of the medium, at 26°C for 48 h. By this time, the fast growing test bacteria attained the stationary growth phase. Subsequently, the five target microorganisms were inoculated on the other half by streaking a line perpendicularly to the test bacteria and parallel to each other. The cultures were grown at 37 °C in the same position as described before. The presence of growth inhibition was observed after 24 h. Target organisms grown in MA medium were used as controls.
To confirm the positive results obtained with the preliminary assay, a liquid screening assay was performed based on Graça et al. (2013). Of the 57 bacteria that showed antimicrobial activity only 48 were assayed as nine lost viability (#91_37, #91_40, #91_43, #91_45, #118_14, #118_23, #118_27, #118_33, and #118_39). The bacteria were grown in MB for 3 and 5 days at 25°C and 200 r.p.m. At days 3 and 5, an aliquot of 2 mL of the culture was centrifuged and the supernatant was 0.22 μm filtered and stored at −20°C—aqueous extract. Another aliquot of 2 mL of culture was mixed with 1.8 mL acetone +0.2 mL DMSO, incubated for 1 h after which the upper phase was collected and stored at −20°C—acetone extract. In 96-well plates, duplicates of 10 μL of each extract were assayed against 90 μL of each target microorganism cultivated in LB medium (≈ 2.5 × 105 CFUs/mL). Additionally, 100 μL of each target microorganism were cultivated to allow the assessment of the total growth of the microorganisms. As positive controls, amphotericin B (0.19; 0.39; 0.78; and 1.56 μg/ml) and rifampicin (62.5; 125; 250; and 500 mg/ml) were used against C. albicans and E. coli, respectively. LB medium was used as negative control. In order to assess the possible interference of the solvents acetone+DMSO present in the extracts, both C. albicans and E. coli were grown in the presence of the same concentration used for the bioactivity assays. As no significant variation was induced by the solvents used to obtain the extracts, these values will not be considered in the calculation of bioactivities. The plates were incubated at 37°C for 24 h at 250 r.p.m. The optical density (OD) of the cultures was measured at 600 nm in a Multiskan GO plate reader (Thermo Scientific). The growth of the target microorganisms (%) was calculated by the following equation: (ODcultures with extract – ODLB medium)/ODpositive control × 100. Variability in the positive controls was of about 10%, reason why only growth inhibitions of equal or more than 25% were considered.
Search for PKS-I and NRPS genes
The presence of genes involved in the production of secondary metabolites was screened in all the isolates obtained. The degenerate primers MDPQQRf (5′-RTRGAYCCNCAGCAICG-3′) and HGTGTr (5′-VGTNCCNGTGCCRTG-3′) (Kim et al., 2005) were used to amplify the β-ketosynthase (KS) domain fragment within the Type I polyketide synthase genes PKS-I. For the amplification of NRPS, primers MTF2 [5′- GCNGG(C/T)GG(C/T)GCNTA(C/T)GTNCC-3′(AGGAYVP, core motif I)] and MTR [5′- CCNCG(AGT)AT(TC)TTNAC(T/C)TG-3′(QVKIRG, core motif V)] (Neilan et al., 1999) were used. The PCR mixture contained 1x PCR buffer, 0.8 units of Go Taq DNA Polymerase (Promega), 0.2 mM of each dNTPs, 0.1 μM of each primer and 1 μL genomic DNA as template. The PCR profile consisted of an initial denaturing step of 5 min at 95°C, 11 cycles of 1 min at 95°C, 30 s at 60°C and 1 min at 72°C, with the annealing temperature reduced by 2°C per cycle, followed by 30 cycles of 95°C for 1 min, 40°C for 30 s and 72°C for 1 min with a final extension of 10 min at 72°C (Kim et al., 2005). The PCR was carried out in a MyCycler™ Thermo Cycler (Bio-Rad) and the amplicons were visualized in a Roti-Safe (Roth) stained 1.2% agarose gel.
Results
16S rRNA gene analysis identification
A total of 83 isolates were selected after enrichment procedure from the three specimens of E. deficiens. All assayed media allowed the isolation of bacterial strains although marine agar was the medium that provided the highest number of isolates and broad diversity from the three sponges. Curiously, M17 medium that is designed to grow fastidious lactic streptococci, allowed the growth of several Firmicutes and Actinobacteria. The majority of the strains obtained in this study after the enrichment belong to Proteobacteria (58%), followed by Actinobacteria (23%) and Firmicutes (19%) (Table 1; Figure 1). Within the Proteobacteria, the most abundant class was the Gammaproteobacteria with 46 isolates and the genus Pseudoalteromonas (27 isolates). Within the Actinobacteria, the genus Microbacterium was represented by 13 isolates. Of the selected strains and based on a 16S rRNA gene 97% threshold, 10 different phylotypes were from sponge #91, five phylotypes from sponge #118 and only one phylotype from sponge #66.
Table 1.
Affiliation, bioactivity and presence of the PKS-I and NRPS genes of E. deficiens bacterial isolates.
| Isolate | Similaritya | Related straina | Phylum/Class | Genera | Activities in solid assays | Activities in liquid assays | PKS-I | NRPS |
|---|---|---|---|---|---|---|---|---|
| #91_43 | 99.8 | Cellulomonas sp. R-32740; AM403591 | Actinobacteria | Cellulomonas | CA, VA | ND | ND | ND |
| #91_17 | 98.9 | Dermacoccus sp. Ellin185; AF409027 | Actinobacteria | Dermacoccus | CA, VA | N | ND | D |
| #91_34 | 100 | Microbacterium esteraromaticum; 2122; EU714337 | Actinobacteria | Microbacterium | CA | CA | ND | ND |
| #91_42 | 99.9 | Microbacterium foliorum; 150; EU714333 | Actinobacteria | Microbacterium | N | D | ND | |
| #91_19.1 | 99.8 | Microbacterium foliorum; 720; EU714376 | Actinobacteria | Microbacterium | N | ND | D | |
| #91_29 | 99.9 | Microbacterium foliorum; Bjc15-C14; JX464206 | Actinobacteria | Microbacterium | CA | CA | ND | ND |
| #91_36.2 | 99.2 | Microbacterium foliorum; BJC15-C1; JX401513 | Actinobacteria | Microbacterium | CA, VA | CA | ND | ND |
| #91_37 | 100 | Microbacterium foliorum; BJC15-C1; JX401513 | Actinobacteria | Microbacterium | CA, BS, VA | ND | ND | ND |
| #91_40 | 99.4 | Microbacterium foliorum; Bjc15-C14; JX464206 | Actinobacteria | Microbacterium | CA, VA | ND | D | ND |
| #91_41 | 99.4 | Microbacterium foliorum; S2-157; JQ660074 | Actinobacteria | Microbacterium | N | ND | ND | |
| #91_31 | 99.9 | Microbacterium hydrocarbonoxydans; 3084; EU714352 | Actinobacteria | Microbacterium | CA | CA | ND | ND |
| #91_38 | 99.7 | Microbacterium hydrocarbonoxydans; 3517; EU714368 | Actinobacteria | Microbacterium | N | D | ND | |
| #91_45 | 99.7 | Microbacterium hydrocarbonoxydans; 3517; EU714368 | Actinobacteria | Microbacterium | CA | ND | ND | ND |
| #91_47 | 99.7 | Microbacterium hydrocarbonoxydans; HR73; JF700446 | Actinobacteria | Microbacterium | N | ND | ND | |
| #91_35 | 99.8 | Microbacterium phyllosphaerae (T); DSM 13468; P 369/06; AJ277840 | Actinobacteria | Microbacterium | CA, BS, VA | CA | D | ND |
| #91_20 | 97.0 | Rhodococcus hoagii; CUB1156; AJ272469 | Actinobacteria | Rhodococcus | N | ND | ND | |
| #91_36.1 | 100 | Rhodococcus equi; type strain: DSM20307; X80614 | Actinobacteria | Rhodococcus | EC, CA, VA | N | D | D |
| #91_48 | 99.9 | Rhodococcus hoagii; type strain: DSM20307; X80614 | Actinobacteria | Rhodococcus | N | D | ND | |
| #91_54 | 99.9 | Rhodococcus qingshengii; KUDC1814; KC355321 | Actinobacteria | Rhodococcus | CA, VA | CA | ND | D |
| #91_51 | 99.9 | Brevundimonas vesicularis (T); LMG 2350 (T); AJ227780 | Alphaproteobacteria | Brevundimonas | N | ND | D | |
| #91_16 | 99.0 | Paracoccus sphaerophysae strain Zy-3; NR117441 | Alphaproteobacteria | Paracoccus | CA, EC | CA | ND | D |
| #118_42 | 100 | Bacillus amyloliquefaciens; BAC3048; HM355639 | Firmicutes | Bacillus | N | ND | D | |
| #118_43 | 100 | Bacillus amyloliquefaciens; BAC3048; HM355639 | Firmicutes | Bacillus | N | ND | D | |
| #91_46 | 100 | Bacillus aryabhattai; D27; FR750269 | Firmicutes | Bacillus | N | ND | ND | |
| #91_19.2 | 100 | Bacillus flexus; FJ584305 | Firmicutes | Bacillus | N | ND | ND | |
| #91_39 | 100 | Bacillus flexus; FJ584305 | Firmicutes | Bacillus | CA, VA | CA | ND | ND |
| #91_53 | 99.8 | Bacillus sp. CCBAU 10722; EF377323 | Firmicutes | Bacillus | CA, VA | N | ND | ND |
| #91_28 | 99.9 | Bacillus sp. G2DM-19; DQ416802 | Firmicutes | Bacillus | N | ND | ND | |
| #91_8 | 99.7 | Bacillus sp. PEB04; GU213160 | Firmicutes | Bacillus | CA | CA | ND | ND |
| #118_1 | 100 | Enterococcus sp. T004_8; JQ739635 | Firmicutes | Enterococcus | N | ND | D | |
| #118_4 | 99.8 | Enterococcus faecalis; AF039902 | Firmicutes | Enterococcus | VA | CA | ND | D |
| #118_3 | 99.9 | Enterococcus faecalis; GIMC503:NBS-2; JF728295 | Firmicutes | Enterococcus | CA, VA | CA | ND | D |
| #118_19 | 99.9 | Enterococcus faecalis; Y18293 | Firmicutes | Enterococcus | EC, CA, VA | CA | ND | D |
| #118_7 | 100 | Enterococcus faecalis; ZZ18; HM776212 | Firmicutes | Enterococcus | VA | CA | ND | ND |
| #91_5 | 99.8 | Paenibacillus glucanolyticus; KSI 1323; KC113143 | Firmicutes | Paenibacillus | N | ND | D | |
| #91_7 | 99.8 | Paenibacillus sp. JAM-FM32; AB526335 | Firmicutes | Paenibacillus | N | ND | D | |
| #91_24 | 100 | Paenibacillus sp. JAM-FM32; AB526335 | Firmicutes | Paenibacillus | CA, VA | CA | ND | D |
| #118_24 | 98.2 | Citrobacter koseri ATCC BAA-895; CP000822 | Gammaproteobacteria | Citrobacter | N | ND | D | |
| #118_38 | 99.7 | Citrobacter koseri ATCC BAA-895; CP000822 | Gammaproteobacteria | Citrobacter | N | ND | ND | |
| #118_39 | 95.0 | Citrobacter koseri ATCC BAA-895; CP000822 | Gammaproteobacteria | Citrobacter | CA | ND | ND | ND |
| #118_20 | 99.7 | bacterium NLAE-zl-H279; JX006497 | Gammaproteobacteria | Proteus | EC, CA | CA | ND | ND |
| #118_13 | 99.9 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | CA | EC, CA | ND | D |
| #118_14 | 99.6 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | EC, CA, VA | ND | ND | — |
| #118_23 | 99.7 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | CA | ND | ND | ND |
| #118_27 | 100 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | CA | ND | ND | ND |
| #118_30 | 100 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | N | ND | ND | |
| #118_34 | 100 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | N | ND | ND | |
| #118_40 | 99.9 | Proteus mirabilis; ATCC 29906T; AF008582 | Gammaproteobacteria | Proteus | N | ND | ND | |
| #118_5 | 99.7 | Proteus mirabilis; IFS10; AB272366 | Gammaproteobacteria | Proteus | CA | CA | ND | ND |
| #118_25 | 99.8 | Proteus mirabilis; LH-52; JN861767 | Gammaproteobacteria | Proteus | N | ND | ND | |
| #118_33 | 99.3 | Proteus mirabilis; LH-52; JN861767 | Gammaproteobacteria | Proteus | CA | ND | D | ND |
| #66_2 | 97.9 | Bacterium K2-82; AY345483 | Gammaproteobacteria | Pseudoalteromonas | N | D | ND | |
| #91_3 | 99.9 | Bacterium Antarctica-11; EF667985 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | ND | ND |
| #91_9 | 99.9 | Bacterium Antarctica-11; EF667985 | Gammaproteobacteria | Pseudoalteromonas | CA | CA | ND | ND |
| #91_12 | 99.9 | Bacterium Antarctica-11; EF667985 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | D | ND |
| #91_13 | 99.9 | Bacterium Antarctica-11; EF667985 | Gammaproteobacteria | Pseudoalteromonas | EC, CA, VA | CA | ND | ND |
| #91_23 | 99.9 | Bacterium Antarctica-11; EF667985 | Gammaproteobacteria | Pseudoalteromonas | CA | CA | ND | ND |
| #66_11 | 99.3 | Pseudoalteromonas issachenkonii (T); KMM 3549; F13; AF316144 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | EC, CA | D | D |
| #91_22 | 98.6 | Pseudoalteromonas sp. 12; DQ642815 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | ND | ND |
| #66_17 | 100 | Pseudoalteromonas sp. 8007; AM111068 | Gammaproteobacteria | Pseudoalteromonas | EC, CA, VA | CA | D | D |
| #66_6 | 99.8 | Pseudoalteromonas sp.; AR0307; AB019947 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | EC, CA | ND | ND |
| #66_7 | 99.8 | Pseudoalteromonas sp.; AR0307; AB019947 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | ND | ND |
| #66_16 | 99.9 | Pseudoalteromonas sp.; AR0307; AB019947 | Gammaproteobacteria | Pseudoalteromonas | N | D | D | |
| #91_10.1 | 99.9 | Pseudoalteromonas sp.; AR0307; AB019947 | Gammaproteobacteria | Pseudoalteromonas | CA | EC, CA | ND | ND |
| #91_10.2 | 99.9 | Pseudoalteromonas sp.; AR0307; AB019947 | Gammaproteobacteria | Pseudoalteromonas | EC, CA, VA | CA | ND | ND |
| #66_10 | 98.6 | Pseudoalteromonas sp. ARCTIC-P16; AY573035 | Gammaproteobacteria | Pseudoalteromonas | VA | CA | D | D |
| #66_1 | 96.4 | Pseudoalteromonas sp. BSi20325; DQ520886 | Gammaproteobacteria | Pseudoalteromonas | CA | N | ND | ND |
| #66_8 | 99.6 | Pseudoalteromonas sp. BSi20325; DQ520886 | Gammaproteobacteria | Pseudoalteromonas | VA | CA | ND | D |
| #66_18 | 97.9 | Pseudoalteromonas sp. BSi20325; DQ520886 | Gammaproteobacteria | Pseudoalteromonas | N | ND | D | |
| #91_26 | 98.0 | Pseudoalteromonas sp. P1; EF627987 | Gammaproteobacteria | Pseudoalteromonas | EC | CA | ND | ND |
| #66_20 | 98.9 | Pseudoalteromonas sp. SM9913; CP001796 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | EC, CA | D | D |
| #66_13 | 100 | Pseudoalteromonas sp. YASM-1; DQ173038 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | D | D |
| #91_21 | 97.0 | Pseudoalteromonas tetraodonis; Do-17; AB257325 | Gammaproteobacteria | Pseudoalteromonas | CA | CA | ND | ND |
| #66_12 | 98.4 | Pseudoalteromonas tetraodonis; SSA930; KC534452 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | D | D |
| #66_3 | 97.5 | Pseudoalteromonas sp. LOB-15; DQ412067 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | CA | D | ND |
| #91_11 | 95.2 | Uncultured bacterium; 11A-6; FJ998316 | Gammaproteobacteria | Pseudoalteromonas | CA | CA | ND | ND |
| #91_27 | 96.6 | Uncultured Pseudoalteromonas sp.; DVBSW_J357; KF722314 | Gammaproteobacteria | Pseudoalteromonas | CA | EC | ND | ND |
| #66_9 | 98.9 | Uncultured Pseudoalteromonas sp.; CI66; FJ695586 | Gammaproteobacteria | Pseudoalteromonas | CA, VA | EC, CA | ND | D |
| #91_50 | 100 | Pseudomonas oryzihabitans; SFK5; KC335294 | Gammaproteobacteria | Pseudomonas | CA, VA | EC, CA | ND | ND |
| #118_11 | 100 | Psychrobacter celer; B_IV_3L25; JF710994 | Gammaproteobacteria | Psychrobacter | VA | CA | ND | D |
| #118_2 | 99.8 | Psychrobacter celer; 91; JF710993 | Gammaproteobacteria | Psychrobacter | VA | CA | ND | D |
| #118_17 | 99.9 | Psychrobacter celer; 91; JF710993 | Gammaproteobacteria | Psychrobacter | EC, CA, VA | CA | ND | ND |
| #118_18 | 99.9 | Psychrobacter sp. 2CpBB14; JN602232 | Gammaproteobacteria | Psychrobacter | N | ND | D |
CA, Candida albicans; EC, Escherichia coli; VA, Vibrio anguillarum; BS, Bacillus subtilis; N, no activity; D, detected; ND, not detected.
Based on SeqMatch (Ribossomal Database Project) (Cole et al., 2014).
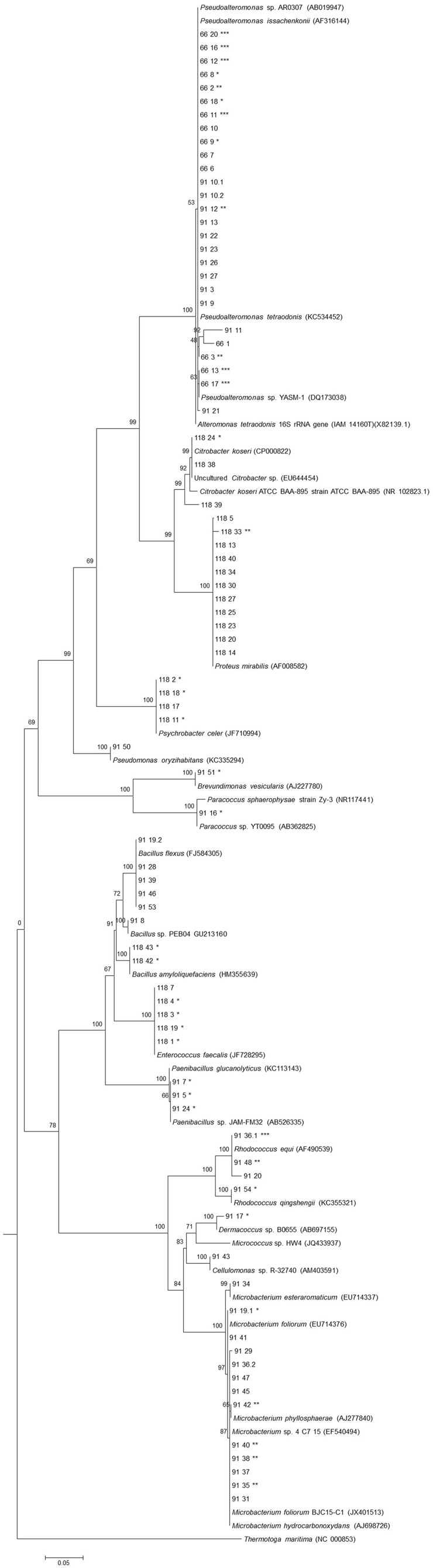
Figure 1.
Phylogenetic 16S rRNA gene tree generated by maximum-likelihood analysis based in General Time Reversible model and Gamma distributed with Invariant sites (G+I) indicating the relationship of the heterotrophic bacteria isolated from three E. deficiens specimens. Thermatoga maritima was used as outgroup. Bar – 0.05 substitutions per 100 nucleotides. *Presence of NRPS genes; **Presence of PKS-I genes; ***Presence of both genes.
Bioactivity of the bacterial isolates
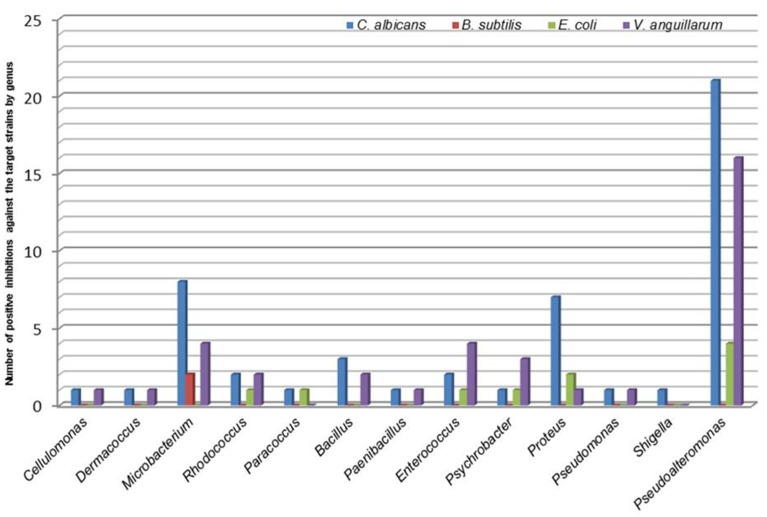
Through the co-culture diffusion pre-screening assay, 68.7% (57 bacteria) of the isolated bacteria showed antimicrobial activity against one or more of the target microorganisms tested. The highest number of positive hits were found in Gammaproteobacteria (63.2%) and Actinobacteria (21.1%) followed by Firmicutes (14.0%) and Alphaproteobacteria (1.8%). Of the thirteen genera showing antimicrobial bioactivity, Pseudoalteromonas (42.1%), Microbacterium (14.0%) and Proteus (12.3%) presented the highest positive hits. The analysis of bioactivity by test bacteria (Table 1) revealed a high number of strains with antifungal activity against C. albicans (87.7%—50 bacteria) and antibiotic against V. anguillarum (63.2%—36 bacteria) and, in lower number, against E. coli (17.5%—10 bacteria) and B. subtilis (3.5%—2 bacteria). None of the isolates produced effective bioactive compounds against the Gram positive B. cereus. Some bioactive bacteria were able to produce secondary bioactive metabolites against two or more target microorganisms. Microbacterium was the only genus able to produce bioactive compounds effective against B. subtilis (Figure 2).
Figure 2.
Relation between the hits of bioactive genera and the target organisms studied in the preliminary screening.
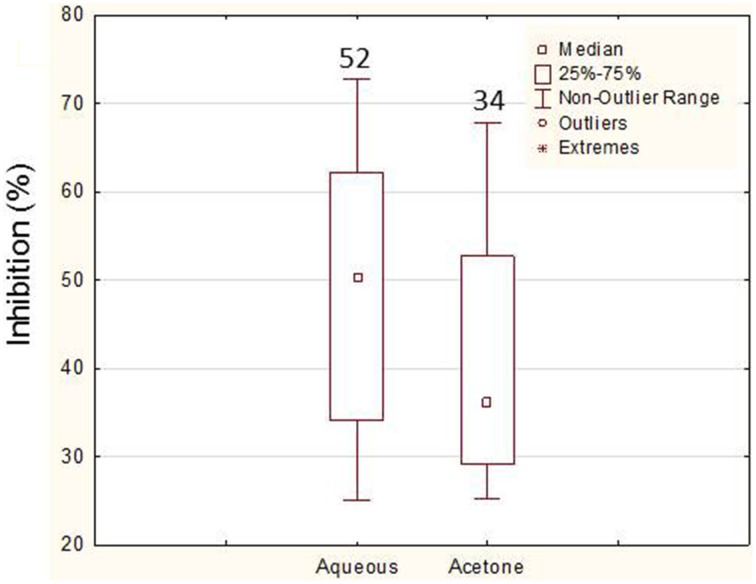
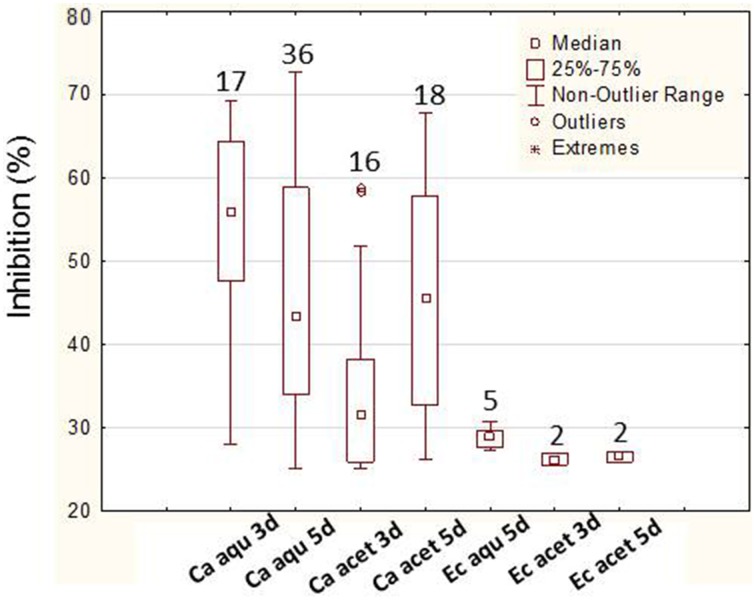
In order to confirm the positive results obtained in the preliminary assay, a liquid screening assay was performed which allowed a more precise evaluation of the bioactivities. In this screening, 92% (45 bacteria) of the bacteria assayed showed bioactivity. Again, more bacteria showed bioactivity against C. albicans (80%—36 bacteria), only 4% (2 bacteria) showed activity against E. coli and 16% (7 bacteria) against both. A total of 86.6% bacteria (n = 39) showed activity in their aqueous extracts (87% against C. albicans and 13% against both C. albicans and E. coli) while only 62.2% bacteria (n = 28) showed bioactivity in acetone extracts. The levels of inhibition obtained (maximum value and median) with the aqueous extracts (3 and 5 days) were higher than the ones with acetone (Figure 3). The analysis of the bioactive extracts also showed higher levels of inhibition against C. albicans than against E. coli in which inhibition levels were very low (Figure 4). Furthermore, it was possible to verify in the assays against C. albicans that the 3 days acetone extracts were less effective than the aqueous and 5 days extracts. Although the median of 5 days aqueous extracts was not so high comparatively to the 3 days aqueous or the 5 days acetone extracts, the maximum value as well as the number of bioactive extracts were higher. No activity was obtained with 3 days aqueous extracts against E. coli.
Figure 3.
Comparison of the levels of inhibition (higher than 25%) between the aqueous and the acetone extracts from Erylus deficiens isolates in the liquid assays. The numbers above each box represent the number of bioactive extracts.
Figure 4.
Levels of inhibition (higher than 25%) of 3 and 5 days and aqueous (aqu) and acetone (acet) extracts from Erylus deficiens isolates against C. albicans (Ca) and E. coli (Ec). No result is shown for aqueous, 3 days extracts against E. coli due to the absence of inhibition. The numbers above each box represent the number of bioactive extracts.
Search for PKS-I and NRPS genes
The β-ketosynthase domain fragment within the Type I polyketide synthase genes PKS-I was potentially amplified from 17 E. deficiens isolates (Table 1). Ten bacteria phylogenetically affiliated to Pseudoalteromonas from sponges #91 and #66, four to Microbacterium and two to Rhodococcus from sponge #91 and one to Proteus from sponge #118 amplified probable PKS-I sequences. The potential amplification of the motifs I and V of non-peptide synthetase adenylate-forming enzymes was observed in thirty strains (Table 1)—10 Pseudoalteromonas from sponge #66, four Enterococcus from sponge #118, three Paenibacillus from sponge #91, three Psycrobacter from sponge #118, two Bacillus from sponge #118, two Rhodococcus from sponge #91 and one Dermacoccus, one Microbacterium, one Brevundimonas and one Paracoccus, from sponge #91 and one Citrobacter and Proteus from sponge #118. A relatively high number of the bioactive isolates amplified for the searched genes and some for both genes. However, other genes should be behind the detected bioactivity in non-amplifying isolates.
Discussion
Since one of the most effective methods used in the discovery of natural products is the cultivation of new microbial strains which may represent novel important chemotypes (Fenical and Jensen, 2006; Haber and Ilan, 2013), our prime goal was to obtain bacterial isolates for screening antimicrobial activities.
The isolation of bacterial strains from E. deficiens after an enrichment procedure allowed the selection of a high number of fast growing heterotrophic bacteria, mainly belonging to Proteobacteria. Similarly, Kamke et al. (2010) also observed that one of the most abundant taxon in both libraries of Ancorina alata and Polymastia sp. was the class Gammaproteobacteria. Proteobacteria was the most abundant phylum in the phylogenetic distribution of sponge-isolated bacteria (Webster and Taylor, 2012) namely from another Erylus species, E. discophorus (Graça et al., 2013). Furthermore, species of Firmicutes and Actinobacteria, both phyla commonly present in sponges (Webster and Taylor, 2012), were also obtained.
Almost no bacterium was common among the three specimens as evidenced by the analysis of the 16S rRNA gene sequences (Figure 1). A possible explanation for this result may be the use of sponge portions from different structural parts of the sponge body that are known to be inhabited by different microbial communities (Thiel et al., 2006). The group of Pseudoalteromonas was the only one shared by specimens #91 and #66 which could be explained by sponges sampling site's proximity. This genus is often isolated from marine macroorganisms like sponges (Ivanova et al., 2002; Lau et al., 2005).
Several of the bioactive genera isolated in this study have been described as possessing antimicrobial activity. Pseudoalteromonas is a well-known bioactive compounds producer Hentschel et al., 2001; Bowman, 2007; Thomas et al., 2008; Sivasubramanian et al., 2011; Chen et al., 2012b; Vynne et al., 2012 as well as Bacillus (Bernal et al., 2002; Bhatta and Kapadnis, 2010; Jamal and Mudarris, 2010; Kamat and Kerkar, 2011; Kumar et al., 2012; Graça et al., 2013). Bioactivity has also been found in Pseudomonas (Kumar et al., 2005; Isnansetyo and Kamei, 2009; Graça et al., 2013), Enterococcus faecalis (Gútiez et al., 2013), Paenibacillus (Chung et al., 2000; Tupinambá et al., 2008; Wen et al., 2011; Graça et al., 2013) and Microbacterium (Lang et al., 2004; Graça et al., 2013). Antibacterial activity produced by Psychrobacter has been previously found (Kanagasabhapathy et al., 2008; Kennedy et al., 2009; Rojas et al., 2009; Flemer et al., 2011) but not antifungal activity. Rhodococcus (Chiba et al., 1999; Riedlinger et al., 2004; Lo Giudice et al., 2007; Hong et al., 2009; Abdelmohsen et al., 2010, 2014) and Paracoccus (Lo Giudice et al., 2007; Deng et al., 2011) showed antimicrobial activity. Dermacoccus was shown to produce antimicrobial, antitumor and antiprotozoal activity (Pospísil et al., 1998; Goodfellow and Fiedler, 2010). No antimicrobial activity has been detected in the genera Cellulomonas and Proteus. Okada et al. (2006) verified that the marine sponge Erylus placenta collected off Hachijo Island exhibited a broad spectrum of activity against several strains of yeasts and a fungus. This result is in agreement with the high levels of bioactivity against C. albicans obtained in our E. deficiens isolates.
The two preliminary and liquid assays allowed the confirmation of a great number of positive bacterial hits against C. albicans (72%—36 bacteria) but dissimilar results were obtained for E. coli and B. subtilis. In fact, these assays were not always coincident in the bioactivities obtained (Table 1), which could be due to differences in the cultivation (solid vs. liquid) and the extraction methods as previously observed (Bode et al., 2002; Graça et al., 2013; Haber and Ilan, 2013). In the diffusion assay there is always a potential problem with the diffusion of the bioactive molecules as well as the establishment of a concentration gradient. However, celerity and simplicity favor the use of this qualitative method (Valgas et al., 2007). Additionally, interaction between target and test microorganisms could induce a quorum-sensing response with antimicrobial production (Haber and Ilan, 2013). The assays using liquid broths have the advantage of being quantitative, simple, rapid, reproducible, and inexpensive and can be performed in a high throughput way. Furthermore, this method facilitates the upscale of the process in the case of a positive hit (avoiding constrains caused by the use of solid media).
The aqueous extracts presented higher bioactivities than the organic ones. This may be indicative of a hydrophilic nature of the bioactive molecules which could be proteins or sugars. With the acetone extraction, proteins would be precipitated and removed from the extract that could justify the decrease in bioactivity.
The genomic screening of bioactive potential is a valuable complement to the search for new bioactive molecules. The presence of PKS-I or NRPS genes was confirmed in some positive antimicrobial bacteria but non-bioactive bacteria also possessed these genes. This may indicate their ability to produce compounds with different types of activities like against other bacteria or even anticancer (Davidson et al., 2001). The PKS genes were previously found in the genera Pseudoalteromonas (Kennedy et al., 2009; Zhu et al., 2009; Chau et al., 2012) and Rhodococcus (Ayuso-Sacido and Genilloud, 2005; Abdelmohsen et al., 2014). However, to our knowledge, the present study is the first to detect PKS genes in the genera Proteus and Microbacterium. Regarding the NRPS genes, they have been previously detected in Pseudoalteromonas (Zhu et al., 2009; Chen et al., 2012a), Enterococcus (in the database: Biocyc.org1), Paenibacillus (Wu et al., 2011), Bacillus (Luo et al., 2014), Rhodococcus (Bosello et al., 2013), Dermacoccus (Pathom-aree et al., 2006), Microbacterium (Wu et al., 2012), Citrobacter (Siezen and Khayatt, 2008), and Proteus (Himpsl et al., 2010) but as far as perceived not in Psycrobacter, Brevundimonas and Paracoccus.
Of the confirmed bacteria that showed bioactivity against C. albicans, strain #91_11 belonging to the class Gammaproteobacteria is probably a novel species of Pseudoalteromonas based on the 16S rRNA gene (96.2% to P. tetraodonis, Figure 1). Furthermore, and based on the same gene similarity, other isolates with non-confirmed bioactivity (#66_1, #91_27, and #118_39) are also potential new taxa of Gammaproteobacteria.
Our results show a great antifungal potential of bacteria associated with E. deficiens. Curiously, in a previous work with heterotrophic bacteria of another Erylus species, E. discophorus (Graça et al., 2013), no antifungal activity was obtained. The bioactivity found was essentially against Bacillus subtilis which was almost absent in this work. Both works evidence for the production of bioactive compounds of bacteria associated with Erylus sponges which should be further studied regarding isolation and identification of the molecules accountable for the bioactivity.
Conclusions
The isolation and phylogenetic analyses of the culturable fraction of the bacterial community isolated from E. deficiens revealed a biodiversity with putative new taxa of Gammaproteobacteria that should be further studied regarding genome analysis and taxonomic characterization.
A great number of Erylus isolates were found to produce antibacterial and high antifungal bioactivity against pathogenic and environmental strains. As far as we know, this is the first report of antimicrobial production in the genera Cellulomonas and Proteus. Furthermore, PKS and NRPS genes are potentially associated with several Erylus isolates, namely the genera Proteus, Microbacterium for PKS-I and Psycrobacter, Brevundimonas, and Paracoccus for NRPS for which these genes have not been previously described. Our results showed the biotechnological potential of bacterial diversity associated with E. deficiens, inspiring further studies for the search of new leading compounds.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the European Regional Development Fund (ERDF) through the COMPETE—Operational Competitiveness Programme and national funds through FCT—Foundation for Science and Technology, under the projects PEst-C/MAR/LA0015/2013, PEst-OE/QUI/UI0612/2014 and PTDC/QUI-QUI/098053/2008. AG acknowledges an ERAMUS fellowship. JB was financed by FCT (PhD grant SFRH/BD/35933/2007). JX research was partially funded by a FCT postdoctoral fellowship (grant no. SFRH/BPD/62946/2009). We also thank Dr. Anabela Castro for providing some of the target organisms and Universidade Lusófona (Portugal), for the opportunity to participate in the Luso Expedição 2008.
Footnotes
References
- Abdelmohsen U. R., Pimentel-Elardo S. M., Hanora A., Radwan M., Abou-El-Ela S. H., Ahmed S., et al. (2010). Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated Actinomycetes. Mar. Drugs 8, 399–412. 10.3390/md8030399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen U. R., Yang C., Horn H., Hajjar D., Ravasi T., Hentschel U. (2014). Actinomycetes from red sea sponges: sources for chemical and phylogenetic diversity. Mar. Drugs 12, 2771–2789. 10.3390/md12052771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. L., Hooper J. N. A. (2001). A revision of Australian Erylus (Porifera: demospongiae: Astrophorida: Geodiidae) with a tabular review of worldwide species. Invertebr. Taxon. 15, 319–340 10.1071/IT00028 [DOI] [Google Scholar]
- Afiyatullov S. S., Kalinovsky A. I., Antonov A. S., Ponomarenko L. P., Dmitrenok P. S., Aminin D. L., et al. (2007). Isolation and structures of erylosides from the Carribean sponge Erylus goffrilleri. J. Nat. Prod. 70, 1871–1877. 10.1021/np070319y [DOI] [PubMed] [Google Scholar]
- Antonov A. S., Kalinovsky A. I., Dmitrenok P. S., Kalinin V. I., Stonik V. A., Mollo E., et al. (2011). New triterpene oligoglycosides from the Caribbean sponge Erylus formosus. Carbohydrate Res. 346, 2182–2192. 10.1016/j.carres.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Antonov A. S., Kalinovsky A. I., Stonik V. A., Afiyatullov S. S., Aminin D. L., Dmitrenok P. S., et al. (2007). Isolation and structures of erylosides from the Caribbean sponge Erylus formosus. J. Nat. Prod. 70, 169–178. 10.1021/np060364q [DOI] [PubMed] [Google Scholar]
- Ayuso-Sacido A., Genilloud O. (2005). New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49, 10–24. 10.1007/s00248-004-0249-6 [DOI] [PubMed] [Google Scholar]
- Bergmann W., Feeney R. J. (1950). The isolation of a new thymine pentoside from sponges. J. Am. Chem. Soc. 72, 2809–2810 10.1021/ja01162a543 [DOI] [Google Scholar]
- Bergmann W., Feeney R. J. (1951). Contributions to the study of marine products. XXXII The nucleosides of sponges. I. J. Org. Chem. 16, 981–987 10.1021/jo01146a023 [DOI] [Google Scholar]
- Bernal G., Illanes A., Ciampi L. (2002). Isolation and partial purification of a metabolite from a mutant strain of Bacillus sp. with antibiotic activity against plant pathogenic agents. Electron. J. Biotechnol. 5, 717–3458 10.2225/vol5-issue1-fulltext-4 [DOI] [Google Scholar]
- Bhatta D. R., Kapadnis B. P. (2010). Production optimization and characterization of bioactive compound against Salmonella bacillus subtilis KBB isolated from Nepal. Sci. World 8, 19–29 10.3126/sw.v8i8.3841 [DOI] [Google Scholar]
- Blunt J. W., Copp B. R., Keyzers R. A., Munroa M. H. G., Prinsep M. R. (2013). Marine natural products. Nat. Prod. Rep. 30, 237–323. 10.1039/C2NP20112G [DOI] [PubMed] [Google Scholar]
- Bode H. B., Bethe B., Höfs R., Zeeck A. (2002). Big effects from small changes: possible ways to explore Nature' s chemical diversity. Chembiochem 3, 619–627. [DOI] [PubMed] [Google Scholar]
- Bondoso J., Albuquerque L., Nobre M. F., Lobo-da-Cunha A., da Costa M. S., Lage O. M. (2011). Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium. Int. J. Syst. Evol. Microbiol. 61, 2844–2850. 10.1099/ijs.0.027474-0 [DOI] [PubMed] [Google Scholar]
- Bosello M., Zeyadi M., Kraas F. I., Linne U., Xie X., Marahiel M. A. (2013). Structural characterization of the heterobactin siderophores from Rhodococcus erythropolis PR4 and elucidation of their biosynthetic machinery. J. Nat. Prod. 76, 2282–2290. 10.1021/np4006579 [DOI] [PubMed] [Google Scholar]
- Bowman J. P. (2007). Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 5, 220–241. 10.3390/md504220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull A. T., Stach J. E. M. (2007). Marine actinobacteria: new opportunities for natural product search and discovery. Trends. Microbiol. 15, 491–499. 10.1016/j.tim.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Carballeira N. M., Negrón V. (1991). Identification and characterization of two new methylicosadienoic acids from Erylus formosus. J. Nat. Prod. 54, 305–309. 10.1021/np50073a041 [DOI] [PubMed] [Google Scholar]
- Carmely S., Roll M., Loya Y., Kashman Y. (1989). The structure of eryloside, A., a new antitumor and antifungal 4-methylated steroidal glycoside from the sponge Erylus lendenfeldi. J. Nat. Prod. 52, 167–170. 10.1021/np50061a022 [DOI] [PubMed] [Google Scholar]
- Chau R., Kalaitzis J. A., Neilan B. A. (2012). Genome of an octopus-derived Pseudoalteromonas reveals unprecedented natural product biosynthesis gene clusters. Planta Med. 78, CL53 10.1055/s-0032-1320288 [DOI] [Google Scholar]
- Chen W., Zhu P., He S., Jin H., Yan X. (2012a). Nonribosomal peptides synthetases gene clusters and core domain in Pseudoalteromonas sp. NJ631. Wei Sheng Wu Xue Bao. 52, 1531–1539. [PubMed] [Google Scholar]
- Chen Y., Guillemin G. J. (2009). Kynurenine pathway metabolites in humans: disease and healthy states. Int. J. Tryptophan. Res. 2, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Kuo J., Sung P. J., Chang Y. C., Lu M. C., Wong T. Y., et al. (2012b). Isolation of marine bacteria with antimicrobial activities from cultured and field-collected soft corals. World J. Microbiol. Biotechnol. 28, 3269–3279. 10.1007/s11274-012-1138-7 [DOI] [PubMed] [Google Scholar]
- Chiba H., Agematu H., Kaneto R., Terasawa T., Sakai K., Dobashi K., et al. (1999). Rhodopeptins (Mer-N1033), novel cyclic tetrapeptides with antifungal activity from Rhodococcus sp. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 52, 695–699. 10.7164/antibiotics.52.695 [DOI] [PubMed] [Google Scholar]
- Chung Y. R., Kim C. H., Hwang I., Chun J. (2000). Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Microbiol. 50, 1495–1500. 10.1099/00207713-50-4-1495 [DOI] [PubMed] [Google Scholar]
- Cohen-Bazire G., Sistrom W. R., Stanier R. Y. (1957). Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell Physiol. 49, 25–68. 10.1002/jcp.1030490104 [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Fish J. A., Chai C., McGarrell D. M., Sun Y., et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucl. Acids Res. 41, D633–D642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S. K., Allen S. W., Lim G. E., Anderson C. M., Haygood M. G. (2001). Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 67, 4531–4537. 10.1128/AEM.67.10.4531-4537.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. S., Zhao L. F., Xu L., Kong Z. Y., Zhao P., Qin W., et al. (2011). Paracoccus sphaerophysae sp. nov., a siderophore-producing, endophytic bacterium isolated from root nodules of Sphaerophysa salsula. Int. J. Syst. Evol. Microbiol. 61, 665–669. 10.1099/ijs.0.021071-0 [DOI] [PubMed] [Google Scholar]
- Elyakov G. B., Kuznetsova T., Mikhailov V. V., Maltsev I. I., Voinov V. G., Fedoreyev S. A. (1991). Brominated diphenyl ethers from a marine bacterium associated with the sponge Dysidea sp. Experientia 47, 632–633 10.1007/BF01949894 [DOI] [Google Scholar]
- Fenical W., Jensen P. R. (2006). Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2, 666–673. 10.1038/nchembio841 [DOI] [PubMed] [Google Scholar]
- Fischbach M. A., Walsh C. T. (2006). Assembly-line enzymology for polyketides and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496. 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- Flemer B., Kennedy J., Margassery L. M., Morrissey J. P., O'Gara F., Dobson A. D. (2011). Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp. J. Appl. Microbiol. 112, 289–301. 10.1111/j.1365-2672.2011.05211.x [DOI] [PubMed] [Google Scholar]
- Fouad M., Al-trabeen K., Badran M., Wray V. (2004). New steroidal saponins from the sponge Erylus lendenfeldi. ARKIVOC xiii, 17–27. [Google Scholar]
- Fusetani N., Sata N., Matsunaga S. (1993). Isolation and structure elucidation of erylusamine, B., a new class of marine natural products, which blocked an IL-6 receptor, from the marine sponge Erylus placenta Thiele. Tetrahedron. Lett. 34, 4067–4070 10.1016/S0040-4039(00)60617-2 [DOI] [Google Scholar]
- Gaspar H., Cachatra V., Frade S., Monteiro V., Neng N., Xavier J., et al. (2012). Targeting new inhibitors of enzymes involved in neurodegenerative diseases by HTS of Portuguese marine sponges, in 9th IUPAC International Symposium on Bimolecular Chemistry and 8th International Symposium for Chinese Medicinal Chemistry (Beijing: ), Book of abstracts, P60. [Google Scholar]
- Goodfellow M., Fiedler H. P. (2010). A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie van Leeuwenhoek 98, 119–142. 10.1007/s10482-010-9460-2 [DOI] [PubMed] [Google Scholar]
- Graça A. P., Bondoso J., Gaspar H., Xavier J. R., Monteiro M. C., de la Cruz M., et al. (2013). Antimicrobial activity of heterotrophic bacterial communities from the marine sponge Erylus discophorus (Astrophorida, Geodiidae). PLoS ONE 8:e78992. 10.1371/journal.pone.0078992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulavita N. K., Wright A. E., Kelly-Borges M., Longley R. E., Yarwood D., Sills M. A. (1994). Eryloside E from an Atlantic sponge Erylus goffrilleri. Tetrahedr. Lett. 35, 4299–4302 10.1016/S0040-4039(00)73338-7 [DOI] [Google Scholar]
- Gútiez L., Gómez-Sala B., Recio I., Del Campo R., Cintas L. M., Herranz C., et al. (2013). Enterococcus faecalis strains from food, environmental, and clinical origin produce ACE-inhibitory peptides and other bioactive peptides during growth in bovine skim milk. Int. J. Food Microbiol. 166, 93–101. 10.1016/j.ijfoodmicro.2013.06.019 [DOI] [PubMed] [Google Scholar]
- Haber M., Ilan M. (2013). Diversity and antimicrobial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J. Appl. Microbiol. 116, 519–532. 10.1111/jam.12401 [DOI] [PubMed] [Google Scholar]
- Hentschel U., Schmid M., Wagner M., Fieseler L., Gernert C., Hacker J. (2001). Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35, 305–312. 10.1111/j.1574-6941.2001.tb00816.x [DOI] [PubMed] [Google Scholar]
- Himpsl S. D., Pearson M. M., Arewång C. J., Nusca T. D., Sherman D. H., Mobley H. L. T. (2010). Proteobactin and a yersiniabactin-related siderophore mediate iron acquisition in Proteus mirabilis. Mol. Microbiol. 78, 138–157. 10.1111/j.1365-2958.2010.07317.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmuth T., Piel J. (2009). Polyketide synthases of bacterial symbionts in sponges—evolution-based applications in natural products research. Phytochemistry 70, 1841–1849. 10.1016/j.phytochem.2009.04.010 [DOI] [PubMed] [Google Scholar]
- Hong K., Gao A.-H., Xie Q.-Y., Gao H., Zhuang L., Lin H. P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. 10.3390/md7010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. N. A., Van Soest R. W. (2002). Class demospongiae sollas, 1885, in Systema Porifera A Guide to the Classification of Sponges, eds Hooper J. N. A., Van Soest R. W. M., Willenz P. (New York, NY: Springer; ), 15–51. [Google Scholar]
- Imhoff J. F., Labes A., Wiese J. (2011). Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol. Adv. 29, 468–482. 10.1016/j.biotechadv.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Isnansetyo A., Kamei Y. (2009). Bioactive substances produced by marine isolates of Pseudomonas. J. Ind. Microbiol. Biotech. 36, 1239–1248. 10.1007/s10295-009-0611-2 [DOI] [PubMed] [Google Scholar]
- Ivanova E. P., Shevchenko L. S., Sawabe T., Lysenko A. M., Svetashev V. I., Gorshkova N. M., et al. (2002). Pseudoalteromonas maricaloris sp. nov., isolated from an Australian sponge, and reclassification of [Pseudoalteromonas aurantia] NCIMB 2033 as Pseudoalteromonas flavipulchra sp. nov. Int. J. Syst. Evol. Microbiol. 52, 263–271. [DOI] [PubMed] [Google Scholar]
- Jamal M. T., Mudarris M. S. A. (2010). Separation of YbdN bioactive protein from Bacillus subtilis isolated from the Red Sea algae Sargassum sp. with bioactivity against antibiotic resistant bacterial pathogens. Mar. Sci. 21, 53–64 10.4197/Mar.21-1.2 [DOI] [Google Scholar]
- Jenke-Kodama H., Dittmann E. (2009). Bioinformatic perspectives on NRPS/PKS megasynthases: advances and challenges. Nat. Prod. Rep. 26, 874–883. 10.1039/b810283j [DOI] [PubMed] [Google Scholar]
- Kalinin V. I., Ivanchina N. V., Krasokhin V. B., Makarieva T. N., Stonik V. A. (2012). Glycosides from marine sponges (Porifera, Demospongiae): structures, taxonomical distribution, biological activities and biological roles. Mar. Drugs 10, 1671–1710. 10.3390/md10081671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat T., Kerkar S. (2011). Bacteria from Salt Pans: a potential resource of antibacterial metabolites. Rec. Res. Sci. Tech. 3, 46–52. [Google Scholar]
- Kamke J., Taylor M. W., Schmitt S. (2010). Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 4, 498–508. 10.1038/ismej.2009.143 [DOI] [PubMed] [Google Scholar]
- Kanagasabhapathy M., Sasaki H., Nagata S. (2008). Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan. World J. Microbiol. Biotechnol. 24, 2315–2321 10.1007/s11274-008-9746-y [DOI] [Google Scholar]
- Kennedy J., Baker P., Piper C., Cotter P. D., Walsh M., Mooij M. J., et al. (2009). Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish Waters. Mar. Biotechnol. 11, 384–396. 10.1007/s10126-008-9154-1 [DOI] [PubMed] [Google Scholar]
- Khan S. T., Musarrat J., Alkhedhairy A. A., Kazuo S. (2014). Diversity of bacteria and polyketide synthase associated with marine sponge Haliclona sp. Ann. Microbiol. 64, 199–207 10.1007/s13213-013-0652-7 [DOI] [Google Scholar]
- Kim T. K., Mary J., Garson M. J., Fuerst J. A. (2005). Marine actinomycetes related to the 'Salinospora' group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 7, 509–518. 10.1111/j.1462-2920.2005.00716.x [DOI] [PubMed] [Google Scholar]
- Kubanek J., Fenical W. (2001). New antifeedant triterpene glycosides from the Caribbean sponge Erylus formosus. Nat. Prod. Lett. 15, 275–285. 10.1080/10575630108041292 [DOI] [PubMed] [Google Scholar]
- Kubanek J., Pawlik J., Eve T., Fenical W. (2000). Triterpene glycosides defend the Caribbean reef sponge Erylus formosus from predatory fishes. Mar. Ecol. Prog. Ser. 207, 69–77 10.3354/meps207069 [DOI] [Google Scholar]
- Kumar A., Kumar S., Kumar S. (2005). Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem. Eng. J. 22, 151–159 10.1016/j.bej.2004.09.006 [DOI] [Google Scholar]
- Kumar S. N., Siji J. V., Ramya R., Nambisan B., Mohandas C. (2012). Improvement of antimicrobial activity of compounds produced by Bacillus sp. associated with a Rhabditid sp.(entomopathogenic nematode) by changing carbon and nitrogen sources in fermentation media. J. Microb. Biotech. Food Sci. 1, 1424–1438. [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laatsch H. (2008). Antibase 2008—The Natural Compound Identifier. Weinheim: Wiley-VCH. [Google Scholar]
- Lage O. M., Bondoso J. (2011). Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol. 78, 366–375. 10.1111/j.1574-6941.2011.01168.x [DOI] [PubMed] [Google Scholar]
- Lane D. J. (1991). 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley and Sons; ), 115–175. [Google Scholar]
- Lang S., Beil W., Tokuda H., Wicke C., Lurtz V. (2004). Improved production of bioactive glucosylmannosyl-glycerolipid by sponge-associated Microbacterium species. Mar. Biotechnol. 6, 152–156. 10.1007/s10126-003-0009-5 [DOI] [PubMed] [Google Scholar]
- Lau S. C. K., Tsoi M. M. Y., Li X., Dobretsov S., Plakhotnikova Y., Wong P. K., et al. (2005). Pseudoalteromonas spongiae sp. nov., a novel member of the γ-Proteobacteria isolated from the sponge Mycale adhaerens in Hong Kong waters. Int. J. Syst. Evol. Microbiol. 55, 1593–1596. 10.1099/ijs.0.63638-0 [DOI] [PubMed] [Google Scholar]
- Lo Giudice A., Brilli M., Bruni V., de Domenico M., Fani R., Michaud L. (2007). Bacterium-bacterium inhibitory interactions among psychrotrophic bacteria isolated from Antarctic seawater (Terra Nova Bay, Ross Sea). FEMS Microbiol. Ecol. 60, 383–396. 10.1111/j.1574-6941.2007.00300.x [DOI] [PubMed] [Google Scholar]
- Luo C., Liu X., Zhou H., Wang X., Chen Z. (2014). Identification of four NRPS gene clusters in Bacillus subtilis 916 for four families of lipopeptides biosynthesis and evaluation of their intricate functions to the typical phenotypic features. Appl. Environ. Microbiol. 10.1128/AEM.02921-14 [DOI] [Google Scholar]
- Martins A., Vieira H., Gaspar H., Santos S. (2014). Marketed marine natural products in the pharmaceutical and cosmoceutical industries: tips for success. Mar. Drugs 12, 1066–1101. 10.3390/md12021066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilan B. A., Dittmann E., Rouhiainen L., Bass R. A., Schaub V., Sivonen K., et al. (1999). Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181, 4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Matsunaga S., Van Soest R. W. M., Fusetani N. (2006). Sokodosides, steroid glycosides with an isopropyl side chain, from the marine sponge Erylus placenta. J. Org. Chem. 71, 884–4888. 10.1021/jo060653j [DOI] [PubMed] [Google Scholar]
- Pathom-aree W., Stach J. E. M., Ward A. C., Horikoshi K., Bull A. T., Goodfellow M. (2006). Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10, 181–189. 10.1007/s00792-005-0482-z [DOI] [PubMed] [Google Scholar]
- Pospísil S., Benada O., Kofronova O., Petricek M., Janda L., Havlicek V. (1998). Kytococcus sedentarius (formerly Micrococcus sedentarius) and Dermacoccus nishinomiyaensis (formerly Micrococcus nishinomiyaensis) produce monensins, typical Streptomyces cinnamonensis metabolites. Can. J. Microbiol. 44, 1007–1011. 10.1139/w98-081 [DOI] [PubMed] [Google Scholar]
- Riedlinger J., Reicke A., Zänher H., Krismer B., Bull A. T., Maldonado L. A., et al. (2004). Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 57, 271–279. 10.7164/antibiotics.57.271 [DOI] [PubMed] [Google Scholar]
- Rojas J. L., Martín J., Tormo J. R., Vicente F., Brunati M., Ciciliato I., et al. (2009). Bacterial diversity from benthic mats of Antarctic lakes as a source of new bioactive metabolites. Mar. Genomics 2, 33–41. 10.1016/j.margen.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Romero F., Espliego F., Baz J. P., de Quesada T. G., Grávalos D., la Calle F., et al. (1997). Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora. I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 50, 734–737. 10.7164/antibiotics.50.734 [DOI] [PubMed] [Google Scholar]
- Sandler J., Forsburg S., Faulkner J. (2005). Bioactive steroidal glycosides from the marine sponge Erylus lendenfeld. Tetrahedron 61, 1199–1206 10.1016/j.tet.2004.11.039 [DOI] [Google Scholar]
- Sarà M., Vacelet J. (1973). Écologie des Démosponges, in Traité de Zoologie, Spongiaires, ed Grassé P. (Paris: Masson et Cie; ), 462–576. [Google Scholar]
- Sata N., Asai N., Matsunaga S., Fusetani N. (1994). Erylusamines, IL-6 receptor antagonists, from the marine sponge Erylus placenta. Tetrahedron 50, 1105–1110 10.1016/S0040-4020(01)80821-8 [DOI] [Google Scholar]
- Shin J., Lee H. S., Woo L., Rho J. R., Seo Y., Cho K. W., et al. (2001). New triterpenoid saponins from the sponge Erylus nobilis. J. Nat. Prod. 64, 767–771. 10.1021/np010047d [DOI] [PubMed] [Google Scholar]
- Siezen R. J., Khayatt B. I. (2008). Natural products genomics. Microb. Biotechnol. 1, 275–282. 10.1111/j.1751-7915.2008.00044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian K., Ravichandran S., Vijayapriya M. (2011). Antagonistic activity of marine bacteria Pseudoalteromonas tunicata against microbial pathogens. Afr. J. Microbiol. Res. 5, 562–567. [Google Scholar]
- Stackebrandt E., Goebel B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16s rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- Stead P., Hiscox S., Robinson P. S., Pike N. B., Sidebottom P. J., Roberts A. D., et al. (2000). Eryloside F, a novel penasterol disaccharide possessing potent thrombin receptor antagonist activity. Bioorg. Med. Chem. Lett. 10, 661–664. 10.1016/S0960-894X(00)00063-9 [DOI] [PubMed] [Google Scholar]
- Takada K., Nakao Y., Matsunaga S., van Soest R. W. M., Fusetani N. (2002). Nobiloside, a new neuraminidase inhibitory triterpenoidal saponin from the marine sponge Erylus nobilis. J. Nat. Prod. 65, 411–413. 10.1021/np010480n [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular Evolutionary Genetic Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. W., Radax R., Steger D., Wagner M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. 10.1128/MMBR.00040-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N. L., Müller W. E. G. (2004). Biotechnological potential of marine sponges. Curr. Sci. 86, 1506–1512. [Google Scholar]
- Thiel V., Neulinger S. C., Staufenberger T., Schmaljohann R., Imhoff J. F. (2006). Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol. Ecol. 59, 47–63. 10.1111/j.1574-6941.2006.00217.x [DOI] [PubMed] [Google Scholar]
- Thomas T., Evans F. F., Schleheck D. (2008). Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS ONE 3:e3252. 10.1371/journal.pone.0003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T. R. A., Kavlekar D. P., LokaBharathi P. A. (2010). Marine drugs from sponge-microbe association-a review. Mar. Drugs 8, 1417–1468. 10.3390/md8041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupinambá G., Da Silva A. J. R., Alviano C. S., Souto-Padron T., Seldin L., Alviano D. S. (2008). Antimicrobial activity of Paenibacillus polymyxa SCE2 against some mycotoxin-producing fungi. J. Appl. Microbiol. 105, 1044–1053. 10.1111/j.1365-2672.2008.03844.x [DOI] [PubMed] [Google Scholar]
- Valgas C., Machado de Souza S., Smania E. F. A., Smanis A., Jr. (2007). Screening methods to determine antibacterial activity of natural products. Braz. J. Microbial. 38, 369–380 10.1590/S1517-83822007000200034 [DOI] [Google Scholar]
- Van Altena I., van Soest R. W. M., Roberge M., Andersen R. J. (2003). Trisphaerolide A, a novel polyketide from the Dominican sponge Erylus trisphaerus. J. Nat. Prod. 66, 561–563. 10.1021/np0205147 [DOI] [PubMed] [Google Scholar]
- Vynne N. G., Mansson M., Gram L. (2012). Gene sequence based clustering assists in dereplication of Pseudoalteromonas luteoviolacea strains with identical inhibitory activity and antibiotic production. Mar. Drugs 10, 1729–1740. 10.3390/md10081729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. (2006). Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 33, 545–551. 10.1007/s10295-006-0123-2 [DOI] [PubMed] [Google Scholar]
- Webster N. S., Taylor M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. 10.1111/j.1462-2920.2011.02460.x [DOI] [PubMed] [Google Scholar]
- Wen Y., Wu X., Teng Y., Qian C., Zhan Z., Zhao Y., et al. (2011). Identification and analysis of the gene cluster involved in biosynthesis of paenibactin, a catecholate siderophore produced by Paenibacillus elgii B69. Environ. Microbiol. 13, 2726–2737. 10.1111/j.1462-2920.2011.02542.x [DOI] [PubMed] [Google Scholar]
- Williams P. G. (2009). Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol. 27, 45–52. 10.1016/j.tibtech.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Wu H., Chen W., Wang G., Dai S., Zhou D., Zhao H., et al. (2012). Culture-dependent diversity of Actinobacteria associated with seagrass (Thalassia hemprichii). Afr. J. Microbiol. Res. 6, 87–94. [Google Scholar]
- Wu X. C., Qian C. D., Fang H. H., Wen Y. P., Zhou J. Y., Zhan Z. J., et al. (2011). Paenimacrolidin, a novel macrolide antibiotic from Paenibacillus sp. F6-B70 active against methicillin-resistant Staphylococcus aureus. Microb. Biotechnol. 4, 491–502 10.1111/j.1751-7915.2010.00201.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier J. R., Van Soest R. W. M. (2007). Demosponge fauna of Ormonde and Gettysburg Seamounts (Gorringe Bank, Northeast Atlantic): diversity and zoogeographical affinities. J. Mar. Biol. Assoc. 87, 1643–1653 10.1017/S0025315407058584 [DOI] [Google Scholar]
- Zhang H., Lee Y. K., Zhang W., Lee H. K. (2006). Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie van Leeuwenhoek 90, 159–169. 10.1007/s10482-006-9070-1 [DOI] [PubMed] [Google Scholar]
- Zhu P., Zheng Y., You Y., Yan X., Shao J. (2009). Molecular phylogeny and modular structure of hybrid NRPS/PKS gene fragment of Pseudoalteromonas sp. NJ6-3-2 isolated from marine sponge Hymeniacidon perleve. J. Microbiol. Biotechnol. 19, 229–237. [PubMed] [Google Scholar]