Abstract
Objectives
The goal of this study is to develop free-breathing high spatiotemporal resolution DCE liver MRI using non-Cartesian parallel imaging acceleration, and quantitative liver perfusion mapping.
Materials and Methods
This study is HIPAA-compliant, IRB approved, and written informed consent was obtained from all participants. Ten healthy subjects and five patients were scanned on a Siemens 3T Skyra scanner. A stack-of-spirals trajectory was undersampled in-plane with a reduction factor of 6, and reconstructed using 3D through-time non-Cartesian GRAPPA. High resolution 3D images were acquired with a true temporal resolution of 1.6~1.9 seconds, while the subjects were breathing freely. A dual-input single-compartment model was used to retrieve liver perfusion parameters from DCE-MRI data, which were co-registered using an algorithm designed to reduce the effects of dynamic contrast changes on registration. Image quality evaluation was performed on spiral images and conventional images from five healthy subjects.
Results
Images with a spatial resolution of 1.9×1.9×3 mm3 were obtained with whole liver coverage. With an imaging speed of better than 2 sec/volume, free-breathing scans were achieved, and dynamic changes in enhancement were captured. The overall image quality of free-breathing spiral images was slightly lower than conventional long breath-hold Cartesian images, but provided clinical acceptable or better image quality. The free-breathing 3D images were registered with almost no residual motion in liver tissue. Following the registration, quantitative whole liver 3D perfusion maps were obtained and the perfusion parameters are all in good agreement with the literature.
Conclusions
This high spatiotemporal resolution free-breathing 3D liver imaging technique allows voxel-wise quantification of liver perfusion.
Keywords: liver perfusion, through-time spiral GRAPPA, DCE MRI
INTRODUCTION
The cornerstone of lesion characterization in liver MRI is a dynamic contrast enhanced (DCE) series, in which breath-hold images are obtained prior to, and at multiple time-points after contrast. However, even with the state-of-the-art parallel imaging techniques and fast pulse sequences, the breath-holds in current scanning protocols continue to be long and difficult for patients. A significant proportion of patients (especially sicker individuals) cannot provide the requisite breath-holds, which results in motion-corrupted examinations.1 To address this problem, several techniques based on a parallel imaging technique called Controlled Aliasing in Parallel Imaging Results in Higher Acceleration (CAIPIRINHA) have been recently developed to further expedite the data acquisition.2–5 Another current limitation is that due to the need for long breath-holds, only a limited number of frames (typically four or five) are obtained to characterize the liver enhancement time course. These few time-points are insufficient for quantitative analysis of liver perfusion parameters and therefore image interpretation is entirely qualitative. An ideal solution to these limitations would be a free-breathing true DCE examination coupled with appropriate biophysical modeling that allows estimates of quantitative clinically relevant parameters such as arterial and portal fractions, and time to peak enhancement.6–8
However, quantitative DCE-MRI in the liver poses several challenges. First, it is necessary to have sufficient spatial resolution to identify small lesions (the goal here was less than 2 mm in-plane spatial resolution, and less than 3 mm through plane), which is particularly challenging as large volume coverage is required for imaging the entire liver. Second, very high temporal resolution is required (less than 2 sec per frame) for accurate arterial input function (AIF) characterization and perfusion model fitting.9 For these reasons, thick slice imaging or partial organ coverage are often utilized.8 Finally, an imaging period of 3~5 minutes is required to adequately cover the contrast dynamics for the application of quantitative liver perfusion modeling. While most standard MR liver exams involve breath-holding to limit motion artifacts, this strategy is not feasible for such a long acquisition window in perfusion imaging. To resolve this problem, the typical imaging protocol consists of multiple acquisitions/breath-holds spanning the period of contrast enhancement.7,10 These breath-holding spells can be difficult to reproduce, exhaust the patient and result in motion degradation of images. Missing data between breath-holds, particularly in the early, fast evolving phase of arterial and liver enhancement, could potentially lead to inaccurate fitting of the perfusion model.
A solution to overcome all of these problems is to apply fast MR imaging techniques to perform scans during free-breathing and at a resolution comparable to a traditional study. Recently, a golden-angle radial acquisition combined with compressed sensing and parallel imaging technique has been developed to provide high-quality liver DCE images during free breathing. However, this technique requires further improvement in the temporal resolution (currently 3~4 sec per frame) to accurately characterize the AIF.11,12 One way to achieve high spatial and temporal resolution is to use view-sharing techniques, but these methods lead to broad temporal footprints in the images, which in turn affect model fitting, particularly the accuracy of the AIF characterization. Recently, a non-Cartesian parallel imaging technique known as through-time non-Cartesian GRAPPA has been developed and has provided robust acceleration in real-time cardiac imaging.13 In this study, we examined the feasibility of adopting this approach in high spatiotemporal resolution hepatic DCE imaging without view sharing.
Another important obstacle for the application of free-breathing MR scans in quantitative liver perfusion studies is the need to accurately register acquired images to each other. The non-rigid motion due to respiration, bowel peristalsis and patient movement, could result in inaccuracies in free-breathing quantitative perfusion analysis.14,15 Thus reliable registration of 3D volumes acquired in different phases of the respiratory cycle is needed, if robust quantitative analysis is desired.
In this work, an undersampled spiral acquisition was coupled with the 3D through-time non-Cartesian GRAPPA reconstruction to obtain whole liver free-breathing high spatiotemporal resolution 3D images. The reconstructed images were aligned using an automatic non-rigid body registration step to account for respiratory motion. Finally, a quantitative dual-input single-compartment model was applied to the data to obtain robust quantitative liver perfusion parameter mapping.
MATERIALS AND METHODS
The prospective study was HIPAA compliant and conducted with approval of the local institutional review board. From March 2013 till April 2014, ten asymptomatic subjects (M:F, 7:3; age, 20.9 ± 1.3 years) were recruited for this study. Five patients (M:F, 2:3; age 68.0 ± 0.7 years) who underwent liver examination as part of clinical care at our institution were also included in this study. Of these patients, four had metastatic adenocarcinoma with 7 total lesions and one had a solitary sclerosing hemangioma. The liver pathologies for all the patients were conclusively established with image-guided biopsies. Written consent was obtained from all volunteers before the MRI examination.
3D Dynamic MR Acquisition
All MRI studies with fast spiral acquisitions were performed on a Siemens 3T Skyra scanner using up to 30 coil elements. For each subject, coronal unenhanced T2-weighted images were first obtained to identify the location of liver and position the imaging volumes. Contrast-enhanced T1-weighted 3D volumes were then acquired using an interleaved variable-density stack-of-spirals spoiled gradient echo sequence with fat suppression.13 A total of 48 in-plane spiral interleaves were required to meet the Nyquist criterion. To accelerate the acquisition, data were undersampled in-plane such that 8 spiral interleaves were acquired for each slice (reduction factor 6). A total of 100 to 120 volumes were acquired with a temporal resolution of 1.6 to 1.9 seconds per 3D volume. The total imaging time was approximately 3.5 min of free breathing acquisition. After the 5th volume was acquired, 0.1 mmol/kg Gadobenate (Multihance, Bracco Diagnostics, Princeton, NJ) was injected at 3 mL/s followed by 20 mL of saline flush. Other imaging parameters are listed in Table 1 (left column). In order to calculate the GRAPPA weights for image reconstruction, a separate calibration scan of 3 fully sampled 3D volumes (free breathing, ~40 s) was also acquired post-contrast at the end of the perfusion exam for better SNR.
Table 1.
Imaging parameters for both spiral and conventional Cartesian acquisitions.
| DCE Spiral | Cartesian Scan (normal subjects) | Cartesian Scan (patients) | |
|---|---|---|---|
| FOV (mm) | 360 ~ 440 | 360 ~ 440 | 350 ~ 400 |
| Matrix size | 192 ~ 240 | 192 ~ 240 | 256 ~ 288 |
| In-plane resolution (mm) | 1.9 | 1.9 | 1.2 ~ 1.4 |
| TR/TE (msec) | 4.5-4.7/0.6 | 4.4/1.9 | 3.1-5.4/1.5-2.5 |
| Flip angle (Degree) | 15 | 9 | 10 |
| Number of slices | 60 | 60 | 48 ~ 75 |
| Slice thickness (mm) | 3 | 3 | 3~4 |
| Parallel imaging | 6 | 2 | 2 |
| Acquisition time per volume (s) | 1.6 ~ 1.9 | 14 ~ 18 | 17 ~ 19 |
For the purpose of image quality comparison, conventional Cartesian T1-weighted fat-suppressed images were also acquired from five of the ten healthy volunteers immediately after the fast spiral acquisition (post-contrast). For each subject, two separate scans, one with breath-hold (BH-VIBE) and the other with free breathing (FB-VIBE), were acquired at the same spatial resolution as the spiral acquisitions. All the subjects were able to hold their breath during the breath-hold examinations and detailed imaging parameters are listed in Table 1 (middle column).
Four of the five patients (except one patient who had CT examinations) enrolled in this study also completed their standard clinical MRI examinations during a separate visit. The exams were performed on a Siemens 1.5T scanner or a Philips 3T scanner. Conventional multi-phase DCE imaging was performed using 3D breath-hold T1-weighted gradient-echo sequence and the imaging parameters are listed in Table 1 (right column).
Through-Time Spiral GRAPPA Reconstruction
All conventional Cartesian images were reconstructed online and saved as DICOM files for post-processing. The undersampled spiral images were reconstructed offline using the through-time spiral GRAPPA reconstruction algorithm in MATLAB (The Mathworks, Natick, MA). This algorithm for 2D spiral acquisitions has been described in detail previously.13 To summarize, spiral data is undersampled to accelerate data acquisition, and the missing spiral arms are reconstructed using GRAPPA weights estimated using a hybrid through-time/through-k-space scheme. To calculate the GRAPPA weights for any local geometry, the fully sampled k-space data from similar geometry in a dataset acquired through time can be combined to provide better image reconstruction. For the 3D acquisition, this similar geometry exists not only in different frames but also in different partitions within the same 3D frame, thereby making it possible to combine both to calculate the GRAPPA weights.16 In this study, a GRAPPA kernel of size 2 × 3 was used in the spiral arm × readout direction. A k-space data block including 8 kernels with similar geometry was used to provide enough information for calibration. Following the GRAPPA reconstruction, gridding and image reconstruction was performed using a non-uniform Fast Fourier Transform (NUFFT) toolbox.17 All the image reconstruction was performed on a desktop computer (Intel Xeon E3-1270 quad-core CPUs at 3.4 GHz and 16GB of RAM) and the total reconstruction time for one 3D volume of 60 slices was approximately 2.5 min.
Image Registration
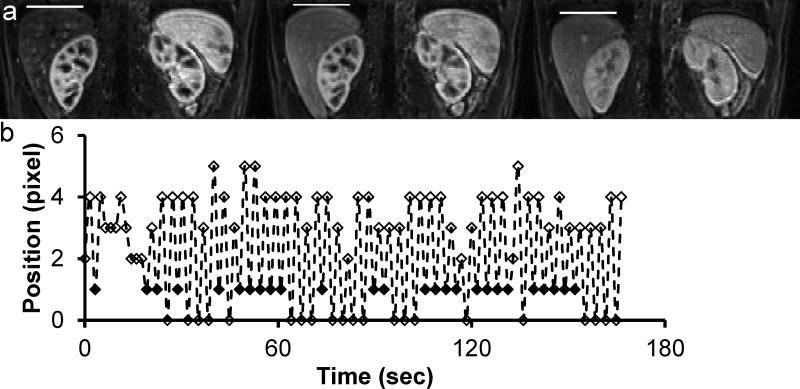
The reconstructed images were registered to eliminate respiratory motion between different volumes. In addition to respiratory motion, the image intensity variation between frames due to contrast dynamics is an obstacle for the application of most image registration methods. Typically, either a pre- or post-contrast volume is used as a reference for the registration of all the time series. In this study, multiple volumes from the same respiration level were selected and used as multiple references. Since the liver movement due to respiration is a quasiperiodic motion predominantly in the craniocaudal direction, an edge detection method was used to automatically measure the craniocaudal motion as shown in Figure 1a. All time frames measured where the liver is in the same position (approximately 20-25% of the total frames in each exam) formed a reference list (Figure 1b) and the remaining frames were registered to their nearest temporal neighbor within this list, minimizing the contrast change between the two volumes in each registration step. Non-rigid registration was then performed using FMRIB's Non-linear Image Registration Tool.18 The image registration was performed on the same computer as image reconstruction, and the average time to register one frame to its reference was approximately 20 min with a single thread.
Figure 1.
Measurement of liver motion in craniocaudal direction. (a) A simple edge-detection method was used to automatically find the top of liver (white bar) at different contrast enhancement levels. (b) Motion waveform from the measurement. The solid diamonds represent the selected references/time frames from the same respiratory level.
Data Processing
After image registration, perfusion quantification over the whole liver was performed on a voxel-by-voxel basis for all subjects using in-house developed Matlab software. Regions of interest were drawn manually in the abdominal aorta and portal vein to measure the arterial and portal input functions (AIF and PIF). To minimize in-flow artifacts, slices near the end of the imaging slab were selected for the AIF characterization. Pixelwise perfusion mapping was then performed for liver parenchyma without any spatial filtering, so the true spatial resolution from the acquired images was preserved in the perfusion analysis.
Signal intensity was first converted to the tissue longitudinal relaxation time T1 using the signal equation for the spoiled gradient echo pulse sequence as following:
| (1) |
where M0 is equilibrium longitudinal magnetization and θ is the flip angle. To estimate M0, signal intensities from five pre-contrast volumes were averaged and applied to Eq. 1. Contrast concentration was further calculated from the longitudinal relaxation time using the following relationship
The following fixed parameters were obtained from the literature: T1,blood 1800 msec (3T); T1,liver 800 msec (3T); and r1 (relaxivity for Gadobenate at 3T), 6.3 s−1mM−1.19,20 For the arterial and portal input functions, a fractional hematocrit level of 0.4 was used to convert them from blood concentration to plasma concentration.
Liver perfusion parameters were subsequently retrieved from the DCE-MRI data using a dual-input single-compartment model21
where CL(t), Ca(t) and Cp(t) represent the contrast concentration in liver tissue, aorta and portal vein, respectively; k1a represents the aortic inflow rate constant, k1p represents the portal venous inflow rate constant and k2 represents the outflow rate constant; τa and τb represent the transit time for aorta and portal vein, respectively. Liver perfusion parameters, such as arterial fraction (AF = k1a/(k1a + k1p)), distribution volume (DV = (k1a + k1p)/k2) and mean transit time (MTT = 1/k2) were further calculated, and the perfusion maps for the whole liver were generated on a voxel-byvoxel basis.
Image Quality Analysis
Image quality analysis was performed by two independent radiologists (14 and 4 years of experience in abdominal MRI) using the delayed enhanced images acquired from five healthy volunteers. For each volunteer, two conventional Cartesian T1-weighted 3D volumes (BH-VIBE and FB-VIBE) and the last volume in the free-breathing spiral acquisition (FB-SPIRAL) were presented to the radiologists in a random order. The radiologists, who were blinded to the type of images, were asked to rate the images on a scale of 1-5 (5 = excellent image quality, 4 = good, 3 = acceptable, 2 = poor, and 1 = unacceptable) for motion artifact, image blurring, liver edge sharpness, clarity of vessels, and overall image quality.
Statistical Analysis
A two-tailed Student's t test was used for the comparison of liver perfusion parameters obtained from ten healthy subjects and four patients with metastatic adenocarcinoma. For image qualitative analysis, statistical analysis was performed using a randomized block ANOVA with Tukey adjustment for multiple pairwise comparisons. Adjusted P values less than 0.05 were considered statistically significant.
Results
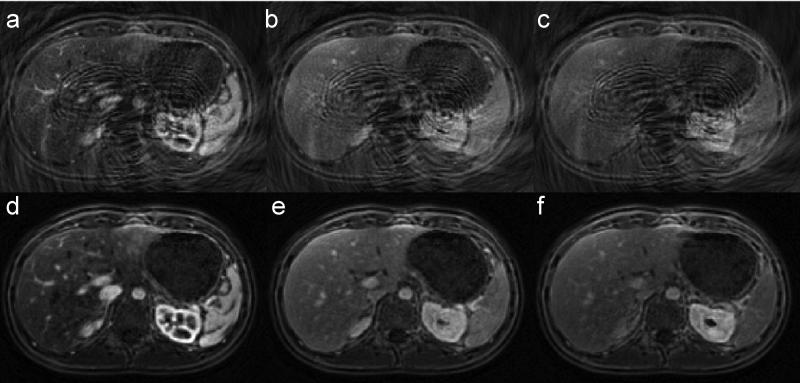
Figure 2 shows representative zero-filled undersampled images and through-time GRAPPA reconstructed images acquired from a normal subject at the arterial phase (~20 sec after contrast injection), portal phase (~70 sec) and equilibrium phase (~180 sec). Movie E1 (online) depicts the rapid contrast dynamics and periodic abdominal motion due to respiration over a 3.5 min scan.
Figure 2.
Representative undersampled (a-c) and through-time GRAPPA reconstructed (d-f) spiral images acquired at arterial phase (a&d), portal phase (b&e) and equilibrium phase (c&f) from a healthy volunteer after contrast injection.
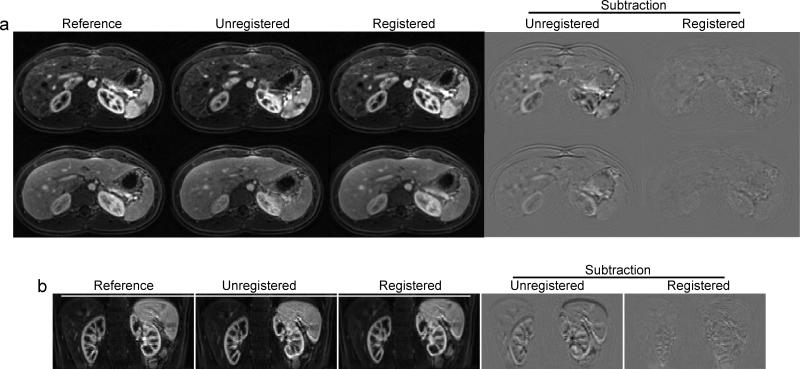
The effect of using image registration to reduce respiratory motion in liver is demonstrated in Figure 3 and Movie E2 (online) in axial and reformatted coronal views. With the automatic edge detection method, about 20-25% of the volumes from each subject were automatically detected by the algorithm to originate from the same position in the respiratory cycle, and thus were effectively pre-registered. After registering the remaining frames to the nearest “pre-registered” neighbor, almost no residual motion in liver tissue was observed, as can be seen from subtracted images before and after the registration (right two columns in Figure 3).
Figure 3.
Representative reference, unregistered and registered images in both axial (a) and reformatted coronal (b) views from a healthy subject. The subtraction images before and after image registration are also shown in the last two columns. Robust image registration with different contrast enhancement is demonstrated with representative images at both arterial phase and portal phase.
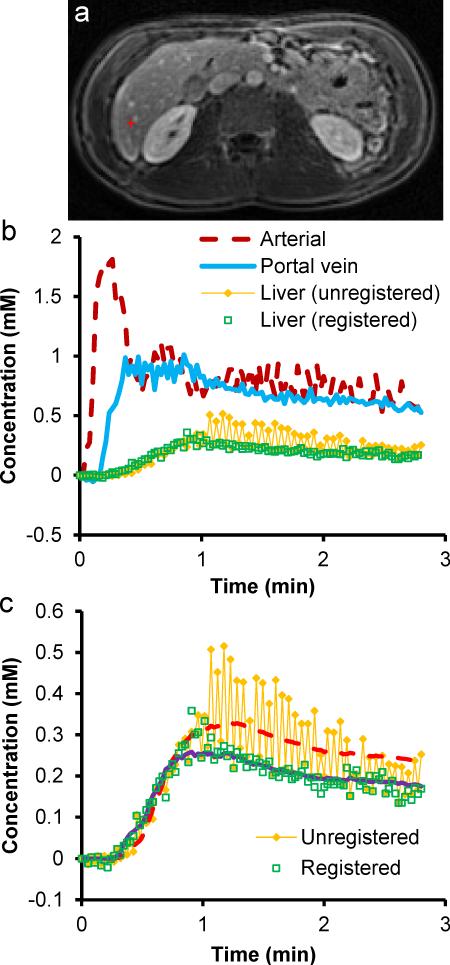
Figure 4 shows representative time courses of contrast concentration from the aorta, portal vein, and hepatic parenchyma (single voxel), and the model fit to the parenchymal time course from a healthy subject. The time course from the same liver voxel without registration is also plotted as a comparison. The model fit after registration yielded an arterial fraction (AF), distribution volume (DV) and mean transit time (MTT) of 18.8%, 24.2%, and 4.6 s, respectively, while the same quantities prior to registration were 13.5%, 35.2%, and 10.6 s, respectively. The perfusion parameters after registration are all in good agreement with published literature for CT and MRI of the liver.7,8,22
Figure 4.
(a) T1-weighted image showing a liver pixel close to a small vessel. (b) Concentration-time curves in the aorta, portal vein and the selected single-voxel liver tissue shown in (a). (c) Measured and fitted data of liver tissue using a dual-input single-compartment model. After image registration, the concentration-time curve is much smoother and the residual error from the fitting is lower. Substantial changes in arterial fraction, distribution volume and mean transit time were also observed before and after registration.
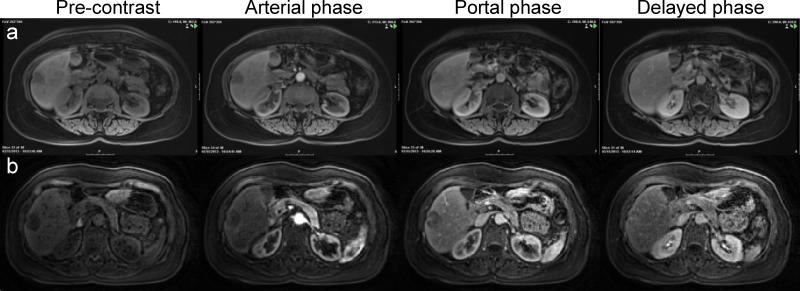
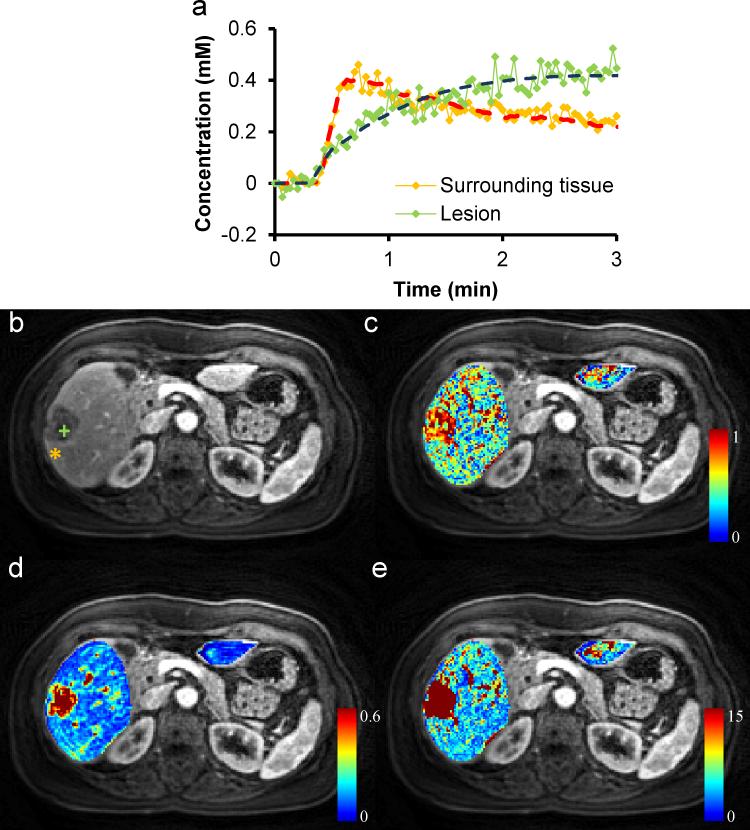
Five patients were scanned using the novel DCE technique as well as traditional MRI scans as representative clinical examples. Traditional and corresponding free-breathing DCE images for one patient with biopsy proven metastatic breast adenocarcinoma are shown in Figure 5. Perfusion modeling for the same patient is presented in Figure 6 and the lesion showed AF, DV, and MTT of 67.5%; 40.4%; and 99.8 s, respectively, all clearly different from surrounding parenchyma in the parametric maps. The mean AF, DV and MTT measured from all 7 lesions of the four patients with adenocarcinoma are presented in Table 2, and the numbers are statistically different as compared to those from surrounding liver (P < 0.05) and also compared to the healthy subjects (P < 0.05).
Figure 5.
Representative pre-contrast and post-contrast images acquired at arterial phase, portal phase and equilibrium phase of a subject with metastatic breast cancer. (a) Clinical standard images acquired with long breath-hold of ~18 sec/volume. (b) Free-breathing images acquired with 1.9 sec/volume.
Figure 6.
Liver perfusion maps for the subject with metastatic breast cancer. (a) Representative concentration-time curves of both lesion and normal surrounding tissue as shown in the T1-weighted image (b). Corresponding liver perfusion maps of (c) arterial fraction, (d) distribution volume, and (e) mean transit time.
Table 2.
Liver perfusion parameters for ten normal volunteers and four patients with metastatic adenocarcinoma (7 total lesions).
| Arterial fraction (%) | Distribution volume (%) | Mean transit time (sec) | ||
|---|---|---|---|---|
| Normal (n=10) | 25.0 ± 4.3 | 29.4 ± 8.3 | 8.8 ± 6.1 | |
| Met AdenoCA (n = 7) | Lesion | 90.8 ± 10.8*† | 46.1 ± 12.1*† | 51.9 ± 23.4*† |
| Surrounding tissue | 38.1 ± 12.3 | 23.6 ± 10.4 | 8.0 ± 5.1 | |
Note. --- Data are means ± standard deviations.
P<0.05 as compared with the normal group
P<0.05 as compared with surrounding tissue.
n: number of volunteers in the normal group and number of lesions in the patient group.
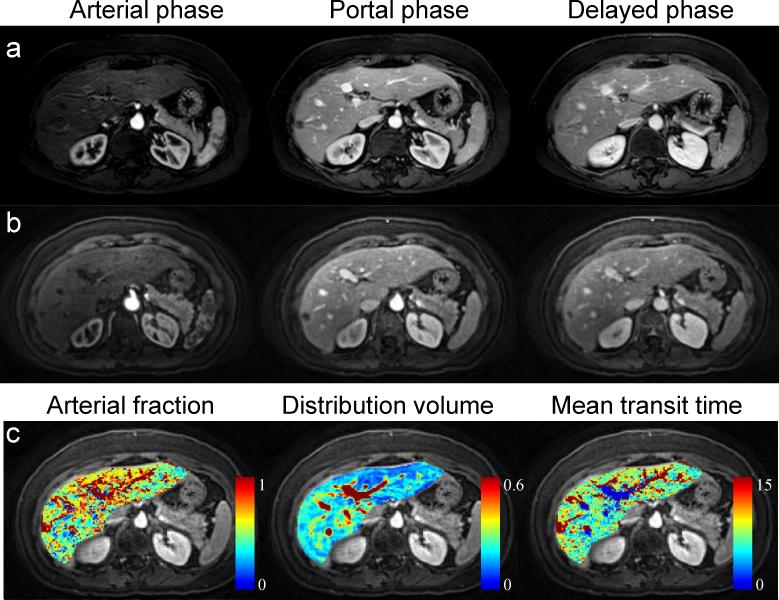
The DCE images and perfusion maps from another patient who had a biopsy proven sclerosing hemangioma are shown in Figure 7. In the lesion of this patient, the AF, DV and MTT were 99.4%, 21.0%, and 27.6 s, respectively, again markedly different from surrounding tissue, and demonstrating nearly 100% arterial fraction as expected from hemangiomata.23,24
Figure 7.
Liver perfusion images for the subject with sclerosing hemangioma. (a) Clinical standard images acquired with long breath-hold of ~18 sec/volume. (b) Free-breathing images acquired with 1.9 sec/volume. (c) Liver perfusion maps.
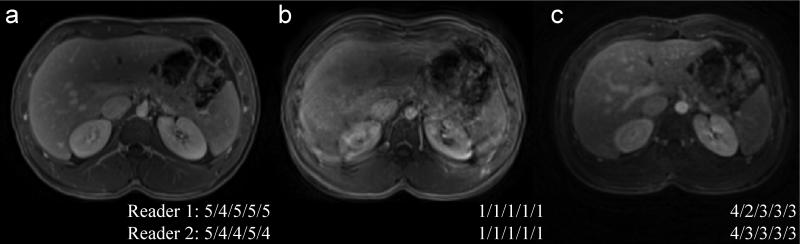
The summary of image quality analysis for the free-breathing spiral images and two types of conventional Cartesian images is presented in Table 3. An example of image evaluation from one of the subjects is shown in Figure 8. Both BH-VIBE and FB-SPIRAL had significantly higher scores as compared to FB-VIBE in all categories (adjusted P < 0.01). BH-VIBE had significantly less image blurring than FB-SPIRAL (adjusted P = 0.025), but no statistical difference was found in motion artifacts, liver edge sharpness, vessel clarity, or overall image quality (though the ratings were higher for the long breath-hold VIBEs) between these two. All FB-SPIRAL images were rated as having acceptable or better overall image quality by both readers.
Table 3.
Mean image quality scores of the post-contrast spiral and Cartesian T1-weighted images from five normal subjects.
| Motion artifacts | Image blurring | Liver edge sharpness | Clarity of vessels | Overall quality | |
|---|---|---|---|---|---|
| BH-VIBE | 4.8 ± 0.3 | 3.9 ± 0.2 | 4.1 ± 0.7 | 4.1 ± 1.0 | 4.2 ± 0.4 |
| FB-SPIRAL | 4.4 ± 0.4 | 3.0 ± 0.4* | 3.8 ± 0.6 | 3.9 ± 0.5 | 3.5 ± 0.5 |
| FB-VIBE | 1.4 ± 0.5*† | 1.4 ± 0.5*† | 1.4 ± 0.6*† | 1.5 ± 0.7*† | 1.4 ± 0.5*† |
Note. --- Data are means ± standard deviations.
Tukey-adjusted P < 0.05 as compared with BH-VIBE
Tukey-adjusted P < 0.05 as compared with FB-SPIRAL
Figure 8.
Representative images from a healthy subject for image quality analysis: (a) 18-sec breath-hold VIBE (BH-VIBE); (b) 18-sec free-breathing VIBE (FB-VIBE); (c) 1.9-sec free-breathing spiral (FB-SPIRAL). The scores from both readers for motion artifact, image blurring, liver edge sharpness, clarity of vessels, and overall image quality are shown as inset.
Discussion
There has been significant recent interest in rapid 3D liver perfusion imaging.12,25–28 Several acquisition and reconstruction techniques have been utilized, including parallel imaging, compressed sensing, and view sharing. In the present study, we demonstrate high spatiotemporal resolution 4D liver DCE imaging with quantitative dual-input single tissue compartment perfusion modeling, using a stack-of-spirals acquisition and through-time non-Cartesian GRAPPA acceleration in combination with non-rigid body image registration. An important characteristic of the presented method is that since no data sharing across frames is needed, the acquisition time of the images represents the true temporal footprint, ensuring high data fidelity. High spatiotemporal resolution (1.9×1.9×3 mm3 × 1.6-1.9s) allows rapid dynamic imaging of the whole liver during free breathing and voxel-wise quantification of multiple perfusion parameters in the liver.
As can be seen from Figures 5, 7, and 8 and Table 3, despite full liver coverage at high spatiotemporal resolution without view sharing, the image quality of free-breathing spiral images (FB-SPIRAL) is much better than free-breathing Cartesian acquisitions, and minimally compromised in comparison to the long breath-hold clinical standard (BH-VIBE) when there is a successful breath-hold for the latter. Mild differences in appearance between the pre-contrast images in Figure 5a and 5b could relate to differences in field strength, as the clinical exam for this patient was at 1.5T. Almost no residual aliasing artifacts are seen after the 3D through-time non-Cartesian GRAPPA reconstruction. All free-breathing spiral images were rated as acceptable or better image quality and are significantly superior to the free-breathing Cartesian images in all categories (Table 3). Slightly lower overall image quality as compared to standard BH-VIBE images was observed which is likely due to the mild image blurring artifacts originating from the spiral readout. It is well-known that spiral imaging with prolonged readout time is susceptible to blurring due to off-resonance effects. To minimize this artifact in this study, a spiral trajectory with 48 interleaves was designed to fully cover the k-space in one slice and the readout time of each spiral interleaf was only ~3 msec. This short spiral readout combined with an effective fat suppression led to only mild image blurring without adversely influencing image sharpness or vessel clarity (Table 3).
The high imaging acquisition speed (~2 seconds per volume) achieved in this study results in minimal motion artifacts despite the free-breathing nature of the acquisition (Table 3). This creates the possibility of eliminating breath-holding in pre- and post-contrast imaging of the abdomen. This could be particularly important in populations in which breath-holding is difficult or impossible, such as sedated patients, children, and patients who are intrinsically short of breath. Through-time non-Cartesian GRAPPA acceleration has been used previously to accelerate renal images for perfusion modeling.29 However, the imaging volume to be encoded was significantly larger for the liver. Thus the imaging speed required for free-breathing liver coverage with good image quality was more challenging, and the spiral acquisition was adopted as it had been previously shown to provide greater scan efficiency.29
Image registration is critical for accurate quantitative analysis in abdominal DCE-MRI.14,15 The complex contrast dynamics in the abdomen result in a rapidly changing appearance of the solid organs, particularly the kidneys, spleen, and liver. These contrast variations pose a challenge for image registration methods, as no single image/frame can serve as a reference for the entire data set. Many methods have been developed to address this issue. For example, Lausch et al. have proposed a floating reference image scheme in which each registered frame is used as a reference for the subsequent unregistered frame.15 While this strategy does allow registration of images despite the changing appearance of the organs, it is subject to error propagation, especially for long dynamic acquisitions. In the current study, the registration problem was solved by using multiple volumes that are automatically detected as stationary with respect to one another as references for the non-rigid registration of the remaining frames. This strategy is easy to implement and can easily be adapted to many other different image registration methods. The efficiency of image registration is also improved because image registration is not performed on stationary volumes, and moreover these volumes are not deformed for registration. Image fidelity is completely retained in these frames, which are thus ideal for radiologist review.
As can be seen from Movie E1 (online), complex contrast dynamics in the abdominal solid organs can be captured with the free-breathing approach, with no data gaps due to resting periods between scans. Multi-parametric perfusion maps were obtained for normal liver parenchyma as well as for liver lesions, with normal values in good agreement with the literature.7,8,22 While the patient examples provided here illustrate the possibility of qualitative lesion characterization, this is a small feasibility study. More extensive evaluation in the setting of various liver lesions is needed and will be performed in the future to evaluate the ability of parametric mapping to differentiate various liver lesions from one another. Existing literature has already shown that quantitative perfusion analysis may be helpful in predicting therapeutic response in metastatic lesions such as neuroendocrine tumors.8,10,28,30 Furthermore, it has been suggested that quantitative perfusion imaging, if performed with high spatiotemporal resolution, may be useful in imaging follow-up of patients with cirrhosis and for early detection of hepatocellular carcinoma (HCC), both extremely common worldwide.25 While the model utilized here is that introduced by Materne et al.,21 any desired liver model can be theoretically applied to the high spatiotemporal resolution 3D data obtained for this study.
This method requires a free breathing calibration scan for the through-time spiral GRAPPA reconstruction to calculate GRAPPA weights and reconstruct the missing spiral interleaves in the undersampled data, in a manner very similar to the autocalibration scan in Cartesian GRAPPA.31 However, unlike Cartesian GRAPPA, this calibration is not incorporated into the undersampled scan. This calibration scan is also performed in a free-breathing manner and adds minimal time (approximately 1 minute) to the overall examination and thus is easily incorporated into the imaging routine. This additional acquisition does not affect the actual signal intensity or SNR of the reconstructed dynamic images.
A potential limitation of the proposed method is the long post-processing time for image reconstruction and registration. In this proof-of-concept study, all the image reconstruction work was performed off-line using MATLAB. However, non-Cartesian GRAPPA reconstruction using C++ and parallel computing have been recently implemented.32 Initial findings indicate that the reconstruction time is reduced to ~10 seconds per 3D volume using C++ and a single GPU card. The speed of image registration can also be improved significantly using parallel computing. Based on the proposed registration algorithm, the overall calculation time can be reduced by a factor equal to the number of threads or number of cores utilized in the computation.
It will be important to compare the perfusion results obtained from the proposed free breathing approach to other methods. Since there is no existing gold standard method for liver perfusion quantification using MRI, definitive validation is difficult to obtain. In this study, the perfusion results acquired from normal volunteers, including the arterial fraction, distribution volume and mean transit time, are similar to that reported previously.7,8,22 The increased arterial fraction in patients with hepatic metastases is also consistent with previous findings.8 Future work will be undertaken to compare the quantitative perfusion parameters such as arterial fraction to the results obtained from other image modalities such as CT.
In conclusion, we demonstrate the feasibility of a high spatiotemporal resolution 3D liver imaging technique based on a stack-of-spirals acquisition, 3D through-time non-Cartesian GRAPPA acceleration and non-rigid image registration. This technique allows fast imaging of the whole liver during free breathing and voxel-wise quantification of liver perfusion parameters.
Supplementary Material
Acknowledgments
Conflicts of interest and sources of funding: supported by Siemens Healthcare and NIH grants 1R01DK098503, R00EB011527, 1R01HL094557, and 2KL2TR000440.
References
- 1.Hanley J, Debois MM, Mah D, et al. Deep inspiration breath-hold technique for lung tumors: the potential value of target immobilization and reduced lung density in dose escalation. Int. J. Radiat. Oncol. Biol. Phys. 1999;45(3):603–611. doi: 10.1016/s0360-3016(99)00154-6. [DOI] [PubMed] [Google Scholar]
- 2.Wright KL, Harrell MW, Jesberger JA, et al. Clinical evaluation of CAIPIRINHA: Comparison against a GRAPPA standard. J. Magn. Reson. Imaging. 2014;39(1):189–194. doi: 10.1002/jmri.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riffel P, Attenberger UI, Kannengiesser S, et al. Highly accelerated T1-weighted abdominal imaging using 2-dimensional controlled aliasing in parallel imaging results in higher acceleration: a comparison with generalized autocalibrating partially parallel acquisitions parallel imaging. Invest. Radiol. 2013;48(7):554–561. doi: 10.1097/RLI.0b013e31828654ff. [DOI] [PubMed] [Google Scholar]
- 4.Michaely HJ, Morelli JN, Budjan J, et al. CAIPIRINHA-Dixon-TWIST (CDT)-volume-interpolated breath-hold examination (VIBE): a new technique for fast time-resolved dynamic 3-dimensional imaging of the abdomen with high spatial resolution. Invest. Radiol. 2013;48(8):590–597. doi: 10.1097/RLI.0b013e318289a70b. [DOI] [PubMed] [Google Scholar]
- 5.Park YS, Lee CH, Kim IS, et al. Usefulness of controlled aliasing in parallel imaging results in higher acceleration in gadoxetic acid-enhanced liver magnetic resonance imaging to clarify the hepatic arterial phase. Invest. Radiol. 2014;49(3):183–188. doi: 10.1097/RLI.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 6.Pandharipande P V, Krinsky GA, Rusinek H, et al. Perfusion imaging of the liver: current challenges and future goals. Radiology. 2005;234(3):661–673. doi: 10.1148/radiol.2343031362. [DOI] [PubMed] [Google Scholar]
- 7.Hagiwara M, Rusinek H, Lee VS, et al. Advanced liver fibrosis: diagnosis with 3D whole-liver perfusion MR imaging--initial experience. Radiology. 2008;246(3):926–934. doi: 10.1148/radiol.2463070077. [DOI] [PubMed] [Google Scholar]
- 8.Koh TS, Thng CH, Lee PS, et al. Hepatic metastases: in vivo assessment of perfusion parameters at dynamic contrast-enhanced MR imaging with dual-input two-compartment tracer kinetics model. Radiology. 2008;249(1):307–320. doi: 10.1148/radiol.2483071958. [DOI] [PubMed] [Google Scholar]
- 9.Sourbron S. Technical aspects of MR perfusion. Eur. J. Radiol. 2010;76(3):304–313. doi: 10.1016/j.ejrad.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki K, Orton MR, Davidson RL, et al. Neuroendocrine tumor liver metastases: use of dynamic contrast-enhanced MR imaging to monitor and predict radiolabeled octreotide therapy response. Radiology. 2012;263(1):139–148. doi: 10.1148/radiol.12110770. [DOI] [PubMed] [Google Scholar]
- 11.Feng L, Grimm R, Tobias Block K, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn. Reson. Med. 2014;72(3):707–717. doi: 10.1002/mrm.24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandarana H, Feng L, Block TK, et al. Free-breathing contrast-enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden-angle radial sampling. Invest. Radiol. 2013;48(1):10–16. doi: 10.1097/RLI.0b013e318271869c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seiberlich N, Lee G, Ehses P, et al. Improved temporal resolution in cardiac imaging using through-time spiral GRAPPA. Magn. Reson. Med. 2011;66(6):1682–1688. doi: 10.1002/mrm.22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melbourne A, Atkinson D, White M, et al. Registration of dynamic contrast-enhanced MRI using a progressive principal component registration (PPCR). Phys. Med. Biol. 2007;52(17):5147–5156. doi: 10.1088/0031-9155/52/17/003. [DOI] [PubMed] [Google Scholar]
- 15.Lausch A, Ebrahimi M, Martel A. IEEE International Symposium on Biomedical Imaging from Nano to Macro. Vol. 2011. IEEE; 2011. Image registration for abdominal dynamic contrast-enhanced magnetic resonance images. pp. 561–565. [Google Scholar]
- 16.Wright K, Lee G, Ehses P, et al. Three-dimensional through-time radial GRAPPA for renal MR angiography. J. Magn. Reson. Imaging. 2014;40(4):864–874. doi: 10.1002/jmri.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fessler JA. On NUFFT-based gridding for non-Cartesian MRI. J. Magn. Reson. 2007;188(2):191–195. doi: 10.1016/j.jmr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkinson M, Beckmann CF, Behrens TEJ, et al. FSL. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Pintaske J, Martirosian P, Graf H, et al. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1.5, and 3 Tesla. Invest. Radiol. 2006;41(3):213–221. doi: 10.1097/01.rli.0000197668.44926.f7. [DOI] [PubMed] [Google Scholar]
- 20.Stanisz GJ, Odrobina EE, Pun J, et al. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med. 2005;54(3):507–12. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 21.Materne R, Smith AM, Peeters F, et al. Assessment of hepatic perfusion parameters with dynamic MRI. Magn. Reson. Med. 2002;47(1):135–142. doi: 10.1002/mrm.10045. [DOI] [PubMed] [Google Scholar]
- 22.Van Beers BE, Leconte I, Materne R, et al. Hepatic perfusion parameters in chronic liver disease: dynamic CT measurements correlated with disease severity. Am. J. Roentgenol. 2001;176(3):667–673. doi: 10.2214/ajr.176.3.1760667. [DOI] [PubMed] [Google Scholar]
- 23.Li G-W, Chen Q-L, Jiang J-T, et al. The origin of blood supply for cavernous hemangioma of the liver. Hepatobiliary Pancreat. Dis. Int. HBPD INT. 2003;2(3):367–370. [PubMed] [Google Scholar]
- 24.Jang H-J, Kim TK, Lim HK, et al. Hepatic Hemangioma: Atypical Appearances on CT, MR Imaging, and Sonography. Am. J. Roentgenol. 2003;180(1):135–141. doi: 10.2214/ajr.180.1.1800135. [DOI] [PubMed] [Google Scholar]
- 25.Brodsky EK, Bultman EM, Johnson KM, et al. High-spatial and high-temporal resolution dynamic contrast-enhanced perfusion imaging of the liver with time-resolved three-dimensional radial MRI. Magn. Reson. Med. 2014;71(3):934–941. doi: 10.1002/mrm.24727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KW, Lee JM, Jeon YS, et al. Free-breathing dynamic contrast-enhanced MRI of the abdomen and chest using a radial gradient echo sequence with K-space weighted image contrast (KWIC). Eur. Radiol. 2013;23(5):1352–60. doi: 10.1007/s00330-012-2699-4. [DOI] [PubMed] [Google Scholar]
- 27.Xu B, Spincemaille P, Chen G, et al. Fast 3D contrast enhanced MRI of the liver using temporal resolution acceleration with constrained evolution reconstruction. Magn. Reson. Med. 2013;69:370–381. doi: 10.1002/mrm.24253. [DOI] [PubMed] [Google Scholar]
- 28.Sourbron S, Reiser MF, Zech CJ. Combined quantification of liver perfusion and function with dynamic gadoxetic acid –enhanced MR imaging. Radiology. 2012;263(3):874–883. doi: 10.1148/radiol.12110337. [DOI] [PubMed] [Google Scholar]
- 29.Wright KL, Chen Y, Saybasili H, et al. Quantitative high-resolution renal perfusion imaging using 3-dimensional through-time radial generalized autocalibrating partially parallel acquisition. Invest. Radiol. 2014;49(10):666–674. doi: 10.1097/RLI.0000000000000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh TS, Thng CH, Hartono S, et al. Dynamic contrast-enhanced MRI of neuroendocrine hepatic metastases: A feasibility study using a dual-input two-compartment model. Magn. Reson. Med. 2011;65(1):250–260. doi: 10.1002/mrm.22596. [DOI] [PubMed] [Google Scholar]
- 31.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn. Reson. Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 32.Saybasili H, Herzka DA, Seiberlich N, et al. Real-time imaging with radial GRAPPA: Implementation on a heterogeneous architecture for low-latency reconstructions. Magn. Reson. Imaging. 2014;32(6):747–758. doi: 10.1016/j.mri.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.