Abstract
To gain insights into the interrelationships among childhood undernutrition, the gut microbiota, and gut mucosal immune/barrier function, we purified bacterial strains targeted by IgA from the fecal microbiota of two cohorts of Malawian infants and children. IgA responses to several bacterial taxa, including Enterobacteriaceae, correlated with anthropometric measurements of nutritional status in longitudinal studies. The relationship between IgA responses and growth was further explained by enteropathogen burden. Gnotobiotic mouse recipients of an IgA+-bacterial consortium purified from the gut microbiota of undernourished children exhibited a diet-dependent enteropathy characterized by rapid disruption of the small intestinal and colonic epithelial barrier, weight loss and sepsis that could be prevented by administering two IgA-targeted bacterial species from a healthy microbiota. Dissection of a culture collection of 11 IgA-targeted strains from an undernourished donor, sufficient to transmit these phenotypes, disclosed that Enterobacteriaceae interacted with other consortium members to produce enteropathy. These findings indicate that bacterial targets of IgA responses have etiologic, diagnostic, and therapeutic implications for childhood undernutrition.
Introduction
Childhood undernutrition is a devastating and pervasive global health problem that is not due to food insecurity alone, but rather reflects a number of intra- and intergenerational factors (1-3). The gut microbiota is one factor being explored to better understand the pathogenesis of undernutrition and to develop more effective treatment and prevention strategies. The gut microbiota operates as a metabolic ‘organ’ that performs functions required for healthy growth, including vitamin biosynthesis and biotransformation of food ingredients (4). Recent studies in Malawi and Bangladesh have shown that the normal pattern of microbiota assembly is disrupted in children with undernutrition: this immaturity persists after commonly used therapeutic food interventions are applied, leading to the proposal that disrupted microbiota development imperils healthy postnatal growth (5, 6).
The microbiota co-develops with the immune system during postnatal life (7, 8), impacting healthy mucosal barrier function and the risk for, and evolution of, enteropathogen infections (9, 10), another contributing factor in undernutrition. Immunoglobulin A (IgA) is secreted into the gut where it functions by binding bacterial, food and other antigens, preventing their direct interaction with the host via immune exclusion (11). Many features of IgA, including its role in mediating host-microbiota homeostasis, its responsiveness to transient antigenic stimulation, and its stability within the intestinal tract make it an attractive molecule to identify microbes that directly interact with the gut mucosal immune system (12-14). Whereas secretory IgA concentrations in serum, nasal washings, tears, saliva and duodenal aspirates have been studied in children with undernutrition (15-17), little is known about the identities of the bacterial taxa targeted by IgA in the gut or other body habitats, or about the role of IgA-targeted bacteria in the pathogenesis of this disease or diseases. Here, we delineate interrelationships among diet, the gut microbiota, gut mucosal barrier function and enteropathogen infection in childhood undernutrition by isolating, identifying and then functionally characterizing the gut bacterial targets of host IgA responses in members of two cohorts of Malawian children.
Results
Identifying bacterial targets of IgA responses in gnotobiotic mice colonized with fecal microbiota from twins discordant for kwashiorkor
As part of ongoing efforts to better understand how the gut microbiota contributes to undernutrition, we transplanted fecal microbiota samples collected from a 21-month-old Malawian monozygotic twin pair discordant for kwashiorkor (a form of severe acute undernutrition), into separate groups of adult male germ-free C57BL/6J mice (6) (see twin pair 57 in table S1). Mice were fed a sterile macro- and micronutrient deficient diet, designed to represent the diets of the donor population, starting one week prior to gavage with the human microbiota, or were maintained on a nutritionally-sufficient mouse chow low in fat and rich in plant polysaccharides (‘standard diet’). Transplantation is an efficient and reproducible process with capture of the majority of species-level bacterial taxa in recipient animals (6). Consistent with our prior report (6), two weeks after transplantation, mice ‘humanized’ with microbiota from the co-twin with kwashiorkor and fed the Malawian diet (‘KM’ mice) had lost significantly more weight than animals fed the same diet but that were colonized with microbiota from the healthy co-twin (‘HM’ mice). Mice fed the standard diet lost significantly less weight than their counterparts fed the deficient Malawian diet, regardless of their donor microbiota (‘KS’ and ‘HS’ mice, respectively) (fig. S1A, which includes results of statistical tests).
We identified IgA-bound components of the transplanted microbiota and subsequently purified these organisms, in a viable form, from each of the four groups of humanized gnotobiotic animals (KM, KS, HM and HS) by fluorescence-activated cell sorting (FACS) (14, 18-22) (see Methods and fig. S2A-G for details about the sorting protocol). Our goal was to first analyze fecal microbiota samples recovered from these groups of gnotobiotic mice using this method (which we call BugFACS), then to characterize the role of components of the enriched IgA+ microbial consortia in mediating disease and/or promoting health through transfer to germ-free animals, and finally to apply BugFACS directly to human fecal samples obtained from cohorts of Malawian infants and children with varying degrees of undernutrition.
IgA-bound bacteria were recovered from fecal samples collected from members of each of the four groups of humanized gnotobiotic mice 13-16 days after they had been colonized. Samples from individual mice were independently subjected to BugFACS followed by bacterial 16S rRNA gene sequencing of the “input”, the IgA+, and IgA- fractions. The differential representation of each taxon between the IgA+ and IgA- fractions was expressed in the form of an IgA index (fig. S1D,E). For a given taxon, the value of the IgA index can range from a maximum of 1.0 (taxon detected exclusively in the IgA+ fraction) to a minimum of -1.0 (present only in the IgA- fraction). The input fraction served to define how a taxon's representation in the IgA+ and IgA- fractions corresponded to its overall abundance within the fecal sample and as an internal control to validate enrichment of IgA+ bacteria. The IgA index value was calculated for each taxon present in a given sample to give a relative measure of targeting; the index does not provide an absolute quantitation of the IgA response since the total amount of IgA bound to each taxon is not determined. Data obtained about a given taxon in each group of mice subjected to a given treatment were combined to compute a mean IgA index value and a P-value for its enrichment in the IgA+ or IgA- fraction (see Methods, fig. S2, and fig. S3A,B for a more detailed description and validation of this approach).
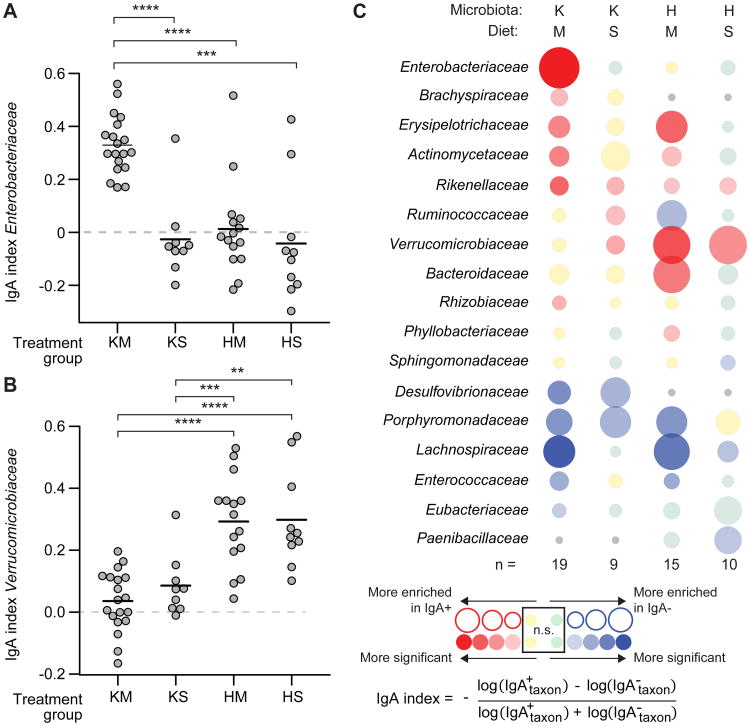
The results revealed significant enrichment for members of the family-level taxon Enterobacteriaceae in the IgA+ fraction purified from the fecal microbiota of KM mice (two independent experiments, n=19 mice; 19 fecal samples analyzed; P<0.0001, paired Wilcoxon rank-sum test; Fig. 1A and table S2). Neither HM mice that had received the healthy co-twin's microbiota and were fed the Malawian diet (n=15), nor KS mice that had received the kwashiorkor co-twin's microbiota but were fed standard mouse chow (n=9), developed a significant IgA response to Enterobacteriaceae, despite its presence in the microbiota of all treatment groups (Fig. 1A, fig. S1B). In contrast, the most prominent IgA response in mice colonized with fecal microbiota from the healthy co-twin (HM and HS groups) was to Akkermansia muciniphila, the only representative of the family Verrucomicrobiaceae present (Fig. 1B, fig. S1C, table S2). Among taxa that were present in all four groups of mice, Enterobacteriaceae was the only family-level taxon whose targeting by IgA was significantly greater in KM mice compared to all three of the other groups of animals (see Fig. 1A which includes results of statistical tests). Erysipelotrichaceae, a member of the Firmicutes, was a target of the IgA response in both groups fed the Malawian diet, regardless of the source of their human microbiota (Fig. 1C, table S2).
Fig. 1. IgA responses in humanized gnotobiotic colonized with the fecal microbiota of twins discordant for kwashiorkor.
Diet- and microbiota-associated differences in IgA responses to bacterial taxa present in gnotobiotic mice containing transplanted microbiota from kwashiorkor or healthy co-twins from discordant pair 57. Separate groups of mice received fecal microbiota from either the kwashiorkor (K) or healthy (H) co-twin and were fed an irradiated (sterile) prototypic Malawian (M) diet (KM or HM groups) or a control nutrient-sufficient standard (S) mouse chow (KS and HS groups). Fecal samples collected from recipient mice 13-16 days after gavage of the human donor microbiota were analyzed by BugFACS. Results shown are from two independent experiments. (A) Enterobacteriaceae were significantly enriched in the IgA+ fraction prepared from the fecal microbiota of KM mice compared to all other groups of animals, indicating the microbiota and diet specificity of the gut mucosal immune response. Each data point represents a fecal microbiota sample from a different animal. (B) Mice colonized with the microbiota from the healthy co-twin had an IgA response to Verrucomicrobiaceae that was significantly greater than the responses of mice harboring microbiota from the sibling with kwashiorkor. ** P < 0.01; *** P < 0.001; ****, P < 0.0001 (Wilcoxon rank-sum test). (C) ‘Bubble plot’ depicting IgA responses (defined by the IgA index) to different family-level bacterial taxa. Each column represents a different group of gnotobiotic mice while each row shows the relative enrichment of a given family-level taxon in the IgA+ or IgA- fraction. The color of the circle represents the direction of enrichment, with darker colors indicating greater significance as determined by paired Wilcoxon test (threshold for significantly enriched, P < 0.05). The diameter of a given circle represents the average magnitude of enrichment for a given taxon in the fecal microbiota of members of a given treatment group. n, number of gnotobiotic mice analyzed per treatment group (see table S2 for IgA indices for members of each family-level taxon shown). ns, not significant. Gray circles indicate that a taxon was not observed within a given treatment group.
Transmissible diet-dependent pathologic phenotypes
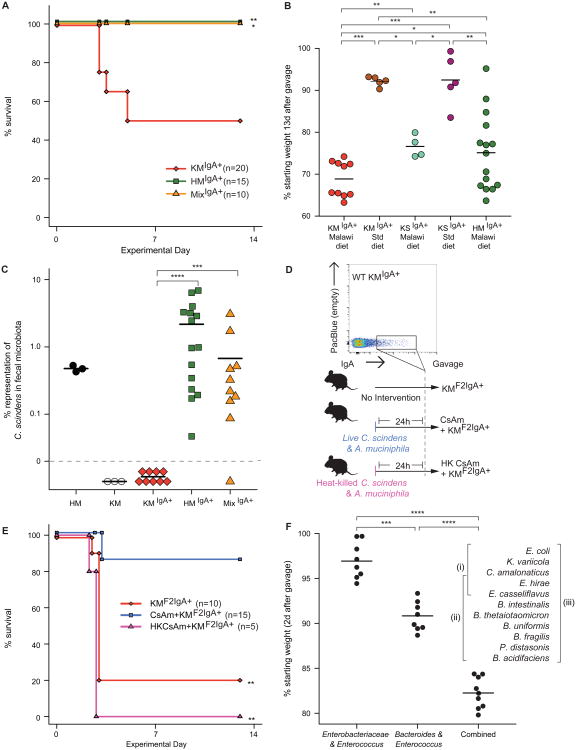
We performed a microbial adoptive transfer experiment to functionally characterize the IgA+ consortia recovered from mice colonized with either the kwashiorkor or healthy co-twins' microbiota (KM and HM groups). We focused on the IgA-enriched fraction and a positive selection approach, realizing that the IgA-negative fraction could, in principle, contain a subpopulation of those strains present in the IgA positive population, and understanding that other, IgA-negative strains could also contribute to disease pathogenesis. Germ-free adult C57BL/6J mice were started on the Malawian diet one week before colonization. One gnotobiotic isolator contained mice gavaged with 105 sorted IgA+ microbial cells derived from the fecal microbiota of KM mice (KMIgA+ group). Another isolator contained mice gavaged with 105 microorganisms from the IgA+ fraction of HM mouse fecal microbiota (HMIgA+ group). A third gnotobiotic isolator contained mice gavaged with a mixture of 5 × 104 IgA+ microbes from KM mice and 5 × 104 IgA+ microbes from HM mice so that the total number of microbes administered was also 105 per mouse (MixIgA+) (fig. S1F; see fig. S3B,C for Hellinger distance comparisons between the three sorted fractions and documentation of colonization with the IgA+ microbes).
KMIgA+ mice fared poorly over the two weeks following colonization, with 50% dying within 5 days (Fig. 2A; two independent experiments; n=20 animals). In contrast, 100% of the HMIgA+ mice survived for the full course of the experiment despite being maintained on the same Malawian diet as the KM group and receiving the same number of sorted microbes (n=15; two independent experiments). Remarkably, 100% of the MixIgA+ group also survived over the entire course of the experiment (n=10, two independent experiments; Fig. 2A). Moreover, MixIgA+ mice lost less weight than KMIgA+ animals over the first several days after colonization (fig. S4), although they experienced more weight loss than HMIgA+ animals at later time points [loss: 28.3±1.5% (mean±s.e.m.) versus 20.7±2.3% 12 days after gavage; P=0.04, unpaired Wilcoxon rank-sum test].
Fig. 2. Bacterial targets of IgA responses to kwashiorkor and healthy co-twin microbiota, introduced into germ-free mice.
Adult germ-free C57BL/6J mice were fed an irradiated Malawian diet starting one week prior to gavage with IgA+ fractions. These fractions were purified from fecal microbiota obtained from KM or HM mice 42 days after they had been colonized with the respective co-twin's microbiota. Following gavage, the recipient mice were maintained on the Malawian diet. (A) KMIgA+ mice (n=20) experienced significantly greater mortality than HMIgA+ mice (n=15). Mice that received an equivalent number of cells from IgA+ consortia purified from HM and KM mouse fecal microbiota did not exhibit mortality during the course of the experiment (MixIgA+, n=10). *, P<0.05; **, P<0.01 compared to KMIgA+ group (Fisher's Exact test). Results represent data from two independent experiments. (B) Impact of diet. KMIgA+ mice fed a Malawian diet lost more weight over a 2-week period following colonization than did animals colonized with the same IgA+ consortium but fed a standard nutrient-sufficient mouse chow. Mice receiving the IgA+ consortium purified from the fecal microbiota of mice harboring the same family 57 kwashiorkor donor microbiota but fed a standard mouse chow (KSIgA+ mice) lost less weight than did KMIgA+ mice, regardless of whether they were fed the Malawian diet or a standard mouse chow. *, P<0.05; **, P<0.01; ***, P<0.001 (Wilcoxon rank-sum test). (C) Clostridium scindens was present in the fecal microbiota of HM, HMIgA+ and MixIgA+ mice, but was not detectable in the microbiota of KM or KMIgA+ animals. ***, P< 0.001; ****, P< 0.0001 (Wilcoxon rank-sum test). (D) Experimental design of a follow-up experiment where three groups of adult germ-free male mice were gavaged with an IgA+ consortium purified by BugFACS from the fecal microbiota of surviving KMIgA+ mice. All recipients [second generation (F2) KMF2IgA+ mice] were fed the Malawian diet. The first group of these KMF2IgA+ mice received no intervention (n=10). Another group received an equal mixture of live C. scindens and A. muciniphila by gavage 24 h before introduction of the IgA+ consortium purified from KMIgA+ mice (CsAm + KMF2IgA+, n=15). A third group was gavaged with heat-killed C. scindens and A. muciniphila 24 h prior to introduction of the IgA+ consortium (heat-killed CsAm + KMF2IgA+, n=5). (E) CsAm + KMF2IgA+ mice exhibited significantly reduced mortality compared to either KMF2IgA+ or heat-killed CsAm + KMF2IgA+ animals. **, P< 0.01 (Fisher's Exact test). (F) Effects of colonizing germ-free mice fed a Malawian diet with different components of the 11 OTU culture collection, generated from KMF2IgA+ microbiota, on weight. Data for individual mice in each treatment group are plotted. ****, P<0.0001 (Wilcoxon rank-sum test).
These phenotypes were diet-dependent. Lethality in recipients of the IgA+ consortium purified from KM mice was prevented when they were fed nutrient-sufficient chow rather than the nutrient-deficient Malawian diet. In addition, if the IgA+ fraction was derived from mice that had been colonized with the intact kwashiorkor microbiota, but fed a standard chow diet, the recipient KSIgA+ mice exhibited significantly less weight loss (P<0.01, Wilcoxon rank-sum test; Fig. 2B), and somewhat reduced mortality (20%; P=0.3, Fisher's exact test) even while consuming the Malawian diet (n=5 mice/treatment group). These results emphasize the transmissible effects of diet and the relative severity of the KMIgA+-induced phenotype.
Lethality phenotype transmissible by the KMIgA+ consortium is ameliorated by IgA-targeted taxa from the healthy co-twin's microbiota
16S rRNA analysis was performed using fecal samples obtained from KMIgA+, HMIgA+ or MixIgA+ mice 13 days after they had been inoculated with an IgA+ consortium. Clostridium scindens was the only species-level taxon that satisfied our criteria of being present in the fecal microbiota of mice colonized with the healthy co-twin's microbiota (HM group) or its derivatives (HMIgA+ and MixIgA+ animals), but not in mice colonized with the kwashiorkor co-twin's microbiota (KM group), or its derivatives (‘first generation’ KMIgA+ animals) (Fig. 2C). C. scindens is part of a consortium of members of Clostridia that induces colonic regulatory T cells and tolerogenic responses in mice (23, 24).
We subsequently tested whether C. scindens and A. muciniphila could prevent the lethality caused by the purified KMIgA+ consortium. A. muciniphila was selected because (i) it induced a significantly more robust IgA response in mice colonized with the healthy co-twin's compared to the kwashiorkor co-twin's fecal microbiota (Fig. 1B), and (ii) its presence has been associated with healthy, non-inflamed gut mucosa in humans [abundance is reduced in the microbiota of patients with inflammatory bowel disease (25)]. A. muciniphila also improves barrier function in a mouse model of diet-induced obesity (26). An equal mixture of C. scindens and A. muciniphila (abbreviated CsAm) was introduced into mice 24 h prior to gavage with 105 bacteria from the IgA+ consortium purified from the feces of animals previously colonized with a KMIgA+ consortium. These recipient mice represented ‘second generation’ KMIgA+ animals (KMF2IgA+) (Fig. 2D). Control groups consisted of animals gavaged with heat-killed (HK) CsAm 24h prior to introduction of the KMIgA+ consortium, and KMF2IgA+ mice that received no intervention. KMF2IgA+ mice had high mortality (80% within 4 days of gavage) while mice that received live CsAm experienced significantly less mortality (14%; P<0.01, Fisher's exact test) compared to mice that were gavaged with an equivalent number of heat-killed CsAm (100% mortality), or that received no intervention (two independent experiments examining the protective effect of the CsAm consortium in mice harboring the KMIgA+ or KMF2IgA+ communities; n=5-15 mice/treatment group/experiment; effect on KMF2IgA+ mice shown in Fig. 2E).
We analyzed serum cytokine profiles in ten KMIgA+ mice that survived until the end of the experiment (day 13) and five moribund KMIgA+ animals that had to be sacrificed on days 3-4. There was an increase in a number of cytokines in the moribund group, including granulocyte-colony stimulating factor (G-CSF), interleukin-10 (IL-10), IL-12p40, IL-1β, IL-6, KC (a mouse IL-8 homolog), and monocyte chemotactic protein-1 (MCP-1) (fig. S5A). This cytokine signature is similar to that reported in mouse models of sepsis due to cecal ligation and puncture (27). Follow-up flow cytometry of mice surviving to day 13 disclosed an increase in B cells (TCR-β-B220+) and activated CD4+ T cells (CD44hiCD62Llow) within the mesenteric lymph nodes (MLN) but not the spleens of KMIgA+ compared to HMIgA animals, consistent with increased delivery of microbes from the gut to MLN and gut barrier dysfunction in KMIgA+ animals (fig. S5B,C).
Enterobacteriaceae are a necessary but not sufficient component of an 11 strain KMIgA+ derived cultured bacterial consortium that disrupts gut barrier integrity
We transplanted cecal contents of KMF2IgA+ mice, harvested at the time of sacrifice from two moribund KMF2IgA+ donors (samples pooled prior to gavage) into recipient germ-free animals fed the Malawian diet to directly test if there was a transmissible effect on barrier function. Recipients exhibited rapid weight loss within 2d after gavage [21.1±1.9% reduction (mean±SD); n=15 animals], and were sacrificed at that time point. Cultures of homogenates prepared from their spleens revealed that they had a polymicrobial sepsis dominated by members of Enterobacteriaceae (three morphologically and biochemically distinct strains of E. coli and a single strain of Klebsiella variicola). Whole genome sequencing of the E. coli strains disclosed known virulence factors, but their representation was not characteristic of that reported for enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC) or enteroaggregative E. coli (EAEC) isolates (see ‘57A Mouse Isolates’ in table S3).
We then performed a histopathologic analysis of the small intestines and colons of these mice, as well as animals belonging to three control groups: one that had received the CsAm consortium one day prior to gavage with KMF2IgA+ cecal microbiota, another that had been monocolonized with a strain of E. coli recovered from the spleens of previously treated KMF2IgA+ mice (57A isolate A7 in table S3), and a third group that had been maintained germ-free on the Malawian diet. The proximal colons of untreated recipients of KMF2IgA+ cecal microbiota exhibited a severe and broadly distributed phenotype with 100% penetrance (n=13 mice surveyed), manifest by atrophic crypts and surface cuff epithelial cells with disrupted intercellular cohesion (‘discohesion’) (note that differentiating post-mitotic epithelial cells normally migrate out of colonic crypts to form a surface cuff epithelium that surrounds each crypt orifice; fig. S6A,B). There were no mitotic figures or apoptotic bodies in the remnant crypts, indicating a profound inhibition of proliferation of epithelial cell progenitors. Interestingly, these changes occurred in the absence of substantial infiltration of innate or adaptive immune cells (i.e., neutrophils and lymphocytes) (fig. S6B). This phenotype was specific for mice colonized with KMF2IgA+cecal microbiota; it was not observed in germ-free mice maintained on the same Malawian diet, or in mice monocolonized with the splenic isolate 57A.A7 (n=5 animals surveyed/treatment group; fig. S6C-E). Pre-treatment with the CsAm consortium one day prior to colonization with KMF2IgA+ cecal microbiota substantially ameliorated the phenotype; six of eight animals did not show evidence of severe epithelial damage (crypt and surface epithelial cells had normal morphology; crypts contained mitotic figures and abundant differentiated goblet cells; the surface cuff epithelium was intact and its cells were cohesive) (fig. S6F,G). The crypt phenotype observed in KMF2IgA+ mice resembled that seen in germ-free mice with dextran sodium sulfate (DSS)-induced colitis (28). However, the surface epithelial discohesion is not a feature of the DSS injury model (28). Equally remarkable is the fact that this profound barrier disruption was observed without prior chemical insults or genetic manipulations of the host (10, 22).
There was no evidence of crypt atrophy in the small intestine: instead, the major defect in all mice that received KMF2IgA+ cecal microbiota was a ‘hobnailed’ pattern along the length of the villus that was associated with sloughing of epithelial cells into the lumen (we use the term hobnailed to refer to projection of an epitheilal cell above the upper apical boundary of its neighboring cell; see fig. S7A). This phenotype was not observed in the small intestines of germ-free or E. coli monocolonized controls (fig. S7B). In CsAm pre-colonized mice, the hobnailed pattern of the epithelium was less pronounced and there was no evidence of cellular extrusion along the length of villi.
The villus epithelial phenotype was reminiscent of a pattern observed in children with congenital deficiency of Epithelial Cell Adhesion Molecule (EpCAM1) (29, 30). EpCAM is normally localized to the lateral cell borders of villus epithelial cells. The only exception occurs at the tips of villi, where cytoplasmic aggregates of EpCAM can be readily observed. EpCAM functions as a binding partner for multiple cell adhesion molecules (31). Congenital EpCAM1 deficiency is a rare autosomal recessive disorder that presents with refractory diarrhea in early childhood, gut barrier dysfunction, and mortality (32). Mice colonized with the cecal microbiota of KMF2IgA+ donors exhibited mislocalization of EpCAM with abundant cytoplasmic aggregates of immunoreactive protein apparent in epithelial cells distributed from the bases to apices of villi (fig. S7C,D). Small intestinal crypts appeared normal. This generalized cytoplasmic aggregation of EpCAM was not observed in the small intestinal epithelium of germ-free or E. coli monocolonized controls or in CsAm+KMF2IgA+ mice (fig. S7E-H). These results indicate that this cell adhesion molecule is a biomarker of the barrier disruption produced by the KMF2IgA+ consortium.
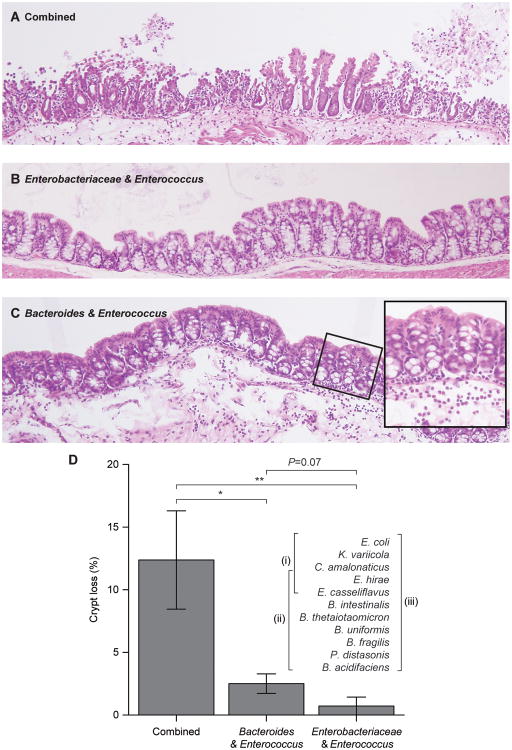
To identify which member or members of the KMF2IgA+ consortium mediate the weight loss and barrier disruption phenotypes, we generated a clonally arrayed bacterial culture collection from this IgA-selected community consisting of 11 operational taxonomic units (OTUs), grouped based on strains sharing ≥97%ID identity in the V4 region of their 16S rRNA genes. We sequenced at least two representatives of each OTU (see table S3 for details of the draft genome assemblies and their content of known virulence factors). Groups of adult male C57BL/6J germ-free mice were colonized with (i) all 11 OTUs, (ii) the collection minus the three 97% ID OTUs assigned to Enterobacteriaceae, or (iii) the three members of Enterobacteriaceae plus two Enterococcus strains assigned to E. hirae and E. casseliflavus (note that these two members of Enterococcus were also present in the other two consortia) (n=8-9 animals/group). Within two days after a single gavage, animals with the complete group of 11 OTUs had lost significantly more weight than the other two groups (P<0.0001; Wilcoxon rank-sum test), while mice colonized with only the Enterobacteriaceae and Enterococcus strains exhibited the least weight loss (Fig. 2F). 16S rRNA sequencing of fecal samples obtained from animals in all three groups confirmed that their communities had the expected content of strains (fig. S8A). The weight loss phenotype correlated with colonic histopathology: crypt loss was greatest in mice harboring all 11 OTUs while those colonized with the consortium that lacked the Enterobacteriaceae strains exhibited an intermediate level of crypt loss compared to the other two treatment groups (Fig. 3A-D). Examination of the small intestines demonstrated the characteristic hobnailing pathology in mice receiving the complete, 11 OTU consortium (present in 6 of 9 animals) while this phenotype was absent in the groups receiving the other two consortia (P < 0.01, Fisher's Exact test).
Fig. 3. Identifying bacterial strains that transmit gut barrier disruption phenotypes.
Adult germ-free mice consuming the prototypic Malawian diet were gavaged with all 11 OTUs contained in the clonally arrayed culture collection generated from the cecal microbiota of KMF2IgA+ mice, or two subsets of the culture collection: a consortium of the five strains belonging to Enterobacteriaceae (E. coli, K. variicola, C. amalonaticus) and Enterococcus (E. hirae and E. casseliflavus) or a consortium of 8 strains that included all but the three strains of Enterobacteriaceae in the collection (see table S3 for details about the genome sequences of these organisms, including their virulence factor content). All animals were sacrificed 2 days after gavage and hematoxylin and eosin stained sections of their proximal colons were prepared. (A) Colonization with the 11 OTU consortium produced generalized disruption of the colonic epithelium with marked loss of crypts. (B) The epithelium and crypt numbers were preserved in mice harboring the 5-strain consortium. (C) The 8-strain consortium lacking members of Enterobacteriaceae did not produce the epithelial disruption seen with the entire 11-strain consortium and crypts were largely preserved. However, there was an associated neutrophil infiltrate in the lamina propria (highlighted in inset). (D) Quantification of crypt number per unit area of the colonic epithelium. *, P<0.05; **, P<0.01 (Wilcoxon rank-sum test).
Together, these results establish that the strains of Enterobacteriaceae represented in the culture collection were not sufficient to produce the transmitted weight loss and gut barrier disruption phenotypes; rather, the phenotypes appeared to be mediated by interactions involving additional members of the cultured consortium. This latter conclusion is consistent with a 16S rRNA enumeration showing that the proportional representation of Enterobacteriaceae in the fecal microbiota of KMF2IgA+ animals was not significantly changed by treatment with A. muciniphila and C. scindens (fig. S8B,C).
Translation to two Malawian cohorts of infants and children
Our approach does not exclude the possibility that strains other than these 11 OTUs, including IgA negative strains present in KM microbiota from which the 11 OTUs were recovered, could contribute to disease. To further explore the relationship between IgA targeting of gut bacteria and childhood undernutrition, we subsequently performed a direct analysis of fecal samples collected from two Malawian populations; one composed of twins, the other a singleton birth cohort. Our goals were to characterize the interrelationships between IgA responses to different bacterial taxa, enteropathogen burden, and nutritional status. The twin cohort consisted of 11 same-gender Malawian pairs who became discordant for kwashiorkor during their first three years of life, plus eight same-gender pairs who remained concordant for healthy status during this period (n=134 samples analyzed; see fig. S3D,E for data confirming staining specificity and reproducibility of the BugFACS procedure when applied directly to human fecal samples). In accordance with prevailing clinical practices in Malawi (6), both co-twins in discordant pairs had been treated with a peanut-based ready-to-use therapeutic food (RUTF) for four weeks (table S1 for subject characteristics). The second group consisted of participants enrolled in the Lungwena Child Nutrition Intervention Study #5 (LCNI-5) that was designed to examine the effects of a lipid-based nutrient supplement on stunting (33). The 165 samples analyzed from LCNI-5 were obtained from children at six and/or 18 months of age who manifested varying degrees of stunting [defined by length-for- age Z scores (LAZ)] and wasting [defined by weight-for-height Z scores (WHZ), see table S5 for clinical characteristics].
A strong IgA response to Enterobacteriaceae is indicative of a pathogenic community
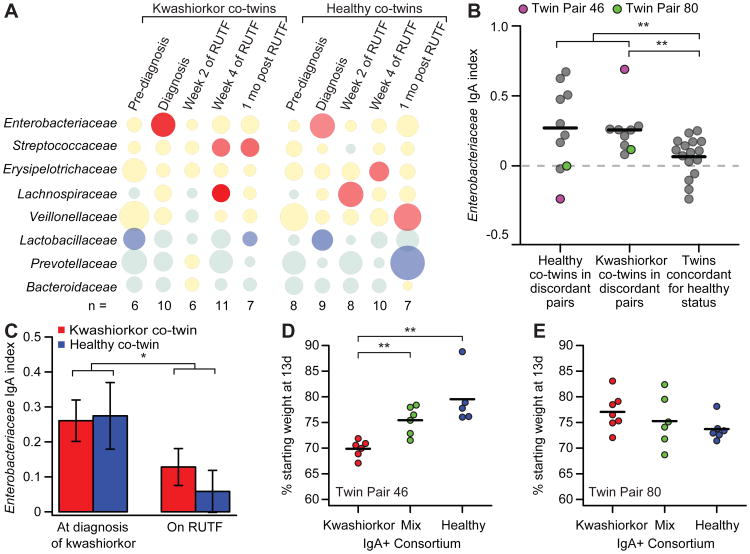
Samples obtained from members of discordant twin pairs at the time when one co-twin developed kwashiorkor had significantly more positive IgA indices for Enterobacteriaceae than did members of concordant healthy pairs (P<0.01, Wilcoxon rank-sum test; Fig. 4A,B; see table S4A for the IgA indices for all members of Enterobacteriaceae as well as IgA indices for other family- and species-level bacterial taxa identified in the BugFACS analysis). During RUTF treatment, the IgA response to Enterobacteriaceae decreased significantly in both kwashiorkor and healthy co-twins in discordant pairs (Fig. 4C). This latter finding provided another demonstration of the effect of a nutrient sufficient diet on reducing mucosal immune responses to members of this bacterial family (see Fig. 1A for a comparison of the IgA index for Enterobacteriaceae in KM versus KS mice).
Fig. 4. Enterobacteriaceae are targeted by the gut mucosal IgA response in children from the Malawian twin study.
(A) IgA responses were defined by BugFACS of fecal samples obtained from 11 twin pairs discordant for kwashiorkor. Data from five time points are shown. The first column represents samples taken 1.2±0.6 months prior to diagnosis of kwashiorkor (“Pre-diagnosis”). The second column represents samples taken at the time of diagnosis (“Diagnosis”). The third column and fourth columns are samples taken 2 and 4 weeks after initiation of treatment with RUTF, while the fifth column represents fecal microbiota characterized 1 month after the completion of RUTF therapy. Data represent mean values for the indicated number (n) of kwashiorkor co-twins and healthy co-twins whose fecal samples were available, and are presented in the form of a bubble plot. See table S1 and table S4 for clinical characteristics and details of the datasets, including IgA indices for individual taxa identified as present within each family-level taxonomic group for each individual fecal sample analyzed. (B) At the time of diagnosis, the IgA index for Enterobacteriaceae was significantly higher in co-twins with kwashiorkor in discordant pairs than in twin pairs concordant for healthy status (data from twins concordant for healthy status represent the averaged IgA indices of an individual's fecal specimens obtained between 6 and 24 months of age in order to allow for comparison with discordant twins of varying ages at the time of diagnosis). Purple and green circles highlight IgA indices for co-twins in discordant pairs 46 and 80 who were used for microbial adoptive transfer experiments (see panels D and E). **, P<0.01 (Wilcoxon rank-sum test). (C) Treatment of kwashiorkor co-twins in the discordant pairs shown in panel A with RUTF resulted in a significant decrease in the IgA index score for Enterobacteriaceae. *, P<0.05 (Wilcoxon rank-sum test). Data represent the average IgA index scores for samples obtained 2 and 4 weeks after initiation of RUTF treatment. Mean values ± SEM are plotted. (D) Germ-free mice colonized with a BugFACS-purified IgA+ consortium from the kwashiorkor child in twin pair 46 lost more weight than mice colonized with either the IgA+ consortium purified from the fecal microbiota of his healthy co-twin or a mixture of the two IgA+ populations. **, P<0.01 (Wilcoxon Rank sum). (E) Colonization of germ-free mice with KwashIgA+, HealthyIgA+ or MixIgA+ consortia prepared from discordant twin pair 80 whose members had similar IgA index values for Enterobacteriaceae (see panel B) did not exhibit significant differences in weight loss (n=5-7 mice/treatment group). All mice were fed the Malawian diet starting 1 week before gavage with the IgA+ consortia. Body weights at the time of sacrifice 13 days post-gavage were used to plot the data shown (each mouse represented by a circle).
We cultured purified IgA+ consortia from kwashiorkor co-twins in five of the discordant pairs under aerobic conditions, to recover and characterize Enterobacteriaceae that were the targets of their gut mucosal IgA responses. Sequencing full-length 16S rRNA gene amplicons established that members of Escherichia dominated these cultures. Whole genome shotgun sequencing of the dominant cultured morphotypes followed by screening of the contigs against the 2,311 genes in the Virulence Factor Database (34) disclosed that the three sequenced isolates from kwashiorkor co-twin 46's IgA+ consortium and the two sequenced isolates from kwashiorkor co-twin 26's IgA+ consortium were enteroaggregative E. coli (EAEC, see table S3). The two sequenced strains from kwashiorkor co-twin 138 and the two strains from kwashiorkor co-twin 207 were enterotoxigenic E. coli (ETEC), while the kwashiorkor co-twin from twin pair 80 harbored an atypical enteropathogenic E. coli (EPEC) strain (see table S3 for virulence factors present in these sequenced isolates).
We selected fecal microbiota samples, collected at the time of diagnosis with kwashiorkor, from two of these discordant twin pairs for follow-up functional studies: twin pair 46 where the degree of IgA targeting of Enterobacteriaceae was substantially higher in the kwashiorkor co-twin compared to his healthy co-twin, and twin pair 80 where there were negligible differences in IgA indices for Enterobacteriaceae between the undernourished and healthy siblings (Fig. 4B). IgA+ consortia purified directly from the kwashiorkor co-twin's fecal microbiota, from the healthy co-twin's microbiota, or a 1:1 mixture of the kwashiorkor and healthy co-twin's IgA+ microbes were transplanted into separate groups of germ-free animals (n=5-7 animals/treatment group/donor microbiota; total of 6 groups). All recipient mice were fed the representative Malawian diet. Mice receiving the IgA+ consortium recovered from the kwashiorkor co-twin's microbiota from twin pair 46 lost significantly more weight than did mice receiving his healthy co-twin's IgA+ consortium or the IgA+ mix (P<0.01 for comparison of Kwashiorkor IgA+ consortium to Mix IgA+ and Healthy IgA+ consortia; Wilcoxon rank-sum test; Fig. 4D). In contrast, there was no significant difference in weight loss between recipients of the healthy co-twin's IgA+, kwashiorkor co-twin's IgA+ or Mix IgA+ consortia recovered from twin pair 80 (Fig. 4E).
We used Random Forests, a machine-learning algorithm, to identify taxa that were most associated with weight phenotypes among the six different treatment groups. Analysis of 16S rRNA datasets generated from fecal microbiota samples collected one day prior to sacrifice from mice colonized with the various combinations of IgA+ consortia purified from twin pair 46 and 80 donor microbiota (n=36 samples from 36 animals) disclosed that Enterobacteriaceae were most discriminatory, with highest relative abundances in the fecal microbiota of animals exhibiting the greatest weight loss (fig. S9). The results provided additional evidence that a high positive IgA index to Enterobacteriaceae is indicative of a community capable of transmitting disease, at least in a subset of this cohort.
Identifying interrelationships between IgA responses to bacterial taxa, enteropathogen burden, and nutritional status in the LCNI-5 singleton cohort
Recent reports have used multiplex PCR assays to characterize the relationship between enteropathogen burden and diarrheal disease in children living in low-income countries (35, 36). The LCNI-5 singleton cohort, sampled at 6 and 18 months of age, and exhibiting a broad range of nutritional phenotypes (anthropometric scores), provided an opportunity to systematically assess the extent to which enteropathogen burden correlates with nutritional status and IgA responses. An expanded TaqMan array card (35) was used to assay fecal samples collected from 114 individuals for 39 genes that are known markers for bacterial and viral pathogens, helminths and protozoans. Pathogen carriage rates were very high in this population (see table S6A,B for summary and individual subject results, respectively). The total number of virulence factors detected within an individual's fecal microbiota negatively correlated with nutritional status at 18 months. Bacterial virulence factors were the major contributors [when these factors were considered alone, Spearman rho value of -0.37 for WHZ (P=0.005), rho value of -0.29 for WAZ (P=0.032); correlation was not significant for LAZ]. No significant correlations between WHZ or WAZ and bacterial pathogen burden were identified at 6 months of age although a significant correlation existed between LAZ and viral pathogen burden (table S6A; rho -0.23, P=0.019).
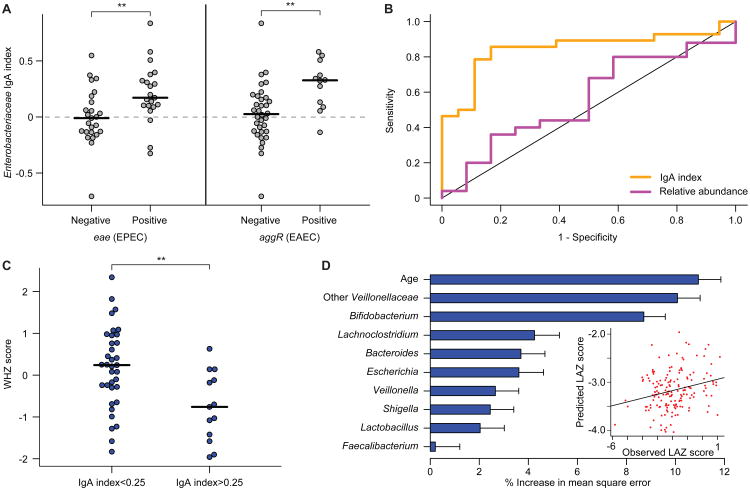
Using this molecular diagnostic method, we examined whether IgA responses (table S4B) correlated with enteropathogen representation in the fecal microbiota of LCNI-5 participants. The most statistically significant relationship involved Enterobacteriaceae: the IgA index to this family was significantly higher when EPEC or EAEC was present (as judged by a positive test for eae and aggR, respectively) (P<0.01, Wilcoxon rank-sum test; Fig. 5A; the relationship was not significant for ETEC). The Receiver Operating Characteristic (ROC) curve presented in Fig. 5B showed that an IgA index cutoff of ≥0.05 had a specificity and sensitivity of 83% and 86%, respectively, for the presence of EPEC (eae+) or EAEC (aggR+). In contrast, the relative abundance of Enterobacteriaceae defined by 16S rRNA analysis of the input fecal sample used for BugFACS did not correlate with the presence of EPEC or EAEC (P>0.05, binomial logistic regression; Fig. 5B).
Fig. 5. Relationships among IgA indices, enteropathogen burden and nutritional status in 18 month-old Malawian children from the LCNI-5 cohort.
(A) IgA indices for Enterobacteriaceae were significantly higher in children that harbored EPEC and EAEC in their microbiota. Each circle represents results from an individual child. **, P<0.01 (Wilcoxon rank-sum test). (B) Receiving Operating Characteristic (ROC) curves for detection of EPEC (eae) and/or EAEC (aggR) using either Enterobacteriaceae relative abundance (purple, defined by V2-16S rRNA sequencing) or Enterobacteriaceae IgA index (orange). Samples were excluded where Enterobacteriaceae were not detected by 16S rRNA sequencing. The correlation to the presence of eae or aggR was significant for IgA index (P < 0.01; binomial logistic regression) but not relative abundance. (C) 18 month-old children from the LCNI-5 cohort with an IgA index value greater than 0.25 for Enterobacteriaceae had lower WHZ scores than did children with an index value less than 0.25. **, P<0.01 (unpaired Student's t-test). (D) Feature importance scores of bacterial taxa that are predictive of LAZ scores were generated by training a sparse Random Forests model using age and genus-level IgA index data from 134 fecal samples collected from the 11 kwashiorkor discordant twin pairs and the eight concordant healthy pairs enrolled in the Malawi Twin Study. To build the model, we included genus-level taxa (features) that had an IgA index value greater than 0.05 or less than -0.05 in 30% of all fecal samples (to remove genera that were only rarely seen and/or had very little enrichment in either the IgA+ or IgA- fractions). The IgA indices for the 25 taxa that satisfied this criterion were regressed against LAZ, and feature importance scores for each genus-level taxon were defined (mean±SD values shown). Shown are the nine genus-level taxa with mean importance scores greater than 1.5% that were incorporated into a 10 feature sparse model, which also included the chronological age of a child. The R2 value of 0.23 represents the goodness of fit of the model when applied to the twin training set, as defined using out-of-bag predictions. The plot in the inset shows that application of this model to 165 fecal samples collected from 6- and 18-month old singleton children in the LCNI-5 study predicted LAZ scores that correlated significantly with their actual LAZ measurements (Spearman's rho=0.2, P=0.009).
In addition to insights about the presence or absence of these enteropathogens, a positive IgA index to Enterobacteriaceae provided additional information about health status. First, we tested a range of cutoffs for the IgA index and found that a value of 0.25 best discriminated healthy from undernourished 18 month-old children in LCNI-5 [P<0.01 (WHZ) and P<0.05 (WAZ); unpaired Student's t-test; Fig. 5C]. Second, combining a positive test for eae (EPEC) and an IgA index >0.25 revealed a statistically significant relationship to LAZ scores (P<0.05; Wilcoxon rank-sum test; table S7). Third, turning to 6 month-old infants, we observed that a positive test for eae and an IgA index >0.25 was associated with better (more positive) LAZ and WAZ scores compared to infants with IgA indices <0.25 (P<0.01, Wilcoxon rank-sum test; table S7). Fourth, 6-month old children with high IgA indices (regardless of pathogen carriage) had significantly better LAZ scores at 18 months compared to those with an index <0.25 (P=0.0001, Wilcoxon rank-sum; n=20 and 25, respectively). In addition, the worsening of LAZ scores between 6 and 18 months of age was significantly greater in children with the lower IgA indices for Enterobacteriaceae [-0.58±0.13 (mean±s.e.m.) in children with IgA indices <0.25 versus -0.15±0.14 in children with indices >0.25, P=0.03; table S7]. In light of the high carriage rates of EPEC and EAEC at 6 months (93% for eae or aggR), these data suggest that a robust IgA response in children harboring these enteropathogens may be protective against growth faltering.
Finally, to further characterize the relationship between IgA response and nutritional status, we used Random Forests to identify taxa whose IgA targeting was correlated with LAZ scores in the Malawian twin cohort. Nine genus-level taxa with mean feature importance scores greater than 1.5% were incorporated into a ten feature sparse model that also included the chronologic age of a child (Fig. 5D). This Random Forests-based model was then applied to the LCNI-5 cohort; the results revealed that the model's predicted LAZ scores correlated with observed LAZ measurements (Spearman's rho=0.23; P=0.009). In addition to members of Enterobacteriaceae, taxa belonging to two groups in the Firmicutes, the family Veillonellaceae and the genus Lachnoclostridium (of which C. scindens is a member), were major contributors to the accuracy of prediction (Fig. 5D). As with Enterobacteriaceae, there was an age effect with Veillonellaceae; IgA indices for this family significantly and positively correlated with LAZ at 6 months (Spearman's rho=0.25; P=0.017) but not at 18 months.
Discussion
Integrating our gnotobiotic mouse data with data from two Malawian cohorts, we observed that children with undernutrition harbored disease-promoting/producing (dysbiotic) gut microbial communities that were characterized by disrupted normal postnatal assembly (immaturity) (5, 6), targeting of specific bacterial taxa by the IgA response, and increased enteropathogen burden. These targets of the gut mucosal IgA responses, prominently represented by members of Enterobacteriaceae and associated enteropathogenic strains, transmitted a weight loss phenotype and disrupted gut barrier function in gnotobiotic mice through mechanisms that were not solely mediated by Enterobacteriaceae, but rather required interactions among members of this family and other IgA-targeted components of the microbiota. Moreover, we identified IgA targeted bacterial taxa (C. scindens, A. muciniphila) that promoted health by ameliorating the effects of KMIgA+ consortia.
While our gnotobiotic mouse studies focused on the contributions of IgA targeted bacterial taxa to disease pathogenesis, a similar approach could be used to delineate the role of IgA negative taxa. The ability to incorporate immunoselected components of human gut communities from children with and without undernutrition, and their diets, into gnotobiotic mouse models will enable further exploration of a proposed pathogenic mechanism where progression to a disease-promoting/producing microbiota reflects a complex intersection and interplay of factors. These factors include (i) personal histories of exposures to microbial reservoirs (encompassing strains acquired from family members as well as other elements of an individual's environment), (ii) the effects of macro-and micronutrient deficient diets on immune responses to acquired strains, (iii) the degree to which functional maturation of the microbiota is compromised, and (iv) the extent to which these communities disrupt gut epithelial barrier integrity, thereby impairing net nutrient absorption and increasing susceptibility to bacterial invasion and systemic infection. Determining the extent to which our findings are generalizable will require studies of additional populations from disparate geographic locations with distinct cultural traditions and nutritional practices. Furthermore, understanding how pathological community structures develop will require characterization of the temporal pattern of evolution IgA responses to gut bacteria in reference groups of children from these populations who have healthy growth phenotypes.
The capacity to recover organisms that are targets of an IgA response in clonally arrayed culture collections, and then introduce these organisms into gnotobiotic mice fed different diets provides a potentially powerful tool in a discovery platform designed to further characterize the combinatorial interactions that are required to effect pathology and to identify disease-modifying as well as disease-preventing next generation probiotics that are naturally occurring gut organisms adapted to the gut ecosystems of individuals living in areas where childhood undernutrition is rampant.
Finally, our studies suggest that performing time series studies of IgA responses to gut bacteria during early childhood in healthy versus undernourished individuals could be used to characterize the normal co-development of mucosal immunity/barrier function and the microbiota, stratify populations at risk for or with already manifest undernutrition, define the efficacy (including the durability) of current and future therapeutic interventions, and perhaps better define the optimal timing of vaccinations in populations where undernutrition is prevalent [e.g. (37, 38)].
Materials and Methods
Study Design
Human study protocols were approved by the College of Medicine Research Ethics Committee of the University of Malawi, the Ethics Committee at the Pirkanmaa Hospital District, Finland, and the Human Research Protection Office of Washington University School of Medicine.
Malawi Twin Study
This clinical trial has been described in a prior publication (6). Briefly, twins were recruited from five villages in southern Malawi: Makhwira, Mitondo, M'biza, Chamba, and Mayaka. A team of American and local health care workers visited each site on a monthly basis, measured height and weight, and screened children for pitting edema of the lower extremities. Fecal specimens were collected every three months from twin pairs who remained healthy. In twin pairs where one co-twin developed kwashiorkor, both co-twins were switched to a peanut-based Ready-to-Use Therapeutic Food (RUTF). Sampling of fecal specimens was increased to bi-weekly while children were receiving RUTF. Fecal specimens were flash frozen in liquid nitrogen shortly after they were produced and stored at -80°C prior to analysis.
LCNI-5 Study
Details of this study may be found at clinicaltrials.gov (ID: NCT00524446) (33). Infants were sampled at 6 months of age prior to any nutritional intervention while samples obtained at 18 months were from members of group D (corn-soy blend which had no statistically significant effect on growth phenotypes).
All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee. Assignment of mice to microbial and diet treatment groups was not blinded since the different diets and the different microbial consortia used were deliberately assembled and administered by the same investigators who performed the animal husbandry and collected the phenotypic data. However, immunostained slides were evaluated in a blinded manner. The numbers of mice assigned to each treatment group in each experiment, and the number of independent repetitions of a given experimental design are described in the text and below, as are the statistical tests used to determine the significance of observed differences.
Gnotobiotic mouse experiments
All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee.
Diets
The Malawian diet was based on food consumption patterns in the catchment area, and consisted primarily of corn flour, mustard greens, yellow onions and tomatoes purchased from Whole Foods Market (stores located in St. Louis, MO). Details of how the diet was assembled/cooked are described in an earlier publication (6). Batches of diet were aliquoted (500 g or 750 g) into plastic bags that were vacuum-sealed, and then placed in a second bag and sterilized by gamma irradiation (Steris Co, Libertyville, IL). Food was stored for up to 6 months at 4°C. Nutritional analyses were performed by N.P. Analytical Labs (St. Louis, MO).
Adult germ-free male C57 BL/6J mice were maintained on a strict 12 h light cycle (lights on at 0600) in flexible plastic gnotobiotic isolators (Class Biologically Clean Ltd., Madison, WI). Mice were weaned onto an autoclaved, standard mouse chow diet low in fat and rich in plant polysaccharides (B&K Universal, East Yorkshire, U.K; diet 7378000). Seven days before introduction of gut microbes by gavage, animals were switched to the Malawian diet or, in the case of controls, maintained on the mouse chow.
Aliquots (approximately 100-200 mg) of previously frozen fecal samples, obtained from members of discordant twin pairs at the time of diagnosis of kwashiorkor in one of the co-twins, were suspended in 1 mL of sterile PBS supplemented with 0.1% cysteine under anaerobic conditions, vortexed vigorously for 5 min at room temperature, placed on ice, and then allowed to undergo gravity sedimentation for 5-10 min. A 200 μL aliquot of this suspension was subsequently introduced into germ-free mice by gavage (see ref. 6 for details of sample preparation, handling and gavage protocols).
Sample preparation for BugFACS
Mouse fecal pellets (∼10-50 mg wet weight), frozen at -80°C immediately after collection, were suspended in sterile PBS by vortexing, placed on ice, and allowed to sediment by gravity for 5-10 min. A 200 μL aliquot of the resulting clarified fecal suspension was passed through a 70 μm sterile nylon mesh filter into a new, sterile tube. Bacteria that passed through the filter were then pelleted by centrifugation (10,000 × g for 1 min at 24°C). The cell-free supernatant was removed; the pellet was washed by resuspension in an additional 1 mL of PBS, and again centrifuged. The resulting pellet was resuspended in 100μL of PBS containing a 1:50 dilution of polyclonal goat anti-mouse IgA conjugated to DyLight649 (Abcam) and incubated on ice for 30 min. The suspension was then washed with 1 mL of sterile PBS, pelleted again by centrifugation, and 200 μL of 0.9%NaCl/0.1 M HEPES buffer (pH 7.2) containing a 1:4000 dilution of SytoBC (Invitrogen/Life Technologies) was added.
Human fecal samples (20-100 mg aliquots), previously frozen at -80°C, were processed as above and stained with a goat anti-human IgA antibody conjugated to DyLight649 (Abcam; note that there was no significant relationship between the weight of the fecal aliquot and IgA staining efficiency).
Samples were sorted using a FACSAria III (BD Biosciences) instrument in a laminar flow bio-containment hood (BioProtect IV Safety Cabinet, Baker Co, Sanford, ME). Fecal samples were analyzed without the use of a neutral density filter to allow the maximum degree of sensitivity for small particles. Threshold settings were set to the minimal allowable voltage for SSC. The gating strategies used to collect different bacterial populations are shown in fig. S1. We collected 20,000-50,000 events from the IgA+ population and a minimum of 100,000 events from the IgA- and ‘input’ populations into sterile tubes (the terms “events” and “microbes” are used interchangeably). Each fraction (typically 50-200 μL) was stored at -20°C prior to PCR and sequencing of bacterial 16S rRNA genes.
Multiple precautions were taken to minimize potential contamination of sorted fractions: (i) freshly autoclaved PBS was used for sheath fluid; (ii) the flow cytometer was sterilized according to the manufacturer's recommended protocol; (iii) the sheath fluid filter was replaced routinely; and (iv) 16S rRNA analysis was performed on samples collected from the flow cytometer droplet stream before and after every sort, thus permitting identification of any sequences that did not originate from the sorted sample.
A simplified in vitro system using a monoclonal IgA antibody generated against B. thetaiotaomicron (39) verified that this approach could be used to isolate and quantify IgA bound bacteria (fig. S2A-G). Analysis of fecal samples collected from adult conventionally-raised wild-type C57BL/6J mice revealed robust IgA targeting to members of the microbiota, while analysis of fecal communities obtained from C57BL/6J Rag1-/- animals that lack B- and T-cells produced no IgA staining above background, confirming the specificity of the BugFACS procedure (fig. S3A).
Bacterial 16S rRNA gene analyses
DNA was prepared from fecal samples by bead beating, followed by phenol-chloroform extraction. Bacterial V2-16S rRNA gene amplicons were generated using barcoded primers. PCR was performed using either 5prime HotMaster Mix or Invitrogen High Fidelity Platinum Taq according to the manufacturer's protocols and cycling conditions described in ref. (40).
In the case of BugFACS, we added 1 μL of the IgA+, IgA-, or ‘input’ fractions directly to PCR master mix containing PCR primers (three replicate 20 μL reactions/sample). Cycling conditions consisted of 95°C for 10 min followed by 30 to 35 cycles of 95°C for 20 s, 52°C for 20 s and 65°C for 60 s. ‘No-template’ controls were run with every sample to ensure that there was no contamination of the barcoded primers or reagents. PCR products were subjected to gel electrophoresis to confirm the presence of amplicons, quantified (Qubit, Invitrogen), and barcoded amplicons were pooled from the different samples for subsequent multiplex pyrosequencing (454 Titanium FLX chemistry; see table S1 and table S4 for information about the datasets).
Pyrosequencer reads were de-multiplexed and clustered based on 97% nucleotide sequence identity (ID) against the Greengenes reference operational taxonomic unit (OTU) database (41). De novo OTUs were generated from unmatched sequences using the UCLUST method in QIIME version 1.5 (42). Data were filtered so that each sample had at least 1000 reads and each 97%ID OTU had to be observed at least twice across all samples. We assigned taxonomy to OTUs using RDP 2.4 trained on a custom database derived from (i) sequence data downloaded from the Greengenes ‘Isolated named strains 16S' database and (ii) modified phylogeny from the NCBI taxonomy database (41, 43). Data were rarefied to a depth of 1000 reads per sample for analyses that required the use of 97%ID OTU level taxonomy.
In calculating the IgA index, we first summarized taxonomy to phylum, class, order, family, genus and species levels. The threshold used for designating that a taxon was called ‘Present’ was ≥0.1% relative abundance in either the IgA+, IgA- or input fractions. A pseudocount (equal to 0.001, which was the lower limit of detection of fractional representation in the community) was then added to every taxon detected in both the IgA+ and IgA- fractions generated from every fecal sample. The sum of the fractional abundance of a given taxon and the pseudocount was log transformed before performing a paired Wilcoxon test (using R (44)) comparing the significance of differences in abundances of that taxon in the IgA+ to IgA- fractions prepared from a group of samples. If a taxon was not detected in a given sample, that sample was excluded from analysis of that taxon. IgA index values of zero represent samples in which a taxon was detected in equal proportions in both the IgA+ and IgA- fractions, or was detected in the ‘input’ fraction but not the IgA+ or IgA- fractions.
The following criteria were used to define sequences as arising from ‘contaminants’ from the flow cytometer: (i) after OTU picking and taxonomy assignments, contaminating OTUs had to constitute >1% of the pre- and post-sort sheath flow-through sequences; (ii) these OTUs had to be detected on at least three separate sorting runs; (iii) if an OTU met criteria (i) and (ii) and was assigned to a genus, all OTUs with the same genus-level assignment were removed from our analysis of BugFACS data. Samples with fewer than 1000 reads after removing contaminating sequences were not included in the analysis.
Transplantation of IgA+ microbial populations into gnotobiotic mice
When sorting gut microbes for subsequent transplantation of the IgA+ population into mice, we maintained the viability of bacteria by minimizing exposure to oxygen as follows: (i) all sample preparation steps, including staining with SytoBC and the DyLight 649-conjugated goat anti-mouse IgA or goat anti-human IgA, were performed in an anaerobic chamber (Coy Lab Products, Grass Lake, MI; atmosphere composed of 75% N2/20% CO2/5% H2); (ii) all buffers used during the preparation and staining steps (PBS and 0.1 M HEPES/0.9% NaCl) were supplemented with 0.1% cysteine HCl; (iii) all buffers were stored in the anaerobic chamber for a minimum of 48 h before use; (iv) plasticware used for preparing samples was stored under anaerobic conditions for a minimum of 3 d prior to use; (v) for steps requiring centrifugation outside of the anaerobic chamber, microbes were first placed in sealed 2 mL screwtop tubes (Axygen) before exiting the chamber, and then returned to the chamber prior to additional processing.
Fecal pellets were harvested from KM and HM mice 42 d after they had been gavaged with the corresponding human co-twin's fecal microbiota. We combined filtered fecal suspensions from three mice in the KM or in the HM group to generate a pooled microbiota that was subsequently used to purify IgA+ consortia that were introduced into germ-free recipients to produce KMIgA+ and HMIgA+ animals. Similarly, KMF2IgA+ fractions were generated from the combined fecal supernatants from five KMIgA+ mice. (KMF2IgA+ animals are the third generation of mice harboring a microbiota derived from one of the Malawian co-twins, and the second generation of mice receiving an IgA-enriched consortium).
Note that microbes were sorted using SSC, or FSC and SSC thresholds set at the minimum possible setting of 200. Once a sufficient number of events was collected, we centrifuged the sorted microbes (10,000 × g; 5 min; 4°C), aspirated the supernatant, and resuspended the microbial cell pellet in a volume of PBS/0.1% cysteine sufficient to capture 100,000 events in 200 μL. Sorted bacteria were sealed in tubes in the anaerobic chamber, brought to the gnotobiotic facility (10 min trip), introduced into the isolator after sterilization of the tube's surface (Clidox), and then gavaged into germ-free mouse recipients.
Other assays of mucosal immune function
We vaccinated KM and HM mice with oral cholera toxin and ovalbumin (45) on days 21, 28 and 35 after gavage of human donor microbiota in one experiment and on day 21 in a repeat experiment. Each dose of vaccine contained 10 μg of cholera toxin and 10 mg of hen egg ovalbumin (Sigma) dissolved in sodium bicarbonate pH 8.0. Vaccines were mixed and filter sterilized by passage through a 0.22 μm-diameter filter then administered in a volume of 200 μL by gavage. No reproducible differences were observed between KM and HM mice as judged by serum ELISA.
Serum cytokine analysis
Sera were collected from mice at the time of sacrifice by retro-orbital phlebotomy, and frozen at -80°C until use. Serum cytokine analysis was performed using a MAGPIX plate reader (Bio-Rad) and Bio-Plex Pro Assay kit according to the manufacturer's instructions.
Histochemical and immunohistochemical analyses
The small intestine and colon were flushed with and fixed overnight with 4% paraformaldehyde in PBS, then transferred to 70% ethanol for storage. The fixed small intestine was cut open along its cephalocaudal axis, and rolled up with the villi pointing outwards. These ‘Swiss rolls’ were secured by embedding in 2% agar. The material was then paraffin embedded, and cut into 5 μm-thick sections that were stained with hematoxylin and eosin. The proximal third of the colon was also opened along its cephalocaudal axis and pinned lumen-side up on paraffin; multiple colons from animals in the same treatment group were stacked one upon the other, secured with agar, and the stack embedded in paraffin prior sectioning and staining.
Small and large intestine samples were examined, in a blinded fashion, for epithelial disruption, crypt number, and neutrophilic infiltrate (in the crypt or lamina propria). The hobnailing pattern of small intestinal epithelium was also noted when present.
Cellular patterns of EpCAM localization were defined by immunostaining. De-paraffinized sections of small intestine were incubated with a 1:500 dilution of a rabbit polyclonal anti-human EpCAM (kindly supplied by Mark Udey, Dermatology Branch, Center for Cancer Research, NIH) followed by 1:500 dilution of donkey anti-rabbit AlexaFluor 594 IgG1 (Invitrogen). Immunostained slides were evaluated in a blinded manner and scored for the presence and location of apical cytoplasmic aggregates in the epithelium below the villus tip.
‘Probiotic’ interventions
Akkermansia muciniphila BAA-835 and Clostridium scindens 35704 were obtained from ATCC (Manassas, VA). Both strains were grown overnight at 37°C in gut microbiota medium (GMM (46)) under strict anaerobic conditions. Organisms harvested in stationary phase were mixed in equal numbers, pelleted by centrifugation (10,000 × g; 1 min, 24°C), and resuspended in PBS with 0.1% cysteine. As many of these manipulations as possible were conducted in an anaerobic Coy chamber (see above). Tubes containing bacteria were sealed when they had to be transported out of the Coy chamber (e.g. for centrifugation and to the gnotobiotic facility for gavage into mice). A separate aliquot of the culture was autoclaved (121°C for 20 min) to kill the organisms, including spores associated with C. scindens, and was then introduced into separate groups of animals as a heat-killed control.
Generation and characterization of arrayed culture collections
In vitro culture of IgA+ fractions prepared from the fecal microbiota of children with kwashiorkor
Fecal specimens from kwashiorkor co-twins in discordant pairs, collected at the time of their diagnosis, were subjected to BugFACS under non-reducing conditions. 105 bacteria from the IgA+ gate were collected, plated onto LB agar plates, and incubated overnight at 37°C under aerobic conditions. Colonies were picked into LB broth (Difco), and grown overnight to stationary phase. DNA was harvested from stationary phase cultures (phenol-chloroform), and purified (QIAquick PCR Purification kit, Qiagen). Full-length amplicons from bacterial 16S rRNA genes were generated using primers 8F and 1391R and ReddyMix master mix (Thermo; cycling conditions were 95°C for 10 min, followed by 35 cycles of 95°C for 45 s, 56°C for 45 s then 72°C for 2 min). Amplicons were purified (QIAquick PCR Purification kit), and sequenced (Sanger method) in both directions using the amplification primers (Retrogen, San Diego). Sequences were assembled using BioEdit (47) and referenced against the GreenGenes database (41) to obtain species-level identification. In the case of twin pair 57, fecal samples were plated directly onto GMM and grown under aerobic conditions overnight (46).
For bacterial genome sequencing, purified genomic DNA was sheared twice for 10 min on ice using a Bioruptor XL sonicator (Diagenode; Denville, NJ) set to ‘High’. Libraries (500 bp) were prepared for each of the strains listed in Supplemental Data Table 3 for sequencing on the Illumina MiSeq instrument (paired-end 250 nt reads). Multiplex sequencing of all strains was achieved using an 8 base pair barcode for each individual strain (48). Sequences were assembled using MIRA (49) (version 4.0.2) and the resulting contigs were used for comparison to a library of known virulence factors31. Whole genome assemblies were compared to publically available sequenced isolates (50).
Generation of clonally arrayed culture collections from the cecal contents of KMF2IgA+ mice
Cecal contents were harvested 2 days after colonization of germ-free mice with a KMF2IgA+ microbiota (mice were fed the Malawian diet 7 days prior to and following gavage). Homogenized cecal contents were spread onto GMM agar plates and incubated for 48 h at 37 °C under anaerobic conditions. The contents of the plates were recovered with a sterile tissue culture cell scraper and resuspended in sterile PBS supplemented with 15% glycerol and 0.1% cysteine. Aliquots were sealed in glass vials under anaerobic conditions and stored at -80°C until use. Clonally arrayed culture collections were generated by first diluting an aliquot of the scraped culture collection onto GMM agar plates. After two days of anaerobic growth at 37°C, 376 individual colonies were picked into four 96 deep-well plates each containing liquid GMM medium and grown for 48 h under anaerobic conditions. Aliquots from each well of the arrayed culture collection that exhibited growth were used to generate glycerol stocks. Genomic DNA was extracted from the remaining contents of each well and used to generate V4 16S rRNA amplicons for sequencing. Whole genome sequencing was subsequently performed on two isolates from each OTU represented in the arrayed culture collection as described above.
Separate groups of mice were gavaged with a pool generated from all wells containing Enterobacteriaceae strains, or a pool containing all non-Enterobacteriaceae strains or a pool containing both the “Enterobacteriaceae” and “Non-Enterobacteriaceae” components of the arrayed culture collection. Our strategy of pooling of multiple independent isolates from the arrayed culture collection was intended to adequately represent strain level diversity in the recipient animals.
FACS analysis of B-cell and T-cell populations recovered from gnotobiotic mice
Mesenteric lymph nodes (MLNs) and spleens were collected at the time of sacrifice and kept at 4°C until processing. All tissue processing was conducted in sterile PBS supplemented with 0.1% bovine serum albumin (Sigma). Tissues were mechanically homogenized and passed through a nylon filter (mesh diameter, 70 μm; BD Falcon). Erythrocytes were removed from splenic samples using ammonium chloride lysis (ACK Lysing Buffer, Gibco). Lymphocyte surface markers were stained using the following antibodies: PerCP-Cy5.5 anti-TCR-β, V500 anti-B220, and FITC anti-CD8 from BD biosciences, plus APC anti-CD4, PE anti-CD44, and PE-Cy7 anti-CD62L from eBioscience. We also stained for FoxP3 using an eFluor450 labeled antibody (eBioscience) with Fixation/Permeabilization Buffer followed by permeabilization/staining with 0.01% saponin/0.009% sodium azide (eBioscience) supplemented with 1% rat serum (Sigma-Aldrich).
Using Random Forests to predict LAZ scores as a function of IgA index values in the Malawian twin and LCNI-5 cohorts
Our training dataset came from the Malawian Twin cohort. To build the model, we included only those genus-level bacterial taxa that had an IgA index value greater than 0.05 or less than -0.05 in 30% of all fecal samples; 25 satisfied this criterion, which was designed to remove taxa that were only rarely seen and/or were only minimally enriched in either the IgA+ or IgA- fractions. Next, we regressed the IgA indices for these 25 taxa against LAZ scores using the Random Forests package as implemented in R (51). Random Forests is a supervised learning method that allows the user to assess the contribution of taxa (“features”) to the accuracy of the overall regression. Feature importance scores are quantified as the percent increase in mean squared error when values (in this case IgA indices) belonging to a feature are randomly permuted. We determined feature importance scores for each genus-level taxon using 1000 iterations with 500 trees/iteration. We also included chronologic age as an additional feature. We then incorporated only those taxa (and chronologic age of the subject in months) that yielded mean importance scores greater than 1.5% to produce a sparse model (52) that explained 18% of the variance relating to LAZ in the training set. We used this sparse model to predict LAZ scores from an individual's IgA index in the LCNI-5 cohort.
Enteropathogen detection by qPCR
Extracted nucleic acids were assayed with TaqMan Array cards as described (35) to detect a panel of enteropathogens, including several belonging to Enterobacteriaceae, such as Enteroaggregative E. coli (EAEC, targeting aggR, aaiC, and aatA genes), Enteropathogenic E. coli (EPEC, targeting eae and bfpA), Enterotoxigenic E. coli (ETEC, targeting both heat stable and heat labile toxin genes), Shiga toxin-producing E. coli (targeting stx1 and stx2), Enteroinvasive E. coli (EIEC, targeting ipaH), Salmonella spp. (targeting ttr), and Shigella spp. (targeting ipaH). Extraction blanks were included to exclude lab contamination. A quantification cycle (Cq) cutoff of 35 was applied to determine positivity based on the lower limit of detection of the assays. Copy numbers of each positive target were calculated from standard curves and normalized to the combined extraction and amplification efficiency derived from the quantities of an extrinsic control (Phocine Herpes Virus) that was incorporated into both the extraction and detection steps.
Statistical Methods
Statistical analyses were performed in R version 3.1.1. A P-value < 0.05 was considered significant. Bargraphs are presented as mean values with error bars representing the standard error of the mean except where otherwise noted. Horizontal bars shown with individual data points represent sample means unless otherwise noted. Specific statistical test are noted in the figure legends.
Supplementary Material
Fig. S1. Characterization of gnotobiotic mice receiving intact uncultured fecal microbiota from discordant twins and the experimental design for subsequent transplantation of IgA+ consortia purified from their fecal microbiota into germ-free recipients.
Fig. S2. Validation of BugFACS using an artificial two-member bacterial community.
Fig. S3. Control experiments validating the specificity of BugFACS.
Fig. S4. Combining HMIgA+ and KMIgA+ consortia attenuates early weight loss in recipient gnotobiotic animals.
Fig. S5. Serum cytokine profiles in KMIgA+ animals and a comparison of immunophenotypes in recipients of KMIgA+, HMIgA+ and MixIgA+ consortia.
Fig. S6. KMF2IgA+ microbiota transmits a colonic epithelial discohesion phenotype to recipient gnotobiotic mice
Fig. S7. IgA-targeted bacterial taxa in the kwashiorkor microbiota disrupt small intestinal epithelial barrier in recipient gnotobiotic mice.
Fig. S8. Representation of bacterial taxa within the gut microbiota of gnotobiotic mice colonized with IgA+ consortia.
Fig. S9. Heatmap of bacterial family-level taxa represented in the fecal microbiota of gnotobiotic mice sampled 12 d after receiving IgA+consortia from discordant twin pairs 46 or 80.
Table S1. Clinical characteristics of Malawian twin donors.
Table S2. IgA indices determined from BugFACS analysis of fecal samples obtained from mice colonized with the fecal microbiota of co-twins discordant for kwashiorkor (pair 57).
Table S3. Characteristics of the genome sequences and content of virulence factors of Enterobacteriaceae isolates obtained from in vitro culture of IgA+ consortia or from spleens of mice colonized with KMF2IgA+ microbiota (originating from donor 57A)
Table S4. IgA indices and relative abundances of family and genus level bacterial taxa in two cohorts of Malawian infants
Table S5. Clinical characteristics of individuals from the Lungwena Child Nutrition Intervention Study #5 (LCNI-5).
Table S6. Multiplex quantitative PCR analysis of enteropathogen burden in members of the LCNI-5 cohort
Table S7. Subgroup analysis of LCNI-5 cohort
Acknowledgments
We thank David O'Donnell, Maria Karlsson, and Sabrina Wagoner for help with gnotobiotic husbandry, Su Deng, Janaki Guruge, and Marty Meier for their superb technical assistance, and Sathish Subramanian, Philip Ahern plus other members of the Gordon lab for their many helpful comments during the course of this study.
Funding: This work was supported by grants from the Bill & Melinda Gates Foundation and the NIH (R01 DK30292, J.I.G). A.L.K is the recipient of a NIH Ruth Kirschstein National Research Service Award (F32 DK091044); part of his salary was also supported by a grant from Danone. J.D.P. is a member of the Washington University Medical Scientist Training Program (NIH GM007200).
Footnotes
Author Contributions: A.L.K., J.D.P. and J.I.G. designed the experiments. S.R. and C.H. helped develop flow cytometry protocols for enriching IgA-coated bacteria. M.J.M and I.T. designed, enrolled and collected specimens from Malawian twins. K.M., P.A., K.D. directed the clinical study design, enrollment, plus clinical data and sample collection for LCNI-5. E.H. developed protocols for TaqMan array-based measurements of pathogens. A.L.K., J.D.P., S.R., T.Y., T.L., T.S., and J.L., performed experiments. A.L.K., J.D.P., and J.I.G. analyzed the data. A.L.K., J.D.P., and J.I.G. wrote the paper.
Competing financial interests: J.I.G is co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. A provisional patent application entitled “Method of isolating and characterizing microorganisms that are targets of host immunity” (Non-Provisional Patent Application Serial No. 14/398,087) has been filed by Washington University related to the techniques described in this paper. The other authors declare that they have no competing interests.
Data and Materials Availability: 16S rRNA datasets are available through the European Nucleotide Archive (ENA Study Accession Number PRJEB5989). Reads from shotgun sequencing datasets generated from cultured bacterial strains and assembled contigs are available through ENA Accession Numbers PRJEB8296 to PRJEB8306.
References
- 1.Levels and Trends in Child Malnutrition, UNICEF-WHO-The World Bank Joint Child Malnutrition Estimates. 2012 URL: http://www.who.int/nutgrowthdb/estimates/en/
- 2.Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, de Onis M, Ezzati M, Grantham-McGregor S, Katz J, Martorell R, Uauy R. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;382:427–451. doi: 10.1016/S0140-6736(13)60937-X. [DOI] [PubMed] [Google Scholar]
- 3.Ruel MT, Alderman H. Nutrition-sensitive interventions and programmes: how can they help to accelerate progress in improving maternal and child nutrition? Lancet. 2013;382:536–551. doi: 10.1016/S0140-6736(13)60843-0. [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewedacross age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokes CR, Soothill JF, Turner MW. Immune exclusion is a function of IgA. Nature. 1975;255:745–746. doi: 10.1038/255745a0. [DOI] [PubMed] [Google Scholar]
- 12.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R, 3rd, McCoy KD, Macpherson AJ. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beatty DW, Napier B, Sinclair-Smith CC, McCabe K, Hughes EJ. Secretory IgA synthesis in Kwashiorkor. J Clin & Lab Immunol. 1983;12:31–36. [PubMed] [Google Scholar]
- 16.Sirisinha S, Suskind R, Edelman R, Asvapaka C, Olson RE. Secretory and serum IgA in children with protein-calorie malnutrition. Pediatrics. 1975;55:166–170. [PubMed] [Google Scholar]
- 17.Reddy V, Raghuramulu N, Bhaskaram C. Secretory IgA in protein-calorie malnutrition. Arch Dis Child. 1976;51:871–874. doi: 10.1136/adc.51.11.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994;16:270–279. doi: 10.1002/cyto.990160312. [DOI] [PubMed] [Google Scholar]
- 19.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–3913. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amor KB, Breeuwer P, Verbaarschot P, Rombouts FM, Akkermans AD, De Vos WM, Abee T. Multiparametric flow cytometry and cell sorting for the assessment of viable, injured, and dead bifidobacterium cells during bile salt stress. Appl Environ Microbiol. 2002;68:5209–5216. doi: 10.1128/AEM.68.11.5209-5216.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 24.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 26.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu S, Ashok M, Li J, Li W, Yang H, Wang P, Tracey KJ, Sama AE, Wang H. Spermine protects mice against lethal sepsis partly by attenuating surrogate inflammatory markers. Mol Med. 2009;15:275–282. doi: 10.2119/molmed.2009.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci USA. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strnad J, Hamilton AE, Beavers LS, Gamboa GC, Apelgren LD, Taber LD, Sportsman JR, Bumol TF, Sharp JD, Gadski RA. Molecular cloning and characterization of a human adenocarcinoma/epithelial cell surface antigen complementary DNA. Cancer Res. 1989;49:314–317. [PubMed] [Google Scholar]
- 30.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol. 1994;125:437–446. doi: 10.1083/jcb.125.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladwein M, Pape UF, Schmidt DS, Schnolzer M, Fiedler S, Langbein L, Franke WW, Moldenhauer G, Zoller M. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Experimental Cell Res. 2005;309:345–357. doi: 10.1016/j.yexcr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Sivagnanam M, Mueller JL, Lee H, Chen Z, Nelson SF, Turner D, Zlotkin SH, Pencharz PB, Ngan BY, Libiger O, Schork NJ, Lavine JE, Taylor S, Newbury RO, Kolodner RD, Hoffman HM. Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology. 2008;135:429–437. doi: 10.1053/j.gastro.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangani C, Maleta K, Phuka J, Cheung YB, Thakwalakwa C, Dewey K, Manary M, Puumalainen T, Ashorn P. Effect of complementary feeding with lipid-based nutrient supplements and corn-soy blend on the incidence of stunting and linear growth among 6- to 18-month-old infants and children in rural Malawi. Matern Child Nutr. 2013 doi: 10.1111/mcn.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40:D641–645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AK, Houpt ER. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. The Lancet Infectious Diseases. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 37.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qadri F, Bhuiyan TR, Sack DA, Svennerholm AM. Immune responses and protection in children in developing countries induced by oral vaccines. Vaccine. 2013;31:452–460. doi: 10.1016/j.vaccine.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host & Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The R Development Core Team. R Foundation for Statistical Computing; Vienna: 2011. R: A language and environment for statistical computing. URL http://www.R-project.org/ [Google Scholar]
- 45.Fahlen-Yrlid L, Gustafsson T, Westlund J, Holmberg A, Strombeck A, Blomquist M, MacPherson GG, Holmgren J, Yrlid U. CD11c (high)dendritic cells are essential for activation of CD4+ T cells and generation of specific antibodies following mucosal immunization. J Immunol. 2009;183:5032–5041. doi: 10.4049/jimmunol.0803992. [DOI] [PubMed] [Google Scholar]
- 46.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 48.Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med. 2013;5:184ra160. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chevreux B, Wetter T, Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics. 1999;99:45–56. [Google Scholar]
- 50.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 52.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Characterization of gnotobiotic mice receiving intact uncultured fecal microbiota from discordant twins and the experimental design for subsequent transplantation of IgA+ consortia purified from their fecal microbiota into germ-free recipients.
Fig. S2. Validation of BugFACS using an artificial two-member bacterial community.
Fig. S3. Control experiments validating the specificity of BugFACS.
Fig. S4. Combining HMIgA+ and KMIgA+ consortia attenuates early weight loss in recipient gnotobiotic animals.
Fig. S5. Serum cytokine profiles in KMIgA+ animals and a comparison of immunophenotypes in recipients of KMIgA+, HMIgA+ and MixIgA+ consortia.
Fig. S6. KMF2IgA+ microbiota transmits a colonic epithelial discohesion phenotype to recipient gnotobiotic mice
Fig. S7. IgA-targeted bacterial taxa in the kwashiorkor microbiota disrupt small intestinal epithelial barrier in recipient gnotobiotic mice.
Fig. S8. Representation of bacterial taxa within the gut microbiota of gnotobiotic mice colonized with IgA+ consortia.
Fig. S9. Heatmap of bacterial family-level taxa represented in the fecal microbiota of gnotobiotic mice sampled 12 d after receiving IgA+consortia from discordant twin pairs 46 or 80.
Table S1. Clinical characteristics of Malawian twin donors.
Table S2. IgA indices determined from BugFACS analysis of fecal samples obtained from mice colonized with the fecal microbiota of co-twins discordant for kwashiorkor (pair 57).
Table S3. Characteristics of the genome sequences and content of virulence factors of Enterobacteriaceae isolates obtained from in vitro culture of IgA+ consortia or from spleens of mice colonized with KMF2IgA+ microbiota (originating from donor 57A)
Table S4. IgA indices and relative abundances of family and genus level bacterial taxa in two cohorts of Malawian infants
Table S5. Clinical characteristics of individuals from the Lungwena Child Nutrition Intervention Study #5 (LCNI-5).
Table S6. Multiplex quantitative PCR analysis of enteropathogen burden in members of the LCNI-5 cohort
Table S7. Subgroup analysis of LCNI-5 cohort