Abstract
The unique proline isomerase Pin1 is pivotal for protecting against age-dependent neurodegeneration in Alzheimer’s disease (AD), with its inhibition providing a molecular link between tangle and plaque pathologies. Pin1 is oxidatively modified in human AD brains, but little is known about its regulatory mechanisms and pathological significance of such Pin1 modification. In this paper, our determination of crystal structures of oxidized Pin1 reveals a series of Pin1 oxidative modifications on Cys113 in a sequential fashion. Cys113 oxidization is further confirmed by generating antibodies specifically recognizing oxidized Cys113 of Pin1. Furthermore, Pin1 oxidation on Cys113 inactivates its catalytic activity in vitro, and Ala point substitution of Cys113 inactivates the ability of Pin1 to isomerize tau as well as to promote protein turnover of tau and APP. Moreover, redox regulation affects Pin1 subcellular localization and Pin1-mediated neuronal survival in response to hypoxia treatment. Importantly, Cys113-oxidized Pin1 is significantly increased in human AD brain comparing to age-matched controls. These results not only identify a novel Pin1 oxidation site to be the critical catalytic residue Cys113, but also provide a novel oxidative regulation mechanism for inhibiting Pin1 activity in AD. These results suggest that preventing Pin1 oxidization might help to reduce the risk of AD.
Keywords: Alzheimer’s disease, Pin1, Oxidation, Tau, APP
Introduction
Proline-directed protein phosphorylation (pSer/Thr-Pro) is a central signaling mechanism in diverse cellular processes. Certain pSer/Thr-Pro motifs in polypeptides exist in two completely distinct conformations, cis and trans, the conversions of which are markedly slowed upon phosphorylation, but yet are specifically catalyzed by the unique peptidyl-prolyl cis/trans isomerase Pin1 (Lu & Zhou, 2007; Lee et al., 2011b; Liou et al., 2011). This striking substrate specificity results from the unique N-terminal WW domain and C-terminal PPIase domain of Pin1 (Lu & Zhou, 2007; Lee et al., 2011b; Liou et al., 2011). The WW domain binds only to specific pSer/Thr-Pro-motifs and targets Pin1 close to its substrates, where the PPIase domain isomerizes specific pSer/Thr-Pro motifs and induces conformational changes in proteins (Lu & Zhou, 2007; Lee et al., 2011b; Liou et al., 2011). Importantly, such Pin1-induced conformational changes following phosphorylation control various protein functions, including their catalytic activity, phosphorylation status, protein interaction, subcellular location, and/or protein stability (Lu & Zhou, 2007; Lee et al., 2011b; Liou et al., 2011). Not surprisingly, due to its vast protein targets, Pin1 is important in many cellular processes involving Pro-directed phosphorylation, including the cell cycle, cell signaling, transcription and splicing, DNA damage responses, germ cell development and neuronal survival (Lu & Zhou, 2007; Girardini et al., 2011; Lee et al., 2011b; Liou et al., 2011; Yuan et al., 2011). Significantly, these Pin1-induced conformational changes after phosphorylation can profoundly impact diverse cellular processes, especially in aging and Alzheimer’s disease (AD) (Lu et al., 1999, Zhou et al., 2000; Ryo et al., 2001; Liou et al., 2002; Atchison et al., 2003; Liou et al., 2003; Lu et al., 2003; Butterfield et al., 2006a; Lee et al., 2009; Lee et al., 2011b).
These cis and trans conformation-specific functions and their regulation by Pin1 have been directly demonstrated by the development of cis and trans conformation-specific antibodies (Nakamura et al., 2012). Pin1 protein levels were shown to be especially low in vulnerable neurons or degenerative neurons in AD (Liou et al., 2003), suggesting a neuroprotective role for Pin1 (Lu et al., 2003). Indeed, in normal brains, Pin1 is mainly expressed in the nucleus in most neurons at unusually high levels and is in the soluble fraction (Lu et al., 1996; Lu et al., 1999; Ryo et al., 2001; Wulf et al., 2001; Thorpe et al., 2004). However, in AD brains, Pin1 co-localizes and co-purifies with intracellular neurofibrillary tangles (NFTs), resulting in depletion of soluble Pin1 (Lu et al., 1999; Thorpe et al., 2001; Ramakrishnan et al., 2003; Thorpe et al., 2004). Direct evidence for this notion has come from determining Pin1 expression in human brains and analyzing the neuronal phenotypes of Pin1 knockout (KO) mice. Neurons in different subregions of the hippocampus are known to have differential vulnerability to AD neurodegeneration (Pearson et al., 1985; Hof & Morrison, 1991; Arriagada et al., 1992; Davies et al., 1992; Thal et al., 2000). Moreover, Pin1 expression inversely correlates with the predicted neuronal vulnerability in normally aged brains and also with actual neurofibrillary degeneration in AD (Liou et al., 2003; Pastorino et al., 2006). Pin1 KO mice develop progressive age-dependent neuropathy characterized by motor and behavioral deficits, tau hyperphosphorylation, tau filament formation, Aβ pathology and neuronal degeneration (Liou et al., 2003; Pastorino et al., 2006; Cancino et al., 2013). These phenotypes resemble many aspects of AD neurons and those in many tau/APP-related transgenic mice (Liou et al., 2003; Pastorino et al., 2006; Cancino et al., 2013). Finally, transgenic overexpression of Pin1 in postnatal neurons is able to suppress tau hyperphosphorylation, tangle formation and neurodegeneration induced by overexpression of human wild-type tau (Lim et al., 2008). Thus, Pin1 is pivotal for protecting against age-dependent tau- and Aβ-related pathologies and neurodegeneration in AD. However, little is known about how Pin1 activity is inhibited in AD.
Oxidative stress has been implicated in the pathogenesis and progression of AD, manifested by protein oxidation, lipid peroxidation, DNA oxidation, advanced glycation and products, and reactive oxygen species (ROS) formation (Markesbery, 1997; Butterfield et al., 2001; Butterfield & Lauderback, 2002; Butterfield et al., 2010). ROS itself can facilitate different kinds of protein oxidation (Stadtman & Berlett, 1997). Our previous studies have shown that Pin1 is oxidized in human AD brains and the level of Pin1 oxidization is elevated in AD (Sultana et al., 2006), suggesting a possible link between Pin1 oxidation and Pin1 inhibition in AD. However, the regulatory mechanisms and pathological significance of Pin1 oxidative modification remain unknown.
In this study, we treated Pin1 protein crystals with various concentrations of H2O2 and identified a series of Pin1 oxidative modifications on Cys113 in a sequential fashion, eventually leading to inactivation of Pin1 catalytic activity. To confirm these results, we generated antibodies specifically recognizing oxidized Cys113 Pin1, and found that Pin1 was oxidized on Cys113 in vitro and in vivo and that such Pin1 oxidization was induced by treatment with H2O2 or diamine. In confirmation of the significance of Pin1 Cys113 oxidation in tauopathy, APP processing and Aβ production in cells, we found that Pin1, but not Pin1 C113A point mutant, promoted protein turnover of tau and APP, and increased neuronal survival in hypoxia. Of note, the levels of oxidized Pin1 was significantly increased in human AD and MCI brains, as compared with age-matched controls. These results not only identify a novel Pin1 oxidation site, but provide a novel oxidative regulation mechanism for Pin1 enzymatic activity in AD.
Results
Structural basis for the inhibitory function of Pin1 Cys113 oxidation
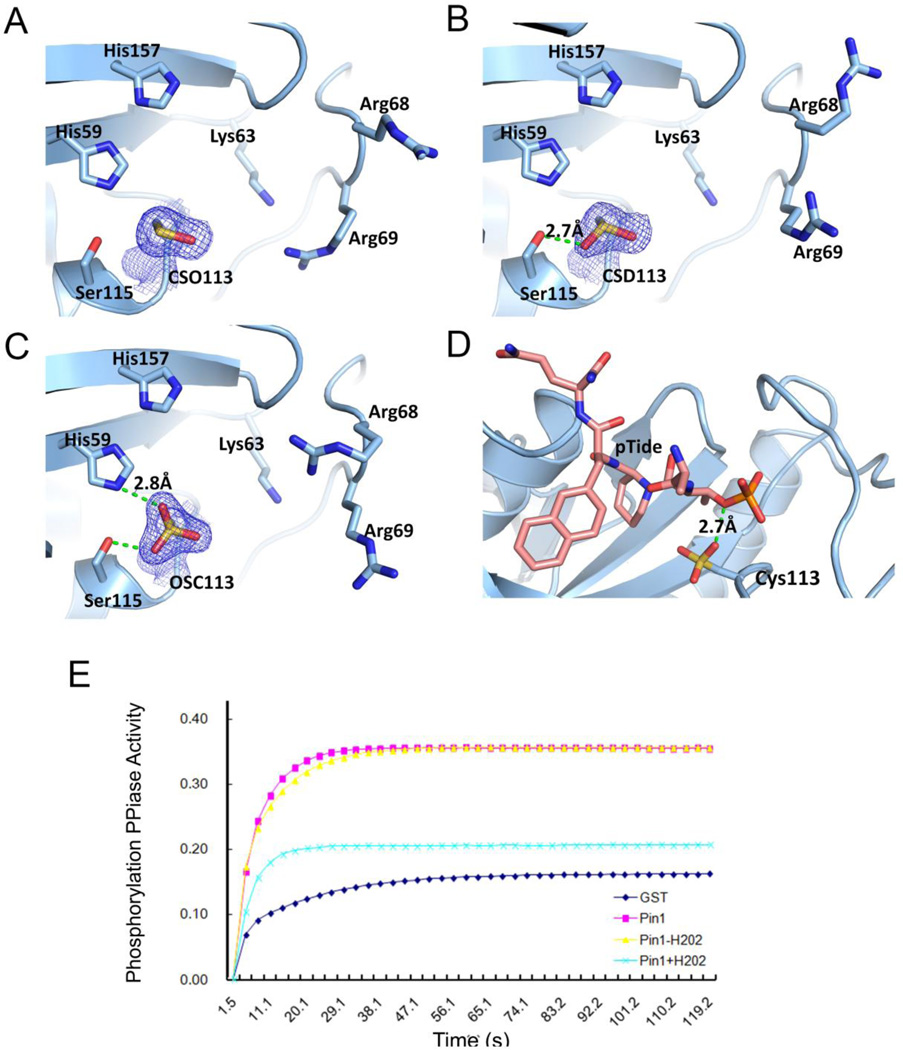
Although our previous proteomic analysis showed that Pin1 is oxidized in human AD brains and Pin1 oxidization in vitro inhibits its catalytic activity (Sultana et al., 2006), it is not known where Pin1 is oxidized and how this oxidative modification affects Pin1 catalytic activity. In order to mimic the oxidized environment of AD brains, we treated Pin1 crystals with various concentrations of H2O2 while altering exposure time. This dose-dependent strategy can reveal the susceptibility of residues upon oxidation damage. With low dosage of H2O2 treatment, Pin1 crystals consistently diffracted to high resolution of 1.7 Å. Careful examination of the Pin1 structure showed that the only residue undergoing modification is Cys113 (Table 1). The other cysteine residue in Pin1 (Cys57) exhibits no additional density. Preference for oxidation site is affected by three factors: thiolnucleophilicity of cysteine residue, local environment and the proximity of the target thiol to the reactive oxygen species. Hydrogen network calculation has shown that Cys113 has a perturbed pKa that is 3 units less than usual (Barman & Hamelberg, 2014), making this residue much more susceptible for sulfenic modification. Interestingly, the position of the oxygen addition follows a consistent sequential fashion influenced by the local environment. The first oxygen atom generates the s-hydroxy cysteine (cysteine sulfenic acid), which potentially could be reversibly reduced. The oxygen atom is always added at the position that is most exposed to the solvent (Fig. 1A). With higher concentrations of H2O2 and prolonged exposure, a second oxygen atom was observed as sulfinic acid (Fig. 1B). The position of the second oxygen atom is stabilized by an intramolecular hydrogen bond with Ser115 (2.7 Å). Finally, the extended treatment of H2O2 saturated the Cysteine with the third oxygen atom added as sulfonic form (Fig. 1C), intra-molecularly hydrogen bonding to His59 (2.9Å). More exposure to H2O2 result in the total loss of diffraction of Pin1 crystals, suggesting that the structural integrity of the protein is compromised upon high dose of oxidation.
Table 1.
X-ray Data Collection and Refinement Statistics
| Pin1 with sulfenic | Pin1 with sulfinic | Pin1 with sulfonic | |
|---|---|---|---|
| Protein | Cys113 | Cys113 | Cys113 |
| PDB | 4U84 | 4U85 | 4U86 |
| Data Collection statistics | |||
| Source | ALS 5.0.2 | ALS 5.0.2 | ALS 5.0.2 |
| Wavelength (Å) | 1.00 50-1.78 |
1.00 50-1.70 |
1.00 50-1.60 |
| Resolution (Å) | (1.81-1.78)* | (1.73-1.70) | (1.63-1.60) |
| Space group | P3121 | P3121 | P3121 |
| Unit Cell | 68.90, 68.90, | 68.77, 68.77, | 69.07, 69.07, |
| a, b, c (Å) | 79.76 | 79.60 | 79.73 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Data cutoff | F>0 | F>0 | F>0 |
| Molecular per asymmetric unit | 1 | 1 | 1 |
| Number of unique reflections | 21461 | 24448 | 19744 |
| Redundancy | 5.2 (3.6) | 5.3 (3.9) | 5.2 (3.7) |
| Completeness (%) | 98.6 (96.6) | 99.1 (91.5) | 99.7 (99.4) |
| I/σ (I) | 23 (1.5) | 36 (1.7) | 32 (1.9) |
| Rsym (%) | 7.5 | 7 | 7.8 |
| Refinement statistics | |||
| Refinement | |||
| Resolution limit (Å) | 59.67-1.78 | 59.56-1.70 | 59.82-1.60 |
| No. reflections (test) | 20060 (1414) | 22968 (1231) | 28112 (1499) |
| Rwork/Rfree (%)# | 19.7/22.4 | 20.5/24.5 | 19.8/23.1 |
| No. atoms | 1296 | 1291 | 1282 |
| protein | 1176 | 1166 | 1167 |
| Water | 101 | 102 | 98 |
| PEG | 19 | 23 | 17 |
| B-factors (Å2) | 28.9 | 26.1 | 27.0 |
| protein | 28.3 | 25.5 | 26.4 |
| Water | 35.4 | 33.0 | 33.7 |
| PEG | 31.7 | 28.0 | 30.6 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.03 | 0.03 | 0.03 |
| Bond angles (°) | 2.37 | 2.5 | 2.4 |
| Ramachandran plot (%) | |||
| Most favored regions | 95.2 | 93.5 | 95.2 |
| Additional allowed regions | 4.8 | 6.5 | 4.8 |
| Generously allowed regions | 0 | 0 | 0 |
| Disallowed regions | 0 | 0 | 0 |
| MolProbity score^ | 96% | 94% | 92% |
| Bad Rotamer | 1.57% | 1.60% | 0.80% |
| Clashscore | 96% | 94% | 96% |
Highest resolution shell is shown in parenthesis.
Rfree is calculated with 5% of the data randomly omitted from refinement.
MolProbity score combines the clashscore, rotamer, and Ramachandran evaluations into a single score, normalized to be on the same scale as X-ray resolution. 100th percentile is the best among structures of comparable resolution; 0th percentile is the worst. For clashscore the comparative set of structures was selected in 2004, for MolProbity score in 2006
Fig. 1.
Structural basis for the inhibitory function of Pin1 Cys113 oxidation.
A) 2Fo-Fc map of Pin1 with Cys113 oxidized to sulfenic acid. Important active site residues are labeled. Cys113 subject to oxidation is shown in Italic. Contour level of 2Fo-Fc map is 1σ.
B) 2Fo-Fc map of Pin1 with Cys113 oxidized to sulfinic acid; The hydrogen bond is represented by a dashed green line.
C) 2Fo-Fc map of Pin1 with Cys113 oxidized to sulfonic acid.
D) Superimposition of oxidized Pin1 structures with structure of Pin1 bound with pTide (PDB 2ITK). Cysteine sulfonic acid is shown by sticks.
E) Oxidation of Pin1 abolishes the pSer-Pro specific PPIase activity. Purified GST-fusion proteins were produced and treated with H2O2 or vehicle, then followed by assaying their catalytic activity towards a pSer-Pro containing peptide.
After the identification of the target site of oxidation on Pin1, we sought to understand if the oxidation of Cys113 would affect the binding affinity of Pin1 to its target. Superimposition of the various oxidized forms of Pin1 with the complex structure of Pin1 with a high affinity substrate-mimicking peptidemimetic (PTide: Ac-Phe-Phos.Thr-Pip-Nal-Gln-NH2) showed that the oxidized Cys113 in Pin1 does not cause steric clash with peptide inhibitor (Fig. 1D). Instead, a hydrogen bond can be formed between the phosphate group of the ligand and oxygen atom (Fig. 1D). Indeed, oxidized Pin1 still effectively recognized the high affinity ligand, as shown by oxidized Pin1 being pulled by the biotinylated Pin1 trapping peptide pTide beads (Fig. 2B).
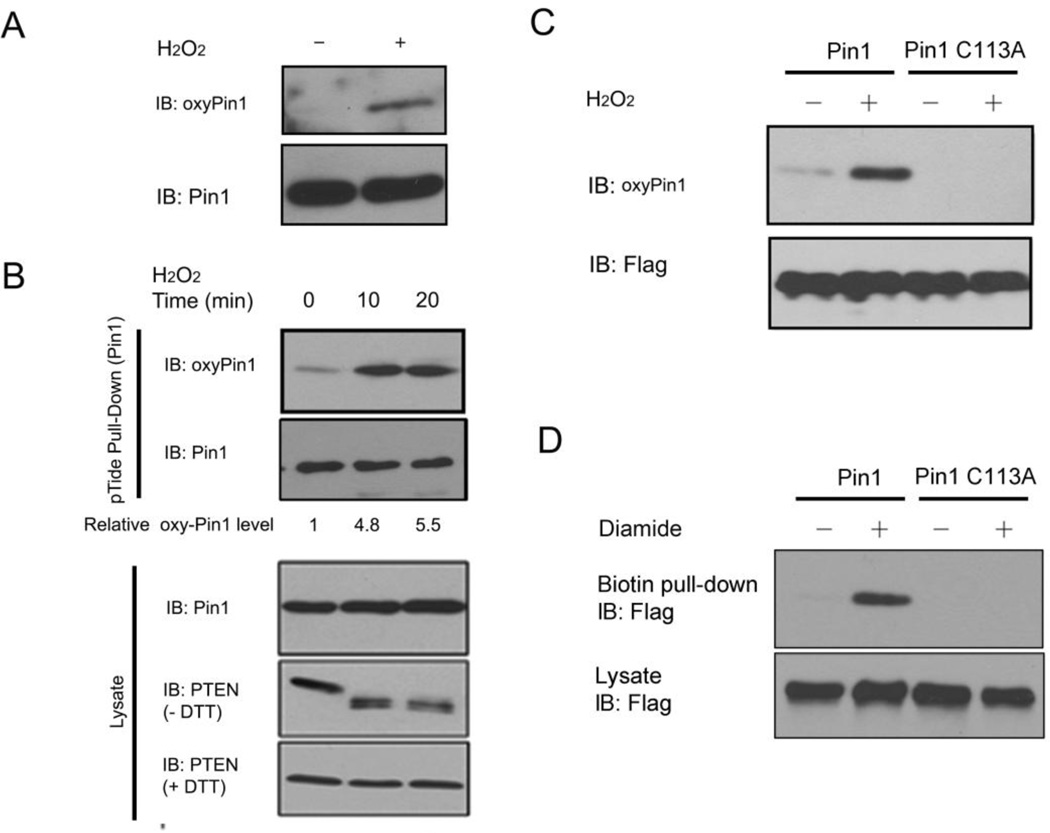
Fig. 2.
Pin1 is oxidized on Cys113 in vitro and in cells.
A) Validation of oxyPin1 antibodies in vitro. Purified recombinant Pin1 was treated with H2O2 or vehicle, followed by immunoblot analysis with oxyPin1 or Pin1 antibodies.
B) Validation of Pin1 oxidation on Cys 113 in cells. Cells were treated with H2O2 and followed by pull-down with biotin-pTide beads before subjecting to immunoblotting with oxyPin1, Pin1 and PTEN antibodies.
C) Pin1 is oxidized on Cys113. Cells were co-transfected with Flag-Pin1 or its mutants and then treated with H2O2. Cell extracts were pulled down with biotin-pTide beads and immunoblotted by oxyPin1 and Flag antibodies.
D) Pin1 is oxidized on Cys113 after diamine treatment. Cells were co-transfected with Flag-Pin1 or its mutants and then treated with 250 µM Diamine for 15 min. Oxidized Pin1 were labeled with biotin-maleimide, and cell extracts were purified with streptavidin-sepharose beads, and immunoblotted by oxyPin1 and Flag antibodies.
However, the oxidation of the free thiol on Cys113 can potentially compromise its role in catalytic reaction, the mutation of which abolishes most of the isomerase reaction (Ranganathan et al., 1997). To test if oxidation inhibits the enzymatic activity of Pin1, we oxidized purified recombinant Pin1 with H2O2, followed by assaying its PPIase activity. Indeed, upon oxidation, Pin1 showed little phospho-specific PPIase activity once when compared to pre-treated Pin1 (Fig. 1E). Our results thus indicate a model in which that the oxidative damage on Pin1 occurs to a specific residue at Cys113, which abolishes its catalytic activity even though it can still effectively bind to substrate peptide and therefore, trap the substrate.
Identification of Pin1 Cys113 oxidation in vitro and in cells in response to oxidative stress
To confirm oxidation of Pin1 on Cys113 in cells, we generated antibodies specifically recognizing oxidized Cys113 in Pin1 (oxyPin1) using an antibody-based method for the monitoring of Pin1’s oxidative state (Persson et al., 2005). We found that the antibodies readily detected robust signal when recombinant Pin1 protein was treated with H2O2, but not in vehicle treatment (Fig. 2A). Having demonstrated Pin1 Cys113 oxidation in vitro, we assessed Pin1 oxidation in cells. By treating cells with H2O2 for various times and subsequently pulling down endogenous Pin1 with the high affinity (1.1 nM) Pin1 trapping peptide pTide beads (Wildemann et al., 2006), followed by immunoblotting analysis with oxyPin1 antibodies. Indeed, H2O2 treatment resulted in a time-dependent increase in oxyPin1 (Fig.2B). To further confirm the oxidation site(s) on Cys113, we generated a Pin1 C113A mutant (Cys113 residue to Ala), and showed that the antibodies failed to recognize the Pin1 C113A mutant, in contrast to wild-type Pin1 (Fig. 2C). These results indicate that Cys113 is a Pin1 oxidation site, and its Pin1 oxidation is increased after H2O2 stimulations. To confirm Pin1 oxidization under a different condition, we treated cells expressing Flag-Pin1 or its mutant C113A, with diamide, a chemical reagent well known to induce protein oxidization (Anastasiou et al., 2011), and lysed cells under denaturing conditions in the presence of maleimide to block reduced cysteine in proteins, followed by reduction of oxidized cysteine and labeling with biotin-maleimide for the detection of Pin1 oxidation with streptavidin, as described previously (Anastasiou et al., 2011). Using this method, we also detected biotin-labeled Flag-Pin1 in lysates of diamide-treated cells, but not in Flag-Pin1 C113A mutant cells (Fig. 2D). Together with our results in Figure 1, we propose that the C113A mutated Pin1 will not only abolish Pin1 catalytic activity but cannot undergo the oxidation event. These results also indicate that Pin1 is oxidized on Cys113 in cells.
Pin1 Cys113 oxidation inhibits its ability to increase protein turnover of Tau and APP in neuroblastoma cells
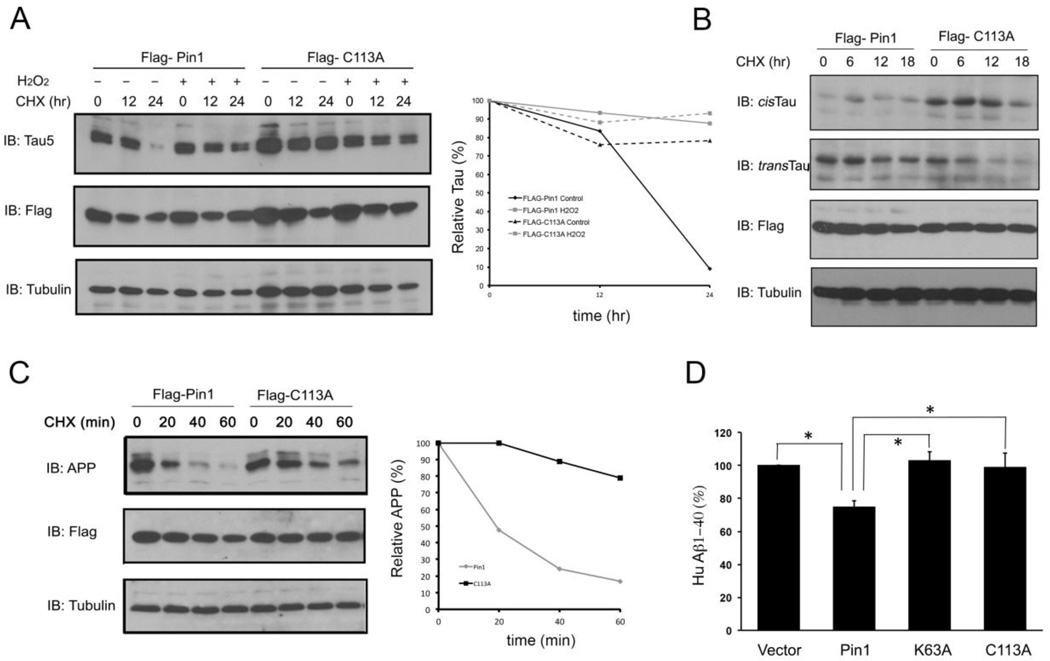
Given that Pin1 is oxidized on the critical active site Cys113 in cells, and such oxidization abolishes Pin1 phospho-specific PPIase activity in vitro, a critical question is whether Pin1 oxidization affects Pin1 cellular functions. Since one of the well-characterized Pin1 functions in protecting against AD is its ability to promote protein turnover of tau and APP, we asked whether Pin1 Cys113 oxidation affects its ability to regulate tau and APP protein turnover (Driver et al., 2014). To clarify this, we first co-transfected tau with Flag-Pin1 or Flag-Pin1 C113A mutant into SH-SY5Y neuroblastoma cells, then added cyclohexmide to inhibit de novo tau synthesis in the presence of H2O2 treatment. Oxidation almost completely inhibited the ability of Pin1 to reduce tau stability in wild-type Pin1, but not its C113A mutant-expressing cells (Fig. 3A). To demonstrate whether Pin1, but not C113A mutant, indeed catalyze cis to trans isomerization of the pT231-Pro motif in tau, we assayed cis and trans pT231-tau conformations using conformation-specific antibodies, as described previously (Nakamura et al., 2012). Overexpression of Pin1, but not its C113A mutant, significantly promoted cis to trans isomerization of pT231-tau (Fig. 3B). These results confirm our previous findings that cis p-tau is much more stable than trans, and that Pin1 catalyzes cis to trans isomerization to reduce tau protein stability (Nakamura et al., 2012). It also indicates that Cys113 is critical for the ability of Pin1 to catalyze cis to trans isomerization of pT231-tau.
Fig. 3.
Oxidation of Pin1 on Cys113 inhibits its cellular function to promote tau and APP protein turnover in neurons.
A) Pin1 Cys113 oxidation inhibits the Pin1 ability to promote tau turnover. Cells were co-transfected with tau or Flag-Pin1 or its mutants with tau and then treated with cycloheximide (100 mg/ml) in the presence or absence of H2O2 for the indicated times, followed by immunoblot analysis with anti-Flag, tau or tubulin antibodies, tau levels were semi-quantitated using tubulin as a loading control.
B) Cells were co-transfected with Flag-Pin1 or its mutants and then treated with cycloheximide (100 mg/ml) for the indicated times, followed by immunoblot analysis with anti-Flag, tau, cis-tau, trans-tau or tubulin antibodies.
C) Cells were co-transfected with Flag-Pin1 or its mutants with APP and then treated with cycloheximide (100 mg/ml) for the indicated times, followed by immunoblot analysis with anti-Flag, APP or tubulin antibodies. APP levels were semi-quantitated using tubulin as a loading control.
D) Cells were co-transfected with Flag-Pin1 or its mutants and then treated with cycloheximide (100 mg/ml) for the indicated times The levels of secreted Aβ 1–40 were measured by sandwich ELISA and data were normalized against the vector control. Results shown are mean ± SEM, n = 3. *, p <0.05.
To examine the role of Cys113 in the ability of Pin1 to regulate APP and Aβ production, we co-transfected APP with Flag-Pin1 or Flag-Pin1 C113A mutant into SH-SY5Y cells, followed by assaying APP stability and Aβ production. Overexpression of wild-type Pin1 reduced APP protein stability and Aβ production (Fig. 3C, D), as shown previously (Pastorino et al., 2006; Ma et al., 2012a; Pastorino et al., 2012). However, the C113A Pin1 mutant displayed little activity to reduce APP protein stability and Aβ production, with activity similar to the catalytic inactive mutant K63A mutant (Fig. 3C, D). These results provide the first evidence for the essential role of the active site residue Cys113 in Pin1’s regulation of tau and APP protein stability in cell models, and suggest that Pin1 oxidation would inhibit the ability of Pin1 to prevent tau- and APP-related pathologies in AD.
Redox regulation affects Pin1 subcellular localization and Pin1-mediated neuroblastoma cells survival
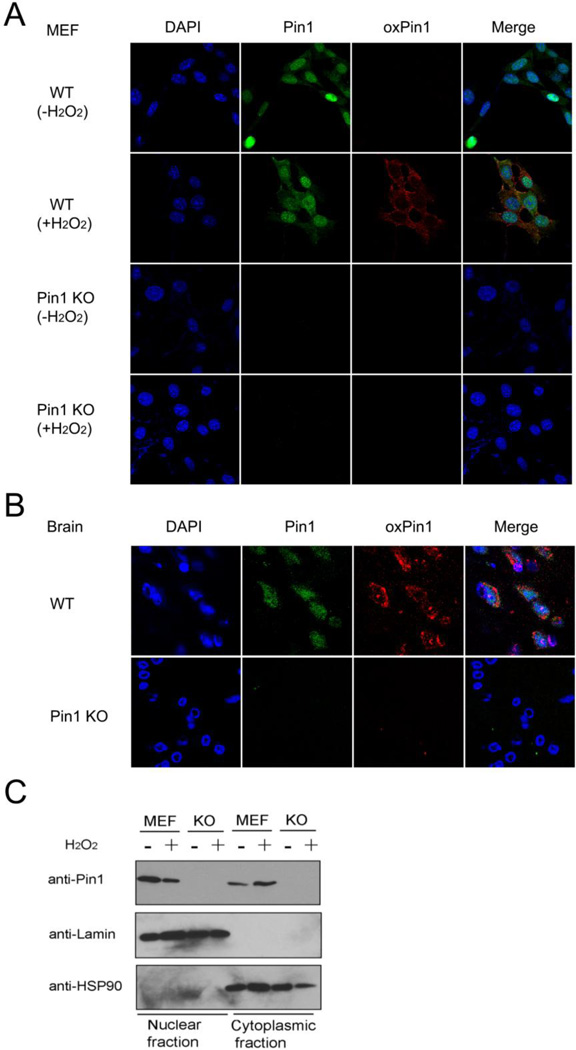
Given that the oxidation of Pin1 on Cys113 impairs its catalytic activity and cellular function, an important question is whether this oxidation has any biological significance or pathological consequence. We first examined whether Pin1 oxidization affects its subcellular localization using immunostaining analysis with anti-oxidized Cys113 antibody or Pin1 antibody. Indeed, after H2O2 treatment, Pin1 cytosolic localization was significantly increased (Fig. 4A and 4B). These results were further confirmed by subcellular fractionation experiments showing that H2O2 treatment reduced levels of nuclear Pin1, but increased levels of cytoplasmic Pin1 (Fig.4C). These results suggest that Pin1 oxidation impairs Pin1 nuclear localization. Interestingly, it has been previously shown that Pin1 is relocalized from the nucleus to the cytoplasm in AD brains (Lu et al., 1996; Lu et al., 1999, Ryo et al., 2001; Wulf et al., 2001; Thorpe et al., 2004).
Fig. 4.
Redox regulation impairs Pin1 subcellular localization.
A and B) H2O2 treatment reduces Pin1 nuclear localization and increases cytoplasmic localization. WT or Pin1 KO cells or mouse brain tissues were immunostained with anti-Pin1 (green), anti-oxyPin1 (red) antibodies and DAPI (blue).
C) H2O2 treatment reduces Pin1 nuclear localization. WT or Pin1 KO cells were harvested and nuclear/cytoplasm fractions isolated, followed by detecting Pin1 protein using anti-Pin1 antibodies.
Next, we searched for signals that induce Pin1 oxidation. Recent evidence has shown that H2O2 acts as an intracellular messenger associated with important signaling pathways in diverse physiological conditions such as hypoxic microenvironments (Haskew-Layton et al., 2010). Moreover, it has been shown that hypoxia-inducible transcription factor-1 (HIF-1) controls a large percentage of hypoxic responses and involves in neurodegenerative disorders. Given that Pin1 is an important regulator of the HIF-1 activity (Jalouli et al., 2014; Lonati et al., 2014), we next examined whether Pin1 oxidation affects its function in cell proliferation and survival in neurons.
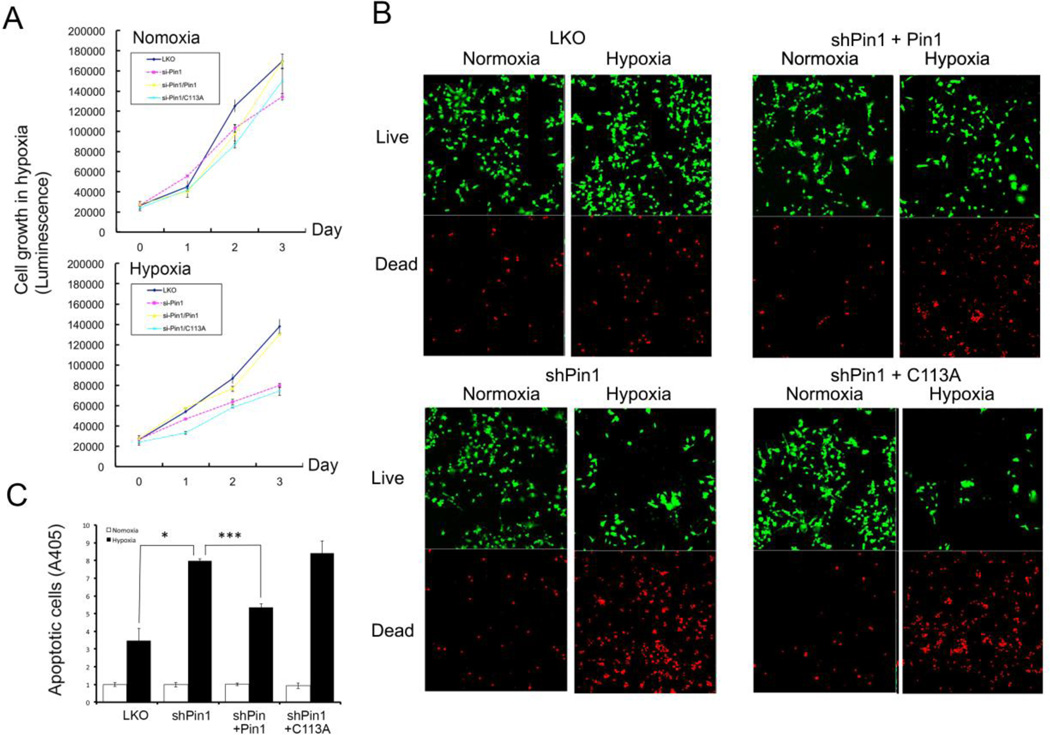
To address this question, we generated SH-SY5Y vector control, Pin1 knockdown cells, and Pin1 shRNA cells stably re-expressing shRNA-resistant Pin1 or Pin1 C113A, followed by assaying cell proliferation using MTT assay. We showed that Pin1 was required for cell proliferation in SH-SY5Y cells and that cell growth was more sensitive in response to the hypoxia treatment in Pin1 knockdown cells and Pin1 C113A mutant cells (Fig.5A).
Fig. 5.
Redox regulation of Pin1-mediated neuronal survival.
A) Oxidation Pin1 on Cys113 is involved in neuronal survival in response to hypoxia treatment. Vector control or Pin1 knockdown cells or Pin1 knockdown cells re-expressing Pin1 or Pin1 mutant were subjected to hypoxia treatment, followed by MTT assay.
B) Oxidation Pin1 on Cys113 is involved in neuronal survival in response to hypoxia treatment. Vector control or Pin1 knockdown cells or Pin1 knockdown cells re-expressing Pin1 or Pin1 mutant were incubated first in the presence 1% oxygen for 24 hr and then assayed for apoptosis using Live/Dead Cell Assay.
C) Oxidation Pin1 on Cys113 is involved in hypoxia-induced cell apoptosis. Vector control or Pin1 knockdown cells or Pin1 knockdown cells re-expressing Pin1 or Pin1 mutant were incubated first in the presence 1% oxygen for 24 hr, then assayed for apoptosis using Cell Death-Detection ELISA. Results shown are mean ± SEM, n = 3. *, p <0.05 and ***, p <0.001.
Consistent with our MTT assay results, we found that cell viability was decreased in the live/dead assay (Fig. 5B), and a similar result was shown to display higher apoptosis in SH-SY5Y Pin1 knockdown or Pin1 C1113A stably expressing cells (Fig. 5C). These results not only indicate that Pin1 is required for cell proliferation and survival under the condition of hypoxia, but Cys113 is critical for Pin1-mediated cell survival under oxidative stress.
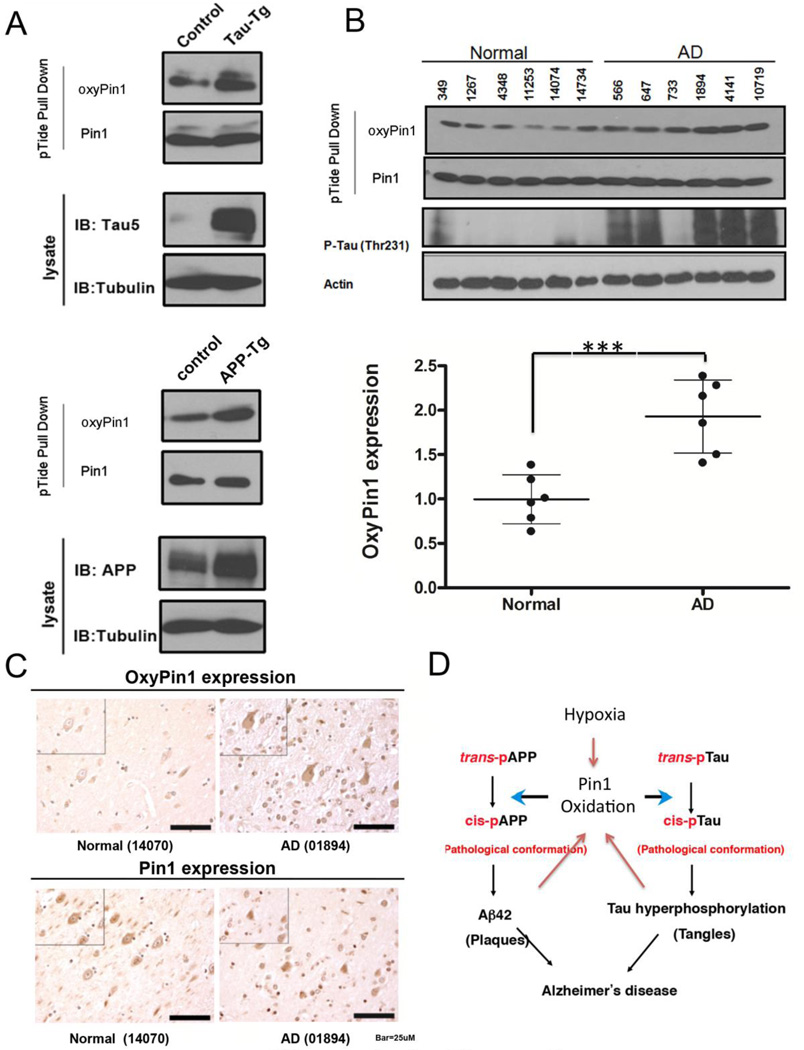
Pin1 Cys113 oxidation is increased both in AD mouse models and human AD samples
To examine Pin1 oxidation in response to oxidative damage in mouse models of AD, we used the Pin1 trapping pTide to isolate Pin1 from tau-transgenic (tg), APP-tg mice and their WT littermates at 18 months old, followed by immunoblotting with oxyPin1 antibodies. As expected, we found that Pin1 Cys113 oxidation was elevated in the brains of both tau and APP transgenic mouse models (Fig. 6A), which are known to have elevated oxidative stress (Chou et al., 2011; Lv et al., 2014). To determine the role of Pin1 oxidation in AD, we measured levels of Pin1 Cys113 oxidation in human AD hippocampus and age-matched controls by using biotinylated pTide to pull down Pin1, followed by immunoblotting with anti-oxyPin1 antibodies. Consistent with previous studies (Kim et al., 2014), phosphorylated Thr231 of tau was highly overexpressed in the brains of AD. More importantly, oxidized Pin1 levels were significantly increased in AD brains, as compared to age-matched controls (p<0.05) (Fig. 6B). Increased Pin1 oxidation was further confirmed by immunohistochemical staining on AD brain sections (Fig. 6C). Thus, Pin1 Cys113 oxidation is increased in human AD brains and AD mouse models.
Fig. 6.
Pin1 Cys113 oxidation is elevated in human AD brains and AD mouse models.
A) Pin1 Cys113 oxidation is increased in brains of tau-Tg mice, APP-Tg mice. Brain tissues were prepared from tau-Tg mice, APP-Tg mice and their age-matched control mice, then subjected to immunoblotting with oxyPin1, Pin1, tau, P-tau (231) or actin antibodies.
B) Pin1 Cys113 oxidation is increased in brains of human AD patients. Brain tissues were prepared from 6 AD patients and 6 age-matched controls, then subjected to immunoblotting with oxyPin1, Pin1, tau, P-tau (231) or actin antibodies. Densitometry values for oxyPin1 were expressed as the mean ± standard error (***, p <0.001 vs. normal control; ANOVA/Dunnett’s test).
C) Paraffin-embedded brain sections from AD patients and controls were immunostained to compare the levels of Pin1 and oxyPin1. Original magnification, Å~40 (main photographs); Å~60 (insets); Scale bar =100 µm.
D) A model by which Pin1 oxidation inhibits its function to regulate tau and APP in both physiological and pathological AD conditions.
Discussion
Functional inhibition of Pin1 contributes to the development of AD by inducing tau and Aβ pathologies and neurodegeneration in an age-dependent manner. However, not much is known about how Pin1 activity is inhibited in AD. In this report, we identified a new oxidation site, Cys113 in the Pin1 catalytic domain, and linked it to the pathobiology of AD. Our crystal structural analysis showed that Pin1 is oxidized on the critical active site Cys113 in a sequential fashion within oxidative environments created by H2O2. The oxidation of active site Cys113 does not prohibit the binding of substrate at the PPIase domain but totally abolishes the isomerase activity. The oxidation of Cys113 greatly reduces Pin1-dependent protein stability of tau and APP in neurons. Moreover, redox regulation impairs Pin1 Cys113 subcellular localization and Pin1-mediated neuronal survival. These results suggest that Pin1 is required for cell proliferation and survival under hypoxic conditions, and that Cys113 is critical for such Pin1-mediated cell survival. Moreover, Cys113 oxidation of Pin1 is elevated in AD mouse models and human AD samples. These results have demonstrated that the oxidation on the active site Cys113 residue causes Pin1 inactivation and mislocalization, contributing to AD pathology.
In contrast to most other PPIases (Hunter, 1998; Fischer & Aumuller, 2003), emerging evidence suggests that Pin1 function may be regulated at multiple levels in neurons. Indeed, in normal brains, Pin1 is mainly expressed in most neurons at unusually high levels and is in the soluble fraction (Lu et al., 1996; Lu et al., 1999; Ryo et al., 2001; Wulf et al., 2001; Thorpe et al., 2004). However, in AD brains, cytoplasmic Pin1 co-localizes and co-purifies with NFTs, resulting in depletion of soluble Pin1 (Lu et al., 1999; Thorpe et al., 2001; Ramakrishnan et al., 2003; Thorpe et al., 2004). Increasing evidence also suggests that Pin1 is subject to post-translational modifications. Pin1 is oxidatively modified, which inhibits its PPIase activity (Sultana et al., 2006). Moreover, the oxidized Pin1/total Pin1 levels is elevated in the early stage of AD pathology (Butterfield et al., 2006b). Oxygen glucose deprivation can trigger partial inhibition of Pin1 enzymatic activity and also increase Ser16 phosphorylation (Lonati et al., 2014). Our work has recently identified an essential role of DAPK1 in regulating PPIase activity of Pin1 on Ser71 in aberrant tau protein regulation and function, providing a link between DAPK1 and Pin1 in regulating age-dependent neurodegeneration (Lee et al., 2011a; Kim et al., 2014). Moreover, a SNP preventing Pin1 inhibition is associated with delayed onset of human AD, preventing its suppression by AP4 (Ma et al., 2012b). However, it will remain to be a major challenge to elucidate the significance and regulation of Pin1 post-translational modifications.
Although Pin1 has been shown to be activated by multiple mechanisms during oncogenesis, much less is known about whether and how Pin1 enzymatic activity is regulated in AD. Here we showed that Pin1 Cys113 oxidation abolishes its PPIase activity. Two reaction mechanisms for prolyl isomerase activity of Pin1 have been proposed, both of which place Cys113 in a pivotal role in mediating the catalytic reaction. In one model, the thiolated Cys113 would act as a nucleophile and attack the carbonyl carbon of the peptidyl-prolyl bond of the substrate and form a covalent bond (Ranganathan et al., 1997). The oxidation of the Cys113 prevents the nucleophilic, attacking by eliminating the thiolated cysteine, therefore abolishing isomerase activity. This mechanism is reminiscent of the redox regulation of tyrosine phosphatase in which the nucleophile cysteine is reversibly oxidized to turn on and off the activity for signal transduction (Lo Conte & Carroll, 2013). The alternative mechanism using kinetic isotope effect does not support the formation of a covalent bond intermediate between Pin1 and substrate. Instead, Cys113 is proposed to destabilize the pseudo double bond character of the peptidyl-prolyl bond of the substrate and facilitate isomerization, in a non-covalent fashion (Mercedes-Camacho et al., 2013). It was also suggested that an extended hydrogen-bonding network formed by Cys113-Serl 15-His59-His157-Thr152 plays a central role for the destabilization effect (Barman & Hamelberg, 2014). Since the reaction of isomerization is highly dependent on the environment of the active site to destabilize the substrate and promote a twisted amide transition state, the oxidation of cysteine can interfere with this process. In both scenarios, the oxidation of Cys113 will not support the isomerase reaction mediated by human Pin1, which has been confirmed by our Pin1 PPIase assay in vitro and Pin1 cellular function to regulate tau and APP protein stability in the neuron.
The transcription factor, HIF-1, is a critical mediator and its activation by hypoxia involves O2-dependent posttranslational modifications and nuclear translocation (Zepeda et al., 2013). The interaction between Pin1 and HIF-1 allows for the activation of specific HIF-1-dependent genes through p42/44 MAPK pathway activation, providing an important link between HIF-1, Pin1 activity and VEGF expression in cancer cells (Jalouli et al., 2014). However, several lines of evidence suggest that hypoxic conditions may also play an important role in AD progression (Ogunshola & Antoniou, 2009; Bulbarelli et al., 2012; Lonati et al., 2014). Intracellular oxidative stress is produced in hypoxia by formation of ROS and results in a process that can damage cell structure such as lipids, membranes, proteins and DNA (Zepeda et al., 2013). It has been reported that hypoxia can facilitate AD pathogenesis by up-regulating APP processing and Aβ production by increasing BACE1 gene expression (Sun et al., 2006). Moreover, Aβ impairs mitochondria redox activity and increases the generation of ROS (Yatin et al., 1999; Kadowaki et al., 2005; Calkins & Reddy, 2011). Several studies also indicate that Aβ-induced oxidative stress leads to apoptotic neuronal cell death (Behl et al., 1994; Mattson & Goodman, 1995; Pillot et al., 1999; Yatin et al., 1999; Calkins & Reddy, 2011). Interestingly, in neurons subjected to OGD, the binding and catalyzing of HIF-1 isomerization by Pin1 are partially inhibited playing a central role in GSK-3β-mediated proteasomal degradation of HIF-1 (Farr et al., 2014; Lonati et al., 2014). Consistent with this idea is the pivotal role of Pin1 in AD. Pin1 interacts with a number of proteins in a phosphorylation-dependent and mitosis-specific manner (Lu, 2004; Wulf et al., 2005; Butterfield et al., 2006a). Interestingly, many of these mitotic phosphoproteins such as tau and APP also have well documented roles in AD (Lu, 2004). Indeed, Pin1 knockout or inhibition has been shown to play an important role in the development of AD, both in mouse models and human patients (Liou et al., 2003; Lu, 2004; Pastorino et al., 2006; Sultana et al., 2006). In contrast, a genetic variation preventing Pin1 inhibition delays age of onset of late onset of AD (Ma et al., 2012b). This is especially exciting given our current findings that Pin1 is oxidized in the brain of human AD and tau and APP-transgenic mouse models. Given elevated oxidative stress in both APP- and tau-transgenic mice (Chou et al., 2011; Lv et al., 2014), these results suggest that inhibition of Pin1 enzymatic activity by hypoxia or other mechanisms might contribute to age-dependent tau- and Aβ-pathologies and neurodegeneration in AD in a positive feedback loop (Fig. 6D). Our recent studies show that upon the hypoxia treatment, neurons robustly produce cis p-tau, which disrupts the microtubule network, interrupts mitochondrial transport in neurites, spreads to other neurons, and leads to massive apoptosis. Given that these cis and trans conformation-specific functions and their regulation by Pin1 (Nakamura et al., 2012). Together these findings not only support our results but provide a molecular mechanism involved in this neuroprotective action of Pin1 on hypoxia-mediated SH-SY5Y cell death. However, further studies are required to elucidate molecular mechanisms regulating Pin1 activity and its coordination with various cell signaling, and how its deregulation contribute to age-dependent neurodegeneration in AD.
Materials and Methods
Plasmids
The expression constructs for wild type and Pin1 mutantsPin1 mutants in which the Cys residue 113 were each replaced by an Ala residue, was generated by site-directed mutagenesis, and then subcloned to pLenti6/V5-GW/lacZ vector as previously described (Lee et al., 2011a; Chen et al., 2013).
PPIase assay
The PPIase activity of Pin1 and oxidized Pin1 were determined using the protease free PPIase activity assay with the substrate Suc-Ala-pSer-Pro-Phe-pNA, Suc-Ala-Glu-Pro-Phe-pNA or Suc-Ala-Ala-Pro-Phe-pNA (50 µM) in 35 mM HEPES pH 7.8 at 10°C, as described previously (Yaffe et al., 1997).
Crystallization and H2O2 treatment of Pin1
The human Pin1 R14A was constructed and purified using previous reported procedure (Jez et al., 2000). Briefly, the Pin1 R14A was overexpressed using E. coli BL21 (DE3) strain at 16°C overnight induced by isopropyl-β-D-thiogalactopyranoside (IPTG). After elution from Ni-NTA (Invitrogen NY) chromatography purification, the N-terminal polyhistidine tag was removed by thrombin protease (Novegen Germany) during the overnight dialysis. The protein was further purified by gel filtration superdex75 (GE Healthcare) in 20mM HEPES 7.5 and 50mM NaCl. Pin1R14A was crystallized with hanging drop vaporization tray mixed 1µl of ~10 mg ml−1 protein solution and 1µl of crystallization buffer (50mM HEPES pH7.5, 1% PEG400, 1.3–1.5 M Ammonium Sulfate). The crystals appeared after three days of incubation at 4°C. Mature crystals were then treating with addition of 1%, 5% or 10% of H2O2 for 30 min to 16 hrs. The oxidization experiment was quenched by harvesting the crystals for data collection.
Diffraction Data Collection and Structure Determination
X-Ray diffraction data were collected from the Advanced Light Source (Berkeley, CA) synchrotron radiation beamlines 5.0.2. Data were processed and scaled using the HKL2000 software suite (Otwinowski & Minor, 1997). Data collection statistics are summarized in Table 1. Molecular replacement was used to determine the structure of oxidized Pin1 R14A with Pin1 R14A (PDB 2ITK) as search model by the program Phaser from the CCP4 package suite (1994). Structures were refined by Rafmac5 (1994) and by iterative model building in COOT (Emsley & Cowtan, 2004; Emsley et al., 2010). The final structures are validated by PROCHECK (Laskowski et al., 1993) and MolProbity (Chen et al., 2010). Refinement statistics are summarized in Table 1. Molecular figures were generated using PyMOL (Schrodinger, 2010). The coordinates and structure factors of Pin1 oxidized states were deposited to Protein Data Bank with codes, 4U84, 4U85 and 4U86.
Protein stability assay
For protein stability assay, cells were transfected stably or transiently with expression plasmids as indicated. Cycloheximide (100 µg/ml) was added to the media to block new protein synthesis. Cells were harvested at each time point, and total lysates were analyzed by immunoblot with anti-tau, anti-APP, anti-Pin1, anti-tubulin antibodies. The blots were scanned and semi-quantitated by using the software NIH image 1.6.2, as described previously (Lee et al., 2009). The results from at least three independent experiments are plotted such that the protein levels at 0 h time point is set at 1.
Establishment of stable cell lines
SH-SY5Y cells were infected with Pin1, and its mutants or control constructs and stable lines were selected using 5 µg/ml of blastidine, as described previously (Lee et al., 2011a; Chen et al., 2013). To overexpress Pin1 and its variants constructs in SH-SY5Ycells, cells were sequentially infected with Pin1 lentiviruses or control vectors, followed by selection. Stable cell clones or pools were checked for protein expression by immunoblotting analysis with various antibodies to confirm the expected protein expression. We maintained stable cell lines continuously in culture, splitting on every fourth day and seeding at the concentration of 6x105 cells per 10 cm culture dish.
Sample preparation
Hippocampus from AD (n = 6) and age-matched controls were individually homogenized separately in Media-I [10 mMHepes buffer (pH 7.4), 137 mMNaCl, 4.6 mMKCl, 1.1 mM KH2PO4, 0.1 mM EDTA, 6 mM MgSO4, leupeptin (0.5 mg/ml), pepstatin (0.7 µg/ml), type II S soybean trypsin inhibitor (0.5 µg/ml), and phenylmethylsulfonyl fluoride (40 µg/Ml)]. These homogenates were centrifuged at 3,000 g for 10 min to remove unbroken cells and nuclear fraction. Protein concentration in the supernatant was determined by the BCA assay (Pierce Chemical, Rockford, IL, USA).
Immunoprecipitation of Pin1
For the immunoprecipitation, 250 µg of the samples were first precleared with protein A/G-agarose beads for an hour at 4 °C. Samples were then incubated overnight with anti-Pin1 antibody (Stressgen, CA). The antigen-antibody-protein A/G complex was centrifuged at 1000 g for 5 min and the resultant pellet was washed five times with IP buffer [phosphate-buffered saline (PBS) containing 0.05% NP-40 and the protease inhibitors leupeptin (4 µg/ml final concentration), pepstatin (4 µg/ml final concentration), and aprotinin (5 µg/ml final concentration), adjusted to pH 8]. The final pellet was suspended in deionized water. Proteins were resolved on SDS-PAGE, followed by immunoblotting on a nitrocellulose membrane (Bio-Rad).
Immunodetection of oxidized Cys-113 (Oxy-Cys 113) of Pin1
The membranes were blocked with 3% bovine serum albumin (BSA) in PBST for 1 h at room temperature, followed by incubated with anti-Oxy-Cys 113 polyclonal antibody (1:1000) for 2 h at room temperature. Following the primary antibody incubation, the membranes were washed three times in Wash Blot for 5 min each and incubated with ECL Plex Cy 5 Dye conjugated anti-rabbit antibody (GE Healthcare, Piscataway, NJ, USA) for hour in dark at room temperature. The membranes were washed in Wash Blot three times for 5 min each and the membrane was scanned using Storm860 phosphoimager (GE Healthcare). For the detection of Pin1 levels the membrane were stripped using stripping buffer (100mM 2-mercaptoethanol, 2% (w/v) sodium dodecyl sulphate, 62.4 mMTris-HCl, pH 6.7), and probed with anti Pin1 antibody (Stressgen, CA), following by incubation with anti-rabbit IgG alkaline phospohotase (ALP)-linked secondary antibody (GE Healthcare, Piscataway, NJ, USA), and developed using BCIP and NBT, and membranes were scanned using a MicrotekScanmaker 4900 scanner.
Image analysis
The images were saved as Tiff files in grayscale mode and the intensity of the oxidized Cys113 and Pin1 were quantified using ImageQuant (GE Healthcare) analysis software.
Statistical analysis
Raw values were exported to Microsoft Excel and the specific oxidation of Cys-113 was determined by dividing the intensity on the blot probed with anti-Cys 113 by total amount of Pin1. The final result was normalized to percentage control values and analyzed by Student’s t tests. A value of p =/< 0.05 was considered statistically significant.
Highlights.
Crystal structures of oxidized Pin1 reveals a series of Pin1 oxidative modifications on Cys113.
Pin1 Cys113 oxidation inactivates its ability to isomerize and regulate tau and APP.
Redox regulation affects Pin1 subcellular localization and Pin1-mediated neuronal survival.
Pin1 Cys113 oxidation is significantly increased in human AD brains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
X.Z.Z. and K.P.L. are inventors of Pin1 technology, which was licensed by BIDMC to Pinteon Therapeutics. Both Dr. Lu and Dr. Zhou own equity in, and consult for, Pinteon. Dr. Lu also serves on its Board of Directors. Their interests were reviewed and are managed by BIDMC in accordance with its conflict of interest policy.
References
- Bailey SM. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada PV, Marzloff K, Hyman BT. Distribution of Alzheimer-type pathologic changes in nondemented elderly individuals matches the pattern in Alzheimer's disease. Neurology. 1992;42:1681–1688. doi: 10.1212/wnl.42.9.1681. [DOI] [PubMed] [Google Scholar]
- Atchison FW, Capel B, Means AR. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development. 2003;130:3579–3586. doi: 10.1242/dev.00584. [DOI] [PubMed] [Google Scholar]
- Barman A, Hamelberg D. Cysteine-mediated dynamic hydrogen-bonding network in the active site of Pin1. Biochemistry. 2014;53:3839–3850. doi: 10.1021/bi5000977. [DOI] [PubMed] [Google Scholar]
- Behl C, Davis JB, Lesley R, Schubert D. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell. 1994;11:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Bulbarelli A, Lonati E, Brambilla A, Orlando A, Cazzaniga E, Piazza F, Ferrarese C, Masserini M, Sancini G. Abeta42 production in brain capillary endothelial cells after oxygen and glucose deprivation. Molecular and cellular neurosciences. 2012;49:415–422. doi: 10.1016/j.mcn.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Abdul HM, Opii W, Newman SF, Joshi G, Ansari MA, Sultana R. Pin1 in Alzheimer's disease. Journal of neurochemistry. 2006a;98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Drake J, Pocernich C, Castegna A. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends in molecular medicine. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Galvan V, Lange MB, Tang H, Sowell RA, Spilman P, Fombonne J, Gorostiza O, Zhang J, Sultana R, Bredesen DE. In vivo oxidative stress in brain of Alzheimer disease transgenic mice: Requirement for methionine 35 in amyloid beta-peptide of APP. Free radical biology & medicine. 2010;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free radical biology & medicine. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol Dis. 2006b;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Calkins MJ, Reddy PH. Amyloid beta impairs mitochondrial anterograde transport and degenerates synapses in Alzheimer's disease neurons. Biochimica et biophysica acta. 2011;1812:507–513. doi: 10.1016/j.bbadis.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancino GI, Miller FD, Kaplan DR. p73 haploinsufficiency causes tau hyperphosphorylation and tau kinase dysregulation in mouse models of aging and Alzheimer's disease. Neurobiol Aging. 2013;34:387–399. doi: 10.1016/j.neurobiolaging.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH, Wei S, Li FA, Chen RH, Zhou XZ, Shih HM, Lu KP. SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer research. 2013;73:3951–3962. doi: 10.1158/0008-5472.CAN-12-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JL, Shenoy DV, Thomas N, Choudhary PK, Laferla FM, Goodman SR, Breen GA. Early dysregulation of the mitochondrial proteome in a mouse model of Alzheimer's disease. Journal of proteomics. 2011;74:466–479. doi: 10.1016/j.jprot.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Davies DC, Horwood N, Isaacs SL, Mann DM. The effect of age and Alzheimer's disease on pyramidal neuron density in the individual fields of the hippocampal formation. Ada Neuropathol (Berl) 1992;83:510–517. doi: 10.1007/BF00310028. [DOI] [PubMed] [Google Scholar]
- Driver JA, Zhou XZ, Lu KP. Regulation of protein conformation by Pin1 offers novel disease mechanisms and therapeutic approaches in Alzheimer's disease. Discovery medicine. 2014;17:93–99. [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr SA, Ripley JL, Sultana R, Zhang Z, Niehoff ML, Platt TL, Murphy MP, Morley JE, Kumar V, Butterfield DA. Antisense oligonucleotide against GSK-3beta in brain of SAMP8 mice improves learning and memory and decreases oxidative stress: Involvement of transcription factor Nrf2 and implications for Alzheimer disease. Free radical biology & medicine. 2014;67:387–395. doi: 10.1016/j.freeradbiomed.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Aumuller T. Regulation of peptide bond cis/transisomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol. 2003;148:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, Mano M, Rosato A, Crook T, Scanziani E, Means AR, Lozano G, Schneider C, Del Sal G. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Haskew-Layton RE, Payappilly JB, Smirnova NA, Ma TC, Chan KK, Murphy TH, Guo H, Langley B, Sultana R, Butterfield DA, Santagata S, Alldred MJ, Gazaryan IG, Bell GW, Ginsberg SD, Ratan RR. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proc Natl Acad Sci U S A. 2010;107:17385–17390. doi: 10.1073/pnas.1003996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Morrison JH. Neocortical neuronal subpopulations labeled by a monoclonal antibody to calbindin exhibit differential vulnerability in Alzheimer's disease. Exp Neurol. 1991;111:293–301. doi: 10.1016/0014-4886(91)90096-u. [DOI] [PubMed] [Google Scholar]
- Hunter T. Prolyl isomerase and nuclear function. Cell. 1998;92:141–143. doi: 10.1016/s0092-8674(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Jalouli M, Dery MA, Lafleur VN, Lamalice L, Zhou XZ, Lu KP, Richard DE. The prolyl isomerase Pin1 regulates hypoxia-inducible transcription factor (HIF) activity. Cellular signalling. 2014;26:1649–1656. doi: 10.1016/j.cellsig.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Kadowaki H, Nishitoh H, Urano F, Sadamitsu C, Matsuzawa A, Takeda K, Masutani H, Yodoi J, Urano Y, Nagano T, Ichijo H. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell death and differentiation. 2005;12:19–24. doi: 10.1038/sj.cdd.4401528. [DOI] [PubMed] [Google Scholar]
- Kim BM, You MH, Chen CH, Lee S, Hong Y, Hong Y, Kimchi A, Zhou XZ, Lee TH. Death-associated protein kinase 1 has a critical role in aberrant tau protein regulation and function. Cell death & disease. 2014;5:el237. doi: 10.1038/cddis.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, Lu KP. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell. 2011a;42:147–159. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev Mol Med. 2011b;13:e21. doi: 10.1017/S1462399411001906. [DOI] [PubMed] [Google Scholar]
- Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, Balastik M, Pastorino L, Wulf G, Zhou XZ, Lu KP. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nature cell biology. 2009;11:97–105. doi: 10.1038/ncb1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Balastik M, Lee TH, Liou YC, Sun A, Finn G, Pastorino L, Lee VM-Y, Lu KP. Pin1 has opposite effects on wild-type and P301L tau stability and tauopathy. J. Clin. Invest. 2008;118:1877–1889. doi: 10.1172/JCI34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y-C, Sun A, Ryo A, Zhou XZ, Yu Z-X, Huang H-K, Bronson R, Bing G, Li X, Hunter T, Lu KP. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424:556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, Uchida T, Hunter T, Lu KP. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. The Journal of biological chemistry. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonati E, Brambilla A, Milani C, Masserini M, Palestini P, Bulbarelli A. Pin1, a new player in the fate of HIF-1 alpha degradation: an hypothetical mechanism inside vascular damage as Alzheimer's disease risk factor. Frontiers in cellular neuroscience. 2014;8:1. doi: 10.3389/fncel.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lu KP, Liou YC, Vincent I. Proline-directed phosphorylation and isomerization in mitotic regulation and in Alzheimer's disease. BioEssays. 2003;25:174–181. doi: 10.1002/bies.10223. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- Lv C, Liu X, Liu H, Chen T, Zhang W. Geniposide Attenuates Mitochondrial Dysfunction and Memory Deficits in APP/PS1 Transgenic Mice. Current Alzheimer research. 2014;11:580–587. doi: 10.2174/1567205011666140618095925. [DOI] [PubMed] [Google Scholar]
- Ma SL, Pastorino L, Zhou XZ, Lu KP. Prolyl isomerase Pin1 promotes amyloid precursor protein (APP) turnover by inhibiting glycogen synthase kinase-3beta (GSK3beta) activity: novel mechanism for Pin1 to protect against Alzheimer disease. The Journal of biological chemistry. 2012a;287:6969–6973. doi: 10.1074/jbc.C111.298596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SL, Tang NL, Tam CW, Lui VW, Lam LC, Chiu HF, Driver JA, Pastorino L, Lu KP. A PIN1 polymorphism that prevents its suppression by AP4 associates with delayed onset of Alzheimer's disease. Neurobiol Aging. 2012b;33:804–813. doi: 10.1016/j.neurobiolaging.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free radical biology & medicine. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Goodman Y. Different amyloidogenic peptides share a similar mechanism of neurotoxicity involving reactive oxygen species and calcium. Brain research. 1995;676:219–224. doi: 10.1016/0006-8993(95)00148-j. [DOI] [PubMed] [Google Scholar]
- Mercedes-Camacho AY, Mullins AB, Mason MD, Xu GG, Mahoney BJ, Wang X, Peng JW, Etzkorn FA. Kinetic isotope effects support the twisted amide mechanism of Pin1 peptidyl-prolyl isomerase. Biochemistry. 2013;52:7707–7713. doi: 10.1021/bi400700b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell. 2012;149:232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunshola OO, Antoniou X. Contribution of hypoxia to Alzheimer's disease: is HIF1 alpha a mediator of neurodegeneration? Cellular and molecular life sciences: CMLS. 2009;66:3555–3563. doi: 10.1007/s00018-009-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pastorino L, Ma SL, Balastik M, Huang P, Pandya D, Nicholson L, Lu KP. Alzheimer's disease-related loss of Pin1 function influences the intracellular localization and the processing of AbetaPP. Journal of Alzheimer's disease: JAD. 2012;30:277–297. doi: 10.3233/JAD-2012-111259. [DOI] [PubMed] [Google Scholar]
- Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson L, Lu KP. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- Pearson RC, Esiri MM, Hiorns RW, Wilcock GK, Powell TP. Anatomical correlates of the distribution of the pathological changes in the neocortex in Alzheimer disease. Proc Natl Acad Sci USA. 1985;82:4531–4534. doi: 10.1073/pnas.82.13.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C, Kappert K, Engstrom U, Ostman A, Sjoblom T. An antibody-based method for monitoring in vivo oxidation of protein tyrosine phosphatases. Methods. 2005;35:37–43. doi: 10.1016/j.ymeth.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Pillot T, Drouet B, Queille S, Labeur C, Vandekerchkhove J, Rosseneu M, Pincon-Raymond M, Chambaz J. The nonfibrillar amyloid beta-peptide induces apoptotic neuronal cell death: involvement of its C-terminal fusogenic domain. Journal of neurochemistry. 1999;73:1626–1634. doi: 10.1046/j.1471-4159.1999.0731626.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P, Dickson DW, Davies P. Pin1 colocalization with phosphorylated tau in Alzheimer's disease and other tauopathies. Neurobiol Dis. 2003;14:251–264. doi: 10.1016/s0969-9961(03)00109-8. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- Ryo A, Nakamura N, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nature Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chemical research in toxicology. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Markesbery WR, Zhou XZ, Lu KP, Butterfield DA. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: A redox proteomics analysis. Neurobiol Aging. 2006;27:918–925. doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W. Hypoxia facilitates Alzheimer's disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai DR, Rub U, Schultz C, Sassin I, Ghebremedhin E, Del Tredici K, Braak E, Braak H. Sequence of Abeta-protein deposition in the human medial temporal lobe. J Neuropathol Exp Neurol. 2000;59:733–748. doi: 10.1093/jnen/59.8.733. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Morley SJ, Rulten SL. Utilizing the peptidyl-prolyl cis-trans isomerase pin1 as a probe of its phosphorylated target proteins. Examples of binding to nuclear proteins in a human kidney cell line and to tau in Alzheimer's diseased brain. J Histochem Cytochem. 2001;49:97–108. doi: 10.1177/002215540104900110. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Mosaheb S, Hashemzadeh-Bonehi L, Cairns NJ, Kay JE, Morley SJ, Rulten SL. Shortfalls in the peptidyl-prolyl cis-trans isomerase protein Pin1 in neurons are associated with frontotemporal dementias. Neurobiol Dis. 2004;17:237–249. doi: 10.1016/j.nbd.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Wildemann D, Erdmann F, Alvarez BH, Stoller G, Zhou XZ, Fanghanel J, Schutkowski M, Lu KP, Fischer G. Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem. 2006;49:2147–2150. doi: 10.1021/jm060036n. [DOI] [PubMed] [Google Scholar]
- Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: Is there an underlying theme? Nature Cell Biol. 2005;7:435–441. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Lu KP. Pin1 is overexpressed in breast cancer and potentiates the transcriptional activity of phosphorylated c-Jun towards the cyclin D1 gene. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld J, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- Yatin SM, Varadarajan S, Link CD, Butterfield DA. In vitro and in vivo oxidative stress associated with Alzheimer's amyloid beta-peptide (1–42) Neurobiol Aging. 1999;20:325–330. doi: 10.1016/s0197-4580(99)00056-1. discussion 339–342. [DOI] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, Tsai CH, Chen HY, Chiang CT, Lai CK, Lu LT, Chen CH, Gu DL, Pu YS, Jou YS, Lu KP, Hsiao PW, Shih HM, Chen RH. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20:214–228. doi: 10.1016/j.ccr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Zepeda AB, Pessoa A, Jr, Castillo RL, Figueroa CA, Pulgar VM, Farias JG. Cellular and molecular mechanisms in the hypoxic tissue: role of HIF-1 and ROS. Cell biochemistry and function. 2013;31:451–459. doi: 10.1002/cbf.2985. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin 1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]