Background: Δ9-Tetrahydrocannabinol (THC) and cannabidiol (CBD) modulate endocannabinoid tone in vivo through unknown mechanisms.
Results: THC and CBD bind to fatty acid-binding proteins (FABPs) and reduce endocannabinoid metabolism. Neither THC nor CBD inhibit human fatty acid amide hydrolase activity.
Conclusion: FABPs are intracellular transporters of THC and CBD.
Significance: These findings identify a new mechanism by which phytocannabinoids influence endocannabinoid signaling.
Keywords: Anandamide (N-Arachidonoylethanolamine) (AEA), Cannabinoid, Endocannabinoid, Fatty Acid Binding Protein, Lipid Trafficking, Lipid Transport, Δ-tetrahydrocannabinol (THC), Cannabidiol (CBD), Cannabinoid Transporters, Fatty Acid Amide Hydrolase (FAAH)
Abstract
Δ9-Tetrahydrocannabinol (THC) and cannabidiol (CBD) occur naturally in marijuana (Cannabis) and may be formulated, individually or in combination in pharmaceuticals such as Marinol or Sativex. Although it is known that these hydrophobic compounds can be transported in blood by albumin or lipoproteins, the intracellular carrier has not been identified. Recent reports suggest that CBD and THC elevate the levels of the endocannabinoid anandamide (AEA) when administered to humans, suggesting that phytocannabinoids target cellular proteins involved in endocannabinoid clearance. Fatty acid-binding proteins (FABPs) are intracellular proteins that mediate AEA transport to its catabolic enzyme fatty acid amide hydrolase (FAAH). By computational analysis and ligand displacement assays, we show that at least three human FABPs bind THC and CBD and demonstrate that THC and CBD inhibit the cellular uptake and catabolism of AEA by targeting FABPs. Furthermore, we show that in contrast to rodent FAAH, CBD does not inhibit the enzymatic actions of human FAAH, and thus FAAH inhibition cannot account for the observed increase in circulating AEA in humans following CBD consumption. Using computational molecular docking and site-directed mutagenesis we identify key residues within the active site of FAAH that confer the species-specific sensitivity to inhibition by CBD. Competition for FABPs may in part or wholly explain the increased circulating levels of endocannabinoids reported after consumption of cannabinoids. These data shed light on the mechanism of action of CBD in modulating the endocannabinoid tone in vivo and may explain, in part, its reported efficacy toward epilepsy and other neurological disorders.
Introduction
Cannabis sativa contains more than 100 known cannabinoids, including Δ9-tetrahydrocannabinol (THC)3 and cannabidiol (CBD). THC and CBD are mainly formed from marijuana when their corresponding acids are decarboxylated upon heating (1). THC is thought to act primarily via the activation of specific cannabinoid receptors, CB1 and CB2 (2). Unlike THC, CBD does not serve as a CB receptor agonist and binds to CB receptors with very low affinity (3). CBD is non-psychoactive, however, its pharmacological use has been shown to exert anti-inflammatory, anti-convulsant, anxiolytic, and neuroprotective effects in a CB receptor-independent manner (2, 4, 5). The mechanism of action of CBD is still a matter of some debate, however, there is evidence that its anxiolytic, neuroprotective, and antidepressant effects are mediated by its ability to activate 5-HT1A receptors (6–8).
THC and CBD are hydrophobic and are transported in the blood by lipoproteins and albumin (9, 10). However, intracellular transporters for phytocannabinoids have not been described. In theory these hydrophobic compounds may be transported from the cell membrane through the aqueous milieu by soluble intracellular carriers. The fatty acid-binding proteins (FABPs) have been shown to be intracellular transporters for the endocannabinoid anandamide (AEA) and other N-acylethanolamines (11–13). AEA requires transport from the membrane to intracellular fatty acid amide hydrolase (FAAH) for inactivation (11, 14, 15). Inhibitors of the FABPs have recently been shown to raise brain levels of AEA by limiting its transport into the cell to FAAH (11, 12, 16). Similar observations of heightened blood and brain anandamide concentrations following consumption of phytocannabinoids have been made, however, the mechanism of action is unclear (17). The mammalian brain expresses three FABP subtypes: FABP3, FABP5, and FABP7 (18). Here, we examine whether cannabinoids also bind to the FABPs and, as such, act as inhibitors of anandamide inactivation.
EXPERIMENTAL PROCEDURES
Chemicals
AEA was from Cayman Chemical (Ann Arbor, MI), THC, CBD, and fatty acid-free BSA were from Sigma, [14C]AEA (arachidonoyl-[1-14C]ethanolamide, 60 mCi/mmol) was provided by the Drug Supply Program at the National Institute on Drug Abuse.
Cell Culture and Transfections
Wild-type and FABP5 shRNA-expressing HeLa cells (12) were grown in DMEM supplemented with 10% fetal bovine serum, 100 units/ml of penicillin/streptomycin, 2 mm l-glutamine, and 1 mm sodium pyruvate. Transfections of human FAAH were carried out using the GenJet Plus reagent (SignaGen, Rockville, MD) according to the manufacturer's instructions.
In Vitro Binding of Ligands to FABPs
Binding analysis of THC and CBD to FABPs was carried out as described (14).
In Silico Studies
Initial Docking
THC and CBD were docked using DOCK version 6.5 (19). Energy grids were first generated using the grid program for the three FABP serotypes: FABP3 (Protein Data Bank (PDB) code 3RSW, 2.6-Å resolution), FABP5 (PDB code 1B56, 2.05-Å resolution), and FABP7 (PDB code 1FE3, 2.8-Å resolution) (20, 21). Likewise, energy grids were generated for the FAAH variants: rat FAAH (PDB code 3QJ8, 2.9-Å resolution), humanized FAAH (PDB code 2WAP, 2.8-Å resolution), and mutant FAAH (a variant created by introducing mutations F160L and M463V in 2WAP with UCSF Chimera v1.9) (22–24). Each ligand molecule was then flexibly docked to each of the respective FABP or FAAH grids (DOCK FLX protocol) (25) and the single lowest energy pose was retained. To generate comparative energy scores, the CBD pose generated in rat FAAH was fixed anchor docked to humanized FAAH and F160L/M463V mutant FAAH (DOCK FAD protocol) (25) and the single lowest energy pose was retained for each respective FAAH.
Simulation Setups and Molecular Dynamics
MD simulations were adapted from the methodology carried out in Ref. 27. Each MD trajectory (2 ns) was performed using NAMD 2.9 (28) employing charmrun to assign multiple processors (∼10) on the Seawulf Cluster at Stony Brook University. Preparation of each the initial MD coordinates started with docked complexes, which were subsequently stripped of crystallographic waters and prepared using the AMBER8 suite (leap, antechamber) (29). AMBER8 was used to assign force field parameters and solvate each complex in a periodic box containing ∼5000 waters (∼60 Å3). Force fields were assigned independently using AMBER8: FF99SB (30) for the protein, GAFF for the ligand, and TIP3P (31) for the water. Prior to running each simulation, a nine-step equilibration of three minimization steps (2000 steps of steepest decent) and seven molecular dynamics runs (50 ps each) as described in Ref. 32 were performed to equilibrate the system before the production run (2 ns) (32).
MD simulations were carried out in the NPT ensemble using Langevin dynamics at a constant temperature of 310 K and constant pressure of 1.01325 bar (33). All MD simulations employed a 1-fs time step that required the use of SHAKE (34). Additional key parameters included particle mesh Ewald (35) to compute long-range electrostatics (1.5 Å grid spacing) and a 10-Å cutoff (8 Å smoothing switch) for non-bonded interactions. Each MD frame is composed of 1000 fp (1 ps) and was run for a total of 2 ns.
MMGBSA Calculations
Free energies of binding (ΔGbind Calculated) were carried out using the MM-GBSA method (36, 37). All MM-GBSA calculation were carried out post production. For each complex, AMBER8 (p-traj) was employed to strip all explicit water from each of the 2-ns MD trajectories. A single point calculation was carried out using AMBER8 (sander) for each ps (2000 ps in 2 ns) with the following parameters: AMBER radii (mbondi2), Dielectric constants (1 and 78.5), and GB model (igb = 5) (38).
Following Equation 1, the MM-GBSA free energy of binding (ΔGbind) was calculated using the non-bonded intermolecular protein-ligand interactions (ΔGcoul and ΔGvdw), the change in hydration energy (ΔGhyd), and the temperature multiplied by the change in solute vibrational, rotational, and translational entropy (TΔS).
Specifically, ΔGcoul and ΔGvdw were calculated from the change in energy for all nonbonded intermolecular protein-ligand interactions (Coulombic and van deer Waals) as the receptor and ligand go from a free state to a bound state (complex) shown in Equation 2.
The ΔΔGhyd was calculated from the ΔGhyd = Gpolar + Gnonpolar for each species (complex, receptor, and ligand) as shown in Equation 3.
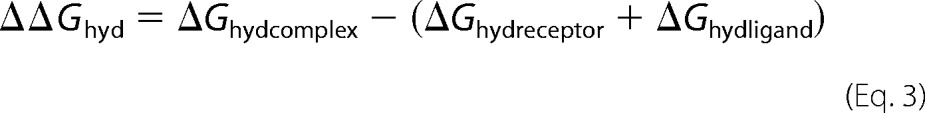 |
Furthermore, Gpolar was calculated using the GB method and molecular solvent accessible surface (SASA) was used to calculate Gnonpolar = γ × SASA + β using standard constants (γ = 0.00542 kcal mol−1 Å−2 and β = 0.82 kcal/mol) (39). Entropy (TΔS) was calculated using quazi-harmonic normal mode analysis of energy minimized structures (see Entropy Calculation) at 310 K (37).
Calculation of Entropy (Normal Mode) Using Amber
Entropy calculations were carried out using Amber 12.0 (MMPBSA.py) (40). All dcd trajectories (NAMD trajectory files) were first converted back into mdcrd files using Amber 12.0 (ptraj). Then each mdcrd file consisting of 1000 frames (1 ps/frame) were loaded into MMPBSA.py version 12.0. Key parameters included: nmstartframe = 1; nmendframe = 1000; nminterval = 10; nmode_igb = 1; nmode_istrng = 0.1; drms = 0.0001; maxcyc = 100000; and NMODE frames = 100. Calculations were performed on 10 processors. Total entropy (TΔS) of the system was calculated from a difference in total energy (translation energy, rotational Energy, and vibrational energy) of the system as shown in Equation 4. Entropy calculations were carried out at 310 K.
Enzyme Assays
FAAH activity in homogenates was determined as described previously (11).
AEA Uptake
Inhibition of AEA uptake into cells was performed as described (11). HeLa cells were preincubated with 20 μm THC, CBD, or vehicle (0.1% ethanol in DMEM supplemented with 0.15% BSA) for 15 min followed by incubation with 100 nm [14C]AEA for 5 min. AEA uptake was quantified as previously described (11).
Immunohistochemistry
Brain sections were blocked in 1% serum or 1% nonfat dry milk in PBS-T (0.3% Trition X-100 in PBS) and then incubated with anti-human/mouse serum albumin primary antibody (MAB1455 from R&D Systems used at 1:20) and anti-macrophage/microglia Iba1 (rabbit polyclonal 019-19741 from WakoUSA used at 1:1000) overnight at 4 °C. Incubation with Alexa Fluor 488 and Alexa Fluor 555-conjugated secondary antibodies at 1:1000 was performed at room temperature for 1 h, followed by washing with PBS and mounting using Fluoromount-G with DAPI (Southern Biotech). 4–8 brain sections were imaged per biological replicate.
Statistics
Results represent mean ± S.E. of at least three independent experiments performed in triplicate. Statistical significance was determined using two-tailed unpaired t tests against controls or by one-way analysis of variance with post-hoc Dunnett's test compared with vehicle controls (significance set at p < 0.05) on the raw data, as indicated. Analysis was performed using GraphPad (version 6.0, Prism).
RESULTS
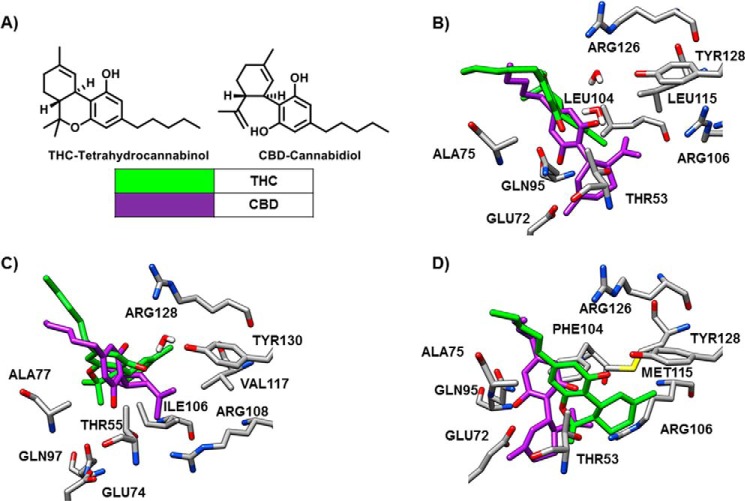
Computational methods were initially employed to determine the likelihood of FABPs being carriers for cannabinoids. THC and CBD displayed energy scores consistent with appreciable binding to FABP3, FABP5, and FABP7, the main FABPs that are expressed in the brain. This analysis showed tighter binding of CBD to FABP3, FABP5, and FABP7 (ΔGbinding = −23.64 ± 2.31, −21.02 ± 0.27, and −22.94 ± 2.25 kcal/mol) than did THC (ΔGbinding = −15.50 ± 3.22, −15.28 ± 0.52, and −15.54 ± 3.06 kcal/mol) (Table 1). THC and CBD were accommodated in the pockets of the FABPs (Fig. 1). Conspicuously, THC binds deeper into the pocket of FABP7 than FABP3 and FABP5. Noticeably, the binding pose is unique for CBD in FABP5 (Fig. 1).
TABLE 1.
Energy scoring for THC and CBD binding to FABP3, FABP5, and FABP7
| Trajectory | ΔEVDW (A) | ΔEcoul (B) | ΔGpolar (C) | ΔGnonpolar (D) | MMGBSA (A+B+C+D) | TΔS (E) | ΔGbind (A+B+C+D-E) |
|---|---|---|---|---|---|---|---|
| THC FABP3 | −42.81 ± 2.59 | −7.29 ± 1.03 | 18.84 ± 0.67 | −4.03 ± 0.03 | −35.29 ± 3.54 | −19.78 ± 0.83 | −15.50 ± 3.22 |
| CBD FABP3 | −49.20 ± 1.45 | −14.56 ± 1.51 | 21.15 ± 0.88 | −3.88 ± 0.05 | −46.48 ± 2.33 | −22.85 ± 0.55 | −23.64 ± 2.31 |
| THC FABP5 | −44.54 ± 1.33 | −7.29 ± 1.29 | 19.27 ± 0.67 | −4.07 ± 0.07 | −36.63 ± 1.85 | −21.35 ± 1.37 | −15.28 ± 0.52 |
| CBD FABP5 | −45.60 ± 1.06 | −13.75 ± 2.91 | 21.66 ± 1.44 | −3.98 ± 0.03 | −41.67 ± 0.81 | −20.43 ± 0.65 | −21.02 ± 0.27 |
| THC FABP7 | −45.08 ± 1.26 | −8.18 ± 3.14 | 21.01 ± 1.23 | −4.09 ± 0.02 | −36.34 ± 3.12 | −20.81 ± 0.75 | −15.54 ± 3.06 |
| CBD FABP7 | −48.43 ± 0.53 | −15.78 ± 1.42 | 24.15 ± 1.23 | −3.91 ± 0.04 | −43.98 ± 1.23 | −21.03 ± 1.10 | −22.94 ± 2.25 |
FIGURE 1.
Structures of THC and CBD and the superimposed predicted binding pose for each ligand in FABP3, FABP5, and FABP7. A, chemical structures of THC and CBD. B, THC appears sterically crowded in the binding pocket of FABP3 with water-mediated hydrogen bonding seen with TYR128, a key amino acid in the binding of fatty acids. THC instead adopts a direct hydrogen bond with Ala75 (2.6 Å). CBD remains in hydrogen bonding distance to Tyr128 (5.0 Å), whereas making closer hydrogen bonds through its 1′-OH with Thr53 (2.4 Å), Gln95 (2.2 Å), and Glu72 (3.9 Å). Water-mediated hydrogen bonding with Tyr128, similar to THC, is critical for binding. C, THC shows a long-range water mediated hydrogen bonding with Tyr130 (5.3 Å), a key amino acid residue critical to FA binding. The Ile106 and Val117 residues in FABP5 provide less steric hindrance compared with Leu104 and Leu115 of FABP3, allowing THC to penetrate further into the binding pocket. CBD adopts a unique orientation in FABP5, compared with similar binding poses in FABP3 and FABP7, blocking Tyr129 by making a strong hydrogen bond with Arg128 (2.1 Å). The 1′-OH of CBD forms hydrogen bonding with Thr55. D, THC forms a strong hydrogen bonding directly with Tyr128 (2.6 Å), a key amino acid residue critical to fatty acid binding, whereas providing a weaker interaction with Arg126 (4.1 Å). The Phe104 and Met115 residues in FABP7 provide even less steric hindrance compared with similar residues in FABP3 and FABP5, allowing THC to penetrate even further into the binding pocket, which is now without water. CBD adopts a similar binding mode as in FABP3, providing long range interactions between Thr53 (2.8 Å) and Tyr128 (5.3 Å), whereas additionally providing hydrogen bonding with Ala75 (2.3 Å) and Glu72 (2.1 Å) via the 1′-OH.
In vitro binding studies of THC and CBD, using a fluorescent NBD-stearate displacement assay, indicated that these two cannabinoids bind to FABP3, FABP5, and FABP7 with low micromolar affinities (Table 2). In addition, for FABP5 there was agreement with the in silico results showing that CBD bound more tightly than did THC, however, a significant difference in binding affinities was not observed for FABP3 and FABP7. Interestingly, THC and CBD bind the FABPs as well or nearly as well as the endocannabinoids and a variety of other compounds including various transport inhibitors as discussed below. However, they do not bind as well as many of the fatty acids including oleic acid that binds with nearly 50-fold higher affinity for some of the FABPs (see Table 2).
TABLE 2.
Binding affinities of THC, CBD, AEA, 2-arachidonoylglycerol (2-AG), and oleic acid (OA) to human FABPs
The Ki values were determined by NBD-stearate fluorescence displacement assays as described under “Experimental Procedures.” Values are averages ± S.E. of at least three independent experiments.
| Compound | Ki |
||
|---|---|---|---|
| FABP3 | FABP5 | FABP7 | |
| μm | |||
| CBD | 1.69 ± 0.20 | 1.88 ± 0.10 | 1.52 ± 0.39 |
| THC | 1.98 ± 0.36 | 3.14 ± 0.26 | 1.04 ± 0.13 |
| AEA | 3.07 ± 0.28 | 1.26 ± 0.18 | 0.80 ± 0.06 |
| 2-AG | 1.65 ± 0.22 | 1.45 ± 0.21 | 0.20 ± 0.03 |
| OA | 0.04 ± 0.00 | 0.56 ± 0.14 | 0.04 ± 0.00 |
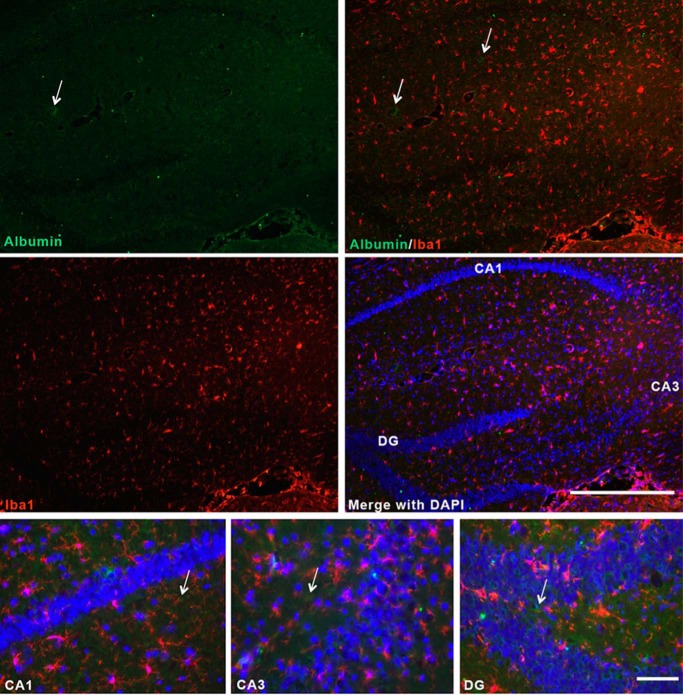
Because albumin is a known carrier of the cannabinoids in the blood and was proposed to function as an intracellular endocannabinoid-binding protein (9, 15), we examined whether it could function in a similar capacity as an intracellular transporter in the brain by initially examining its expression pattern in the mouse brain. One group has previously reported that microglial cells synthesize albumin (41). Contrary to this report, we did not find any albumin expression in microglia (Fig. 2). In the normal mouse brain albumin expression did not co-localize with the macrophage/microglial marker Iba1, but was rather detected in perivascular areas (Fig. 2, arrows). Very low levels of expression could be detected in the cytoplasm of neurons in the hippocampus after extensive incubation with the primary antibody (not shown). These results indicate that albumin is likely not a major intracellular transporter of the cannabinoids in the brain.
FIGURE 2.
Albumin is not expressed by microglia in the normal mouse brain. Hippocampal brain sections were obtained from C57Bl6 mice and incubated with anti-albumin and anti-Iba1 antibodies. The sections were incubated with the primary antibody overnight at 4 °C. Images were visualized with Alexa Fluor 488- and Alexa Fluor 555-conjugated secondary antibodies. Nuclei were labeled with DAPI. n = 4. Scale bars, 500 μm (upper panels, low magnification images) and 100 μm (lower panels, high magnification images). Arrows point to perivascular albumin staining.
We next determined whether FABPs serve as intracellular carriers for THC and CBD. HeLa cells are a suitable cell type to examine interactions of exogenous cannabinoids with FABPs and their effects upon endocannabinoid transport because these cells conveniently express a single FABP subtype, FABP5 (12, 14), AEA uptake is coupled to its breakdown by FAAH (42, 43). Therefore, HeLa cells, which lack FAAH (44), were transfected with human FAAH and AEA uptake was subsequently examined.
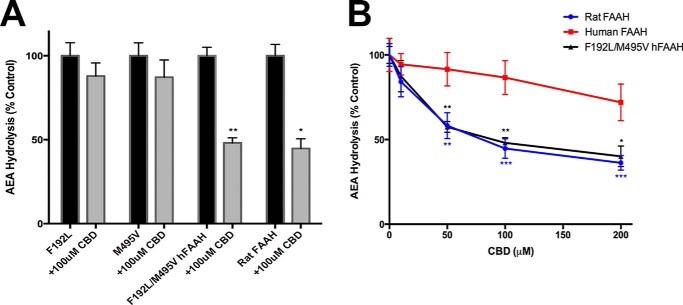
Both THC and CBD inhibited AEA uptake, with CBD being slightly more efficacious (Fig. 3A). The greater potency of CBD as an inhibitor of AEA uptake mirrored its higher affinity (lower Ki) for FABP5 (Table 2). As expected, FABP5 knockdown inhibited AEA uptake (Fig. 3A) (14). Importantly, THC and CBD did not reduce AEA uptake in cells bearing a knockdown of FABP5 (Fig. 3A). Furthermore, THC and CBD did not reduce the proportion of intracellular AEA that is hydrolyzed following uptake, suggesting that the cannabinoids block the delivery of AEA to FAAH but do not affect AEA hydrolysis by FAAH (Fig. 3B). In support of this, we found that CBD and THC did not inhibit AEA hydrolysis by human FAAH in cell homogenates (Fig. 3C). This result was surprising in light of previous studies demonstrating robust inhibition of rodent FAAH by CBD (17, 45–47). Therefore, we examined whether CBD inhibits rat and mouse FAAH. In agreement with previous studies, CBD inhibited both rat and mouse but not human FAAH (Fig. 3D), suggesting a species specificity that may underlie these effects (14, 44).
FIGURE 3.
THC and CBD inhibit AEA uptake by targeting FABP5. A, [14C]AEA uptake by human FAAH-transfected WT and FABP5 shRNA-expressing HeLa cells following a 5-min incubation in the presence or absence of 20 μm THC, CBD, or vehicle-only control. *, p < 0.05 versus control. B, ratio of [14C]AEA hydrolysis to total uptake in vector-transfected (gray) or human FAAH-transfected WT or FABP5 shRNA HeLa cells following a 5-min incubation in the presence or absence of 20 μm THC, CBD, or vehicle-only control. C, [14C]AEA hydrolysis in homogenates of human FAAH-transfected HeLa cells in the absence or presence of 20 μm THC or CBD. D, [14C]AEA hydrolysis in homogenates of rat, mouse, or human FAAH-transfected cells in the presence or absence of 20 μm CBD. *, p < 0.05.
To further investigate this species specificity, we employed site-directed mutagenesis to interconvert key residues within the active site of human FAAH to the homologous residues found in rat FAAH. Previous work has identified six residues that differ between the active sites of human and rat FAAH (F192L, Y194F, T377A, N435S, V491I, and M495V) (Fig. 4) (48). Of these, Phe192 and Met495 were identified as likely candidates to alter interactions with CBD based on the crystal structures of human and rat FAAHs with other bound ligands (23). These two residues appeared to restrict the entry of CBD into the active site of human FAAH. Therefore, we mutated these residues in human FAAH to generate F192L, M495V, as well as FAAH bearing the double mutant. Interestingly, the F192L and M495V mutants did not significantly alter FAAH inhibition by CBD. In contrast, human FAAH carrying mutations at both sites was robustly inhibited by CBD, with a similar inhibitor sensitivity profile as seen in rat FAAH (Fig. 5, A and B).
FIGURE 4.
Multiple sequence alignment of human and rodent FAAHs. Non-homologous residues that reside within the active site are marked with an asterisk (*). Sequences were aligned using the ClustalW program and edited with the BOXSHADE (version 3.21) software.
FIGURE 5.
F192L/M495V human FAAH mutant confers sensitivity to inhibition by CBD. A, [14C]AEA hydrolysis in homogenates of rat or mutant FAAH-transfected cells in the presence or absence of 100 μm CBD. B, CBD inhibitor sensitivity profile in homogenates of human, rat, or F192L/M495V hFAAH-transfected cells. Data represent the mean ± S.E. for 3 replicates per group. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus vehicle control.
Computational methods were subsequently employed to characterize the interactions of CBD within the active site of FAAH. CBD was individually docked to human FAAH, rat FAAH, or the F192L/M495V mutant human FAAH, and predicted free energy scores were obtained (ΔGbinding = −26.55 kcal/mol, −36.84 kcal/mol, and −30.46 kcal/mol, respectively). Consistent with the observed biochemical data, CBD was predicted to bind human FAAH with a lower affinity than either rat FAAH or the human FAAH F192L/M495V mutant.
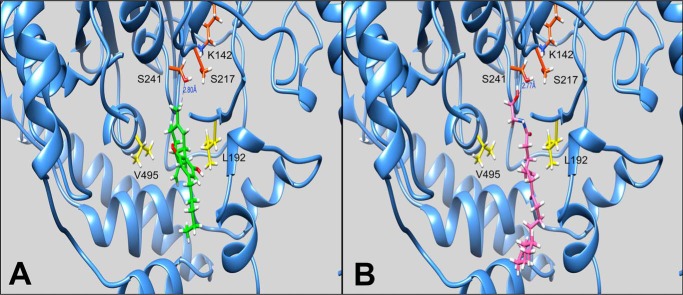
The membrane access channel of FAAH is a cavity of hydrophobic and aromatic residues that connect the catalytic core of the protein to the membrane-associated region, thought to allow lipid substrates to directly access the site of catalysis from the lipid bilayer (49). Substrate catalysis by amidase signature enzymes, such as FAAH, is mediated by an unusual Ser-Ser-Lys catalytic triad (Ser241-Ser217-Lys142 in FAAH). Crystal structures of FAAH inhibitors in complex with FAAH reveal these compounds residing within the membrane access channel cavity (49, 50). We performed in silico docking of CBD to FAAH to investigate the preferred binding pose of this cannabinoid. Like other FAAH inhibitors, CBD was predicted to reside within the membrane access channel cavity, coming within 2.8 Å of Ser241 of the catalytic core (Fig. 6A). AEA was subsequently docked to rat FAAH for comparison and the lowest energy pose was found to be in a highly similar location to that of CBD (Fig. 6B), further suggesting the possibility of CBD acting as a competitive inhibitor of FAAH.
FIGURE 6.
Predicted binding poses of CBD and AEA by in silico docking within rat FAAH. A, lowest energy pose of CBD (green) within rat FAAH. Depicted residues are the Ser-Ser-Lys catalytic triad (red) and two relevant active site residues that differ between rodent and human species (yellow). A peripheral section of the protein has been removed for visibility. B, lowest energy pose of AEA (pink) within rat FAAH. Residues were labeled as in A.
Inspection of the predicted geometry of CBD within the active site reveals a twisted conformation of the compound (Fig. 7). Although THC and CBD are isomers, the pyran ring of THC confers added rigidity to the molecule. The reduced structural flexibility of THC should not allow for the same twisted conformation as observed with CBD and likely accounts for the inability of THC to inhibit FAAH activity from either species. Superimposed images of CBD in rat (red), human (blue), or F192L/M495V mutant (purple) FAAHs reveal similar modes of binding (Fig. 7). CBD penetrates ∼1 Å less deeply into the active site of human FAAH and has a less favorable predicted free energy of binding, which may account for it being less potent as an inhibitor in human FAAH in comparison to rat FAAH in our enzyme assays.
FIGURE 7.
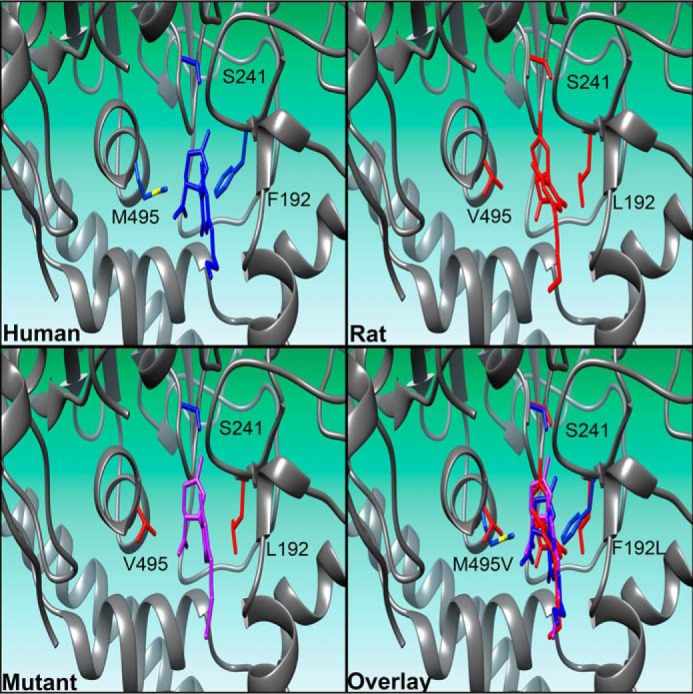
Predicted binding poses of CBD by computational docking within human, rat, or F192L/M495V mutant FAAHs. Poses of CBD within human, rat, and F192L/M495V human mutant FAAHs reveal similar modes of binding, however, CBD penetrates into the active site of human FAAH slightly less deeply. Compound and residues of human FAAH are shown in blue, compound and residues of rat FAAH are shown in red, compound docked to the F192L/M495V mutant FAAH are shown in purple. A portion of the protein in the depictions has been removed for clarity.
DISCUSSION
In the present study, we identified FABPs as the first intracellular carriers for phytocannabinoids and showed that THC and CBD modulate AEA uptake by competing with AEA for binding to FABPs. Therefore, THC and CBD may potentiate AEA signaling by reducing its catabolism. Unexpectedly, in contrast to previous reports demonstrating robust inhibition of rodent FAAH by CBD, our findings clearly indicate that FAAH does not serve as a target for CBD in humans. Therefore, the recent observation linking CBD administration to elevated serum AEA levels in humans is unlikely to be attributable to FAAH inhibition, and points to modulation of AEA transport proteins such as FABPs (see schematic in Fig. 8).
FIGURE 8.
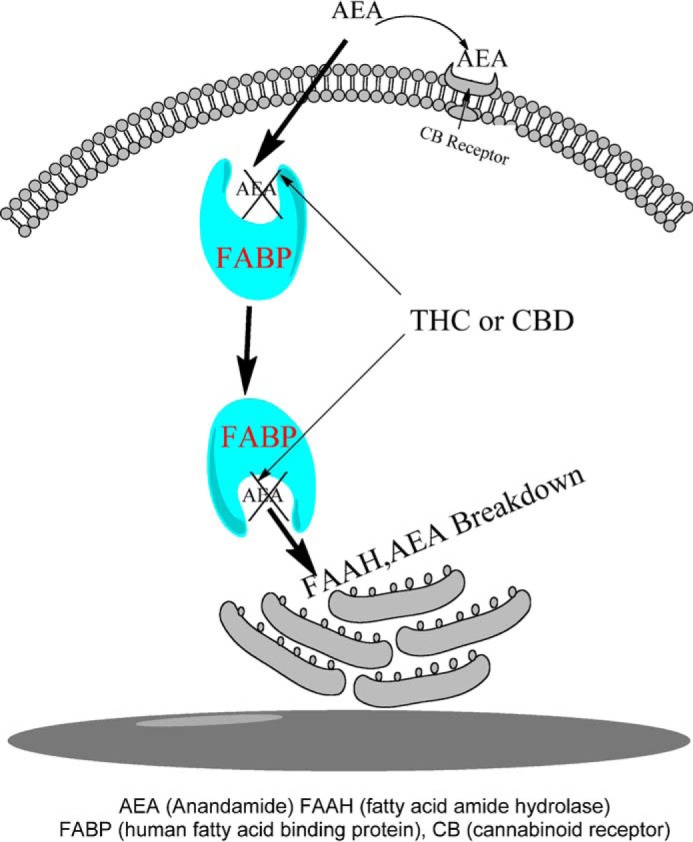
Conceptual schematic of phytocannabinoids modulating endocannabinoid metabolism via FABP inhibition in human cells. Endogenous AEA is shuttled through the aqueous environment of the cytoplasm to intracellularly localized FAAH for catabolism. THC and CBD compete for binding to FABPs, thereby reducing the rate of anandamide breakdown and raising AEA levels.
In silico modeling demonstrated that THC and CBD interact with FABP3, FABP5, and FABP7 and that both ligands reside within the FABP binding pocket in a manner similar to that shown for AEA and 2-AG by x-ray crystallography (51). In vitro experiments confirmed that these cannabinoids bind the FABPs, all with a low micromolar affinity (1.04–3.14 μm). Interestingly, these affinities are only slightly lower than those observed with SBFI26, a truxillic acid compound recently identified as a specific inhibitor of the FABPs (Ki for FABP3, FABP5, and FABP7 = 3.86 ± 0.70, 0.93 ± 0.08, and 0.38 ± 0.04, respectively) (16). However, unlike SBFI26, THC and CBD showed pronounced affinity for FABP3. These two cannabinoids also bind with similar affinities as endocannabinoids AEA and 2-AG (Table 2). The relative similarity in binding affinity between the phytocannabinoids and endocannabinoids suggests that competition for these binding sites may exist. Our results show that the FABPs carry anandamide and that it can be displaced by CBD and THC in milieu of tighter binding of non-esterified fatty acids that, we surmise, are not saturating.
Endocannabinoid signaling is believed to be controlled through a balance of on demand synthesis and prompt catabolism (52–55). AEA is inactivated through hydrolysis by intracellular FAAH (56, 57). We chose to employ HeLa cells because they do not express FAAH, and this permitted us to use cells transfected with human, rat, or mouse FAAH. The uptake of AEA is unique compared with other neurotransmitter systems in that AEA does not require a transmembrane transporter. FAAH enzyme activity maintains an inward concentration gradient that drives uptake and this process requires FABPs as carriers (11, 13, 14).
Conveniently, HeLa cells only express a single FABP subtype, FABP5 (12). Our results indicate that CBD and THC inhibit AEA inactivation by targeting FABPs. This is supported by the observations that both phytocannabinoids were without effect in cells bearing a knockdown of FABPs. Furthermore, we did not observe inhibition of human FAAH by either of these cannabinoids.
Previous reports have shown that CBD, but not THC, inhibits mouse and rat FAAH activity (17, 45–47), which we have confirmed here. Unexpectedly, we are the first to report that CBD does not inhibit human FAAH, even at high concentrations. We explored the species-specific differences and found two key residues within the active site of human FAAH that render it resistant to inhibition by CBD.
In addition to exogenous cannabinoids, FABPs bind to endogenous N-acylethanolamines related to AEA (12). Therefore, it is not surprising that previous studies have demonstrated that N-acylethanolamines that do not themselves activate cannabinoid receptors, potentiate endocannabinoid signaling (58–60). A recent study reported that CBD alleviates the psychotic symptoms of schizophrenia, possibly in part by elevating levels of AEA and N-acylethanolamines in humans (17). Another report showed that administration of a single oral dose of 20 mg of THC to 30 healthy volunteers resulted in higher circulating concentrations of anandamide, 2-AG, palmitoyl ethanolamide, and oleoylethanolamide (61). Therefore, it is tempting to speculate that phytocannabinoids modulate the endocannabinoid tone by targeting FABPs in humans (Fig. 8). Indeed, inhibition of FABPs was recently shown to elevate brain levels of AEA in mice (16, 62). This proposed mechanism of AEA modulation in humans is highly dependent on the pharmacological concentrations of cannabinoids in the brain following consumption. To our knowledge there is little data on this front, however, rats administered a low dose (1 mg/kg) of THC or CBD rapidly attained micromolar brain and plasma concentrations of the parent compound and/or their metabolites (63). The reported brain cannabinoid concentrations are above the Ki for the FABPs, indicating the feasibility of a FABP-dependent mechanism of action in modulating endocannabinoid levels.
Following ingestion, THC is biotransformed primarily to the psychoactive metabolite 11-OH-THC, which is further oxidized to the non-psychoactive 11-nor-9-carboxy-THC (THC-COOH). This inactivation pathway primarily occurs by the cytochrome P-450s in the liver, however, extrahepatic metabolism in other tissues such as the brain, intestines, and lungs may also partially contribute to THC metabolism (64). Phase II metabolism involves glucuronidation of THC-COOH to form soluble adducts that are excreted in urine and feces (10, 64). Phase I metabolism of CBD is similar to that of THC, with primary oxidation at the C-9 position to the alcohol and carboxylic acid, however, little data are available describing its phase II metabolism (64, 65). CBD has been reported to inhibit THC metabolite formation in vitro and some groups have found that co-administration of THC and CBD partially inhibits the formation of select THC metabolites in human subjects (66–68).
Metabolic reactions such as phase I hydroxylation by the cytochrome P-450s and phase II glucuronidation occur at the endoplasmic reticulum and, similar to endocannabinoids, phytocannabinoids likely require a carrier for translocation from the cell membrane to the endoplasmic reticulum. In light of our current study, we postulate that FABPs may play a role in trafficking phytocannabinoids through the cytosol to their respective intracellular enzymes, although it is possible that other identified carriers such as HSP70 may also be involved (15).
Cannabinoids have shown promise in treating several neurological disorders, including addiction, anxiety, schizophrenia, and epilepsy, as well as other conditions such as glaucoma, pain, and nausea (4, 5, 17, 69–71). Sativex, an oromucosal spray containing ∼1:1 ratio of THC:CBD, has shown promise in treating neuropathic pain and spasticity in patients with multiple sclerosis (26, 72, 73). The efficacy of CBD in the treatment of these various medical conditions suggests that both cannabinoid receptor-dependent and -independent mechanisms of action are at play in conferring such therapeutic benefits. Further studies characterizing the in vivo effects of phytocannabinoids are warranted, and the FABPs may emerge as targets of great therapeutic potential in the future of cannabinoid-based pharmacotherapies.
Acknowledgments
We thank Dr. Kyungmin Ji for her immunostaining work and Dr. Robert Rizzo who generously made his facilities available for the computational analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants DA032232, DA035923, DA035949, DA016419, DA026593, GM007518, GM008444, and NS042168.
- THC
- Δ9-tetrahydrocannabinol
- CBD
- cannabidiol
- FABP
- fatty acid-binding protein
- 2-AG
- 2-arachidonoylglycerol
- AEA
- arachidonoylethanolamide
- FAAH
- fatty acid amide hydrolase
- PDB
- Protein Data Bank
- MD
- molecular dynamics.
REFERENCES
- 1. Happyana N., Agnolet S., Muntendam R., Van Dam A., Schneider B., Kayser O. (2013) Analysis of cannabinoids in laser-microdissected trichomes of medicinal Cannabis sativa using LCMS and cryogenic NMR. Phytochemistry 87, 51–59 [DOI] [PubMed] [Google Scholar]
- 2. Howlett A. C., Barth F., Bonner T. I., Cabral G., Casellas P., Devane W. A., Felder C. C., Herkenham M., Mackie K., Martin B. R., Mechoulam R., Pertwee R. G. (2002) International Union of Pharmacology: XXVII. classification of cannabinoid receptors. Pharmacol. Rev. 54, 161–202 [DOI] [PubMed] [Google Scholar]
- 3. Pertwee R. G. (2008) The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Δ9-tetrahydrocannabinol, cannabidiol and Δ9-tetrahydrocannabivarin. Br. J. Pharmacol. 153, 199–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Izzo A. A., Borrelli F., Capasso R., Di Marzo V., Mechoulam R. (2009) Non-psychotropic plant cannabinoids: new therapeutic opportunities from an ancient herb. Trends Pharmacol. Sci. 30, 515–527 [DOI] [PubMed] [Google Scholar]
- 5. Mechoulam R., Parker L. A. (2013) The endocannabinoid system and the brain. Annu. Rev. Psychol. 64, 21–47 [DOI] [PubMed] [Google Scholar]
- 6. Zanelati T. V., Biojone C., Moreira F. A., Guimarães F. S., Joca S. R. (2010) Antidepressant-like effects of cannabidiol in mice: possible involvement of 5-HT1A receptors. Br. J. Pharmacol. 159, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayakawa K., Mishima K., Nozako M., Ogata A., Hazekawa M., Liu A. X., Fujioka M., Abe K., Hasebe N., Egashira N., Iwasaki K., Fujiwara M. (2007) Repeated treatment with cannabidiol but not Δ9-tetrahydrocannabinol has a neuroprotective effect without the development of tolerance. Neuropharmacology 52, 1079–1087 [DOI] [PubMed] [Google Scholar]
- 8. Gomes F. V., Resstel L. B., Guimarães F. S. (2011) The anxiolytic-like effects of cannabidiol injected into the bed nucleus of the stria terminalis are mediated by 5-HT1A receptors. Psychopharmacology 213, 465–473 [DOI] [PubMed] [Google Scholar]
- 9. Fanali G., Cao Y., Ascenzi P., Trezza V., Rubino T., Parolaro D., Fasano M. (2011) Binding of Δ9-tetrahydrocannabinol and diazepam to human serum albumin. IUBMB life 63, 446–451 [DOI] [PubMed] [Google Scholar]
- 10. Huestis M. A. (2005) Pharmacokinetics and metabolism of the plant cannabinoids, Δ9-tetrahydrocannabinol, cannabidiol and cannabinol. Handb. Exp. Pharmacol. 168, 657–690 [DOI] [PubMed] [Google Scholar]
- 11. Kaczocha M., Glaser S. T., Deutsch D. G. (2009) Identification of intracellular carriers for the endocannabinoid anandamide. Proc. Natl. Acad. Sci. U.S.A. 106, 6375–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaczocha M., Vivieca S., Sun J., Glaser S. T., Deutsch D. G. (2012) Fatty acid-binding proteins transport N-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J. Biol. Chem. 287, 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Björklund E., Blomqvist A., Hedlin J., Persson E., Fowler C. J. (2014) Involvement of fatty acid amide hydrolase and fatty acid binding protein 5 in the uptake of anandamide by cell lines with different levels of fatty acid amide hydrolase expression: a pharmacological study. PloS One 9, e103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berger W. T., Ralph B. P., Kaczocha M., Sun J., Balius T. E., Rizzo R. C., Haj-Dahmane S., Ojima I., Deutsch D. G. (2012) Targeting fatty acid binding protein (FABP) anandamide transporters: a novel strategy for development of anti-inflammatory and anti-nociceptive drugs. PLoS One 7, e50968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oddi S., Fezza F., Pasquariello N., D'Agostino A., Catanzaro G., De Simone C., Rapino C., Finazzi-Agrò A., Maccarrone M. (2009) Molecular identification of albumin and Hsp70 as cytosolic anandamide-binding proteins. Chem. Biol. 16, 624–632 [DOI] [PubMed] [Google Scholar]
- 16. Kaczocha M., Rebecchi M. J., Ralph B. P., Teng Y. H., Berger W. T., Galbavy W., Elmes M. W., Glaser S. T., Wang L., Rizzo R. C., Deutsch D. G., Ojima I. (2014) Inhibition of fatty acid binding proteins elevates brain anandamide levels and produces analgesia. PloS One 9, e94200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leweke F. M., Piomelli D., Pahlisch F., Muhl D., Gerth C. W., Hoyer C., Klosterkötter J., Hellmich M., Koethe D. (2012) Cannabidiol enhances anandamide signaling and alleviates psychotic symptoms of schizophrenia. Transl. Psychiatry 2, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Furuhashi M., Hotamisligil G. S. (2008) Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balius T. E., Mukherjee S., Rizzo R. C. (2011) Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons. J. Comput. Chem. 32, 2273–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balendiran G. K., Schnutgen F., Scapin G., Borchers T., Xhong N., Lim K., Godbout R., Spener F., Sacchettini J. C. (2000) Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem. 275, 27045–27054 [DOI] [PubMed] [Google Scholar]
- 21. Hohoff C., Börchers T., Rüstow B., Spener F., van Tilbeurgh H. (1999) Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry 38, 12229–12239 [DOI] [PubMed] [Google Scholar]
- 22. Min X., Thibault S. T., Porter A. C., Gustin D. J., Carlson T. J., Xu H., Lindstrom M., Xu G., Uyeda C., Ma Z., Li Y., Kayser F., Walker N. P., Wang Z. (2011) Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH). Proc. Natl. Acad. Sci. U.S.A. 108, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahn K., Johnson D. S., Mileni M., Beidler D., Long J. Z., McKinney M. K., Weerapana E., Sadagopan N., Liimatta M., Smith S. E., Lazerwith S., Stiff C., Kamtekar S., Bhattacharya K., Zhang Y., Swaney S., Van Becelaere K., Stevens R. C., Cravatt B. F. (2009) Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem. Biol. 16, 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 25. Mukherjee S., Balius T. E., Rizzo R. C. (2010) Docking validation resources: protein family and ligand flexibility experiments. J. Chem. Inf. Model. 50, 1986–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iskedjian M., Bereza B., Gordon A., Piwko C., Einarson T. R. (2007) Meta-analysis of cannabis based treatments for neuropathic and multiple sclerosis-related pain. Curr. Med. Res. Opin. 23, 17–24 [DOI] [PubMed] [Google Scholar]
- 27. Huang Y., Rizzo R. C. (2012) A water-based mechanism of specificity and resistance for lapatinib with ErbB family kinases. Biochemistry 51, 2390–2406 [DOI] [PubMed] [Google Scholar]
- 28. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Wang B., Pearlman D. A., Crowley M., Brozell S., Tsui V., Gohlke H., Mongan J., Hornak V., Cui G., Beroza P., Schafmeister C., Caldwell J. W., Ross W. S., Kollman P. A. (2004) AMBER8, University of California, San Francisco [Google Scholar]
- 30. Harnak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006) Comparison of multiple amber force fields and development of improved protein backbone ParaMETERS. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 32. Balius T. E., Rizzo R. C. (2009) Quantitative prediction of fold resistance for inhibitors of EGFR. Biochemistry 48, 8435–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feller S. E., Zhang Y., Pastor R. W., Brooks B. R. (1995) Constant pressure molecular dynamics simulations: the langevin piston method. J. Chem. Phys. 103, 4613–4621 [Google Scholar]
- 34. Ryckaert J.-P., Ciccotti G., Berendsen H. J. C. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 35. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 36. Srinivasan J., Cheatham T. E. I., Cieplak P., Kollman P. A., Case D. A. (1998) Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J. Am. Chem. Soc. 120, 9401–9409 [Google Scholar]
- 37. Kollman P. A., Massova I., Reyes C., Kuhn B., Huo S. H., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D. A., Cheatham T. E., 3rd (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 [DOI] [PubMed] [Google Scholar]
- 38. Onufriev A., Bashford D., Case D. A. (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–394 [DOI] [PubMed] [Google Scholar]
- 39. Sitkoff D., Sharp K. A., Honig B. (1994) Accurate calculation of hydration free-energies using macroscopic solvent models. J. Phys. Chem. 98, 1978–1988 [Google Scholar]
- 40. Case D. A., Darden T., Cheatham T. E. I., Simmerling C., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B. P., Hayik S., Roitberg A., Seabra G., Swails J., Gotz A. W., Kolossvary I., Wong K. F., Paesani F., Vanicek J., Wolf R. M., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P. A. (2012) AMBER12, University of California, San Francisco, CA [Google Scholar]
- 41. Ahn S. M., Byun K., Cho K., Kim J. Y., Yoo J. S., Kim D., Paek S. H., Kim S. U., Simpson R. J., Lee B. (2008) Human microglial cells synthesize albumin in brain. PloS One 3, e2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Day T. A., Rakhshan F., Deutsch D. G., Barker E. L. (2001) Role of fatty acid amide hydrolase in the transport of the endogenous cannabinoid anandamide. Mol. Pharmacol. 59, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 43. Deutsch D. G., Glaser S. T., Howell J. M., Kunz J. S., Puffenbarger R. A., Hillard C. J., Abumrad N. (2001) The cellular uptake of anandamide is coupled to its breakdown by fatty-acid amide hydrolase. J. Biol. Chem. 276, 6967–6973 [DOI] [PubMed] [Google Scholar]
- 44. Kaczocha M., Glaser S. T., Chae J., Brown D. A., Deutsch D. G. (2010) Lipid droplets are novel sites of N-acylethanolamine inactivation by fatty acid amide hydrolase-2. J. Biol. Chem. 285, 2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bisogno T., Hanus L., De Petrocellis L., Tchilibon S., Ponde D. E., Brandi I., Moriello A. S., Davis J. B., Mechoulam R., Di Marzo V. (2001) Molecular targets for cannabidiol and its synthetic analogues: effect on vanilloid VR1 receptors and on the cellular uptake and enzymatic hydrolysis of anandamide. Br. J. Pharmacol. 134, 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ligresti A., Moriello A. S., Starowicz K., Matias I., Pisanti S., De Petrocellis L., Laezza C., Portella G., Bifulco M., Di Marzo V. (2006) Antitumor activity of plant cannabinoids with emphasis on the effect of cannabidiol on human breast carcinoma. J. Pharmacol. Exp. Ther. 318, 1375–1387 [DOI] [PubMed] [Google Scholar]
- 47. De Petrocellis L., Ligresti A., Moriello A. S., Allarà M., Bisogno T., Petrosino S., Stott C. G., Di Marzo V. (2011) Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br. J. Pharmacol. 163, 1479–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mileni M., Johnson D. S., Wang Z., Everdeen D. S., Liimatta M., Pabst B., Bhattacharya K., Nugent R. A., Kamtekar S., Cravatt B. F., Ahn K., Stevens R. C. (2008) Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc. Natl. Acad. Sci. U.S.A. 105, 12820–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bracey M. H., Hanson M. A., Masuda K. R., Stevens R. C., Cravatt B. F. (2002) Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 298, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 50. Mileni M., Garfunkle J., DeMartino J. K., Cravatt B. F., Boger D. L., Stevens R. C. (2009) Binding and inactivation mechanism of a humanized fatty acid amide hydrolase by α-ketoheterocycle inhibitors revealed from cocrystal structures. J. Am. Chem. Soc. 131, 10497–10506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanson B., Wang T., Sun J., Wang L., Kaczocha M., Ojima I., Deutsch D., Li H. (2014) Crystallographic study of FABP5 as an intracellular endocannabinoid transporter. Acta Crystallogr. D Biol. Crystallogr. 70, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Di Marzo V., Melck D., Bisogno T., De Petrocellis L. (1998) Endocannabinoids: endogenous cannabinoid receptor ligands with neuromodulatory action. Trends Neurosci. 21, 521–528 [DOI] [PubMed] [Google Scholar]
- 53. Marsicano G., Goodenough S., Monory K., Hermann H., Eder M., Cannich A., Azad S. C., Cascio M. G., Gutiérrez S. O., van der Stelt M., López-Rodriguez M. L., Casanova E., Schütz G., Zieglgänsberger W., Di Marzo V., Behl C., Lutz B. (2003) CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science 302, 84–88 [DOI] [PubMed] [Google Scholar]
- 54. Basavarajappa B. S. (2007) Neuropharmacology of the endocannabinoid signaling system: molecular mechanisms, biological actions and synaptic plasticity. Curr. Neuropharmacol. 5, 81–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Howlett A. C., Reggio P. H., Childers S. R., Hampson R. E., Ulloa N. M., Deutsch D. G. (2011) Endocannabinoid tone versus constitutive activity of cannabinoid receptors. Br. J. Pharmacol. 163, 1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cravatt B. F., Giang D. K., Mayfield S. P., Boger D. L., Lerner R. A., Gilula N. B. (1996) Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 384, 83–87 [DOI] [PubMed] [Google Scholar]
- 57. Deutsch D. G., Chin S. A. (1993) Enzymatic synthesis and degradation of anandamide, a cannabinoid receptor agonist. Biochem. Pharmacol. 46, 791–796 [DOI] [PubMed] [Google Scholar]
- 58. Costa B., Comelli F., Bettoni I., Colleoni M., Giagnoni G. (2008) The endogenous fatty acid amide, palmitoylethanolamide, has anti-allodynic and anti-hyperalgesic effects in a murine model of neuropathic pain: involvement of CB(1), TRPV1 and PPARγ receptors and neurotrophic factors. Pain 139, 541–550 [DOI] [PubMed] [Google Scholar]
- 59. García Mdel C., Adler-Graschinsky E., Celuch S. M. (2009) Enhancement of the hypotensive effects of intrathecally injected endocannabinoids by the entourage compound palmitoylethanolamide. Eur. J. Pharmacol. 610, 75–80 [DOI] [PubMed] [Google Scholar]
- 60. Ben-Shabat S., Fride E., Sheskin T., Tamiri T., Rhee M. H., Vogel Z., Bisogno T., De Petrocellis L., Di Marzo V., Mechoulam R. (1998) An entourage effect: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur. J. Pharmacol. 353, 23–31 [DOI] [PubMed] [Google Scholar]
- 61. Walter C., Ferreirós N., Bishay P., Geisslinger G., Tegeder I., Lötsch J. (2013) Exogenous Δ(9)-tetrahydrocannabinol influences circulating endogenous cannabinoids in humans. J. Clin. Psychopharmacol. 33, 699–705 [DOI] [PubMed] [Google Scholar]
- 62. Yu S., Levi L., Casadesus G., Kunos G., Noy N. (2014) Fatty acid-binding protein 5 (FABP5) regulates cognitive function both by decreasing anandamide levels and by activating the nuclear receptor peroxisome proliferator-activated receptor β/δ (PPARβ/δ) in the brain. J. Biol. Chem. 289, 12748–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alozie S. O., Martin B. R., Harris L. S., Dewey W. L. (1980) 3H-Δ9-Tetrahydrocannabinol, 3H-cannabinol and 3H-cannabidiol: penetration and regional distribution in rat brain. Pharmacol. Biochem. Behav. 12, 217–221 [DOI] [PubMed] [Google Scholar]
- 64. Huestis M. A. (2007) Human cannabinoid pharmacokinetics. Chem. Biodiversity 4, 1770–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bergamaschi M. M., Barnes A., Queiroz R. H., Hurd Y. L., Huestis M. A. (2013) Impact of enzymatic and alkaline hydrolysis on CBD concentration in urine. Anal. Bioanal. Chem. 405, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hunt C. A., Jones R. T., Herning R. I., Bachman J. (1981) Evidence that cannabidiol does not significantly alter the pharmacokinetics of tetrahydrocannabinol in man. J. Pharmacokinetics Biopharmaceutics 9, 245–260 [DOI] [PubMed] [Google Scholar]
- 67. Nadulski T., Pragst F., Weinberg G., Roser P., Schnelle M., Fronk E. M., Stadelmann A. M. (2005) Randomized, double-blind, placebo-controlled study about the effects of cannabidiol (CBD) on the pharmacokinetics of Δ9-tetrahydrocannabinol (THC) after oral application of THC versus standardized cannabis extract. Therap. Drug Monitoring 27, 799–810 [DOI] [PubMed] [Google Scholar]
- 68. McArdle K., Mackie P., Pertwee R., Guy G., Whittle B., Hawksworth G. (2001) Selective inhibition of Δ(9)-tetrahydrocannabinol metabolite formation by cannabidiol in vitro. Toxicology 168, 133–134 [Google Scholar]
- 69. Meiri E., Jhangiani H., Vredenburgh J. J., Barbato L. M., Carter F. J., Yang H. M., Baranowski V. (2007) Efficacy of dronabinol alone and in combination with ondansetron versus ondansetron alone for delayed chemotherapy-induced nausea and vomiting. Curr. Med. Res. Opin. 23, 533–543 [DOI] [PubMed] [Google Scholar]
- 70. Tomida I., Azuara-Blanco A., House H., Flint M., Pertwee R. G., Robson P. J. (2006) Effect of sublingual application of cannabinoids on intraocular pressure: a pilot study. J. Glaucoma 15, 349–353 [DOI] [PubMed] [Google Scholar]
- 71. Devinsky O., Cilio M. R., Cross H., Fernandez-Ruiz J., French J., Hill C., Katz R., Di Marzo V., Jutras-Aswad D., Notcutt W. G., Martinez-Orgado J., Robson P. J., Rohrback B. G., Thiele E., Whalley B., Friedman D. (2014) Cannabidiol: pharmacology and potential therapeutic role in epilepsy and other neuropsychiatric disorders. Epilepsia 55, 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Langford R. M., Mares J., Novotna A., Vachova M., Novakova I., Notcutt W., Ratcliffe S. (2013) A double-blind, randomized, placebo-controlled, parallel-group study of THC/CBD oromucosal spray in combination with the existing treatment regimen, in the relief of central neuropathic pain in patients with multiple sclerosis. J. Neurol. 260, 984–997 [DOI] [PubMed] [Google Scholar]
- 73. Collin C., Davies P., Mutiboko I. K., Ratcliffe S., and Sativex Spasticity in MS Study Group (2007) Randomized controlled trial of cannabis-based medicine in spasticity caused by multiple sclerosis. Eur. J. Neurol. 14, 290–296 [DOI] [PubMed] [Google Scholar]