Background: The telomerase gene hTERT is tightly repressed in normal human cells.
Results: KLF2 binds to the hTERT promoter in resting T cells, resulting in hTERT gene repression.
Conclusion: KLF2 is a novel factor that represses hTERT gene expression in human resting T cells.
Significance: KLF2 contributes to human-specific repression of the hTERT gene in resting T cells.
Keywords: Krüppel-like Factor 2 (KLF2), Telomerase Reverse Transcriptase (TERT), Transcription, Transcription Repressor, Zinc Finger, T-cells
Abstract
In normal human T cells, telomerase activity is strictly regulated. T cells are thought to express telomerase to avoid replicative senescence, unlike most normal somatic cells with definite replicative lifespan. T cells in blood and tissues are usually in a state of quiescence without expression of the limiting catalytic subunit of telomerase, human telomerase reverse transcriptase (hTERT). In contrast to activation, repression of hTERT transcription has not been studied well. Our previous studies have found an hTERT promoter element with repressive function. Here we identified KLF2, which represses hTERT transcription by binding to the putative promoter element. KLF2 and hTERT exhibited reciprocal mRNA expression patterns in primary human T cells. In activated T cells, KLF2 binding to the hTERT promoter was eliminated, relieving the repression of hTERT transcription found in resting T cells. Our results suggest that KLF2 is involved in strict repression of hTERT expression through binding to the promoter in primary human T cells.
Introduction
In contrast to stem cells, which prevent cellular senescence by elongation of the telomeres at cell division, most somatic cells in humans express very low levels of telomerase, which limits the capacity for cell division (1–3). Normal T lymphocytes, however, are exceptional in that they allow high-level expression of telomerase in association with cell proliferation, when activated during immune reactions (4–7). Escape from senescence may be important for generation and maintenance of memory T cells with long lifespan (7, 8). Human telomerase is a ribonucleoprotein complex composed of human telomerase reverse transcriptase (hTERT)2 and human telomerase RNA component (hTERC) (9). Transcriptional expression of the catalytic subunit hTERT has been identified as a rate-limiting step of telomerase activity. Subsequently, T cell growth has been shown to be closely associated with induction of hTERT expression; in particular, IL-2 seems to deliver signals to induce hTERT transcription (10, 11). Molecular mechanisms by which transcription of the hTERT gene is activated in T cells have been studied, and several enhancer elements and their associated factors have been identified (12–14).
hTERT expression is silenced in most human T cells in the peripheral blood and lymph nodes, as T cells are in resting phase in those tissues (5, 6). Strict repression of hTERT transcription may be associated with human T cells to avoid accidental immortalization during their increased long lifespan. It remains unknown whether an active inhibitory mechanism is involved in hTERT repression in T cells. Such a question has yet to be addressed. We as well as others previously demonstrated that the hTERT promoter carries a novel element (+9-+30) involved in transcriptional repression of this gene (10, 15, 16). In this study, we sought to understand how this novel element functions in the transcriptional expression of hTERT in human T cells and found that Krüppel-like factor 2 (KLF2, also known as LKLF) binds this element, resulting in transcriptional repression in human T cells.
EXPERIMENTAL PROCEDURES
Cells and Culture
Primary human T cells were isolated from peripheral blood leukocytes with the Pan T cell isolation kit II (Miltenyi Biotec). Collection of peripheral blood leukocytes was performed with Internal Review Committee approval from the Tokyo Medical and Dental University. The human T leukemia cell lines Jurkat and Kit 225 were cultured in RPMI 1640 medium containing 10% FCS with or without IL-2 (1 nm, Shionogi). Phytohemagglutinin (Sigma) and concanavalin A (Sigma) were added at concentrations of 10 and 3 μg/ml, respectively.
Antibodies
Anti-KLF2 antibody (AB4137) was obtained from Millipore. Anti-NF-κB p65 (sc-372X), anti-c-Myc (sc-764X), anti-KLF2 (sc-28675), anti-p21 (sc-469), anti-Sp1 (sc-14027), and anti-β-tubulin (sc-9104) antibodies and normal rabbit IgG (sc-2027) were from Santa Cruz Biotechnology.
Plasmids
Luciferase reporter plasmids, pTERT(−281)-L, pTERT(−281)-S, pGL3-Prom/43×3, and p21(−827)-Luc, were previously described elsewhere (10, 16, 17). KLF2 expression plasmids (pENTR-KLF2 and pGST-KLF2 for expressing GST fused to KLF2) were generated by insertion of human KLF2 cDNA into pENTR-HA and pGEX-4T-2 (GE Healthcare), respectively. pGL3-hTERT/(+9-+30)×3 was generated by insertion of three tandem repeats of the +9-+30 fragment of the hTERT promoter into the BglII site of pGL3-promoter vector (Promega).
Yeast One-hybrid Screening
Three tandem repeats of the 22-bp sequences (+9-+30 from the transcription start site) in the hTERT promoter were used by the MATCHMAKER one-hybrid system (K-1603-1, Clontech) with the Human Leukocyte MatchMakerTM cDNA library (Clontech). Library plasmids were isolated from yeast transformants, and the nucleotide sequences were determined.
RT-PCR
First strand cDNA was synthesized from total RNA extracted using ISOGEN (Nippon Gene). RT-PCR was performed with the following primer sets: KLF2 forward, 5′-gcgcccccagccttcggtctct-3′ and KLF2 reverse, 5′-catgtgcagcgccaggtgat-3′; GAPDH forward, 5′-ggagtccactggcgtcttca-3′ and GAPDH reverse, 5′-gaggggccatccacagtctt-3′; PPIB forward, 5′-ggccgggtgatctttggtctcttc-3′ and PPIB reverse, 5′-cccggctgtctgtcttggtgctct-3′; and KLF4 forward, 5′-accggccggctgcacacgact-3′ and KLF4 reverse, 5′-agcgggcgaatttccatccacagc-3′. Real-time RT-PCR for hTERT was performed with LightCyclerTM (Roche Diagnostics) using the LightCycler FastStart DNA MasterPLUS HybProbe (Roche Diagnostics) and LightCycler primer and probe sets (4410726, Roche Diagnostics). Triplicate samples were normalized to the amount of 18 S rRNA.
Transfection and Reporter Assay
Cells were transfected with plasmids by the DEAE-dextran method and cultured for 40 h. Luciferase assays were performed using the Luciferase Assay System (Promega). For primary human T cells, plasmids were introduced by electroporation with the IngenioTM electroporation solution (Takara Bio). All assays were performed at least three times in duplicate.
ChIP Assay
Cells were cross-linked with 1% formaldehyde and then sonicated 10 times for 30 s with 30-s intervals on ice. Cleared lysates of sonicated chromatin were incubated with antibodies overnight at 4 °C. Antibody-bound complexes were captured by incubation with Dynabeads protein A (Invitrogen). After washing with radioimmunoprecipitation assay buffer, pellets were de-cross-linked for DNA purification. DNA fragments were subjected to PCR with the following primer sets: hTERT forward, 5′-ttcaccttccagctccgcctcctc-3′ and hTERT reverse, 5′-ccccaggcgccgcacgaacg-3′; pGL3 promoter forward, 5′-caaaataggctgtccccagt-3′ and pGL3 promoter reverse, 5′-cctcggcctctgcataaata-3′.
EMSA
The 30-bp fragment (+1-+30) from the hTERT promoter was used as a probe. The probe was labeled with [32P]dATP (3000 Ci/mmol; PerkinElmer) using the Klenow enzyme or with biotin. Recombinant GST-KLF2 and GST produced in the Escherichia coli BL21 strain were purified using glutathione-Sepharose 4 Fast Flow (GE Healthcare) according to the manufacturer's protocol. These proteins were incubated with the probe. In supershift assays, anti-KLF2 or anti-p65 antibody was added to the reaction mixture before the addition of the probe. In competition assays, unlabeled wt fragment or unlabeled mutated fragment was used at 10-fold molar excess. Nucleotide sequences of the DNA fragments used for competition assays are as follows: WT fragment, 5′-ctcctcgcggcgcgagtttcaggcagcgct-3′ and mutated fragment, 5′-ctcctcgcatagttagtttcaggcagcgct-3′ (the underlined nucleotides are substitutions). A portion of reaction mixture was separated by 5% non-denaturing polyacrylamide gel electrophoresis. Probes were detected with a BAS 1500 imaging analyzer (Fujifilm) or the LightShift chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer's protocol.
Preparation of Nuclear and Cytoplasmic Extract
Nuclear and cytoplasmic fractions were prepared using the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific) according to the manufacturer's protocol and then subjected to Western blotting.
Western Blotting
For whole cell extracts, cells were lysed in the radioimmunoprecipitation assay buffer containing 10 mm Tris (pH 7.4), 1% Triton, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 5 mm EDTA, and protease inhibitor cocktail (Roche Diagnostics). The cell extracts were separated by SDS-PAGE and blotted onto Immobilon-P membranes (Atto). The membranes were blocked with 5% skim milk solution and further incubated with specific antibodies. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare). Detection was performed with the ECL Plus Western blotting detection system (GE Healthcare) according to the manufacturer's protocol.
Immunofluorescence Staining
Cells were fixed in PBS with 4% paraformaldehyde on a poly-l-lysine-coated glass slide and then permeabilized with 0.5% Triton X-100 in PBS and treated with rabbit serum to block nonspecific binding of antibodies. Cells were incubated with anti-KLF2 antibody for 30 min at room temperature, and then FITC-conjugated anti-rabbit IgG antibody (Molecular Probes) was added. DNA in the nuclei was detected with DAPI. Cells were examined with a confocal laser scanning fluorescence microscope (FV10i, Olympus).
siRNAs
Accell siRNA for KLF2 (E-006928 (mixture of #1-#4), A-006928-14 (#1), A-006928-15 (#2), A-006928-16 (#3), A-006928-17 (#4)), KLF4 (E-005089), PPIB (D-001920), and non-targeting control (D-001910) were purchased form Thermo Fisher Scientific, and introduced into primary human T cells. In brief, primary T cells were stimulated with mitogens for 3 days. After being washed with PBS three times, the cells were subjected to transfection with siRNA in Accell siRNA delivery medium (Thermo Scientific) containing 2% FCS for 3 days, and then RNA was isolated to perform RT-PCR and real-time PCR. For Western blotting, cells were cultured for 4 days, and then cellular proteins were isolated.
Statistical Analysis
A paired t test was performed for all statistical analyses. Statistical significances were considered at p values < 0.05.
RESULTS
A Nuclear Factor Bound to the Repressive Element in the hTERT Promoter
Our previous studies demonstrated that the hTERT promoter carries a repressive element downstream of the transcription initiation site (+9-+30) (Fig. 1A) (10, 16). This repressive element was used in the yeast one-hybrid assay as a probe to screen for a factor that regulates hTERT gene expression. The screening recovered 69 clones from the Human Leukocyte MatchMakerTM cDNA library. Out of the 69 clones, 12 were derived from the gene encoding KLF2 (Table 1).
FIGURE 1.
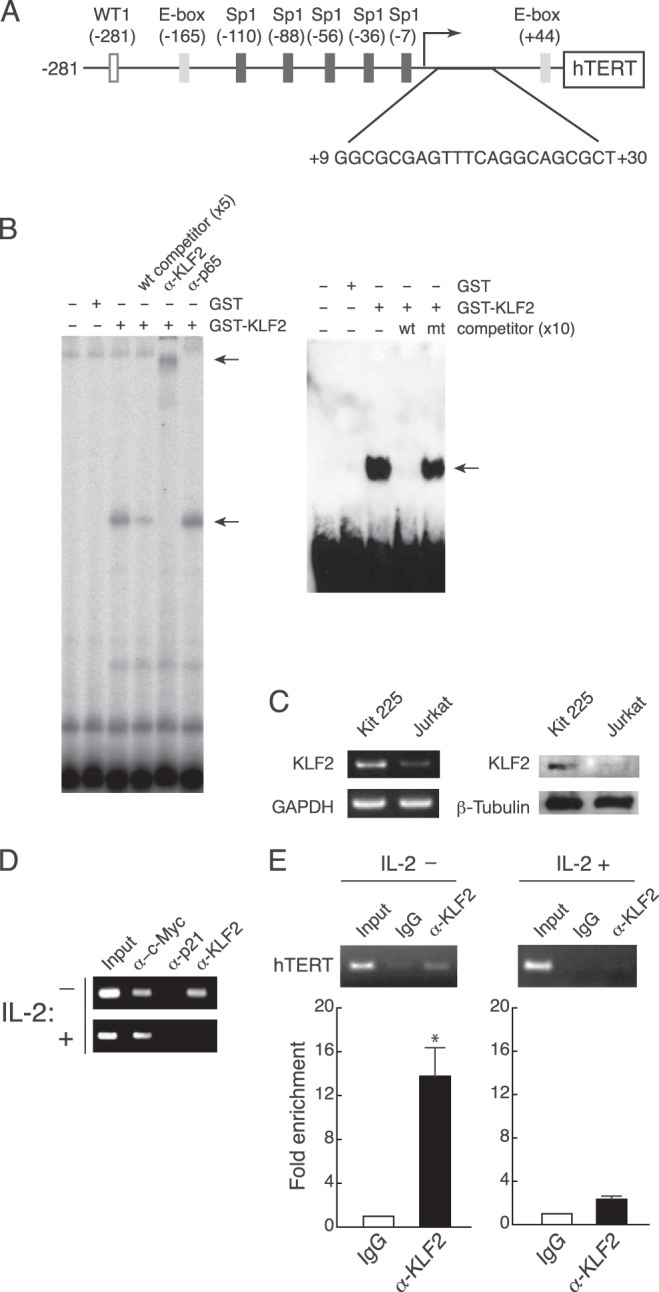
Binding of KLF2 to an hTERT promoter element. A, in addition to nucleotide sequences encompassing the element (10, 16), previously identified elements are shown (12, 13). B, recombinant GST-KLF2 was incubated with the 32P-labeled probe (+1-+30) in the absence or presence of 5-fold excess of cold competitor, anti-KLF2 antibody, or anti-p65 antibody (left). For competition assays, the unlabeled wild-type fragment (wt) or unlabeled mutated fragment (mt) at 10-fold excess was mixed with the biotin-labeled probe and GST-KLF2 (right). Arrows indicate specific binding and an antibody-dependent super-shifted band. Experiments were performed in triplicate, and representative data are shown. C, expression of KLF2 in Kit 225 and Jurkat cells. KLF2 expression was examined by RT-PCR with primers specific to KLF2 and GAPDH (left) and Western blotting with antibodies specific to KLF2 and β-tubulin (right). Kit 225 cells were cultured in the presence of IL-2 for sample preparation. Experiments were performed in triplicate, and representative data are shown. D and E, ChIP assays in Kit 225 cells (D) and primary T cells (E). Kit 225 cells were transfected with the hTERT promoter reporter plasmid pGL3-Prom/43×3 and cultured with (+) or without (−) IL-2. Primary T cells were cultured with (+) or without (−) IL-2. Chromatin lysates were prepared from those cells and subjected to immunoprecipitation by antibodies followed by PCR with primers specific to the hTERT promoter. Experiments were performed in triplicate, and representative data are shown. Real-time PCR products were quantified and presented as relative amounts to rabbit IgG-mediated products by measuring SYBR Green I fluorescence (E, lower panel). Mean values ± S.E. of triplicate examinations are shown. *, p = 0.0007.
TABLE 1.
Molecules interacting with the hTERT promoter element (+9-+30)
Molecules that gave more than two independent colonies in the yeast one-hybrid screening are shown.
| Names of molecules | Number of colonies |
|---|---|
| Krüppel-like factor 2 (KLF2) | 12 |
| Proteasome 26S subunit, non-ATPase, 4 (PSMD4) | 7 |
| V-rel reticuloendotheliosis viral oncogene homolog A (Rel A) | 4 |
| Interferon regulatory factor 8 (IRF8) | 3 |
| Mitogen-activated protein kinase kinase kinase 2 (MAP3K2) | 2 |
Binding of KLF2 to the Element in Vivo and in Vitro
KLF2 binding to the element in vitro was examined by EMSA. The GST-KLF2 chimera protein bound to a probe containing DNA sequences from the repressive element in the hTERT promoter (Fig. 1B). The complex formation was abolished in the presence of the unlabeled wild-type competitor, whereas the mutated competitor did not alter the binding. The addition of anti-KLF2 antibody, but not anti-p65 antibody, showed a supershift band. KLF2 binding to the element was investigated in vivo by studies of ChIP assays in the human T cell line Kit 225 and primary human T cells. Kit 225 cells are positive for KLF2 (Fig. 1C) and, interestingly, Kit 225 cells require IL-2 to grow and depletion of IL-2 causes growth arrest at the G1 phase of the cell cycle (10). Kit 225 cells were transfected with pGL3-Prom/43×3 carrying three tandem repeats of the 43-bp fragment (+9-+51) with the repressive element and a Myc binding site (E-box) in the hTERT promoter and subjected to transient ChIP assays. KLF2 binding to the exogenous element was seen only in Kit 225 cells cultured in the absence of IL-2, indicating that KLF2 binds to the element in resting Kit 225 cells (Fig. 1D). In contrast, c-Myc bound to the exogenous fragment irrespective of growth phase of Kit 225 cells. Consistent with the results of transient ChIP assays, the endogenous element was enriched in resting primary T cells, when anti-KLF2 antibody was used in the ChIP assays (Fig. 1E). Primary T cells stimulated with IL-2 showed little or no KLF2 bound to the element (Fig. 1E), similar to the results with growing Kit 225 cells. These results indicate that KLF2 binding to the hTERT promoter is inversely associated with T cell activation.
Inverse Correlation between KLF2 Expression and hTERT Expression
Contrary to KLF2 binding, hTERT mRNA level was profoundly higher in activated primary human T cells rather than in resting cells (Fig. 2A), which is consistent with previous studies (10, 11, 18). To gain insights into the mechanism of KLF2-mediated regulation of hTERT gene expression, the properties of KLF2 expression were examined in terms of kinetics in resting and activated primary T cells. When primary T cells were activated with phytohemagglutinin and concanavalin A, KLF2 mRNA expression was transcriptionally down-regulated (Fig. 2B). In parallel with the result of the transcription, the protein level of KLF2 was also reduced upon the stimulation with mitogens (Fig. 2C). Interestingly, depletion of mitogens recovered expression of KLF2 mRNA and KLF2 protein (Fig. 2, B and C). Inversely, expression of hTERT mRNA was increased in response to the stimulation, and mitogen-depleted cells weakly transcribed the hTERT gene (Fig. 2B), as also shown previously (10).
FIGURE 2.
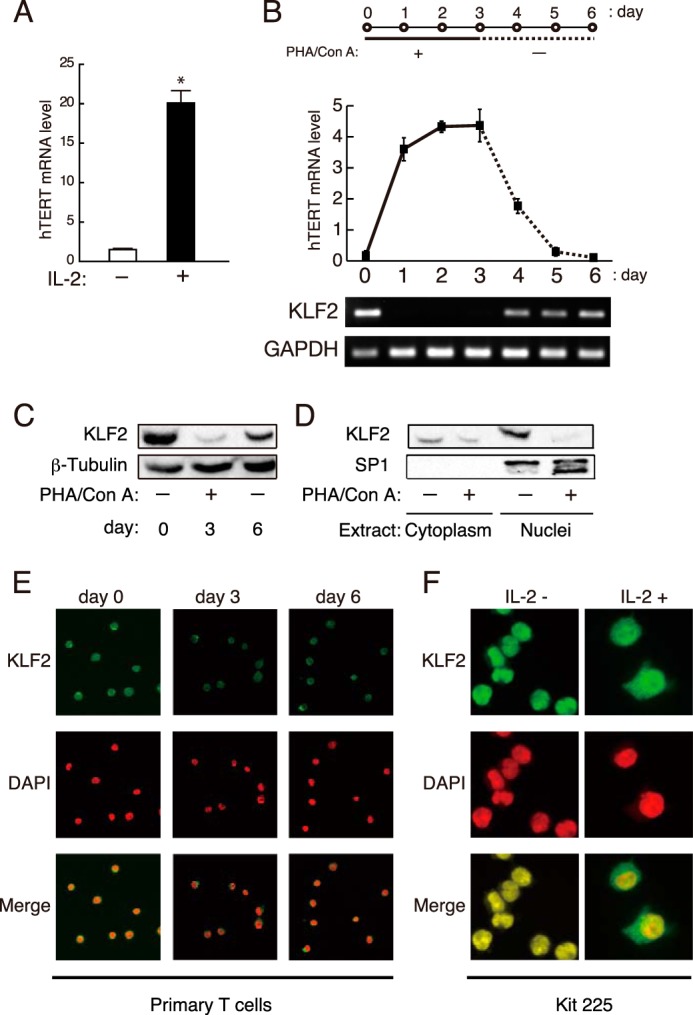
KLF2 expression in human T cells. A, peripheral blood leukocytes were stimulated with mitogens and cultured with or without IL-2. Levels of hTERT mRNA were determined by real-time RT-PCR. Mean values ± S.E. of four samples are shown. *, p = 0.00017. B, primary human T cells were cultured with mitogens for 3 days (days 0–3) and further cultured without mitogens (days 4–6) (upper). Cells were harvested every day until day 6 to determine the expression of hTERT and 18 S rRNA by real-time RT-PCR. Results are shown as a ratio of hTERT mRNA to 18 S rRNA (middle). Mean values ± S.E. of four samples are shown. KLF2 and GAPDH expression was monitored by RT-PCR (lower). PHA, phytohemagglutinin; Con A, concanavalin A. C and D, primary T cells were cultured with mitogens for 3 days and further cultured without mitogens for 3 days. Extracts from whole cells (days 0, 3, and 6) (C) and nuclear and cytoplasmic fractions (day 3) (D) were subjected to Western blotting with antibodies specific to KLF2, SP1, and β-tubulin. Experiments were performed in triplicate, and representative data are shown. E, primary human T cells on days 0, 3, and 6 as shown in panel B were subjected to immunostaining with anti-KLF2 antibody and DAPI. F, Kit 225 cells cultured with (+) or without (−) IL-2 were subjected to immunostaining with anti-KLF2 antibody and DAPI. Fluorescence images were observed with a confocal laser-scanning microscope. All experiments were performed at least three times, and representative data are shown.
To characterize the functional properties of KLF2 in hTERT regulation, nuclear and cytoplasmic fractions from resting and activated primary T cells were subjected to Western blot analysis for determining subcellular distribution of KLF2. KLF2 predominantly located in the nucleus of resting cells (Fig. 2D). Upon activation, the amount of KLF2 protein in the nucleus was dramatically reduced, consistent with the down-regulation of KLF2 in activated cells (Fig. 2D). Immunostaining of primary T cells with anti-KLF2 antibody also illustrated the presence of KLF2 predominantly in the nucleus of resting cells (days 0 and 6), whereas asynchronously activated cells (day 3) demonstrated that KLF2 was also found in the cytoplasm (Fig. 2E). Changes in KLF2 localization were also seen with Kit 225 cells, although the protein level of KLF2 does not change in Kit 225 cells irrespective of cell cycle phase. In resting Kit 225 cells, KLF2 was located in the nucleus, but growing Kit 225 cells cultured with IL-2 contained KLF2 in the cytoplasm and nucleus (Fig. 2F). Thus we suggest that KLF2 activity in growing T cells may be reduced in two ways: repression of its transcription and subcellular localization of the protein from the nucleus to the cytoplasm. These results imply that KLF2 may repress hTERT gene transcription by direct binding to the promoter in resting T cells and that this repression may be released in activated T cells, resulting in induction of hTERT gene expression.
Repression of hTERT Transcription by KLF2
The ability of KLF2 to repress hTERT gene expression was assessed by modulation of KLF2 expression. The KLF2 expression plasmid (pENTR-KLF2) was transfected along with luciferase reporter plasmids carrying the hTERT promoter into the Jurkat human T cell line, which lacks endogenous KLF2 expression (Fig. 1C). The p21WAF1/Cip1 promoter, which is a cell cycle inhibitor and is positively regulated by KLF2 (19), was activated by ectopic KLF2 expression in luciferase assays (Fig. 3A). Under such conditions, the wild-type vector pTERT(−281)-L containing the repressive element was significantly repressed by ectopic expression of KLF2, whereas a mutant pTERT(−281)-S lacking the element was not affected (Fig. 3A). Primary T cells exhibited similar results; ectopic introduction of the KLF2 expression plasmid into activated T cells induced reduction in endogenous hTERT mRNA expression (Fig. 3B). Conversely, inhibition of KLF2 expression with a mixture of four siRNA for KLF2 in primary resting T cells specifically increased the level of endogenous hTERT mRNA (Fig. 3C). Each siRNA species was examined for its effect on expression of KLF2 and hTERT. Among them, two (#1 and #4) showed reduction of KLF2 mRNA and protein levels (Fig. 3D). Reduction of KLF2 was associated with up-regulation of hTERT expression, indicating that KLF2 knockdown led the hTERT gene to down-regulation in its expression. It is intriguing to note that ablation of KLF4, which is similar to KLF2 in terms of functional properties (20), did not recover hTERT mRNA expression (Fig. 3C). Our data suggest that KLF2 contributes to strict regulation of the hTERT transcription in resting human T cells.
FIGURE 3.
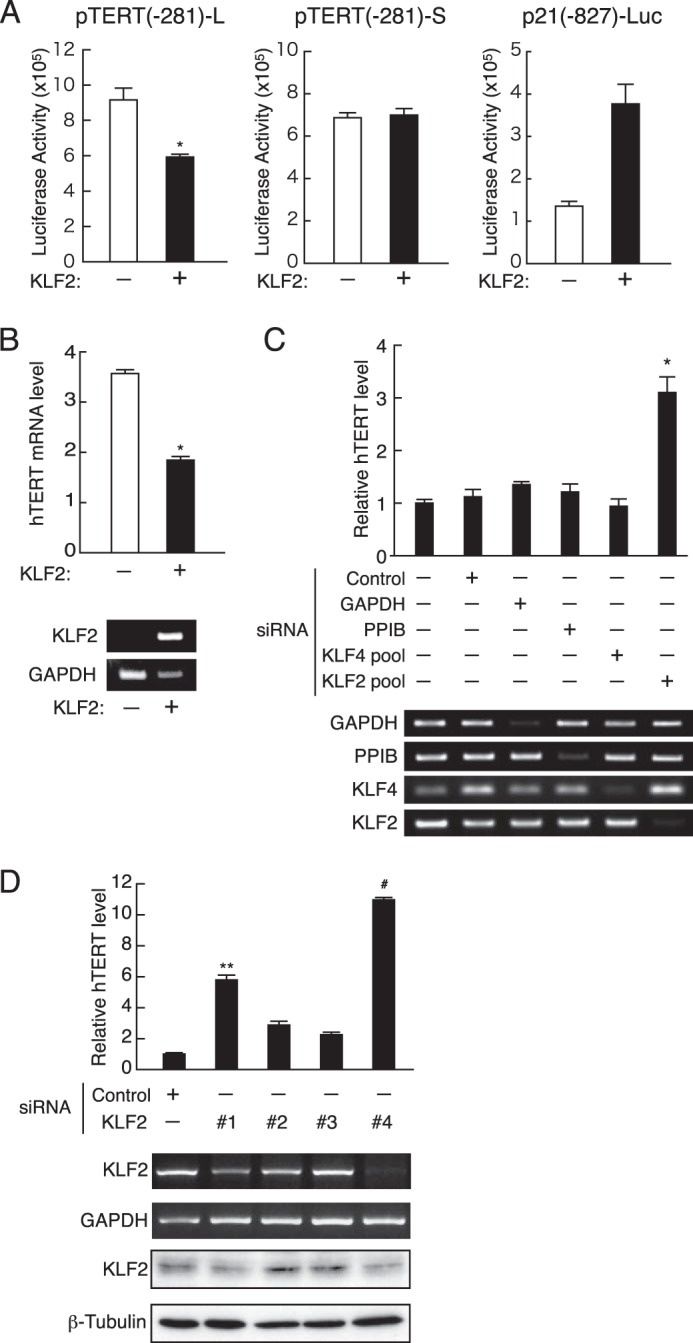
KLF2-mediated repression of the hTERT promoter. A, Jurkat cells were transfected with the hTERT promoter reporter plasmids (pTERT(−281)-L or pTERT(−281)-S) along with the KLF2 expression plasmid pENTR-KLF2. Luciferase activity was determined at 40 h after transfection. The p21-luc plasmid carries the p21WAF1/Cip1 promoter, which is shown to be activated by KLF2. Mean values ± S.E. of four samples are shown. *, p = 0.0082. B, peripheral blood leukocytes were cultured with mitogens for 2 days and then transfected with pENTR-HA or pENTR-KLF2 by electroporation. hTERT mRNA levels were determined by real-time RT-PCR with RNA prepared from cells 48 h after transfection. KLF2 and GAPDH expression was detected by RT-PCR. Mean values ± S.E. of four samples are shown. *, p = 0.00002. C and D, after culture with mitogens for 72 h, primary T cells were transfected with non-targeting siRNA or siRNA specific for various target genes and cultured in Accell siRNA delivery medium containing 2% FCS without mitogens. RNA and cellular protein were harvested at 72 and 96 h, respectively. hTERT mRNA levels were determined by real-time RT-PCR. Results are shown as the ratio of hTERT mRNA to 18 S rRNA. mRNA expression of KLF2, KLF4, PPIB, and GAPDH was examined by RT-PCR. Whole cell lysates were subjected to Western blotting with antibodies specific to KLF2 and α-tubulin. Representative data are shown. Mean values ± S.E. of four samples are shown. *, p = 0.0004, **, p = 0.0009, and #, p = 0.00008.
DISCUSSION
The major finding of the present study is that KLF2 represses hTERT gene expression via binding to the hTERT promoter region. Our results indicate that KLF2 binds to the hTERT promoter in resting T cells, which do not express hTERT, whereas KLF2 binding is not seen in activated T cells, which do express hTERT.
hTERT expression is generally associated with extension of the replicative lifespan of T cells to avoid shortening of telomere during immune responses. In the peripheral blood, T cells mostly stay in a phase of growth quiescence with little or no expression of hTERT. Immune stimulation induces activation and proliferation of T cells along with transient hTERT expression. If hTERT is constitutively expressed, undesirable immortalization may occur. Thus hTERT expression must be under stringent regulation in human T cells. Multiple lines of evidence indicate that hTERT expression is linked with the cell cycle of human T cells (5, 7, 10, 13, 16).
Several elements in the hTERT promoter, including two E-boxes, have been shown to be involved in the activation and repression of hTERT transcription through the binding of transcription factors (c-Myc, Mad, Sp1, E2F1, USF, WT1, MZF-2, VDR, ER, ER81, and ETS) (13). Mutagenesis approaches in our previous studies demonstrated that two distinct elements in the hTERT promoter contribute to repression of the promoter in resting T cells. One is an E-box at +44-+49, which has been suggested to independently exert repressive effects on transcription in our previous study (10). In addition, we and others have observed that the Myc binding site (E-box at +44-+49) functioned as an activator (12, 16), but its molecular mechanisms are still controversial as described in a recent study (21). The other (+9-+30) is examined in this study for its molecular mechanism underlying hTERT repression in resting cells. We identified the KLF2 as a repressor of the hTERT gene in resting T cells. KLF2 is a zinc finger type transcription factor expressed in the lung, endothelial cells, lymphocytes, and cell lines derived from such tissues (22). In mouse, KLF2 plays an important role in naive T cell trafficking through induction of various molecules (23–25). In addition, growth arrest is induced by forced expression of KLF2 in human T cells via induction of p21WAF1/Cip1 and reduction of c-myc transcription (19, 26). However, our previous study showed that overexpression of p21WAF1/Cip1 did not change hTERT promoter activity in human T cells, whereas overexpression of p21WAF1/Cip1 decreased E2F activity, presumably resulting in cell growth arrest (16). Thus our results suggest that KLF2 directly represses hTERT expression, irrespective of p21WAF1/Cip1 expression.
The recovery of KLF2 expression appeared incomplete after depletion of mitogens on day 6 (Fig. 2C). We assume two possible hypotheses. One is that 3 days are not enough for as much accumulation of KLF2 as on day 0. This notion may be supported in part by data that KLF2 mRNA levels increase gradually after mitogen depletion (Fig. 2B). The other is related to degradation mechanism of KLF2. The E3 ligase WWP1 is shown to catalyze ubiquitination of KLF2, leading its degradation (27). Under our experimental conditions, WWP1 might be active shortly after induction of cell resting by depletion of mitogens.
In mouse, even normal somatic tissues and cells express robust levels of telomerase activity (28). Notably, the repressive element shown in this study is not conserved in the mouse TERT promoter (15). Introduction of the human repressive element into the mouse TERT promoter showed significant repression of the promoter. These results indicate that human and mouse differ in the molecular mechanism of telomerase regulation. Our finding of the involvement of KLF2 in telomerase regulation may be specific to human T cells. A recent study showed that KLF2 is not required for quiescence in post-activated mouse T cells (29), supporting the difference in KLF2 function between human and mouse. Our results indicate that KLF2 regulates hTERT gene expression even in post-activated human T cells. Consequently, hTERT expression returns to the background level after diminishing activation (Fig. 2B).
The KLF/specificity protein (Sp) family of transcription factors consists of 25 members and regulates diverse biological processes including proliferation, differentiation, growth, development, survival, and responses to external stress (22). Among them, KLF2, KLF4, and KLF5 show structural and functional similarity. ChIP-on-chip assays indicate that KLF2, KLF4, and KLF5 share many common gene targets in ES cells (20). Interestingly, the present study suggests that KLF2 has a unique function in regulation of the hTERT gene, which is not shared by KLF4 (Fig. 3C). Our findings clearly show a novel role of KLF2 in hTERT gene expression in human T cells.
Acknowledgments
We thank R. Weinberg for an hTERT promoter clone and N. Enomoto and H. Shimizu for technical assistance. We also thank J. McCubery, L. Preston, and P. Varga-Weisz for critical reading of the manuscript and members of our laboratory for discussions and support.
Note Added in Proof
Table 1 was inadvertently omitted from the version of this article that was published as a Paper in Press on February 18, 2015.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Japan Society for Promotion of Science.
- hTERT
- human telomerase reverse transcriptase
- KLF2
- Krüppel-like factor 2
- PPIB
- peptidylprolyl isomerase B.
REFERENCES
- 1. Harley C. B., Futcher A. B., Greider C. W. (1990) Telomeres shorten during aging of human fibroblasts. Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 2. Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., Weinberg R. A., Stewart S. A., Hahn W. C. (2003) Telomerase maintains telomere structure in normal human cells. Cell 114, 241–253 [DOI] [PubMed] [Google Scholar]
- 3. Deng Y., Chan S. S., Chang S. (2008) Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer 8, 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hiyama K., Hirai Y., Kyoizumi S., Akiyama M., Hiyama E., Piatyszek M. A., Shay J. W., Ishioka S., Yamakido M. (1995) Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 155, 3711–3715 [PubMed] [Google Scholar]
- 5. Buchkovich K. J., Greider C. W. (1996) Telomerase regulation during entry into the cell cycle in normal human T cells. Mol. Biol. Cell 7, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weng N. P., Levine B. L., June C. H., Hodes R. J. (1996) Regulated expression of telomerase activity in human T lymphocyte development and activation. J. Exp. Med. 183, 2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hodes R. J., Hathcock K. S., Weng N. P. (2002) Telomeres in T and B cells. Nat. Rev. Immunol 2, 699–706 [DOI] [PubMed] [Google Scholar]
- 8. Akbar A. N., Vukmanovic-Stejic M. (2007) Telomerase in T lymphocytes: use it and lose it? J. Immunol. 178, 6689–6694 [DOI] [PubMed] [Google Scholar]
- 9. Nugent C. I., Lundblad V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev. 12, 1073–1085 [DOI] [PubMed] [Google Scholar]
- 10. Matsumura-Arioka Y., Ohtani K., Hara T., Iwanaga R., Nakamura M. (2005) Identification of two distinct elements mediating activation of telomerase (hTERT) gene expression in association with cell growth in human T cells. Int. Immunol. 17, 207–215 [DOI] [PubMed] [Google Scholar]
- 11. Kawauchi K., Ihjima K., Yamada O. (2005) IL-2 increases human telomerase reverse transcriptase activity transcriptionally and posttranslationally through phosphatidylinositol 3′-kinase/Akt, heat shock protein 90, and mammalian target of rapamycin in transformed NK cells. J. Immunol. 174, 5261–5269 [DOI] [PubMed] [Google Scholar]
- 12. Poole J. C., Andrews L. G., Tollefsbol T. O. (2001) Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT). Gene 269, 1–12 [DOI] [PubMed] [Google Scholar]
- 13. Janknecht R. (2004) On the road to immortality: hTERT upregulation in cancer cells. FEBS Lett. 564, 9–13 [DOI] [PubMed] [Google Scholar]
- 14. Ducrest A. L., Szutorisz H., Lingner J., Nabholz M. (2002) Regulation of the human telomerase reverse transcriptase gene. Oncogene 21, 541–552 [DOI] [PubMed] [Google Scholar]
- 15. Horikawa I., Chiang Y. J., Patterson T., Feigenbaum L., Leem S. H., Michishita E., Larionov V., Hodes R. J., Barrett J. C. (2005) Differential cis-regulation of human versus mouse TERT gene expression in vivo: Identification of a human-specific repressive element. Proc. Natl. Acad. Sci. U.S.A. 102, 18437–18442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hara T., Matsumura-Arioka Y., Ohtani K., Nakamura M. (2008) Role of human T-cell leukemia virus type I Tax in expression of the human telomerase reverse transcriptase (hTERT) gene in human T-cells. Cancer Sci. 99, 1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Funato N., Ohtani K., Ohyama K., Kuroda T., Nakamura M. (2001) Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol. Cell Biol. 21, 7416–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu K., Schoonmaker M. M., Levine B. L., June C. H., Hodes R. J., Weng N. P. (1999) Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 96, 5147–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu J., Lingrel J. (2004) KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene 23, 8088–8096 [DOI] [PubMed] [Google Scholar]
- 20. Jiang J., Chan Y.-S., Loh Y.-H., Cai J., Tong G.-Q., Lim C.-A., Robson P., Zhong S., Ng H.-H. (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 21. Zhao Y., Cheng D., Wang S., Zhu J. (2014) Dual roles of c-Myc in the regulation of hTERT gene. Nucleic Acids Res. 42, 10385–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaczynski J., Cook T., Urrutia R. (2003) Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carlson C. M., Endrizzi B. T., Wu J., Ding X., Weinreich M. A., Walsh E. R., Wani M. A., Lingrel J. B., Hogquist K. A., Jameson S. C. (2006) Krüppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 [DOI] [PubMed] [Google Scholar]
- 24. Bai A., Hu H., Yeung M., Chen J. (2007) Krüppel-like factor 2 controls T cell trafficking by activating l-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J. Immunol. 178, 7632–7639 [DOI] [PubMed] [Google Scholar]
- 25. Odumade O. A., Weinreich M. A., Jameson S. C., Hogquist K. A. (2010) Krüppel-like factor 2 regulates trafficking and homeostasis of γδ T cells. J. Immunol. 184, 6060–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuo C. T., Veselits M. L., Leiden J. M. (1997) LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science 277, 1986–1990 [DOI] [PubMed] [Google Scholar]
- 27. Zhang X., Srinivasan S. V., Lingrel J. B. (2004) WWP1-dependent ubiquitination and degradation of the lung Krüppel-like factor, KLF2. Biochem. Biophys. Res. Commun. 316, 139–148 [DOI] [PubMed] [Google Scholar]
- 28. Prowse K. R., Greider C. W. (1995) Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. U.S.A. 92, 4818–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takada K., Wang X., Hart G. T., Odumade O. A., Weinreich M. A., Hogquist K. A., Jameson S. C. (2011) Krüppel-like factor 2 is required for trafficking but not quiescence in postactivated T cells. J. Immunol. 186, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]


