Background: Copper levels stimulate trafficking and hyperphosphorylation of the copper transporter ATP7B.
Results: Hyperphosphorylation can occur prior to exit from the TGN and requires intact N and C terminus but is not required for apically directed trafficking.
Conclusion: Copper-stimulated hyperphosphorylation of ATP7B occurs on the C terminus.
Significance: Hyperphosphorylation and apically directed trafficking are independent events.
Keywords: Copper, Copper Transport, Phosphoproteomics, Phosphorylation, Protein Kinase, Trafficking
Abstract
The Wilson disease protein ATP7B exhibits copper-dependent trafficking. In high copper, ATP7B exits the trans-Golgi network and moves to the apical domain of hepatocytes where it facilitates elimination of excess copper through the bile. Copper levels also affect ATP7B phosphorylation. ATP7B is basally phosphorylated in low copper and becomes more phosphorylated (“hyperphosphorylated”) in elevated copper. The functional significance of hyperphosphorylation remains unclear. We showed that hyperphosphorylation occurs even when ATP7B is restricted to the trans-Golgi network. We performed comprehensive phosphoproteomics of ATP7B in low versus high copper, which revealed that 24 Ser/Thr residues in ATP7B could be phosphorylated, and only four of these were copper-responsive. Most of the phosphorylated sites were found in the N- and C-terminal cytoplasmic domains. Using truncation and mutagenesis, we showed that inactivation or elimination of all six N-terminal metal binding domains did not block copper-dependent, reversible, apical trafficking but did block hyperphosphorylation in hepatic cells. We showed that nine of 15 Ser/Thr residues in the C-terminal domain were phosphorylated. Inactivation of 13 C-terminal phosphorylation sites reduced basal phosphorylation and eliminated hyperphosphorylation, suggesting that copper binding at the N terminus propagates to the ATP7B C-terminal region. C-terminal mutants with either inactivating or phosphomimetic substitutions showed little effect upon copper-stimulated trafficking, indicating that trafficking does not depend on phosphorylation at these sites. Thus, our studies revealed that copper-dependent conformational changes in the N-terminal region lead to hyperphosphorylation at C-terminal sites, which seem not to affect trafficking and may instead fine-tune copper sequestration.
Introduction
ATP7B is a P1B-type ATPase that transports the essential element copper across cellular membranes. Mutations in ATP7B cause Wilson disease (OMIM 277900), a condition of copper toxicity that can lead to liver failure and/or neurological disturbances (1, 2). The ATP7B protein carries out two main functions: it delivers copper to cuproenzymes during their biosynthesis, and it exports copper to maintain cellular homeostasis as a mechanism for protecting the cell from oxidative damage caused by excess copper (3). Cellular copper levels affect the intracellular location of ATP7B such that it resides in the trans-Golgi network (TGN)2 in low copper to fulfill its biosynthetic role. In elevated copper, ATP7B moves via vesicles to the apical surface to rid the cell of excess copper (4–8).
Copper-dependent trafficking of ATP7B is also associated with increased phosphorylation on Ser/Thr and was previously termed hyperphosphorylation (9–13). Previous studies showed that copper-dependent TGN exit of ATP7B correlated with its hyperphosphorylation (9). ATP7B phosphorylation also occurs during its biosynthesis; these modifications have been referred to as “basal” phosphorylation and seem to affect protein stability, lifetime, and/or quality control (9, 12–14). Mass spectrometry (MS) analysis of recombinant ATP7B isolated from Sf9 cells revealed that the loop between metal binding domain 3 (MBD3) and MBD4 as well as 10 other N-terminal (N-term) peptides were phosphorylated following in vitro radiolabeling in the presence of mammalian cell extract (15). Recent large scale MS proteomics analyses have identified still more phosphorylated ATP7B residues (see Table 1 and Refs. 16–21). Most of these residues were identified when either cells or tissues were exposed to basal copper, making their copper dependence equivocal.
TABLE 1.
Summary of phosphosite identifications in ATP7B
H, human; M, mouse. Square brackets indicate ambiguities.
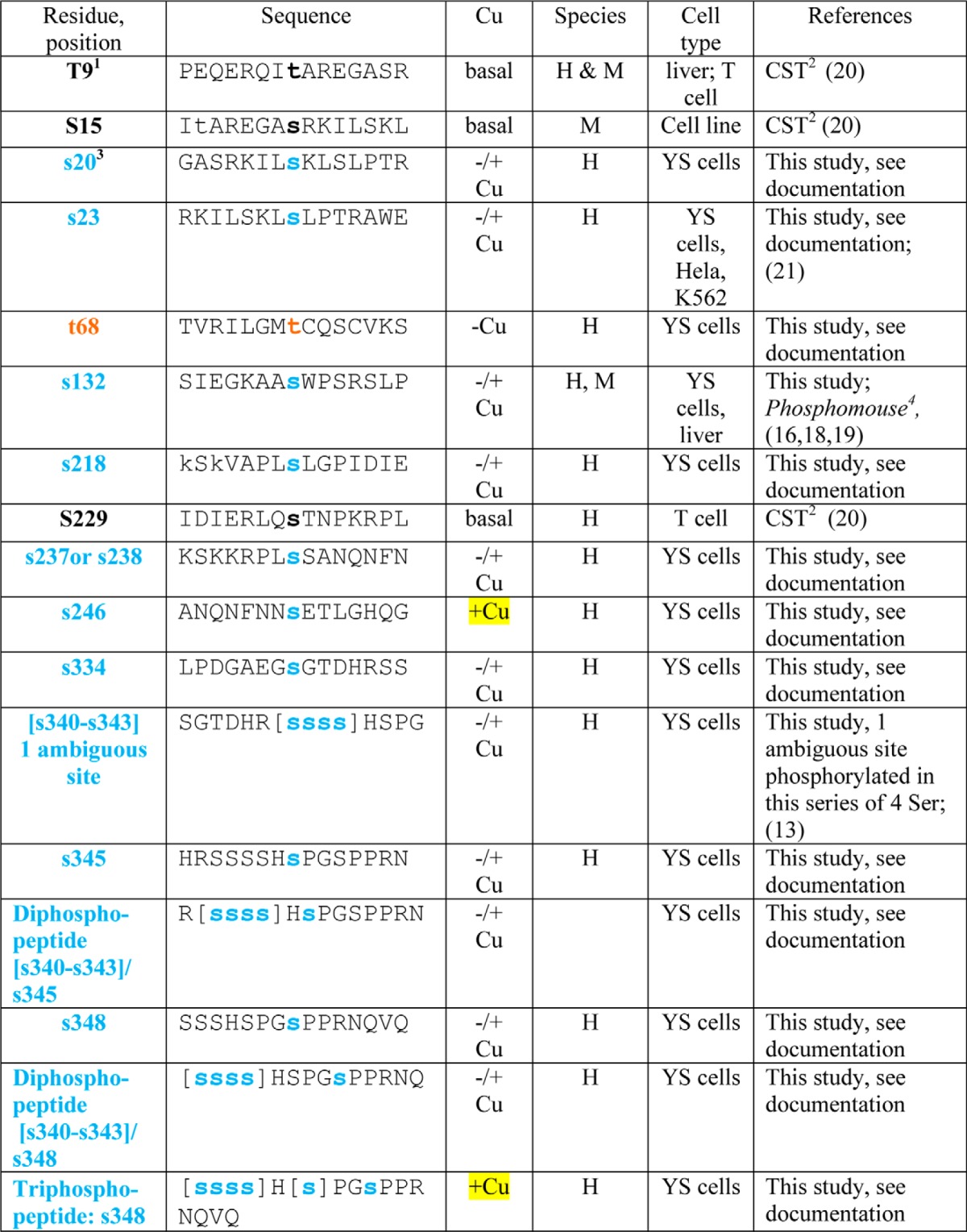
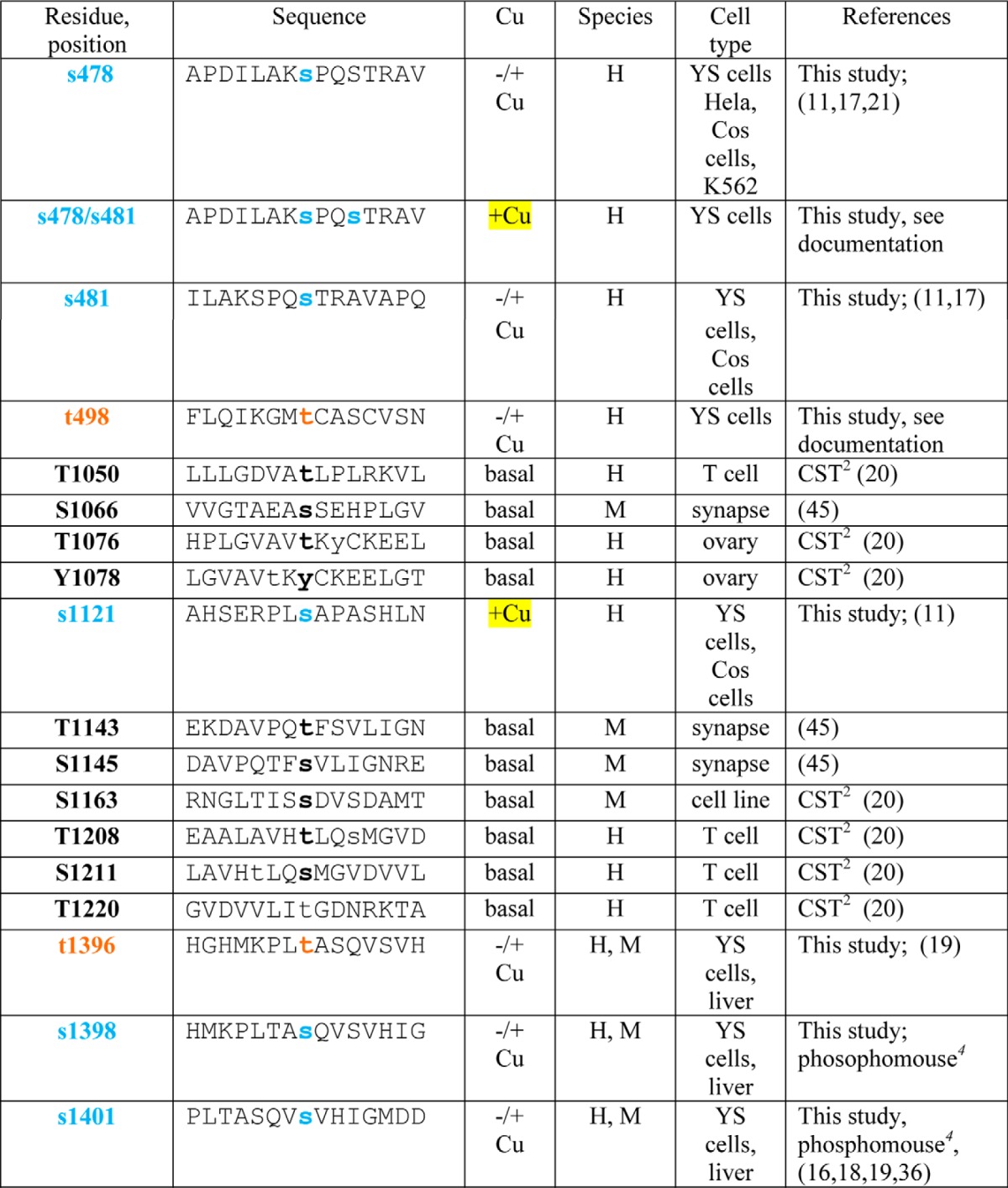
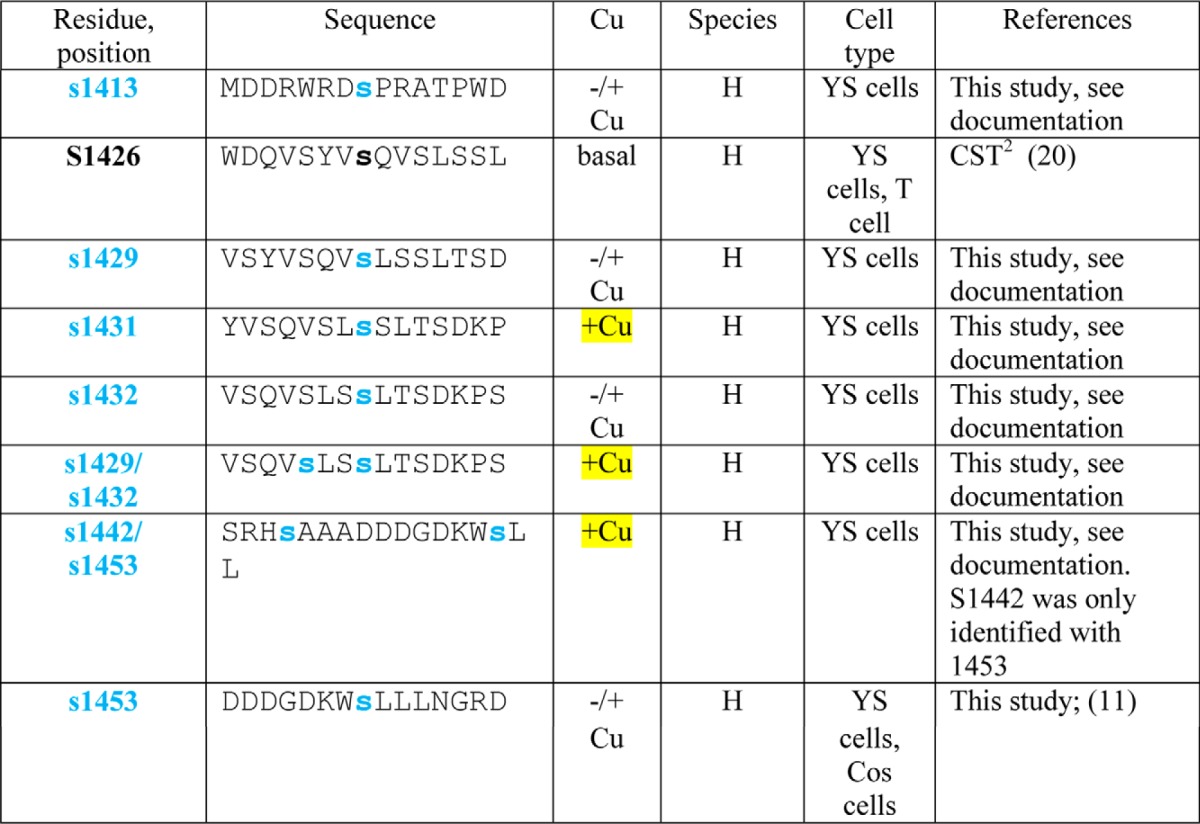
1 Black Ser and Thr residues were identified as referenced.
2 CST, Cell Signaling Technology. PhosphoSitePlus (PSP) is an online systems biology resource providing comprehensive information and tools for the study of protein post-translational modifications including phosphorylation, ubiquitination, acetylation, and methylation.
3 Blue (Ser) and brown (Thr) residues were identified in this study; the reference is provided if identified in abother study. Documentation for all previously unidentified sites as well as di- and triphosphopeptides is shown in the supplemental data. Phosphosites found only in copper-treated cells are highlighted in yellow.
4 Phosphorylation site identification in mouse tissues was performed by Gygi and co-workers (16).
More recently, four unique serines (Ser-478, Ser-481, Ser-1121, and Ser-1453) were identified following in vitro phosphorylation of microsomes isolated from COS-1 cells expressing myc-tagged ATP7B, and three of these residues (Ser-478, Ser-471, and Ser-1153) were substrates for kinase(s) in those cells (11). An ATP7B mutant with all four of these Ser changed to Ala exhibited defective TGN exit, suggesting that phosphorylation at these sites is required for TGN exit (12). The conformation of a different Ser cluster (Ser-340 and Ser-341), located in the loop between MBD3 and MBD4 also appears to be important for TGN exit in HEK293TRex cells (13). Inactivating mutations (Ser-340 and Ser-341 to Ala) did not reduce basal phosphorylation of ATP7B. Moreover, all mutations at this cluster, whether to Ala/Gly (inactivating), Asp (phosphomimic), or Thr (phosphosite), had similar effects on trafficking; i.e. they all showed increased ATP7B in vesicles. The conclusion of this study was that phosphorylation does not initiate ATP7B trafficking per se but rather maintains the protein in a trafficking-permissive state following a copper-induced conformational change (13).
Overall, the relationship between copper-stimulated Ser/Thr hyperphosphorylation and copper-dependent trafficking remains enigmatic, and the ATP7B residues phosphorylated physiologically in high copper remain undetermined. In this study, we used comparative MS-based proteomics to determine ATP7B phosphorylation under conditions of low and high copper in fibroblasts cells. We then used mutagenesis of selected sites in conjunction with expression in hepatic cells to determine the physiological consequences of copper-stimulated phosphorylation. We found little evidence to suggest that copper-stimulated apical trafficking requires phosphorylation, leaving the physiological consequence of this post-translational modification yet to be determined.
EXPERIMENTAL PROCEDURES
Generation of ATP7B Mutants
Full-length wild-type ATP7B fused at its N terminus to green fluorescent protein (GFP) (WTATP7B; cataloging designation, pLB1080 (22)) and the N-terminal mutant 1–63 Δ1–4MBD (cataloging designation YG36 (23)) in pAdLOX (24) were described previously. The QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used with YG36 as a template to create plasmids with substitutions in N-term metal binding domains (Table 2; all nicknames used throughout are in parentheses): 1–63 Δ1–4MBD + C499S/C502S/C575S/C578S (MBD5&6 C>S). The template pLB1080 was used to create plasmids with C-terminal Ser/Thr phosphomimetic substitutions (Table 2; T1396D, S1398D, and S1401D (3-mimetic); 3-mimetic + S1429D, S1431D, S1432D, T1434E, and S1435D (8-mimetic); and 8-mimetic + S1442D (9-mimetic)). LB1080 was also used to create plasmids with C-terminal Ser/Thr to Ala substitutions: T1396A, S1398A, and S1401A (3 S/T>A); 3 S/T>A + S1413A, T1417A, S1423G, S1426G, S1429A, S1431A, S1432A, S1435A, S1438A, and S1442A (13 S/T>A/G); 13 S/T>A/G + S1116A and A1121A (13 S/T>A/G + N). Each construct in Table 2 encodes GFP at the N terminus, and the locations of the C-terminal substitutions are shown in Fig. 6. All primers were from Integrated DNA Technologies (Coralville, IA). Sequences of all mutated regions in each construct were verified, and several constructs were sequenced in their entirety.
TABLE 2.
Summary of mutant constructs used in this study
V, vesicles; ap, apical; ND, not determined; *, increased presence of vesicles.
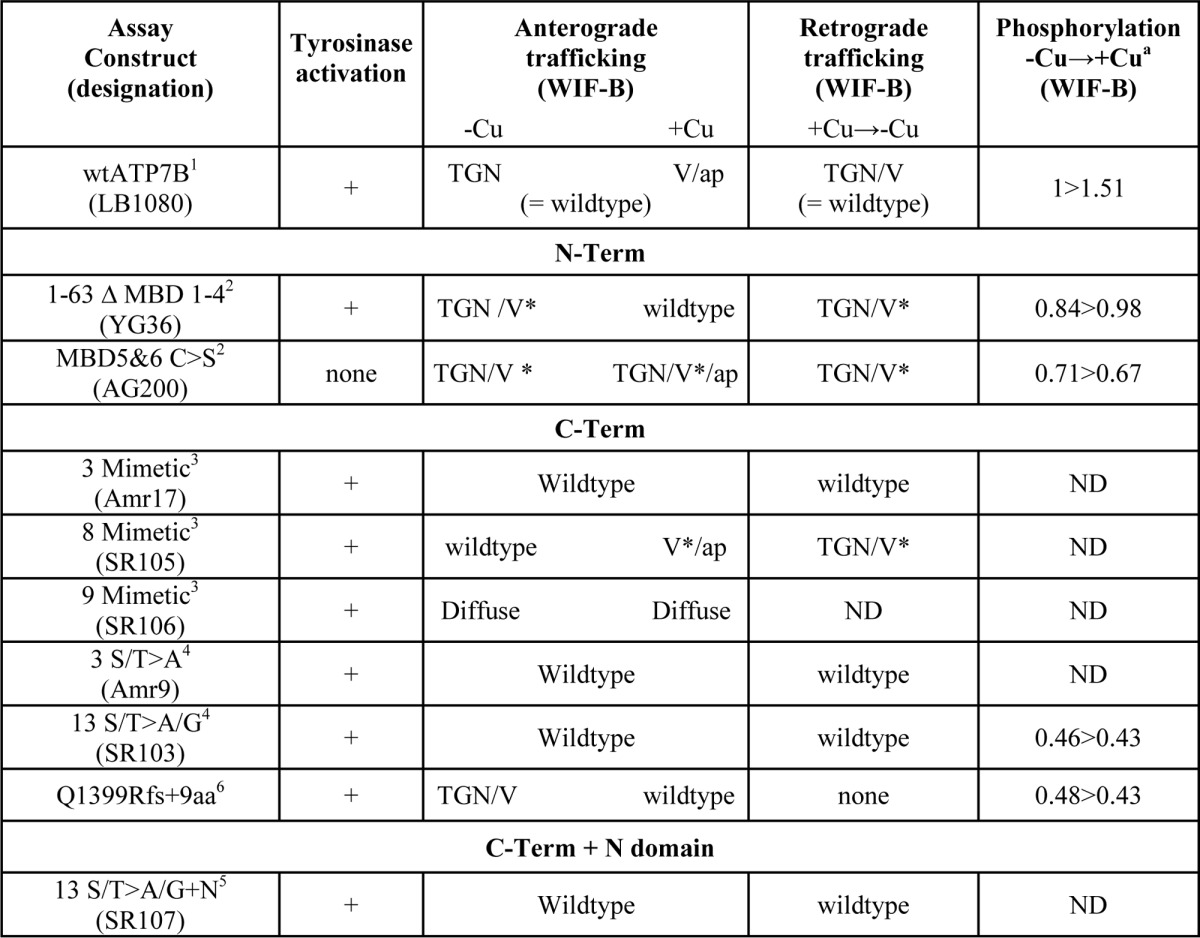
a Specific activities are all relative to WTATP7B in chelator (−Cu); changes in specific activities in the presence of copper (+Cu) are shown.
FIGURE 6.
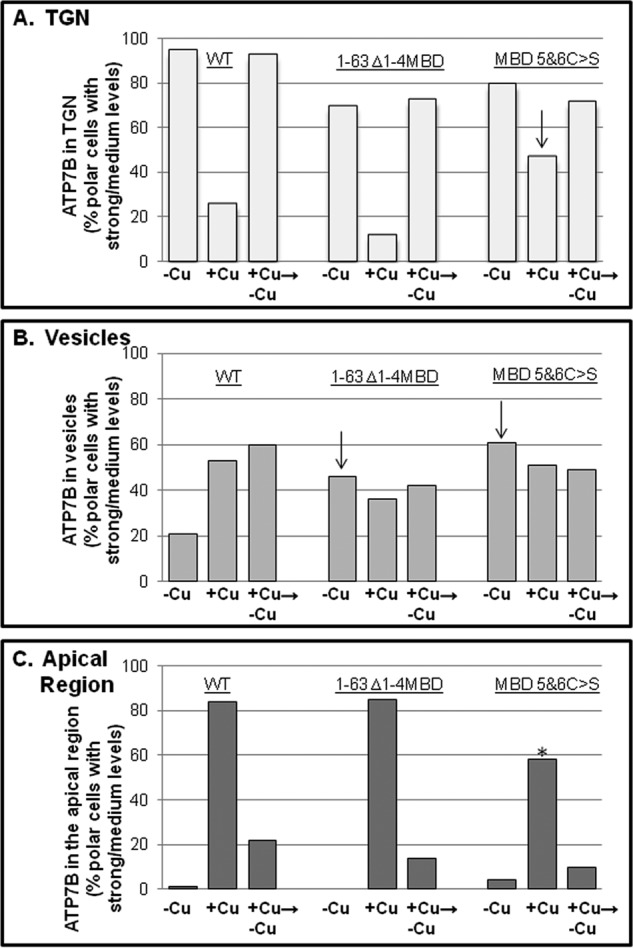
N-term metal binding domain mutant trafficking phenotypes. GFP-WTATP7B (from three independent experiments) and the two N-terminal mutants (from two independent experiments performed in parallel) shown in Fig. 5 were expressed and evaluated for copper-directed trafficking. Wide field images of 50–100 cells for each treatment were evaluated, and graphs illustrate the percentage of cells with a medium/strong presence in TGN (A), vesicles (B), or apical region (C) under the three copper conditions. Totals exceed 100% because some cells showed a mixed phenotype. Arrows indicate where and in which copper condition the mutants differ from WT. The 1–63 Δ1–4MBD C>S mutant has a smaller fraction of cells with a strong/medium level at the apical region (*).
All constructs were packaged into adenoviruses and purified as described (25). To verify that packaged viruses encoded the expected mutation and were not cross-contaminated, adenoviral DNA was purified from infected 293A cell pellets, PCR-amplified, and sequenced as described (5). All sequencing was performed by The Johns Hopkins University DNA Sequencing Facility.
Cell Culture and Adenoviral Infection
Two immortalized derivatives of SV40-transformed Menkes-null fibroblasts, designated YS and YST, were cultured as described previously (4, 23). For protein expression and immunoprecipitation studies, YS cells were plated in 10-cm tissue culture dishes with or without six glass coverslips (22 × 22 mm) at densities of 1.5 × 106 and 7.5 × 105 for infection 2 or 3 days later, respectively.
WIF-B cells were plated in 10-cm tissue culture dishes with or without six glass coverslips (22 × 22 mm) at a density of 5 × 105, cultured as described (26), and used ∼9–11 days later when maximal polarity had been achieved. YS and WIF-B cells on coverslips were infected as described previously (5) except that titers were further reduced so that the predominant GFP signal was detected with antibody (see below). YS and WIF-B cells in dishes were infected with ∼750 and 500–700 virus particles/cell, respectively.
Characterization and Copper-responsive Trafficking Assays of GFP-ATP7B Mutants (5)
Each ATP7B construct was tested for its copper transport activity in YST cells co-transfected with the construct and apotyrosinase, and then assayed 12–16 h later for tyrosinase activity as described previously (4, 23). Briefly, for YS cells, 16–24 h after infection, GFP-ATP7B was staged in the TGN by culturing cells in basal medium containing 10–25 μm TTM, 90 μg/ml cycloheximide, and 10 mm HEPES, pH 7.5 for 3–4 h. To promote trafficking out of the TGN, YS cells were cultured in basal medium for 16–20 h and then in basal medium containing 100–200 μm CuCl2, 90 μg/ml cycloheximide, and 10 mm HEPES, pH 7.5 for 3–4 h. For WIF-B cells, ATP7B and GFP-ATP7B were maintained in the TGN with 10 μm BCS for 16–20 h, and then fresh basal medium with 10 μm BCS, 50 μg/ml cycloheximide, and 10 mm HEPES, pH 7.5 was added for 1.5 h. To promote trafficking out of the TGN, WIF-B cells were cultured in basal medium containing 10 μm BCS for 16–20 h, rinsed twice in basal medium, and then changed to basal medium containing 10–20 μm CuCl2, 50 μg/ml cycloheximide, and 10 mm HEPES, pH 7.5 for 1.5 h. Retrograde assays were performed in parallel to anterograde trafficking assays as follows. Infected WIF-B cells were cultured in medium with 10 μm BCS for 16–24 h, rinsed twice with 10 μm CuCl2, and then incubated in medium with 10 μm CuCl2 for 1 h. The same cells were rinsed twice in 10 μm BCS and cultured for 2 h followed by fresh medium with 10 μm BCS + 50 μg/ml cycloheximide for an additional hour. All coverslips were then subsequently fixed and processed as described below (26).
Indirect Immunofluorescence
Primary and secondary antibodies used for microscopy were as described previously (5). To detect low levels of GFP-ATP7B, chicken anti-GFP (Abcam) and goat anti-chicken secondary antibody conjugated to Alexa Fluor 488 from Invitrogen were used. Labeled cells were analyzed using a 63× PLAN-APO, 1.4 numerical aperture oil immersion objective on a Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss, Germany). Images of 10–25 polar cells were acquired using Volocity software (PerkinElmer Life Sciences). Images of 8–20 polar cells were also acquired using a 100× PLAN-APO, 1.4 numerical aperture oil immersion objective on an LSM 510 META confocal microscope (Carl Zeiss) or a 63× objective on an LSM 700 confocal microscope (Carl Zeiss). Z-section imaging was done at 0.47-μm intervals with an overlap of 0.08 μm. Zen 2012 software was used for image capture, processing, and analysis. We focused on cells that expressed low levels of exogenous protein and assessed the distribution of the GFP protein relative to the organelle marker Golgin 97 (for YS cells) or TGN38 (for WIF-B cells) and either the apical cyst (26) or aminopeptidase N. Experiments were performed two or more times.
Evaluation of Copper-directed Trafficking
To assess the trafficking phenotypes of the mutants shown in Figs. 5 and 10, WIF-B cells expressing GFP-WTATP7B or GFP mutants were fixed and labeled with antibodies to GFP, TGN38, or the apical marker aminopeptidase N. Polar cells expressing low to moderate levels of GFP-ATP7B were scored for the presence of the protein in vesicles, the TGN, or apical region. The scores given were: not present (0), weak (+), “medium” (++), or “strong” (+++). The number of cells with medium to strong presence in the TGN, vesicles, or apical region was tallied. For each mutant, 50–100 ATP7B-expressing polar cells were evaluated from two independent experiments for each condition.
FIGURE 5.
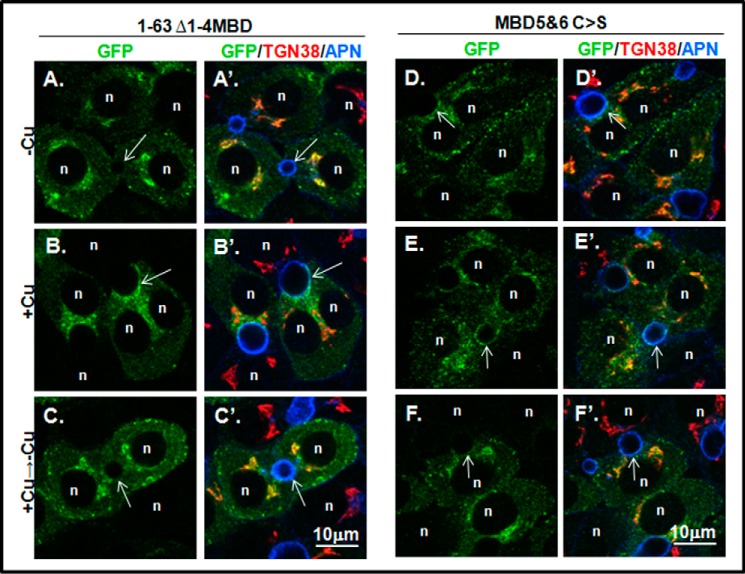
Deletion/mutation of N-terminal metal binding domains does not disrupt copper-responsive appearance at the apical domain. A–F′, WIF-B cells expressing the GFP-ATP7B constructs shown in A were fixed, stained, and imaged as in Fig. 3. Images on the left (B–E) show the GFP channel (green); images on the right (B′–E′) show overlays with the TGN (red) and apical (blue) markers. Cells were incubated overnight in 10 μm BCS (−Cu; top row) or BCS followed by 10–20 μm CuCl2 for 1–1.5 h (+Cu; middle row); some were then washed and treated with 10 μm BCS to initiate retrograde trafficking (+Cu → −Cu; bottom row). n, nucleus; arrows, apical surface; APN, aminopeptidase N.
FIGURE 10.
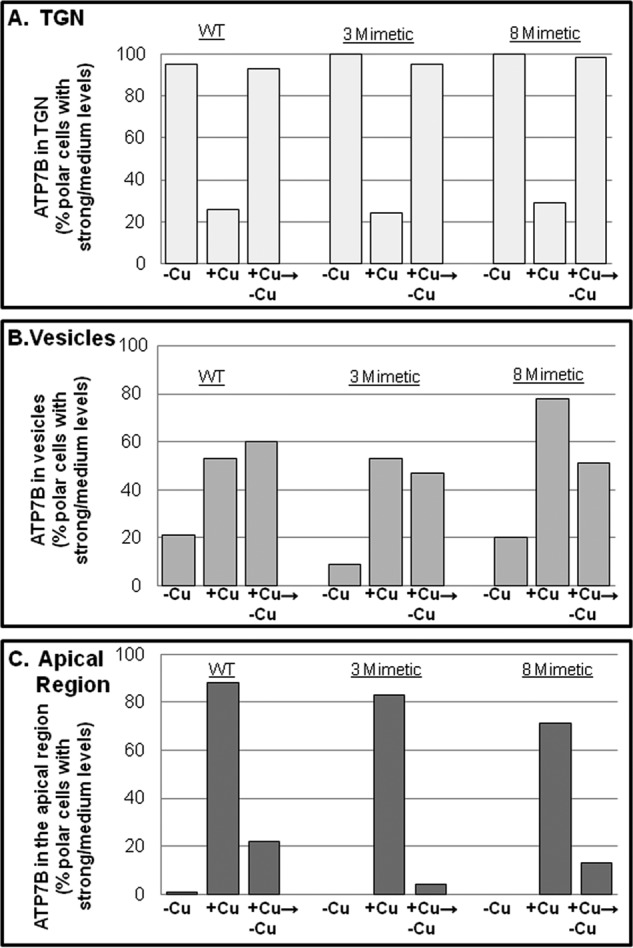
C-term phosphomimetic mutant trafficking phenotypes. GFP-WTATP7B (from three independent experiments; also shown in Fig. 5) and the two C-terminal mutants (from two independent experiments performed in parallel) shown in Fig. 9 were expressed and evaluated for copper-directed trafficking. Wide field images of 50–100 cells for each treatment were evaluated, and graphs illustrate the percentage of cells with a medium/strong presence in TGN (A), vesicles (B), or apical region (C) under the three copper conditions. Totals exceed 100% because some cells showed a mixed phenotype. The 8-mimetic shows somewhat more vesicles in the +copper (+Cu) condition than either the 3-mimetic or WT.
ATP7B Staging for Biochemical Assays
Endogenous or exogenous ATP7B was staged by manipulating copper levels in cells grown in 10-cm dishes as described above. Prior to cell lysis and ATP7B purification, localization of GFP-ATP7B was examined by removing a coverslip from each dish, fixing the cells, and evaluating them by fluorescence microscopy.
Metabolic Labeling
To correlate the subcellular distribution of ATP7B with phosphorylation of specific sites and the total level of phosphorylation, copper levels were also manipulated as described above except that phosphate-free DMEM containing 5% dialyzed fetal bovine serum with 10 mm HEPES, pH 7.5 (labeling medium) was substituted for basal medium. Incubations for YS cells were as follows. Phosphate starvation was for 2 h with or without TTM in labeling medium. Radiolabeling was for 2 h in labeling medium with TTM or copper (100–200 μm CuCl2), ∼0.1–0.3 mCi/ml 32P (PerkinElmer Life Sciences), and 90 μg/ml cycloheximide for an additional 2 h. The medium was removed, cells were rinsed in Hanks' balanced salt solution containing 0.1% bovine serum albumin, and then the “chase” was initiated by addition of labeling medium with cycloheximide and either TTM or copper for 1.5 h. Chelator and copper treatments in labeling medium were for 5.5 and 3.5 h, respectively. Incubations for WIF-B cells were as follows. Phosphate starvation was for 2 h with BCS in labeling medium. Radiolabeling was for 2 h in labeling medium with BCS, 50 μg/ml cycloheximide, and ∼0.1–0.3 mCi/ml 32P for an additional 2 h. The medium was removed, cells were rinsed in Hanks' balanced salt solution containing 0.1% bovine serum albumin, and then the chase was initiated by addition of labeling medium with either BCS or copper and cycloheximide for 1.5h. Chelator and copper treatments were for 5.5 and 1.5 h, respectively. Prior to cell lysis and ATP7B purification, localization of GFP-ATP7B was examined by removing a coverslip from each dish, fixing the cells, and evaluating them by fluorescence microscopy. The cells in dishes were rinsed twice in ice-cold PBS, and then 500 μl of ice-cold lysis buffer B (0.5% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, and HALT protease and phosphatase inhibitors (Thermo Scientific, Rockford, IL)) was added.
Immunoprecipitation of Endogenous ATP7B and GFP-ATP7B
All steps were performed on ice. YS and WIF-B cells (∼107/dish and 0.5–0. 8 × 107/dish, respectively) were scraped into 500 μl of lysis buffer B. Cell extracts were triturated with a 1-ml syringe and 28-gauge needle (four times) and incubated for a total of 30 min, and then insoluble material was sedimented at 16,000 × g for 10 min. The supernatants were diluted in dilution buffer B (150 mm NaCl, 10 mm Tris-HCl, pH7.5, 0.5 mm EDTA, and phosphatase inhibitors to yield 0.25% Nonidet P-40) and incubated with control agarose resin (Thermo Scientific, Rockford, IL) at 4 °C for 0.5 h on a rotating wheel to remove nonspecific radiolabeled material. The beads were sedimented, and the supernatant was added to either anti-ATP7B (Ab 3985 (4)) cross-linked to Protein A gel (Seize X Immunoprecipitation kit, Pierce) or GFP-Trap-A (ChromoTek GmbH, Martinsried, Germany) and incubated at 4 °C for 2 h on a rotating wheel. The beads were sedimented and rinsed twice with 0.25% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl, pH 7.5, and 0.5 mm EDTA and twice in dilution buffer B. Endogenous ATP7B or GFP-ATP7B was eluted with lithium dodecyl sulfate sample buffer (Invitrogen) plus 0.1 m dithiothreitol by boiling for 5 min; the released proteins were then separated by SDS-PAGE (4–12% gradient NuPAGE or Bolt gels, Invitrogen) and transferred to nitrocellulose. For analysis of kinase-mediated phosphorylation, the radiolabeled [32P]ATP7B was measured by exposure of the dried membrane to a phosphor screen (BAS III, Fujifilm, Tokyo, Japan), which was imaged using the PharosFX scanner (Bio-Rad) and analyzed using Quantity One software (Bio-Rad). To determine specific activity, the nitrocellulose membrane was incubated with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE) and then rabbit anti-GFP followed by anti-rabbit IRDye 800cw as described (4). The radioactive signal was normalized to the amount of protein in each sample; levels of 32P incorporation in the copper-treated cells were normalized to that in the chelator-treated cells.
Phosphoprotein Stain
For each treatment, GFP-ATP7B was purified from ∼5 dishes of fibroblasts (see “Large Scale GFP-ATP7B Purification”), separated by SDS-PAGE, then fixed, washed, and stained with ProQ Diamond as recommended by the supplier (Invitrogen/Molecular Probes, Eugene OR). ProQ Diamond is a non-covalent fluorescent dye that is reported to detect phosphoserine-, phosphothreonine-, and phosphotyrosine-containing proteins displayed on SDS-PAGE gels. After destaining, the gel bands were imaged with the PharosFX Plus laser-based gel scanner using a 532 nm excitation laser and 605-nm bandpass emission filter and analyzed using Quantity One software. To determine total protein, the same gels were then stained with SYPRO Ruby fluorescent protein stain (Invitrogen/Molecular Probes) according to the manufacturer's instructions. This stain replaces the ProQ Diamond stain. Protein bands were also imaged with the PharosFX Plus laser-based gel scanner using a 488 nm excitation laser and a 605-nm bandpass emission filter and then analyzed using Quantity One software.
λ-Phosphatase Treatment
GFP-ATP7B immobilized on GFP-Trap-A was placed in protein metallophosphatase buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT, and 0.01% Brij 35); 30 μl of protein metallophosphatase buffer + 1 mm MnCl2 were added to duplicate tubes. For phosphatase treatment, 2 μl of λ-protein phosphatase (New England Biolabs, Ipswich, MA) was added to one tube, and buffer only was added to a second tube. To monitor the λ-protein phosphatase, 2 μl was added to protein metallophosphatase buffer + 1 mm MnCl2. All samples were incubated for 1 h at 30 °C. The reaction was stopped with 4× lithium dodecyl sulfate sample buffer and 0.1 m DTT, and the samples were boiled for 5 min. The polypeptides were separated by SDS-PAGE, and the gels were stained with the ProQ Diamond Stain and SYPRO Ruby stain.
Large Scale GFP-ATP7B Purification
Pilot studies revealed that GFP-ATP7B was soluble in a variety of detergents including 2% CHAPS and 0.5% Nonidet P-40. Initially, lysis buffer A (2% CHAPS, 50 mm NaCl, and 10 mm HEPES, pH 7.4 with phosphatase inhibitors) was used for to purify GFP-ATP7B from fibroblasts. Because we obtained variable yields and impurities with lysis buffer A, we used lysis buffer B for later experiments. For purifications, 400 μl of lysis buffer was added to each of 5–10 100-mm dishes, and the cell lysates were scraped, triturated first with a 1-ml syringe and 25-gauge needle (four times) and then with a 28-gauge needle (four times), pooled, and incubated for a total of 30 min. The insoluble material in ∼1-ml aliquots was sedimented (35,000 rpm for 10 min in a TLA-55 rotor (Beckman Coulter, Brea, CA)). The supernatants were pooled and diluted with either dilution buffer A (50 mm NaCl and 10 mm HEPES, pH 7.4 to yield 1% CHAPS) or dilution buffer B (150 mm NaCl, 10 mm Tris-HCl, pH 7.5, and 0.5 mm EDTA to yield 0.25% Nonidet P-40) and then added to GFP-Trap-A, which was used essentially as recommended by the manufacturer. The samples were incubated at 4 °C for 2 h on a rotating wheel. The beads were sedimented and rinsed twice with 0.25% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl, pH 7.5, and 0.5 mm EDTA and twice in dilution buffer B. GFP-ATP7B was eluted twice with 100 μl of 0.5% SDS by boiling for 5 min, and then the pooled SDS-eluted sample was lyophilized. The lyophilized proteins were resuspended in 1× lithium dodecyl sulfate sample buffer and separated by SDS-PAGE (4–12% gradient NuPAGE or Bolt gels), the gels were stained with Simply Blue (Bio-Rad), and the GFP-ATP7B band was cut from the gel and stored at −80 °C for MS analysis.
LTQ Orbitrap Velos Mass Spectrometer Analysis
The excised gel bands were destained, reduced in 5 mm DTT at 60 °C for 45 min, alkylated with 20 mm iodoacetamide at room temperature for 20 min in the dark, and proteolyzed with 10 ng/μl trypsin (Promega, Madison, WI) overnight at 37 °C or chymotrypsin (Roche Applied Science) as described previously (27). Dry peptides were resuspended in 12 μl of 0.1% formic acid; 8 μl of this was loaded on a 75-μm × 2.5-cm trap packed with Magic AQ C18 (5-μm, 100-Å material) and then fractionated by reverse-phase HPLC using a 5–45% acetonitrile and 0.1% formic acid gradient from 0 to 45 min and then up to 90% at 48 min with a flow rate of 300 nl/min. LC-MS/MS analysis was performed on a 2D nanoLC system (Eksigent, Dublin, CA) coupled on line to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Data were acquired in data-dependent mode in which a full MS scan was followed by MS/MS scans of collision-induced dissociation of the 10 most intense ions. Precursor and fragment ions were analyzed at 30,000 and 7,500 resolution, respectively.
Peptide Identifications and Analysis
Peptide sequences were identified using Proteome Discoverer 1.3 (Thermo Scientific, Waltham, MA) software configured with Mascot to search the mass spectrometry data against NCBI Reference Sequences 40 or version 2012. The following criteria were used in the peptide searches: human species; variable modifications, Met oxidation and Asp or Glu deamidation; and fixed modification, Cys carbamidomethylation; additional searches, Thr and Tyr phosphorylation or N-acetylhexosamine on Ser and Thr. Mass tolerances on precursor and fragment masses were 15 ppm and 0.03 Da, respectively, and a 1% false discovery rate cutoff was used to filter the peptide search results. Mascot search results were imported into Scaffold (version 3.6.5) to validate the fragmentation spectrum-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (28). Protein probabilities were assigned by the Protein Prophet algorithm (29). Finally, peptide data were researched using PEAKS (version 7) as additional validation of phosphosites and to obtain the annotated mass spectra fragmentation images reported in the supplemental data (30).
RESULTS
Copper-dependent Phosphorylation Is Phosphatase-sensitive and Can Occur in the TGN
We first confirmed that the behavior of GFP-WTATP7B in the human skin fibroblast YS cell line (23) corresponds with previous studies that demonstrated an association between copper-dependent trafficking and kinase-mediated hyperphosphorylation of ATP7B (9–13). We used recombinant adenovirus to express GFP-WTATP7B in the YS cell line and then exposed the cells to either a cell-permeant copper chelator (−Cu), which promoted GFP-WTATP7B retention in the TGN (Fig. 1A, left upper panel), or copper (+Cu), which shifted the GFP-WTATP7B localization to vesicles and the plasma membrane of many cells (Fig. 1A, left lower panel). To test for GFP-WTATP7B hyperphosphorylation in the presence of copper, we metabolically labeled YS cells with 32P in the presence of cycloheximide, a protein synthesis inhibitor. Cycloheximide was included to ensure that we were studying immunoprecipitated GFP-WTATP7B that had been phosphorylated post-translationally in the TGN or in a post-TGN compartment.
FIGURE 1.
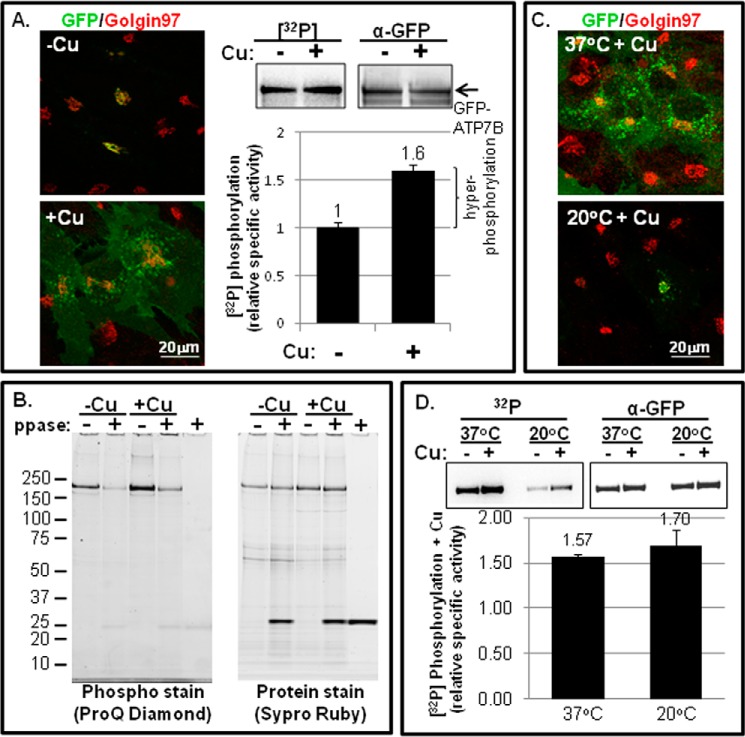
Copper-stimulated ATP7B trafficking and Ser/Thr phosphorylation in fibroblasts. A, YS cells were incubated overnight in 25 μm TTM (−Cu) or overnight in basal medium followed by 1.5 h in 100–200 mm CuCl2 (+Cu). Cells were then fixed and labeled with antibodies to GFP (green) and Golgin 97 (left panels; single confocal images are shown), or they were metabolically labeled with [32P]orthophosphate followed by ATP7B immunoprecipitation and gel electrophoresis (right panels; representative autoradiogram of 32P incorporation and corresponding immunoblot of protein are shown). The bar graph (bottom right) shows the relative specific activities ±copper. The bracket indicates the amount of hyperphosphorylated ATP7B in the presence of copper relative to the basal phosphorylation observed in the presence of TTM (n = 13). B, phosphorylated GFP-WTATP7B was isolated from YS cells exposed to either 25 μm TTM (−Cu) or 100–200 μm copper (+Cu) and then treated with phosphatase or vehicle (ppase; + or −). Following immunoprecipitation and electrophoresis, phosphorylated GFP-ATP7B was detected using ProQ Diamond (left gel), and then the gel was reprobed for total protein using SYPRO Ruby stain (right gel). C, cells equilibrated at 37 (top panel) or 20 °C (bottom panel) were exposed to copper for 1.5 h, then fixed, and stained as described in A. D, YS cells were equilibrated at 37 or 20 °C and then metabolically labeled with [32P]orthophosphate followed by ATP7B immunoprecipitation as described in A. Gels on the left are autoradiograms; gels on the right are immunoblots for total ATP7B protein. The bar graph shows the specific activity (+Cu) normalized to chelator-treated cells (−Cu). The mean of three independent experiments is shown; error bars represent S.E.
Our results indicate that GFP-WTATP7B is basally phosphorylated in the presence of chelator and that copper enhances phosphorylation by ∼50% (Fig. 1A, right). We used the non-radioactive ProQ Diamond staining technique to establish that phospho-ATP7B is sensitive to λ-phosphatase, indicating that Ser, Thr, and/or Tyr are the modified residues (Fig. 1B), consistent with previous work (9).
To distinguish whether hyperphosphorylation was dependent upon elevated copper per se or rather was a downstream consequence of its copper-dependent redistribution into vesicles, we incubated GFP-WTATP7B-infected YS cells in basal medium at either 37 or 20 °C before adding copper; the latter temperature blocks exit from the TGN (31, 32). Fig. 1C shows that GFP-WTATP7B expressed in cells maintained at 37 °C and treated with copper showed normal trafficking, whereas in cells maintained at 20 °C plus copper, GFP-WTATP7B remained in the TGN. We repeated the metabolic labeling in conjunction with immunoprecipitation using cells maintained at both temperatures. Although the specific activity of phospho-GFP-WTATP7B was reduced at 20 °C as would be expected, when phosphorylation amounts were normalized to the their respective basal values, the magnitude of increased phosphorylation due to copper remained similar at both temperatures; i.e. copper enhanced GFP-WTATP7B phosphorylation by ∼50% (Fig. 1D). Thus, the cellular machinery responsible for copper-dependent phosphorylation can engage ATP7B even when ATP7B localization is restricted to the TGN.
Four Unique Phosphoresidues, Four Diphosphopeptides, and One Triphosphopeptide Were Phosphorylated upon Exposure of ATP7B to Copper
We used MS to catalog all phosphorylated residues on ATP7B and to determine which were phosphorylated independently of copper and which were copper-dependent. Large scale cultures of adenovirally infected YS cells were grown to obtain ample quantities of GFP-WTATP7B. Cultures were treated such that ATP7B was staged either in the TGN with a cell-permeant copper chelator or in vesicles plus plasma membrane with copper. Following immunoprecipitation, efficient recovery of GFP-WTATP7B was demonstrated by immunoblotting and protein staining (Fig. 2, A and B). Recovered GFP-WTATP7B was digested with trypsin, and the peptides were analyzed by MS (see “Experimental Procedures” for details). Of the 179 potential cytosolic Ser/Thr residues identified by in silico amino acid analysis, a total of 24 Ser/Thr residues were phosphorylated, and 10 of those were identified previously (Table 1). The identifications are supported by the ions in the fragmentation spectra (supplemental data); a table shows the ion mass for each newly identified phosphorylated residue (supplemental data).
FIGURE 2.
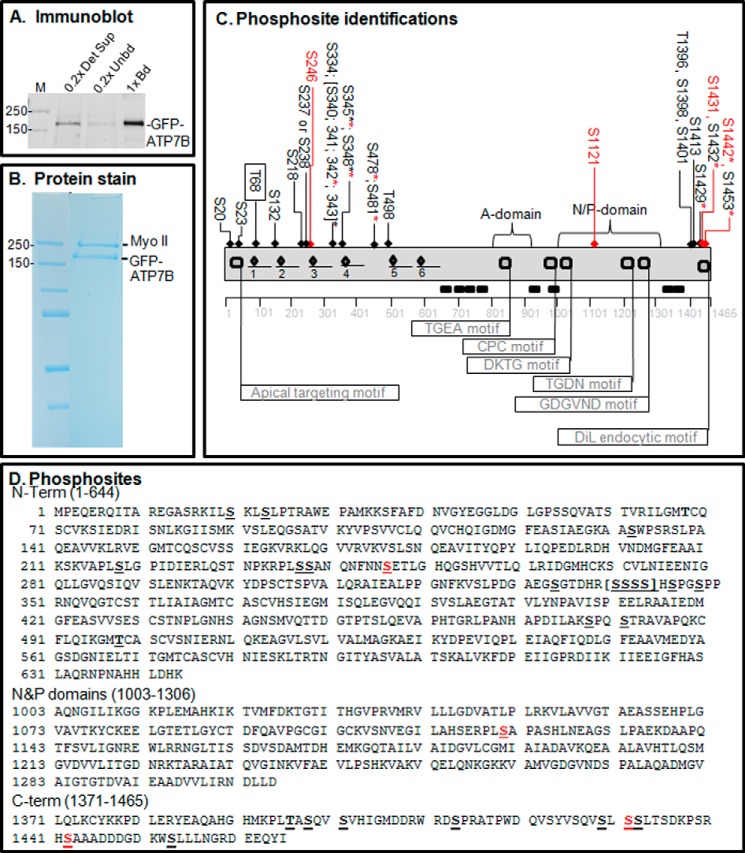
Proteomics analysis of copper-dependent and copper-independent phosphorylation. A, YS cells expressing GFP-WTATP7B were extracted in detergent, and the ATP7B was immunoprecipitated. The gel shown is an immunoblot of the starting extract (0.2× Det Sup; left), depleted supernatant (0.2× Unbd; middle) and immunoprecipitated ATP7B protein (1× bound (Bd); right); molecular mass markers are indicated in kDa (far left). B, GFP-WTATP7B was immunoprecipitated, separated by SDS-PAGE, and then stained in-gel with Simply Blue. Gel bands of five independent samples for each treatment were used for MS analysis, and three and four independent MS analyses were performed for ±copper-treated cells, respectively. C, schematic depiction of phosphosites identified by proteomics in the context of the ATP7B domain structure (flags show the locations of characterized motifs). An underline shows the location of the six metal binding domains, and diamonds indicate the CXXC motifs that bind copper. All of the sites identified with >95% confidence are indicated across the top of the diagram; those highlighted in red were found only in the high copper condition. The boxed site near the N terminus was found only in the chelation condition. Asterisks indicate residues found as part of a diphosphopeptide or triphosphopeptide. See the supplemental data for documentation of the mass spectroscopy data; PEAKS analysis showed that ATP7B sequence coverage was 84%. D, phosphosite identifications mapped onto the ATP7B primary sequence. Red Ser residues were hyperphosphorylated in the presence of copper. DiL, dileucine.
Most of the modified residues were located on the cytoplasmic N and C termini, and most were present under both conditions (Fig. 2, C and D, and Table 1). The modified sites fell into three general categories: 1) present only in chelator-treated cells (Fig. 2C, Thr-68 only, boxed); 2) present in both chelator- and copper-treated cells (19 Ser/Thr with black arrows); and 3) present only in copper-treated cells (Fig. 2C, the four Ser/Thr with red arrows). This approach revealed that Ser-246, Ser-1121, Ser-1431, and Ser-1442 were copper-responsive sites. Additionally, we found two N-term and two C-term diphosphopeptides as well as one triphosphopeptide present only in copper-treated cells (Table 1 and supplemental data). Thus, in addition to copper-dependent N-terminal phosphorylation previously validated (13), our phosphoproteomics study shows that ATP7B is also hyperphosphorylated on C-terminal Ser/Thr in response to copper.
Copper-dependent ATP7B Phosphorylation in Hepatic Cells
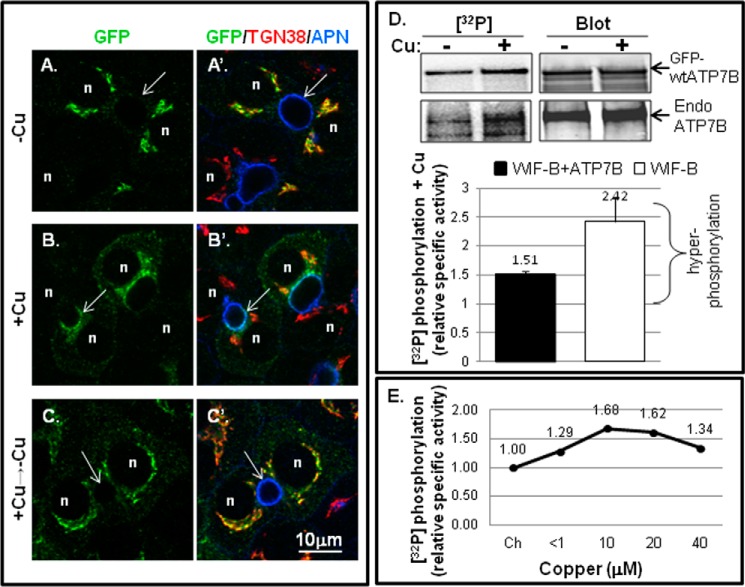
To verify that basal and copper-dependent hyperphosphorylation occurred in a more physiologically relevant cell type, we turned to the hepatic WIF-B cell line (6, 26). GFP-WTATP7B was expressed in WIF-B cells and then exposed to chelator, which promoted its retention in the TGN (Fig. 3, A and A′). Copper treatment (10 μm CuCl2 for 1 h) promoted ATP7B exit from the TGN and its appearance in vesicles and the apical plasma membrane (Fig. 3, B and B′). Upon subsequent removal of copper by chelation, GFP-WTATP7B returned to the TGN (Fig. 3, C and C′), demonstrating an itinerary comparable with that of endogenous ATP7B in WIF-B (6). We next tested whether GFP-WTATP7B in WIF-B cells was hyperphosphorylated to a degree similar to that in YS cells after exposure to copper (Fig. 3D). Metabolic labeling indicated that GFP-WTATP7B was basally phosphorylated when copper was chelated and was enhanced by ∼50% after addition of copper (Fig. 3D). In Fig. 3E, we show the dose-response of copper on ATP7B phosphorylation in WIF-B cells expressing GFP-WTATP7B. Copper-dependent hyperphosphorylation of ATP7B occurred at 1 μm CuCl2, and maximal hyperphosphorylation was produced at 10–20 μm copper (Fig. 3E). Copper treatment for 1.5 h enhanced total phosphorylation of endogenous ATP7B by ∼2-fold.
FIGURE 3.
Copper-stimulated ATP7B trafficking and Ser/Thr phosphorylation in WIF-B cells. WIF-B cells were incubated overnight in 10 μm BCS (−Cu) to stage endogenous ATP7B or GFP-WTATP7B in the TGN. For the +copper (+Cu) condition, cells were incubated in 10–20 μm CuCl2 for 1–1.5 h. A–C′, WIF-B cells infected with GFP-WTATP7B were fixed; stained with anti-GFP (green), TGN38 (red), and aminopeptidase N (APN) (blue); and imaged by confocal microscopy. Single plane images show copper-dependent trafficking. n, nucleus; arrows, apical surface. D, representative autoradiogram of 32P incorporation and corresponding immunoblot of immunoprecipitated endogenous (GFP-wtATP7B; top gel bands) and (Endo ATP7B; bottom gel bands) from metabolically labeled WIF-B cells ±copper (top). The bar graph (bottom) shows the mean relative specific activity normalized to chelator-treated cells. n = 7 for GFP-ATP7B (black bar; left); n = 3 for endogenous ATP7B (white bar; right). Error bars represent S.E. E, dose response of hyperphosphorylation to varying copper concentrations. <1, the copper level in basal medium. The relative specific activity of 32P-labeled GFP-WTATP7B in WIF-B cells is shown normalized to the value obtained in chelator (Ch).
Having confirmed that basal hyperphosphorylation and copper-dependent hyperphosphorylation of GFP-ATP7B were similar in YS and hepatic WIF-B cells and identified the N and C termini as sites of GFP-ATP7B basal and hyperphosphorylation in YS cells, we next used deletion and/or mutagenesis of selected phosphorylation sites in conjunction with GFP-ATP7B expression in hepatic cells to determine whether hyperphosphorylation was essential for its copper-dependent trafficking.
The N-term Metal Binding Domains Are Required for Copper-stimulated Phosphorylation but Not Basal Phosphorylation
The extreme N-terminal part of ATP7B consists of a motif involved in apical targeting (amino acids 1–63) (4) followed by six MBD repeats, each containing a CXXC motif that binds copper (Fig. 2C) (33). We previously described the 1–63 Δ1–4MBD mutant, which lacks MBD1–4 but exhibits near normal trafficking when treated with 200 μm copper (23). Interestingly, the 1–63 Δ1–4MBD mutant lacks nine of the 12 N-term phosphoresidues found through MS (Fig. 4A), which raises the possibility that phosphorylation of this mutant might be diminished. To eliminate all copper binding of the N-term, we generated recombinant adenovirus expressing 1–63 Δ1–4MBD in which the cysteines in the two remaining MBDs (5 and 6) were changed to Ser, a zero volume substitution that would likely preserve the ferredoxin-like folds of MBDs 5 and 6 (designated 1–63 Δ1–4MBD C>S; Fig. 4A) (34). This mutant failed to supply copper to metallate and thus activate tyrosinase, indicating the requirement of MBDs 5 and 6 for copper transport activity (Fig. 4A).
FIGURE 4.
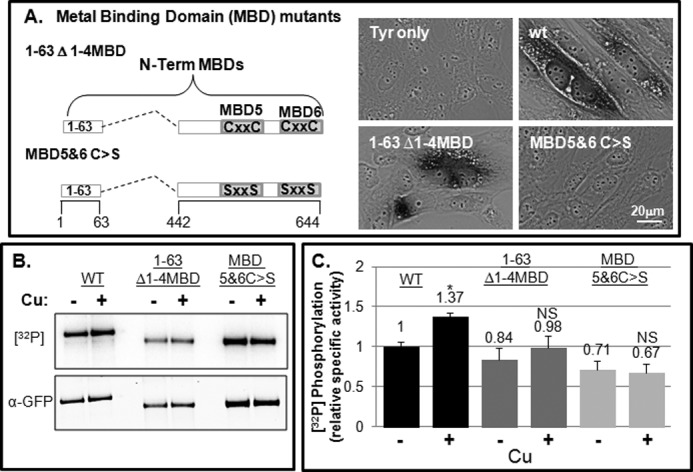
N-term MBD mutants exhibit basal phosphorylation and not copper-dependent hyperphosphorylation. A, schematic showing N-terminal MBD mutants (right). YST cells were co-transfected with pTyrosinase alone (Tyr) or with various ATP7B constructs as indicated. The black reaction product indicates the presence of copper-dependent tyrosinase activity (left). B and C, GFP-WTATP7B and the two N-terminal mutants were expressed and evaluated for phosphorylation as described under “Experimental Procedures.” B, autoradiogram and blot showing 32P incorporation and protein levels of the three constructs. C, bar graph showing the mean relative specific phosphorylation of GFP-WTATP7B, 1–63 Δ1–4MBD, and 1–63 Δ1–4MBD C>S normalized to the −copper (−Cu) condition. n = 4. Error bars, mean ± S.E.; *, p < 0.05 using two-tailed t test; NS, not significant.
We next used metabolic labeling to determine whether deletion of the N-term phosphorylation sites altered either basal or copper-stimulated phosphorylation of ATP7B. Both the 1–63 Δ1–4MBD and 1–63 Δ1–4MBD C>S mutants were expressed in WIF-B cells and assayed. Despite the fact that nine of the 12 N-terminal phosphorylation sites we identified by MS were deleted in both mutants (Fig. 2), these mutant proteins exhibited only a modest reduction in basal phosphorylation as measured by [32P]orthophosphate incorporation (∼84 and ∼71% of WT, respectively) (Fig. 4, B and C). This result indicates that other regions of the protein must be basally phosphorylated. Following the addition of copper, the 1–63 Δ1–4MBD mutant showed a modest but not significant increase in [32P]orthophosphate incorporation, whereas the Δ1–4MBD C>S mutant exhibited no copper-stimulated hyperphosphorylation (Fig. 4C). Because serines were added to this mutant, it is possible that they are substrates for kinases in cells. However, if this were so, the relative specific activity of the 1–63 Δ1–4MBD C>S mutant would likely be greater than for 1–63 Δ1–4MBD when in fact it was lesser (Fig. 4C, right bars).
Deletion/Mutation of N-terminal Metal Binding Domains Does Not Disrupt Copper-responsive Appearance at the Apical Region
Next we tested the effect of the two N-term mutants on copper-dependent trafficking. In the presence of chelator, both the 1–63 Δ1–4MBD and the 1–63 Δ1–4MBD C>S mutants localized in the TGN and in vesicles distributed throughout the cell, indicating that TGN retention was somewhat compromised (Figs. 5, A and D, and 6, A and B). Addition of copper promoted the appearance of the 1–63 Δ1–4MBD C>S mutant at the apical region (Figs. 5, B and E, and 6C). By counting the number of polar cells that showed apical targeting of each MBD mutant, we determined that both mutants could be trafficked to the apical region in a copper-dependent manner, although the 1–63 Δ1–4MBD C>S mutant trafficking was less robust (Fig. 5C). Following removal of copper by treatment with chelator for 3 h, both mutants again localized to the TGN and in vesicles, showing that apical trafficking was reversible (Figs. 5, C and F, and 6).
Compared with constructs with a complete N terminus, both mutants showed an increased presence in vesicles under the chelator condition, consistent with the idea that interdomain contacts are important for TGN retention and can be disrupted by copper (Fig. 5 and Refs. 13 and 35). However, our results clearly demonstrate that the N-term MBDs are not essential for ATP7B copper-dependent trafficking to the apical region in hepatic cells beyond their previously established importance in regulating TGN exit and retention. They also indicate that copper-sensitive delivery of ATP7B to the apical domain still occurs even when all copper binding at MBDs has been eliminated, suggesting the existence of additional copper-sensing sites in ATP7B.
Thus, although inactivation of all MBDs did not abolish copper-dependent apical delivery in hepatic cells, it drastically reduced copper-dependent phosphorylation. One interpretation of this result is that copper-stimulated phosphorylation sites are within the deleted MDB1–4 region in the N-term. Alternatively, our data are also consistent with a model in which copper binding to the N-term MBDs produces conformational changes in the ATP7B structure that in turn increase the accessibility of residues in the C terminus to kinases. We thus explored the effects of direct mutation of the C-term phosphorylation sites that we (Fig. 2) and others (11, 16, 18, 19, 36) identified by MS.
C-term Ser/Thr Are Not Required for Copper Transport Activity
We showed previously that the C terminus has a number of critical roles for ATP7B function (5). In that report, we showed that various deletions of the C terminus (Fig. 7A, first four constructs) had little effect on copper-responsive anterograde trafficking; however, the mutants were unable to return to the TGN presumably due to their lack of the C-term tri-Leu motif that interacts with cellular machinery that mediates TGN retrieval (5). Because the deletions encompassed most of the C-term Ser/Thr residues identified in our MS analysis, we used site-directed mutagenesis to create mutants with either negative charge substitutions (Asp/Glu), termed phosphomimetics, or inactivating (Ala/Gly) substitutions (Fig. 7A). We documented that each of the mutants was able to transport copper as demonstrated by the black reaction product indicating active tyrosinase (Fig. 7, B–G). Although the 9-mimetic mutant showed some activity in this qualitative assay (Fig. 7D), we found a large portion of it was retained in the endoplasmic reticulum, and thus we did not study it further.
FIGURE 7.
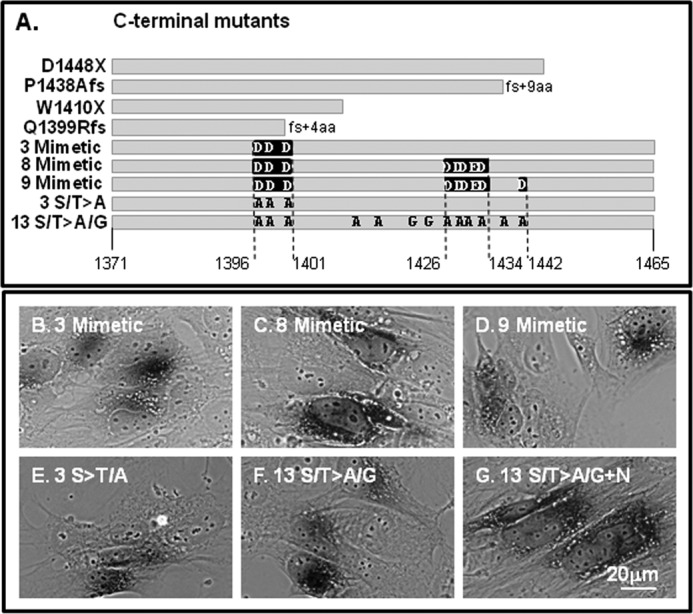
ATP7B C-terminal mutants transport copper. A, diagrams of the various C-terminal mutants used in this study. The gray bars indicate the length of the C-terminal sequence. Negative charge point mutations (phosphomimetic) are indicated by black boxes; Ala/G mutations (inactivating) are shown as black letters. B–G, positive tyrosinase reaction product was observed for each construct with point mutations. The construct shown in G shows the 13 mutant with two additional sites inactivated (S1116A/S1121A present in the N domain). aa, amino acids.
C-term Ser/Thr Are Required for Basal and Copper-stimulated Phosphorylation
To determine whether inactivation of C-term Ser/Thr is responsible for either basal or copper-stimulated phosphorylation, we studied two mutants, one with approximately two-thirds of the C terminus deleted (Q1399Rfs; Fig. 7A), and one with 13 of the 15 C-term Ser/Thr inactivated by substitution with Gly/Ala (13 S/T>A/G; Fig. 7A). The 13 S/T>A/G mutant was generated for two reasons: 1) nine of the 15 C-term Ser/Thr were found to be phosphorylated in our MS study, and 2) in silico analysis revealed that multiple Ser/Thr residues are potential recognition sites for a variety of kinases, which highlights the possibility for kinase promiscuity, which could complicate our analysis. Both mutants were expressed in WIF-B cells, radiolabeled with [32P]orthophosphate, and then immunoprecipitated. Both exhibited reduced basal phosphorylation when copper was chelated, and following the addition of copper, neither became hyperphosphorylated (Fig. 8, A and B). Both findings are consistent with the MS result (Fig. 2), which indicated that nine C-term residues are phosphorylated, two of which were found only in the presence of copper. Thus, both the N-term MBDs and C-term Ser/Thr are required for copper-stimulated phosphorylation.
FIGURE 8.
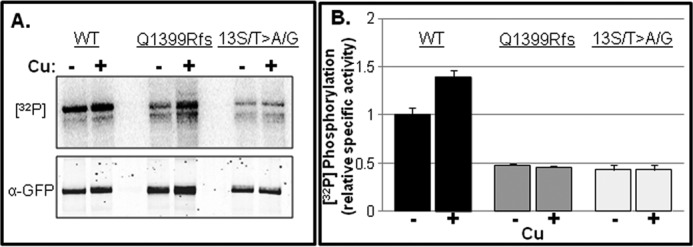
The C terminus is a major region for basal and copper-responsive hyperphosphorylation. GFP-WTATP7B and two C-term mutants, Q1399R and 13 S/T>A/G, were expressed in WIF-B cells, metabolically labeled with [32P]orthophosphate, and treated as for Fig. 3. A, representative autoradiogram showing 32P incorporation (top) and an immunoblot (bottom) of immunoprecipitated GFP proteins. B, bar graph showing the mean specific radioactivity relative to the WT −copper (−Cu) condition. Error bars, mean ± S.E. n = 3.
Negative Charge Substitutions of Eight Ser/Thr in the C terminus Did Not Affect Trafficking
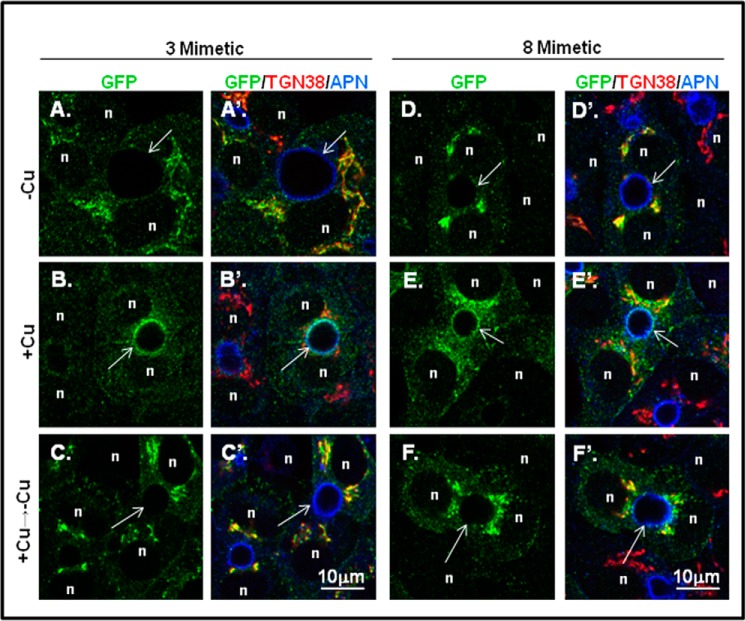
To explore the possibility that C-terminal phosphorylation alters ATP7B trafficking, we first examined the trafficking behavior of the two phosphomimetic mutants, 3-mimetic and 8-mimetic (Fig. 7A), in WIF-B cells. Both mutants showed robust co-localization with the TGN marker when copper was chelated, indicating that charge-mimicking C-term phosphorylation does not promote TGN exit (Fig. 9, A, A′, D, and D′). Upon addition of copper, both mutants exhibited normal copper-directed trafficking to the apical region (Fig. 9, B, B′, E, and E′), which was documented by counting the number of polar ATP7B-expressing cells (Fig. 10C). Following removal of copper by treatment with chelator for 3 h, both mutants again localized to the TGN and in vesicles (Figs. 9, C, C′, F, and F′, and 10). Although we observed normal copper-responsive trafficking for the mutant with eight negative charged substitutions (T1396D/S1398D/S1401D/S1429D/S11431D/S1432D/T1434E/S1435D), there was a modest increase of this mutant in vesicles in the presence of copper (Fig. 10). These results show that increased fixed negative charges on the C terminus does not alter the itinerary of ATP7B.
FIGURE 9.
Introduction of eight negative charges at putative C-term phosphorylation sites does not disrupt copper-directed trafficking. GFP-ATP7B mutants with three or eight negative charges (3- and 8-mimetic) were expressed in WIF-B cells, treated under various copper conditions, and then stained as in Fig. 3. Single confocal planes are shown. n, nucleus; arrows, apical surface; APN, aminopeptidase N.
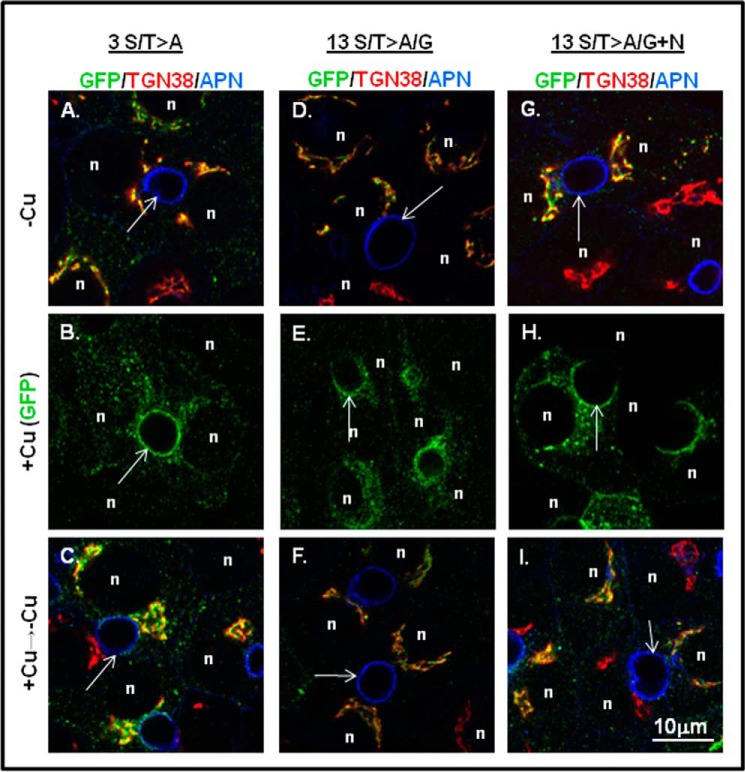
Inactivation of C-term Ser/Thr Did Not Alter GFP-ATP7B Trafficking in WIF-B Cells
To rule out the requirement of copper-stimulated hyperphosphorylation for copper-dependent trafficking of ATP7B, we examined trafficking of the 13 S/T>A/G mutant, which is not hyperphosphorylated in response to copper (Fig. 8). In the presence of chelator (−Cu), both the 3 S/T>A with T1396A, S1398A, and S1401A substitutions and the 13 S/T>A/G mutants localized in the TGN (Fig. 11, A and D). Addition of copper promoted the appearance of both mutants at the apical region in WIF-B cells (Fig. 11, B and E). Following removal of copper by treatment with chelator for 3 h, both mutants again localized to the TGN and in vesicles (Fig. 11, C and F). Thus, neither copper-stimulated hyperphosphorylation of the C terminus nor the full complement of basal phosphorylation is required for copper-responsive trafficking of ATP7B.
FIGURE 11.
Introduction of 13 inactivating residues (Ala or Gly) in the C terminus does not disrupt copper-responsive trafficking. GFP-ATP7B mutants with three (left column) or 13 (middle column) Ala/Gly inactivating mutations were expressed in WIF-B cells, treated under various copper conditions, and then stained as in Fig. 3. Single confocal planes are shown. n, nucleus; arrows, apical surface; APN, aminopeptidase N. The right column shows the 13 S/T>A/G mutant with two additional sites inactivated (S1116A/S1121A present in the N domain).
Previous studies showed that Ser-1121 in the flexible loop of the nucleotide binding domain was phosphorylated in response to copper (11, 12); our mass spectroscopy analysis confirmed this finding. Because copper has been shown to alter interdomain interactions that may be functionally relevant for trafficking (35), we used site-directed mutagenesis to inactivate Ser-1121 as well as the closely associated site Ser-1116 located in the nucleotide (N) domain (13 S/T>A/G+N). The 13 S/T>A/G+N mutant was expressed in WIF-B cells, and its copper-responsive trafficking was examined. In the presence of chelator, the S/T>A/G+N mutant showed a robust TGN localization (Fig. 11G). Addition of copper promoted its TGN exit and a robust signal at the apical plasma membrane (Fig. 11H). Following removal of copper by treatment with chelator for 3 h, the 13 S/T>A/G+N mutant again localized to the TGN and vesicles (Fig. 11I). Thus, phosphorylation of Ser-1121 in the N domain and the 13 Ser/Thr in the C terminus are not required for trafficking in hepatic cells. Taken together, our analysis definitively shows that copper, but not copper-stimulated phosphorylation, is required for ATP7B to reach the apical region where copper can be exported. Our data also show that communication between the N and C termini of ATP7B promotes its copper-dependent hyperphosphorylated state, and we speculate that this regulates the rate or extent of copper transport.
DISCUSSION
We report the most comprehensive phosphoproteomics analysis of ATP7B to date. Comparison with the PhosphoSitePlus® post-translational modification resource, which compiles the results from all proteomics studies, shows that 14 of the 24 sites phosphosites we found have not been identified previously. Fourteen additional phosphosites appear in that database, making a total of 38 phosphomodifications on ATP7B that are supported by experimental evidence (Table 1). Some of the ATP7B sites may be cell type-specific because a number of sites were identified only in T-cells, isolated synapses, or ovary (Table 1). The PhosphoSitePlus post-translational modification resource also reports that six lysines were either acetylated or ubiquitinated. Our own MS studies also revealed a number of such modifications, raising the possibility that they may be important for ATP7B trafficking and/or function.
Our phosphoproteomics in fibroblasts demonstrates that multisite phosphorylation of ATP7B occurs overwhelmingly on the N and C termini, and surprisingly, it occurs in both high and low copper. We did, however, identify one triphosphopeptide, four unique phosphoresidues, and four diphosphopeptides that were found only in high copper, which is consistent with the increase in [32P]orthophosphate incorporation after addition of copper in hepatic cells. Amino acid sequence alignments reveal that the C-term sites (1429/1432 and 1442/1453) in two diphosphopeptides are conserved in mammals (5). Taken together, we identified a complex pattern of Ser/Thr phosphorylation on APT7B.
We show that copper-stimulated trafficking and phosphorylation could be experimentally separated, and we report several novel findings pertaining to the relationship between the two processes. We first showed that hyperphosphorylation in response to copper occurs even when ATP7B localization is restricted to the TGN. This result demonstrates that there are kinases(s) located in the TGN that phosphorylate ATP7B preferentially when copper is high. It also argues against a model in which the phosphorylation occurs downstream of trafficking only after ATP7B has exited the TGN in vesicles. How cellular copper levels promote ATP7B hyperphosphorylation is unknown. Most models envision a copper-dependent conformational change in ATP7B that in turn affects access of kinase(s) to ATP7B phosphosites. Recent studies demonstrate that copper can directly activate Ras/mitogen protein kinase signaling as well BRAF signaling (37, 38), but inhibitors of Erk2 did not block ATP7B 32P incorporation (12). There are additional indirect mechanisms by which one can envision physiologic changes in copper levels impinging on signaling systems that in turn regulate TGN-associated kinases in hepatocytes (e.g. phorbol 12-myristate 13-acetate (12)). A previous study implicated the TGN-associated kinase PKD in ATP7B regulation (12). Although ATP7B trafficking from the TGN was reported to be reduced by mutation of four serines to alanines in that study, there was no direct evidence that inhibition of PKD altered ATP7B trafficking (12). Perhaps PKD promotes hyperphosphorylated ATP7B carrier vesicle fission. Thus, the kinase(s) responsible for hyperphosphorylation in the TGN remains unknown.
The distal N terminus of ATP7B (amino acids 1–63) contains a targeting motif that is required for apical targeting in response to copper (4). Six N-terminal MBDs, each capable of binding copper, lie downstream of amino acids 1–63. When all copper binding by MBDs had been eliminated, we could still observe some copper-stimulated TGN exit and reversible trafficking to the apical domain of WIF-B cells. Perhaps deletion of MBD1–4 alters the N-term structure in such a manner that the mutants are permissive for TGN exit but are only competent to reach the apical surface following the addition of copper. Thus, our studies reached the unexpected conclusion that the competence of all six MBDs to bind copper was not required to observe copper-dependent apical targeting, suggesting the existence of another copper-sensing domain(s) in ATP7B (39, 40) that regulates the apical targeting step in polarized cells. Our studies extend previous ones that have suggested that copper binding to MBDs triggers conformational changes that propagate structural changes to other domains of ATP7B and trigger its TGN exit via vesicles (13). We suggest an additional role for copper-stimulated conformational changes: perhaps those structural changes expose both the apical targeting motif and C-terminal Ser/Thr to TGN-resident kinases in a copper-dependent manner.
The C-terminal tail of ATP7B is also implicated in two trafficking steps: TGN exit and return (5). Our discovery that phosphorylation occurs at 60% of the sites in the tail domain is consistent with our finding that residues in the C terminus are required for basal phosphorylation. Importantly, we show that increased fixed negative charges on the C terminus do not alter the itinerary of ATP7B, but we did not address the question of whether the dynamics of apical trafficking could be altered by the negative charges.
Because we found that phosphorylation of the C terminus is required for copper-stimulated phosphorylation of the N terminus, we speculated that conformational changes promoted by hyperphosphorylation regulated ATP7B trafficking. So we eliminated all hyperphosphorylation by changing 13 of the serines in the C terminus to Ala/Gly in the context of the full-length protein with all six functional N-terminal MBDs. Despite its total lack of hyperphosphorylation, the copper-dependent trafficking of this mutant was indistinguishable from wild type. When eight of these same residues were instead changed to negative charges to mimic the hyperphosphorylated state, ATP7B trafficking to the apical region was largely unaffected. Overall, by a variety of criteria, copper-dependent trafficking and hyperphosphorylation appear to be coincident but independent events. Our study found little evidence to link copper-dependent hyperphosphorylation to intracellular trafficking. Our results do not exclude an effect of phosphorylation on trafficking per se, but they do argue against those ATP7B molecules uniquely modified by copper-enhanced phosphorylation functioning as critical initiators of TGN exit or trafficking from the TGN.
Multisite phosphorylation has been shown to dynamically regulate the localization, activity, and expression of the voltage-gated sodium channel and potassium channel in mammalian brain (41). In contrast, multiple phosphorylations on the C terminus of photoexcited rhodopsin are required for its inactivation (for a review, see Ref. 42). These examples highlight the idea that multisite phosphorylation is another paradigm for the regulation of membrane protein trafficking and function. Perhaps copper-stimulated phosphorylation fine-tunes the rate or extent of copper transport as indicated by studies in yeast (43) and mammalian liver (44). Thus, the precise roles of both basal and copper-stimulated hyperphosphorylation remain enigmatic.
Supplementary Material
Acknowledgments
We thank Drs. Michael Schell and Svetlana Lutsenko for critical discussions and reading; L. Nyasae for assistance with the figures; L. DeVine of the Mass Spectrometry and Proteomics Facility, Johns Hopkins University School of Medicine and S. Rao, A. Murthy, and T. Sharma for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 DK-072084 and P01 GM-067166.
- TGN
- trans-Golgi network
- MBD
- metal binding domain
- N-term
- N-terminal
- C-term
- C-terminal
- TTM
- tetrathiomolybdate
- BCS
- bathocuproinedisulfonic acid
- N
- nucleotide.
REFERENCES
- 1. Ferenci P. (2006) Regional distribution of mutations of the ATP7B gene in patients with Wilson disease: impact on genetic testing. Hum. Genet. 120, 151–159 [DOI] [PubMed] [Google Scholar]
- 2. Weiss K. H., Stremmel W. (2012) Evolving perspectives in Wilson disease: diagnosis, treatment and monitoring. Curr. Gastroenterol. Rep. 14, 1–7 [DOI] [PubMed] [Google Scholar]
- 3. Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Function and regulation of human copper-transporting ATPases. Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 4. Braiterman L., Nyasae L., Guo Y., Bustos R., Lutsenko S., Hubbard A. (2009) Apical targeting and Golgi retention signals reside within a 9-amino acid sequence in the copper-ATPase, ATP7B. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G433–G444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braiterman L., Nyasae L., Leves F., Hubbard A. L. (2011) Critical roles for the COOH terminus of the Cu-ATPase ATP7B in protein stability, trans-Golgi network retention, copper sensing, and retrograde trafficking. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G69–G81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nyasae L. K., Schell M. J., Hubbard A. L. (2014) Copper directs ATP7B to the apical domain of hepatic cells via basolateral endosomes. Traffic 15, 1344–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Berghe P. V., Klomp L. W. (2010) Posttranslational regulation of copper transporters. J. Biol. Inorg. Chem. 15, 37–46 [DOI] [PubMed] [Google Scholar]
- 8. Polishchuk R., Lutsenko S. (2013) Golgi in copper homeostasis: a view from the membrane trafficking field. Histochem. Cell Biol. 140, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vanderwerf S. M., Cooper M. J., Stetsenko I. V., Lutsenko S. (2001) Copper specifically regulates intracellular phosphorylation of the Wilson's disease protein, a human copper-transporting ATPase. J. Biol. Chem. 276, 36289–36294 [DOI] [PubMed] [Google Scholar]
- 10. Vanderwerf S. M., Lutsenko S. (2002) The Wilson's disease protein expressed in Sf9 cells is phosphorylated. Biochem. Soc. Trans. 30, 739–741 [DOI] [PubMed] [Google Scholar]
- 11. Pilankatta R., Lewis D., Adams C. M., Inesi G. (2009) High yield heterologous expression of wild-type and mutant Cu+-ATPase (ATP7B, Wilson disease protein) for functional characterization of catalytic activity and serine residues undergoing copper-dependent phosphorylation. J. Biol. Chem. 284, 21307–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pilankatta R., Lewis D., Inesi G. (2011) Involvement of protein kinase D in expression and trafficking of ATP7B (copper ATPase). J. Biol. Chem. 286, 7389–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasan N. M., Gupta A., Polishchuk E., Yu C. H., Polishchuk R., Dmitriev O. Y., Lutsenko S. (2012) Molecular events initiating exit of a copper-transporting ATPase ATP7B from the trans-Golgi network. J. Biol. Chem. 287, 36041–36050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta A., Bhattacharjee A., Dmitriev O. Y., Nokhrin S., Braiterman L., Hubbard A. L., Lutsenko S. (2011) Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein. Proc. Natl. Acad. Sci. U.S.A. 108, 5390–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartee M. Y., Ralle M., Lutsenko S. (2009) The loop connecting metal-binding domains 3 and 4 of ATP7B is a target of a kinase-mediated phosphorylation. Biochemistry 48, 5573–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grimsrud P. A., Carson J. J., Hebert A. S., Hubler S. L., Niemi N. M., Bailey D. J., Jochem A., Stapleton D. S., Keller M. P., Westphall M. S., Yandell B. S., Attie A. D., Coon J. J., Pagliarini D. J. (2012) A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab. 16, 672–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson-Grady J. T., Haas W., Gygi S. P. (2013) Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods 61, 277–286 [DOI] [PubMed] [Google Scholar]
- 20. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J., Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 [DOI] [PubMed] [Google Scholar]
- 22. Braiterman L. T., Murthy A., Jayakanthan S., Nyasae L., Tzeng E., Gromadzka G., Woolf T. B., Lutsenko S., Hubbard A. L. (2014) Distinct phenotype of a Wilson disease mutation reveals a novel trafficking determinant in the copper transporter ATP7B. Proc. Natl. Acad. Sci. U.S.A. 111, E1364–E1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Y., Nyasae L., Braiterman L. T., Hubbard A. L. (2005) NH2-terminal signals in ATP7B Cu-ATPase mediate its Cu-dependent anterograde traffic in polarized hepatic cells. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G904–G916 [DOI] [PubMed] [Google Scholar]
- 24. Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M. L. (1997) Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71, 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bastaki M., Braiterman L. T., Johns D. C., Chen Y. H., Hubbard A. L. (2002) Absence of direct delivery for single transmembrane apical proteins or their “secretory” forms in polarized hepatic cells. Mol. Biol. Cell 13, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ihrke G., Neufeld E. B., Meads T., Shanks M. R., Cassio D., Laurent M., Schroer T. A., Pagano R. E., Hubbard A. L. (1993) WIF-B cells: an in vitro model for studies of hepatocyte polarity. J. Cell Biol. 123, 1761–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shevchenko A., Wilm M., Vorm O., Jensen O. N., Podtelejnikov A. V., Neubauer G., Shevchenko A., Mortensen P., Mann M. (1996) A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24, 893–896 [DOI] [PubMed] [Google Scholar]
- 28. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 29. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 30. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by MS/MS. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 31. Griffiths G., Fuller S. D., Back R., Hollinshead M., Pfeiffer S., Simons K. (1989) The dynamic nature of the Golgi complex. J. Cell Biol. 108, 277–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ladinsky M. S., Wu C. C., McIntosh S., McIntosh J. R., Howell K. E. (2002) Structure of the Golgi and distribution of reporter molecules at 20°C reveals the complexity of the exit compartments. Mol. Biol. Cell 13, 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huster D., Lutsenko S. (2003) The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson's disease protein. J. Biol. Chem. 278, 32212–32218 [DOI] [PubMed] [Google Scholar]
- 34. Banci L., Bertini I., Cantini F., Rosenzweig A. C., Yatsunyk L. A. (2008) Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry 47, 7423–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsivkovskii R., MacArthur B. C., Lutsenko S. (2001) The Lys1010–Lys1325 fragment of the Wilson's disease protein binds nucleotides and interacts with the N-terminal domain of this protein in a copper-dependent manner. J. Biol. Chem. 276, 2234–2242 [DOI] [PubMed] [Google Scholar]
- 36. Christensen G. L., Kelstrup C. D., Lyngsø C., Sarwar U., Bøgebo R., Sheikh S. P., Gammeltoft S., Olsen J. V., Hansen J. L. (2010) Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol. Cell. Proteomics 9, 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brady D. C., Crowe M. S., Turski M. L., Hobbs G. A., Yao X., Chaikuad A., Knapp S., Xiao K., Campbell S. L., Thiele D. J., Counter C. M. (2014) Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turski M. L., Brady D. C., Kim H. J., Kim B. E., Nose Y., Counter C. M., Winge D. R., Thiele D. J. (2012) A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol. Cell. Biol. 32, 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cater M. A., La Fontaine S., Mercer J. F. (2007) Copper binding to the N-terminal metal-binding sites or the CPC motif is not essential for copper-induced trafficking of the human Wilson protein (ATP7B). Biochem. J. 401, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borjigin J., Payne A. S., Deng J., Li X., Wang M. M., Ovodenko B., Gitlin J. D., Snyder S. H. (1999) A novel pineal night-specific ATPase encoded by the Wilson disease gene. J. Neurosci. 19, 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baek J. H., Cerda O., Trimmer J. S. (2011) Mass spectrometry-based phosphoproteomics reveals multisite phosphorylation on mammalian brain voltage-gated sodium and potassium channels. Semin. Cell Dev. Biol. 22, 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arshavsky V. Y. (2002) Rhodopsin phosphorylation: from terminating single photon responses to photoreceptor dark adaptation. Trends Neurosci. 25, 124–126 [DOI] [PubMed] [Google Scholar]
- 43. Valverde R. H., Britto-Borges T., Lowe J., Einicker-Lamas M., Mintz E., Cuillel M., Vieyra A. (2011) Two serine residues control sequential steps during catalysis of the yeast copper ATPase through different mechanisms that involve kinase-mediated phosphorylations. J. Biol. Chem. 286, 6879–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cardoso L. H., Britto-Borges T., Vieyra A., Lowe J. (2014) ATP7B activity is stimulated by PKCϵ in porcine liver. Int. J. Biochem. Cell Biol. 54, 60–67 [DOI] [PubMed] [Google Scholar]
- 45. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.