Background: Ubiquitous and conserved Hsp70 heat shock proteins play key roles in protein folding.
Results: DnaK, a model Hsp70, forms a specific dimer, and mutations on the dimer interfaces compromise both chaperone activity and Hsp40 interaction.
Conclusion: Dimerization of DnaK is required for the efficient interaction with Hsp40.
Significance: Our studies shed light on the molecular mechanism of the Hsp70 chaperone machinery.
Keywords: 70-Kilodalton Heat Shock Protein (Hsp70), Chaperone DnaJ (DnaJ), Chaperone DnaK (DnaK), Heat Shock Protein (HSP), Protein Folding, Protein-Protein Interaction, Hsp40
Abstract
Highly conserved molecular chaperone Hsp70 heat shock proteins play a key role in maintaining protein homeostasis (proteostasis). DnaK, a major Hsp70 in Escherichia coli, has been widely used as a paradigm for studying Hsp70s. In the absence of ATP, purified DnaK forms low-ordered oligomer, whereas ATP binding shifts the equilibrium toward the monomer. Recently, we solved the crystal structure of DnaK in complex with ATP. There are two molecules of DnaK-ATP in the asymmetric unit. Interestingly, the interfaces between the two molecules of DnaK are large with good surface complementarity, suggesting functional importance of this crystallographic dimer. Biochemical analyses of DnaK protein supported the formation of dimer in solution. Furthermore, our cross-linking experiment based on the DnaK-ATP structure confirmed that DnaK forms specific dimer in an ATP-dependent manner. To understand the physiological function of the dimer, we mutated five residues on the dimer interface. Four mutations, R56A, T301A, N537A, and D540A, resulted in loss of chaperone activity and compromised the formation of dimer, indicating the functional importance of the dimer. Surprisingly, neither the intrinsic biochemical activities, the ATP-induced allosteric coupling, nor GrpE co-chaperone interaction is affected appreciably in all of the mutations except for R56A. Unexpectedly, the interaction with co-chaperone Hsp40 is significantly compromised. In summary, this study suggests that DnaK forms a transient dimer upon ATP binding, and this dimer is essential for the efficient interaction of DnaK with Hsp40.
Introduction
Hsp70 heat shock proteins are a class of abundant molecular chaperones that are most frequently induced in response to various environmental stresses, such as elevated temperature, radiation, and inflammation, and to alleviate protein denaturation damages caused by such stresses. At the same time, under normal conditions, Hsp70s also play multiple essential roles in maintaining cellular protein homeostasis (proteostasis) by assisting in protein folding, assembly, translocation into organelles, and degradation (1–6). The central roles of Hsp70s in proteostasis inevitably link them to many human diseases, especially various cancers and neurodegenerative diseases; thus they are potential targets for treating these diseases (7–11).
The importance of Hsp70s is also demonstrated by their ubiquitous expression and high degree of conservation. Except for a number of archaea, Hsp70s have been found in all organisms examined and in all cellular compartments of eukaryotes. Hsp70s are highly conserved with more than 40% pairwise sequence identity even between the most divergent members, indicating an overall conserved molecular mechanism (1, 5, 6, 10, 12).
All Hsp70s contain two functional domains, the nucleotide-binding domain (NBD)6 at the N terminus and the substrate-binding domain (SBD) at the C terminus (1, 3, 6). Each domain has an essential intrinsic activity. The NBD binds adenine nucleotides ATP and ADP and also possesses an ATPase activity. A number of crystal structures of the isolated NBD have revealed the structural basis of nucleotide binding (13–17). The SBD is the site for peptide substrate binding and is further divided into α and β subdomains (18). SBDβ forms a single binding site for peptide substrates, whereas SBDα functions as a lid covering the peptide-binding site on the SBDβ. Previous biochemical and structural studies demonstrated that Hsp70s prefer stretches of hydrophobic segments, which normally fold inside native proteins (18–21). A short and highly conserved linker segment, the inter-domain linker, connects NBD and SBD (22, 23). Although the binding functions of NBD and SBD are independent, Hsp70 chaperone activity is strictly dependent on the tight coupling of these two domains upon ATP binding, which leads to modulation of the two intrinsic activities (1, 5, 24–26). ATP binding dramatically reduces the affinity for peptide substrates by increasing both the rate of binding and especially the rate of dissociation (27, 28). At the same time, peptide substrate binding stimulates the ATP hydrolysis rate (28). Thus, this ATP-induced allosteric coupling ensures that the energy from ATP hydrolysis is efficiently used to regulate peptide substrate binding and release, thereby conferring efficient chaperone activity. In summary, the two intrinsic activities and ATP-induced allosteric coupling are at the heart of the Hsp70 chaperone activity.
Purified Hsp70s have been shown to form low-ordered oligomers in the ADP-bound and nucleotide-free states (29–33). It is well accepted that this oligomerization is due to the binding of the inter-domain linker by the SBD from another molecule of Hsp70 in the absence of peptide substrates (34, 35). In contrast, in the presence of ATP, Hsp70s mainly form monomers, and it was proposed that this monomeric Hsp70-ATP is the active form (22, 35). Recently, a small amount of dimer was observed for DnaK, an Escherichia coli Hsp70 (36, 37); however, little is known about the functional meaning of this dimer.
Up to now, two classes of co-chaperones have been discovered for Hsp70s that dramatically facilitate their chaperone activity as follows: Hsp40s and nucleotide-exchange factors (NEFs) (1, 2, 6, 38–40). Although NEFs facilitate the exchange from ADP to ATP after ATP hydrolysis, the essential and functionally conserved Hsp40 co-chaperones facilitate all biological processes associated with Hsp70s by specifically enhancing the ATP hydrolysis rate of Hsp70s (39, 41, 42). At the same time, many Hsp40s bind unfolded or partially folded polypeptides directly to prevent their aggregation. Thus, it was proposed that Hsp40s also bring polypeptide substrates to Hsp70s. Although Hsp40s are less conserved than Hsp70s, all share a conserved J-domain named after the well studied E. coli Hsp40, DnaJ. The signature J-domain has been shown to be critical for stimulating the ATPase activity of Hsp70s. A number of biochemical and structural studies suggest that the bottom cleft of the NBD and inter-domain linker of Hsp70s are crucial for interacting with the J-domain (16, 43–48). However, because of the transient nature of Hsp70-Hsp40 interaction, the molecular mechanism and the exact interacting site of this interaction remain a mystery.
Consistent with the well established role of Hsp40s in stimulating the ATP hydrolysis rate of Hsp70s, Hsp40s interact robustly with Hsp70s only in the ATP-bound state. Hsp40s are divided into three classes. Class I and II are the canonical Hsp40s. Both of these classes form a stable dimer. As described above, Hsp70s were believed to function mainly as monomer in the ATP-bound state. The Hsp70-Hsp40 interaction is not symmetrical. DnaK, the major Hsp70 in E. coli, has been used extensively as a model to study Hsp70s due to its availability in protein purification. Recently, we solved a crystal structure of an intact DnaK in the ATP-bound state (37), which is supposed to be the active state for interacting with Hsp40 co-chaperones. Interestingly, DnaK-ATP packs as a dimer in the asymmetric unit with large buried surface area, suggesting a functional role of the dimer in Hsp70 chaperone activity. In this study, after mutating the dimer interface in our DnaK-ATP structure, we found that four out of five dimer mutations abolished the in vivo chaperone activity of DnaK and compromised the formation of dimer, indicating the functional importance of the DnaK-ATP dimer in chaperone activity. Our following biochemical analysis suggested an essential role of this dimer in efficiently interacting with Hsp40 co-chaperones.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The DnaK-T199A/L′3,4 (where L3,4 indicates loop3,4) proteins used for crystallization and native gel analysis were purified as described previously (37).
All the mutant DnaK proteins used in this work were full-length DnaK overexpressed from a dnak expression plasmid pBB46 (ampR) in a dnak deletion strain BB205 (camR kanR) (49). A His6 tag was added at the C terminus of DnaK to facilitate protein purification (44). Wild-type (WT) DnaK protein was used as positive control. The mutations were introduced by QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). All the DnaK proteins were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside in LB medium for 5 h at 30 °C. Cell pellets were resuspended in ice-cold 2× PBS (20 mm Na2HPO4, 1.76 mm KH2PO4, pH 7.4, 274 mm NaCl, and 5.4 mm KCl) and sonicated in an ice-water bath. The supernatant of the cell lysate after centrifugation at 12,000 × g for 1 h was applied to a HisTrap column. The eluted DnaK protein from the HisTrap column was further purified on a HiTrap Q column. Before being flash-frozen in liquid nitrogen, the final protein was concentrated to >10 mg/ml in a buffer containing 10 mm Hepes-KOH, pH 7.5, 50 mm KCl, and 1 mm DTT.
The DnaJ and GrpE proteins were purified as described previously (44). Briefly, the open reading frames of DnaJ and GrpE were cloned into a pSMT3 vector (a generous gift from Dr. Lima) as a Smt3 fusion protein with an N-terminal hexahistidine tag (50). After the HisTrap column, the hexahistidine tag and Smt3 were removed by Ulp1 protease. Both proteins were further purified on a Superdex 75 16/60 size-exclusion column.
The DnaK-BCCP fusion protein used for Biacore assay was expressed using the pSMT3 vector as DnaJ and GrpE and purified on a HisTrap column first, and the Q column after the hexahistidine tag and Smt3 were removed by Ulp1 protease.
Analytical Ultracentrifugation
Sedimentation velocity experiments with DnaK proteins were carried out with a Beckman Optima XL-I analytical ultracentrifuge (Beckman Coulter Inc.) at 20 °C. DnaK proteins were dialyzed with buffer A (25 mm Hepes-KOH, pH 7.5, 150 mm KCl, 10 mm Mg(OAc)2, and 1 mm DTT) with 2 mm ATP (ATP samples) or 100 μm ADP (ADP samples) for more than 4 h in the cold room and diluted to 0.25 or 1 mg/ml after determining the protein concentrations with Bio-Rad Protein Assay using the WT DnaK protein without nucleotides as a standard. Protein samples were loaded in the ultracentrifugation cells with 2-sector carbon-filled Epon centerpieces using the dialysis buffer as references. Then the samples were centrifuged at 25,000 rpm, and scans using Rayleigh interference optical system were collected at 5-min intervals and analyzed using the SEDFIT and SEDPHAT programs.
Cross-linking with Glutaraldehyde
DnaK proteins were diluted to 4 mg/ml using buffer B (25 mm Hepes-KOH, pH 7.5, 150 mm KCl, 10 mm Mg(OAc)2, and 10% glycerol) with addition of 2 mm DTT and 2 mm ATP and incubated on ice for 1 h. Glutaraldehyde was freshly dilute to 0.00625, 0.0125, and 0.025% with the same buffer and added to DnaK protein in 1:1 volume ratio to start cross-linking reaction. After incubating on ice for 30 min, 5 μl of 0.5 m Tris-HCl, pH 7.5, was added to a 20-μl cross-linking reaction to quench glutaraldehyde, and proteins were separated on SDS-PAGE.
Disulfide Cross-linking with Copper-Phenanthroline
All the DnaK proteins were diluted to 5 mg/ml in buffer B with addition of 5 mm DTT and incubated for 2 h on ice to make sure all the introduced cysteine residues are fully reduced. DTT was quickly removed on a spin column pre-equilibrated with buffer B. After determining protein concentration, each DnaK protein was diluted to 0.5 mg/ml using buffer B. 100 μm CuSO4 and 200 μm 1,10-phenanthroline were mixed freshly and added to the indicated proteins to start oxidation. After incubating for 1 h on ice, the oxidation reactions were stopped by adding EDTA to a final concentration of 5 mm. Proteins were separated on SDS-PAGE.
Site-directed Mutagenesis, Growth Test, and Western Blot Analysis of DnaK Expression Level
All the cysteine and dimer mutations in DnaK were introduced into a dnak expression plasmid pBB46 using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). The in vivo function of the dimer mutations were tested as described previously (49, 51). To test the expression level of the dimer mutants, overnight cultures of fresh transformants were diluted to LB medium containing 50 μg/ml ampicillin, 25 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 20 μm isopropyl 1-thio-β-d-galactopyranoside. After shaking at 30 °C for 3 h, samples were taken, and equal amount of E. coli cells were loaded onto SDS-PAGE. Western blot analysis with an anti-DnaK antibody, mAb 8E2/2 (Calbiochem) was performed as described previously (51).
Fluorescence Anisotropy Peptide Substrate Binding Assay
NR peptide (sequence NRLLLTG) with fluorescein labeled at the N terminus, F-NR, was ordered from NeoBioscience with greater than 95% purity. When determining the affinity for peptide binding, 20 nm F-NR peptide was incubated with serial dilutions of DnaK proteins in buffer C (25 mm Hepes-KOH, pH 7.5, 150 mm KCl, 10 mm Mg(OAc)2, 10% glycerol, and 1 mm DTT) for over 3 h to allow binding to reach equilibrium. Fluorescence anisotropy measurements were read on a Beacon 2000 instrument (Invitrogen). Dissociation constants (Kd) were determined by fitting anisotropy data to a one-site binding equation using PRISM (GraphPad).
For determining the release of bound F-NR peptide upon addition of ATP, 5 μm DnaK proteins were incubated with F-NR peptide (20 nm final concentration) to allow binding to reach equilibrium as described above. Fluorescence anisotropy was determined. ATP was added to final concentration of 2 mm. After incubating for 2 min, the anisotropy was read to determine the release of F-NR.
Single-turnover ATPase Assay
Single-turnover ATPase assay was carried out as described before (44, 52). Briefly, 20 μg of DnaK protein was incubated with 25 μCi of [α-32P]ATP (NEG503H250UC, 3000 Ci/mmol; PerkinElmer Life Sciences) in buffer C in the presence of 20 μm unlabeled ATP for 5 min on ice. The DnaK-ATP complex was quickly isolated from free ATP on a spin column pre-equilibrated with buffer C. We started each reaction by mixing equal volumes of the DnaK-ATP complex with NR peptide, GrpE, or DnaJ in the indicated concentrations. After stopping the reactions at the indicated time points, ATP was separated from ADP using PEI-cellulose thin layer chromatography plates (Sigma). The amounts of radioactive ATP and ADP were quantified after visualized with a Typhoon phosphorimaging system (GE Healthcare). First-order rate equation by nonlinear regression (GraphPad Prism) was used to determine the rate of ATP hydrolysis (kcat).
Tryptophan Fluorescence Assay
The measurements for tryptophan fluorescence were performed as described before (37, 44). Briefly, DnaK proteins were diluted to 1 μm in buffer C in the presence either 2 mm ATP or ADP. Emission spectra were collected from 310 to 400 nm with excitation wavelength at 295 nm.
Surface Plasmon Resonance (SPR) Analysis of DnaK-DnaJ Interaction
SPR analysis was carried on a Biacore T200 system (GE Healthcare). When using DnaK proteins as the analyte, a DnaJ-BCCP fusion protein was expressed in BL21(Gold), and cell lysate was prepared by sonication (43, 45). We immobilized this DnaJ-BCCP fusion on an SA sensor chip for about 200 response units. All the SPR assays were performed at 25 °C with Biacore buffer (25 mm Hepes-KOH, pH 7.5, 100 mm KCl, 10 mm Mg(OAc)2, 1 mm DTT, 0.003% P20, and 1 mm ATP). Each DnaK protein was diluted to 2 μm with Biacore buffer and injected for 5 min at 30 μl/min. The first channel on the sensor was coated with biotin and used as background control. The data were plotted after subtracting this background control.
When using DnaJ as the analyte, purified DnaK-BCCP fusion protein was immobilized on a SA sensor chip for about 200 response units. Serial dilutions of purified DnaJ protein were injected and analyzed as described above.
Luciferase Refolding Assay
After diluting to a final concentration of 100 nm in the presence of 3 μm of the indicated DnaK protein in buffer D (25 mm Hepes-KOH, pH 7.5, 100 mm KOAc, 10 mm Mg(OAc)2, 2 mm DTT, and 3 mm ATP), purified firefly luciferase (Promega) was denatured by incubating at 42 °C for 20 min. The heat-denatured luciferase was diluted into refolding reaction mixtures containing 3 μm DnaK, 0.33 μm GrpE, and various concentrations of DnaJ in buffer D to start the refolding reaction. After incubating at room temperature for 30 min, a 2-μl refolding reaction was mixed with 50 μl of luciferase substrate (Promega), and luciferase activity was measured in a luminometer (Berthold LB9507). The activity of unheated luciferase in buffer D was read and set as 100%.
RESULTS
DnaK, an E. coli Hsp70, Forms Specific Dimers in the Presence of ATP
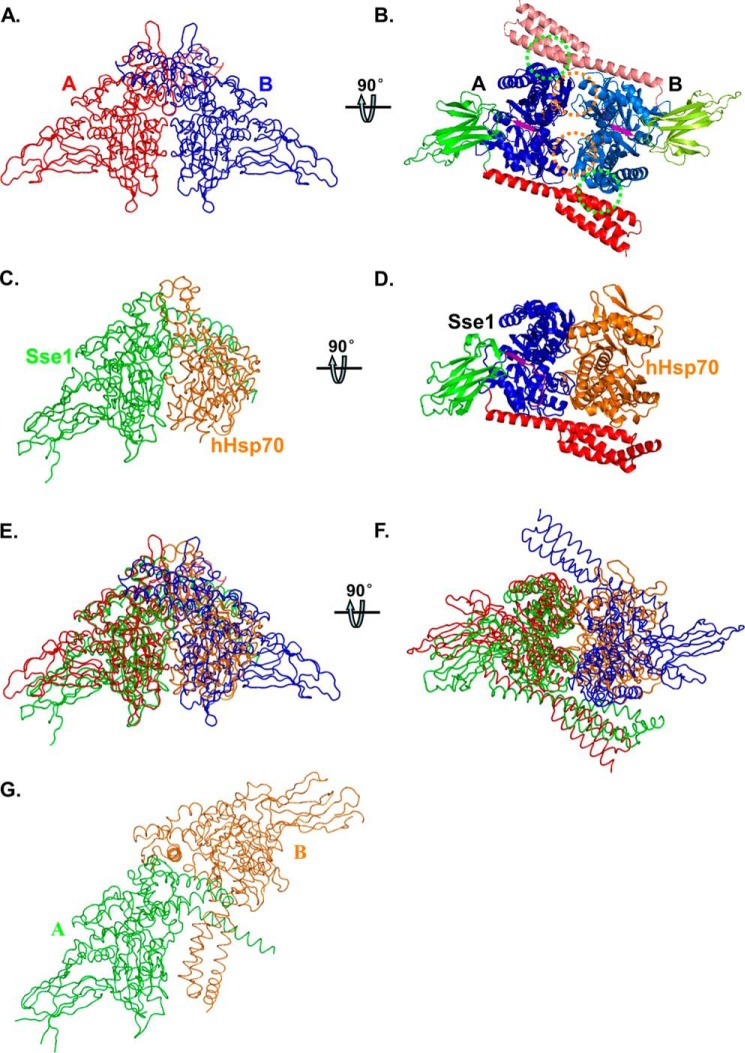
Recently, we solved a crystal structure of an intact DnaK in complex with ATP, DnaK-ATP (37). There are two molecules per asymmetric unit in the crystal (Fig. 1, A and B). The interfaces between the two protomers are extensive with high shape complementarity, suggesting DnaK may form a dimer of functional importance in solution. There are mainly two interfaces, NBD-NBD′ and NBD-SBD′. The NBD-NBD′ interface shares high similarity to that of the Sse1-hHsp70 complex (Fig. 1, A–F) (53). Sse1 is an Hsp110 chaperone from yeast. Hsp110s, distant homologs of Hsp70s, have been shown to function as nucleotide-exchange factors for Hsp70 (54–58). In contrast, this DnaK-ATP dimer is quite different from the Sse1-ATP dimer in the crystal structure that we solved previously (Fig. 1, A and G) (51).
FIGURE 1.
Structure of the DnaK-ATP dimer. A and B, worm and ribbon diagrams of the DnaK-ATP homodimer, respectively. A, view has the 2-fold axis vertical and the NBD-NBD′ interface along the line of sight. The two DnaK protomers A and B are colored in red and blue in A, respectively. B, view is rotated 90° from A about a horizontal axis to view the DnaK dimer along the 2-fold axis from below. The DnaK protomers are colored based on domain as follows: NBD (blue for protomer A and marine for protomer B), inter-domain linker (purple), SBDβ (green for protomer A and lemon green for protomer B), and SBDα (red for protomer A and salmon for protomer B). NBD-NBD′ contacts are highlighted with orange circles, and the SBDa-NBD′ contacts are highlighted with green circles. C and D, worm and ribbon diagrams of the Sse1-hHsp70(NBD) heterodimer (Protein Data Bank code 3D2F), respectively. C, Sse1 is colored green and hHsp70(NBD) colored orange. The NBD of Sse1 is superimposed onto the NBD of DnaK protomer A in A, thereby providing an (A)-equivalent view of this complex. D, Sse1 is colored based on domain as in A, and hHsp70(NBD) is colored orange. The view is rotated relative to that in C just as B is rotated relative to A. E, superposition of A and C. F, superposition of B and D in worm diagrams. G, worm diagram of the Sse1 dimer (Protein Data Bank code 2QXL). The two protomers are colored in green and orange, respectively. The NBD of Sse1 protomer A is superimposed onto the NBD of DnaK protomer A in A, thereby providing an (A)-equivalent view of this complex.
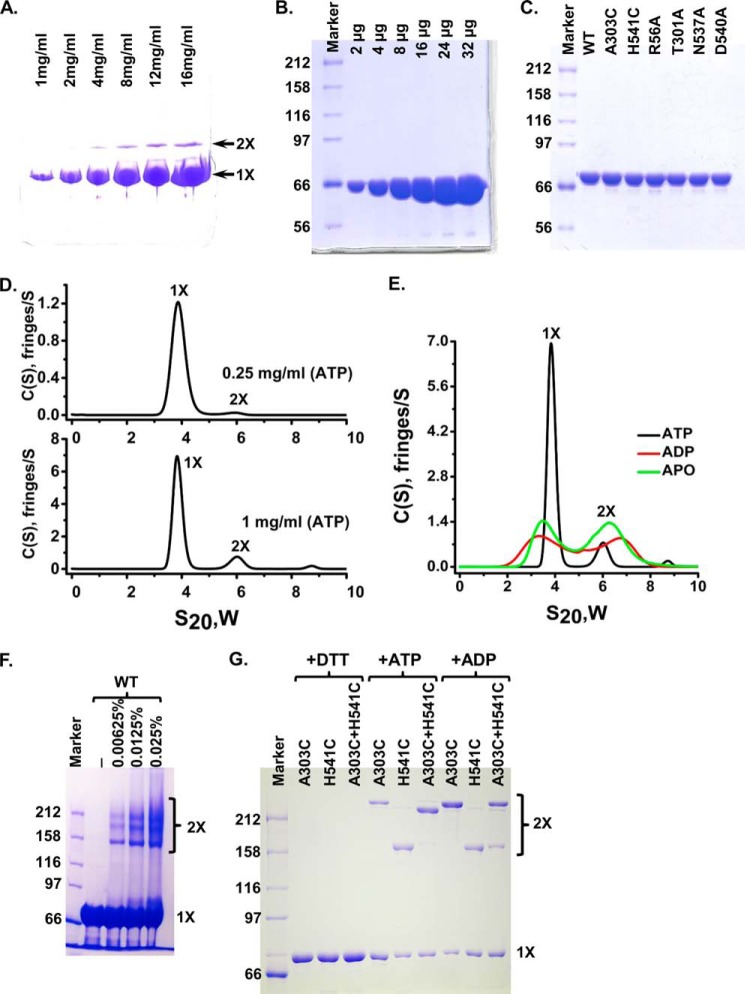
Consistent with the crystallographic dimer, our previous analysis with the crystallization construct DnaK-T199A/L′3,4 using analytical ultracentrifugation (AUC) suggested that DnaK forms a weak dimer in the presence of ATP in solution (37). Moreover, on native gels a weaker band with a higher molecular weight was observed with this construct in the presence of ATP (Fig. 2A), although this protein was purified to high purity (Fig. 2B), indicating the formation of a small amount of dimer. To test whether wild-type (WT) DnaK forms dimer in solution in the presence of ATP like the crystallization construct, we purified WT DnaK to high purity (Fig. 2C) and carried out AUC analysis using sedimentation velocity experiments. As shown in Fig. 2D, at 1 mg/ml, WT DnaK mainly existed as a monomer and dimer in solution with about 14% total protein as dimer. The amount of dimer is concentration-dependent and seems in constant equilibrium with the monomer. At 0.25 mg/ml, the lowest concentration of DnaK that we could use to get reliable results with interference measurement for AUC, the amount of dimer is reduced to about 3% of total protein. In contrast, in the absence of nucleotide (apo-form) or in the presence of ADP, there were similar amounts of monomer and dimer at 1 mg/ml, and the s20,w values for both the monomer and the dimer were different from those in the presence of ATP (Fig. 2E), suggesting that the shapes of the monomer and dimer of the apo-form or ADP-bound form are different from those in the presence of ATP. Furthermore, when the WT DnaK protein was treated with cross-linking reagent glutaraldehyde, several bands with molecular weight in the dimer range were observed (Fig. 2F), further supporting that WT DnaK protein forms a small amount of dimer in the presence of ATP. Consistent with our observations, a recent report from Gestwicki and co-workers (36) demonstrated that DnaK protein forms a small amount of dimer in the presence of saturating concentrations of ATP using gel filtration.
FIGURE 2.
DnaK forms specific dimer in solution in the presence of ATP. A, native PAGE analysis of DnaK-T199A/L′3,4. 2 μl of DnaK-T199A/L′3,4 with indicated concentrations were loaded onto an 8–25% gradient PhastGel (GE Healthcare). The positions of the monomer (1×) and dimer (2×) are indicated by arrows. B, DnaK-T199A/L′3,4 was purified to high purity. Equal amounts of DnaK-T199A/L′3,4 for each lane from A were loaded onto SDS-PAGE. There are no apparent contaminants in the purified DnaK-T199A/L′3,4 protein. C, mutant DnaK proteins were purified to a similar purity as that of the WT DnaK. Each mutant protein was loaded and separated on SDS-polyacrylamide gel. WT DnaK protein purified to high purity was used for comparison. D, AUC analysis of the WT DnaK protein in the presence of ATP. Sedimentation velocity experiments were carried out with WT DnaK protein at 0.25 (top panel) and 1 mg/ml (bottom panel). The positions of the monomer and dimer are labeled as 1× and 2×, respectively. E, AUC analysis of the WT DnaK protein in the presence of different nucleotides. Sedimentation velocity experiments were performed on WT DnaK protein at 1 mg/ml in the presence of ATP, ADP, or in the absence of nucleotide (apo-form). F, glutaraldehyde cross-linking. WT DnaK protein was treated with 0.00625, 0.0125, and 0.025% glutaraldehyde and separated on SDS-PAGE. The concentrations of glutaraldehyde were labeled on the top of the gel. The positions of the monomer (1×) and dimer (2×) are labeled on the right. G, disulfide bond formation between A303C and H541C. A303C, DnaK-C15A/A303C; H541C, DnaK-C15A/H541C; A303C+H541C, DnaK-C15A/A303C and DnaK-C15A/H541C were mixed in 1:1 ratio. Oxidation with copper-phenanthroline was carried out in the presence of ATP (+ATP) or ADP (+ADP). Samples in the presence of ATP were treated with DTT as loading controls (+DTT). The positions of the monomer (1×) and dimer (2×) are labeled on the right.
To further confirm the formation of a specific dimer in solution in the presence of ATP, we have mutated two residues individually on the NBD-SBD′ interface to cysteine: Ala-303 and His-541. In addition, the only endogenous cysteine (Cys-15) was mutated to alanine to prevent nonspecific disulfide bond formation. If DnaK forms a specific dimer as observed in the crystal, then the formation of a disulfide bond between A303C and H541C should be dependent on the nucleotide, i.e. dimer formation under oxidizing conditions should occur in ATP but not in ADP. We purified these two mutant DnaK proteins in the presence of the reducing agent DTT as follows: C15A/A303C and C15A/H541C (Fig. 2C). Oxidation with copper-phenanthroline was performed directly after removing the DTT. Each of the two mutant proteins form a disulfide-bonded band on SBD-Polyacrylamide gel when oxidized regardless of nucleotide (Fig. 2G), suggesting disulfide bond formation of A303C-A303C and H541C–H541C, respectively, because both A303C and H541C are on the surface of DnaK and readily accessible to form a disulfide bond. After treating with DTT, these bands disappeared. When these two mutant proteins were mixed together in the presence of ADP, we observed two disulfide-bonded bands that ran at the same positions as the A303C–A303C and H541C–H541C disulfide-bonded bands. In contrast, in the presence of ATP, these two mutant proteins mainly formed a new disulfide-bonded band running at a different position from that of either A303C–A303C or H541C–H541C disulfide-bonded band, suggesting a specific disulfide-bond formation between A303C and H541C. Taken together, DnaK forms specific dimer in solution as seen in our reported crystal structure only in the presence of ATP.
Dimerization of DnaK Is Essential for Function
After confirming DnaK forms specific dimers in the presence of ATP in solution, we tested the in vivo functional importance of this dimer. As described above, there are two dimer interfaces: NBD-NBD′ and NBD-SBD′. A number of hydrogen bonds and hydrophobic interactions are featured on each interface (Figs. 1, A and B, and 3, A and B). On the NBD-NBD′ interface, Arg-56 (NBD) forms a hydrogen bond with Glu-272 (NBD′), and Glu-28 (NBD) forms two hydrogen bonds with Arg-345 (NBD′). On the NBD-SBD′ interface, Asn-537 (SBD′) forms two hydrogen bonds with the main-chain atoms of both Gln-277 and Ala-303 from NBD; Asp-540 forms a hydrogen bond with Thr-301; and Arg-536 forms hydrophobic interactions with Ala-303 and Thr-301.
FIGURE 3.
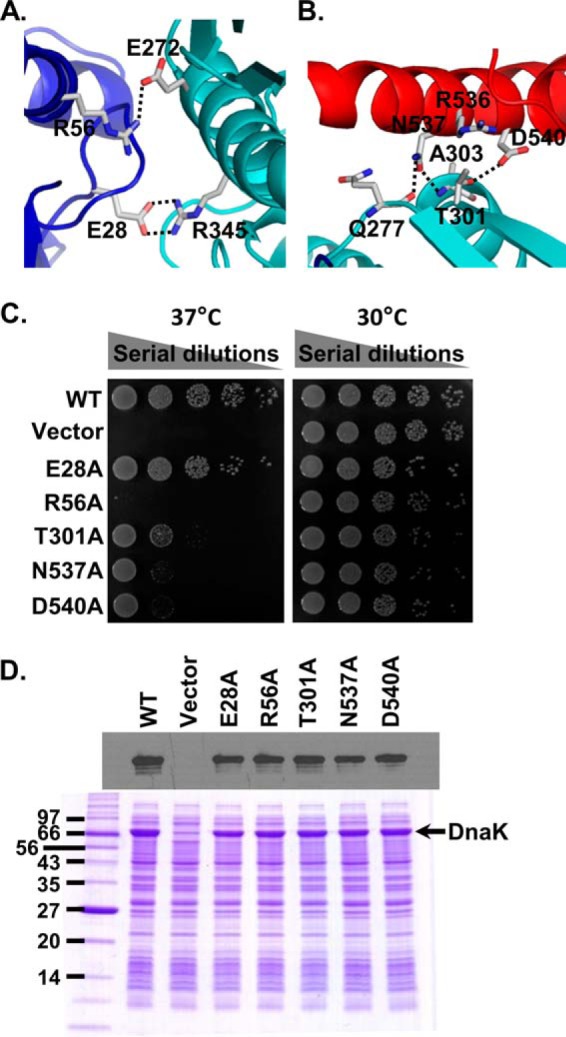
DnaK dimer is essential for in vivo chaperone activity. A, contacts between the NBDs in the DnaK dimer. The NBDs of protomer A and B are colored blue and cyan, respectively. Viewpoint is as in Fig. 1B. B, contacts between NBD′ (cyan) and the partner SBDα (red) in the DnaK dimer. Viewpoint is as in Fig. 1B. C, growth test of the dimer mutants in DnaK. Serial dilutions of fresh cultures carrying indicated dimer mutations were spotted on LB agar plates and grew for 1 overnight at 30 and 37 °C. We used WT DnaK and empty vector as positive and negative controls, respectively. D, protein expression levels of the dimer mutants. Equal amounts of E. coli cultures carrying each dimer mutant or the WT DnaK were loaded onto SDS-PAGE. The empty vector was used as negative control. The top panel is the Western blot analysis with an anti-DnaK antibody. The bottom panel is a SDS-polyacrylamide gel stained with Coomassie Blue.
To test the importance of the dimer, we mutated five key residues one at a time on the dimer interface: Glu-28, Arg-56, Thr-301, Asn-537, and Asp-540. We mutated each residue to alanine to disrupt the observed hydrogen bond interactions on the dimer interface. To test the in vivo function of these mutants, we took advantage of the observation that DnaK is required for growth at elevated temperature such as 37 °C but not at 30 °C (49, 59, 60). After introducing these mutants into a dnak deletion strain, we analyzed growth at 37 °C using 30 °C as a control. WT DnaK and empty vector were employed as positive and negative controls, respectively. Except for the E28A mutant, all of the alanine mutants grew much less than that of the WT with R56A almost identical to the empty vector, the negative control, indicating a dramatic impairment of their in vivo chaperone function (Fig. 3C). Both Western blot and SDS-polyacrylamide gel showed that the mutant proteins had comparable levels of expression as that of WT DnaK (Fig. 3D).
To confirm dimerization is compromised in the R56A, T301A, N537A, and D540A mutants that conferred a significant growth defect, we first introduced these dimer mutations into the crystallization construct DnaK-T199A/L′3,4. On native gels, purified proteins carrying these mutations demonstrated a significantly less amount of the dimer band than that of the WT control (Fig. 4A, although all these proteins were purified to high purity (Fig. 4B), suggesting these mutations compromise the formation of dimer. Moreover, we purified the R56A, T301A, N537A, and D540A dimer mutants in the regular DnaK construct as the WT DnaK. All the purified proteins have a similar purity as that of the WT DnaK (Fig. 2C). AUC analysis using sedimentation velocity experiments on these mutant proteins in the presence of ATP showed significantly less dimer than that of the WT DnaK (Fig. 4C). In contrast, all the mutant proteins behaved similarly as the WT DnaK in the presence of ADP (Fig. 4D). Thus, these dimer mutations specifically compromise the dimer formation in the presence of ATP. At the same time, when the purified mutant proteins were treated with glutaraldehyde, there was less amount of dimer bands than that of the WT DnaK (Fig. 4E), especially for T301A, N537A, and D540A. Thus, both the AUC and glutaraldehyde cross-linking analyses further confirmed that the formation of DnaK dimer is compromised by these dimer mutations. Consistent with these observations, when T301A and N537A/D540A were introduced into the cysteine mutants A303C and H541C, the specific disulfide-bonded dimer band was significantly less than that of the cysteine mutants A303C and H541C by themselves (Fig. 4, F and G). Taken together, the mutations R56A, T301A, N537A, and D540A indeed compromise the formation of DnaK dimer. In summary, the dimer observed in the DnaK-ATP crystal structure is functionally important.
FIGURE 4.
Dimer mutations R56A, T301A, N537A, and D540A compromise the formation of DnaK dimer. A, native gel analysis of the dimer mutations in the DnaK-T199A/L′3,4 construct. 2 μl of each protein at 12 mg/ml was loaded onto an 8–25% gradient PhastGel (GE Healthcare). The positions of the monomer (1×) and dimer (2×) are indicated by arrows. The DnaK-T199A/L′3,4 protein (WT) was used as control. B, all the dimer mutants in the DnaK-T199A/L′3,4 construct were purified to a similar purity as that of the DnaK-T199A/L′3,4 (WT). 24 μg of each protein were loaded onto SDS-PAGE. C and D, AUC analysis of the dimer mutants in the presence of ATP (C) and ADP (D), respectively. Each mutant protein (at 1 mg/ml) was subjected to sedimentation velocity experiment as in Fig. 2D. The WT DnaK was used as control. The positions of the monomer and dimer are labeled as 1× and 2×, respectively. E, glutaraldehyde cross-linking. The mutant DnaK proteins were treated with 0.00625, 0.0125, and 0.025% glutaraldehyde and separated on SDS-PAGE as in Fig. 2F. The concentrations of glutaraldehyde were labeled at the top of the gel. The positions of the monomer (1×) and dimer (2×) are labeled on the right. F, dimer mutants T301A and N537A/D540A compromise the formation of the disulfide bond between A303C and H541C. T301A and N537A/D540A were introduced into the cysteine mutants A303C and H541C. Purified proteins were oxidized with increasing concentrations of copper-phenanthroline (50, 100, and 200 μm; labeled at the top of the gel) in the presence of ATP as described in Fig. 2G. The positions of the monomer (1×) and dimer (2×) are labeled on the right. G, cysteine mutant DnaK proteins used in F were purified to the similar purity as that of the WT DnaK. Each DnaK protein was loaded and separated on an SDS-polyacrylamide gel.
Intrinsic Biochemical Activities of DnaK Are Largely Intact in the Dimer Interface Mutants
To dissect the functional role of this dimer in the DnaK chaperone cycle, we analyzed the intrinsic biochemical activities of the purified dimer mutants R56A, T301A, N537A, and D540A to tease out what activities are affected. First, we looked at the intrinsic ATPase activity using a single-turnover ATPase assay. As shown in Table 1, three of the four dimer mutants, T301A, N537A, and D540A, have intrinsic ATPase activity close to the WT level. The intrinsic ATPase activity of R56A is about 3–4 times greater than that of the WT. Arg-56 is on the surface of NBD. How this mutant affects the intrinsic ATPase rate remains unclear.
TABLE 1.
The two intrinsic activities of DnaK
The two intrinsic activities of DnaK are as follows: ATPase and peptide substrate binding activities.
| ATPase activity kcat | Peptide binding Kd | |
|---|---|---|
| ×10−2/min | μm | |
| WT | 1.04 ± 0.076 | 1.37 ± 0.04 |
| R56A | 3.61 ± 0.260 | 1.25 ± 0.06 |
| T301A | 0.89 ± 0.089 | 1.28 ± 0.06 |
| N537A | 0.91 ± 0.094 | 0.72 ± 0.07 |
| D540A | 0.99 ± 0.109 | 0.75 ± 0.06 |
Next, we determined the peptide substrate binding activity using fluorescence polarization assay. For this assay, we used a model peptide NR with a fluorescein label at the N terminus, F-NR. The intrinsic peptide binding affinity for the WT DnaK is 1.37 ± 0.04 μm in this study (Table 1), consistent with previously published results (44). Both R56A and T301A have affinity close to that of the WT DnaK. Both N537A and D540A showed slightly higher affinity than that of the WT. In the isolated SBD structure of DnaK (representing the ADP-bound and nucleotide-free states), Asn-537 and Asp-540 (on SBDα) form hydrogen bonds with the main chain of Mer-404 and side chain of Glu-467 on SBDβ, respectively (Fig. 5, A and B). These interactions are thought to stabilize the SBDα lid in covering the peptide-binding pocket on SBDβ. Removing the whole SBDα lid reduces the peptide binding affinity severalfold (61). However, like the N537A and D540A mutants in this study, a previous work reported that the D540A and K548A double mutant of DnaK did not show an appreciable defect in peptide substrate binding, but it lost chaperone activity in refolding luciferase (62), suggesting that the hydrogen bonds between Asp-540 and Glu-467 have a minor role in the ADP-bound and nucleotide-free states. Moreover, mutating Glu-467 to cysteine did not show an obvious effect in chaperone activity (Fig. 5C). In summary, neither of the intrinsic biochemical activities of DnaK is appreciably affected in the dimer mutants T301A, N537A, and D540A, suggesting the dimer has little role in these intrinsic activities.
FIGURE 5.
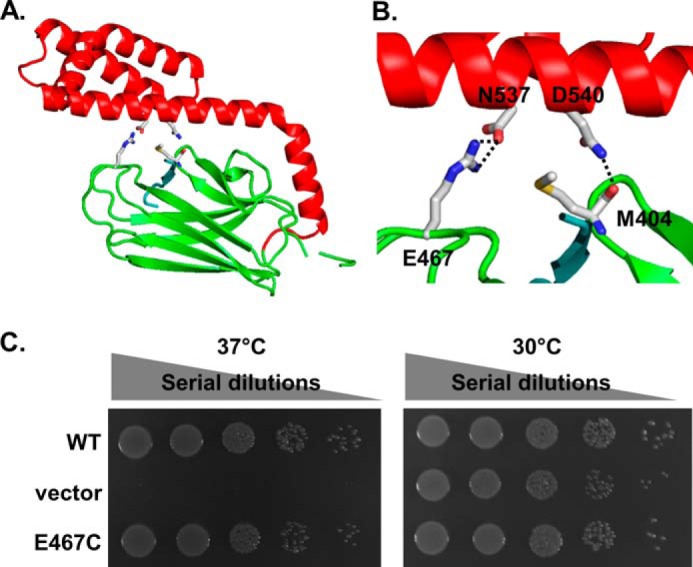
Hydrogen bonds formed between Asn-537 and Glu-467 are not essential for DnaK's chaperone activity. A, ribbon diagram of the isolated DnaK SBD structure. SBDβ and SBDα are colored in green and red, respectively. The bound NR peptide is shown in cyan. Asn-537, Asp-540, Glu-467, and Met-404 are highlighted in stick representation. B, close-up view of the hydrogen bonds formed between Asn-537 and Glu-467 and between Asp-540 and the main chain of Met-404. Coloring is the same as in A. C, E467C mutation has little effect on DnaK's in vivo chaperone activity. The growth test was done the same way as that of Fig. 3C.
Dimer Mutations Have Little Influence on the ATP-induced Allosteric Coupling of DnaK
In each protomer of the DnaK-ATP structure, the SBDα has an elongated conformation, and only the N-terminal end of the first long α-helix binds its NBD. The rest of SBDα protrudes out of the monomer structure and forms dimer contacts with the NBD from the other protomer (Fig. 1B). It seems that this SBDα-NBD′ dimer contacts stabilizes the DnaK-ATP protomer conformation. Thus, disrupting dimer formation may influence ATP-induced allosteric coupling. To test this hypothesis, we analyzed the ATP-induced allosteric coupling in the dimer mutants. We used three different approaches. The first one was the well established tryptophan fluorescence assay (26). DnaK has a single tryptophan in the NBD, Trp-102. The intrinsic fluorescence emission spectrum of Trp-102 is sensitive to the nucleotide-bound states of DnaK; relative to the nucleotide-free and the ADP-bound states, ATP binding induces a blue shift. As expected, we observed an ∼6-nm blue shift for the WT DnaK (Fig. 6A). When we examined the dimer mutants, all showed a similar amount of blue shift, suggesting the ATP-induced allosteric coupling is largely intact.
FIGURE 6.
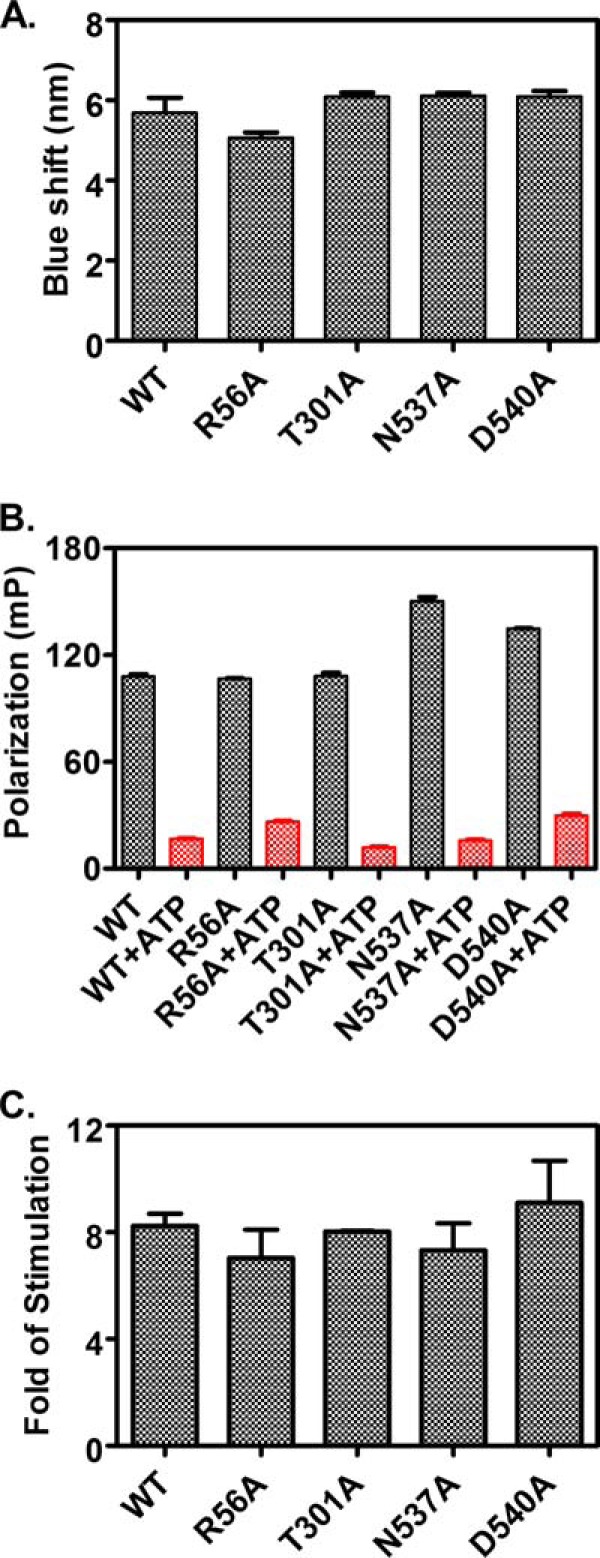
Tests of the ATP-induced allosteric coupling in the dimer mutant proteins. A, ATP-induced tryptophan fluorescence shift. The blue shift for each protein was calculated as the wavelength difference of the maximal emission between the samples incubated with ATP and ADP. B, ATP-induced bound peptide substrate release. F-NR was incubated with 5 μm each of DnaK protein for more than 3 h to allow binding to reach equilibrium. Fluorescence anisotropy was measured for binding in the absence of ATP (black bars). Then ATP was added to a final concentration of 2 mm and incubated for 2 min. The resulting anisotropy measurements represent the binding in the presence of ATP (red bars). C, NR peptide stimulation of the intrinsic ATPase activity of DnaK. Fold of stimulation in the presence of 40 μm NR peptide was calculated by setting the intrinsic ATPase activity rate (kcat) as 1.
Second, we tested the ATP-induced release of bound peptide substrate due to the significantly reduced affinity of DnaK for peptide substrates upon ATP binding. We used the aforementioned fluorescence polarization assay with F-NR peptide. Consistent with the tryptophan fluorescence results, all the dimer mutants released bound F-NR to a similar extent as that of WT DnaK (Fig. 6B).
To further confirm these results, we also determined the stimulation of the intrinsic ATPase activity upon peptide substrate binding. We utilized the single-turnover ATPase assay described above and included 40 μm NR peptide as substrate. Consistent with previous results (44), 40 μm NR peptide stimulated the ATPase activity of the WT DnaK protein about 8-fold (Fig. 6C). All the dimer mutant proteins showed a similar fold simulation as that of the WT DnaK, although R56A has higher intrinsic ATPase activity.
Taken together, the above three assays supported that the dimer mutations have little effect on the ATP-induced allosteric coupling, indicating most likely the DnaK dimer is not important for the ATP-induced allosteric coupling.
Dimer Mutants T301A, N537A, and D540A Have Normal Interaction with the Nucleotide-exchange Factor Co-chaperone GrpE
Because neither the intrinsic activities nor the ATP-induced allosteric coupling showed appreciable defects in the dimer mutants, we analyzed co-chaperone interactions. DnaK has two co-chaperones, DnaJ and GrpE. We first tested the GrpE interaction using the aforementioned single-turnover ATPase assay. GrpE facilitates the bound nucleotide release from DnaK (40). When incubated with the DnaK-ATP complex in the single-turnover ATPase assay, GrpE speeds up the release of bound [32P]ATP before it can be hydrolyzed. In the presence of a high concentration of unlabeled ATP, this released [32P]ATP has little chance to rebind to DnaK for hydrolysis. Thus, on the surface, it seems that GrpE inhibits the hydrolysis of [32P]ATP by DnaK. Consistent with previous observations with Mge1, a GrpE homolog in yeast mitochondria (63), GrpE blocks the ATP hydrolysis of DnaK in the presence of 250 μm unlabeled ATP (Fig. 7A). Simply adding 250 μm unlabeled ATP only reduced the ATP hydrolysis moderately because DnaK binds ATP quite tightly before hydrolysis. When we looked the dimer mutants, T301A, N537A, and D540A behaved quite similarly to that of the WT DnaK (Fig. 7, C–E), suggesting that these dimer mutations have little influence in interacting with GrpE. These results are consistent with the observation that Thr-301, Asn-537, and Asp-540 are far away from the DnaK-GrpE interface (17).
FIGURE 7.
ATP release activity of GrpE in the single-turnover ATPase assay. DnaK-[32P]ATP complexes were incubated with 250 μm unlabeled ATP (purple diamonds) or 3 and 6 μm GrpE together with 250 μm unlabeled ATP (green triangles and blue upside-down triangles). Samples were taken at the indicated times, and the fraction of ATP converted to ADP was calculated after quantification. The samples of the DnaK-[32P]ATP complexes incubated with buffer were used as control (red-filled circles). A, WT DnaK; B, DnaK-R56A; C, DnaK-T301A; D, DnaK-N537A; and E, DnaK-N540A.
Arg-56 has been shown to be on the interface with GrpE (17). However, a previous study suggested R56A still forms stable complex with GrpE and retained significant GrpE nucleotide-exchange activity (64). The NBD is made of two big lobes, lobe I and II. Between the lobes is the nucleotide-binding pocket. Arg-56 (on the surface of lobe I) was hypothesized to form a salt bridge with Glu-264 from lobe II and thus play a role in controlling the release rate of bound nucleotide, although such a salt bridge was not observed in any published DnaK structures (17, 37, 65). Consistent with these observations, in our assays, R56A binds ATP less tightly than that of WT DnaK because ATP hydrolysis is significantly reduced even in the presence of only unlabeled ATP (Fig. 7B). Nevertheless, GrpE further inhibited the ATP hydrolysis to the similar extent as that of the WT DnaK, indicating R56A still can interact with GrpE significantly.
All the Dimer Mutants Showed Significantly Reduced Interaction with the Hsp40 Co-chaperone
Finally, we tested the Hsp40 co-chaperone DnaJ interaction using the well established SPR and the aforementioned single-turnover ATPase assays. In the SPR assay, we immobilized DnaJ to a sensor chip using a previously characterized DnaJ construct with the BCCP fused at the C terminus (43, 45, 66). When WT DnaK was passed through the sensor chip, a strong signal was observed in the presence of ATP (Fig. 8, A and B), indicating a robust interaction, which is consistent with previous observations (43–45, 66, 67). In contrast, when the dimer mutant proteins were applied, the response signals for T301A, N537A, and D540A were about 25% of the WT DnaK, and the R56A has an even lower signal (Fig. 8B), suggesting a significant defect in the interaction of the dimer mutants with Hsp40 co-chaperone DnaJ. This defect was further confirmed with the single-turnover ATPase assay. It is well established that DnaJ markedly stimulates the intrinsic ATPase activity of DnaK by directly accelerating the ATP hydrolysis rate. Consistent with previous observations, DnaJ increased the ATPase rate of WT DnaK more than 60-fold when 0.6 μm DnaJ was included in the single-turnover ATPase assay (Fig. 8C). All the dimer mutants showed significant reduction in the stimulation by DnaJ, supporting the DnaJ interaction defect observed in the SPR assay. Thus, the DnaK dimer is required for efficient interaction with Hsp40 co-chaperone DnaJ.
FIGURE 8.
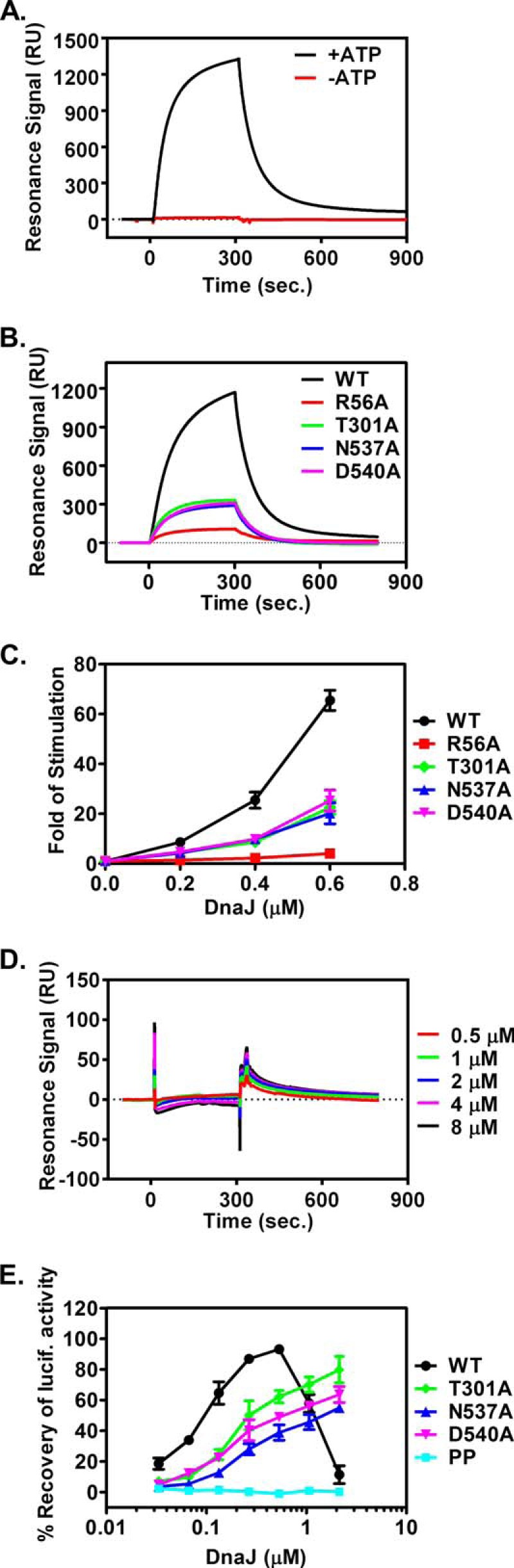
DnaJ interaction is compromised in the dimer mutant proteins. A and B, SPR analysis of the DnaK-DnaJ interaction with DnaJ immobilized. 2 μm DnaK proteins were injected over a sensor chip with DnaJ immobilized on the surface. Resonance signals after subtracting the background binding to a control channel were recorded over time. All the analyses in B were carried out in the presence of ATP. C, DnaJ stimulation of DnaK's ATPase activity. Various concentrations of DnaJ were incubated with DnaK-[32P]ATP complexes in the single-turnover ATPase assay. The hydrolysis rate, kcat, for each concentration of DnaJ was determined, and the fold of stimulation was calculated by setting the intrinsic ATPase rate as 1. D, SPR analysis with DnaK immobilized on a sensor chip. DnaJ protein was injected as analyte. The concentrations of DnaJ were labeled on the right. E, DnaJ dependence in refolding the heat-denatured firefly luciferase. Various concentrations of DnaJ were included in the refolding reactions of denatured firefly luciferase. The refolding activity was calculated by setting the undenatured luciferase activity as 100%.
Moreover, a DnaK-BCCP fusion protein with BCCP fused to the C terminus of DnaK was purified and immobilized on a sensor chip at low density; thus, the DnaK molecules on the sensor chip were almost all monomers, and immobilization on the chip prevents monomer-dimer equilibrium. When serial dilutions of DnaJ protein were passed through the sensor chip as analyte, no appreciable interaction was observed (Fig. 8D), further supporting the above observation that the DnaK dimer is involved in efficient interaction with DnaJ.
As chaperone machinery, DnaK, DnaJ, and GrpE can refold heat-denatured luciferase (35). The efficiency of the refolding activity depends on the concentration of DnaJ; the activity reaches maximum at relatively low DnaJ concentration, whereas high concentrations of DnaJ inhibit refolding activity (68). In our hands, the maximal refolding efficiency was achieved at around 0.5 μm DnaJ with the DnaK concentration at 3 μm (Fig. 8E). Because all of the dimer mutant proteins showed a significant defect in interacting with DnaJ in both the SPR and single-turnover ATPase assay, and this DnaJ interaction defect is the major defect that we could detect for the T301A, N537A, and D540A mutants, the refolding activities of these mutants should require a higher DnaJ concentration to reach maximal efficiency. In other words, a higher DnaJ concentration may be able to compensate for the chaperone activity defect of these dimer mutants if the DnaJ interaction defect is the major defect of these mutants. As expected, T301A, N537A, and D540A have lower refolding activity than that of the WT DnaK at DnaJ concentrations lower than 0.5 μm, the optimal DnaJ concentration for WT DnaK (Fig. 8E), consistent with the in vivo chaperone activity defect in supporting E. coli growth as shown above (Fig. 3C). In contrast, when DnaJ concentration is above 0.5 μm, the refolding activities of the three dimer mutant proteins are getting higher with increasing concentrations of DnaJ instead of lower for the WT DnaK, suggesting the optimal DnaJ concentrations for these dimer mutants are higher than that of the WT DnaK. We did not test R56A because it also has defects in both ATPase activity and binding nucleotide. As a negative control, we tested the DnaK-PP mutant (DnaK-G461P/G468P), whose major defect is locking DnaK in the ATP-bound state (37). Thus, its refolding activity should not have higher dependence on DnaJ concentration as those of the dimer mutants. As expected, the refolding activity is always low, supporting the DnaJ dependence of the dimer mutants is specific. In summary, the higher optimal DnaJ concentration required for T301A, N537A, and D540A in refolding activity supports the hypothesis that the major defect of these dimer mutants is in their interaction with DnaJ. Taken together, the DnaK dimer observed in our DnaK-ATP structure is important for the efficient interaction with the DnaJ Hsp40 co-chaperone.
DISCUSSION
Our recently solved DnaK-ATP structure suggested that DnaK forms a dimer in the ATP-bound state. In this study, we have demonstrated that DnaK forms specific dimers in solution as seen in the crystal structure only in the presence of ATP, and this dimer is important for the chaperone activity by providing efficient interaction with DnaJ Hsp40 co-chaperones. Thus, we proposed the following refined chaperone cycle for DnaK (Fig. 9). In the ATP-bound state, DnaK dimerizes (Fig. 9, step 1), which provides the DnaJ dimer with contact surfaces. With the polypeptide substrate-bound, the DnaJ dimer binds to the bottom of the DnaK dimer, and thus brings the polypeptide substrate to DnaK for binding (Fig. 9, step 2). With both DnaJ and polypeptide substrate-bound, DnaK hydrolyzes ATP to ADP. DnaJ then dissociates from DnaK. DnaK becomes a monomer in the ADP-bound state with the polypeptide-binding channel closed and SBDα covering the polypeptide-binding site (Fig. 9, step 3). This monomeric DnaK-ADP is ideal for GrpE to interact and facilitate the exchange of ADP for ATP. Upon ATP-ADP exchange, DnaK returns to the ATP-bound state, and the NBD and SBD form extensive contacts, which lead to radical conformational changes in SBD and the opening of the peptide-binding pocket (Fig. 9, step 4). Subsequently, bound polypeptide substrate is released either to fold or to enter another round of the chaperone cycle.
FIGURE 9.

Proposed model of the DnaK chaperone cycle. DnaK domain coloring is the same as protomer A in Fig. 1B. The domains of DnaJ are as follows: J-domain (yellow), substrate binding domain (pink), and dimerization domain (cyan). J-domain has been shown to be the site that binds Hsp70s. Polypeptide substrates are highlighted in black.
DnaK and other Hsp70s have been shown to mainly form monomer in the ATP-bound state. The dimer observed in our study is a small population, and it may be transient. The transient nature of this dimer is also consistent with the transient nature of the Hsp70-Hsp40 interaction. Under stress conditions, Hsp70 level increases, which will increase the dimer concentration for more efficient interaction with Hsp40s and thus result in more efficient chaperone activity in refolding.
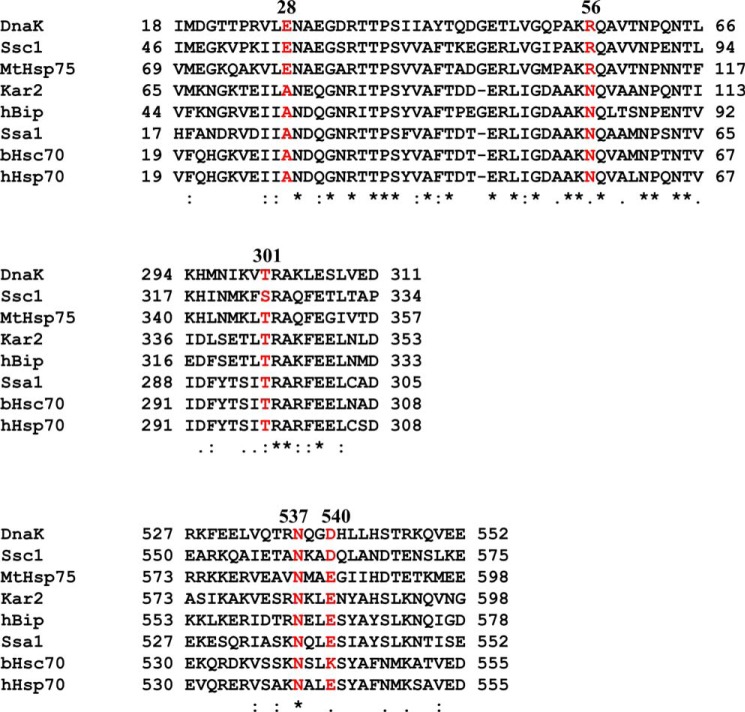
On the NBD-NBD′ interface, Glu-28 and Arg-R56 are not conserved in Hsp70s (Fig. 10), which may be consistent with their phenotype; E28A mutant has not effect on growth. and R56A affects other aspects of the biochemical activities. In contrast, the NBD-SBDα′ dimer interface is quite conserved; Thr-301 and Asn-537 are highly conserved, and Asp-540 is moderately conserved. They seem to play a more important role in the dimer formation than the NBD-NBD′ interface. Based on the conservation, it is possible that the formation and function of dimer are conserved in other Hsp70s. Hsp40s have been traditionally divided into three classes as follows: I, II, and III. Class I (DnaJ-like Hsp40s) and II have the J-domain at the N terminus and form stable dimers in solution. In contrast, class III Hsp40s do not always form dimers, such as auxilin (69, 70). It is possible that the Hsp70-ATP dimer is only needed for working with class I and II Hsp40s.
FIGURE 10.
Sequence alignment of segments containing key dimer contacts. Residues corresponding to Glu-28, Arg-56, Thr-301, Asn-537, and Asp-540 in DnaK are highlighted in red.
The NBD surface that mediates the NBD-NBD′ interface seems a busy surface. It is also involved in interacting with various NEFs, including Hsp110 and GrpE (17, 53, 58, 71). In the ATP-bound state, this interface seems involved in dimer formation. Once ATP is hydrolyzed, the dimer dissociates, and then this NBD surface is available for Hsp110 or GrpE to bind and facilitate nucleotide exchange in Hsp70s. Formation of the dimer may play a role in preventing NEFs from interacting with Hsp70s in the ATP-bound state and releasing bound ATP, which seems unnecessary for the Hsp70 chaperone cycle, although it is observed in the single-turnover ATPase assay.
Acknowledgments
We thank Drs. Lois Greene, Wayne Hendrickson, Elizabeth Craig, Diomedes Logothetis, Young-Jai You, and Leon Avery for critically reading the manuscript and providing insightful suggestions. We thank Drs. Carlos Escalante, John Burgner, and Francisco Zarate-Perez for help with AUC analysis. We thank Drs. Ruifeng Qi and Xinping Xu for technical support in the initial phase of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01GM098592 (to Qinglian Liu). This work was also supported by startup funds from the Virginia Commonwealth University School of Medicine (to Qinglian Liu), New Scholar Award in Aging Grant AG-NS-0587-09 from the Ellison Medical Foundation (to Qinglian Liu), and Grant-in-aid Award 11GRNT7460003 from the American Heart Association (to Qinglian Liu).
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- NEF
- nucleotide-exchange factor
- AUC
- analytical ultracentrifugation
- SPR
- surface plasmon resonance
- BCCP
- biotin carboxyl carrier protein.
REFERENCES
- 1. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 3. Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 4. Bukau B., Deuerling E., Pfund C., Craig E. A. (2000) Getting newly synthesized proteins into shape. Cell 101, 119–122 [DOI] [PubMed] [Google Scholar]
- 5. Bukau B., Horwich A. L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 [DOI] [PubMed] [Google Scholar]
- 6. Young J. C. (2010) Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans C. G., Chang L., Gestwicki J. E. (2010) Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 53, 4585–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brodsky J. L., Chiosis G. (2006) Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 9. Murphy M. E. (2013) The HSP70 family and cancer. Carcinogenesis 34, 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muchowski P. J., Wacker J. L. (2005) Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 11. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 12. Boorstein W. R., Ziegelhoffer T., Craig E. A. (1994) Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 38, 1–17 [DOI] [PubMed] [Google Scholar]
- 13. Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 14. Sriram M., Osipiuk J., Freeman B., Morimoto R., Joachimiak A. (1997) Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5, 403–414 [DOI] [PubMed] [Google Scholar]
- 15. Wisniewska M., Karlberg T., Lehtiö L., Johansson I., Kotenyova T., Moche M., Schüler H. (2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70–2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS One 5, e8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P., Lafer E. M., Sousa R. (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 18. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 [DOI] [PubMed] [Google Scholar]
- 21. Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M. E. (1994) Specificity of DnaK-peptide binding. J. Mol. Biol. 235, 848–854 [DOI] [PubMed] [Google Scholar]
- 22. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vogel M., Mayer M. P., Bukau B. (2006) Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 281, 38705–38711 [DOI] [PubMed] [Google Scholar]
- 24. Swain J. F., Gierasch L. M. (2006) The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 16, 102–108 [DOI] [PubMed] [Google Scholar]
- 25. Saibil H. R. (2008) Chaperone machines in action. Curr. Opin. Struct. Biol. 18, 35–42 [DOI] [PubMed] [Google Scholar]
- 26. Buchberger A., Theyssen H., Schröder H., McCarty J. S., Virgallita G., Milkereit P., Reinstein J., Bukau B. (1995) Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270, 16903–16910 [DOI] [PubMed] [Google Scholar]
- 27. Schmid D., Baici A., Gehring H., Christen P. (1994) Kinetics of molecular chaperone action. Science 263, 971–973 [DOI] [PubMed] [Google Scholar]
- 28. Flynn G. C., Chappell T. G., Rothman J. E. (1989) Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 [DOI] [PubMed] [Google Scholar]
- 29. Benaroudj N., Batelier G., Triniolles F., Ladjimi M. M. (1995) Self-association of the molecular chaperone HSC70. Biochemistry 34, 15282–15290 [DOI] [PubMed] [Google Scholar]
- 30. Osipiuk J., Georgopoulos C., Zylicz M. (1993) Initiation of λ DNA replication. The Escherichia coli small heat shock proteins, DnaJ and GrpE, increase DnaK's affinity for the λP protein. J. Biol. Chem. 268, 4821–4827 [PubMed] [Google Scholar]
- 31. Liu Q., D'Silva P., Walter W., Marszalek J., Craig E. A. (2003) Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300, 139–141 [DOI] [PubMed] [Google Scholar]
- 32. Schönfeld H. J., Schmidt D., Schröder H., Bukau B. (1995) The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J. Biol. Chem. 270, 2183–2189 [DOI] [PubMed] [Google Scholar]
- 33. Carlino A., Toledo H., Skaleris D., DeLisio R., Weissbach H., Brot N. (1992) Interactions of liver Grp78 and Escherichia coli recombinant Grp78 with ATP: multiple species and disaggregation. Proc. Natl. Acad. Sci. U.S.A. 89, 2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang Y. W., Sun Y. J., Wang C., Hsiao C. D. (2008) Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J. Biol. Chem. 283, 15502–15511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schröder H., Langer T., Hartl F. U., Bukau B. (1993) DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson A. D., Bernard S. M., Skiniotis G., Gestwicki J. E. (2012) Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress Chaperones 17, 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 39. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan C. Y., Lee S., Cyr D. M. (2003) Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J., Qian X., Sha B. (2009) Heat shock protein 40: structural studies and their functional implications. Protein Pept. Lett. 16, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suh W. C., Burkholder W. F., Lu C. Z., Zhao X., Gottesman M. E., Gross C. A. (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. U.S.A. 95, 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar D. P., Vorvis C., Sarbeng E. B., Cabra Ledesma V. C., Willis J. E., Liu Q. (2011) The four hydrophobic residues on the Hsp70 inter-domain linker have two distinct roles. J. Mol. Biol. 411, 1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis J. E., Voisine C., Craig E. A. (1999) Intragenic suppressors of Hsp70 mutants: interplay between the ATPase- and peptide-binding domains. Proc. Natl. Acad. Sci. U.S.A. 96, 9269–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greene M. K., Maskos K., Landry S. J. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. U.S.A. 95, 6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gässler C. S., Buchberger A., Laufen T., Mayer M. P., Schröder H., Valencia A., Bukau B. (1998) Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. U.S.A. 95, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wisén S., Bertelsen E. B., Thompson A. D., Patury S., Ung P., Chang L., Evans C. G., Walter G. M., Wipf P., Carlson H. A., Brodsky J. L., Zuiderweg E. R., Gestwicki J. E. (2010) Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem. Biol. 5, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burkholder W. F., Zhao X., Zhu X., Hendrickson W. A., Gragerov A., Gottesman M. E. (1996) Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc. Natl. Acad. Sci. U.S.A. 93, 10632–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mossessova E., Lima C. D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 51. Liu Q., Hendrickson W. A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Q., Krzewska J., Liberek K., Craig E. A. (2001) Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 276, 6112–6118 [DOI] [PubMed] [Google Scholar]
- 53. Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 54. Easton D. P., Kaneko Y., Subjeck J. R. (2000) The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5, 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shaner L., Morano K. A. (2007) All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hendrickson W. A., Liu Q. (2008) Exchange we can believe in. Structure 16, 1153–1155 [DOI] [PubMed] [Google Scholar]
- 59. Georgopoulos C., Tilly K., Drahos D., Hendrix R. (1982) The B66.0 protein of Escherichia coli is the product of the dnaK+ gene. J. Bacteriol. 149, 1175–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Craig E. A. (1985) The heat shock response. CRC Crit. Rev. Biochem. 18, 239–280 [DOI] [PubMed] [Google Scholar]
- 61. Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., Zuiderweg E. R. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303 [DOI] [PubMed] [Google Scholar]
- 62. Fernández-Sáiz V., Moro F., Arizmendi J. M., Acebrón S. P., Muga A. (2006) Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J. Biol. Chem. 281, 7479–7488 [DOI] [PubMed] [Google Scholar]
- 63. Miao B., Davis J. E., Craig E. A. (1997) Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J. Mol. Biol. 265, 541–552 [DOI] [PubMed] [Google Scholar]
- 64. Brehmer D., Rüdiger S., Gässler C. S., Klostermeier D., Packschies L., Reinstein J., Mayer M. P., Bukau B. (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8, 427–432 [DOI] [PubMed] [Google Scholar]
- 65. Kityk R., Kopp J., Sinning I., Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 [DOI] [PubMed] [Google Scholar]
- 66. Suh W. C., Lu C. Z., Gross C. A. (1999) Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 274, 30534–30539 [DOI] [PubMed] [Google Scholar]
- 67. Mayer M. P., Laufen T., Paal K., McCarty J. S., Bukau B. (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289, 1131–1144 [DOI] [PubMed] [Google Scholar]
- 68. Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jiang R. F., Greener T., Barouch W., Greene L., Eisenberg E. (1997) Interaction of auxilin with the molecular chaperone, Hsc70. J. Biol. Chem. 272, 6141–6145 [DOI] [PubMed] [Google Scholar]
- 70. Ungewickell E., Ungewickell H., Holstein S. E., Lindner R., Prasad K., Barouch W., Martin B., Greene L. E., Eisenberg E. (1995) Role of auxilin in uncoating clathrin-coated vesicles. Nature 378, 632–635 [DOI] [PubMed] [Google Scholar]
- 71. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]