Background: The mechanism of HCC resistant to Fas-mediated apoptosis is not clearly understood.
Results: ORP8 triggered HCC cell apoptosis via relocation of cytoplasmic Fas to the cell plasma membrane and FasL up-regulation.
Conclusion: ORP8 increases sensitivity of HCC cells to Fas-mediated apoptosis.
Significance: New insights reveal that ORP8 may serve as a potential target for HCC therapy.
Keywords: Apoptosis, Endoplasmic Reticulum Stress (ER Stress), Liver Cancer, MicroRNA (miRNA), Protein Translocation, Fas, ORP8
Abstract
Human hepatoma (HCC) has been reported to be strongly resistant to Fas-mediated apoptosis. However, the underlying mechanisms are poorly understood. In this study the function of oxysterol-binding protein-related protein 8 (ORP8) in human hepatoma cells apoptosis was assessed. We found that ORP8 is down-regulated, whereas miR-143, which controls ORP8 expression, is up-regulated in clinical HCC tissues as compared with liver tissue from healthy subjects. ORP8 overexpression triggered apoptosis in primary HCC cells and cell lines, which coincided with a relocation of cytoplasmic Fas to the cell plasma membrane and FasL up-regulation. Co-culture of HepG2 cells or primary HCC cells with Jurkat T-cells or T-cells, respectively, provided further evidence that ORP8 increases HCC cell sensitivity to Fas-mediated apoptosis. ORP8-induced Fas translocation is p53-dependent, and FasL was induced upon ORP8 overexpression via the endoplasmic reticulum stress response. Moreover, ORP8 overexpression and miR-143 inhibition markedly inhibited tumor growth in a HepG2 cell xenograft model. These results indicate that ORP8 induces HCC cell apoptosis through the Fas/FasL pathway. The role of ORP8 in Fas translocation to the plasma membrane and its down-regulation by miR-143 offer a putative mechanistic explanation for HCC resistance to apoptosis. ORP8 may be a potential target for HCC therapy.
Introduction
Apoptosis plays a key role in various forms of physiological cell death and disease states (1). Resistance to apoptosis is a characteristic of malignant cells. Thus, one of the main goals for cancer therapy is to overcome of tumor cell resistance to apoptosis (2). The best-characterized apoptotic pathway is that mediated by the surface molecule Fas/Fas ligand (FasL)3 system. Fas/APO-1 (or CD95) is a 45-kDa cell surface glycoprotein that transduces a cellular death signal for apoptosis in various normal and neoplastic cells (3). Engagement of Fas by FasL or anti-Fas antibodies results in trimerization of Fas and recruitment of the adapter protein Fas-associating protein with death domain (FADD) followed by caspase activation and apoptosis (1). The Fas/FasL route is one of the key apoptotic pathways in the liver (4, 5). In normal human hepatocytes, the Fas receptor is constitutively expressed, and Fas-mediated apoptosis may occur in an autocrine or paracrine fashion via binding of soluble FasL (6). However, it is important to note that not all Fas-bearing cells are susceptible to Fas-mediated killing. Unlike normal hepatocytes, which are susceptible to Fas-mediated apoptosis, human hepatoma cells have been reported to be strongly resistant to Fas-mediated apoptosis (7, 8). A low level of Fas expression was implicated in hepatoma treatment resistance (9). However, the resistance to apoptosis remains unchanged in Fas-positive hepatocellular carcinoma (HCC) cell lines (8). The majority of noncancerous specimens expressed Fas both on the cell surface and in the cytoplasm, whereas the majority of HCC specimens expressed Fas only in the cytoplasm, with the cell apoptosis rate being higher in tissue with surface Fas expression than that with cytoplasmic expression only (10). This suggests the importance of the subcellular location of Fas in HCC apoptosis.
ORP8 is a member of the oxysterol-binding protein-related protein family that contains a single C-terminal transmembrane domain targeting the protein to the endoplasmic reticulum (ER). Our previous study indicated that ORP8 decreases cholesterol efflux in macrophages by suppressing ABCA1 expression, implying that it may play a role in the development of atherosclerotic lesions (11). In hepatic cells ORP8 functions as a negative regulator of intracellular cholesterol (12). Other roles have been suggested for ORP8, including the inhibition of cell migration through interaction with nucleoporin Nup62 (13) and mediation of oxysterol interference of the cell cycle through interaction with Astrin/SPAG5 in HepG2 cells (14). ORP8 has previously been reported to be regulated by miR-143 in mice (15). Quantitative mass spectrometric analysis of hepatic protein expression in mice overexpressing miR-143 revealed a miR-143-dependent down-regulation of ORP8. Reduced ORP8 expression in cultured liver cells impaired the ability of insulin to induce AKT activation, revealing a mechanism of AKT regulation that depends on ORP8 (15).
In the present study we show that the expression of ORP8 protein is significantly down-regulated in HCC tissues, whereas miR-143 is up-regulated as compared with normal liver tissues. The relationship between ORP8 expression/miR-143 inhibition and HCC cell apoptosis and its role in a HepG2 xenograft tumor model was assessed. The results suggest that ORP8 triggers HCC cells apoptosis via increasing Fas plasma membrane localization and FasL expression and may thus be a potential target for HCC therapy.
EXPERIMENTAL PROCEDURES
cDNA Constructs and Transfection
Human ORP8 cDNA (accession number NM_001003712), ORP5 cDNA (accession number NM_020896), and truncated ORP8 cDNA (ORP8–1 without ORD, ORP8–2 without PHD) were inserted into the XbaI site of pcDNA4HisMaxC (Invitrogen) to obtain constructs expressing proteins fused with an N-terminal Xpress epitope tag. Transient transfections of cultured cells were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For primary HCC cell transfection, cells were electroporated by using of Neon® Transfection System (Life Technology).
Antibodies and Other Reagents
Rabbit antibodies against human ORP8 were produced and affinity-purified. Anti-caspase-8, anti-cleaved caspase-8, anti-caspase-3, anti-cleaved caspase-3, anti-FasL, anti-Chop, and anti-p65 polyclonal antibody were purchased from Cell Signaling Technology. Anti-phospho-PERK, anti-PERK, anti-phospho-eIF2α, and anti-Fas polyclonal antibody was purchased from Santa Cruz Biotechnology. Anti-Xbp-1s was purchased from BioLegend. Anti-ATF4 and Anti-β-actin monoclonal antibody were purchased from Proteintech. XpressTM monoclonal antibody, AlexaFluor-488 goat anti-mouse IgG, AlexaFluor-543 goat anti-rabbit IgG, and oligonucleotide primers were obtained from Invitrogen. p53 inhibitor PFT-a was from Sellect.
Tissue Specimens and Cell Lines
This study was approved by the institutional ethics committee of Jinan University and was performed in accordance with the Declaration of Helsinki. After patients scheduled to undergo surgery provided written informed consent, explanted hepatoma tissue was obtained during the surgery. Healthy liver tissues were obtained from donors in the Adult-to-Adult Living Donor Liver Transplantation Program who had provided written informed consent. Primary liver cells, HepG2 cells, and Huh7 cells were maintained in DMEM containing 10% FBS, 100 units/ml of penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2. LO2 cells and Jurkat T-cells were cultured in RPMI 1640 containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2. Fresh leukocytes were isolated from human peripheral blood of healthy donors after obtaining written informed consent and maintained in RPMI 1640 containing 10% FBS.
Quantitative Real-time PCR
Total RNA was isolated with TRIzol reagent according to the manufacturer's instructions. For mRNA quantitative PCR, RNA samples were reverse-transcribed using random hexamer primers in the presence of RNase inhibitor (Takara Bio, Shiga, Japan). For miRNA quantitative PCR, RNA samples were directly reverse-transcribed using the One Step PrimeScript miRNA cDNA synthesis kit (Takara Bio). qRT-PCR was performed with SYBR Premex EX Taq (Takara Bio) using the 7300 Sequence Detection System (Life Technologies/Applied Biosystems, Carlsbad, CA). Primers used are shown in Table 1. Relative quantification analysis was performed using the ΔΔCt method, with actin and RUN48 as endogenous controls for mRNA and miRNA, respectively. Relative gene expression was presented as the ratio of the target gene to reference control.
TABLE 1.
The oligonucleotide primers used
qPCR, quantitative PCR.
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| ORP8 qPCR | GAACAGGGAGATTTTGAATCA | TCCTGTGAGTGGATCAAGTTC |
| FasL qPCR | TGCCTTGGTAGGATTGGGC | GCTGGTAGACTCTCGGAGTTC |
| Fas qPCR | GCTGGGCATCTGGACCCTCCTACCT | CAGTCACTTGGGCATTAACACTT |
| Chop qPCR | GCCTTTCTCCTTTGGGACACTGTCCAGC | CTCGGCGAGTCGCCTCTACTTCCC |
| Bip qPCR | CCTGGGTGGCGGAACCTTCGATGTG | CTGGACGGGCTTCATAGTAGACCGG |
| Actin qPCR | GGCATCCTCACCCTGAAGTA | AGGTGTGGTGCCAGATTTTC |
| miR-143–3p qPCR | GCCCTGAGATGAAGCACTGTAGCTC | Provided by kit |
| RUN48 qPCR | AGTGATGATGACCCCAGGTAACTC | Provided by kit |
miR-143 Mimic and Inhibitor Transfection
HepG2 and Huh7 cells (5 × 104 cells/500 μl/well) were cultured in a 24-well plate and transfected with miR-143 inhibitor (100 nm) (Qiagen, Shanghai, China) or control RNA, and LO2 cells (5 × 104 cells/500 μl/well) were transfected with miR-143 mimic (100 nm; Qiagen) or control RNA by using Lipofectamine 2000 (Invitrogen) for 48 h. The cells were collected and lysed for Western blot analysis.
Western Blot Analysis
Cellular total protein samples were mixed with sample loading buffer, boiled for 10 min, and subjected to SDS-PAGE. Western blot analysis was conducted as described elsewhere (11). Proteins were quantified by densitometry using WCIF ImageJ, and the data were normalized to β-actin.
RNA Interference
One day before transfection, cells were seeded on 6-well plates at 30–50% confluency and then transfected with siORP8, siPERK, sip65, or control non-targeting siRNA (siNT) for 72 h (siORP8: GAGUGGUCUUGCAAAUUAUdTdT; siNT: UAGCGACUAAACACAUCAAdTdT; siPERK and sip65 were purchased from SantaCruz) by using Lipofectamine 2000 (Invitrogen).
Cell Apoptosis Assays
Propidium iodide staining followed by flow cytometry was used to detect cells with sub-G1 DNA content. Cells were trypsinized, washed twice with PBS, and fixed in 70% ethanol at a density of 0.5 to 1 × 106 cells/ml. The fixed cells were resuspended in propidium iodide staining solution containing 50 μg/ml propidium iodide and incubated at room temperature for 30 min. Fluorescence was measured on a BD Biosciences FACSCalibur instrument. Cells with sub-G1 DNA content were quantified using Cell QUEST software.
Immunohistochemical Analysis
Human hepatoma tissue was fixed with paraformaldehyde and embedded in paraffin. Paraffin-embedded samples were sectioned into 4-μm thick slices, dewaxed, rehydrated, and then pretreated by boiling for 10 min in 0.01 m citrate buffer (pH 6.0) in a microwave oven (750 watts). Sections were incubated overnight with primary antibodies at 4 °C. Subsequently, the sections were incubated with biotinylated secondary antibodies for 30 min followed by 30 min of incubation with ABComplex/HRP. Finally, the sections were stained with 3,30-diaminobenzidine (DAB) and counterstained with hematoxylin. The slides were mounted under glass coverslips and analyzed by light microscopy.
Primary HCC Cell Isolation
Liver tissue was minced (1–4 mm3), washed 3 times in PBS, and transferred to a small flask containing 20 ml of prewarmed 0.25% trypsin solution. After 10 min of incubation at 37 °C, the trypsin was decanted, and 10 ml of fresh prewarmed trypsin was added. The tissue was then triturated 10–15 times with a pipette having a bore of about 3 mm in diameter. The large fragments were allowed to settle, and the supernatant containing single cells was collected and centrifuged at 600 rpm for 3 min at room temperature; the pellet was resuspended and cultured as described above.
Immunofluorescence Microscopy
Cells were seeded onto coverslips and fixed with 4% paraformaldehyde for 30 min at room temperature followed by permeabilization for 5 min with 0.1% Triton X-100 and blocked for 30 min with 10% FBS at room temperature. Cells were then incubated overnight with primary antibodies in 5% FBS at 4 °C. After washing 3 times (10 min each) with PBS, cells were incubated for 30 min with secondary antibody conjugated with fluorochromes at 37 °C. The specimens were analyzed using a Zeiss LSM 510 Meta laser scanning confocal microscope system. For immunofluorescence staining, the sections were blocked with 1% BSA in PBS after microwave antigen retrieval and incubated with the Fas antibody diluted in 1% (w/v) BSA in PBS for 45 min at room temperature. After washing in PBS, the sections were incubated with fluorescently labeled secondary antibody diluted in 1% (w/v) BSA in PBS for 30 min at 37 °C. The sections were mounted in fluorescence mounting medium (Invitrogen) and analyzed with a laser scanning confocal microscope (Zeiss LSM 510 Meta).
RT2 Profiler PCR Array
RT2 ProfilerTM PCR arrays for human apoptosis (PAHS-012A-2) were purchased from SuperArray Biosciences (Shanghai, China), and cDNA was synthesized according to the manufacturer's instructions. Real-time PCR was performed using ABI7300 Sequence Detection System with SYBR Green dye mix in a 96-well RT2 ProfilerTM PCR array containing 84 key genes involved in the apoptosis pathway. The PCR reaction was run as follows: one 10-min cycle at 95 °C followed by 40 cycles of 15 s each at 95 °C and 1 min at 60 °C. Threshold cycle (Ct) data generated from the real-time instrument were analyzed using the SuperArray Biosciences web portal software.
Detection of Fas Localization in Plasma Membrane and Total Fas Expression by Flow Cytometry
For analysis of surface Fas, cells with or without ORP8 overexpression for 36 h were detached using cell dissociation solution (Sigma) and stained with FITC-labeled anti-Fas antibody (BioLegend). For detection of total Fas expression, cells were permeabilized with detergent before staining as described above. Flow cytometric analysis was performed using a BD Biosciences FACSCalibur instrument.
Isolation of Nuclear and Plasma Membranes
Nuclear fractions were isolated using a nuclear/cytosol fractionation kit (Biovision, Milpitas, CA) according to the manufacturer's instructions. For plasma membrane isolation, cells were collected and resuspended in 0.2 mm EDTA in 1 mm NaHCO3 in an approximate ratio of 1 ml per 108 cells and incubated on ice for 30 min to swell the cells. Cells were then homogenized using a Dounce homogenizer. The homogenates were centrifuged for 10 min at 175 × g at 4 °C to remove unbroken cells and nuclei, and the supernatant was centrifuged a second time at 25,000 × g for 30 min at 4 °C to prepare a plasma membrane-enriched microsome fraction. The supernatant was discarded, and the pellets were resuspended in 0.2 m potassium phosphate buffer (pH 7.2). The resuspended membranes then were loaded onto a two-phase system with a polymer mixture containing 6.6% Dextran T500 (GE Healthcare), 6.6% (w/w) poly(ethylene glycol) 3350 (Fisher), and 0.2 m potassium phosphate (pH 7.2). The phases were separated by centrifugation at 1150 × g for 5 min at 4 °C. The upper phase, containing primarily plasma membranes, was collected.
In Vivo Animal Studies
All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86–23, revised 1985) and according to our institutional ethical guidelines for animal experiments. Four-week-old male Balb/C athymic (nu/nu) nude mice were purchased from the animal center of Guangzhou Province (Guangzhou, China) and kept under pathogen-free conditions in the Laboratory Animal Center, Jinan University. The animals were adapted to new conditions for 1 week before the experiments.
Tumor Induction and Measurement
To examine the effect of ORP8 overexpression on tumor growth, aliquots (107 cells/200 μl) of HepG2 cells in PBS were injected subcutaneously into the left and right rear flanks of the same female BALB/c athymic nude mouse at 5 weeks of age. At 19 days after inoculation, tumor volumes were determined and then again every 3 days after intratumoral plasmid or lentivirus treatment (see below). Two bisecting diameters of each tumor were measured with calipers. The tumor volume was estimated according to the following formula: tumor volume (mm3) = ab2/2, where a = the larger diameter and b = smaller diameter.
Intratumoral Plasmid or Lentivirus Treatment
Linear polyethyleneimine was used to achieve efficient in vivo gene transfer into tumor cells. The tumor-bearing mice were injected intratumorally with DNA-linear polyethyleneimine complexes of pcDNA4HismaxC-ORP8 and in vivo-jetPEITM in right flank tumors and complexes of pcDNA4HismaxC and in vivo-jetPEITM in the left flank tumors according to the manufacturer's instructions. The injection was performed every 3 days. The first injection was performed at 19 days after tumor inoculation, and the last injection was performed at 28 days after inoculation (i.e. 3 times over 9 days). The mice were sacrificed on day 28, and the tumors were harvested and weighed. For lentivirus treatment, tumor-bearing mice were injected intratumorally with lenti-shNT or lenti-miR-143 inhibitor at 19 days after tumor inoculation. The mice were sacrificed on day 28, and the tumors were harvested and weighed.
TUNEL Assay
The in situ cell death detection kit (POD; Roche Diagnostics) was used. Four-micrometer histologic sections were cut and processed for TUNEL staining. The slides were stained with 3,3′-diaminobenzidine substrate, counterstained with hematoxylin, mounted under glass coverslip, and analyzed by light microscopy.
Statistical Analyses
The data are expressed as the mean ± S.D. Differences between groups were assessed by one-way analysis of variance or Kruskall-Wallis when data were not normally distributed (SigmaStat Software Version 3.5). For groups with small n values or when the values were not normally distributed, the non-parametric Mann-Whitney U test (SPSS10.0 software package) was used.
RESULTS
ORP8 Protein Is Down-regulated in Human HCC Tissues and Cell Lines
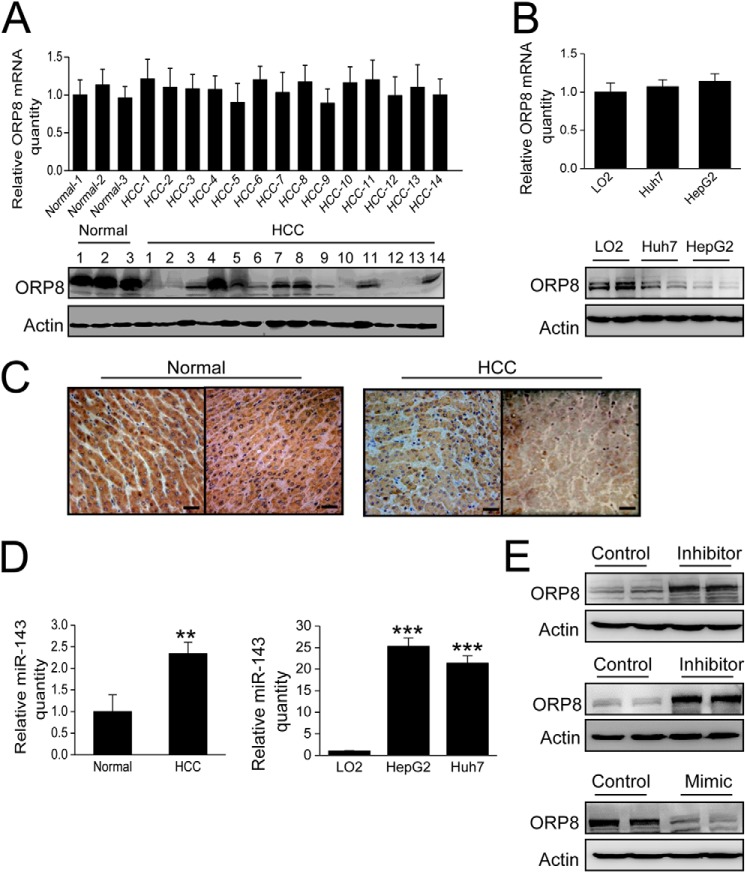
Our previous study demonstrated that ORP8 overexpression in mouse liver significantly reduces the plasma total cholesterol level in vivo (12). Because aberrant elevation of the cholesterol level is associated with various types of cancers (16), a total of 67 clinical HCC samples were analyzed for ORP8 expression for both mRNA and protein expression by qRT-PCR and Western blot. The results indicated no difference in the mRNA expression levels between normal and HCC tissues (Fig. 1A, upper panel, 3 normal liver tissues versus 14 clinical HCC samples are shown). However, compared with the normal liver tissues, ORP8 protein expression was significantly down-regulated in 13 of 14 HCC tissues (Fig. 1A, lower panel). A similar difference in ORP8 expression was observed between the non-cancerous LO2 hepatocyte cell line and the Huh7 and HepG2 HCC cell lines (Fig. 1B). Immunohistochemical staining confirmed the lower level of ORP8 protein detected in HCC tissue as compared with the healthy control (Fig. 1C).
FIGURE 1.
ORP8 protein is down-regulated in HCC. A, qRT-PCR and Western blot analysis of ORP8 mRNA (upper panel) and protein (lower panel) expression in normal and HCC human liver tissue specimens. B, qRT-PCR and Western blot analysis of ORP8 mRNA (upper panel) and protein (lower panel) expression in LO2, Huh7, and HepG2 cell lines. C, the pattern of expression of ORP8 protein in normal and HCC liver tissues by immunohistochemical staining. ORP8 is represented by a dark brown staining pattern (20×, counterstained with hematoxylin. Scale bars, 10 μm). D, summarized data for the relative expression of miR-143 in 3 normal and 14 HCC tissues (left panel), LO2, HepG2 and Huh7 cell lines (right panel). The data were normalized to RUN48 microRNA. E, upper panel, Western blot analysis of ORP8 protein in HepG2 cells transfected with control or an inhibitor of miR-143. Middle panel, Western blot analysis of ORP8 protein in Huh7 cells transfected with control or an inhibitor of miR-143. Lower panel, Western blot analysis of ORP8 protein in LO2 cells transfected with control or mimic of miR-143. The data represent mean ± S.D. from three individual experiments (n = 3; **, p < 0.01; ***, p < 0.001).
The above observation indicated that down-regulation of ORP8 occurs at a post-transcriptional level. ORP8 has previously been reported to be a target of miR-143 in mice (15), and miR-143 was observed to be up-regulated in HCC (17). We, therefore, explored whether the down-regulation of ORP8 in HCC is due to miR-143 deregulation. In contrast to down-regulation of ORP8 protein in HCC, miR-143 was significantly up-regulated compared with normal liver tissue (Fig. 1D, left panel). Similarly, increased miR-143 expression was observed in the HCC cell lines HepG2 and Huh7 but not in the non-cancerous liver cell line LO2 (Fig. 1D, right panel). When we treated HepG2 and Huh7 cells with a miR-143 inhibitor, ORP8 protein expression significantly increased (Fig. 1E, upper and middle panels), whereas treatment with a miR-143 mimic potently inhibited ORP8 expression in LO2 cells (Fig. 1E, lower panel). These results confirmed that ORP8 is a target of miR-143 in human liver cells and that high levels of miR-143 lead to reduced expression of ORP8 protein in HCC.
ORP8 Induces HCC Cell Apoptosis via the Fas-FasL Pathway
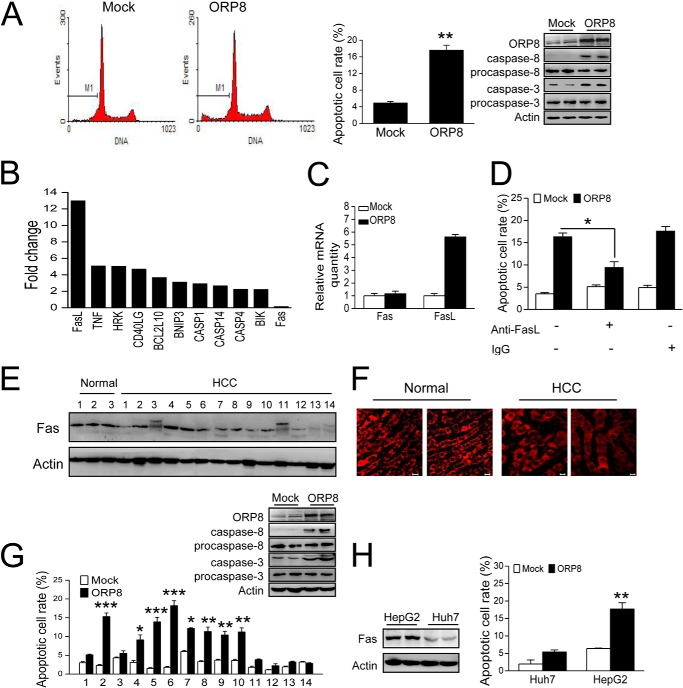
To investigate the role of ORP8 in HCC, we transfected HepG2 cells with ORP8 cDNA. Increased apoptosis was detected in cells overexpressing ORP8 (Fig. 2A). We also analyzed the cleaved caspase-8 and caspase-3 levels by Western blot. Cleavage was clearly enhanced in ORP8-overexpressing cells (Fig. 2A).
FIGURE 2.
ORP8 induces HCC cell apoptosis via the Fas/FasL pathway. A, HepG2 cells were transfected with ORP8 cDNA or empty vector for 36 h. Left panels, apoptosis was evaluated by flow cytometric analysis of DNA content after staining with propidium iodide. Right panel, mean cellular apoptosis rate and Western blot assay showing that caspase-8 and caspase-3 were activated in cells overexpressing ORP8. B, up-regulated genes with a -fold change >2 in HepG2 cells overexpressing ORP8 as compared with controls. C, qRT-PCR analysis of fas and fasl gene expression in HepG2 cells with or without ORP8 overexpression. D, HepG2 cells were transfected with ORP8 cDNA or empty vector for 12 h and then treated with or without 10 μg/ml of anti-FasL neutralizing antibody or control IgG for 24 h with evaluation of the apoptotic rate. E, Western blot analysis of Fas expression levels in normal and HCC liver tissues. F, immunofluorescence analysis indicating Fas localization in HCC and normal liver tissues. Scale bars, 10 μm. G, cellular apoptotic rate in primary HCC cells isolated from patient tissue samples upon ORP8 overexpression for 36 h; Western blots showing that caspase-8 and caspase-3 were activated in ORP8-overexpressing HCC-2 cells. H, Western blot analysis of Fas expression levels in HepG2 and Huh7 cells and the cellular apoptotic rate in cells upon ORP8 overexpression for 36 h are shown. The data represent the mean ± S.D. from three individual experiments (n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
To better understand the molecular apoptotic signature induced by ORP8, we first profiled 84 apoptosis-related mRNAs using a PCR array. The results identified a total of 18 genes that were differentially expressed (16 up-regulated and 2 down-regulated) in cells overexpressing ORP8 as compared with the control based on the cutoff value of 2. The up-regulated genes with a -fold change of >2 are shown (Fig. 2B). Among the identified genes, FasL expression increased the most, a result confirmed by qRT-PCR (Fig. 2C). The Fas/FasL route is one of the key apoptotic pathways in the liver (4, 5). HCC cells have been reported to be strongly resistant to Fas-mediated apoptosis for low expression of Fas (7, 8). Based on the results from the PCR array analysis, we next determined whether ORP8-induced cell apoptosis occurs in tandem with changes in Fas/FasL expression. Fas expression did not change in HepG2 cells overexpressing ORP8 (Fig. 2, B and C), whereas treatment of HepG2 cells with anti-FasL neutralizing antibody significantly reduced the apoptosis induced by ORP8 overexpression (Fig. 2D), indicating that Fas/FasL pathway was involved in HepG2 cell apoptosis induced by ORP8 overexpression.
The Fas expression and cell surface level are relatively low in hepatoma (18, 19). In our study some of HCC samples (HCC-2, HCC-4, HCC-5, and HCC-6) displayed Fas expression at a level similar to normal liver tissues, but it was much lower or undetectable in the other HCC samples (Fig. 2E). Immunofluorescence staining indicated that most Fas localized in the cytoplasm of HCC samples (Fig. 2F). Cells bearing higher Fas expression showed a significantly increased apoptotic rate, whereas cells with lower Fas expression displayed no significant change in apoptosis upon ORP8 overexpression (Fig. 2G). Moreover, caspase-8 and caspase-3 was activated in HCC-2 cells with ORP8 overexpression (Fig. 2G). Huh7 cells expressed relatively lower levels of Fas as compared with HepG2 cells (Fig. 2H, left panel). Both cell lines showed an increased cell death rate, and Huh7 displayed a blunter apoptotic response upon ORP8 overexpression (Fig. 2H, right panel), indicating that ORP8-induced apoptosis is positively related to Fas level.
ORP8 Increases Sensitivity to Fas-mediated Apoptosis by Shifting Fas to the Plasma Membrane in a p53-dependent Manner
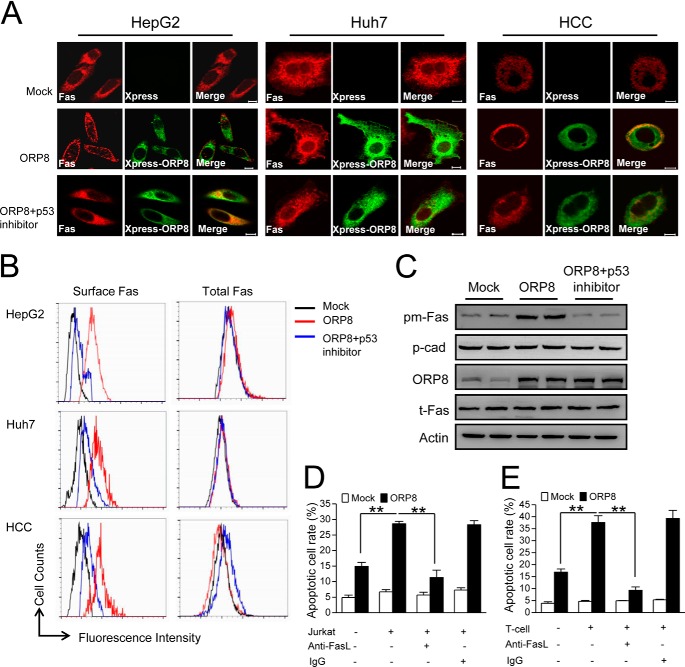
To further confirm the mechanism by which ORP8 functions in HCC cell apoptosis, we analyzed both total and surface Fas expression. The total Fas protein level did not change in ORP8 overexpressing cells (Fig. 3, B and C). In mock-transfected HepG2, Huh7, and primary HCC cells (HCC-2), Fas was localized in the cytoplasm, whereas in cells overexpressing ORP8, Fas was clearly present on the cell surface, indicating transport of Fas from the cytoplasm to the plasma membrane (Fig. 3A). By flow cytometry using anti-Fas antibody, we further confirmed more Fas plasma membrane localization of ORP8 overexpression cells (Fig. 3B). Cell fractionation to isolate plasma membranes followed by Western blot analysis further confirmed the relocation of Fas to the plasma membrane of HepG2 cells overexpressing ORP8 (Fig. 3C). To figure out the downstream regulatory factor involved in ORP8-induced Fas transport, we focused on the p53 protein, which was reported to increase surface Fas expression by Fas transport from cytoplasmic stores (20). As shown in Fig. 3, A–C, p53 inhibition abolished the Fas transport induced by ORP8 overexpression. These data indicate that ORP8 promoted plasma membrane Fas trafficking through the p53.
FIGURE 3.
ORP8 increases sensitivity to Fas-mediated apoptosis by shifting Fas to the plasma membrane. A, confocal microscopy analysis indicating Fas (red staining) localization in HepG2 cells, Huh7 cells, and HCC cells (HCC-2). Fluorescence images were acquired with or without ORP8 (green straining) overexpression for 36 h or ORP8 overexpression plus 50 μm p53 inhibitor PFT-a. Scale bars, 10 μm. B, ORP8 overexpression induced cell surface and total Fas as determined by flow cytometry. C, Western blot analysis of Fas protein in plasma membrane fraction (pm-Fas) and total lysis (t-Fas) from HepG2 cells. P-cadherin (p-cad) was employed as a loading control for plasma membrane fractions. D, HepG2 cells were transfected with ORP8 cDNA or empty vector for 12 h then treated with or without 10 μg/ml anti-FasL neutralizing antibody in the presence or absence of Jurkat T-cells for 24 h, after which apoptosis were evaluated. E, primary HCC cells (HCC-2) were transfected with ORP8 cDNA or empty vector for 12 h and then treated with or without 10 μg/ml of anti-FasL neutralizing antibody in the presence or absence of T-cells for 24 h, and apoptosis was evaluated by flow cytometric analysis. The data represent the mean ± S.D. from three individual experiments (n = 3; **, p < 0.01).
Fas ligand expressed in T-lymphocytes can engage Fas on the cell membrane to trigger target cell apoptosis, which is important for successful immune surveillance and apoptosis of tumor cells (21, 22). Given that ORP8 increases the plasma membrane localization of Fas, it is possible that ORP8 may enhance susceptibility of HCC cells to apoptosis as mediated by T-cells. Accordingly, we co-cultured HepG2 cells with Jurkat T-cells. Co-culture with mock-transfected HepG2 cells did not trigger cell apoptosis. However, apoptosis significantly increased in ORP8-transfected HepG2 cells (Fig. 3D). Moreover, treatment of ORP8-transfected HepG2 cells with anti-FasL neutralizing antibody drastically reduced cell apoptosis (Fig. 3D). These results indicated that ORP8 increases the susceptibility of HepG2 cells to apoptosis via the Fas/FasL pathway. We further co-cultured T-cells with primary HCC cells (HCC-2). Like HepG2 cells, apoptosis in ORP8-transfected HCC cells was significantly greater compared with mock-transfected cells, and the apoptosis could be neutralized by anti-FasL neutralizing antibody (Fig. 3E).
ORP8 Induces ER Stress to Invoke NFκB Activation for FasL Induction
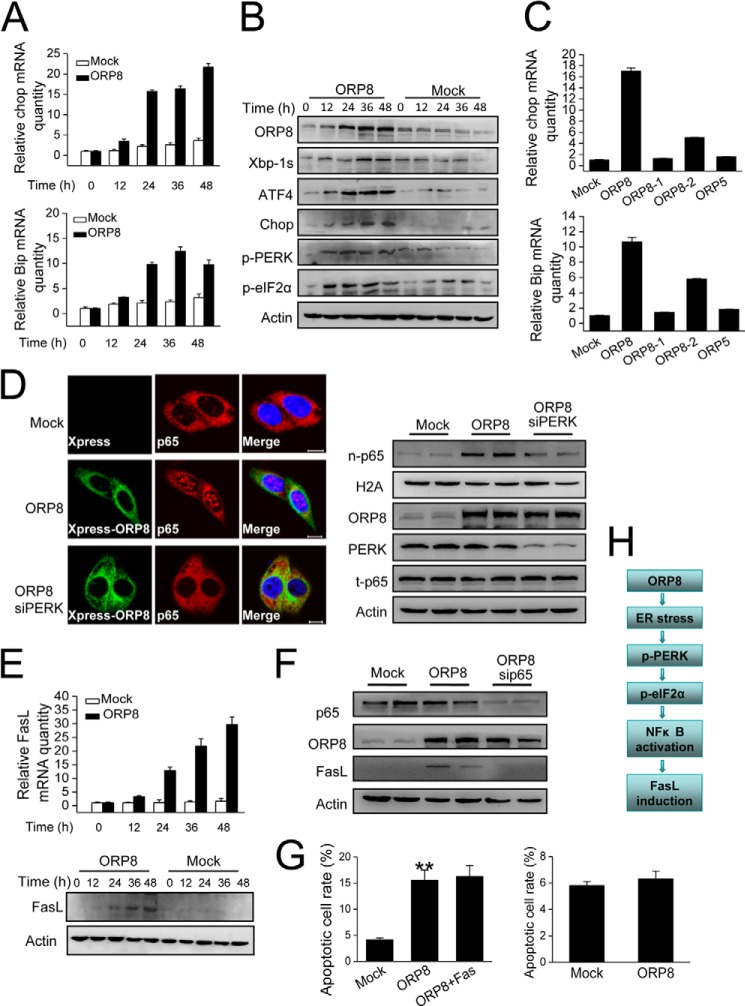
We next focused on the cell signaling events upstream of FasL in HepG2 cells. FasL expression is reported to be regulated by the transcription factor NFκB, which is induced by ER stress (23–25). To determine whether ER stress was involved in ORP8-induced FasL up-regulation, we first examined the expression of Chop and Bip mRNAs, central components involved in ER stress responses (26). qRT-PCR revealed Chop and Bip mRNAs were robustly induced in HepG2 cells after transfection with ORP8 cDNA as compared with mock-treated cells (Fig. 4A). We also examined the protein expression of ER stress markers by Western blot analysis, including Xbp-1s (the form of Xbp-1 derived from its spliced mRNA after ER stress induction), ATF4, Chop, phospho-PERK, and phospho-eIF2α. All of these markers significantly increased in a time-dependent manner over 12–48 h of ORP8 overexpression whereas expression did not change in cells transfected with the empty vector (Fig. 4B). Because ORP8 is localized in the ER membranes (11), we controlled for the possibility that in cells overexpressing ORP8, general ER protein overload induced ER stress; transfection of HepG2 cells with ORP5, another ORP localized in the ER (27), and analysis of Chop and Bip mRNA expression showed no detectable ER stress response (Fig. 4C). A truncated ORP8 cDNA lacking the PHD or ORD domains also failed to induce ER stress (Fig. 4C), further supporting the notion that ORP8 overexpression induces an ER stress response in a specific manner in HepG2 cells.
FIGURE 4.
Mechanistic analysis of ORP8-induced fasl up-regulation in HepG2 cells. A, HepG2 cells were transfected with ORP8 cDNA or empty vector for the indicated times, and relative Chop and Bip mRNA levels were measured by qRT-PCR. B, the ER stress markers indicated were analyzed by Western blot. β-Actin was used as a loading control. C, HepG2 cells were transfected for 18 h with empty vector, full-length ORP8 cDNA (ORP8), ORP8-truncated constructs (ORP8–1 without ORD, ORP8–2 without PHD), or ORP5 cDNA. The mRNA levels of Chop and Bip were determined with qRT-PCR. D, confocal microscopy analysis indicated the localization of the NFκB subunit p65 (red staining) in HepG2 cells. Scale bars, 10 μm. Western blot analysis demonstrated increased p65 protein levels in nuclear fractions (n-p65) of HepG2 cells with ORP8 overexpression; t-p65 indicated the total p65 protein; H2A was used as a loading control for nuclear fractions. E, qRT-PCR (upper panel) and Western blot (lower panel) analyses showed FasL was up-regulated in ORP8-overexpressing HepG2 cells. F, Western blot analysis was performed to determine the protein level of FasL in ORP8 overexpression cells or ORP8 overexpression cells with p65 knockdown. G, left panel, apoptotic cell rate in HepG2 cells upon ORP8 overexpression and in cells with ORP8 plus Fas co-overexpression for 36 h. Right panel, apoptotic cell rate in Jurkat T-cells upon co-culture with HepG2 cells overexpressing ORP8. The data represent the mean ± S.D. from three individual experiments (n = 3; **, p < 0.01). H, flow chart showing the signaling pathway of FasL up-regulation induced by ORP8 overexpression.
Phosphorylation of eIF2α by phosphorylated PERK during ER stress is essential for induction of NFκB transcriptional activity. Upon activation, the heterodimeric p65 subunit translocates from the cytoplasm to the nucleus to regulate gene expression, including that of FasL (28, 29). Cellular localization of NFκB in response to ER stress induced by ORP8 was visualized by immunofluorescence microscopy. Cytoplasmic localization of p65 was readily visible in control cells, whereas in ORP8-overexpressing cells p65 was localized in the nucleus (Fig. 4D). Also cell fractionation followed by Western blot analysis revealed that p65 level in the nucleus was increased in the ORP8-overexpressing cells (Fig. 4D). However, the p65 nucleus localization was abrogated by PERK knockdown (Fig. 4D), indicating that PERK is required for NFκB activation invoked by ORP8 overexpression. Consistent with these results, overexpression of ORP8 resulted in a significant increase in FasL mRNA and protein expression (Fig. 4E), which was inhibited by p65 knockdown (Fig. 4F). Taken together, these data are evidence that ORP8 induced HepG2 cell apoptosis through Fas relocalization and FasL up-regulation. Furthermore, Fas overexpression in ORP8-overexpressing HepG2 cells did not induce a further increase the apoptotic cell rate (Fig. 4G, left panel), suggesting that the amount of FasL induced by ORP8 overexpression was not enough to form Fas-FasL complexes to trigger more cell apoptosis. Accordingly, co-culture with ORP8-transfected HepG2 cells did not trigger Jurkat T-cell apoptosis (Fig. 4G, right panel).
Intratumoral Administration of ORP8 Inhibits Tumor Growth in Vivo
Considering the role of ORP8 in inducing apoptosis of HepG2 cells, we explored ORP8 overexpression as a potential approach for hepatoma treatment by using a HepG2 xenograft model in BALB/c nude mice. At 19 days post-inoculation of HepG2 cells, tumor-bearing mice were injected intratumorally with complexes of ORP8-cDNA/in vivo-jetPEITM (ORP8 group, 8 mice) or complexes of empty vector/in vivo-jetPEITM (mock group, 8 mice). To confirm that the two groups were comparable with regard to tumor size at baseline, we determined tumor volume before the first intratumoral gene delivery, finding no significant difference between groups (mock group = 325.2 mm3; ORP8 group = 347.1 mm3, p = 0.93). Tumor growth was recorded from the time of first injection. ORP8 overexpression led to a marked reduction of tumor growth (Fig. 5A), with successful delivery of ORP8 cDNA by in vivo-jetPEITM confirmed by Western blot analysis (Fig. 5C).
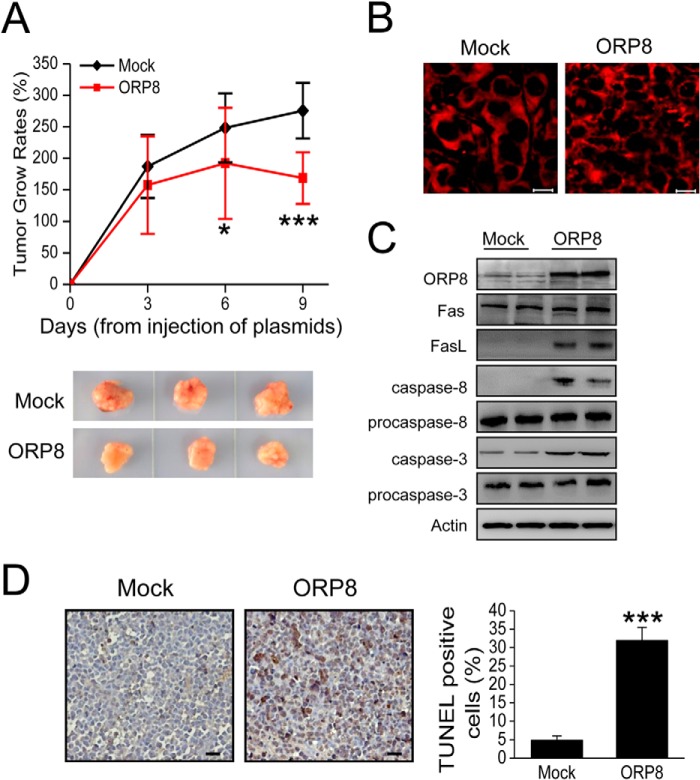
FIGURE 5.
ORP8 suppresses HepG2 xenograft tumor growth in BALB/c nude mice. A, upper panel, the tumor volumes of HepG2 cell xenograft tumor. The results are expressed as tumor relative growth rate to the tumor volume on day 0 when the first intratumoral injections were performed. The data represent the mean ± S.D. (n = 8; *, p < 0.05; ***, p < 0.001). Lower panel, photographs of representative xenograft tumor masses from nude mice. B, confocal microscopy analysis of Fas localization in xenograft tumor tissues. Scale bars, 10 μm. C, Western blot analysis of ORP8, FasL, cleaved caspase-8, and caspase-3 expression in xenograft tumor tissues. D, in situ TUNEL assays in which tumor tissue with ORP8 overexpression exhibits increased apoptosis. Scale bars, 10 μm. Quantification of the percentage of TUNEL-positive cells in control or miR-143 inhibition expressing tumors is shown. Three representative areas with vital tumor tissue were randomly selected.
We observed intratumor Fas localization by confocal microscopy. Fas was evenly distributed in the cytosol in the Mock-transfected tumors, whereas in ORP8-transfected tumors Fas distribution was uneven and concentrated in brightly stained patches (Fig. 5B). The FasL expression was also up-regulated in ORP8-transfected tumors (Fig. 5C). To further investigate whether ORP8 suppressed tumor growth in BALB/c nude mice via apoptosis, we analyzed the cleaved caspase-8 and caspase-3 in the xenograft tumor tissues. The cleaved caspase-8 and caspase-3 in tumor tissues overexpressing ORP8 was higher than that in the mock group (Fig. 5C). We also found more TUNEL-positive staining in sections from ORP8-overexpressing xenograft tumors compared with controls (Fig. 5D). Taken together, these results suggest that ORP8 overexpression inhibits HepG2 xenograft growth in BALB/c nude mice through induction of tumor cell apoptosis.
miR-143 Inhibition Results in HepG2 Cell Apoptosis and Suppresses Tumorigenicity
Based upon the effect of miR-143 in ORP8 expression, we hypothesized that the abolition of miR-143 and restoration of tumor suppressor ORP8 expression in HepG2 cells might have an anti-tumor effect. Notably, the inhibition of miR-143 results in plasma membrane Fas trafficking, FasL up-regulation, caspase-8 and caspase-3 cleaved (Fig. 6A), and cell apoptosis (Fig. 6B) in HepG2 cells. However, these effects were abrogated by ORP8 knockdown (Fig. 6, A and B), indicating that ORP8 mediated the effects of miR-143 inhibition. In addition, we investigated the tumor growth in vivo. Tumor-bearing mice were injected intratumorally with lenti-shNT or lenti-miR-143 inhibitor at 19 days after tumor inoculation. The mice were sacrificed on day 28, and the tumors were harvested and weighed. As expected, miR-143 inhibition led to a significant reduction in tumor growth (Fig. 6C). Furthermore, FasL up-regulation and Fas trafficking were induced in miR-143-inhibited tumor (Fig. 6, D and E). We also found that miR-143 inhibitor-induced inhibition in the tumor growth was through activation of caspase-3 and -8 with more TUNEL-positive cells in sections from miR-143 inhibitor-treated tumors compared with controls (Fig. 6, D and F).
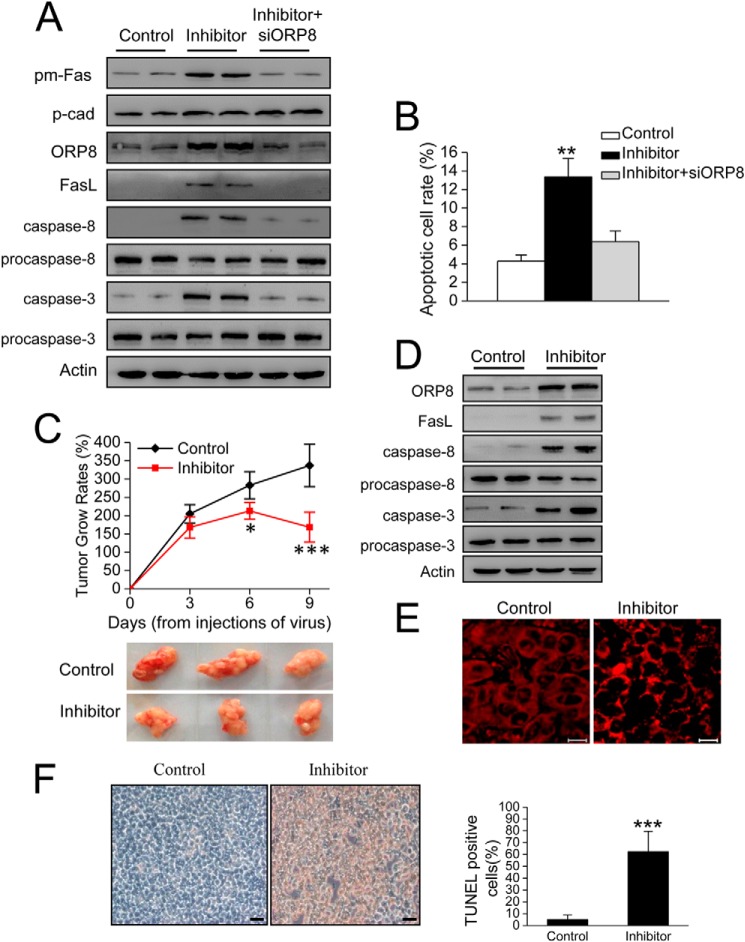
FIGURE 6.
miR-143 inhibition results in HepG2 cell apoptosis and suppresses tumor growth in BALB/c nude mice. A, Western blot shows Fas protein in plasma membrane fraction (pm-Fas) and FasL expression in HepG2 cells. P-cadherin (p-cad) was employed as a loading control for plasma membrane fractions. B, cell apoptosis in HepG2 cells with miR-143 inhibition. C, tumor volumes of HepG2 cell xenograft tumor. The results are expressed as tumor relative growth rate to the tumor volume on day 20 when the first measurements were performed. Data represent the mean ± S.D. (n = 8; **, p < 0.01; ***, p < 0.001). Lower panel, photographs of representative xenograft tumor masses from nude mice. D, Western blot indicated ORP8 and FasL expression and caspases-8 and 3 activation in tumor. E, confocal microscopy analysis of Fas localization in xenograft tumor tissues. Scale bars, 10 μm. F, in situ TUNEL assays in which tumor tissue from miR-143 inhibitor-treated mice exhibits increased apoptosis. Scale bars, 10 μm. Quantification of the percentage of TUNEL-positive cells in control or miR-143 inhibition expressing tumors is shown. Three representative areas with vital tumor tissue were randomly selected.
DISCUSSION
A low level of Fas expression was implicated in hepatoma treatment resistance (9). However, the resistance to apoptosis was reported to remain unchanged in Fas-positive HCC cell lines (8). We observed that most clinical cases of HCC display low Fas protein expression, but some of HCC cases we analyzed appeared to have a normal expression level. These results indicated that mechanisms other than weak Fas expression are involved in hepatoma cell apoptosis resistance.
In this study we found that ORP8 expression in primary human HCC cells and HCC cell lines was significantly diminished. Overexpression of ORP8 in primary HCC cells and cell lines induced apoptosis, which was also replicated in HepG2 xenografts in vivo. These results suggested that down-regulation of ORP8 expression in HCC may protect liver cancer cells from apoptosis.
Binding of Fas to its ligand or Fas antibody on the cell surface leads to the activation of caspase-8, downstream caspases, and cell death (30). In our study Fas expression did not change with ORP8 overexpression in HepG2 cells. Another hepatoma cell line, Huh7, which also is reported to be resistant to Fas-mediated apoptosis (7), expressed relatively low levels of Fas compared with HepG2 and displayed a blunter apoptotic response upon ORP8 overexpression. Thus, ORP8-induced apoptosis may be positively related to Fas expression. Treatment of HepG2 cells with anti-FasL neutralizing antibody significantly reduced the apoptosis induced by ORP8 overexpression, indicating that Fas/FasL pathway mediates ORP8-induced apoptosis. We further found that Fas normally is distributed in the cytoplasmic compartment but localizes to the plasma membrane in ORP8-overexpressing HCC cells. Considering that ORP8 overexpression up-regulates FasL, Fas/FasL should be a main pathway for ORP8-induced HCC cell apoptosis. FasL expressed in T-lymphocytes can engage Fas on the cell membrane to trigger apoptosis (21). ORP8 overexpression in our experiments significantly enhanced the sensitivity of HepG2 cells and primary HCC cells to apoptosis mediated by Jurkat T-cells or T-cells in co-culture. Based on these results, we propose that ORP8 facilitates the translocation of cytoplasmic Fas to the cell surface, after which FasL from T-cells triggers HCC cell apoptosis.
ORP8 was previously shown to localize to the ER and nuclear envelope (11, 12). We found that ORP8 was also distributed to the hepatocyte plasma membrane. Even through we failed to detect a direct interaction of ORP8 with Fas by co-immunoprecipitation (data not shown), the appearance of ORP8 at the plasma membrane may provide a mechanistic clue to its role in Fas transport to this membrane. We further showed that ORP8-mediated Fas translocation was p53-dependent.
We also found that ORP8 overexpression increased XBP-1s, ATF4, Chop, phospho-PERK, and phospho-eIF2α protein levels in a time-dependent manner. These results clearly indicated that overexpression of ORP8 in HepG2 cells induces an ER stress response, which results in FasL up-regulation.
Deregulation of miRNA expression has been implicated in many diseases, including cancer (31). Studies have indicated that miRNAs directly contribute to HCC by targeting many of the critical regulatory gene products modulating apoptosis, cell cycle checkpoints, and growth factor-stimulated responses (32). Perturbation of these pathways can result in malignant transformation and ultimately HCC development. However, the exact nature of this relationship is not fully understood. miR-143 is highly conserved in vertebrates (33). The up-regulation of miR-143 through repression of FNDC3B was observed in an HCC model during tumor metastasis (17). ORP8 has previously been reported to be a major target of miR-143 in mice (15). We explored the effect of miR-143 on the expression of ORP8 in human using miR-143 inhibitor and miR-143 mimic. The results confirmed that ORP8 is also a target of miR-143 in human hepatic cells, which is consistent with ORP8 playing an important role in HCC apoptosis.
The present study provides evidence for a novel function of ORP8 in the induction of apoptosis in hepatoma cells and suggests that ORP8-induced apoptosis occurs through movement of Fas from the cytoplasm to the cell surface and up-regulation of FasL expression. Moreover, the pro-apoptotic effect of ORP8 overexpression and miR-143 inhibition was confirmed in vivo using a HepG2 xenograft model. Thus, the down-regulation of ORP8 expression in HCC, which coincides with elevated expression of miR-143, may contribute to HCC cell escape from immune surveillance by reducing Fas transport to the cell surface. ORP8 may, therefore, be considered as a potential new target for HCC therapy. Finally, the dampening of ORP8 expression by miR-143 in HCC cells raises the possibility that ORP8 may be a participant in the development of HCC.
Acknowledgment
We thank Dr. Vesa Olkkonen for critical reading of this manuscript.
This work was supported by grants from the NSFC (National Natural Science Foundation of China), China (91439122 and 30971104 (to D. Y.)), the National Basic Research Program of China (2012CB517502 (to D. Y.), and Fundamental Research Funds for the Central Universities, China (21610609 (to D. Y.)).
- FasL
- Fas ligand
- HCC
- hepatocellular carcinoma
- ORP8
- oxysterol-binding protein-related protein 8
- ER
- endoplasmic reticulum
- qRT
- quantitative real-time
- miRNA
- microRNA.
REFERENCES
- 1. Pinkoski M. J., Brunner T., Green D. R., Lin T. (2000) Fas and Fas ligand in gut and liver. Am. J. Physiol. Gastrointest. Liver Physiol. 278, G354–G366 [DOI] [PubMed] [Google Scholar]
- 2. Schulze-Bergkamen H., Krammer P. H. (2004) Apoptosis in cancer: implications for therapy. Semin. Oncol. 31, 90–119 [DOI] [PubMed] [Google Scholar]
- 3. Itoh N., Yonehara S., Ishii A., Yonehara M., Mizushima S., Sameshima M., Hase A., Seto Y., Nagata S. (1991) The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 66, 233–243 [DOI] [PubMed] [Google Scholar]
- 4. Galle P. R., Hofmann W. J., Walczak H., Schaller H., Otto G., Stremmel W., Krammer P. H., Runkel L. (1995) Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J. Exp. Med. 182, 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Faubion W. A., Gores G. J. (1999) Death receptors in liver biology and pathobiology. Hepatology 29, 1–4 [DOI] [PubMed] [Google Scholar]
- 6. Wang X., Lu Y., Cederbaum A. I. (2005) Induction of cytochrome P450 2E1 increases hepatotoxicity caused by Fas agonistic Jo2 antibody in mice. Hepatology 42, 400–410 [DOI] [PubMed] [Google Scholar]
- 7. Ogawa K., Yasumura S., Atarashi Y., Minemura M., Miyazaki T., Iwamoto M., Higuchi K., Watanabe A. (2004) Sodium butyrate enhances Fas-mediated apoptosis of human hepatoma cells. J. Hepatol. 40, 278–284 [DOI] [PubMed] [Google Scholar]
- 8. Natoli G., Ianni A., Costanzo A., De Petrillo G., Ilari I., Chirillo P., Balsano C., Levrero M. (1995) Resistance to Fas-mediated apoptosis in human hepatoma cells. Oncogene 11, 1157–1164 [PubMed] [Google Scholar]
- 9. Ito Y., Monden M., Takeda T., Eguchi H., Umeshita K., Nagano H., Nakamori S., Dono K., Sakon M., Nakamura M., Tsujimoto M., Nakahara M., Nakao K., Yokosaki Y., Matsuura N. (2000) The status of Fas and Fas ligand expression can predict recurrence of hepatocellular carcinoma. Brit. J. Cancer 82, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higaki K., Yano H., Kojiro M. (1996) Fas antigen expression and its relationship with apoptosis in human hepatocellular carcinoma and noncancerous tissues. Am. J. pathol. 149, 429–437 [PMC free article] [PubMed] [Google Scholar]
- 11. Yan D., Mäyränpää M. I., Wong J., Perttilä J., Lehto M., Jauhiainen M., Kovanen P. T., Ehnholm C., Brown A. J., Olkkonen V. M. (2008) OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J. Biol. Chem. 283, 332–340 [DOI] [PubMed] [Google Scholar]
- 12. Zhou T., Li S., Zhong W., Vihervaara T., Béaslas O., Perttilä J., Luo W., Jiang Y., Lehto M., Olkkonen V. M., Yan D. (2011) OSBP-related protein 8 (ORP8) regulates plasma and liver tissue lipid levels and interacts with the nucleoporin Nup62. PLoS ONE 6, e21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Béaslas O., Vihervaara T., Li J., Laurila P. P., Yan D., Olkkonen V. M. (2012) Silencing of OSBP-related protein 8 (ORP8) modifies the macrophage transcriptome, nucleoporin p62 distribution, and migration capacity. Exp. Cell Res. 318, 1933–1945 [DOI] [PubMed] [Google Scholar]
- 14. Zhong W., Zhou Y., Li J., Mysore R., Luo W., Li S., Chang M. S., Olkkonen V. M., Yan D. (2014) OSBP-related protein 8 (ORP8) interacts with Homo sapiens sperm-associated antigen 5 (SPAG5) and mediates oxysterol interference of HepG2 cell cycle. Exp. Cell Res. 322, 227–235 [DOI] [PubMed] [Google Scholar]
- 15. Jordan S. D., Krüger M., Willmes D. M., Redemann N., Wunderlich F. T., Brönneke H. S., Merkwirth C., Kashkar H., Olkkonen V. M., Böttger T., Braun T., Seibler J., Brüning J. C. (2011) Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434–446 [DOI] [PubMed] [Google Scholar]
- 16. Dessì S., Batetta B., Pulisci D., Spano O., Anchisi C., Tessitore L., Costelli P., Baccino F. M., Aroasio E., Pani P. (1994) Cholesterol content in tumor tissues is inversely associated with high-density lipoprotein cholesterol in serum in patients with gastrointestinal cancer. Cancer 73, 253–258 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X., Liu S., Hu T., Liu S., He Y., Sun S. (2009) Up-regulated microRNA-143 transcribed by nuclear factor κB enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50, 490–499 [DOI] [PubMed] [Google Scholar]
- 18. Yano H., Fukuda K., Haramaki M., Momosaki S., Ogasawara S., Higaki K., Kojiro M. (1996) Expression of Fas and anti-Fas-mediated apoptosis in human hepatocellular carcinoma cell lines. J. Hepatol. 25, 454–464 [DOI] [PubMed] [Google Scholar]
- 19. Strand S., Hofmann W. J., Hug H., Müller M., Otto G., Strand D., Mariani S. M., Stremmel W., Krammer P. H., Galle P. R. (1996) Lymphocyte apoptosis induced by CD95 (APO-1/Fas) ligand-expressing tumor cells: a mechanism of immune evasion? Nat. Med. 2, 1361–1366 [DOI] [PubMed] [Google Scholar]
- 20. Bennett M., Macdonald K., Chan S. W., Luzio J. P., Simari R., Weissberg P. (1998) Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science 282, 290–293 [DOI] [PubMed] [Google Scholar]
- 21. el-Khatib M., Stanger B. Z., Dogan H., Cui H., Ju S. T. (1995) The molecular mechanism of FasL-mediated cytotoxicity by CD4+ Th1 clones. Cell. Immunol. 163, 237–244 [DOI] [PubMed] [Google Scholar]
- 22. Lowin B., Mattman C., Hahne M., Tschopp J. (1996) Comparison of Fas(Apo-1/CD95)- and perforin-mediated cytotoxicity in primary T lymphocytes. Int. Immunol. 8, 57–63 [DOI] [PubMed] [Google Scholar]
- 23. Pahl H. L., Baeuerle P. A. (1997) The ER-overload response: activation of NF-κB. Trends Biochem. Sci. 22, 63–67 [DOI] [PubMed] [Google Scholar]
- 24. Pahl H. L., Baeuerle P. A. (1995) A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-κB. EMBO J. 14, 2580–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu S. C., Gavrilin M. A., Lee H. H., Wu C. C., Han S. H., Lai M. Z. (1999) NF-κB-dependent Fas ligand expression. Eur. J. Immunol. 29, 2948–2956 [DOI] [PubMed] [Google Scholar]
- 26. Kim I., Xu W., Reed J. C. (2008) Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug. Discov. 7, 1013–1030 [DOI] [PubMed] [Google Scholar]
- 27. Du X., Kumar J., Ferguson C., Schulz T. A., Ong Y. S., Hong W., Prinz W. A., Parton R. G., Brown A. J., Yang H. (2011) A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang H. Y., Wek S. A., McGrath B. C., Scheuner D., Kaufman R. J., Cavener D. R., Wek R. C. (2003) Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell Biol. 23, 5651–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng J., Lu P. D., Zhang Y., Scheuner D., Kaufman R. J., Sonenberg N., Harding H. P., Ron D. (2004) Translational repression mediates activation of nuclear factor κB by phosphorylated translation initiation factor 2. Mol. Cell Biol. 24, 10161–10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X., Cederbaum A. I. (2007) Acute ethanol pretreatment increases FAS-mediated liver injury in mice: role of oxidative stress and CYP2E1-dependent and -independent pathways. Free Radic. Biol. Med. 42, 971–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sassen S., Miska E. A., Caldas C. (2008) MicroRNA: implications for cancer. Virchows Arch. 452, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao X., Yang Z., Li G., Li D., Zhao Y., Wu Y., Robson S. C., He L., Xu Y., Miao R., Zhao H. (2012) The role and clinical implications of microRNAs in hepatocellular carcinoma. Science China 55, 906–919 [DOI] [PubMed] [Google Scholar]
- 33. Trakooljul N., Hicks J. A., Liu H. C. (2010) Identification of target genes and pathways associated with chicken microRNA miR-143. Anim. Genet. 41, 357–364 [DOI] [PubMed] [Google Scholar]