Background: Modulation of histone deacetylase function is crucial for DNA damage-induced cell death and survival. The mechanism of histone deacetylase regulation is poorly understood.
Results: Ubiquitination and de-ubiquitination influence sirtuin 1 (SIRT1) histone deacetylase function during DNA damage-induced cell responses.
Conclusion: Sirtuin function requires DNA damage-induced SIRT1 ubiquitination.
Significance: SIRT1 ubiquitination is a crucial mechanism regulating cell death and survival.
Keywords: Deubiquitylation (Deubiquitination), Histone Deacetylase (HDAC), Post-translational Modification (PTM), Sirtuin, Ubiquitylation (Ubiquitination)
Abstract
Downstream signaling of physiological and pathological cell responses depends on post-translational modification such as ubiquitination. The mechanisms regulating downstream DNA damage response (DDR) signaling are not completely elucidated. Sirtuin 1 (SIRT1), the founding member of Class III histone deacetylases, regulates multiple steps in DDR and is closely associated with many physiological and pathological processes. However, the role of post-translational modification or ubiquitination of SIRT1 during DDR is unclear. We show that SIRT1 is dynamically and distinctly ubiquitinated in response to DNA damage. SIRT1 was ubiquitinated by the MDM2 E3 ligase in vitro and in vivo. SIRT1 ubiquitination under normal conditions had no effect on its enzymatic activity or rate of degradation; hypo-ubiquitination, however, reduced SIRT1 nuclear localization. Ubiquitination of SIRT1 affected its function in cell death and survival in response to DNA damage. Our results suggest that ubiquitination is required for SIRT1 function during DDR.
Introduction
Protein function can be temporally and spatially regulated by post-translational modification such as lysine acetylation. Histone deacetylases, also known as lysine deacetylases, regulate the reversible acetylation of lysine residues. Sirtuin 1 (SIRT1)2 is an NAD+-dependent histone deacetylase that is the human ortholog of the yeast Sir2 protein and founding member of the Class III histone deacetylases. Like most histone deacetylases, SIRT1 plays important roles in many normal and abnormal physiological processes (1) such as caloric restriction-related longevity, metabolism, DNA damage response (DDR), aging, and tumorigenesis (2–5).
DDR is crucial for maintaining genome stability when cells are exposed to endogenous and/or exogenous factors that induce DNA damage. One function of SIRT1 is regulating DDR. SIRT1 is involved in multiple steps of DDR, including damage sensing, signal transduction, DNA repair, and apoptosis (6). SIRT1 plays a role in both cell death and survival by deacetylating several DDR proteins, including p53, Ku70, NBS1, Tip60, hMOF, and PARP-1 (3, 7–14). For example, SIRT1-deacetylated NBS1 triggered DDR when cells were subjected to ionizing radiation (IR), which induces double-strand breaks (12). SIRT1 can target multiple substrates (such as Tip60, p53, and PARP-1) to regulate apoptosis (7–10, 14). SIRT1 deacetylates p53 at multiple lysine residues; K382 deacetylation represses p53-dependent apoptosis in response to cellular damage (7, 8). SIRT1 is believed to inhibit apoptosis induced by certain types of DNA damage, including IR- and etoposide-induced double-strand breaks. However, our previous study suggests that SIRT1 promotes cell death in response to stimuli such as H2O2 (15). Although the importance of SIRT1 in DDR is indisputable, post-translational modification regulation of SIRT1 in DDR has not been fully explored.
Ubiquitin is a small, 76-amino acid protein containing 7 lysine residues, namely Lys-6, -11, -27, -29, -33, -48, and -63 (16). Ubiquitination is the process by which a cysteine in ubiquitin is covalently linked to a lysine in a protein or another ubiquitin. Typical Lys-48-mediated polyubiquitination usually targets the substrate protein to proteasome-mediated degradation. Atypical polyubiquitination, which is mediated by Lys-6, -11, -27, -29, -33, and -63, regulates non-proteolytic functions such as enzymatic activity, protein interaction, and cellular localization. Post-translational modifications such as phosphorylation, acetylation, and ubiquitination are important for proper DDR. For example, p53 protein is maintained at a low level under normal conditions by ubiquitination-mediated degradation involving MDM2 E3 ligase. The stability of p53 increases after DNA damage by the loss of interaction with MDM2 and a subsequent reduction in ubiquitination (17, 18). Histone H2AX ubiquitination is required for its phosphorylation and for recruiting MDC1 and repair proteins to repair DNA damage (19–21). These data indicate that ubiquitination is an important regulatory mechanism in DDR.
A large scale proteomics study identified that SIRT1 was ubiquitinated (22); however, very little is known about the mechanism and regulation of this modification. One study reported that SIRT1 was de-ubiquitinated by USP7 (23). Another study reported that JNK promoted SIRT1 ubiquitination (24). The E3 ligase responsible for SIRT1 ubiquitination, has yet to be identified, and the role of SIRT1 ubiquitination in DDR is unknown. The current study examines the regulation and functional implications of SIRT1 ubiquitination during DDR.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
WT and H363Y Myc-SIRT1, FLAG-SIRT1, glutathione S-transferase (GST)-SIRT1, and FLAG-p53 plasmids were described previously (25, 26). The lysine-to-arginine mutation was generated using the QuikChange Multisite-directed mutagenesis kit (Stratagene). MDM2, FLAG-MDM2, GST-MDM2, and His-ubiquitin (His-Ub) plasmids were described previously (27). Anti-FLAG, anti-hemagglutinin (HA), and anti-tubulin antibodies, recombinant His-ubiquitin, MG132, and chloroquine were purchased from Sigma. Rabbit anti-acetylated-K382-p53 antibody was purchased from Cell Signaling Technology. Anti-c-Myc, anti-lamin A, anti-SIRT1, and anti-PARP-1 antibodies were purchased from Santa Cruz Biotechnology. Anti-ubiquitin antibody was purchased from Abcam. Active UBE1 and UbcH5C were purchased from Millipore.
Cell Culture
HEK293T, U2OS, and H1299 cells were grown in Dulbecco's modified Eagle's medium. PC3, A549, and DU145 were grown in RPMI 1640 medium. All media were supplemented with 10% fetal calf serum, 100 mg/ml streptomycin, and 100 IU/ml penicillin. Control and SIRT1-knockdown (S1-KD) HeLa cell clones were generated and maintained as described previously (26).
Transfection, Transduction, and Cellular Fractionation
Transfections were performed using either Lipofectamine 2000 (Invitrogen) or PolyJetTM (SignaGen Laboratories) according to the manufacturer's instructions. S1-KD cells were generated using RNAi, in which a SIRT1-coding DNA sequence was targeted by a pSuper shRNA construct containing interfering oligonucleotides (5′-GTTGGATGATATGACACTG-3′). To restore SIRT1 expression in HeLa S1-KD cells, the silent mutant coding DNA sequence of SIRT1, in which the GTTGGATGATATGACACTG template sequence was modified to GTTAGACGACATGACCCTC by multisite-directed mutagenesis (Stratagene), was inserted into the pLenti-CMV-neo-DEST vector (Addgene plasmid 17392) using the Gateway recombination strategy (Invitrogen). 293FT cells were transfected with pLenti-CMV-neo-DEST-SIRT1 together with pLP1, pLP2, and pLP3 plasmids to package SIRT1 lentivirus as described previously (28). HeLa S1-KD cells were then transduced by lentivirus harvested from 293FT culture medium in the presence of 8 μg/ml Polybrene. Twenty-four hours post-infection, culture medium was supplemented with 600 μg/ml G418 and 0.5 μg/ml puromycin. Single cell-derived clones were isolated and transferred to new dishes. SIRT1 expression in these stable clones was verified by Western blot analysis. Cellular fractionation was conducted using the NE-PER nuclear and cytoplasmic extraction kit according to the manufacturer's instructions (Pierce).
Immunoprecipitation, Western Blotting, and Immunofluorescence
These assays were performed as described previously (26). Some Western blot results were quantified by densitometric analysis using Image J software (National Institutes of Health).
Ubiquitination Assays
Ubiquitination assays were performed in vivo using either the HA-tagged (29) or the histidine-tagged (27) ubiquitination method. In the HA-Ub method, cells were co-transfected with SIRT1 and HA-Ub. At 36 h post-transfection, cells were resuspended in 100 μl of lysis buffer (2% SDS, 10 mm Tris-HCl (pH 8.0), and 150 mm NaCl) and immediately boiled for 10 min. Cells were sonicated for three pulses of 10 s each, and cell lysates were diluted with 900 μl dilution buffer (10 mm Tris (pH 8.0), 150 mm NaCl, 2 mm EDTA, and 1% Triton). SIRT1 was immunoprecipitated and analyzed for ubiquitination by Western blotting using anti-HA antibody. In the His-Ub method, cells were co-transfected with FLAG-SIRT1 and His-Ub. At 36 h post-transfection, cells were dissolved in lysis buffer (6 m guanidinium-HCl, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl (pH 8.0), 5 mm imidazole, and 10 mm β-mercaptoethanol). His-tagged proteins were captured using Ni2+-nitrilotriacetic acid (NTA) beads and eluted with imidazole. The presence of SIRT1 in the eluted fraction was analyzed by Western blot.
The in vitro ubiquitination assay was performed as described previously (27). GST, GST-MDM2, and GST-SIRT1 were expressed in Escherichia coli and purified using glutathione-agarose beads. Two μg of GST-SIRT1 was incubated with 40 ng of UBE1 (E1), 200 ng UBcH5C (E2), and 2 μg His-Ub together with 500 ng of GST-MDM2 or GST in reaction buffer (40 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 2 mm dithiothreitol, and 2 mm ATP) at 30 °C for 2 h with agitation. The reaction mixture was then subjected to SDS-PAGE and Western blot analysis. Polyubiquitination of GST-SIRT1 was detected using anti-SIRT1 antibody. The membrane was stripped using stripping buffer (Pierce) and re-probed for autoubiquitination of GST-MDM2 using anti-MDM2 antibody.
Deacetylation Assays
For in vivo deacetylation assays, HeLa cells were co-transfected with FLAG-p53 and either WT or mutant SIRT1. At 32 h post-transfection, cells were treated with 400 ng/ml trichostatin A for 4 h. Cells were lysed in NETN buffer (100 mm NaCl, 0.5 mm EDTA, 20 mm Tris (pH 8.0), and 0.5% (v/v) Nonidet P-40) supplemented with 20 mm sodium butyrate. Whole-cell lysates were subjected to SDS-PAGE and Western blotting. p53 acetylation was detected using anti-Ac-K382 p53 antibody (Cell Signaling Technology). In vitro deacetylation assays were performed using a fluorometric assay kit (Fluor-de-Lys®SIRT1 fluorometric drug discovery assay kit, Enzo Life Sciences) according to the manufacturer's instructions. Fluorescence was measured using the EnVision Multilabel Plate Reader (PerkinElmer, excitation 360 nm and emission 460 nm).
Apoptosis and Necrosis Assays
HeLa cells were treated with 5 μm etoposide (Sigma) for 24 h, 100 μm H2O2 for 2 h or left untreated. Twenty-four hours later, cells were detached using EDTA buffer without trypsin and washed with phosphate-buffered saline (PBS). To detect apoptosis and necrosis, cells were stained with 1 μg/ml FITC-labeled annexin V (BD Biosciences) for 15 min in the dark, washed with PBS, and then incubated with 5 μg/ml propidium iodide (Sigma). Apoptosis and necrosis were analyzed using a FACSCalibur flow cytometer (BD Biosciences).
RESULTS
SIRT1 Is Ubiquitinated by MDM2 in Vivo and in Vitro
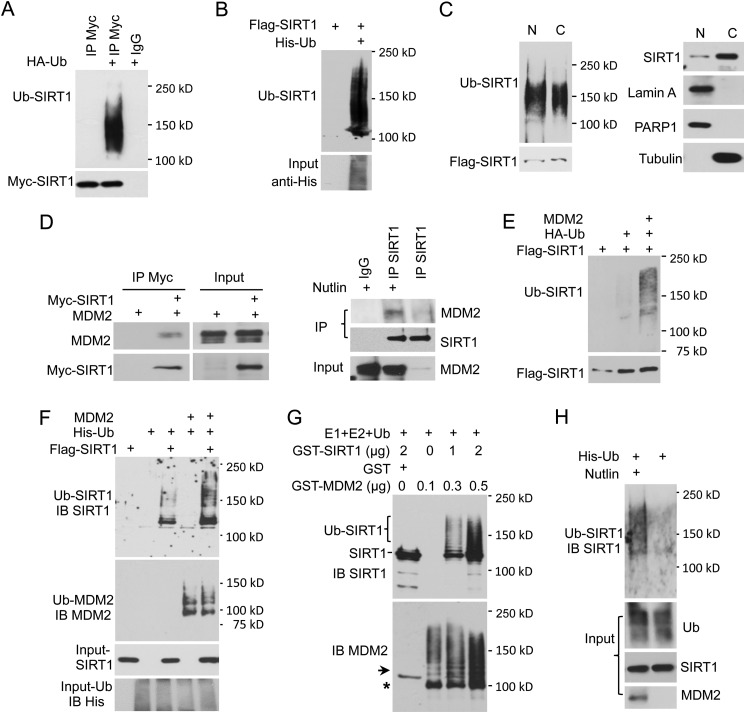
To examine SIRT1 ubiquitination in vivo, 293T cells were co-transfected with Myc-SIRT1 and either HA-tagged Ub or a vector control. Ubiquitination of immunoprecipitated SIRT1 was detected by Western blotting using anti-HA antibody. Our data suggested that SIRT1 was ubiquitinated, as indicated by a protein ladder above the predicted SIRT1 band of 120 kDa (Fig. 1A). To exclude the possibility of contamination from a SIRT1-interacting protein, a stringent guanidinium HCl-urea denaturation method was performed as described previously (27). In these experiments FLAG-SIRT1 was co-expressed with His-tagged Ub, and then ubiquitinated (His-tagged) proteins were denatured by guanidinium HCl and pulled down using nickel resin. The presence of ubiquitinated SIRT1 was detected using an anti-FLAG antibody (Fig. 1B). Because SIRT1 is localized in both the cytoplasm and nucleus (30–33), we examined the localization of ubiquitinated SIRT1 in cellular fractions. We found that ubiquitinated SIRT1 was localized in both compartments (Fig. 1C).
FIGURE 1.
SIRT1 is ubiquitinated. A, 293T cells were transfected with Myc-SIRT1 and either HA-Ub or HA-tag control vector. After 36 h, cells were treated with 50 μm MG132 for 4 h. SIRT1 was immunoprecipitated (IP) using anti-Myc antibody and IgG isotype control and then analyzed for ubiquitination by Western blot using anti-HA antibody. B, cells were transfected with FLAG-SIRT1 and either His-Ub or control vector. Ubiquitinated protein was pulled down by NTA+ beads and resolved by SDS-PAGE. SIRT1 in the pulldown fraction was detected by anti-SIRT1 antibody. C, 293T cells were transfected with FLAG-SIRT1 and HA-Ub, and then cytoplasmic (C) and nuclear (N) extracts were prepared. Expression levels of SIRT1 in cellular fractions were examined by Western blot (right). FLAG-SIRT1 was immunoprecipitated using anti-FLAG antibody. Equal amounts of SIRT1 from both fractions were loaded for ubiquitination analysis (left). D, interaction between MDM2 and SIRT1. 293T cells were co-transfected with Myc-SIRT1 and MDM2. SIRT1 was immunoprecipitated using anti-Myc antibody. Co-immunoprecipitation of MDM2 was detected using anti-MDM2 antibody (left). A549 cells were treated with 7 μm Nutlin overnight or left untreated. Endogenous SIRT1 was pulled down by anti-SIRT1 antibody, and MDM2 co-immunoprecipitation was detected using anti-MDM2 antibody (right). E, SIRT1 is ubiquitinated by MDM2 in vivo. FLAG-SIRT1 was co-expressed with HA-Ub and either MDM2 or vector control. SIRT1 was immunoprecipitated by anti-FLAG antibody and analyzed for ubiquitination using anti-HA antibody. F, FLAG-SIRT1 was co-expressed with His-Ub and MDM2 or control vector. SIRT1 ubiquitination was analyzed as described in B. IB, immunoblot. G, SIRT1 was ubiquitinated by MDM2 in vitro. GST-SIRT1 was incubated with active E1, E2, and ubiquitin in the presence of GST-MDM2 or GST alone. Reaction products were analyzed by Western blot using anti-SIRT1 antibody. The membrane was stripped and re-probed with anti-MDM2 antibody. The arrow indicates residual SIRT1 signal after stripping. The asterisk (*) indicates the band corresponding to non-ubiquitinated GST-MDM2. H, A549 cells were transfected with His-Ub. At 36 h after transfection, cells were treated with 7 μm Nutlin overnight and 50 μm MG132 for 4 h before harvesting. His-Ub-conjugated proteins were pulled down, and ubiquitination of endogenous SIRT1 was examined as described in F.
The MDM2 E3 ubiquitin ligase is responsible for p53 ubiquitination and degradation and plays an important role in regulating cell proliferation, apoptosis, and DDR (34, 35). To determine whether MDM2 modifies SIRT1, we first examined SIRT1 and MDM2 interaction using a co-immunoprecipitation assay. As shown in Fig. 1D, exogenous Myc-SIRT1 co-immunoprecipitated with ectopically expressed MDM2 (left panel). Nutlin, which stabilizes endogenous MDM2, strengthens the endogenous interaction between MDM2 and SIRT1 (right panel). Overexpression of MDM2 dramatically enhanced SIRT1 ubiquitination in vivo (Fig. 1E). Using the guanidinium HCl-urea denaturation method, we still found that SIRT1 was ubiquitinated by MDM2 as detected after high stringency washes of the nickel matrix (Fig. 1F). To examine SIRT1 ubiquitination by MDM2 in vitro, GST-SIRT1 was incubated with either GST-MDM2 or GST in the presence of purified ubiquitin-activating enzyme E1, conjugating enzyme E2, ubiquitin, and ATP. GST-SIRT1 ubiquitination occurred in the presence of GST-MDM2 but not in the presence of GST alone (Fig. 1G). Autoubiquitination of GST-MDM2 was used as a control. To further confirm that MDM2 drives SIRT1 ubiquitination, A549 cells were treated with Nutlin, and ubiquitination of endogenous SIRT1 was examined. Nutlin treatment significantly increased MDM2 protein levels and SIRT1 ubiquitination (Fig. 1H). Together, our results unequivocally demonstrate that MDM2 ubiquitinates SIRT1 both in vivo and in vitro.
SIRT1 Ubiquitination Is Regulated by DNA Damage
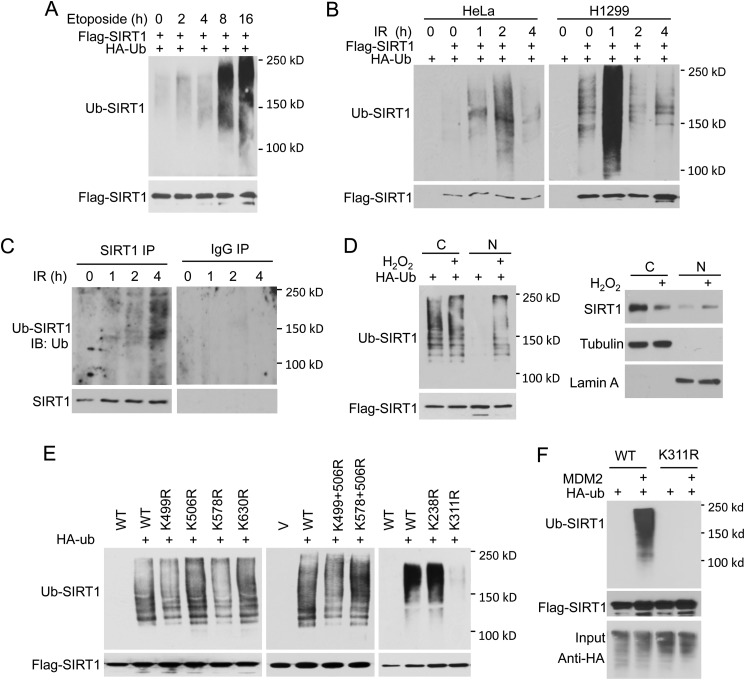
To determine whether SIRT1 ubiquitination is regulated by DNA damage signals, FLAG-SIRT1 was co-expressed in 293T cells with HA-Ub. SIRT1 ubiquitination was analyzed at different time points after treatment with the topoisomerase II inhibitor etoposide, which induces double-strand breaks. SIRT1 ubiquitination significantly increased 2 h after etoposide treatment (Fig. 2A). We examined SIRT1 ubiquitination after exposure to IR. Our data demonstrate that SIRT1 ubiquitination increased dramatically 1 h after IR treatment and then returned to basal levels after 4 h (Fig. 2B). Endogenous SIRT1 ubiquitination was similarly regulated by IR (Fig. 2C). SIRT1 ubiquitination also was affected by H2O2-induced oxidative damage (Fig. 2D). Therefore, SIRT1 ubiquitination is dynamically regulated in response to different types of DNA damaging agents.
FIGURE 2.
SIRT1 ubiquitination is regulated by DNA damage. A–D, DNA damage regulates SIRT1 ubiquitination. FLAG-SIRT1 was co-transfected with HA-Ub. At 36 h post-transfection, cells were treated with 50 μm etoposide, 10 gray IR, or left untreated. FLAG-SIRT1 was immunoprecipitated using anti-FLAG antibody and analyzed for ubiquitination using anti-HA antibody. C, ubiquitination of endogenous SIRT1. Endogenous SIRT1 was immunoprecipitated from HeLa cells using anti-SIRT1 antibody, and ubiquitination was assessed using anti-ubiquitin antibody. D, HeLa cells were transfected with FLAG-SIRT1 and HA-Ub. At 36 h after transfection, cells were treated with 400 μm H2O2 or left untreated. Cytoplasmic (C) and nuclear (N) fractions were extracted and examined for SIRT1 expression (right). Cytoplasmic and nuclear fractions were normalized to contain the same amount of SIRT1, used for immunoprecipitation, and then examined for ubiquitination (left). E and F, WT and lysine-to-arginine mutant of SIRT1 were transfected with or without HA-Ub and along with MDM2 or not. SIRT1 ubiquitination was analyzed as described in Fig. 1A. V, vector control.
We used mass spectrometry (MS) to identify the SIRT1 lysines that were modified by ubiquitination. Together with an earlier report on the global quantification of ubiquitination in cells (22), we and others identified at least six ubiquitinated SIRT1 lysine residues (Lys-238, -311, -499, -506, -578, and -630) (data not shown). To biochemically verify the MS results, a series of lysine-to-arginine SIRT1 mutants was generated and ubiquitination examined as described previously. Mutation of Lsy-311, but not any other candidate lysine, significantly reduced SIRT1 ubiquitination (Fig. 2E). Moreover, MDM2 was unable to ubiquitinate SIRT1 if Lsy-311 was mutated (Fig. 2F). Therefore, although SIRT1 contains multiple ubiquitination sites, we conclude that Lsy-311 is the major SIRT1 lysine that undergoes ubiquitination.
SIRT1 Ubiquitination Affects DDR-induced Cell Death and Survival
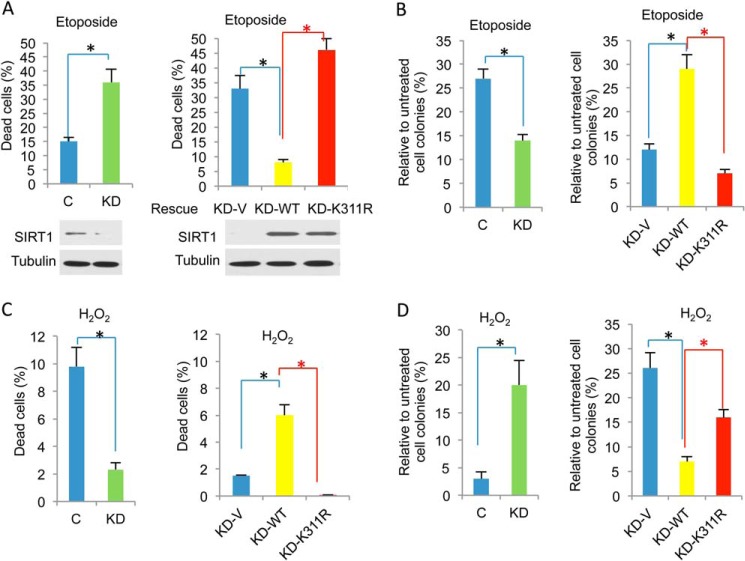
SIRT1 plays important regulatory roles in cell death and survival in response to DNA damage (7, 8). To address the functional implication of SIRT1 ubiquitination during DDR, we produced stable SIRT1 knockdown (KD) HeLa cells and analyzed cell survival rescue after etoposide treatment in the presence of WT or K311R SIRT1. SIRT1 expression was monitored by Western blot analysis. Cell survival was analyzed as described previously (36). SIRT1 KD cells exhibited greater cell death and reduced survival after etoposide treatment than cells treated with the vehicle control (C). Expression of WT SIRT1 (KD-WT), but not K311R SIRT1, reduced apoptosis and increased cell survival after etoposide treatment compared with that of the vector control (KD-V) (Fig. 3, A and B). The role of SIRT1 in cell death and survival depends on the type of DNA damage sustained. For example, SIRT1 is required for H2O2-induced cell death (15, 37, 38). To determine the effect of SIRT1 Lsy-311 ubiquitination in H2O2-induced responses, we treated SIRT1 KD cells with H2O2 and examined the effect of WT and K311R SIRT1 expression on cell death. As expected, SIRT1 knockdown inhibited H2O2-induced cell death and promoted cell survival. WT SIRT1, but not K311R SIRT1, restored SIRT1-induced cell death in response to H2O2 (Fig. 3, C and D). Our results indicate that SIRT1 de-ubiquitination impairs SIRT1 function during DDR-induced cell death and survival.
FIGURE 3.
K311R is defective in regulating cell death and survival during DDR. A, control and SIRT1 KD HeLa cells were treated with 5 μm etoposide for 24 h (left). SIRT1 KD HeLa cells were transfected with WT SIRT1, K311R SIRT1, or vector control (C) and subsequently treated with etoposide (right). Apoptosis and necrosis were examined by flow cytometric analysis using annexin V-PI staining. The percentage of dead cells was determined by measuring the percentage of annexin-positive cells. Levels of SIRT1 and tubulin were assessed by Western blot. B, control, KD, and rescued cells were treated with 5 μm etoposide for 2 h or left untreated. Two weeks later cell colonies were counted and normalized to the untreated control. C, control and SIRT1 KD HeLa cells were treated with 100 μm H2O2 for 2 h (left). SIRT1 KD HeLa cells were transfected with WT SIRT1, K311R SIRT1, or vector control and subsequently treated with H2O2 (right). Cell death was measured by annexin V-PI staining. D, control, KD, and rescued cells were treated with 100 μm H2O2 for 30 min or left untreated. Two weeks later, cell colonies were counted and normalized to the untreated control. All data are presented as the means ± S.E. of at least three independent experiments. A two-tailed unpaired Student's t test was applied to calculate the p value using the GraphPad Prism software. *, p < 0.05 was considered as statistically significant.
Ubiquitination Affects SIRT1 Protein Stability Under DNA Stress
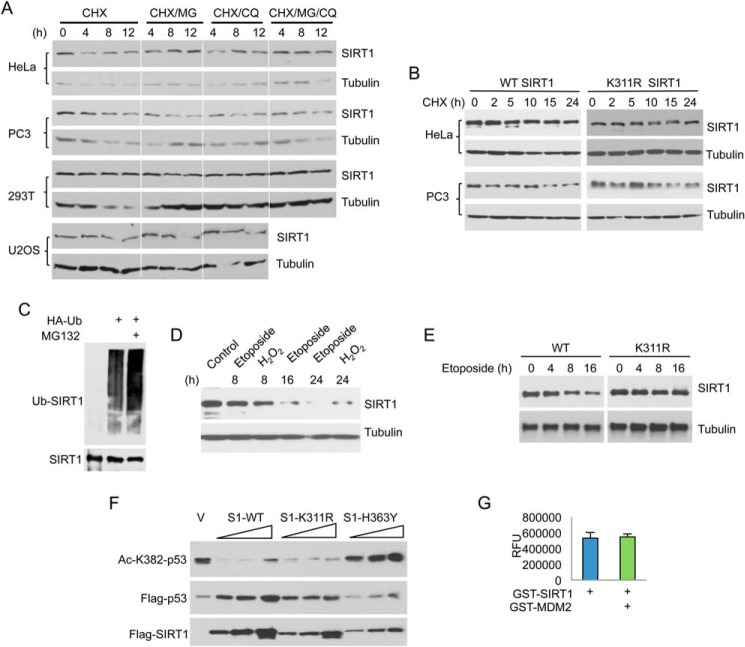
Ubiquitination can facilitate protein degradation mediated by proteasomal, lysosomal, and endoplasmic reticulum mechanisms (39–41). To investigate the molecular mechanisms by which K311R (a de-ubiquitination-mimicking mutation) affects the role of SIRT1 during DDR, we first determined the effect of ubiquitination on SIRT1 protein stability. SIRT1 is a very stable protein because its protein level did not significantly change when cells were treated with 50 μg/ml cycloheximide (CHX) for up to 12 h to halt protein synthesis (Fig. 4, A and B). No significant increase in SIRT1 was observed when cells were subjected to the proteasomal inhibitor MG132 and the lysosomal inhibitor chloroquine (CQ) either alone or together (Fig. 4A). The K311R mutant was just as stable as WT SIRT1 (Fig. 4B). Consistent with the observed SIRT1 stability, MG132 treatment did not significantly increase SIRT1 ubiquitination (Fig. 4C). In contrast, endogenous SIRT1 protein levels decreased dramatically after cells were treated for 16 h with etoposide and H2O2 (Fig. 4D), consistent with the increase in SIRT1 ubiquitination after treatment with etoposide (Fig. 2A) and H2O2 (Fig. 2D). The deacetylation mimic mutant K311R was more stable than WT SIRT1 after treatment with etoposide (Fig. 4E). Therefore, ubiquitination has a limited effect on SIRT1 stability under normal physiological conditions but promotes SIRT1 degradation after exposure to DNA damaging agents.
FIGURE 4.
SIRT1 stability is regulated by DNA damage. A and B, various cell lines were treated with 50 μg/ml cycloheximide (CHX) alone or together with 50 μm MG132 (MG), 20 μm chloroquine (CQ), or both for the indicated times or left untreated. SIRT1 and tubulin protein levels were examined by Western blot. C, ubiquitination of endogenous SIRT1 in the presence or absence of MG132. D, cells were treated with etoposide and H2O2 for the indicated times, and SIRT1 protein level was assessed by Western blot. E, WT and K311R stably transfected HeLa cells were treated with 50 μm etoposide for the indicated times. Protein levels of WT and K311R SIRT1 were examined by Western blot using anti-SIRT1 antibody. F, SIRT1 deacetylase activity in vivo. HeLa cells were co-transfected with FLAG-p53 and a vector control (V) or an increasing amount of WT, K311R, and H353Y SIRT1. Acetylation of p53 at K382 was examined by Western blot using anti-Ac-K382 p53 antibody. Then, the membrane was stripped and reprobed with anti-FLAG antibodies. G, SIRT1 deacetylase activity in vitro. GST-SIRT1 was first ubiquitinated by GST-MDM2 in vitro as described in Fig. 1G. An aliquot of the reaction mixture (containing 0.5 μg SIRT1) was incubated with 100 μm acetylated p53 peptides and 3 mm NAD+ for 1 h at 32 °C. Fluorescence was measured using a fluorometric plate reader, and is presented as relative fluorescence units (RFU).
Lys-311 is located within the enzymatic domain of SIRT1. Therefore, we determined whether SIRT1 de-ubiquitination affects its deacetylase activity. WT, K311R, and H353Y SIRT1 were expressed with FLAG-p53 in HeLa cells. Acetylation of p53 was detected using anti-Ac-K382 p53 antibodies. As shown in Fig. 4F, there was no significant difference in SIRT1 deacetylase activity toward p53 between WT and K311R. In a complementary experiment, GST-SIRT1 was incubated with MDM2 and subjected to an in vitro fluorometric assay to examine SIRT1 deacetylase activity. No significant difference in p53 deacetylation was observed between non-ubiquitinated GST-SIRT1 and ubiquitinated GST-SIRT1 (Fig. 4G).
Ubiquitination Affects SIRT1 Nuclear Localization during DDR
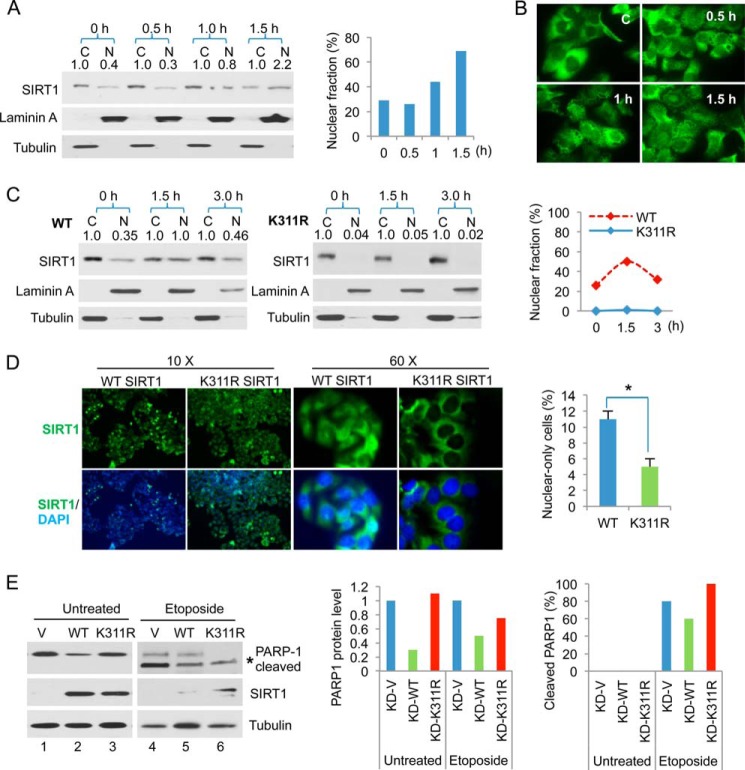
SIRT1 is reported to localize in both the cytoplasm and the nucleus (30–32). Therefore, we determined whether SIRT1 localization is regulated by DNA damage. The prostate cancer cell line DU145, in which SIRT1 resides primarily in the cytoplasm, was treated with a time course of IR. SIRT1 cellular localization was analyzed using immunofluorescence and cell fractionation. We found a small but reproducible increase in nuclear SIRT1 after exposure to IR for 1–1.5 h (Fig. 5, A and B). Next, we assessed whether de-ubiquitination affected SIRT1 cellular localization. Our results showed that WT SIRT1 was localized in the nucleus and cytoplasm, whereas K311R SIRT1 remained mostly cytoplasmic. The nuclear fraction of WT SIRT1 increased at 1.5 h post-IR and returned to approximately basal levels at 3 h post-IR. By contrast, K311R SIRT1 remained in the cytoplasm in response to IR (Fig. 5C). Immunofluorescence staining showed three types of SIRT1 localization patterns: cytoplasmic only, nuclear only, and both cytoplasmic and nuclear. We calculated the percentage of immunostained cells containing only nuclear SIRT1 in WT- and K311R-rescued cells. The results show that SIRT1 is localized in the cytoplasm in most K311R-rescued cells, with few cells showing only nuclear SIRT1 localization (Fig. 5D). We also observed an increase in nuclear SIRT1 localization in H2O2-treated cells (Fig. 2D, right), which is consistent with the increase in nuclear SIRT1 ubiquitination after H2O2 treatment (Fig. 2D, left).
FIGURE 5.
SIRT1 ubiquitination affects subcellular localization. A, DU145 cells were treated with 10 gray IR for the indicated times, and then nuclear (N) and cytoplasmic (C) extracts were prepared for Western blot analysis using antibodies specific for SIRT1, lamin A (nuclear marker), and tubulin (cytoplasmic marker). SIRT1 protein levels were determined by densitometry and are indicated for each fraction. The percentage of SIRT1 in the nucleus relative to total SIRT1 was calculated using the values determined by densitometry. B, DU145 cells were treated as above, and subcellular localization of endogenous SIRT1 was examined by immunofluorescence. C, HeLa cells expressing WT or K311R SIRT1 were treated as in A. The percentage of SIRT1 present in the nucleus was quantified relative to total SIRT1. D, immunofluorescence analysis of SIRT1 localization in WT and K311R-rescued HeLa cells. Nuclei were counterstained with DAPI. 10× and 60× images are shown. The percentage of cells containing only nuclear-localized SIRT1 was calculated and compared between WT and K311R cells. *, p < 0.002. E, SIRT1-KD cells were rescued with WT, K311R, or vector (V) control and treated with 20 μm etoposide for 24 h or left untreated. Because WT SIRT1 rescue substantially reduced PARP-1, 3-fold more WT cell lysate was loaded onto the fifth lane to clearly show PARP-1 cleavage. Total and cleaved PARP-1, SIRT1, and tubulin were examined by Western blot (left). PARP-1 protein levels in WT and K311R SIRT1 cells were normalized with respect to the vector control (middle). The percentage of PARP-1 cleavage was quantified relative to total PARP-1 (right).
We showed that K311R SIRT1 did not inhibit etoposide-induced apoptosis compared with that of WT SIRT1 (Fig. 3A). We explored the underlying molecular mechanism of this observation by examining PARP-1 expression level and protein cleavage because SIRT1 regulates PARP-1 transcription and activity (14). We found that SIRT1 knockdown significantly increased PARP-1 protein levels under normal conditions and after etoposide treatment (data not shown). Rescue of SIRT1-deficient cells with WT SIRT1, but not K311R SIRT1, reduced PARP-1 protein levels. Rescue with WT SIRT1 partially inhibited PARP-1 cleavage, which is an apoptosis marker. However, K311R SIRT1 rescue did not inhibit PARP-1 cleavage (Fig. 5E). These data suggest that K311R SIRT1 deficiency in regulating etoposide-induced apoptosis might be due to its ineffectiveness in regulating PARP-1 cleavage and expression (Fig. 3A).
DISCUSSION
SIRT1 is a multifunctional protein that has important roles in regulating restricted caloric intake-related longevity, insulin sensitivity, metabolism, mitochondrial respiration, DNA damage responses, apoptosis, autophagy, and other physiological activities (2–5). SIRT1 has well characterized functions in regulating the metabolism and multilayer regulation of DDR from damage sensing to cell-fate decision (6). In this study we explored the functional implication of SIRT1 ubiquitination under different DDR scenarios. We discovered that SIRT1 ubiquitination could be induced by different DNA damage stimuli, which results in different cellular responses. For example, SIRT1 knockdown sensitized cells to etoposide-induced apoptosis and reduced cell survival, indicating that SIRT1 protects cells from etoposide-induced DNA damage. In contrast, SIRT1 deficiency inhibited H2O2-induced cell death and facilitated cell survival, suggesting that SIRT1 plays a negative role in H2O2-induced damage response. These differences observed in SIRT1 function may result from differences in SIRT1 substrate activation.
Ubiquitination has both proteolytic and non-proteolytic functions and regulates subcellular localization, enzymatic activity, and protein-protein interactions (39–43). Our results indicate that ubiquitination does not affect SIRT1 degradation under normal conditions. This is consistent with the observation that SIRT1 might be ineffectively ubiquitinated under normal conditions. Treatment with etoposide and H2O2 significantly increases SIRT1 ubiquitination and reduces the level of SIRT1 protein. Therefore, we conclude that SIRT1 ubiquitination regulates protein degradation in response to DDR signaling. In this study we showed that MDM2 drove SIRT1 ubiquitination in vivo and in vitro. In fact, we also examined two other E3 ligases, hMIB1 and Cul4, which were candidates for SIRT1 interaction partners as revealed by mass spectrometry in our screen for SIRT1-interacting proteins, for their abilities to ubiquitinate SIRT1. However, neither could promote SIRT1 ubiquitination (data not shown). MDM2 is barely detectable in 293T and H1299 cells; however, overexpression of HA-Ub alone still promoted SIRT1 ubiquitination, although to a much lower extent than when co-transfected with MDM2 (Fig. 1, A and F). Therefore, it is possible that another as yet identified E3 ligase(s) exists to ubiquitinate SIRT1.
SIRT1 localization in the cytoplasm or nucleus depends on the cellular context (30–33). For example, SIRT1 is localized primarily in the nucleus of normal cells but resides in the cytoplasm of cancerous cells (31). SIRT1 can shuttle between the cytoplasm and nucleus in response to cell stimulation with growth factor (30, 32). Nuclear SIRT1 can interact with and deacetylate nuclear DDR protein substrates such as NBS1, PARP-1, Tip60, and hMOF. Cytoplasmic SIRT1 could access pro-apoptotic substrates such as Bax and caspases. Our data indicate that the K311R SIRT1 mutant shows a higher level of cytoplasmic localization than WT SIRT1 and that K311R-rescued cells exhibit greater apoptosis in response to etoposide-induced DNA damage compared with WT cells. These results are consistent with an earlier report that cytoplasmic localization of SIRT1 promotes apoptosis (30). Thus, we conclude that SIRT1 subcellular localization affects its functions during DDR.
In conclusion, our study reveals that SIRT1 ubiquitination is dynamically regulated by different types of DNA damage and that the ubiquitination response depends on the type of DNA damage. Although the present study focused on Lys-311 ubiquitination, there are several additional SIRT1 lysines that are modified by ubiquitination. Our future studies will determine whether these other SIRT1 lysines serve similar functions.
This work was supported, in whole or in part, by National Institutes of Health grants (to E. S.). This work was also supported by grants from Kaul Foundation (to E. S.) and a fellowship from the James and Esther King Biomedical Research Program (to L. P.).
- SIRT1
- Sirtuin 1
- DDR
- DNA damage response
- MDM2
- mouse double minute 2 homolog
- Ub
- ubiquitin
- KD
- knockdown
- IR
- ionizing radiation
- NTA
- Ni2+-nitrilotriacetic acid.
REFERENCES
- 1. Peng L., Seto E. (2011) Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb. Exp. Pharmacol. 206, 39–56 [DOI] [PubMed] [Google Scholar]
- 2. Finkel T., Deng C. X., Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Satoh A., Stein L., Imai S. (2011) The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb. Exp. Pharmacol. 206, 125–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haigis M. C., Guarente L. P. (2006) Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 5. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gorospe M., de Cabo R. (2008) AsSIRTing the DNA damage response. Trends Cell Biol. 18, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 8. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 9. Tang Y., Luo J., Zhang W., Gu W. (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 [DOI] [PubMed] [Google Scholar]
- 10. Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B. (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeong J., Juhn K., Lee H., Kim S. H., Min B. H., Lee K. M., Cho M. H., Park G. H., Lee K. H. (2007) SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp. Mol. Med. 39, 8–13 [DOI] [PubMed] [Google Scholar]
- 12. Yuan Z., Zhang X., Sengupta N., Lane W. S., Seto E. (2007) SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Chen J. (2010) SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J. Biol. Chem. 285, 11458–11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rajamohan S. B., Pillai V. B., Gupta M., Sundaresan N. R., Birukov K. G., Samant S., Hottiger M. O., Gupta M. P. (2009) SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell. Biol. 29, 4116–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guan D., Lim J. H., Peng L., Liu Y., Lam M., Seto E., Kao H. Y. (2014) Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell Death Dis. 5, e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 17. Jackson S. P., Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 [DOI] [PubMed] [Google Scholar]
- 18. Brooks C. L., Gu W. (2011) p53 regulation by ubiquitin. FEBS Lett. 585, 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu C. Y., Kang H. Y., Yang W. L., Wu J., Jeong Y. S., Wang J., Chan C. H., Lee S. W., Zhang X., Lamothe B., Campos A. D., Darnay B. G., Lin H. K. (2011) Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol. Chem. 286, 30806–30815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., Kimura H., Ikura M., Nishikubo S., Ito T., Muto A., Miyagawa K., Takeda S., Fishel R., Igarashi K., Kamiya K. (2007) DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27, 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mattiroli F., Vissers J. H., van Dijk W. J., Ikpa P., Citterio E., Vermeulen W., Marteijn J. A., Sixma T. K. (2012) RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 [DOI] [PubMed] [Google Scholar]
- 22. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin Z., Yang H., Kong Q., Li J., Lee S. M., Gao B., Dong H., Wei J., Song J., Zhang D. D., Fang D. (2012) USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484–494 [DOI] [PubMed] [Google Scholar]
- 24. Gao Z., Zhang J., Kheterpal I., Kennedy N., Davis R. J., Ye J. (2011) Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 286, 22227–22234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Zhang M., Dong H., Yong S., Li X., Olashaw N., Kruk P. A., Cheng J. Q., Bai W., Chen J., Nicosia S. V., Zhang X. (2009) Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28, 445–460 [DOI] [PubMed] [Google Scholar]
- 26. Peng L., Yuan Z., Ling H., Fukasawa K., Robertson K., Olashaw N., Koomen J., Chen J., Lane W. S., Seto E. (2011) SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 31, 4720–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan Y., Chen J. (2003) MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23, 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choo Y. S., Zhang Z. (2009) Detection of protein ubiquitination. J. Vis. Exp. 30, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin Q., Yan T., Ge X., Sun C., Shi X., Zhai Q. (2007) Cytoplasm-localized SIRT1 enhances apoptosis. J. Cell. Physiol. 213, 88–97 [DOI] [PubMed] [Google Scholar]
- 31. Byles V., Chmilewski L. K., Wang J., Zhu L., Forman L. W., Faller D. V., Dai Y. (2010) Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int. J. Biol. Sci. 6, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. (2007) Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 282, 6823–6832 [DOI] [PubMed] [Google Scholar]
- 33. Mattagajasingh I., Kim C. S., Naqvi A., Yamamori T., Hoffman T. A., Jung S. B., DeRicco J., Kasuno K., Irani K. (2007) SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 35. Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334 [DOI] [PubMed] [Google Scholar]
- 36. Peng L., Ling H., Yuan Z., Fang B., Bloom G., Fukasawa K., Koomen J., Chen J., Lane W. S., Seto E. (2012) SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol. Cell. Biol. 32, 2823–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Furukawa A., Tada-Oikawa S., Kawanishi S., Oikawa S. (2007) H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD+ depletion. Cell Physiol. Biochem. 20, 45–54 [DOI] [PubMed] [Google Scholar]
- 38. Cao C., Lu S., Kivlin R., Wallin B., Card E., Bagdasarian A., Tamakloe T., Wang W. J., Song X., Chu W. M., Kouttab N., Xu A., Wan Y. (2009) SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes. J. Cell Mol. Med. 13, 3632–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chastagner P., Israël A., Brou C. (2006) Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 7, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding Y., Zhang Y., Xu C., Tao Q. H., Chen Y. G. (2013) HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J. Biol. Chem. 288, 8289–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mao Y., Shang Y., Pham V. C., Ernst J. A., Lill J. R., Scales S. J., Zha J. (2011) Polyubiquitination of insulin-like growth factor I receptor (IGF-IR) activation loop promotes antibody-induced receptor internalization and down-regulation. J. Biol. Chem. 286, 41852–41861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. (2008) Control of AMPK-related kinases by USP9X and atypical Lys-29/Lys-33-linked polyubiquitin chains. Biochem. J. 411, 249–260 [DOI] [PubMed] [Google Scholar]
- 43. Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]