Background: Protein disulfide isomerase (PDI) family members are chaperones involved in apoptotic signaling through unclear mechanisms.
Results: Pharmacological inhibition of PDI and PDIA3 activities reduces apoptotic signaling. Purified PDI and PDIA3 proteins induce Bak-dependent mitochondrial outer membrane permeabilization in vitro.
Conclusion: PDI and PDIA3 possess proapoptotic function through inducing Bak oligomerization.
Significance: The data show a novel mechanism of PDI/PDIA3-mediated apoptosis.
Keywords: Apoptosis, B Cell Lymphoma 2 (Bcl-2), Bax, Endoplasmic Reticulum (ER), Protein Disulfide Isomerase, Bak, Mitochondrial Outer Membrane Permeabilization
Abstract
Protein disulfide isomerase (PDI) family proteins are classified as enzymatic chaperones for reconstructing misfolded proteins. Previous studies have shown that several PDI members possess potential proapoptotic functions. However, the detailed molecular mechanisms of PDI-mediated apoptosis are not completely known. In this study, we investigated how two members of PDI family, PDI and PDIA3, modulate apoptotic signaling. Inhibiting PDI and PDIA3 activities pharmacologically alleviates apoptosis induced by various apoptotic stimuli. Although a decrease of PDIA3 expression alleviates apoptotic responses, overexpression of PDIA3 exacerbates apoptotic signaling. Importantly, Bak, but not Bax, is essential for PDIA3-induced proapoptotic signaling. Furthermore, both purified PDI and PDIA3 proteins induce Bak-dependent, but not Bax-dependent, mitochondrial outer membrane permeabilization in vitro, probably through triggering Bak oligomerization on mitochondria. Our results suggest that both of PDI and PDIA3 possess Bak-dependent proapoptotic function through inducing mitochondrial outer membrane permeabilization, which provides a new mechanism linking ER chaperone proteins and apoptotic signaling.
Introduction
Protein disulfide isomerase (PDI)2 is a multidomain and multifunctional enzyme that reconstructs misfolded proteins (1). The PDI gene family currently comprises 21 genes, and PDI proteins are mainly present in the endoplasmic reticulum (ER) and also found in the nucleus, cytosol, mitochondria, and cell membrane (2). PDI (P4HB) is the first described member of the PDI gene family and plays a crucial role in oxidizing, reducing, and isomerizing disulfide bonds (S-S) both in vitro and in vivo (3). Another PDI family member, PDIA3 (also known as ERp57 and ERp60), also primarily localizes in the ER and catalyzes intramolecular disulfide bond formation in proteins (4).
In eukaryotic cells, ER stress responses frequently result in an unfolded protein response to induce up-regulated chaperone expression, such as PDI and PDIA3, to protect against misfolded protein aggregation (2, 5). Loss of PDIs activity has been associated with the pathogenesis of numerous disease states (6). In particular, PDI and PDIA3 prevent apoptotic cell death associated with ER stress and protein misfolding in various in vivo and in vitro models (7–13). The up-regulation of PDIA3 correlates with the accumulation of misfolded prion proteins and suppresses prion neurotoxicity (7), whereas reducing PDIA3 expression in cancer cells increases the apoptotic response to fenretinide (12). In response to hypoxia or transient forebrain ischemia in astrocytes, PDI is up-regulated and protects against apoptotic cell death (10). Inhibition of PDI enzymatic activity sensitizes cells to apoptosis induced by oxidized low-density lipoprotein (11), nitrosative stress (8), and chemotherapy drugs (13). Furthermore, in prion-infected animals, expression of prion protein mutants results in S-nitrosylation of PDI, and blocking cellular nitric oxide synthase inhibits S-nitrosylation of PDI and subsequent cytotoxicity (9).
Given their vital role in protein folding, PDI family members are generally considered to be prosurvival by assisting cells to adapt to the unfolded protein response (14). However, persistent ER stress causes the release of PDI from the ER, initiates apoptosis, and plays a critical role in the pathogenesis of multiple diseases (15–17). A recent study demonstrates that inhibiting PDI activities in rat brain cells suppresses apoptosis induced by misfolded huntingtin and amyloid precursor protein/β amyloid protein (18). This proapoptotic function of PDI is distinct from ER stress-mediated canonical apoptosis pathways (18). Therefore, it has been proposed that the up-regulation of PDI family members possesses a protective function to repair misfolded proteins and restores normal cellular homeostasis at the early stage of ER stress. However, apoptotic cell death pathways are initiated when the PDI accumulates at threshold levels in response to misfolded proteins, similar to p53 expression, which induces apoptosis at extreme levels of DNA damage (19). Recently, more evidence of proapoptotic activities of PDI members has emerged. In human endothelial cells, reducing PDIA3 expression inhibits hyperoxia- or tunicamycin-induced apoptosis by blocking caspase-3 activation and BiP/GRP78 induction (20). Similarly, decreasing or inhibiting activities of PDIA3 and GST pi 1(GSTP1) reduces oxidative processing and S-glutathionylation of Fas, resulting in cell survival (21). Furthermore, a decrease in PDIA3 expression attenuates the influenza A virus burden and reduces subsequent caspase-12 activation and apoptosis in epithelial cells (22). PDI may induce apoptosis through the reversal of iron(III)-mediated caspase-3 inhibition through the formation of iron-sulfur complexes at active-site thiols (23). Likewise, reducing endogenous PDI expression preferentially amends cytotoxicity caused by a mutant prion protein with an extra octa repeat insertion (9).
The proapoptotic functions of PDI family members and the detailed molecular mechanisms of apoptosis mediated by them, especially PDIA3, are not completely known. Therefore, a comprehensive study is necessary to elucidate the proapoptotic signaling of PDI and PDIA3. In this work, we explored whether PDI and PDIA3 directly induce apoptotic signaling and, if so, which mechanism(s) is/are involved. Our data suggest that both PDI and PDIA3 trigger MOMP through activating Bak, providing a new mechanism linking the unfolded protein response and apoptotic signaling.
EXPERIMENTAL PROCEDURES
Reagents
Actinomycin D, Polybrene, and thapsigargin were purchased from Sigma. Tunicamycin was purchased from Enzo (Farmingdale, NY). Propidium iodide was obtained from Invitrogen. Securinine, bacitracin, thiomuscimol, and muscimol were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). DMEM and penicillin/streptomycin were obtained from Mediatech (Manassas, VA), and fetal bovine serum was purchased from Gemini (Broderick, CA). The SensoLyte homogeneous Rh110 caspase-3/7 assay kit was purchased from AnaSpec (San Jose, CA). The antibodies used were anti-actin mAb (Sigma), anti-cytochrome c mAb (Santa Cruz Biotechnology), anti-Bak, N-terminal pAb (Millipore, Billerica, MA), anti-Tom40 pAb (Santa Cruz Biotechnology), anti-PDI pAb (Enzo, Western blot), anti-PDI pAb (Santa Cruz Biotechnology, immunofluorescence), anti-PDIA3 pAb (Enzo, Western blot), and anti-PDIA3 pAb antibodies (Santa Cruz Biotechnology, immunofluorescence). Recombinant human full-length PDI protein with a histidine tag at its N terminus was purchased from ProSpec-Tany (East Brunswick, NJ). Recombinant human full-length PDIA3 protein with a GST tag at its N terminus was purchased from Abnova (Walnut, CA). Recombinant Bcl-2 proteins Bax, hBcl-xL, and htBid were acquired as described previously (24).
Plasmids
Murine Bak cDNA or murine Bax cDNA was cloned into the retroviral expression vector pBABE-IRES-EGFP with the GFP functioning as an indicator expressed from an internal ribosomal entry site (IRES). The cDNAs of human PDI and PDIA3 were obtained from Origene (Rockville, MD) and subcloned into pBABE-IRES-EGFP. Human PDI cDNA or human PDIA3 cDNA was cloned into the retroviral expression vector pBABE-Puro. Murine Bak cDNA or murine Bax cDNA was also cloned into pEGFP-C1 (Clontech, Mountain View, CA). The identity of the plasmids was confirmed by sequencing. Lentiviral PDI shRNA and PDIA3 shRNA plasmids were purchased from Santa Cruz Biotechnology.
Retrovirus and Lentivirus Production
For retrovirus production, the package cell line HEK293T was transfected with the plasmids pBABE-mBak-IRES-EGFP, pBABE-mBax-IRES-EGFP, pBABE-hPDI-IRES-EGFP, pBABE-hPDIA3-IRES-EGFP,pBABE-Puro-hPDI, pBABE-Puro-hPDIA3, or the corresponding empty vector and two retroviral helper plasmids (pUMVC and pMD2.G) using jetPRIME® transfection reagent (Polyplus Transfection, New York, NY). Medium containing retrovirus was collected 48–72 h after transfection. To produce lentivirus, HEK293T cells were transfected with the shRNA plasmids along with the helper plasmids pMDLg/pRRE, pRSV.Rev, and pMDG2.0, with jetPRIME® transfection reagent used as a lipid transport milieu. Lentivirus in the medium was obtained 48–72 h after transfection.
Cell Lines
Bak−/−Bax−/− murine embryonic fibroblast (MEF) cells expressing the empty vector, Bak, or Bax were cultured as described previously (25). MEF cells overexpressing PDI or PDIA3 were generated by infection with the retroviral supernatants containing 10 μg/ml of Polybrene (Sigma) to increase infection efficiency. Over 95% of infected cells were GFP-positive, as measured by flow cytometry (FACScalibur, BD Biosciences, San Jose, CA). Because Bak−/−Bax−/− MEF cells reexpressing Bak or Bax are GFP-positive, retroviral medium obtained from cells transfected with pBABE-Puro-hPDI or pBABE-Puro-hPDIA3 was used to infect respective cells to overexpress PDI or PDIA3. Cell lines stably overexpressing PDI or PDIA3 were acquired by culturing cells in medium supplemented with 1.5 μg/ml puromycin. To generate MEF cells with reduced PDI and PDIA3 expression or vector control, medium containing lentivirus was used to infect MEF cells, and 10 μg/ml of Polybrene was added to the medium. Stable cell lines were obtained by culturing cells in the medium with 1.5 μg/ml puromycin. All MEF cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. MEF cells were cultured in a 5% CO2 tissue culture incubator at 37 °C.
Detecting Bak and Bax Oligomerization on Mitochondria and Cytochrome c Release
Mitochondria were isolated from wild-type and Bak−/−Bax−/− MEF cells reexpressing Bak, Bax, or the empty vector as described previously (26). Purified mitochondria were resuspended in buffer containing 12 mm HEPES (pH 7.5), 1.7 mm Tris-HCl (pH 7.5), 100 mm KCl, 140 mm mannitol, 23 mm sucrose, 2 mm KH2PO4, 1 mm MgCl2, 0.67 mm EGTA, and 0.6 mm EDTA supplemented with protease inhibitors (Complete, Roche Diagnostics). After incubation with purified PDI protein or PDIA3 protein and different recombinant Bcl-2 proteins at 30 °C for 1 h, mitochondrial vesicles were resuspended in 100 μl of 100 mm Na2CO3 (pH 11.3) and put on ice for 30 min to remove proteins loosely associated with mitochondria. Mitochondrial vesicles were then incubated with the chemical cross-linking agent bismaleimidohexane (0.1 mm) for 30 min at room temperature. Treated mitochondrial vesicles were centrifuged at 10,000 × g for 10 min, and vesicles were dissolved in 1× SDS-PAGE loading buffer. Proteins in the vesicle fractions were detected by Western blot analysis. Cytochrome c release experiments were carried out as described previously (24). Tom40 was used as a loading control.
Detection of the Insertion of Bax into Mitochondria
Mitochondria were isolated from Bak−/−Bax−/− MEFs as described previously (25). Purified mitochondria were resuspended in 12 mm HEPES (pH 7.5), 1.7 mm Tris-HCl (pH 7.5), 100 mm KCl, 140 mm mannitol, 23 mm sucrose, 2 mm KH2PO4, 1 mm MgCl2, 0.67 mm EGTA, and 0.6 mm EDTA supplemented with protease inhibitors (Complete, Roche Diagnostics). After incubation with PDI and PDIA3 and different recombinant Bcl-2 proteins at 30 °C for 1 h, mitochondrial vesicles were resuspended in 100 μl of 100 mm Na2CO3 (pH 11.3), and the suspension was put on ice for 30 min to remove proteins loosely associated with mitochondria. Treated mitochondrial vesicles were centrifuged at 10,000 × g for 10 min, and vesicles were dissolved in 1× SDS-PAGE loading buffer. Proteins in the vesicle fractions were detected by Western blot analysis. Tom40 was used as a loading control.
Cell Death Assay
After treatment for the indicated time, cells were collected in the presence of 1 μg/ml propidium iodide, and the percentage of live cells was measured using flow cytometry analysis as described previously (25, 26). The viability of treated cells was calculated as the percentage of the viability of untreated cells.
Caspase-3/7 Assay
Caspase-3/7 activity was detected directly in cells using the SensoLyte® homogeneous Rh110 caspase-3/7 assay kit (AnaSpec). Cells were plated in 96-well white-walled plates, and the assay was carried out according to the instructions of the manufacturer. The fluorescence signal (excitation/emission = 496/520 nm) was measured kinetically over 2 h at 1-min intervals using a Gemini EM microplate spectrofluorimeter (Molecular Devices, Sunnyvale, CA). Data were plotted as relative fluorescence units versus time, and the slope was determined (relative fluorescence units per minute).
Western Blot Analysis
Whole cell lysates of collected cells were prepared by lysing cells in radioimmune precipitation assay lysis buffer (150 mm sodium chloride, 1.0% (v/v) Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris-HCl (pH 8.0)) containing Complete protease inhibitors (Roche). The total protein concentration was measured using a BCA assay (Thermo, Waltham, MA). A 20-μg portion of total protein per sample was separated on a 4–12% BisTris gel (Bio-Rad) and transferred onto a PVDF membrane (Millipore). The membrane filters were incubated with appropriate primary and secondary antibodies and 10% (w/v) nonfat dried skimmed milk (Bio-Rad) in blotting buffer (1× PBS, 0.2% Tween 20). Proteins were detected using the enhanced chemiluminescence detection system (Thermo) as described previously (27).
Immunofluorescence
Bak−/−Bax−/− MEF cells (4 × 104) were plated on 18-mm coverslips in a 12-well tissue culture plate and cultured for 24 h. Cells were transiently transfected with the pEGFP-C1 plasmids carrying Bak cDNA or Bax cDNA using Lipofectamine 2000 transfection reagent (Life Technologies) following the instructions of the manufacturer. Forty-eight hours later, cells were washed twice with 1× Hanks' balanced salt solution followed by fixation with 0.4% paraformaldehyde for 15 min. After three washes with 1× Hanks' balanced salt solution, cells were permeabilized with 0.2% Triton X-100 for 15 min. Cells were then washed with incubation solution (0.02% Triton X-100 and 1.5% FBS in 1× Hanks' balanced salt solution) and incubated with primary antibodies in incubation solution for 3 h at room temperature. After three washes with incubation solution, cells were incubated with Alexa Fluor 594 goat anti-rabbit IgG for 3 h at room temperature and again washed three times with incubation solution. The coverslips were put on slides with mounting medium (Dako, Carpinteria, CA) and mounted onto a Nikon Eclipse Ti confocal microscope. Individual cells were visualized using a PlanApo ×60, 1.42 numerical aperture oil immersion objective and confocal images were acquired.
Statistical Analysis
All experiments were performed in triplicate at least three times independently. Results are presented as mean ± S.D. Statistical analysis was performed using Student's t test. p < 0.05 between groups was considered significant.
RESULTS
Cytotoxic Effects of Apoptotic Stimuli Depend on PDI and PDIA3 Catalytic Activities
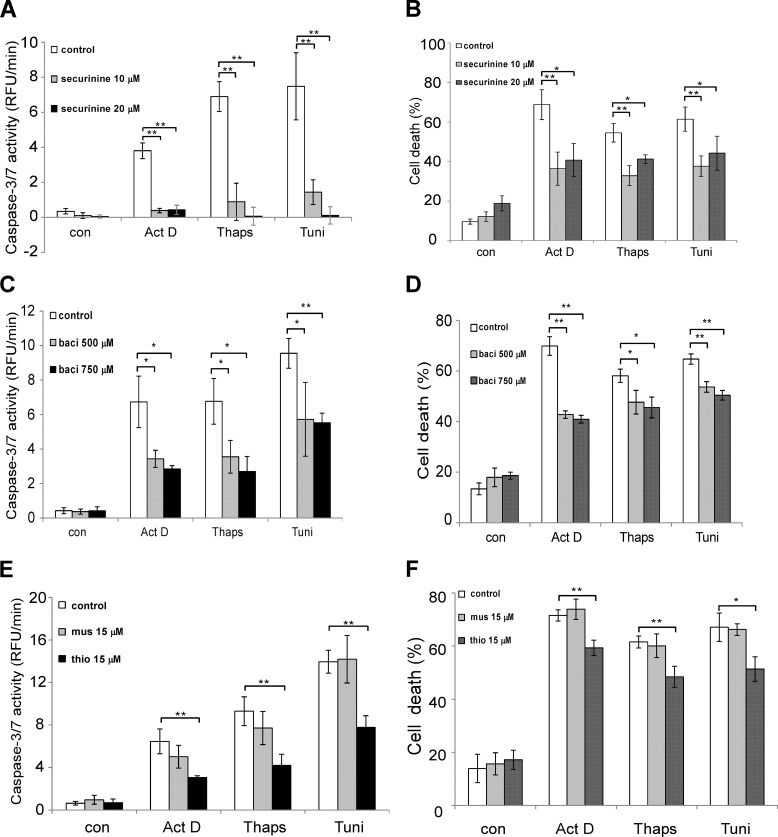
To explore the role of PDI family members in apoptotic signaling, we first investigated whether PDI and PDIA3 catalytic activities to facilitate protein folding are essential to mediate apoptotic responses. Several small molecules have been found to inhibit PDI and PDIA3 catalytic activities and have been used to elucidate the role of the protein folding catalyst in a variety of signaling pathways (18, 21, 28, 29). MEF cells were treated with a range of reagents known to induce apoptosis in the presence or absence of the PDI inhibitor securinine (18). The activity of the effector caspases, caspase-3 and caspase-7, was measured. Lower levels of caspase-3/7 activation were observed in cells preincubated with securinine in all apoptosis paradigms examined (Fig. 1A). Similarly, treatment with securinine resulted in less cell death, consistent with the caspase-3/7 data (Fig. 1B). To further confirm that PDI and PDIA3 catalytic activities are vital to mediate apoptotic signaling, two other PDI and PDIA3 inhibitors, bacitracin and thiomuscimol, were investigated (18, 21, 28, 29). In agreement with the study of securinine, both bacitracin and thiomuscimol alleviated the cytotoxic effects of different apoptotic stimuli in MEF cells (Fig. 1, C–F). Moreover, the thiomuscimol-inactive analog muscimol failed to decrease cell death and caspase-3/7 activation induced by apoptotic stimuli, indicating that the effects of thiomuscimol on cytotoxicity is due to PDI and PDIA3 catalytic activities (Fig. 1, E and F). These data suggest that both PDI and PDIA3 activities to promote protein folding are involved in apoptotic signaling in various apoptotic paradigms.
FIGURE 1.
Inhibiting PDI catalytic activities reduces apoptosis induced by apoptotic stimuli. A, caspase-3/7 activity was measured in wild-type MEFs after 24-h incubation with or without securinine, followed by a treatment with actinomycin D (Act D, 0.4 μg/ml), thapsigargin (Thaps, 0.4 μm), or tunicamycin (Tuni, 0.2 μg/ml) for 24 h. RFU, relative fluorescence unit; con, control. B, MEFs were preincubated with or without 10 or 20 μm securinine for 24 h. MEFs were then treated with actinomycin D (0.4 μg/ml), thapsigargin (0.7 μm), or tunicamycin (0.4 μg/ml) for 24 h, and cell viability was determined using flow cytometry analysis. C, after preincubation with or without 500 or 750 μm bacitracin for 24 h, MEFs were treated with actinomycin D (0.4 μg/ml), thapsigargin (0.4 μm), or tunicamycin (0.2 μg/ml) for 24 h, and caspase-3/7 activity was measured. baci, bacitracin. D, MEFs were preincubated with or without 500 or 750 μm bacitracin for 24 h. MEFs were treated with actinomycin D (0.4 μg/ml), thapsigargin (0.7 μm), or tunicamycin (0.4 μg/ml) for 24 h, and cell viability was measured. E, MEFs were preincubated with 15 μm thiomuscimol or 15 μm muscimol for 24 h. MEFs were then treated with actinomycin D (0.4 μg/ml), thapsigargin (0.4 μm), or tunicamycin (0.2 μg/ml) for 24 h, and caspase-3/7 activity was measured. mus, muscimol; thio, thiomuscimol. F, after incubation with 15 μm thiomuscimol or 15 μm muscimol for 24 h, MEFs were treated with actinomycin D (0.4 μg/ml), thapsigargin (0.7 μm), or tunicamycin (0.4 μg/ml) for 24 h, and cell viability was measured. All data are presented as mean ± S.D. of three different experiments that were carried out in triplicate. *, p < 0.05; **, p < 0.01; unpaired Student's t test.
Reducing PDIA3 Expression Decreases Apoptosis Induced by Apoptotic Stimuli
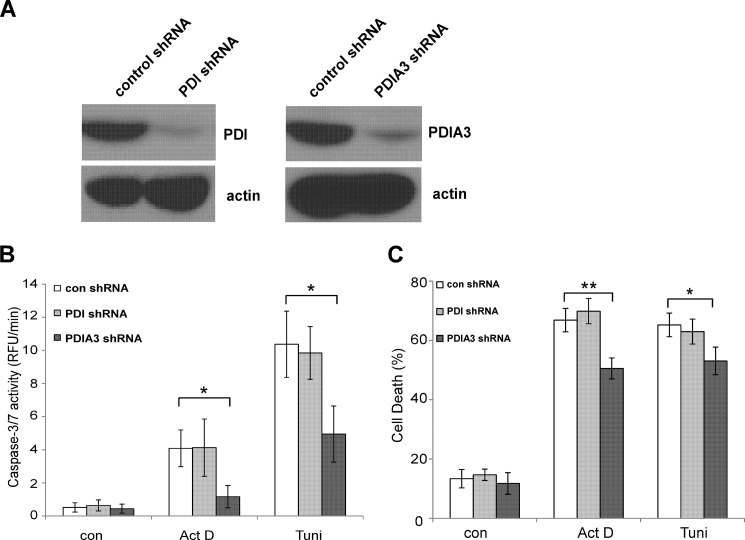
To elucidate the role of individual PDI family members in apoptotic signaling, we examined PDI and PDIA3, two members of PDI family implicated previously in apoptotic regulation (9, 18, 20–23). PDI and PDIA3 expression were stably reduced in MEF cells to a comparable degree using shRNA (Fig. 2A). Less actinomycin D- and tunicamycin-induced cell death and caspase-3/7 activation were observed in MEF cells with decreased PDIA3 expression compared with cells expressing a control shRNA vector (Fig. 2, B and C). In contrast, changes in PDI expression level failed to influence cellular responses to apoptotic stimuli, suggesting that some activity of PDI to mediate apoptotic signaling might be redundant.
FIGURE 2.
Decreasing expression of PDIA3, but not PDI, alleviates apoptotic signaling. A, PDI and PDIA3 expression in MEF cells was stably reduced by shRNA. The expression levels of PDI and PDIA3 were determined by Western blot analysis. B, less apoptosis was detected in MEF cells with reduced PDIA3 expression compared with cells with reduced PDI expression or control vector cells. Caspase-3/7 activity was assessed after 24-h incubation with actinomycin D (Act D, 0.4 μg/ml) or tunicamycin (Tuni, 0.2 μg/ml). RFU, relative fluorescence unit; con, control. C, cell death was measured after 24-h incubation with actinomycin D (0.4 μg/ml) or tunicamycin (0.4 μg/ml) of MEF cells with reduced PDIA1 and PDIA3 expression. Data are mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; unpaired Student's t test.
Overexpression of PDIA3 Exacerbates Apoptosis in Different Apoptotic Paradigms
To further explore the involvement of PDI and PDIA3 in mediating apoptosis, PDI and PDIA3 were stably overexpressed in MEF cells by retroviral infection. Western blot analysis showed that PDI and PDIA3 expression was increased significantly in MEF cells (Fig. 3A). Higher caspase-3/7 activation and cell death were observed in PDIA3-overexpressing cells compared with cells expressing the empty vector upon the treatment with actinomycin D or tunicamycin (Fig. 3, C and D). These data are consistent with the study of cells with reduced PDIA3 expression (Fig. 2), further validating the role of PDIA3 in apoptotic signaling. In accordance with PDI reduction, enhancing PDI expression failed to influence cellular responses to apoptotic stimuli (Fig. 3, B and D).
FIGURE 3.
Overexpression of PDIA3, but not PDI, enhances apoptosis induced by different apoptotic stimuli. A, PDI and PDIA3 were stably overexpressed in MEF cells by retroviral infection. The expression levels of PDI and PDIA3 were determined by Western blot analysis. B, no significant change of caspase3/7 activation was found in MEF cells with higher PDI expression following 24-h treatment with actinomycin D (Act D, 0.1 μg/ml) or tunicamycin (Tuni, 0.1 μg/ml). RFU, relative fluorescence unit. C, actinomycin D (0.1 μg/ml) or tunicamycin (0.1 μg/ml) induced more caspase-3/7 activation in MEF cells with higher PDIA3 expression following 24-h treatment. D, cell death was measured after 24-h incubation with actinomycin D (0.4 μg/ml) or tunicamycin (0.4 μg/ml) of MEF cells with higher PDI and PDIA3 expression. All data are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; unpaired Student's t test.
Decrease in Apoptotic Signaling Mediated by Reducing PDIA3 Expression Is Dependent on Bak
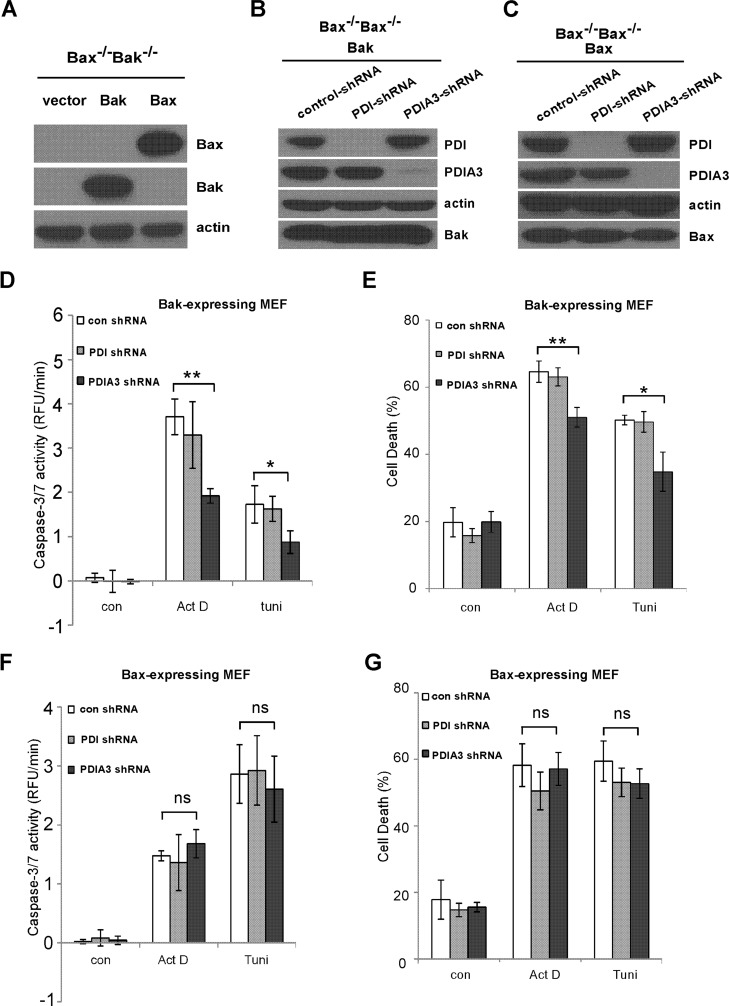
Bax and Bak are the two proapoptotic Bcl-2 family members for the execution of the apoptotic program (30). Although the functions of Bak and Bax are largely overlapping, they display distinct activities in certain apoptotic signaling (30, 31). To investigate the role of Bak and Bax in PDI- and PDIA3-induced apoptotic singling, Bax−/−Bak−/− MEF cell lines stably reexpressing only Bax or Bak were established (Fig. 4A). PDI and PDIA3 expression were then stably reduced in Bax−/−Bak−/− MEF cell lines reexpressing Bak only or Bax only to comparable degrees using shRNA (Fig. 4, B and C). Upon reducing PDIA3 but not PDI expression, less actinomycin D- and tunicamycin-induced caspase-3/7 activation and cell death were observed in Bax−/−Bak−/− MEF cells expressing Bak but not Bax (Figs. 4, D–G), indicating that PDIA3 modulates apoptotic signaling in a Bak-dependent fashion.
FIGURE 4.
PDIA3-induced proapoptotic signaling is dependent on Bak but not Bax. A, stable expression of Bak and Bax in Bak−/−Bax−/− MEF cells was examined by Western blot analysis. B, expression of PDI or PDIA3 was stably reduced by shRNA in Bak−/−Bax−/−MEF cells expressing Bak. C, stable reduction of PDI or PDIA3 expression in Bak−/−Bax−/− MEF cells reexpressing Bax was determined by Western blot analysis. D, upon treatment with actinomycin D (Act D, 0.2 μg/ml) or tunicamycin (tuni, 0.8 μg/ml) for 24 h, less caspase-3/7 activation was detected in Bak-expressing MEF cells with reduced PDIA3 expression. RFU, relative fluorescence unit; con, control. E, actinomycin D (0.4 μg/ml) or tunicamycin (0.8 μg/ml) induced less cell death in Bak−/−Bax−/− MEF cells expressing Bak but not Bax with reduced PDIA3 following 24-h treatment. F, no significant changes in caspase-3/7 activation was detected in Bak−/−Bax−/− MEF cells reexpressing Bax with reduced PDI or PDIA3 expression following 24-h treatment with actinomycin D (0.8 μg/ml) or tunicamycin (0.2 μg/ml). G, cell death was measured in Bax-expressing MEF cells with reduced PDIA1 or PDIA3 expression following 24-h incubation of actinomycin D (0.8 μg/ml) or tunicamycin (0.8 μg/ml). All data are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ns, not significant; unpaired Student's t test.
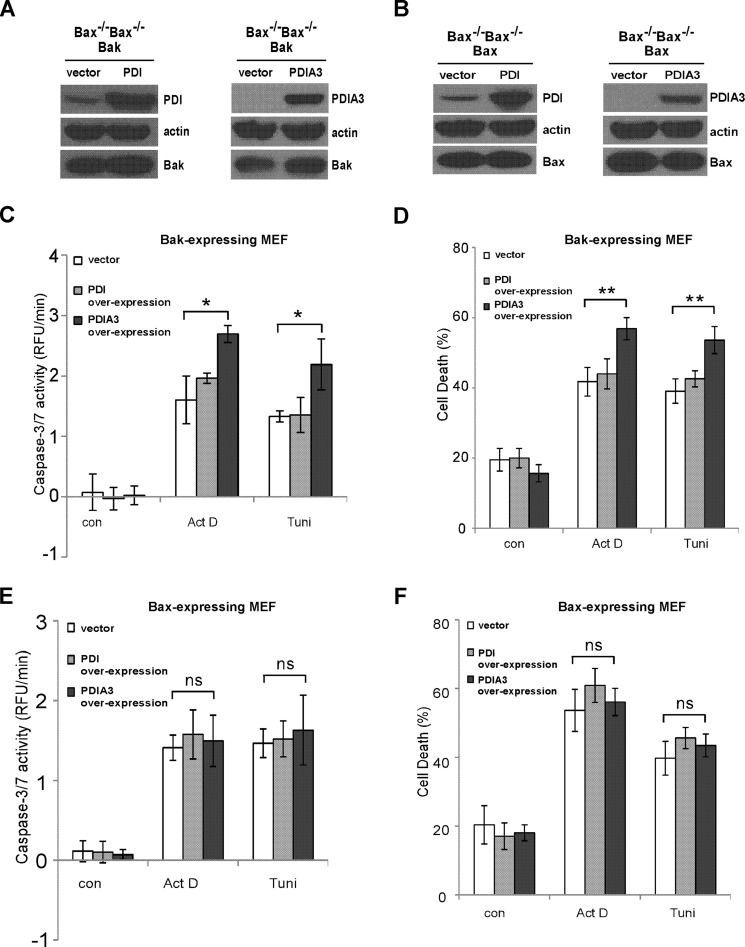
Bak Is Involved in Enhanced Apoptotic Signaling Mediated by PDIA3 Overexpression
To further explore the roles of Bak and Bax in apoptotic singling mediated by PDI and PDIA3, PDI or PDIA3 was stably overexpressed in Bak-expressing or Bax-expressing MEFs, respectively, by retroviral infection. As shown in Fig. 5, A and B, PDI and PDIA3 expression were increased significantly in these two cell lines. Upon treatment with actinomycin D or tunicamycin, more caspase-3/7 activation and cell death were detected in cells overexpressing PDIA3 but not in cells overexpressing PDI in a Bak-dependent manner (Fig. 5, C–F). In agreement with the knockdown experiments (Fig. 4), these data provide more evidence that Bak, but not Bax, is critical for PDIA3-induced apoptotic signaling.
FIGURE 5.
Bak, but not Bax, is involved in PDIA3-mediated proapoptotic signaling. A, PDI and PDIA3 were stably overexpressed in Bak−/−Bax−/− MEF cells expressing Bak by retroviral infection. The expression levels of PDI and PDIA3 were determined by Western blot analysis. B, stable overexpression of PDI and PDIA3 in Bak−/−Bax−/− MEF cells expressing Bax was examined by Western blot analysis. C and D, after treatment with actinomycin D (Act D, 0.1 μg/ml) or tunicamycin (Tuni, 0.4 μg/ml) for 24 h, more caspase-3/7 activation (C) and cell death (D) were detected in Bak-expressing MEF cells with reduced PDIA3 expression but not with reduced PDI expression. RFU, relative fluorescence unit; con, control. E, 24-h treatment with actinomycin D (0.8 μg/ml) or tunicamycin (0.1 μg/ml) induced the same level of caspase-3/7 activation in Bak−/−Bax−/− MEF cells expressing Bax with higher PDIA1 or PDIA3 expression. F, cell death was measured in Bax-expressing MEFs with increased PDIA1 or PDIA3 expression after 24-h incubation of actinomycin D (0.8 μg/ml) or tunicamycin (0.2 μg/ml). All data are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ns, not significant; unpaired Student's t test.
PDI and PDIA3 Induce Mitochondrial Outer Membrane Permeabilization
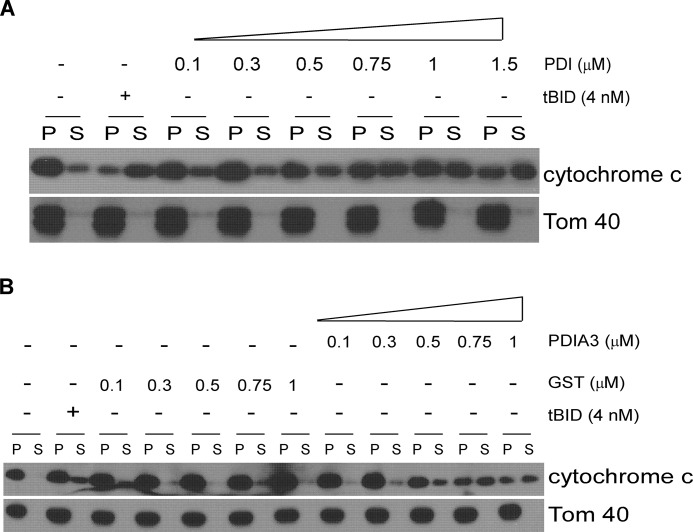
To investigate the detailed molecular mechanisms of apoptosis mediated by a particular PDI family member, we examined the effects of purified recombinant PDI and PDIA3 proteins on MOMP. Mitochondria isolated from wild-type MEFs were incubated with various concentrations of purified PDI protein, purified PDIA3 protein, or purified BH3-only Bcl-2 protein truncated Bid (tBid), which is known to permeabilize the mitochondrial outer membrane (32). The release of cytochrome c from mitochondria was determined as a parameter of MOMP. Like the well documented tBid (32), PDI or PDIA3 was able to induce cytochrome c release from mitochondria in a dose-dependent fashion (Figs. 6, A and B). These data indicate that both PDI and PDIA3 function directly on mitochondria to induce MOMP in vitro.
FIGURE 6.
PDI and PDIA3 induce MOMP in vitro. PDI and PDIA3 cause dose-dependent MOMP. Mitochondria purified from wild-type MEF cells were incubated with recombinant tBid protein or different concentrations of recombinant PDI protein (A) or recombinant PDIA3 protein (B). GST protein was used as a negative control of PDIA3. Cytochrome c in the mitochondrial vesicle fractions (pellet, P) or the released fractions (supernatant, S) was detected by Western blot analysis.
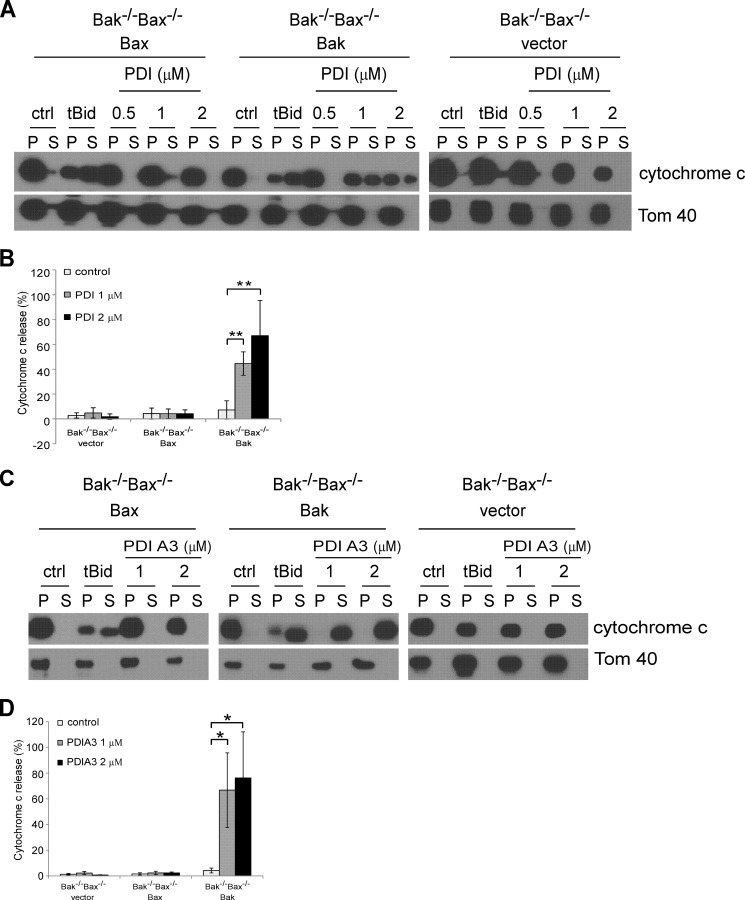
PDI- and PDIA3-induced MOMP Are Dependent on Bak but Not Bax
Bak and Bax are required for MOMP in almost all apoptotic paradigms (33). To investigate whether Bak and Bax are also vital in PDI- and PDIA3-induced MOMP, we examined the influence of PDI and PDIA3 on the integrity of mitochondria containing only Bak, only Bax, or neither of them. Mitochondria were purified from Bax−/−Bak−/− MEFs reexpressing the empty vector, Bak only, or Bax only. After incubation with purified PDI and PDIA3 proteins, mitochondrial membrane integrity was assessed. PDI induced cytochrome c release from mitochondria of Bak-expressing cells but not from those of Bax-expressing cells or the vector control cells (Figs. 7, A and B). By contrast, tBid was able to permeabilize mitochondria from both Bak-expressing and Bax-expressing cells. In agreement with PDI, purified PDIA3 was only able to induce permeabilization of mitochondria with Bak but not those with Bax (Fig. 7, C and D). These data indicate that both PDI and PDIA3 induce MOMP in a Bak- but not Bax-dependent fashion, which is consistent with the cellular study indicating that Bak but not Bax is involved in PDIA3-mediated proapoptotic signaling (Figs. 4 and 5).
FIGURE 7.
Bak, but not Bax, is involved in PDI and PDIA3-induced MOMP. PDI and PDIA3 induce Bak-dependent, but not Bax-dependent, cytochrome c release. A and C, mitochondria isolated from Bak−/−Bax−/− MEF cells expressing the empty vector, Bak, or Bax were incubated with recombinant tBid protein (4 nm), recombinant PDI protein (A), or recombinant PDIA3 protein (C). Cytochrome c in the mitochondrial vesicle fractions (pellet, P) or the released fractions (supernatant, S) was detected by Western blotting. ctrl, control. B, the intensities of cytochrome c in mitochondrial fractions and released fractions shown in A were quantified using ImageJ software (National Institutes of Health). D, the intensities of cytochrome c in mitochondrial fractions and released fractions shown in C were quantified. Cytochrome c release is represented as a percentage of the sum of the protein intensity in the supernatant and pellet. Mean ± S.D. for three independent experiments are shown. *, p < 0.05; **, p < 0.01; unpaired Student's t test.
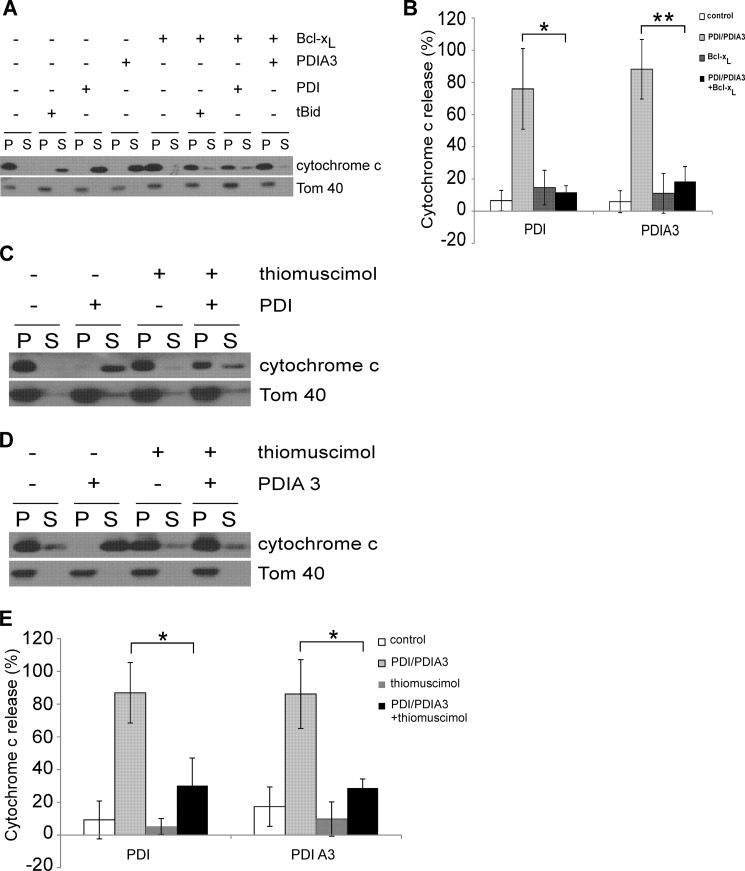
The Permeabilizing Activity of PDI or PDIA3 on the Mitochondrial Membrane Can Be Blocked
Antiapoptotic Bcl-2 protein Bcl-xL has been shown to inhibit apoptosis by blocking MOMP triggered by proapoptotic Bcl-2 protein in vitro (34). We investigated whether MOMP-induced by PDI and PDIA3 proteins was also modulated by Bcl-xL. MOMP assays were carried out using mitochondria isolated from wild-type MEFs (Fig. 8, A and B). The addition of Bcl-xL alone had a minimal effect on cytochrome c release. In the presence of purified PDI or PDIA3 protein, Bcl-xL was able to prevent MOMP, reminiscent of its ability to maintain MOM integrity under similar conditions with tBid (24). Our data suggest that PDI and PDIA3 induce MOMP in a similar fashion as the well studied proapoptotic BH3-only Bcl-2 proteins tBid.
FIGURE 8.
Bcl-xL and inhibiting PDI enzymatic activities regulate the permeabilizing activity of PDI and PDIA3. A, mitochondria purified from wild-type MEF cells were incubated with recombinant PDI protein (2 μm) or recombinant PDIA3 protein (2 μm) with or without recombinant Bcl-xL protein (0.5 μm). Cytochrome c release was determined by Western blot analysis. P, pellet, mitochondrial fraction; S, supernatant, released fraction. B, the intensities of cytochrome c in mitochondrial fractions and released fractions shown in A were quantified. Cytochrome c release is represented as a percentage of the sum of the protein intensity in the supernatant and pellet. C and D, mitochondria purified from wild-type MEF cells were incubated with 2 μm recombinant PDI protein (C) or 2 μm recombinant PDIA3 protein (D) in the absence or presence of thiomuscimol (10 mm). Cytochrome c in the mitochondrial fractions or the released fractions was assessed by Western blot analysis. E, cytochrome c intensities in mitochondrial fractions and released fractions presented in C and D were determined. Cytochrome c release is calculated as a percentage of the sum of the protein intensity in the supernatant and pellet. Data are mean ± S.D. for three independent experiments. *, p < 0.05; **, p < 0.01; unpaired Student's t test.
The best known enzymatic activities of PDI and PDIA3 are oxidizing, reducing, and isomerizing disulfide bonds (1), and our data suggest that PDI and PDIA3 enzymatic activities are involved in apoptotic signaling (Fig. 1). We studied whether their enzymatic activities are essential for their ability to permeabilize mitochondria. Isolated mitochondria from wild-type MEFs were incubated with PDI or PDIA3 proteins with or without the PDI and PDIA3 enzymatic activity inhibitor thiomuscimol (18). Thiomuscimol failed to affect MOMP by itself but blocked MOMP triggered by PDI or PDIA3 (Fig. 8, C–E), providing evidence that the PDI enzymatic activities are essential for its effects on MOM integrity. Overall, our results suggest that permeabilization of mitochondria by PDI or PDIA3 is a controlled biological process.
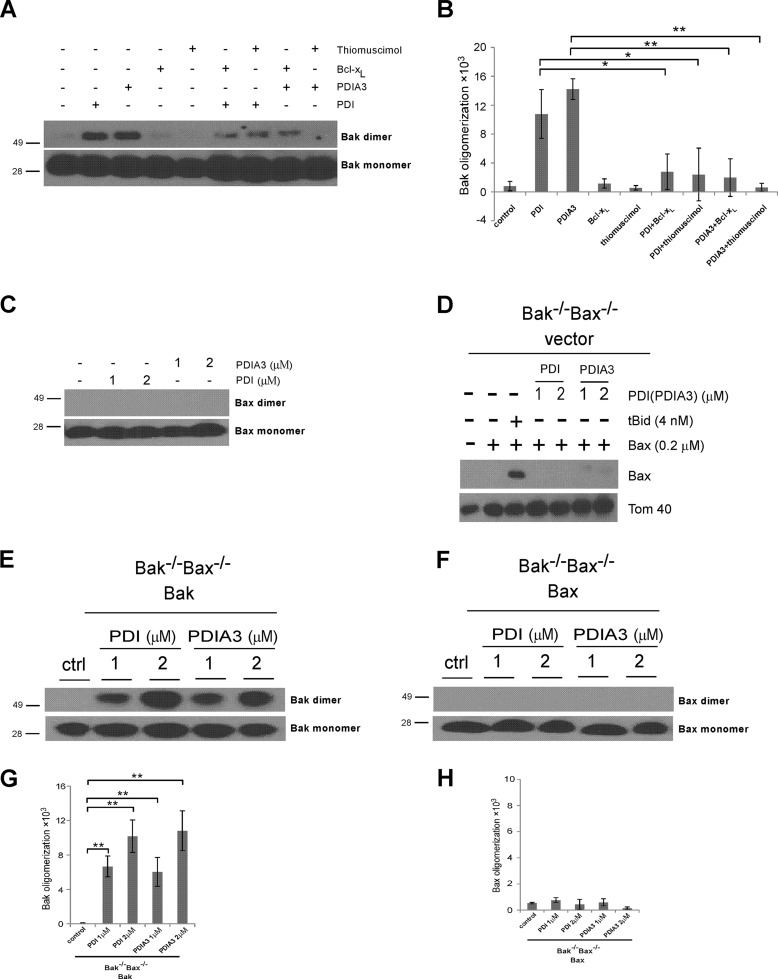
PDI and PDIA3 Induce Bak, but Not Bax, Oligomerization in Mitochondria
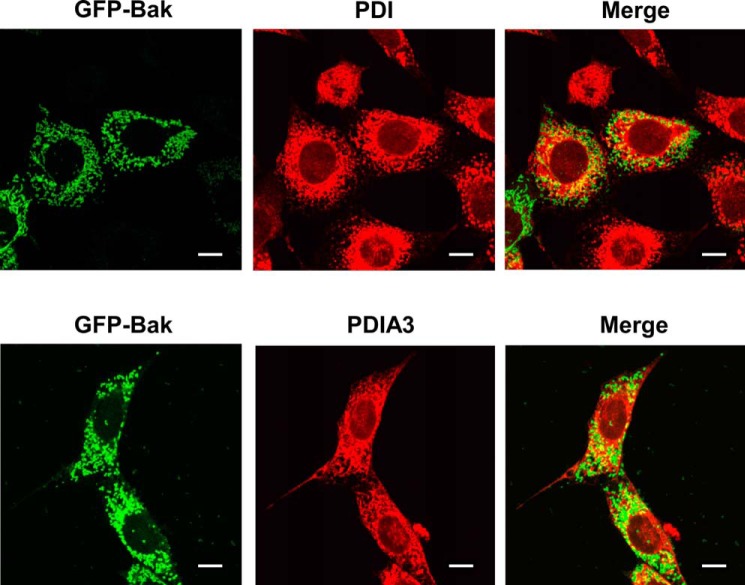
The oligomerization of Bak in the mitochondrial outer membrane is one of the ultimate control points for the mitochondrial apoptosis pathway, which is thought to be responsible for forming permeation pores on mitochondria (32). Because PDI and PDIA3 induce Bak-dependent cytochrome c release, we examined whether they were able to alter Bak protein conformation on mitochondria in a fashion similar to the proapoptotic BH3-only Bcl-2 protein tBid (35). After incubation with purified PDI or PDIA3 protein, mitochondria isolated from wild-type MEF cells were treated with the sulfhydryl-reactive cross-linker bismaleimidohexane to cross-link the oligomerized proteins. Bak oligomerization on mitochondria was increased by both PDI and PDIA3, as indicated by the enhanced intensity of slowly migrating bands recognized by Bak antibodies following treatment (Fig. 9A). Importantly, PDI- or PDIA3-induced oligomerization of Bak was reduced markedly by Bcl-xL, correlating with its influence on MOMP, as shown in Fig. 8. Similar results were obtained using the PDI inhibitor thiomuscimol (Fig. 9). Therefore, Bak oligomerization on mitochondria induced by PDI or PDIA3 is a regulated biological process that depends on its catalytic activities and is modulated by antiapoptotic Bcl-2 proteins. In contrast, PDI and PDIA3 failed to induce detectable Bax oligomerization on mitochondria (Fig. 9C). Furthermore, we isolated mitochondria from Bax−/−Bak−/− MEFs and incubated them with purified recombinant Bax along with tBid, PDI, or PDIA3. Unlike the well documented Bax activator tBid, PDI or PDIA3 failed to induce Bax insertion into mitochondria, providing more evidence that Bax is not involved in PDI- or PDIA3-induced cytochrome c release (Fig. 9D). Moreover, in accordance with PDI- and PDIA3-induced Bak, but not Bax, oligomerization in mitochondria isolated from wild-type MEFs (Fig. 9, A and C), PDI and PDIA3 induced Bak oligomerization in mitochondria from Bax−/−Bak−/− MEF cell lines reexpressing only Bak but failed to trigger Bax oligomerization in mitochondria from Bax-reexpressing cells (Fig. 9, E and F). In addition, immunofluorescent studies demonstrated that both PDI and PDIA3 were partially colocalized with Bak (Fig. 10). In summary, these results reveal a correlation between the assembly of Bak oligomers and the increase in MOM permeability in the presence of PDI or PDIA3 in vitro, suggesting that PDI or PDIA3 permeabilizes the mitochondrial outer membrane by triggering Bak conformational change and subsequent Bak oligomerization on mitochondria.
FIGURE 9.
PDI and PDIA3 induce Bak, but not Bax, oligomerization in mitochondria. A, mitochondria purified from wild-type MEF cells were incubated with the indicated combination of PDI protein (2 μm), PDIA3 protein (2 μm), Bcl-xL protein (0.5 μm), and thiomusciol (10 mm). After treatment with the cross-linking reagent bismaleimidohexane, Bak oligomerization in mitochondria was determined by Western blot analysis. The molecular weights of protein standards are indicated. B, the intensities of Bak dimer in mitochondria shown in A were quantified. The dimerization of Bak levels were normalized to the control sample with buffer incubation only. C, mitochondria isolated from wild-type MEF cells were incubated with the indicated concentrations of PDI or PDIA3 protein. Following the treatment with the cross-linking reagent bismaleimidohexane, Bax oligomerization in mitochondria was determined by Western blot analysis. The molecular weights of protein standards are indicated. D, PDI or PDIA3 failed to induce Bax insertion into mitochondria. Mitochondria purified from Bak−/−Bax−/− MEFs were incubated with Bax and either tBid, PDI, or PDIA3. Levels of Bax inserted into mitochondria were determined by Western blot analysis. E and F, mitochondria purified from Bak−/−Bax−/− MEF cells expressing Bak only (E) or Bax only (F) were incubated with the indicated combination of PDI or PDIA3 protein. Bak or Bax oligomerization in mitochondria was determined by Western blot analysis. ctrl, control. G and H, the intensities of Bak or Bax dimer in mitochondria shown in E and F were quantified. Data are mean ± S.D. for three independent experiments. **, p < 0.01; unpaired Student's t test.
FIGURE 10.
PDI and PDIA3 are partially colocalized with Bak. GFP and Bak fusion proteins were transiently expressed in Bak−/−Bax−/− MEFs. The intracellular distribution of PDI or PDIA3 was detected by immunofluorescent staining. The fluorescence was visualized by confocal microscopy. Scale bars = 10 μm.
DISCUSSION
The mitochondrially mediated apoptotic pathway is the most common form of apoptosis (36). In this pathway, MOMP is recognized to be a “no return” step during apoptotic signaling (37). The induction of MOMP leads to the release of cytochrome c from the mitochondrial intermembrane space, which serves as a cofactor for apoptotic protease-activating factor 1 (APAF-1) to trigger apoptosis (38). Although Hoffstrom et al. (18) have reported that PDI purified from bovine liver is able to cause MOMP in vitro, the PDI preparation contains more than one member of PDI family proteins.3 It is still unclear which particular PDI family member is essential for MOMP. In this study, purified recombinant PDI and PDIA3 proteins were examined, and the evidence that catalytic activities of PDI members are vital for their roles in apoptotic signaling are presented. Both PDI and PDIA3 directly induced MOMP in a fashion similar to the BH3-only Bcl-2 protein tBid in vitro. Importantly, the activity of PDI and PDIA3 on mitochondrial membrane permeability was also diminished by the antiapoptotic Bcl-2 protein Bcl-xL. The permeabilizing activity of PDI and PDIA3 on the mitochondrial membrane is likely attributed to their ability to induce Bak oligomerization.
During MOMP, Bax or Bak oligomerize and insert stably into the mitochondrial outer membrane, which is an important prerequisite for MOMP (32). Bax and Bak are considered functionally redundant because the activation of either of them could induce apoptosis in almost all apoptosis paradigms examined, and loss of Bax or Bak alone fails to provide significant protection against apoptosis (39, 40). However, accumulating evidence also suggests that they might have non-redundant roles in apoptosis induced by certain death stimuli (30, 31, 41–44). Studies show that Bak, but not Bax, is essential for cytochrome c release or apoptosis induced by NBK/BIK (41), Bcl-xS (43), Neisseria gonorrhoeae (Ngo), and cisplatin (31). In this study, we also found that Bak, but not Bax, is essential for PDIA3- induced proapoptotic signaling (Figs. 4 and 5) in cells. Importantly, both PDI- and PDIA3-induced MOMP is Bak-dependent but not Bax-dependent, probably through causing Bak oligomerization on mitochondria (Figs. 7 and 9). These corroborative findings provide the first evidence that proapoptotic ER proteins induce apoptosis exclusively via Bak activation. Indeed, antiapoptotic Bcl-2 proteins can block MOMP through binding to deactivate Bak, which is a prevailing model of Bcl-2 protein interactions inherent to the regulation of MOMP (45). Consistent with this model, Bcl-xL blocked the proapoptotic activities of PDI and PDIA3 in our studies (Figs. 8 and 9). Therefore, we propose that Bcl-xL directly binds to Bak and, subsequently, deactivates Bak to inhibit PDI- and PDIA3-induced Bak oligomerization and MOMP.
It is generally believed that ER stress is an adaptive mechanism to preserve cell function and survival. However, constant ER stress triggers apoptosis, which has a critical role in the development of many diseases (6). Our previous studies indicate that PDI is released from the ER lumen during ER stress (26). Recent studies have also shown that several ER luminal chaperones released from the ER lumen exhibit unique proapoptotic activities in various apoptotic paradigms (46–48). Importantly, this study demonstrates that PDI and PDIA3 protein directly induce MOMP in vitro by selectively activating Bak on mitochondria. It is conceivable that released ER luminal PDIs directly induce Bak-dependent MOMP by catalyzing Bak intermolecular disulfide bond formation. Alternatively, PDIs might accumulate at the mitochondria-associated ER subcompartment to trigger MOMP, as suggested by the studies of human neuronal cells expressing polyglutamine-expanded huntingtin exon 1 (18).
In this work, molecules inhibiting PDI and PDIA3 catalytic activities (securinine, thiomuscimol, or bacitracin) decreased apoptotic cell death induced by different kinds of apoptotic insults (actinomycin D, thapsigargin, and tunicamycin) (Fig. 1). The broad protective effects of PDI and PDIA3 in different apoptosis paradigms suggest that they are involved in a critical step of the canonical intrinsic mitochondrial apoptotic pathway.
Reducing PDIA3 protein expression alleviated apoptotic cell death caused by apoptotic stimuli (Fig. 2), which is consistent with earlier reports (20–22). Furthermore, overexpression of PDIA3 increased caspase-3/7 activation and cell death induced by actinomycin or tunicamycin in MEF cells (Fig. 3, C and D). These corroborative results suggest that PDIA3 exerts its proapoptotic activities in MEF cells. Although purified PDI protein permeabilizes mitochondria as effectively as purified PDIA3 protein in vitro (Fig. 6), reducing or enhancing PDI expression failed to influence the cytotoxic effects of actinomycin or tunicamycin on MEF cells (Figs. 2–5). One possible explanation for this discrepancy is that, in MEF cells, the majority of PDI is retained inside the ER lumen during apoptosis. Therefore, PDI is less likely to translocate to the mitochondrial outer membrane to induce MOMP than PDIA3. As a result, changes in PDI expression levels have a limited influence on cellular responses to apoptotic insults.
In summary, this study reveals novel proapoptotic actions of PDI and PDIA3, two chaperone proteins for reconstructing misfolded proteins. A better understanding of how PDI- and PDIA3-mediated apoptosis pathway may assist in discovering therapeutic approaches for protein misfolding diseases.
Acknowledgments
We thank Dr. John Eaton for intellectual input.
This work was supported, in whole or in part, by National Institutes of Health Grants CA106599, CA175003, and RR018733 (to C. L.). This work was also supported by National Natural Science Foundation of China Grant 81300769 (to H. L.), the James Graham Brown Cancer Center, and the University of Louisville School of Medicine Research Committee (to C. L.).
G. Zhao and C. Li, unpublished data.
- PDI
- protein disulfide isomerase
- ER
- endoplasmic reticulum
- MOMP
- mitochondrial outer membrane permeabilization
- pAb
- polyclonal antibody
- MEF
- mouse embryonic fibroblast
- IRES
- internal ribosomal entry site.
REFERENCES
- 1. Ellgaard L., Ruddock L. W. (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galligan J. J., Petersen D. R. (2012) The human protein disulfide isomerase gene family. Hum. Genomics 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Freedman R. B., Hirst T. R., Tuite M. F. (1994) Protein disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 19, 331–336 [DOI] [PubMed] [Google Scholar]
- 4. Coe H., Michalak M. (2010) ERp57, a multifunctional endoplasmic reticulum resident oxidoreductase. Int. J. Biochem. Cell Biol. 42, 796–799 [DOI] [PubMed] [Google Scholar]
- 5. Turano C., Coppari S., Altieri F., Ferraro A. (2002) Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell. Physiol. 193, 154–163 [DOI] [PubMed] [Google Scholar]
- 6. Mossuto M. F. (2013) Disulfide bonding in neurodegenerative misfolding diseases. Int. J. Cell Biol. 2013, 318319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hetz C., Russelakis-Carneiro M., Wälchli S., Carboni S., Vial-Knecht E., Maundrell K., Castilla J., Soto C. (2005) The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J. Neurosci. 25, 2793–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uehara T., Nakamura T., Yao D., Shi Z. Q., Gu Z., Ma Y., Masliah E., Nomura Y., Lipton S. A. (2006) S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 [DOI] [PubMed] [Google Scholar]
- 9. Wang S. B., Shi Q., Xu Y., Xie W. L., Zhang J., Tian C., Guo Y., Wang K., Zhang B. Y., Chen C., Gao C., Dong X. P. (2012) Protein disulfide isomerase regulates endoplasmic reticulum stress and the apoptotic process during prion infection and PrP mutant-induced cytotoxicity. PloS ONE 7, e38221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka S., Uehara T., Nomura Y. (2000) Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 275, 10388–10393 [DOI] [PubMed] [Google Scholar]
- 11. Muller C., Bandemer J., Vindis C., Camaré C., Mucher E., Guéraud F., Larroque-Cardoso P., Bernis C., Auge N., Salvayre R., Negre-Salvayre A. (2013) Protein disulfide isomerase modification and inhibition contribute to ER stress and apoptosis induced by oxidized low density lipoproteins. Antioxid. Redox Signal. 18, 731–742 [DOI] [PubMed] [Google Scholar]
- 12. Corazzari M., Lovat P. E., Armstrong J. L., Fimia G. M., Hill D. S., Birch-Machin M., Redfern C. P., Piacentini M. (2007) Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. Br. J. Cancer 96, 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lovat P. E., Corazzari M., Armstrong J. L., Martin S., Pagliarini V., Hill D., Brown A. M., Piacentini M., Birch-Machin M. A., Redfern C. P. (2008) Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 68, 5363–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 15. Ryu E. J., Harding H. P., Angelastro J. M., Vitolo O. V., Ron D., Greene L. A. (2002) Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson's disease. J. Neurosci. 22, 10690–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harding H. P., Ron D. (2002) Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes 51, S455–S461 [DOI] [PubMed] [Google Scholar]
- 17. Ni M., Lee A. S. (2007) ER chaperones in mammalian development and human diseases. FEBS Lett. 581, 3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffstrom B. G., Kaplan A., Letso R., Schmid R. S., Turmel G. J., Lo D. C., Stockwell B. R. (2010) Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 6, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helton E. S., Chen X. (2007) p53 modulation of the DNA damage response. J. Cell. Biochem. 100, 883–896 [DOI] [PubMed] [Google Scholar]
- 20. Xu D., Perez R. E., Rezaiekhaligh M. H., Bourdi M., Truog W. E. (2009) Knockdown of ERp57 increases BiP/GRP78 induction and protects against hyperoxia and tunicamycin-induced apoptosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L44–L51 [DOI] [PubMed] [Google Scholar]
- 21. Anathy V., Roberson E., Cunniff B., Nolin J. D., Hoffman S., Spiess P., Guala A. S., Lahue K. G., Goldman D., Flemer S., van der Vliet A., Heintz N. H., Budd R. C., Tew K. D., Janssen-Heininger Y. M. (2012) Oxidative processing of latent Fas in the endoplasmic reticulum controls the strength of apoptosis. Mol. Cell. Biol. 32, 3464–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberson E. C., Tully J. E., Guala A. S., Reiss J. N., Godburn K. E., Pociask D. A., Alcorn J. F., Riches D. W., Dienz O., Janssen-Heininger Y. M., Anathy V. (2012) Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. Am. J. Respir. Cell and Mol. Biol. 46, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sliskovic I., Mutus B. (2006) Reversible inhibition of caspase-3 activity by iron(III): potential role in physiological control of apoptosis. FEBS Lett. 580, 2233–2237 [DOI] [PubMed] [Google Scholar]
- 24. White C., Li C., Yang J., Petrenko N. B., Madesh M., Thompson C. B., Foskett J. K. (2005) The endoplasmic reticulum gateway to apoptosis by Bcl-XL modulation of the InsP3R. Nat. Cell Biol. 7, 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao G., Zhu Y., Eno C. O., Liu Y., Deleeuw L., Burlison J. A., Chaires J. B., Trent J. O., Li C. (2014) Activation of the proapoptotic bcl-2 protein bax by a small molecule induces tumor cell apoptosis. Mol. Cell. Biol. 34, 1198–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X., Olberding K. E., White C., Li C. (2011) Bcl-2 proteins regulate ER membrane permeability to luminal proteins during ER stress-induced apoptosis. Cell Death Differ. 18, 38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eno C. O., Zhao G., Olberding K. E., Li C. (2012) The Bcl-2 proteins Noxa and Bcl-xL co-ordinately regulate oxidative stress-induced apoptosis. Biochem. J. 444, 69–78 [DOI] [PubMed] [Google Scholar]
- 28. Dickerhof N., Kleffmann T., Jack R., McCormick S. (2011) Bacitracin inhibits the reductive activity of protein disulfide isomerase by disulfide bond formation with free cysteines in the substrate-binding domain. FEBS J. 278, 2034–2043 [DOI] [PubMed] [Google Scholar]
- 29. Huber-Ruano I., Pinilla-Macua I., Torres G., Casado F. J., Pastor-Anglada M. (2010) Link between high-affinity adenosine concentrative nucleoside transporter-2 (CNT2) and energy metabolism in intestinal and liver parenchymal cells. J. Cell. Physiol. 225, 620–630 [DOI] [PubMed] [Google Scholar]
- 30. Cartron P. F., Juin P., Oliver L., Martin S., Meflah K., Vallette F. M. (2003) Nonredundant role of Bax and Bak in Bid-mediated apoptosis. Mol. Cell. Biol. 23, 4701–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kepp O., Rajalingam K., Kimmig S., Rudel T. (2007) Bak and Bax are non-redundant during infection- and DNA damage-induced apoptosis. EMBO J. 26, 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chipuk J. E., Green D. R. (2008) How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renault T. T., Chipuk J. E. (2014) Death upon a kiss: mitochondrial outer membrane composition and organelle communication govern sensitivity to BAK/BAX-dependent apoptosis. Chem. Biol. 21, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuwana T., Newmeyer D. D. (2003) Bcl-2-family proteins and the role of mitochondria in apoptosis. Curr. Opin. Cell Biol. 15, 691–699 [DOI] [PubMed] [Google Scholar]
- 35. Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X. (2001) The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933 [PubMed] [Google Scholar]
- 37. Newmeyer D. D., Ferguson-Miller S. (2003) Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112, 481–490 [DOI] [PubMed] [Google Scholar]
- 38. Jiang X., Wang X. (2004) Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 39. Lindsten T., Ross A. J., King A., Zong W. X., Rathmell J. C., Shiels H. A., Ulrich E., Waymire K. G., Mahar P., Frauwirth K., Chen Y., Wei M., Eng V. M., Adelman D. M., Simon M. C., Ma A., Golden J. A., Evan G., Korsmeyer S. J., MacGregor G. R., Thompson C. B. (2000) The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell 6, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimazu T., Degenhardt K., Nur-E-Kamal A., Zhang J., Yoshida T., Zhang Y., Mathew R., White E., Inouye M. (2007) NBK/BIK antagonizes MCL-1 and BCL-XL and activates BAK-mediated apoptosis in response to protein synthesis inhibition. Genes Dev. 21, 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang C., Youle R. J. (2012) Predominant requirement of Bax for apoptosis in HCT116 cells is determined by Mcl-1's inhibitory effect on Bak. Oncogene 31, 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lindenboim L., Kringel S., Braun T., Borner C., Stein R. (2005) Bak but not Bax is essential for Bcl-x (s)-induced apoptosis. Cell Death Differ. 12, 713–723 [DOI] [PubMed] [Google Scholar]
- 44. Madesh M., Zong W. X., Hawkins B. J., Ramasamy S., Venkatachalam T., Mukhopadhyay P., Doonan P. J., Irrinki K. M., Rajesh M., Pacher P., Thompson C. B. (2009) Execution of superoxide-induced cell death by the proapoptotic Bcl-2-related proteins Bid and Bak. Mol. Cell. Biol. 29, 3099–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Llambi F., Moldoveanu T., Tait S. W., Bouchier-Hayes L., Temirov J., McCormick L. L., Dillon C. P., Green D. R. (2011) A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gardai S. J., McPhillips K. A., Frasch S. C., Janssen W. J., Starefeldt A., Murphy-Ullrich J. E., Bratton D. L., Oldenborg P. A., Michalak M., Henson P. M. (2005) Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123, 321–334 [DOI] [PubMed] [Google Scholar]
- 47. Burikhanov R., Zhao Y., Goswami A., Qiu S., Schwarze S. R., Rangnekar V. M. (2009) The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell 138, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Obeid M., Panaretakis T., Joza N., Tufi R., Tesniere A., van Endert P., Zitvogel L., Kroemer G. (2007) Calreticulin exposure is required for the immunogenicity of γ-irradiation and UVC light-induced apoptosis. Cell Death Differ. 14, 1848–1850 [DOI] [PubMed] [Google Scholar]