Background: Centaurin-β2 specifically recognizes Rab35 but not the other 59 Rabs.
Results: The T76S/T81A mutation in the switch II region of Rab35 specifically impaired its centaurin-β2 binding activity.
Conclusion: Thr-76 and Thr-81 are key residues of Rab35 for the specific recognition by centaurin-β2.
Significance: Rab35(T76S/T81A) should be a useful tool for evaluating the functional significance of the Rab35/centaurin-β2 interaction in Rab35-dependent cellular events.
Keywords: Membrane Trafficking, Neurite Outgrowth, Rab, Site-directed Mutagenesis, Small GTPase, Ankyrin
Abstract
The small GTPase Rab35 is a molecular switch for membrane trafficking that regulates a variety of cellular events. We previously showed that Rab35 promotes neurite outgrowth of nerve growth factor-stimulated PC12 cells through interaction with centaurin-β2 (also called ACAP2). Centaurin-β2 is the only Rab35-binding protein reported thus far that exclusively recognizes Rab35 and does not recognize any of the other 59 Rabs identified in mammals, but the molecular basis for the exclusive specificity of centaurin-β2 for Rab35 has remained completely unknown. In this study, we performed deletion and mutation analyses and succeeded in identifying the residues of Rab35 and centaurin-β2 that are crucial for formation of a Rab35·centaurin-β2 complex. We found that two threonine residues (Thr-76 and Thr-81) in the switch II region of Rab35 are responsible for binding centaurin-β2 and that the same residues are dispensable for Rab35 recognition by other Rab35-binding proteins. We also determined the minimal Rab35-binding site of centaurin-β2 and identified two asparagine residues (Asn-610 and Asn-691) in the Rab35-binding site as key residues for its specific Rab35 recognition. We further showed by knockdown-rescue approaches that neither a centaurin-β2 binding-deficient Rab35(T76S/T81A) mutant nor a Rab35 binding-deficient centaurin-β2(N610A/N691A) mutant supported neurite outgrowth of PC12 cells, thereby demonstrating the functional significance of the Rab35/centaurin-β2 interaction during neurite outgrowth of PC12 cells.
Introduction
The Rab family is the largest family within the Ras superfamily of small GTPases and is conserved in all eukaryotes. The members of the Rab family are generally thought to be key players in membrane trafficking, which underlies a variety of cellular events (1–4). Rabs act as switch molecules that cycle between two nucleotide-bound states, a GTP-bound active state and a GDP-bound inactive state, and the cycling is controlled by two regulatory factors, a guanine nucleotide exchange factor, which activates Rabs, and a GTPase-activating protein (GAP),2 which inactivates Rabs (5, 6). The active Rabs drive various steps of membrane trafficking, including vesicle budding from donor membranes, vesicle movement along the cytoskeleton, vesicle docking to acceptor membranes, and vesicle fusion, by recruiting their specific effector molecules (1–4).
The results of recent comprehensive screenings for mammalian Rab effectors have indicated that interactions between Rabs and their effectors are more complicated than previously thought (7, 8), and most Rab isoforms appear to interact with two or more different types of effector molecules. Because mammalian cells and tissues are highly specialized, the presence of multiple Rab effectors may enable a single Rab isoform to control different types (or steps) of membrane trafficking in specialized cells. However, the physiological significance of the presence of multiple Rab effectors and their Rab recognition mechanisms are poorly understood.
Rab35 is one such Rab protein and has been shown to bind various candidate effector molecules, including MICAL-1 (7), MICAL-L1 (7, 9, 10), MICAL-cl (7), OCRL (7, 11), RUSC2 (12), Fascin1 (13), and centaurin-β2 (8, 14), and to be involved in various cellular events, including cytokinesis (11, 15, 16), cell migration (17, 18), phagocytosis (19, 20), immunological synapse formation (21), myelination (22), and neurite outgrowth (10, 14, 23, 24), most likely through regulation of endocytic recycling (25). The pleiotropic roles of Rab35 in membrane trafficking may be attributable to the presence of multiple Rab35 effectors, but the involvement of individual Rab35 effectors in the above cellular events has remained largely unknown. Moreover, almost nothing is known about the structural basis of Rab35·effector complexes, e.g. about the critical amino acid residues for the Rab35/effector interactions. Although identification of a specific amino acid(s) in Rab35 that is exclusively involved in interaction with only one effector molecule would enhance our understanding of the molecular mechanism of Rab35-mediated membrane trafficking, no attempts have ever been made to biochemically analyze Rab35/effector interactions.
In this study, we focused on centaurin-β2 (also called ACAP2), an Arf6-GAP (26) that is required for nerve growth factor (NGF)-induced neurite outgrowth of PC12 cells (8, 14), and analyzed the exclusive specificity of the Rab35/centaurin-β2 interaction with Rab35 by performing deletion and mutation analyses. The results showed that two Thr residues (Thr-76 and Thr-81) in the switch II region of Rab35 are responsible for binding centaurin-β2 and that they are not responsible for Rab35 binding to other effectors. Knockdown-rescue experiments showed that a centaurin-β2 binding-deficient Rab35 mutant, Rab35(T76S/T81A), did not support NGF-induced neurite outgrowth. Based on our findings, we discuss the utility of the Rab35(T76S/T81A) mutant as a tool to investigate the involvement of the Rab35·centaurin-β2 complex in Rab35-dependent cellular events.
EXPERIMENTAL PROCEDURES
Antibodies
Horseradish peroxidase (HRP)-conjugated anti-FLAG tag (M2) mouse monoclonal antibody, anti-FLAG tag antibody-conjugated agarose (Sigma-Aldrich), HRP-conjugated anti-T7 tag mouse monoclonal antibody (Merck Biosciences Novagen, Darmstadt, Germany), anti-centaurin-β2 goat polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-GFP rabbit polyclonal antibody (MBL, Nagoya, Japan), and anti-β-actin mouse monoclonal antibody (Applied Biological Materials, Richmond, British Columbia, Canada) were obtained commercially. Anti-Rab35 antibody was prepared as described previously (14).
Plasmid Construction
Mutant mouse Rab35 expression plasmids carrying a Thr-to-Ser and Thr-to-Ala double mutation at amino acid positions 76 and 81, respectively, named Rab35(T76S/T81A); a swapping mutation in the switch II region between Rab35 (amino acids (AA) 70–76) and Rab5A (AA82–88) named Rab35(S5A) (see Fig. 2A for details); or a short hairpin RNA (shRNA)-resistant mutant named Rab35SR were produced by PCR sewing methods essentially as described previously (27). In brief, PCRs were performed to generate two DNA fragments having overlapping ends into which specific alterations were introduced by using two sets of oligonucleotides, e.g. 5′-CGGATCCATGGCCCGGGACTACGACCA-3′ (Rab35-Met primer; sense) and 5′-ATGGGCCCCCCGATAATAGGAAGA-3′ (Rab35(T76S/T81A)-3′ primer; antisense) and 5′-TCTTCCTATTATCGGGGGGCCCAT-3′ (Rab35(T76S/T81A)-5′ primer; sense) and 5′-TTAGCAGCAGCTTTCTTTCG-3′ (Rab35-stop primer; antisense) (substituted nucleotides are underlined, and stop codons are in bold). After purification of the two DNA fragments, they were combined to generate the fusion product by a second PCR in which the Rab35-Met primer and Rab35-stop primer were used. The resulting Rab35 mutant cDNAs were subcloned into the pEGFP (enhanced green fluorescent protein)-C1 vector (Clontech-Takara Bio Inc., Shiga, Japan), pGBD-C1 vector (28), and pEF-FLAG tag expression vector (29). Mutant mouse centaurin-β2 fragments carrying N610A, N632A, T648A, N691A, T695A/E697A, N724A, S730A, L733A/Y734A, Q764A/Q765A, N610A/N691A, or T648A/N691A mutation(s) were similarly produced by the method described above, and they were subcloned into the pEGFP-C1 vector, pAct2 vector (Clontech-Takara Bio Inc.), and/or pEF-T7 tag expression vector (29). Deletion mutants of centaurin-β2 (AA580–770, AA630–770, AA662–770, AA580–755, AA580–745, AA580–730, AA580–697, and AA630–730; see Fig. 5A for details) were prepared by conventional PCR techniques as described previously (30). cDNAs encoding the ankyrin repeat (ANKR) domain of centaurin-β1 (Centβ1-ANKR; AA548–740) and of centaurin-β5 (Centβ5-ANKR; AA565–833) and full-length Fascin1 were amplified by conventional PCR techniques as described previously (29) and then subcloned into the pAct2 vector and/or pEF-T7 tag expression vector. Deletion mutants of centaurin-β2 (pAct2-Centβ2-ANKR and pEGFP-C1-Centβ2SR(ΔANKR)) were prepared as described previously (8, 14). shRNAs targeting rat Centβ2 (shCentβ2; 19-base target site, 5′-GGGTATCTGTTCAAACGAG-3′) and rat Rab35 (shRab35; 19-base target site, 5′-TATTAGTGGGCAATAAGAA-3′) were also prepared as described previously (14).
FIGURE 2.
Involvement of the switch II region of Rab35 in the binding of Rab35BPs. A, sequence alignment of the switch II regions of mouse Rab5A and Rab35. Amino acid residues that are conserved in the two sequences are shown against a black background. The central parts of their switch II region (dashed line) were swapped, and the Rab35(S5A) mutant contains part of the switch II region of Rab5A. B, the switch II region of Rab35 is required for binding all Rab35BPs except RUSC2. Yeast two-hybrid assays were performed as described in the legend for Fig. 1C. C, the Rab35(S5A) mutant hardly interacted with centaurin-β2 at all in contrast to the weak centaurin-β2 binding activity of the Rab35(T76S/T81A) mutant. Co-immunoprecipitation assays were performed as described in the legend for Fig. 1E. The positions of the molecular mass markers (in kilodaltons) are shown on the left. IP, immunoprecipitation.
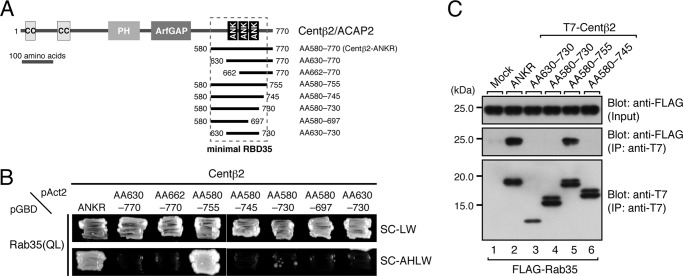
FIGURE 5.
Determination of the minimal Rab35-binding site in centaurin-β2. A, schematic representation of the deletion mutants of the ANKR domain of centaurin-β2 used in this study. AA numbers are shown on both sides of each construct. The minimal RBD35 determined in this study is indicated by enclosure with a broken line. B, Rab35 binding activities of the centaurin-β2 deletion mutants. Note that the ANKR domain of centaurin-β2 alone did not recognize Rab35 at all and that additional N-terminal and C-terminal regions adjacent to the ANKR domain were required for Rab35 binding activity. Yeast cells containing pAct2 plasmid expressing Centβ2-ANKR or each Centβ2-ANKR mutant (AA630–770, AA662–770, AA580–755, AA580–745, AA580–730, AA580–697, or AA630–730) and pGBD plasmid expressing Rab35(QL) were streaked on SC-LW (top panel) and SC-AHLW (bottom panel) and incubated at 30 °C. C, Rab35 binding activity of Centβ2-ANKR mutants (AA630–730, AA580–730, AA580–755, and AA580–745) in COS-7 cells. Co-immunoprecipitation assays were performed as described in the legend for Fig. 1E. The positions of the molecular mass markers (in kilodaltons) are shown on the left. IP, immunoprecipitation; PH, pleckstrin homology domain; ANK, ankyrin repeat.
Co-immunoprecipitation Assays in COS-7 Cells and Immunoblotting
T7-tagged centaurin-β2 (centaurin-β1/2/5-ANKR or Fascin1) and FLAG-tagged Rab35 (wild-type (WT), T76S/T81A, or S5A) were transiently expressed in COS-7 cells, and their associations were evaluated by co-immunoprecipitation assays with anti-FLAG tag antibody-conjugated agarose beads as described previously (29, 31). Proteins bound to the beads were analyzed by 10 or 12.5% SDS-PAGE followed by immunoblotting with HRP-conjugated anti-T7 tag antibody (1:10,000 dilution) and HRP-conjugated anti-FLAG tag antibody (1:10,000 dilution). Immunoreactive bands were visualized by enhanced chemiluminescence (GE Healthcare). The blots shown in this study are representative of at least three independent experiments.
Yeast Two-hybrid Assays
Yeast two-hybrid assays were performed by using pGBD-C1-Rab35(Q67L, Q67L/T76S/T81A, or Q67L/S5A) lacking the C-terminal geranylgeranylation site and pGAD-C1-RUSC2-RUN (12), pAct2-MICAL-1, pAct2-MICAL-cl, pAct2-MICAL-L1, pAct2-OCRL (7), pAct2-Fascin1, or pAct2-Centβ2-ANKR (8) or by using pGBD-C1-Rab35(Q67L or S22N)ΔCys (hereafter simply designated as QL or SN) and pAct2-Centβ2-ANKR(WT or its mutants), pAct2-Centβ1-ANKR, or pAct2-Centβ5-ANKR as described previously (8, 32). The yeast strain, medium, culture conditions, and transformation protocol used were as described previously (28). The assays were performed in duplicate, and the results of one representative set of assays are shown.
Cell Cultures and Transfections
PC12 cell and COS-7 cell cultures and plasmid transfections were performed essentially as described previously (14). Plasmids were transfected into cultured cells 1 day after plating by using Lipofectamine LTX or Lipofectamine 2000 (Invitrogen), each according to the manufacturer's instructions. Because the protein expression level of wild-type and mutant Rab35SR (or centaurin-β2SR) differs slightly even when the same amount of plasmid is transfected into PC12 cells, the amounts of plasmid used for transfection were varied to maintain the total amount of recombinant proteins at the same level (see Figs. 3C and 7C). To avoid overexpression in the knockdown-rescue experiments, we tried to maintain the level of the recombinant proteins at levels that were similar to the level of the endogenous protein (see Figs. 3C and 7C). Under our experimental conditions, the transfection efficiency of the EGFP-expressing plasmids into PC12 cells was ∼40–50%.
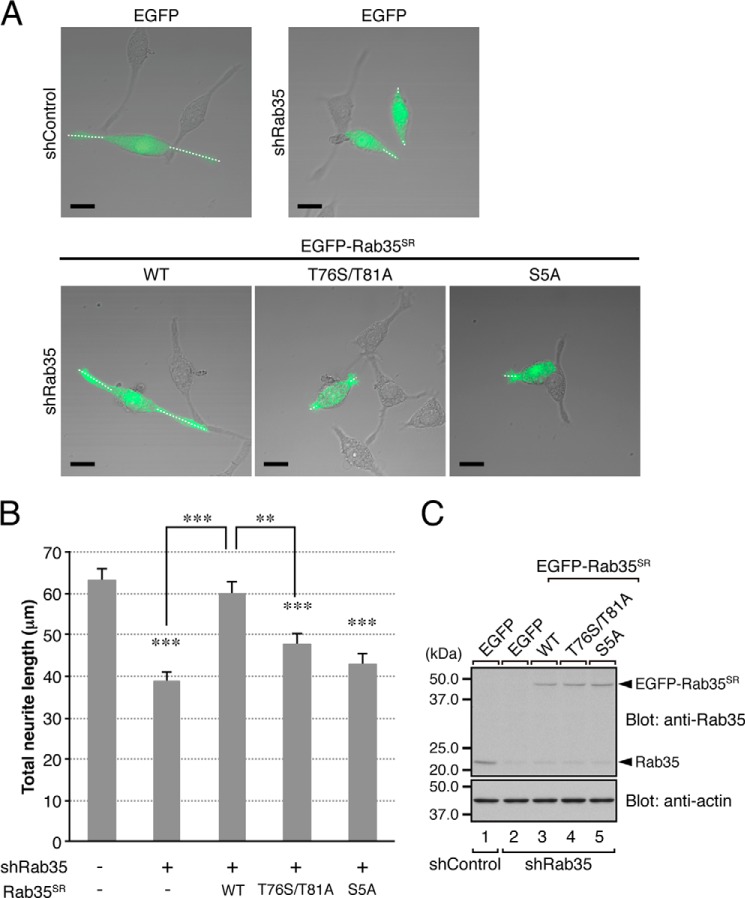
FIGURE 3.
Effect of the T76S/T81A mutation of Rab35 on NGF-induced neurite outgrowth of PC12 cells. A, typical images of PC12 cells (merged bright field images and EGFP fluorescence images) transiently expressing shControl or shRab35 together with pEGFP-C1 (top row) or together with pEGFP-C1-Rab35SR(WT, T76S/T81A, or S5A) (bottom row). The cells were fixed after NGF stimulation for 36 h and examined under a confocal microscope. Note that knockdown of Rab35 in PC12 cells inhibited neurite outgrowth (top right panel), whereas re-expression of Rab35SR(WT) (bottom left panel), but not of Rab35SR(T76S/T81A) (bottom middle panel) or Rab35SR(S5A) (bottom right panel), in shRab35-treated PC12 cells restored neurite outgrowth. Under our experimental conditions, manipulation of Rab35 had no significant effect on the number of neurites (shControl, 2.2 ± 0.06; shRab35, 2.0 ± 0.04; shRab35 + Rab35SR(WT), 2.3 ± 0.06; shRab35 + Rab35SR(T76S/T81A), 2.1 ± 0.05; and shRab35 + Rab35SR(S5A), 1.9 ± 0.05 (mean ± S.E.)), suggesting that Rab35 is involved in neurite extension rather than in neuritogenesis. Scale bars, 20 μm. B, quantification of total neurite length shown in A (sum of the lengths of the broken white lines in each PC12 cell). The bars represent the means and S.E. of data from three independent experiments (n = 100 cells; more than 30 cells were analyzed in each experiment). **, p < 0.01; ***, p < 0.001 (one-way analysis of variance followed by the Tukey-Kramer test). C, equivalent expression level of EGFP-Rab35SR(WT, T76S/T81A, or S5A) in shRab35-expressing PC12 cells. Cell lysates of PC12 cells expressing shRab35 together with EGFP-Rab35SR(WT, T76S/T81A, or S5A) were subjected to 12.5% SDS-PAGE followed by immunoblotting with anti-Rab35 antibody (top panel; 1:1,000 dilution) and anti-actin antibody (bottom panel; 1:20,000 dilution). The positions of the molecular mass markers (in kilodaltons) are shown on the left.
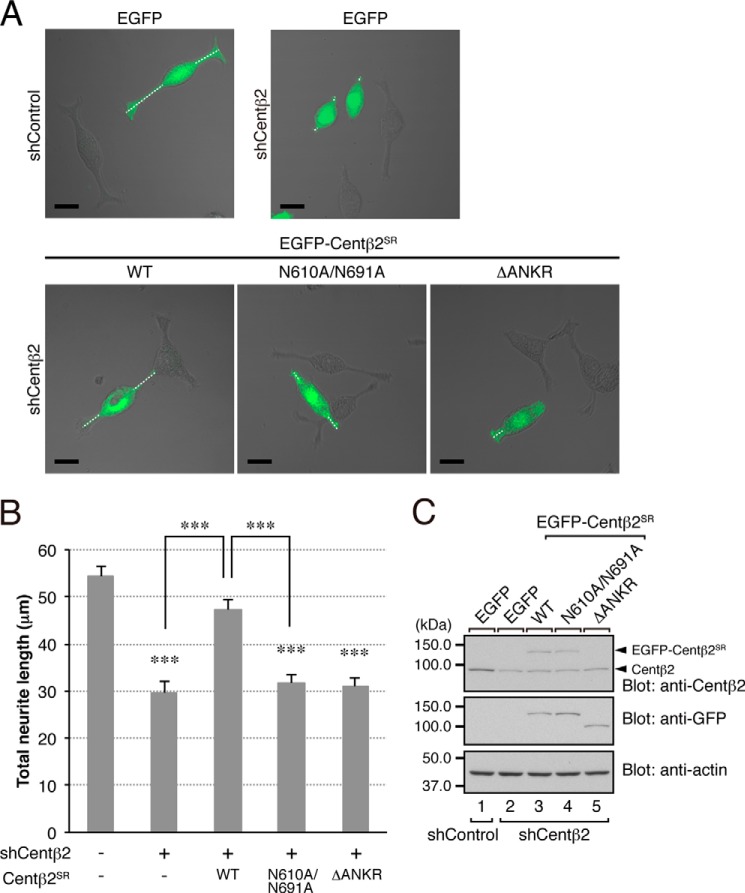
FIGURE 7.
Effect of the N610A/N691A mutation of centaurin-β2 on NGF-induced neurite outgrowth of PC12 cells. A, typical images of PC12 cells (merged bright field images and EGFP fluorescence images) transiently expressing shControl or shCentβ2 together with pEGFP-C1 (top row) or pEGFP-C1-Centβ2SR(WT, N610A/N691A, or ΔANKR) (bottom row). The cells were fixed after NGF stimulation for 36 h and examined under a confocal microscope. Note that knockdown of centaurin-β2 in PC12 cells inhibited neurite outgrowth (top right panel), whereas re-expression of Centβ2SR(WT) (bottom left panel), but not of Centβ2SR(N610A/N691A) (bottom middle panel) or Centβ2SR(ΔANKR) (bottom right panel), in shCentβ2-treated PC12 cells restored total neurite length. Under our experimental conditions, manipulation of centaurin-β2 had no significant effect on the number of neurites (shControl, 2.2 ± 0.05; shCentβ2, 2.0 ± 0.04; shCentβ2 + Centβ2SR(WT), 2.1 ± 0.04; shCentβ2 + Centβ2SR(N610A/N691A), 2.0 ± 0.04; and shCentβ2 + Centβ2SR(ΔANKR), 2.0 ± 0.05 (mean ± S.E.)), suggesting that centaurin-β2 is involved in neurite extension rather than in neuritogenesis, the same as Rab35 is. Scale bars, 20 μm. B, quantification of total neurite length shown in A (sum of the lengths of the broken white lines in each PC12 cell). The bars represent the means and S.E. of data from three independent experiments (n = 100 cells; more than 30 cells were analyzed in each experiment). ***, p < 0.001 (one-way analysis of variance followed by the Tukey-Kramer test). C, equivalent expression level of EGFP-Centβ2SR(WT, N610A/N691A, or ΔANKR) in shCentβ2-expressing PC12 cells. Cell lysates of PC12 cells expressing shCentβ2 together with EGFP-Centβ2SR(WT, N610A/N691A, or ΔANKR) were subjected to 10% SDS-PAGE followed by immunoblotting with anti-Centβ2 antibody (top panel; 1:500 dilution), anti-GFP antibody (middle panel; 1:1,000 dilution), and anti-actin antibody (bottom panel; 1:20,000 dilution). Because our anti-Centβ2 antibody recognized the C-terminal domain of centaurin-β2, it failed to detect EGFP-Centβ2SR(ΔANKR) expression (top panel, lane 5). The positions of the molecular mass markers (in kilodaltons) are shown on the left.
Immunofluorescence Analysis
All of the procedures used to perform the immunofluorescence analyses have been described previously (14)
Neurite Outgrowth Assays
Neurite outgrowth assays were performed essentially as described previously (14). In brief, PC12 cells that had been transfected with pSilencer plasmids together with EGFP-tagged protein-expressing plasmids were treated with 100 ng/ml β-NGF (Merck Biosciences) for 36 h. The transfected cells were identified by EGFP fluorescence, and images of the cells were captured at random with a confocal fluorescence microscope (Fluoview FV1000, Olympus, Tokyo, Japan). The total neurite length of each cell (see Figs. 3A and 7A, broken lines) was measured with MetaMorph software (Molecular Devices, Sunnyvale, CA). The results of the neurite outgrowth assays in this study are reported as means and S.E. of data from three independent experiments (n = 100 cells; more than 30 cells were analyzed in each experiment). Because the response of PC12 cells to NGF often differs according to the number of cell passages and the lot of NGF, we used PC12 cells with a similar passage number and NGF from the same lot in any one set of experiments.
RESULTS
Identification of Critical Residues in Rab35 for Specific Centaurin-β2 Binding by Site-directed Mutagenesis
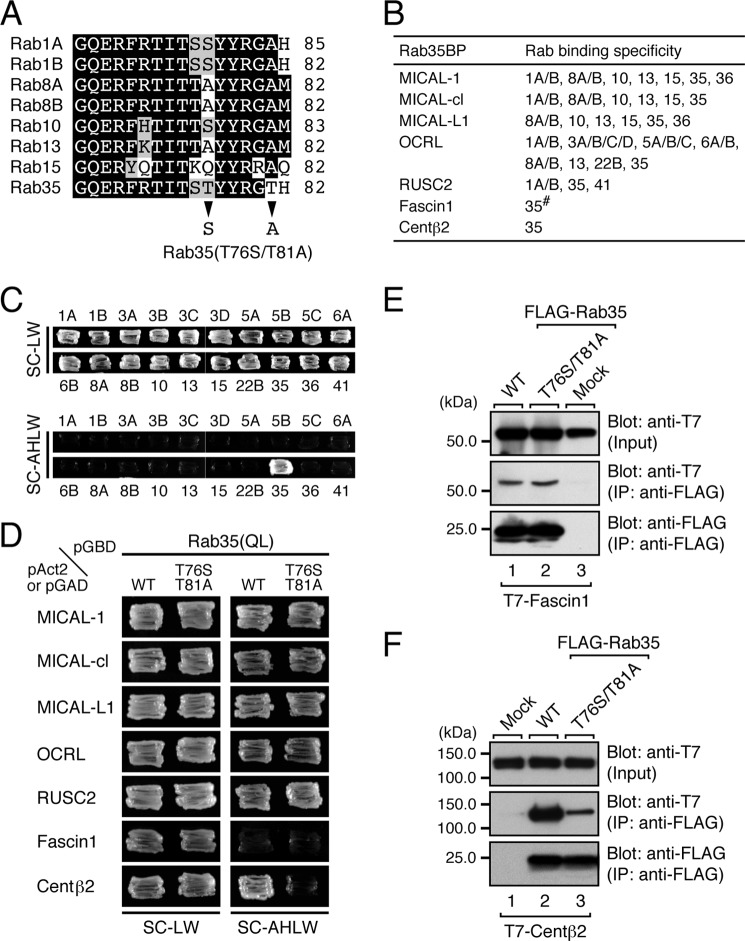
Seven Rab35-binding proteins (Rab35BPs) have been identified thus far (7–13), but their Rab binding specificity is highly diversified, ranging from the exclusive Rab35 binding activity of centaurin-β2 (Fig. 1C) to the multiple Rab binding activity of MICAL family proteins, all of which commonly interact with Rab8A/B, Rab10, Rab13, Rab15, and Rab35, and to the even broader Rab binding activity of OCRL, which interacts with 16 Rabs (summarized in Fig. 1B). In an attempt to identify the structural determinant responsible for the exclusive Rab35 binding specificity of centaurin-β2, we first focused on the switch II region of Rabs that interact with most of the Rab35BPs (Fig. 1A) because several amino acids in the switch II region of certain Rabs have previously been shown to be responsible for specific effector binding (32–38). Because Rab1A/B, Rab8A/B, Rab10, Rab13, Rab15, and Rab35 are phylogenetically similar and belong to the same large branch in the phylogenetic tree (39), the amino acid sequences of their switch II region are highly conserved. Careful inspection of their sequences, however, revealed two Thr residues, one at AA position 76 (Thr-76) and the other at AA position 81 (Thr-81), that are unique to Rab35 (Fig. 1A, arrowheads). To determine whether these two Thr residues are involved in the specific recognition of Rab35 by centaurin-β2, we performed site-directed mutagenesis and generated a Rab35(T76S/T81A) mutant that carries a switch II region of Rab1A/B (Fig. 1A, arrowheads). Intriguingly, the results of the yeast-two hybrid assays showed that the T76S/T81A mutation impaired centaurin-β2 binding activity (Fig. 1D, bottom panel) but had no effect on binding activity toward MICAL-1, MICAL-cl, MICAL-L1, OCRL, or RUSC2 (Fig. 1D, top five panels). By contrast, we were unable to evaluate the effect of the T76S/T81A mutation on Fascin1 binding by yeast two-hybrid assays because even the wild-type Rab35 did not interact with Fascin1 at all under our yeast two-hybrid assay conditions (Fig. 1D, second panel from the bottom). To overcome this problem, we performed co-immunoprecipitation assays in COS-7 cells, and the results showed that both Rab35(WT) protein and Rab35(T76S/T81A) protein did in fact interact with Fascin1 (Fig. 1E). We also investigated the impaired interaction between Rab35(T76S/T81A) and centaurin-β2 by performing co-immunoprecipitation assays in COS-7 cells. As shown in Fig. 1F, the T76S/T81A mutation dramatically decreased the binding activity toward centaurin-β2, although, in contrast to the results of the yeast two-hybrid assays (Fig. 1D), residual binding activity still persisted. These results indicated that both Thr-76 and Thr-81 in the switch II region of Rab35 are critical for recognition by centaurin-β2 but that they are not required for binding to other Rab35BPs.
FIGURE 1.
Identification of critical residues responsible for the specific binding of centaurin-β2 in the switch II region of Rab35 by site-directed mutagenesis. A, sequence alignment of the switch II regions of mouse Rab1A/B, Rab8A/B, Rab10, Rab13, Rab15, and Rab35. Amino acid residues in the sequences that are conserved in more than four switch II regions and that are similar are shown against a black background and a shaded background, respectively. Only two amino acids (arrowheads) in the switch II region of Rab1A/B and Rab35 are different, and we replaced the Thr-76 and Thr-81 of Rab35 with Ser and Ala, respectively, by site-directed mutagenesis (i.e. produced a T76S/T81A mutant, which mimics the switch II region of Rab1A). B, summary of Rab35BPs and their Rab binding specificity. The Rab binding specificity of all of the Rab35BPs except Fascin1 (#) has already been thoroughly investigated by yeast two-hybrid assays (7, 8, 12). C, centaurin-β2 specifically recognized Rab35 but did not recognize other Rabs that interact with MICALs, OCRL, and RUSC2. Yeast cells containing pAct2-cenaurin-β2-ANKR and pGBD-C1-Rabs(QL)ΔCys (7) were streaked on SC-LW (top panels) and SC-AHLW (selection medium; bottom panels) and incubated at 30 °C for 1 day and 1 week, respectively. D, two Thr residues of Rab35, Thr-76 and Thr-81, are critical for binding centaurin-β2 but not for binding other Rab35BPs. Yeast cells containing the pAct2 (or pGAD) plasmid expressing Rab35BP and pGBD plasmid expressing the constitutively active form (Rab35(QL)) of Rab35(WT) or Rab35(T76S/T81A) mutant were streaked on SC-LW (left panels) and SC-AHLW (right panels) and incubated at 30 °C for 1 day and 1 week, respectively. Note that the Rab35 containing the T76S/T81A mutations specifically abrogated binding activity toward centaurin-β2 (bottom right panel). E, the T76S/T81A mutation did not impair Facsin1 binding activity in co-immunoprecipitation assays. T7-tagged Facsin1 and FLAG-tagged Rab35(WT) or Rab35(T76S/T81A) mutant were co-expressed in COS-7 cells, and their associations were analyzed by co-immunoprecipitation assays with anti-FLAG tag antibody-conjugated agarose beads as described previously (29, 31). Co-immunoprecipitated T7-tagged Facsin1 (middle panel) and immunoprecipitated FLAG-tagged Rab35(WT) or Rab35(T76S/T81A) mutant (bottom panel) were detected with HRP-conjugated anti-T7 tag antibody and HRP-conjugated anti-FLAG tag antibody, respectively. Input, of the volume of the reaction mixture used for immunoprecipitation (IP) (top panel). The positions of the molecular mass markers (in kilodaltons) are shown on the left. F, the T76S/T81A mutation dramatically decreased the centaurin-β2 binding activity of Rab35, a finding that was consistent with the results of the yeast two-hybrid assays shown in D. Co-immunoprecipitation assays were performed as described in E.
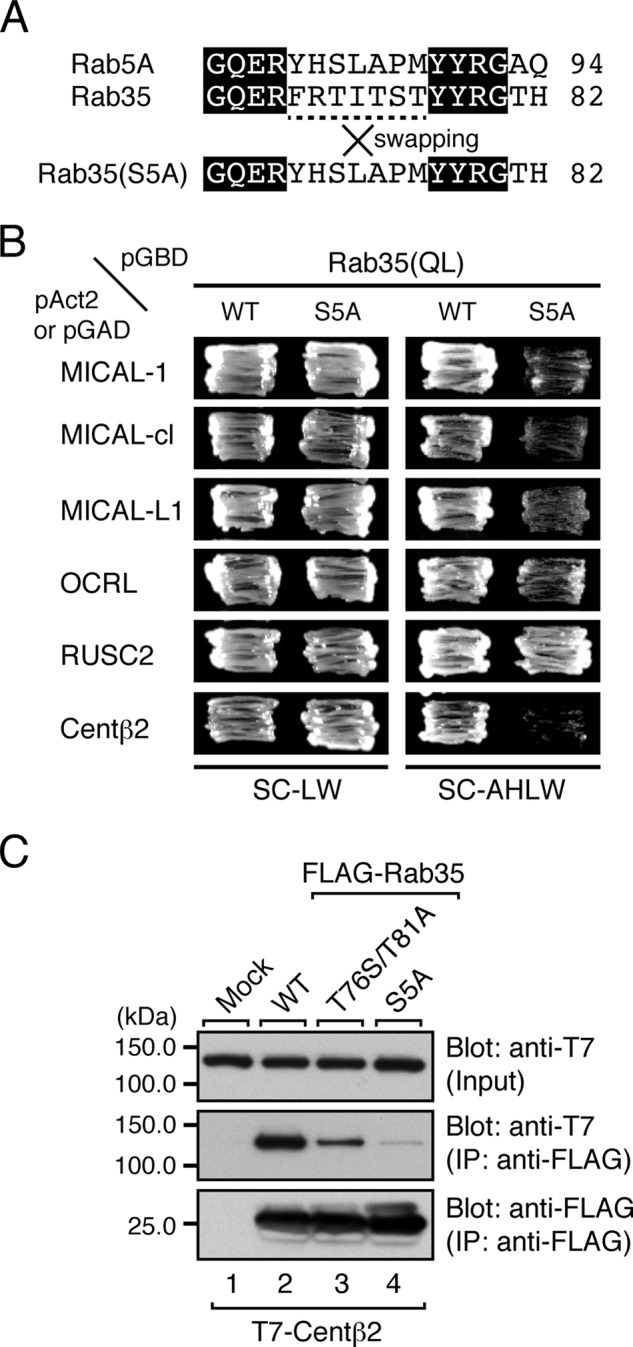
Contribution of the Switch II Sequence in Rab35 to Binding Activity toward Rab35BPs
Next, to evaluate the contribution of the entire switch II region of Rab35 to binding activity toward Rab35BPs, we produced a switch II-swapping mutant of Rab35, named Rab35(S5A), in which the switch II region of Rab35 was replaced by the switch II region of Rab5A, a Rab that is evolutionarily distant from Rab35 (39). The results of the yeast two-hybrid assays showed that the Rab35(S5A) mutant exhibited no binding activity toward centaurin-β2 and exhibited decreased binding activity toward MICAL family members and OCRL, whereas the swapping mutation had no effect on RUSC2 binding activity (Fig. 2B). To further compare the centaurin-β2 binding ability of Rab35(T76S/T81A) and Rab35(S5A), we then performed co-immunoprecipitation assays in COS-7 cells. As shown in Fig. 2C, Rab35(S5A) hardly interacted with centaurin-β2 at all in contrast to the reduced centaurin-β2 binding activity of Rab35(T76S/T81A), suggesting that centaurin-β2 recognizes certain amino acid(s) in the switch II region of Rab35 besides Thr-76 and Thr-81.
Effect of the T76S/T81A Mutation of Rab35 on NGF-induced Neurite Outgrowth of PC12 Cells
Because the T76S/T81A mutation of Rab35 specifically caused a reduction in the centaurin-β2 binding activity without affecting binding activity toward other Rab35BPs (Fig. 1, D and E), the Rab35(T76S/T81A) mutant is expected to be a useful tool for evaluating the physiological significance of the interaction between Rab35 and centaurin-β2 by knockdown-rescue approaches. To apply the Rab35(T76S/T81A) mutant as a tool to evaluate the physiological significance of the Rab35/centaurin-β2 interaction, we focused on NGF-induced neurite outgrowth of PC12 cells because we previously found that Rab35 recruits centaurin-β2 to Arf6-positive recycling endosomes in PC12 cells and that knockdown of either of them with specific shRNAs inhibits neurite outgrowth (14) (Fig. 3, A, upper panels, and B). When we re-expressed an shRNA-resistant form of Rab35(WT) (named Rab35SR(WT)) in Rab35 knockdown cells, NGF-induced neurite outgrowth was almost completely restored (Fig. 3, A, bottom left panel, and B), whereas re-expression of Rab35SR(T76S/T81A) in Rab35 knockdown cells failed to rescue the phenotype (Fig. 3, A, bottom middle panel, and B), indicating that the specific interaction between Rab35 and centaurin-β2 is crucial for neurite outgrowth. Similarly, no rescue effect was observed with Rab35SR(S5A) (Fig. 3, A, bottom right panel, and B), which also lacks centaurin-β2 binding activity (Fig. 2, B and C). The lack of a rescue effect by these two mutants is unlikely to be attributable to their lower protein expression level because equivalent amounts of Rab35SR proteins were expressed under our experimental conditions (Fig. 3C). Although the difference was not statistically significant, the Rab35SR(T76S/T81A)-re-expressing cells tended to possess slightly longer neurites than the Rab35SR(S5A)-re-expressing cells. This difference may be explained by the residual centaurin-β2 binding activity of Rab35(T76S/T81A) as opposed to the almost completely absent centaurin-β2 binding activity of Rab35(S5A) (Fig. 2C), or Rab35(T76S/T81A) may weakly promote neurite outgrowth through interaction with other Rab35BPs, e.g. MICAL-L1 (10).
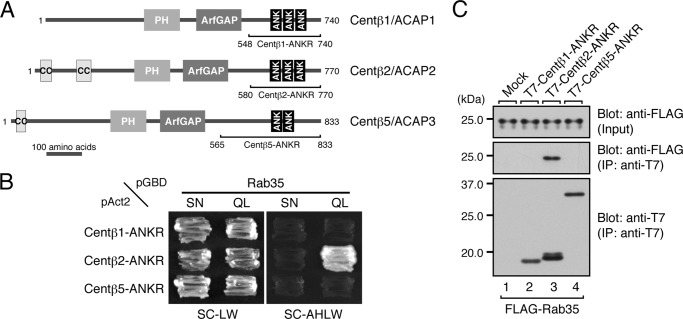
Rab35 Binding Activity of Centaurin-β1/ACAP1 and Centaurin-β5/ACAP3
In the next set of experiments, we turned our attention to centaurin-β2 and attempted to identify the residues that are crucial for Rab35 binding activity. Although we previously showed that a C-terminal ANKR domain of centaurin-β2 functions as a Rab35 effector domain (8, 14), nothing was known about its Rab35 recognition mechanism. To identify the critical residues in the ANKR domain of centaurin-β2 that are responsible for Rab35 binding, we first compared the ANKR domain of ORP1L (i.e. Rab7-binding site) (40) and the ANKR domain of VPS9-ankyrin repeat protein (Varp) (i.e. Rab32/38-binding site) (32, 41) with the centaurin-β2 ANKR domain. Because of their low sequence conservation, however, we were unable to identify shared key residues that are responsible for Rab binding by these three ANKR domains. We then turned our attention to Arf-GAPs because approximately two-thirds of mammalian Arf-GAPs contain ANKR domains and because two of them (42, 43), centaurin-β1 (also called ACAP1) and centaurin-β5 (also called ACAP3), contain a C-terminal ANKR domain similar to the centaurin-β2 ANKR domain (Fig. 4A). If the ANKR domain of centaurin-β1/β5 also serves as a Rab35-binding site, comparison of their sequence was expected to be helpful in identifying key residues responsible for Rab35 binding. To determine whether centaurin-β1 and centaurin-β5 are binding partners of Rab35, we cloned the cDNAs of the mouse ANKR domains of centaurin-β1/β5 (Fig. 4A) and subjected them to yeast two-hybrid assays as described above (Fig. 4B). To our surprise, however, neither the ANKR domain of centaurin-β1 nor the ANKR domain of centaurin-β5 interacted with Rab35 despite their relatively high sequence similarity to the ANKR domain of centaurin-β2, which recognizes a GTP-locked (QL) form of Rab35 but not its GDP-locked (SN) form. We confirmed the lack of interaction between Rab35 and centaurin-β1/β5 by performing co-immunoprecipitation assays in COS-7 cells (Fig. 4C). These results suggested that amino acid residues in the centaurin-β2 ANKR domain that are not conserved in centaurin-β1 or centaurin-β5 were good candidates for key residues responsible for Rab35 binding. Because the original centaurin-β2-ANKR construct contains other regions besides the ANKR domain (Fig. 4A), we wanted to reduce the Rab35-binding region to a minimum before searching for the candidate residues by sequence comparisons between the C-terminal domains of centaurin-β1, -β2, and -β5.
FIGURE 4.
Rab35 binding activity of the centaurin-β2 homologues centaurin-β1 and centaurin-β5. A, schematic representation of the domain organization of Centβ1/ACAP1, Centβ2/ACAP2, and Centβ5/ACAP3. B, Centβ2-ANKR, but not Centβ1-ANKR or Centβ5-ANKR, specifically recognized the active form of Rab35. Yeast cells containing pAct2 plasmid expressing Centβ1-ANKR, Centβ2-ANKR, or Centβ5-ANKR and pGBD plasmid expressing Rab35(QL) (a constitutively active form) or Rab35(SN) (a constitutively negative form) were streaked on SC-LW (left panels) and SC-AHLW (right panels) and incubated at 30 °C. C, Centβ2-ANKR, but not Centβ1-ANKR or Centβ5-ANKR, interacted with Rab35 in COS-7 cells. Co-immunoprecipitation assays were performed as described in the legend for Fig. 1E. The positions of the molecular mass markers (in kilodaltons) are shown on the left. IP, immunoprecipitation; PH, pleckstrin homology domain; ANK, ankyrin repeat.
Determination of the Minimal Rab35-binding Site in Centaurin-β2
To determine the minimal Rab35-binding site (named RBD35), we generated a series of deletion mutants (summarized in Fig. 5A) and evaluated their Rab35 binding activity by yeast two-hybrid assays. The results showed that only the AA580–755 construct strongly interacted with Rab35(QL), the same as the original ANKR construct (AA580–770) did (Fig. 5B), whereas the other deletion constructs did not interact or hardly interacted with Rab35(QL). Similar results were obtained by co-immunoprecipitation assays in COS-7 cells: deletion of the C-terminal 15 amino acids from the original ANKR construct (i.e. AA580–755) had no effect on Rab35 binding activity, whereas deletion of the C-terminal 30 amino acids (i.e. AA580–745) impaired Rab35 binding activity (Fig. 5C). We therefore concluded that the minimal RBD35 in centaurin-β2 is AA580–755 and that additional amino acids in the C-terminal flanking region of the ANKR domain, i.e. AA730–755, are required for high affinity Rab35 binding.
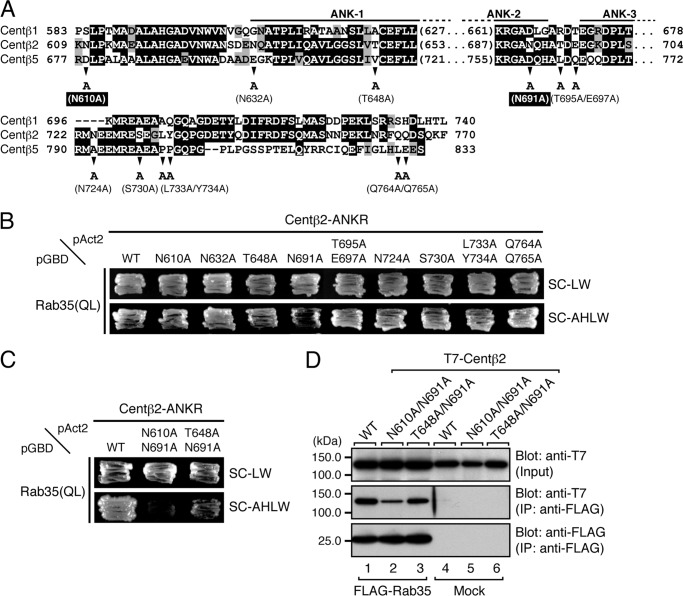
Identification of Critical Residues in the ANKR Domain of Centaurin-β2 That Specifically Recognize Rab35 by Site-directed Mutagenesis
Based on the results shown in Fig. 5B, we searched for candidate residues responsible for Rab35 binding in the minimal RBD35 of centaurin-β2 by performing sequence comparisons among centaurin-β1, -β2, and -β5 (Fig. 6A). The 12 candidate residues conserved in centaurin-β2 alone, i.e. Asn-610, Asn-632, Thr-648, Asn-691, Thr-695, Glu-697, Asn-724, Ser-730, Leu-733, Tyr-734, Gln-764, and Gln-765, were selected, and each was replaced with Ala by site-directed mutagenesis (Fig. 6A, arrowheads). The results of the yeast two-hybrid assays showed that none of the single point mutations dramatically decreased binding activity toward Rab35(QL) (Fig. 6B). However, because the yeast cells expressing the N610A, T648A, or N691A mutant appeared to grow slowly, we generated double mutants of centaurin-β2, i.e. N610A/N691A and T648A/N691A, and assessed their Rab35 binding activity by yeast two-hybrid assays. As shown in Fig. 6C, the N610A/N691A double mutant abrogated binding activity toward Rab35(QL), whereas the T648A/N691A double mutant still exhibited significant Rab35 binding activity. Similar results were obtained by co-immunoprecipitation assays in COS-7 cells; that is the Rab35 binding activity of centaurin-β2(N610A/N691A) mutant was dramatically reduced (Fig. 6D, middle panel, lane 2). These results indicated that both Asn-610 and Asn-691 in the ANKR domain of centaurin-β2 are critical for recognition by Rab35, and we decided to use the N610A/N691A mutant as a Rab35 binding-deficient mutant in the subsequent analysis.
FIGURE 6.
Identification of critical residues responsible for the binding of Rab35 in the ANKR domain of centaurin-β2 by site-directed mutagenesis. A, sequence alignment of the ANKR domains of mouse Centβ1, Centβ2, and Centβ5. Amino acid residues in the sequences that are conserved and that are similar are shown against a black background and a shaded background, respectively. The arrowheads indicate the positions of amino acids that are not conserved between Centβ2 and Centβ1 or Centβ5 and were the focus of the Ala-based site-directed mutagenesis. B, Rab35 binding activity of Centβ2 as determined by yeast two-hybrid assays. Yeast cells containing pAct2 plasmid expressing Centβ2-ANKR(WT) or each Centβ2-ANKR mutant (N610A, N632A, T648A, N691A, T695A/E697A, N724A, S730A, L733A/Y734A, or Q764A/Q765A) and pGBD plasmid expressing Rab35(QL) were streaked on SC-LW (top panel) and SC-AHLW (bottom panel) and incubated at 30 °C. Based on the growth rate of the yeast cells, the N610A, T648A, or N691A mutation in the Centβ2-ANKR appeared to slightly decrease Rab35 binding activity. C, two Asn residues, i.e. Asn-610 and Asn-691, of Centβ2-ANKR are critical for binding Rab35. Yeast two-hybrid assays were performed as described in B. D, the N610A/N691A mutation of Centβ2 dramatically decreased Rab35 binding activity, a finding that was consistent with the results of the yeast two-hybrid assays shown in C. Co-immunoprecipitation assays were performed as described in the legend for Fig. 1E. The positions of the molecular mass markers (in kilodaltons) are shown on the left. IP, immunoprecipitation; ANK1–3, ankyrin repeats 1–3.
Effect of the N610A/N691A Mutation of Centaurin-β2 on NGF-induced Neurite Outgrowth of PC12 Cells
To investigate whether the centaurin-β2(N610A/N691A) mutant is capable of supporting NGF-induced neurite outgrowth, we performed knockdown-rescue experiments as described previously (14). Consistent with our previous finding, knockdown of centaurin-β2 with specific shRNA inhibited neurite outgrowth (Fig. 7, A, upper panels, and B), whereas re-expression of centaurin-β2SR(WT) in centaurin-β2 knockdown cells rescued the phenotype (14) (Fig. 7, A, bottom left panel, and B). By contrast, re-expression of centaurin-β2SR(N610A/N691A) in centaurin-β2 knockdown cells failed to rescue the phenotype (Fig. 7, A, bottom middle panel, and B), the same as the centaurin-β2SR(ΔANKR) mutant, which completely lacked the RBD35 (14) (Fig. 7, A, bottom right panel, and B). The lack of a rescue effect by these two mutants is unlikely to be attributable to their lower protein expression level because equivalent amounts of centaurin-β2SR proteins were expressed in the centaurin-β2 knockdown cells (Fig. 7C). These results taken together indicated that the Rab35/centaurin-β2 interaction is essential for neurite outgrowth.
DISCUSSION
We and others have previously identified a variety of Rab35BPs (7–13), each of which showed extremely different Rab binding specificity (Fig. 1B). Only one of them, centaurin-β2, had been shown to specifically recognize Rab35 (Fig. 1C) (8), but the molecular determinant(s) responsible for the exclusive Rab35 binding specificity of centaurin-β2 had never been investigated. In the present study, we performed site-directed mutagenesis and identified residues in Rab35, i.e. Thr-76 and Thr-81 in the switch II region, and in centaurin-β2, i.e. Asn-610 and Asn-691 in the ANKR domain, that are crucial for the formation of the Rab35·centaurin-β2 complex (Figs. 1 and 6). Because Rab35 is the only Rab isoform that has a Thr residue at each of these positions in the switch II region (Fig. 1A), centaurin-β2 is likely to specifically recognize these two Thr residues. Actually, mutation of these Thr residues resulted in a dramatic reduction in the centaurin-β2 binding ability without affecting binding activity toward any of the other Rab35BPs (Fig. 1, D–F). However, these results do not mean that other Rab35BPs besides centaurin-β2 recognize the switch II region of Rab35. A switch II-swapping analysis indicated that the switch II region of Rab35 also contributes to its recognition by most of the Rab35BPs (Fig. 2). Because RUSC2 normally bound to the Rab35(S5A) mutant, it must recognize some other region of Rab35 besides the switch II region, suggesting that Rab35 interacts with both RUSC2 and one of the other Rab35BPs. Further work will be necessary to determine whether Rab35 interacts with more than one Rab35BP at the same time.
Identification of the minimal RBD35 in centaurin-β2 provided other important information. We previously thought that the ANKR domain of centaurin-β2 was necessary and sufficient for Rab35 binding activity, the same as the ANKR1 domain of VPS9-ankyrin repeat protein that alone recognizes Rab32/38 (32), but the results of our deletion analysis indicated that an N-terminal flanking region and a C-terminal flanking region (AA580–630 and AA730–755, respectively) of the ANKR domain are also required for high affinity Rab35 binding activity (Fig. 5). This observation may well explain why other Arf-GAPs, e.g. centaurin-β1/5, which often have an ANKR domain (42, 43), do not interact with Rab35 because the N-terminal and C-terminal flanking regions of the ANKR domain are not well conserved among Arf-GAPs. A more detailed structural analysis will be needed to evaluate the contribution of the N/C-terminal flanking regions of the ANKR domain to Rab35 binding.
The fact that more than one Rab35BP is simultaneously expressed in a single cell type (10, 14) makes it difficult to determine which Rab35 effector is involved in Rab35-dependent cellular events. Knockdown of a certain Rab35BP is insufficient to show whether a direct interaction between Rab35 and that Rab35BP is involved in cellular events even when the Rab35BP knockdown impairs them. The Rab35SR(T76S/T81A) mutant that we developed in this study, however, is a useful tool for evaluating the functional significance of the Rab35/centaurin-β2 interaction in cellular events. By using this tool in combination with knockdown-rescue approaches, we succeeded in confirming our previous finding that the interaction between Rab35 and centaurin-β2 is essential for neurite outgrowth of PC12 cells because the centaurin-β2 binding-deficient Rab35SR(T76S/T81A) mutant did not support NGF-induced neurite outgrowth of Rab35 knockdown cells (Fig. 3). Similarly, the Rab35 binding-deficient centaurin-β2SR(N610A/N691A) mutant was unable to restore neurite outgrowth of centaurin-β2 knockdown cells (Fig. 7). It will be interesting to apply these tools to investigate the involvement of centaurin-β2 in other Rab35-dependent cellular events in the future. In our preliminary experiments, the Rab35SR(T76S/T81A) mutant was unable to rescue the multinucleation phenotype of HeLaS3 cells induced by Rab35 siRNA (15).3 Because Arf6 is also known to be required for cytokinesis (16), there is a strong possibility that centaurin-β2-mediated functional cross-talk between Rab35 and Arf6 occurs in cytokinesis. Investigation of the possible functional involvement of centaurin-β2 in cytokinesis is now underway in our laboratory.
In conclusion, we performed structure-function analyses of Rab35 and centaurin-β2 by site-directed mutagenesis and identified residues that are crucial for the Rab35/centaurin-β2 interaction. We also showed by knockdown-rescue experiments that centaurin-β2 functions as a Rab35 effector during NGF-induced neurite outgrowth of PC12 cells. The Rab35SR(T76S/T81A) mutant that we developed should be a powerful, easy-to-use tool for probing the involvement of centaurin-β2 in Rab35-dependent cellular events, including cytokinesis (15), cell migration (17, 18), and phagocytosis (19, 20), at the cellular level.
Acknowledgments
We thank Megumi Aizawa for technical assistance, Hotaka Kobayashi for helpful advice, and members of the Fukuda Laboratory for valuable discussions.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, and Technology of Japan (to M. F.).
K. Etoh and M. Fukuda, unpublished observations.
- GAP
- GTPase-activating protein
- AA
- amino acid(s)
- ACAP
- Arf-GAP with coiled coil, ankyrin repeat, and pleckstrin homology domain
- ANKR
- ankyrin repeat
- Arf
- ADP-ribosylation factor
- Centβ
- centaurin-β
- EGFP
- enhanced green fluorescent protein
- MICAL
- molecule interacting with CasL
- OCRL
- oculocerebrorenal syndrome of Lowe
- QL
- Q67L
- Rab35BP
- Rab35-binding protein
- RBD35
- Rab35-binding site
- SC-AHLW
- synthetic complete medium lacking adenine, histidine, leucine, and tryptophan
- SC-LW
- synthetic complete medium lacking leucine and tryptophan
- SN
- S22N
- SR
- shRNA-resistant.
REFERENCES
- 1. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 3. Pfeffer S. R. (2013) Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barr F. A. (2013) Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukuda M. (2011) TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 31, 159–168 [DOI] [PubMed] [Google Scholar]
- 7. Fukuda M., Kanno E., Ishibashi K., Itoh T. (2008) Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 8. Kanno E., Ishibashi K., Kobayashi H., Matsui T., Ohbayashi N., Fukuda M. (2010) Comprehensive screening for novel Rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic 11, 491–507 [DOI] [PubMed] [Google Scholar]
- 9. Rahajeng J., Giridharan S. S., Cai B., Naslavsky N., Caplan S. (2012) MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic 13, 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi H., Fukuda M. (2013) Rab35 establishes the EHD1-association site by coordinating two distinct effectors during neurite outgrowth. J. Cell Sci. 126, 2424–2435 [DOI] [PubMed] [Google Scholar]
- 11. Dambournet D., Machicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., Echard A. (2011) Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13, 981–988 [DOI] [PubMed] [Google Scholar]
- 12. Fukuda M., Kobayashi H., Ishibashi K., Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 [DOI] [PubMed] [Google Scholar]
- 13. Zhang J., Fonovic M., Suyama K., Bogyo M., Scott M. P. (2009) Rab35 controls actin bundling by recruiting fascin as an effector protein. Science 325, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 14. Kobayashi H., Fukuda M. (2012) Rab35 regulates Arf6 activity through centaurin-β2 (ACAP2) during neurite outgrowth. J. Cell Sci. 125, 2235–2243 [DOI] [PubMed] [Google Scholar]
- 15. Kouranti I., Sachse M., Arouche N., Goud B., Echard A. (2006) Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16, 1719–1725 [DOI] [PubMed] [Google Scholar]
- 16. Chesneau L., Dambournet D., Machicoane M., Kouranti I., Fukuda M., Goud B., Echard A. (2012) An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr. Biol. 22, 147–153 [DOI] [PubMed] [Google Scholar]
- 17. Allaire P. D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R. F., McPherson P. S. (2013) Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126, 722–731 [DOI] [PubMed] [Google Scholar]
- 18. Zhu Y., Shen T., Liu J., Zheng J., Zhang Y., Xu R., Sun C., Du J., Chen Y., Gu L. (2013) Rab35 is required for Wnt5a/Dvl2-induced Rac1 activation and cell migration in MCF-7 breast cancer cells. Cell. Signal. 25, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 19. Shim J., Lee S. M., Lee M. S., Yoon J., Kweon H. S., Kim Y. J. (2010) Rab35 mediates transport of Cdc42 and Rac1 to the plasma membrane during phagocytosis. Mol. Cell. Biol. 30, 1421–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egami Y., Fukuda M., Araki N. (2011) Rab35 regulates phagosome formation through recruitment of ACAP2 in macrophages during FcγR-mediated phagocytosis. J. Cell Sci. 124, 3557–3567 [DOI] [PubMed] [Google Scholar]
- 21. Patino-Lopez G., Dong X., Ben-Aissa K., Bernot K. M., Itoh T., Fukuda M., Kruhlak M. J., Samelson L. E., Shaw S. (2008) Rab35 and its GAP EPI64C in T cells regulate receptor recycling and immunological synapse formation. J. Biol. Chem. 283, 18323–18330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyamoto Y., Yamamori N., Torii T., Tanoue A., Yamauchi J. (2014) Rab35, acting through ACAP2 switching off Arf6, negatively regulates oligodendrocyte differentiation and myelination. Mol. Biol. Cell 25, 1532–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chevallier J., Koop C., Srivastava A., Petrie R. J., Lamarche-Vane N., Presley J. F. (2009) Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 583, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 24. Kobayashi H., Etoh K., Ohbayashi N., Fukuda M. (2014) Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biol. Open 3, 803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grant B. D., Donaldson J. G. (2009) Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson T. R., Brown F. D., Nie Z., Miura K., Foroni L., Sun J., Hsu V. W., Donaldson J. G., Randazzo P. A. (2000) ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151, 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 28. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukuda M., Kanno E., Mikoshiba K. (1999) Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 274, 31421–31427 [DOI] [PubMed] [Google Scholar]
- 30. Fukuda M., Kojima T., Aruga J., Niinobe M., Mikoshiba K. (1995) Functional diversity of C2 domains of synaptotagmin family: mutational analysis of inositol high polyphosphate binding domain. J. Biol. Chem. 270, 26523–26527 [DOI] [PubMed] [Google Scholar]
- 31. Fukuda M., Kanno E. (2005) Analysis of the role of Rab27 effector Slp4-a/granuphilin-a in dense-core vesicle exocytosis. Methods Enzymol. 403, 445–457 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K., Ohbayashi N., Maruta Y., Kanno E., Itoh T., Fukuda M. (2009) Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol. Biol. Cell 20, 2900–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eathiraj S., Pan X., Ritacco C., Lambright D. G. (2005) Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Recacha R., Boulet A., Jollivet F., Monier S., Houdusse A., Goud B., Khan A. R. (2009) Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure 17, 21–30 [DOI] [PubMed] [Google Scholar]
- 35. Eathiraj S., Mishra A., Prekeris R., Lambright D. G. (2006) Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364, 121–135 [DOI] [PubMed] [Google Scholar]
- 36. Kukimoto-Niino M., Sakamoto A., Kanno E., Hanawa-Suetsugu K., Terada T., Shirouzu M., Fukuda M., Yokoyama S. (2008) Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure 16, 1478–1490 [DOI] [PubMed] [Google Scholar]
- 37. Fukuda M. (2002) Synaptotagmin-like protein (Slp) homology domain 1 of Slac2-a/melanophilin is a critical determinant of GTP-dependent specific binding to Rab27A. J. Biol. Chem. 277, 40118–40124 [DOI] [PubMed] [Google Scholar]
- 38. Matsui T., Ohbayashi N., Fukuda M. (2012) The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36: Rab36 regulates retrograde melanosome transport in melanocytes. J. Biol. Chem. 287, 28619–28631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pereira-Leal J. B., Seabra M. C. (2001) Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889–901 [DOI] [PubMed] [Google Scholar]
- 40. Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V. M., Neefjes J. (2007) Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor βIII spectrin. J. Cell Biol. 176, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hesketh G. G., Pérez-Dorado I., Jackson L. P., Wartosch L., Schäfer I. B., Gray S. R., McCoy A. J., Zeldin O. B., Garman E. F., Harbour M. E., Evans P. R., Seaman M. N. J., Luzio J. P., Owen D. J. (2014) VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev. Cell 29, 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inoue H., Randazzo P. A. (2007) Arf GAPs and their interacting proteins. Traffic 8, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 43. Kahn R. A., Bruford E., Inoue H., Logsdon J. M., Jr., Nie Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L., Cassel D. (2008) Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 182, 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]