Background: The actin-binding protein profilin-1 is a eukaryotic protein essential for growth, with poorly understood antitumor function.
Results: Profilin-1 antitumor activity requires nuclear localization and is inhibited by Ser-137 phosphorylation.
Conclusion: Profilin-1 has spatially defined functions and is post-translationally regulated.
Significance: Our data support a model to reconcile the seemingly oppositional functions of profilin-1 and may have implications for novel anticancer therapies.
Keywords: Cancer, Cell Compartmentalization, Nucleus, Phosphorylation, Profilin, Proliferation, Apoptosis
Abstract
The actin-binding protein profilin-1 (Pfn1) inhibits tumor growth and yet is also required for cell proliferation and survival, an apparent paradox. We previously identified Ser-137 of Pfn1 as a phosphorylation site within the poly-l-proline (PLP) binding pocket. Here we confirm that Ser-137 phosphorylation disrupts Pfn1 binding to its PLP-containing ligands with little effect on actin binding. We find in mouse xenografts of breast cancer cells that mimicking Ser-137 phosphorylation abolishes cell cycle arrest and apoptotic sensitization by Pfn1 and confers a growth advantage to tumors. This indicates a previously unrecognized role of PLP binding in Pfn1 antitumor effects. Spatial restriction of Pfn1 to the nucleus or cytoplasm indicates that inhibition of tumor cell growth by Pfn1 requires its nuclear localization, and this activity is abolished by a phosphomimetic mutation on Ser-137. In contrast, cytoplasmic Pfn1 lacks inhibitory effects on tumor cell growth but rescues morphological and proliferative defects of PFN1 null mouse chondrocytes. These results help reconcile seemingly opposed cellular effects of Pfn1, provide new insights into the antitumor mechanism of Pfn1, and implicate Ser-137 phosphorylation as a potential therapeutic target for breast cancer.
Introduction
Pfn13 (∼14 kDa) is an actin-binding protein involved in actin polymerization and remodeling. It was the first identified member of the profilin gene family and is ubiquitously expressed. Pfn1 facilitates filamentous actin (F-actin) polymerization by binding and exchanging nucleotides on globular actin (G-actin) and by ushering ATP-bound G-actin to the barbed ends of actin filaments (1–4). Pfn1 is required for cytokinesis (5, 6) and is essential for the development and survival of many eukaryotes including mouse, fly, and yeast (6–8). Besides G-actin, Pfn1 also has a separate binding site for poly-l-proline (PLP) motifs, which are present in many cellular proteins. Whereas some of the known PLP ligands of Pfn1 are actin binding factors (e.g. Ena/VASP, N-WASP, Arp2, and mDia), others are involved in signaling, membrane trafficking, synaptic scaffolding, and nuclear functions (2, 4). Thus Pfn1 may participate in diverse cellular processes depending on its interaction with different PLP ligands. For instance, we have identified huntingtin (Htt), a PLP-containing protein that causes Huntington disease, as a novel Pfn1 ligand. Direct interactions between Htt and Pfn1 inhibit mutant Htt aggregation, thereby implicating Pfn1 as a potential modifier of Huntington disease pathogenesis (9).
Remarkably, despite being essential for cell growth and survival, Pfn1 also has antitumor functions. Its expression is decreased in multiple types of carcinoma (breast, bladder, and pancreas) (10–14), and its ectopic re-expression inhibits the proliferation and survival of several cancer cell lines in vitro and in vivo (12, 14–16). Recently, low Pfn1 expression was correlated with poor outcome of bladder and pancreatic cancer patients (13, 14). However, unlike classic tumor suppressor genes, homozygous deletion and somatic mutations of the PFN1 gene are extremely rare and have not been causally linked to cancer. Although this is consistent with PFN1 being an essential gene, the mechanistic basis of the opposing functions of Pfn1 are completely unknown. On a cellular level, the antitumor effect of Pfn1 has been attributed to cell cycle arrest in G1 phase and sensitization to apoptosis (17). However, at a molecular level, its antitumor function remains poorly understood.
Pfn1 is predominantly cytoplasmic. However, it is also present in the nucleus and, after binding G-actin, is exported back into the cytoplasm by Exportin-6 (18). Nuclear Pfn1 has been functionally implicated in gene expression regulation based on its association with transcriptionally active genes (19), its presence in nuclear speckles/Cajal bodies (20, 21), and its association with nuclear proteins such as the transcription factor p42POP (22) and the pre-mRNA splicing regulatory factor SMN (21). It is also required for actin-dependent RNA synthesis by respiratory syncytial virus (23). However, unlike the well characterized role of cytoplasmic Pfn1 as an actin assembly factor, its nuclear functions are poorly understood.
Recent studies suggest that Pfn1 functions are regulated by phosphorylation. For example, phosphorylation of Pfn1 at Tyr-129 occurs in vascular endothelial cells stimulated with vascular endothelial growth factor, and this is required for efficient actin polymerization at the cell leading edges and for stimulus-induced angiogenesis (24). We originally described Pfn1 phosphorylation on Ser-137 (9, 25) and found that this inhibits Pfn1 binding to the PLP-containing Htt protein and its ability to suppress mutant Htt aggregation (9). Thus, Ser-137 phosphorylation may regulate Pfn1 by controlling its binding to PLP-containing ligands. We have now investigated how Ser-137 phosphorylation affects the tumor inhibitory activities of Pfn1 in the context of breast cancer models. Ser-137 phosphorylation blocks the ability of Pfn1 to inhibit cell cycle progression of breast cancer cells. It also inhibits the proapoptotic activity of Pfn1 and renders tumor cells more resistant to apoptosis in mouse xenografts. Importantly, tumor cell growth inhibition by Pfn1 requires its nuclear localization, whereas cellular proliferation depends on cytoplasmic Pfn1, and both functions are regulated by Ser-137 phosphorylation. Together, our study helps elucidate the antitumor mechanism of Pfn1 and highlights a critical regulatory effect of Ser-137 phosphorylation.
EXPERIMENTAL PROCEDURES
Molecular Cloning
Untagged and Myc-tagged Pfn1 in pcDNA3 were generated previously (9). N-terminally HA-tagged Pfn1 was PCR-amplified and similarly cloned into pcDNA3. For lentiviral constructs, cDNAs encoding untagged Pfn1 were cloned into the pENTR1A vector and subsequently recombined into the pLenti-CMV/TO-Neo-DEST vector (Addgene, #17292) using LR Clonase II according to the manufacturer's protocol (Invitrogen). Pfn1 fused with the nuclear localization (NLS) and export sequences (NES) were PCR-amplified and cloned into the lentiviral pFLRu-NYFP-FH vector (26). Three tandem NLS repeats (DPKKKRKV, adapted from the Clontech pAcGFP1-Nuc) and a single NES sequence (MNLVDLQKKLEELELDEQQ, adapted from the Clontech pCaspase3 sensor vector) were fused to the N terminus of Pfn1 and cloned downstream of YFP in the pFLRu-NYFP-FH vector to generate YFP-NLS-Pfn1 and YFP-NES-Pfn1. To silence Pfn1, a 21-mer sequence (GTGGTTTGATCAACAAGAA) was cloned into the pFLRu-FH vector (26, 27). A second shRNA (TACGTGAATGGGCTGACACTT) in the pLKO.1 vector was obtained from the RNAi consortium at the Genome Institute of Washington University. A control shRNA containing a non-targeting sequence (CAACAAGATGAAGAGCACCAA) was cloned into the pFLRu-FH vector.
Antibodies
Primary antibodies used for Western blots are as follows: mouse anti-HA tag (Covance, MMS-101P), mouse anti-Myc tag (Santa Cruz, sc-40), mouse anti-β-actin (Santa Cruz, sc-47778; Cell Signaling, #3700), mouse anti-α-tubulin (Sigma, T9026; Cell Signaling, #3873), rabbit anti-GFP (Santa Cruz, sc-8334), mouse anti-GFP (Thermo Scientific, PIMA515256), rabbit anti-Pfn1 (Cell Signaling, #3237), rabbit anti-PARP (Cell Signaling, #9532), rabbit anti-cleaved PARP (Cell Signaling, #9541), rabbit anti-cleaved caspase-3 (Cell Signaling, #9661), rabbit anti-Rb (Cell Signaling, #9313). Primary antibodies used for immunofluorescence staining and immunohistochemistry are as follows: rabbit anti-Pfn1 (Cell Signaling, #3237), rabbit anti-cleaved caspase-3 (Cell Signaling, #9661), and mouse anti-Myc-tag (Santa Cruz, sc-40). Secondary antibodies for Western blots are alkaline phosphatase-conjugated (Sigma) and horseradish peroxidase-conjugated anti-rabbit and anti-mouse (Amersham Biosciences) antibodies. For immunofluorescence staining, Alexa Fluor® 488-conjugated goat anti-rabbit IgG was purchased from Invitrogen. For immunohistochemistry, VECTASTAIN Elite ABC kit (rabbit IgG) was purchased from Vector Laboratories.
Cell Culture
All cell lines were purchased from ATCC with the exception of MDA-MB-231 cells stably expressing a tri-modal reporter fusion (firefly luciferase, red fluorescence protein, and HSV1-sr39 truncated thymidine kinase (28)). MDA-MB-231, HEK293, HEK293T, NIH3T3, and immortalized Pfn1−/− chondrocytes cells were all grown in DMEM supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin. For transient transfection of HEK293 and NIH3T3 cells, Lipofectamine 2000 was used according to the manufacturer's protocol (Invitrogen). To generate lentivirus, HEK293T (pFLRu-NYFP-FH-based) or HEK293FT (pLenti-CMV/TO-Neo-DEST-based) cells were co-transfected with vector DNA and packaging plasmids with FuGENE 6 reagent based on manufacturer's protocol (Roche Applied Science). Media containing viruses were collected and clarified 2 days after transfection.
Pulldown Assays
For PLP binding of Pfn1, HEK293 cells grown on a 6-well dish were transfected with HA-tagged Pfn1 constructs (wt, S137A, and S137D) and lysed in 200 μl of buffer containing 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Triton X-100, 1× protease inhibitor mixture (EDTA-free Complete Mini, Roche Applied Science), and 1× phosphatase inhibitor mixture (PhosSTOP, Roche Applied Science). MDA-MB-231 cells stably expressing NLS or NES-tagged Pfn1 (wt, S137A, and S137D) were grown in 10-cm dishes and lysed in 200 μl of radioimmune precipitation assay buffer containing the same protease and phosphatase inhibitors. Cell lysates were centrifuged at 20,000 × g for 5 min at 4 °C, and supernatants were bound to 20 μl of control or PLP-coupled-Sepharose beads for 2 h at 4 °C. Beads were washed 4 times with the lysis buffer and boiled in SDS sample buffer for 5 min. Samples were separated on a 12% SDS-PAGE gel, and bound HA-Pfn1, YFP-NLS-Pfn1, or YFP-NES-Pfn1 were analyzed by Western blot using an anti-HA antibody (1:2000) or anti-GFP antibody (1:2000). For actin binding of NLS or NES-tagged Pfn1, MDA-MB-231 stable cells grown on 10-cm dishes were lysed in 200 μl of radioimmune precipitation assay buffer containing protease and phosphatase inhibitors and bound to ∼20 μl of anti-GFP antibody-conjugated Sepharose beads (Abcam, ab69314) for 2 h at 4 °C. Beads were washed, boiled in SDS sample buffer, and analyzed by SDS-PAGE as above. Co-immunoprecipitated actin was analyzed by Western blot.
Subcellular Fractionation
MDA-MB-231 cells were grown on 10-cm dishes and harvested by trypsinization. Cell pellets were divided into cytoplasmic and nuclear fractions using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific, PI78833) according to the manufacturer's protocol. Cytoplasmic and nuclear fractions were analyzed by Western blots at a 1:5 volume ratio.
Analysis of F-actin/G-actin Ratio
HEK293 cells grown on 60-mm dishes were transfected with pcDNA3 or different Myc-tagged Pfn1 constructs. Two days after transfection cells were harvested and lysed in 200 μl of F-actin stabilization buffer (50 mm PIPES, pH 6.9, 50 mm NaCl, 5 mm MgCl2, 5 mm EGTA, 5% glycerol, 0.1% Triton X-100, 0.1% Nonidet P-40, 0.1% Tween 20, 0.1% β-mercaptoethanol, 1 mm ATP, and 1× EDTA-free Complete Mini protease inhibitor mixture (Roche Applied Science)). Lysates were centrifuged at 150,000 × g for 1 h at 37 °C. Supernatants containing soluble G-actin were collected and boiled in SDS sample buffer. Pellets were resuspended in 200 μl of ice-cold distilled water containing 1 μm cytochalasin D (Calbiochem) and incubated on ice for 1 h to depolymerize F-actin. As a control, F-actin stabilizing Jasplakinolide (0.1 μm) (Calbiochem) was added to cultured cells for 30 min at 37 °C before harvesting and fractionation. After boiling in SDS sample buffer for 5 min, the same volumes of both fractions (G-actin in the supernatant and F-actin in the pellet) were separated on a 12% SDS-PAGE gel, and levels of endogenous actin and overexpressed Pfn1 were analyzed by Western blot using antibodies against β-actin (1:5000) and Myc tag (1:5000).
Immunocytochemistry
Cultured cells were grown on glass coverslips coated with poly-d-lysine and fibronectin (5 μg/ml). For anti-Pfn1 staining, cells were fixed in 4% paraformaldehyde for 30 min at room temperature, washed with PBS, and permeabilized in PBS, 0.1% Triton X-100. After blocking in 2% bovine serum albumin (in PBS, 0.1% Triton X-100) for 1 h, cells were incubated with primary antibody (1:100) at 4 °C overnight, washed, and incubated with Alexa Fluor-conjugated secondary antibodies (1:500) for 2 h at room temperature. For rhodamine-phalloidin staining, cells were fixed with 4% paraformaldehyde, washed, blocked, and incubated for 20 min in 100 nm rhodamine-phalloidin (Cytoskeleton Inc.) along with the secondary antibody. Cells were washed with PBS, 0.1% Triton X-100 following secondary antibody binding, counterstained with DAPI (1 μg/ml), and mounted using the ProLong Gold antifade reagent (Invitrogen). Fluorescent images were acquired using the Zeiss LSM 5 PASCAL confocal system equipped with the following lasers: 405 nm, 488 nm, 543 nm.
Immunohistochemistry
Resected tumors from the xenografted mice were fixed in formalin for 24 h and embedded in paraffin blocks. Slides of 4-μm thickness were sectioned and baked at 60 °C for 30 min before staining. They were deparaffinized in xylenes and hydrated in graded ethanol and deionized H2O. Tissues were subjected to antigen unmasking by microwaving in 10 mm citrate buffer, pH 6.0, for 10 min followed by washing in deionized H2O and PBS. They were treated with 3% hydrogen peroxide for 15 min followed by PBS wash and a 1-h block with 10% normal goat serum in PBS, 0.1% Tween 20. Anti-cleaved caspase-3 antibody (1:1600, in SignalStain diluent, Cell Signaling) was applied and incubated overnight at 4 °C. Slides were washed in PBS, 0.1% Tween 20 and incubated with the SignalStain Boost IHC Detection Reagent (Cell Signaling) for 1 h at room temperature and washed with PBS, 0.1% Tween 20 and PBS. Slides were developed using the 3,3′-diaminobenzidine (DAB) peroxidase substrate kit (Vector Laboratories), counterstained with hematoxylin, and mounted. Bright-field whole-slide imaging was performed using the Olympus NanoZoomer 2.0-HT system (Hamamatsu). Image acquisition and analysis were performed using the NDP.scan 2.5 and NDP.view software, respectively.
Cell Synchronization and DNA Content Analysis
For serum starvation, MDA-MB-231 cells were grown in DMEM media containing 0.1% FBS for 48 h followed by release in DMEM growth medium containing 10% FBS for 24 h. For double thymidine block, MDA-MB-231 cells were cultured in growth medium containing 2 mm thymidine (Sigma) for 18 h followed by 9 h of release in thymidine-free media. They were blocked again with 2 mm thymidine for 16 h followed by various hours of release. Cells were harvested by trypsinization, rinsed with PBS, and fixed in 70% ice-cold ethanol. After storing at −20 °C overnight, cells were pelleted for 5 min at 200 × g, rehydrated in PBS, centrifuged again, and resuspended in staining solution containing 20 μg/ml propidium iodide and 200 μg/ml RNase A in PBS, 0.1% Triton X-100. DNA content was measured using the 488-nm laser of a BD Biosciences flow cytometry analyzer with a Cytek BluFL2 or BluFL3 detector. BluFL2 detector was used for cells expressing untagged Pfn1, whereas BluFL3 detector was used for those expressing YFP and YFP-tagged Pfn1 constructs (to avoid emission overlap between propidium iodide and YFP). Cell cycle analysis was performed using the Watson model of FlowJo.
In Vitro Growth Assays
For two-dimensional cultures, MDA-MB-231 stable cells were seeded at 10,000 cells per well in 12-well dishes, with triplicate wells per stable line. Cells were quantified by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma) viability assay 1 day and 7 days post-seeding. Briefly, they were incubated with 0.5 mg/ml MTT in growth media for 3 h at 37 °C followed by solubilization of MTT formazan for 30 min with 1 ml of isopropyl alcohol containing 4 mm HCl and 0.1% Triton X-100 at room temperature. 200 μl of solvent was transferred to a transparent 96-well plate, and the absorbance at 590 nm was read on a fluorescence plate reader (Tecan Infinite M1000) with 620 nm as the reference wavelength.
Soft Agar Assay
Base layer of 0.6% agar was prepared by diluting 5% agar in DMEM, 10% FBS growth media in 12-well plates (1 ml/well). MDA-MB-231 cells were trypsinized, counted, and mixed with 0.4% agar (diluted in growth media) at 3000 cells/well density and plated as the top layer (1 ml/well). 0.5 ml of growth medium was added to each well after solidification. Triplicate wells were set up for each MDA-MB-231 stable line, and the same experiment was independently conducted three times. 0.005% crystal violet was used to stain colonies for 2 h in the 37 °C incubator before imaging.
Mouse Xenografts
The animal experiment was carried out in strict accordance with the guidelines recommended for care and use of laboratory animals by the National Institutes of Health. The Animal Studies Committee at Washington University (St. Louis, MO) approved all animal protocols used in this study. NOD/SCID female mice were purchased from Charles River and kept under standard institutional care. They were 5 weeks old at the time of surgery. MDA-MB-231 stable cells were trypsinized, washed two times in PBS, and kept on ice before injection. Mice were anesthetized, incised, and injected bilaterally in their 4th mammary glands with 1 × 106 MDA-MB-231 cells per side. The final volume of injection was 50 μl per mammary gland. The incision was sutured, and the mice were monitored for tumor growth weekly via bioluminescence and caliper. For each stable cell line, five mice were injected and kept in the same cage. At 8 weeks post-injection, each mammary tumor was resected, weighed, and split. Half was subjected to rapid snap-freezing for Western blotting, and the remainder was fixed overnight in 10% neutral buffered formalin (Fisher). Tumor samples were embedded in paraffin, and 4-μm sections were prepared for immunohistochemistry.
In Vivo Bioluminescence Imaging
Bioluminescence imaging of xenografted mice was performed using an IVIS Lumina device (Caliper). Imaging and signal quantification were controlled by the acquisition and analysis software LivingImage 3.2 (Caliper) and Igor (Wavemetrics). For primary tumors, mice were imaged 1 day after tumor cell implantation and weekly thereafter for 8 weeks in total. For metastasis, mice were imaged once during the 9th week after primary tumor resection. Before imaging, mice were anesthetized with 2.5% isoflurane in a holding tank and injected intraperitoneally with D-luciferin (Biosynth) at 150 mg/kg in PBS. They were awake for 5 min, anesthetized again with 2.5% isoflurane in the holding tank, and then transferred inside the imaging chamber of the Lumina device with 1.5% isoflurane and O2 flow through a nose cone. 15 min after d-luciferin injection, images were acquired with a fixed series of exposure times and the same binning (1 min, 30 s, 10 s, 5 s, 1 s, and 5 min; binning, low). For image analysis, an oval-shaped region of interest of the same size was used to calculate photon flux of all tumors at all time points. Data were expressed as total photon flux (photons/s).
Statistical Analysis
One way ANOVA analysis was conducted to assess the overall differences of the cell growth rates in two-dimensional cultures and the end-point tumor weights from the mice. Multiple comparisons were conducted using Tukey's method after one-way ANOVA to assess the differences between individual groups. Unpaired Student's t test was used to determine the difference between Pfn1-expressing tumors and those in the control group with regard to bioluminescence signals and tumor volumes. All statistical analyses were performed using Graphpad Prism 5.0 and R (Version 2.15.2). p values <0.05 were considered significant.
RESULTS
Mimicking Ser-137 Phosphorylation Specifically Inhibits Pfn1 Binding to PLPs
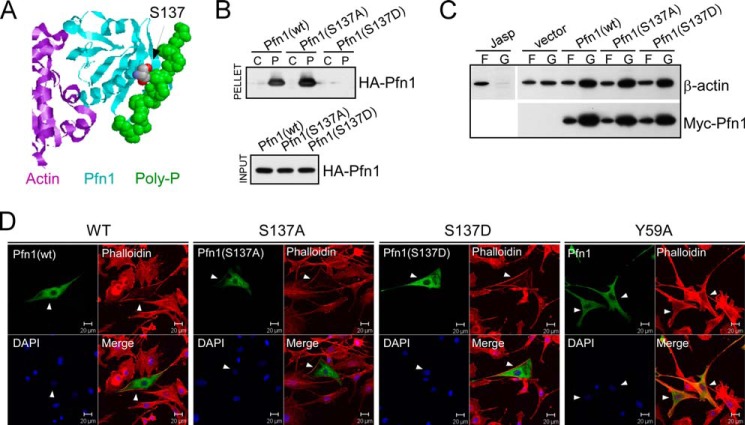
In prior work we found that Ser-137 phosphorylation of Pfn1 blocks its binding to Htt protein, which has two PLP motifs (9). Ser-137 resides within the PLP-binding site of Pfn1 (Fig. 1A). Thus Ser-137 phosphorylation may inhibit general PLP binding via direct steric hindrance or electrostatic repulsion conferred by the negative charge. To test this we transiently expressed wild type, phospho-resistant (S137A), and phosphomimetic (S137D) forms of Pfn1 in HEK293 cells and measured their binding to PLP-conjugated Sepharose beads. The S137D mutation abolished Pfn1 binding to PLPs, whereas the S137A mutation did not (Fig. 1B). Our prior co-immunoprecipitation experiment suggested that Ser-137 phosphorylation does not affect G-actin binding of Pfn1 at physiological salt concentration (9). When transiently overexpressed, wild type Pfn1 and its S137A or S137D mutants similarly decreased the F-actin/G-actin ratio of HEK293 cells (Fig. 1C) and redistributed F-actin (stained by phalloidin) from the cell body to the periphery of NIH3T3 cells (Fig. 1D). An actin-binding Pfn1 mutant (Y59A) (15), however, failed to redistribute F-actin in NIH3T3 cells, suggesting that such an effect of Pfn1 requires actin binding (Fig. 1D). Although the net effects of Pfn1 on F-actin levels and localization depend on cellular context and relative abundance of other actin-regulatory factors (4), our results strongly suggest that Ser-137 phosphorylation does not directly affect Pfn1 G-actin binding. Rather, it appears to be a selective inhibitory event for PLP binding by Pfn1.
FIGURE 1.
Mimicking Ser-137 phosphorylation selectively inhibits PLP binding of Pfn1. A, x-ray diffraction structure of human Pfn1 (middle, cyan) bound with actin (left, purple) and a 16-amino acid PLP peptide derived from the vasodilator-stimulated phosphoprotein (right, green, in space-filling mode) (Protein Data Bank ID 2PAV). Ser-137 of Pfn1, indicated by the arrow, is located next to the PLP peptide but far from actin. B, HEK293 cells were transfected with HA-tagged Pfn1 (wt, S137A, or S137D), and lysates were bound to control (C) or PLP-conjugated (P) Sepharose beads. Anti-HA antibody was used to detect HA-Pfn1 bound to the beads (pellet) and in the lysate (input) by Western blot. C, HEK293 cells transfected with pcDNA3 (vector) or Myc-tagged Pfn1 (wt, S137A, and S137D) were fractionated and analyzed for F-actin (F)/G-actin (G) ratio by Western blot. Jasplakinolide was used to stabilize F-actin before fractionation as a control. Similar expression of all three Myc-Pfn1 proteins was confirmed by Western blotting using a Myc-tag antibody. D, NIH 3T3 cells were transfected with Pfn1 constructs (wt, S137A, S137D, and Y59A) and subsequently co-stained for overexpressed Pfn1 with an anti-Pfn1 antibody and F-actin with rhodamine-phalloidin. Arrowheads point to positive cells which expressed the indicated Pfn1 plasmids.
Mimicking Pfn1 Phosphorylation on Ser-137 Promotes Tumor Growth in Vivo
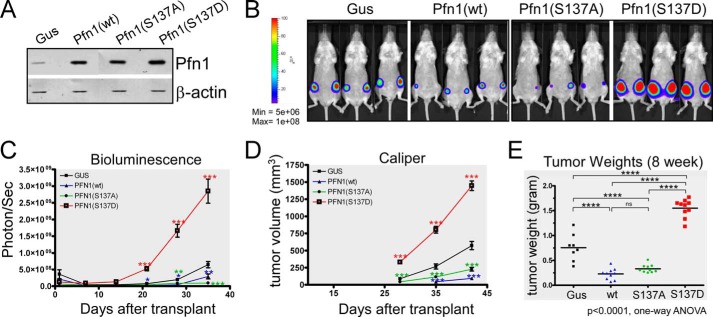
Pfn1 has been found to suppress breast cancer cell growth in mouse xenografts. This requires actin binding (15), but the role of PLP binding has not been clearly defined. Our ability to mimic Ser-137 phosphorylation and selectively block PLP binding by Pfn1 allowed us to test its role in this process. We stably expressed Pfn1(wt), Pfn1(S137A), or Pfn1(S137D) in MDA-MB-231 breast carcinoma cells with the bacterial β-glucuronidase (gus) as a negative control. Western blot showed that exogenous Pfn1 proteins were expressed at levels 2–3-fold above endogenous Pfn1 (Fig. 2A). We injected stable MDA-MB-231 cells into the mammary fat pads of immuno-deficient female NOD/SCID mice and monitored tumor burden weekly with bioluminescence imaging (utilizing stably expressed firefly luciferase (28)) and caliper measurement (feasible 4 weeks post injection). Compared with the Gus control group, mice injected with cells expressing Pfn1(wt) and Pfn1(S137A) exhibited significantly slower tumor growth (Fig. 2, B–D). In contrast, tumors formed by cells expressing Pfn1(S137D) grew significantly faster than all other lines (Fig. 2, B–D). We confirmed these findings by end-point tumor weight measurements (Fig. 2E). Taken together our results suggest Ser-137 phosphorylation via disrupting PLP binding, inhibits Pfn1 antitumor function, and promotes tumor formation in vivo.
FIGURE 2.
Mimicking Pfn1 phosphorylation on Ser-137 promotes tumor growth in vivo. A, Western blot showing stable overexpression of exogenous Pfn1 (wt, S137A, and S137D) over endogenous Pfn1 in MDA-MB-231 cells. gus, a bacterial reporter gene encoding β-glucuronidase as the negative control. β-Actin was blotted as the loading control. B–E, stable MDA-MB-231 cells were injected into the mammary fat pads of 5-week-old female NOD/SCID mice (106 cells per side, 5 mice per group). Tumor cells were monitored 1 day after injection and weekly thereafter using bioluminescence imaging (BLI). Caliper measurements started 4 weeks after injection. B, representative bioluminescence imaging images showing tumor burden at 4 weeks post-injection. C, time course of tumor growth measured by bioluminescence imaging. Total photon flux (photons/s) of 10 s/binning 2 imaging acquisition is shown for each data point. Data are the mean ± S.E. of 10 ROI (region of interest covering each entire tumor) values for each group (2 tumors per mouse, 5 mice per group). D, time course of tumor volume increase measured by caliper. Data are the mean ± S.E. of 10 tumors within each group (2 tumors per mouse, 5 mice per group). p values were based on two-tailed unpaired t test (Pfn1 versus Gus tumors). E, end-point tumor weights at resection. p values were based on one-way ANOVA and multiple comparisons using the Tukey method. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
Mimicking Pfn1 Phosphorylation on Ser-137 Inhibits Tumor Cell Apoptosis in Vivo
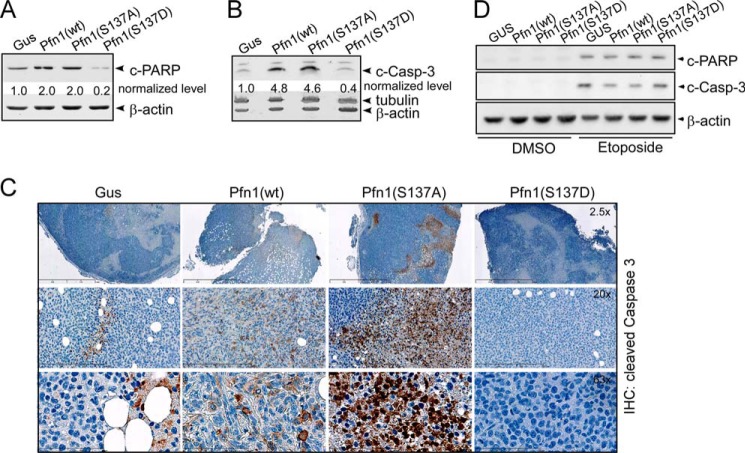
Pfn1 potentiates apoptosis in multiple cancer cell lines (29–31). To test if Ser-137 phosphorylation inhibits the pro-apoptotic activity of Pfn1, we examined the xenograft tumors for cleaved caspase-3 (c-Casp-3) and cleaved PARP (c-PARP) using Western blots. Tumors expressing Pfn1(wt) and Pfn1(S137A) contained significantly higher levels of c-Casp-3 (>4-fold) and c-PARP (2-fold) compared with the Gus control. In contrast, tumors expressing Pfn1(S137D) contained lower levels of both markers compared with the Gus control (80% decrease for c-PARP and 60% decrease for c-Casp-3) (Fig. 3, A and B). The changes in c-Casp-3 levels were further confirmed using immunohistochemistry (Fig. 3C). Tumors expressing Pfn1(wt) and Pfn1(S137A) both contained more c-Casp-3-positive cells than the Gus control tumor. Cells expressing Pfn1(S137A) exhibited more intense staining than those expressing Pfn1(wt), indicating that Pfn1(S137A) might be especially effective in promoting apoptosis. In contrast, we detected many fewer positive cells in Pfn1(S137D)-expressing tumors (Fig. 3C). These data suggest that in vivo Ser-137 phosphorylation not only abolishes the pro-apoptotic activity of Pfn1 but also makes tumors more resistant to apoptosis, causing the fast-growing phenotype of Pfn1(S137D)-expressing tumors. Interestingly, neither c-PARP nor c-Casp-3 was detectable by Western blot when the same stable MDA-MB-231 cells were grown in two-dimensional cultures (Fig. 3D). When triggered with etoposide (a DNA topoisomerase II inhibitor and an apoptotic inducer), both markers appeared. However, Pfn1 expression had no effect on their levels regardless of the phosphorylation status of Ser-137 (Fig. 3D). This indicates that Pfn1 sensitizes tumor cells to apoptosis preferentially in an in vivo microenvironment in response to appropriate stimuli.
FIGURE 3.
Mimicking Pfn1 phosphorylation on Ser-137 inhibits tumor cell apoptosis in vivo. A and B, frozen tumors from the xenografted mice were analyzed for c-PARP and c-casp-3 by Western blot. Protein bands were quantified based on densitometry and normalized against β-actin levels. For simplicity, c-PARP and c-Casp-3 levels within the Gus tumor were arbitrarily set at 1, and those in Pfn1-expressing tumors were calculated accordingly. Tumors from two mice within each group were analyzed (which showed similar results), one of which is shown here. C, immunohistochemical (IHC) analysis of formalin-fixed and paraffin-embedded tumors for c-Casp-3. Nuclei were counterstained with hematoxylin. Shown are representative images at different magnifications. D, stable MDA-MB-231 cells grown in two-dimensional cultures in the absence or presence of 50 μm etoposide (24 h) were analyzed for c-PARP and c-casp-3 levels by Western blot. Etoposide was used as a positive experimental control to induce apoptosis. β-Actin was blotted as the loading control.
Preventing or Mimicking Ser-137 Phosphorylation Both Inhibit Cell Cycle Arrest by Pfn1
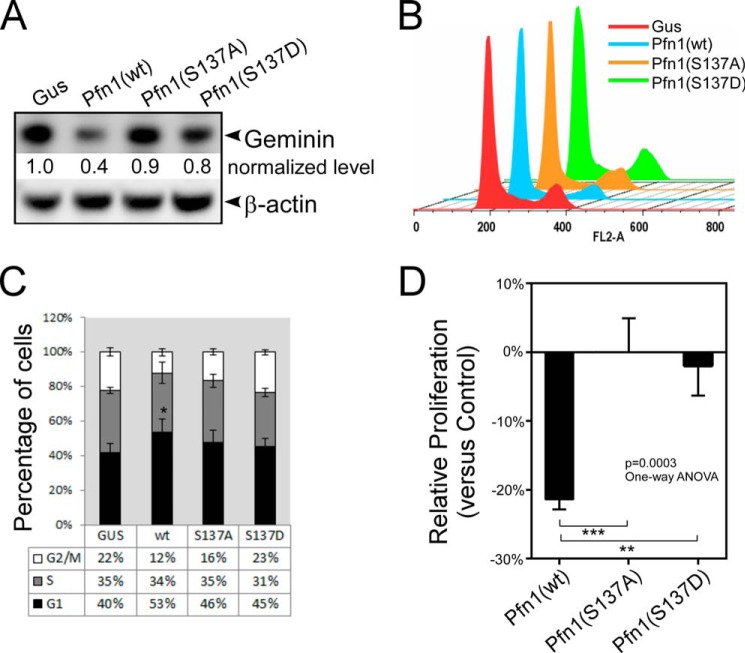
Pfn1 has been reported to cause G1 arrest of the cell cycle in the MDA-MB-231 cells (17). To test if this partially contributes to the antitumor effect of Pfn1 in mouse xenografts, we examined the levels of geminin, a cell cycle marker absent in G1 but present throughout S and G2/M phases. Western blot analysis showed that tumors expressing Pfn1(wt) contained ∼60% less geminin than the Gus control tumors (Fig. 4A), consistent with the Pfn1(wt)-expressing cells being arrested in G1 phase. However, tumors expressing Pfn1(S137A) or Pfn1(S137D) contained similar levels of geminin to the Gus control tumors. DNA content analysis of synchronized cells in two-dimensional cultures confirmed this as only cells expressing Pfn1(wt) were significantly arrested in G1 phase (33% increase in cell number compared with Gus control cells) (Fig. 4, B and C). This is also consistent with the cell growth rates in two-dimensional cultures (under which condition apoptosis of these cells was minimal) as measured by the MTT assay (Fig. 4D). Thus, inhibition of cell proliferation by Pfn1, at least in the MDA-MB-231 cells, may require Ser-137 in a freely phosphorylatable and dephosphorylatable state. We conclude that Pfn1(wt) might inhibit tumor growth through the effects on proliferation and apoptosis together, whereas Pfn1(S137A) functions mainly by sensitization to apoptosis.
FIGURE 4.
Preventing or mimicking Ser-137 phosphorylation both inhibit cell cycle arrest by Pfn1. A, frozen xenograft tumors were analyzed for geminin by Western blot. Protein bands were quantified based on densitometry and normalized against β-actin levels. Relative geminin levels in the Pfn1-expressing tumors over the Gus control tumors were shown. B and C, stable MDA-MB-231 cells expressing Gus or Pfn1 (wt, S137A, or S137D) grown in two-dimensional cultures were serum-starved for 48 h and subsequently released for 24 h. They were labeled with propidium iodide and analyzed for DNA content using flow cytometry. A representative experiment was shown in B. Data in C are the mean ± S.E. of four independent experiments. D, stable MDA-MB-231 cells expressing Gus or Pfn1 (wt, S137A, or S137D) were seeded on plastic and grown for 7 days. They were quantified by MTT labeling on day 1 and day 7, and the day 7/day 1 ratios of Pfn1-expressing cells were normalized against that of Gus control cells to give rise to their relative proliferation rates (y axis: positive % values for increase and negative % values for decrease). Data are the mean ± S.E. of five independent experiments. p values for C and D were based on one-way ANOVA analysis and multiple comparisons using the Tukey method. **, p < 0.01; ***, p < 0.001.
Cell Cycle Arrest and Growth Inhibition by Pfn1 Requires Nuclear Localization
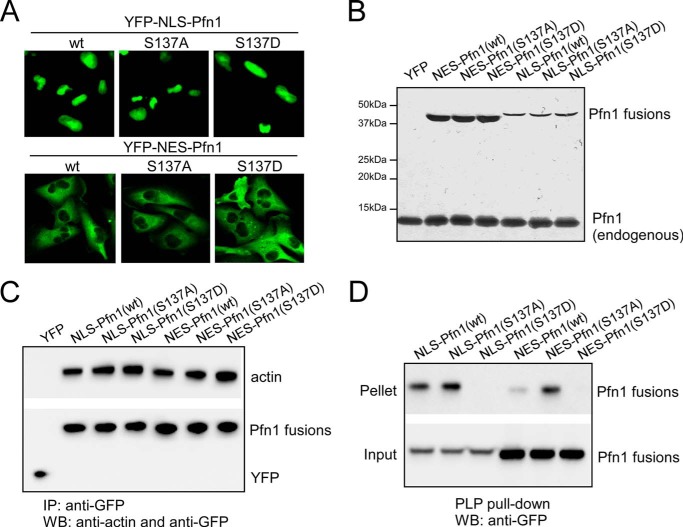
Although predominantly a cytoplasmic protein, a fraction of Pfn1 is present in the nucleus and has been implicated in gene transcription and pre-mRNA processing (19, 20, 23). Thus Pfn1 may have distinct functions depending on its subcellular localization. To test how subcellular localization influences Pfn1 antitumor activity, we fused Pfn1 to YFP along with NLS or NES and stably expressed the fusion proteins (termed NLS-Pfn1 and NES-Pfn1) in MDA-MB-231 cells (Fig. 5A). Western blot showed that NLS-Pfn1 fusions were expressed at levels ∼25% that of endogenous Pfn1, whereas NES-Pfn1 fusions were expressed at levels similar to endogenous Pfn1 (Fig. 5B).
FIGURE 5.
Pfn1 fused to NLS or NES maintains actin and PLP binding abilities. A, subcellular localization of YFP-NLS-Pfn1 and YFP-NES-Pfn1 in stable MDA-MB-231 cells. B, relative expression levels of YFP-NLS-Pfn1 and YFP-NES-Pfn1 in stable MDA-MB-231 cells by Western blot. C, MDA-MB-231 stable cell lysates expressing YFP, NLS-Pfn1, or NES-Pfn1 lacking or containing S137 mutations were immunoprecipitated (IP) with an anti-GFP antibody and analyzed for actin binding by Western blot (WB). D, same cell lysates from C were mixed with PLP-conjugated beads and analyzed for the binding of NLS or NES-tagged Pfn1 by Western blotting using a GFP antibody.
Next, we tested if the NLS- or NES-tagged Pfn1 maintains the structural integrity of the untagged Pfn1 by testing ligand binding. Both NLS- and NES-tagged Pfn1 bind actin and PLPs in the MDA-MB-231 stable cells (Fig. 5, C and D). As for untagged Pfn1, neither S137A nor S137D affected the actin binding of NLS- or NES-tagged Pfn1 (Fig. 5C). However, S137D specifically abolished PLP binding of both NLS- and NES-tagged Pfn1. Of note, S137A subtly increased PLP binding of NLS-Pfn1 and significantly increased PLP binding of NES-Pfn1 (Fig. 5D), consistent with the idea that Ser-137 undergoes a certain degree of phosphorylation in both compartments, resulting in reduced PLP binding.
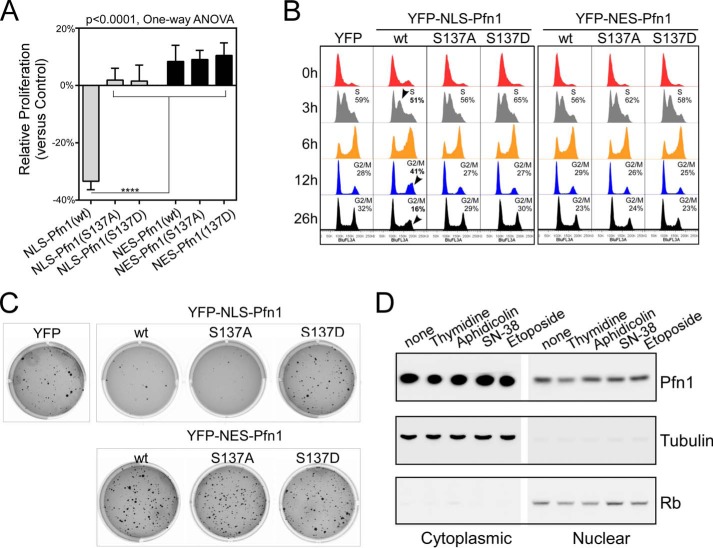
We next compared the in vitro growth rates of the MDA-MB-231 stable cells in two-dimensional cultures (Fig. 6A). NLS-Pfn1 inhibited cell growth compared with the YFP control. NES-Pfn1, however, failed to do so. DNA content analysis of synchronized cells indicated that wild type NLS-Pfn1 arrested cells in G1 phase (Fig. 6B). 3 h after double thymidine block, NLS-Pfn1(wt) caused a 14% decrease of cells in S phase compared with YFP control cells, indicating a delay in G1/S transition. At 6 h, when most YFP control cells completed DNA synthesis, 35% more NLS-Pfn1(wt) cells were in S phase, and 18% fewer were in G2/M phase, reflecting slower S phase entry and completion. By 12 h, most YFP control cells had completed mitosis and reentered G1 phase, whereas 46% more NLS-Pfn1(wt) cells were still in G2/M phase and 20% fewer were in G1 phase. The slower cycling rate of NLS-Pfn1(wt) cells was also reflected at 26 h post-release when 50% fewer were found in G2/M phase compared with YFP cells, presumably due to their delay in G1/S transition in two consecutive cycles. In contrast, no cell cycle arrest and proliferative inhibition was observed in MDA-MB-231 cells expressing NLS-Pfn1(S137A) or NLS-Pfn1(S137D) (Fig. 6, A and B), similar to the behaviors of their untagged counterparts under the same experimental conditions (Fig. 4).
FIGURE 6.
Cell cycle arrest and growth inhibition by Pfn1 requires nuclear localization. A, stable MDA-MB-231 cells were seeded in two-dimensional cultures and quantified by MTT assay at day 1 and day 7. Relative growth rates of Pfn1 fusion-expressing cells over those expressing YFP control were calculated as described in Fig. 4D. Data are the mean ± S.E. of three separate experiments. p values were based on one-way ANOVA analysis and multiple comparisons using the Tukey method. ****, p < 0.0001. B, DNA content analysis of stable MDA-MB-231 cells blocked by double thymidine and released for various lengths of time (3, 6, 12, and 26 h). Arrowheads indicated obvious changes in cell cycle profiles caused by NLS-Pfn1(wt) expression, which have been confirmed by three independent experiments. C, stable MDA-MB-231 cells were grown in 0.4% soft agar for 3 weeks and stained with crystal violet. Shown are representative images from one of three independent experiments, which generated data of similar trends. D, parental MDA-MB-231 cells were subjected to double thymidine (2 mm) block or treated with aphidicolin (5 μg/ml), SN-38 (200 nm), and etoposide (50 μm) for 24 h. Cells were fractionated, and Pfn1 in the cytoplasmic and nuclear fractions was analyzed by Western blot. Tubulin and Rb were used as cytoplasmic and nuclear markers.
Next, we tested if nuclear Pfn1 could inhibit the growth of MDA-MB-231 cells in soft agar (Fig. 6C). Three weeks post-seeding, cells expressing NLS-Pfn1(wt) and NLS-Pfn1(S137A) both formed fewer colonies than the YFP control cells, reminiscent of the effects of untagged Pfn1(wt) and Pfn1(S137A) in mouse xenografts, which both inhibited tumor growth (Fig. 2). Cells expressing NLS-Pfn1(S137D), however, formed a similar number of colonies to the YFP control, consistent with its lack of antitumor activity. NES-Pfn1 did not inhibit colony formation regardless of the phosphorylation state of Ser-137, consistent with its lack of anti-growth effects in two-dimensional culture. Together, these results suggest that profilin-1 functions in the nucleus as an antitumor protein and is subjected to regulation by Ser-137 phosphorylation.
Given the distinct activities of nuclear versus cytoplasmic Pfn1 with regard to tumor cell growth, we tested if their relative distribution within the cell changes as a result of G1/S arrest or apoptotic activation. We treated MDA-MB-231 cells with double thymidine and aphidicolin (DNA polymerase inhibitor) to induce G1/S arrest and DNA damaging agents SN-38 (a metabolite of irinotecan) and etoposide to induce apoptosis. We then separated the cells into cytoplasmic and nuclear fractions and quantified Pfn1 level by Western blot (Fig. 6D). We observed no clear effects of G1/S arrest or DNA damage-induced apoptosis on the relative distribution of Pfn1 between nucleus and cytoplasm. We conclude that although Pfn1 can influence these specific cell events, its subcellular localization is not governed by them per se.
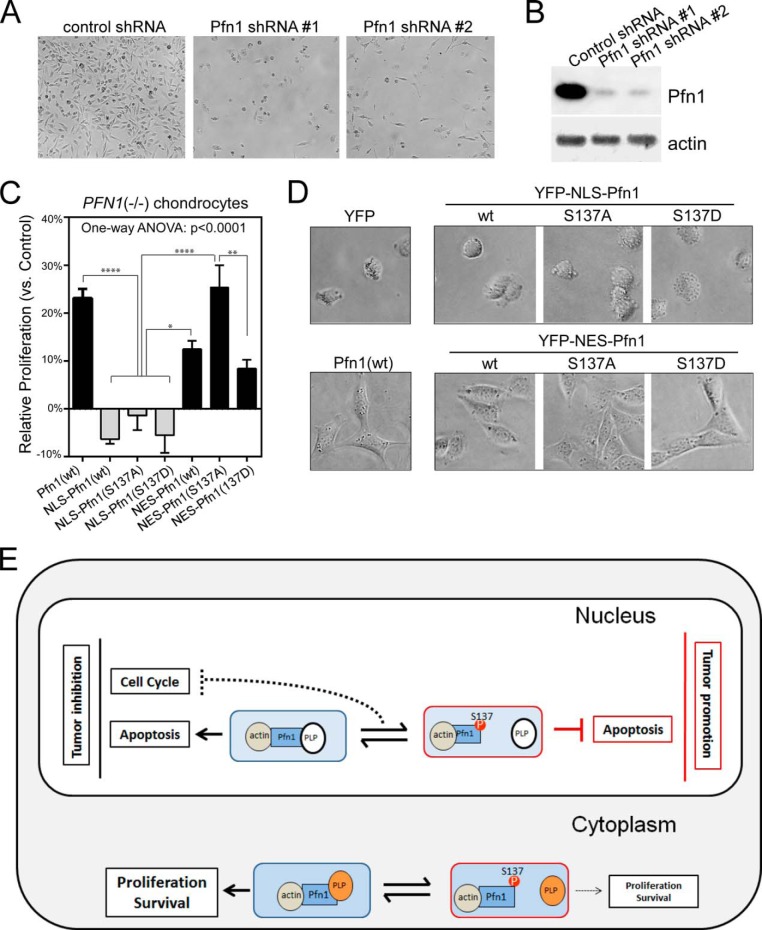
Cytoplasmic Pfn1 Rescues Cellular Defects of PFN1 Null Chondrocytes
Pfn1 is an essential protein for cytokinesis, proliferation, and survival (6–8). In line with this, we observed severe growth inhibition of MDA-MB-231 cells upon acute Pfn1 knockdown by two different shRNAs (Fig. 7, A and B). Systemic knock-out of PFN1 is embryonically lethal in mice (7). Cartilage-specific PFN1 deletion is tolerable, but the mice develop chondrodysplasia, and cultured PFN1 null chondrocytes show abnormally rounded shape, cytokinesis defects due to weakened actin-dependent traction force generation, and slower proliferation (5). We observed that re-expression of wild type Pfn1 in PFN1 null chondrocytes increased their proliferation and normalized their morphology (Fig. 7, C and D). To determine whether the cytoplasmic or nuclear Pfn1 functions were responsible for the rescuing effects, we expressed NLS-Pfn1 or NES-Pfn1. NES-Pfn1 rescued both the morphological and growth defects of PFN1 null cells, whereas NLS-Pfn1 did not (Fig. 7, C and D). This indicates that cytoplasm is the site of action for Pfn1 essential cellular functions, in contrast to its effects on tumor growth, which occur in the nucleus. NES-Pfn1(S137A) more effectively rescued the proliferation defect of PFN1 null cells than NES-Pfn1(wt) and NES-Pfn1(S137D), suggesting that cytoplasmic Pfn1 function may be negatively regulated by Ser-137 phosphorylation.
FIGURE 7.
Cytoplasmic Pfn1 rescues cellular defects of PFN1 null chondrocytes. A, equal numbers of MDA-MB-231 cells were infected with lentiviral shRNAs containing a scrambled sequence (control) or two different Pfn1-targeting sequences (#1 and #2). Cell images were taken at day 7 post-infection. B, Western blot analysis of shRNA infected MDA-MB-231 cells for total Pfn1. Actin was blotted as the loading control. C, Pfn1−/− chondrocytes infected with YFP, YFP-NLS-Pfn1, or YFP-NES-Pfn1 viruses were seeded in two-dimensional cultures and quantified by MTT assay at day 1 and day 4. Relative growth rates of Pfn1-infected cells over YFP-infected control cells were calculated as described in Fig. 5C. Data are the mean ± S.E. of three separate experiments. p values were based on one-way ANOVA analysis and multiple comparisons using the Tukey's method. *, p < 0.05; **, p < 0.01; ****, p < 0.0001. D, morphological differences of virus infected Pfn1−/− chondrocytes grown on plastic. E, A Spatial Confinement model to explain Pfn1's opposing cellular functions and the role of Ser-137 phosphorylation. Pfn1 acts as a tumor suppressor in the nucleus by augmenting apoptosis and blocking cell cycle, whereas it acts in the cytoplasm to support proliferation and survival by facilitating actin polymerization and remodeling. In the nucleus, the pro-apoptotic activity of Pfn1 depends on interaction with actin and an unknown PLP protein. Ser-137 phosphorylation blocks PLP binding, and converts Pfn1 from an apoptotic enhancer to an inhibitor. As for cell cycle inhibition, dynamic association/dissociation of Pfn1 with a different PLP protein might be required (dashed line). In the cytoplasm, Pfn1 interacts with actin and multiple PLP-containing proteins (most are actin-regulatory proteins such as mDia, Ena/VASP, and N-WASP) to support proliferation and survival. Ser-137 phosphorylation inhibits these activities by disrupting Pfn1 binding to PLP.
DISCUSSION
The Opposing Cellular Functions of Pfn1 Are Spatially Defined
Pfn1 is an essential cellular protein with paradoxical antitumor activities. Its systematic deletion is lethal in several eukaryotic organisms (mouse, firefly, and yeast) (6–8), and its genetic knockdown inhibits the growth of cultured mammalian cells (Fig. 7, A and B) (32). Yet, Pfn1 also has anti-proliferative and pro-apoptotic activities, which have been mostly recognized in the context of cancer (Refs. 12 and 15–17 and Figs. 2–4). Recently, Pfn1 has been found to destabilize hypoxia-inducible factor-1α (HIF-1α) and consequently inhibit angiogenesis, indicating the multifaceted nature of its antitumor functions (14, 33). However, until now it has remained puzzling how Pfn1 can simultaneously support and inhibit proliferation within the same cellular context. For instance, we have observed that both overexpression and knockdown of Pfn1 in the MDA-MB-231 cells cause growth inhibition (Figs. 4 and 7). In this paper we have defined the nucleus as the site of action for Pfn1 antitumor function and the cytoplasm as where Pfn1 supports cell proliferation. This forms the basis for a simple “spatial confinement” model, which helps explain the apparent Pfn1 paradox; its opposing cellular functions are compartmentalized between the nucleus and cytoplasm (Fig. 7E).
Although mainly in the cytoplasm, Pfn1 has been detected in nuclear structures and implicated in transcription and RNA splicing (19–23). Thus, nuclear Pfn1 might function as an antitumor protein by regulating gene expression. We speculate that nuclear Pfn1 might partner with actin while executing these nuclear events. This is based on a prior study that actin binding is essential for Pfn1 to suppress tumor growth in a breast cancer xenograft model (15). It is also consistent with the emerging role of nuclear actin as an important regulator of gene expression through association with all three RNA polymerases and chromatin remodeling complexes as well as direct participation in transcription (34–37).
In contrast, cytoplasmic Pfn1 fails to inhibit tumor cell growth but instead rescues morphological and growth defects of PFN1 null cells. Although this finding comes as no surprise given the crucial role of F-actin integrity and dynamics, which are facilitated by Pfn1, in cell division and survival (38–42), it further supports our hypothesis that the opposing functions of Pfn1 are compartmentally separated.
Functional Effects of Ser-137 Phosphorylation and the Importance of PLP Binding
Our data that Ser-137 phosphorylation selectively disrupts PLP binding by Pfn1 and acts as an off-switch for Pfn1 antitumor effects suggest that finding the critical nuclear PLP-containing ligand(s) of Pfn1 will be the most straightforward way to unravel its antitumor mechanisms. Pfn1 has mostly been studied with regard to its cytoplasmic functions, and its nuclear PLP ligands have remained largely unexplored. Only two nuclear PLP proteins have been reported to interact with Pfn1 (transcriptional factor p42POP (22) and pre-mRNA splicing regulatory factor SMN (21)), neither of which has been linked to cancer. Thus, future efforts will be necessary to identify more nuclear PLP ligands of Pfn1 and to test their functional relationships.
Although Pfn1 inhibits tumor growth by influencing both proliferation and apoptosis, these two functions of Pfn1 appear to be differentially regulated by Ser-137 phosphorylation. With regard to apoptosis, our data suggest that Ser-137 phosphorylation “toggles” Pfn1 between a pro-apoptotic and an anti-apoptotic state when tumor cells are triggered by certain in vivo stimuli (Fig. 3). This implies that an unknown PLP-ligand is important for the effect of Pfn1 on apoptosis (Fig. 7E). With regard to cell cycle arrest and proliferative inhibition by Pfn1, we find that both Ala/Asp substitutions at Ser-137 cause loss of function (Figs. 4 and 6). This could be due to complete inactivation of Pfn1. However, given clearly opposite effects of these point mutations in other aspects of Pfn1 function, we favor a model in which dynamic phosphorylation and dephosphorylation of Ser-137 may be required (Fig. 7E). The answer can only come from more definitive future studies.
Interestingly, data using PFN1 null chondrocytes suggest that Ser-137 phosphorylation also inhibits cytoplasmic Pfn1 function. This is consistent with the fact that interactions with several PLP-containing actin-regulatory factors (e.g. mDia, Ena/VASP, and N-WASP) are important for the optimal ability of Pfn1 to promote actin polymerization (2, 4).
Taken together, our data have uncovered the importance of nuclear localization and phosphorylation state of Ser-137 in tumor inhibition by Pfn1. They help reconcile the long-standing paradox regarding the opposing cellular functions of Pfn1 and take us closer to understanding its molecular mechanisms.
Acknowledgments
We thank Dr. Byron Hann for the MDA-MB-231 cell line stably expressing the tri-modal reporter fusion, Dr. Ralph T Bottcher and Dr. Reinhard Fassler for providing the PFN1 null chondrocytes, Dr. Eric Campeau for the pLenti-CMV/TO-Neo-DEST (685–3) destination vector, Dr. Sofia Origanti for scientific advice, Dr. Mingjie Li and Nada Husic for assisting in lentiviral production, Dr. William Eades for assistance with flow cytometry and analysis, Gary London and Dr. Kris Hyrc for assistance with confocal and whole-slide imaging, and Dr. Dyche Mullins and Dr. Brad Zuchero for the PLP-conjugated Sepharose beads. The Hope Center Viral Vectors Core is supported by Neuroscience Blueprint Core National Institutes of Health Grant P30 NS057105 (to Washington University). The Siteman Cancer Center is supported in part by National Institutes of Health Grant NCI Cancer Center Support Grant P30 CA91842. The Molecular Imaging Center is supported in part by National Institutes of Health P50 CA94056 (to Washington University). The Hope Center Alafi Neuroimaging Core is supported by a P30 Neuroscience Blueprint Interdisciplinary Center Core award to Washington University (P30 NS057105). The RNAi consortium at the Genome Institute of Washington University is supported by the Children's Discovery Institute.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS50284 (NINDS; to M. I. D.), R01CA131226 (NCI; to M. I. D.), and R37CA064786, R01CA057621, R01CA140663, U54CA112970, U01CA143233, and U54CA143836 (NCI; to M. B.). This work was also supported by the American Cancer Society Institutional Research Grant (to J. S.), the Muscular Dystrophy Association (to J. S. and M. I. D.), the KOMEN Foundation (to H. P.-W.), the United States Department of Energy (to M. B.), the Office of Biological and Environmental Research and Low Dose Scientific Focus Area (to M. B., contract DE-AC02-05CH1123), the United States Department of Defense (to M. B.), the Hope Center for Neurological Disorders, the Knight-ADRC, the Siteman Cancer Center, and the Molecular Imaging Center.
- Pfn1
- profilin-1
- F-actin
- filamentous actin
- G-actin
- globular actin
- PLP
- poly-l-proline
- Htt
- huntingtin
- NLS
- nuclear localization sequence
- NES
- nuclear export sequence
- PARP
- poly(ADP-ribose) polymerase
- c-PARP
- cleaved PARP
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
- ANOVA
- analysis of variance
- c-Casp-3
- cleaved caspase-3.
REFERENCES
- 1. Birbach A. (2008) Profilin, a multi-modal regulator of neuronal plasticity. Bioessays 30, 994–1002 [DOI] [PubMed] [Google Scholar]
- 2. Jockusch B. M., Murk K., Rothkegel M. (2007) The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 159, 131–149 [DOI] [PubMed] [Google Scholar]
- 3. Schlüter K., Jockusch B. M., Rothkegel M. (1997) Profilins as regulators of actin dynamics. Biochim. Biophys. Acta 1359, 97–109 [DOI] [PubMed] [Google Scholar]
- 4. Witke W. (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14, 461–469 [DOI] [PubMed] [Google Scholar]
- 5. Böttcher R. T., Wiesner S., Braun A., Wimmer R., Berna A., Elad N., Medalia O., Pfeifer A., Aszódi A., Costell M., Fässler R. (2009) Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 28, 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balasubramanian M. K., Hirani B. R., Burke J. D., Gould K. L. (1994) The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 125, 1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Witke W., Sutherland J. D., Sharpe A., Arai M., Kwiatkowski D. J. (2001) Profilin I is essential for cell survival and cell division in early mouse development. Proc. Natl. Acad. Sci. U.S.A. 98, 3832–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verheyen E. M., Cooley L. (1994) Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 120, 717–728 [DOI] [PubMed] [Google Scholar]
- 9. Shao J., Welch W. J., Diprospero N. A., Diamond M. I. (2008) Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell. Biol. 28, 5196–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu N., Zhang W., Yang Y., Liang Y. L., Wang L. Y., Jin J. W., Cai X. M., Zha X. L. (2006) Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 6, 6095–6106 [DOI] [PubMed] [Google Scholar]
- 11. Grønborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell Proteomics 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 12. Janke J., Schlüter K., Jandrig B., Theile M., Kölble K., Arnold W., Grinstein E., Schwartz A., Estevéz-Schwarz L., Schlag P. M., Jockusch B. M., Scherneck S. (2000) Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J. Exp. Med. 191, 1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zoidakis J., Makridakis M., Zerefos P. G., Bitsika V., Esteban S., Frantzi M., Stravodimos K., Anagnou N. P., Roubelakis M. G., Sanchez-Carbayo M., Vlahou A. (2012) Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol. Cell Proteomics 11, 10.1074/mcp.M111.009449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao W., Ji S., Qin Y., Yang J., Xu J., Zhang B., Xu W., Liu J., Shi S., Liu L., Liu C., Long J., Ni Q., Li M., Yu X. (2014) Profilin-1 suppresses tumorigenicity in pancreatic cancer through regulation of the SIRT3-HIF1α axis. Mol. Cancer 13, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wittenmayer N., Jandrig B., Rothkegel M., Schlüter K., Arnold W., Haensch W., Scherneck S., Jockusch B. M. (2004) Tumor suppressor activity of profilin requires a functional actin binding site. Mol. Biol. Cell 15, 1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou L., Jaramillo M., Whaley D., Wells A., Panchapakesa V., Das T., Roy P. (2007) Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br. J. Cancer 97, 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zou L., Ding Z., Roy P. (2010) Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J. Cell. Physiol. 223, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stüven T., Hartmann E., Görlich D. (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 22, 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Söderberg E., Hessle V., von Euler A., Visa N. (2012) Profilin is associated with transcriptionally active genes. Nucleus 3, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skare P., Kreivi J. P., Bergström A., Karlsson R. (2003) Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp. Cell Res. 286, 12–21 [DOI] [PubMed] [Google Scholar]
- 21. Giesemann T., Rathke-Hartlieb S., Rothkegel M., Bartsch J. W., Buchmeier S., Jockusch B. M., Jockusch H. (1999) A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J. Biol. Chem. 274, 37908–37914 [DOI] [PubMed] [Google Scholar]
- 22. Lederer M., Jockusch B. M., Rothkegel M. (2005) Profilin regulates the activity of p42POP, a novel Myb-related transcription factor. J. Cell Sci. 118, 331–341 [DOI] [PubMed] [Google Scholar]
- 23. Harpen M., Barik T., Musiyenko A., Barik S. (2009) Mutational analysis reveals a noncontractile but interactive role of actin and profilin in viral RNA-dependent RNA synthesis. J. Virol. 83, 10869–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan Y., Arif A., Gong Y., Jia J., Eswarappa S. M., Willard B., Horowitz A., Graham L. M., Penn M. S., Fox P. L. (2012) Stimulus-dependent phosphorylation of profilin-1 in angiogenesis. Nat. Cell Biol. 14, 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh S. S., Chauhan A., Murakami N., Styles J., Elzinga M., Chauhan V. P. (1996) Phosphoinositide-dependent in vitro phosphorylation of profilin by protein kinase C. Phospholipid specificity and localization of the phosphorylation site. Recept. Signal. Transduct. 6, 77–86 [PubMed] [Google Scholar]
- 26. Feng Y., Nie L., Thakur M. D., Su Q., Chi Z., Zhao Y., Longmore G. D. (2010) A multifunctional lentiviral-based gene knockdown with concurrent rescue that controls for off-target effects of RNAi. Genomics Proteomics Bioinformatics 8, 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bae Y. H., Ding Z., Zou L., Wells A., Gertler F., Roy P. (2009) Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J. Cell. Physiol. 219, 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ray P., Tsien R., Gambhir S. S. (2007) Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 67, 3085–3093 [DOI] [PubMed] [Google Scholar]
- 29. Yao W., Cai X., Liu C., Qin Y., Cheng H., Ji S., Xu W., Wu C., Chen T., Xu J., Long J., Fang Z., Qu B., Hoth M., Ni Q., Zha X., Yu X. (2013) Profilin 1 potentiates apoptosis induced by staurosporine in cancer cells. Curr. Mol. Med. 13, 417–428 [PubMed] [Google Scholar]
- 30. Yao W., Yu X., Fang Z., Yin P., Zhao C., Li N., Wang L., Li Z., Zha X. (2012) Profilin1 facilitates staurosporine-triggered apoptosis by stabilizing the integrin β1-actin complex in breast cancer cells. J. Cell. Mol. Med. 16, 824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng H., Li J., Liu C., Yao W., Xu Y., Frank T. S., Cai X., Shi S., Lu Y., Qin Y., Liu L., Xu J., Long J., Ni Q. X., Li M., Yu X. J. (2013) Profilin1 sensitizes pancreatic cancer cells to irradiation by inducing apoptosis and reducing autophagy. Curr. Mol. Med. 13, 1368–1375 [DOI] [PubMed] [Google Scholar]
- 32. Ding Z., Lambrechts A., Parepally M., Roy P. (2006) Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 119, 4127–4137 [DOI] [PubMed] [Google Scholar]
- 33. Fan Y., Potdar A. A., Gong Y., Eswarappa S. M., Donnola S., Lathia J. D., Hambardzumyan D., Rich J. N., Fox P. L. (2014) Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat. Cell Biol. 16, 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng B., Han M., Bernier M., Wen J. K. (2009) Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 276, 2669–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Percipalle P., Visa N. (2006) Molecular functions of nuclear actin in transcription. J. Cell Biol. 172, 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jockusch B. M., Schoenenberger C. A., Stetefeld J., Aebi U. (2006) Tracking down the different forms of nuclear actin. Trends Cell Biol. 16, 391–396 [DOI] [PubMed] [Google Scholar]
- 37. Spencer V. A., Costes S., Inman J. L., Xu R., Chen J., Hendzel M. J., Bissell M. J. (2011) Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 124, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stehn J. R., Haass N. K., Bonello T., Desouza M., Kottyan G., Treutlein H., Zeng J., Nascimento P. R., Sequeira V. B., Butler T. L., Allanson M., Fath T., Hill T. A., McCluskey A., Schevzov G., Palmer S. J., Hardeman E. C., Winlaw D., Reeve V. E., Dixon I., Weninger W., Cripe T. P., Gunning P. W. (2013) A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 73, 5169–5182 [DOI] [PubMed] [Google Scholar]
- 39. Takasuka T., Ishibashi S., Ide T. (1987) GC-7 cells are growth arrested by cytochalasin D at two different points in the cell cycle. Exp. Cell Res. 173, 287–293 [DOI] [PubMed] [Google Scholar]
- 40. Senderowicz A. M., Kaur G., Sainz E., Laing C., Inman W. D., Rodríguez J., Crews P., Malspeis L., Grever M. R., Sausville E. A. (1995) Jasplakinolide's inhibition of the growth of prostate carcinoma cells in vitro with disruption of the actin cytoskeleton. J. Natl. Cancer Inst. 87, 46–51 [DOI] [PubMed] [Google Scholar]
- 41. Bousquet P. F., Paulsen L. A., Fondy C., Lipski K. M., Loucy K. J., Fondy T. P. (1990) Effects of cytochalasin B in culture and in vivo on murine Madison 109 lung carcinoma and on B16 melanoma. Cancer Res. 50, 1431–1439 [PubMed] [Google Scholar]
- 42. Odaka C., Sanders M. L., Crews P. (2000) Jasplakinolide induces apoptosis in various transformed cell lines by a caspase-3-like protease-dependent pathway. Clin. Diagn. Lab. Immunol. 7, 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]