Background: The MetNI transporter drives methionine import against its concentration gradient and regulates intracellular methionine levels.
Results: Methionine is a noncompetitive inhibitor of MetNI ATPase activity, binding the transporter at two allosteric sites.
Conclusion: MetNI regulates intracellular methionine concentrations via allosteric regulation.
Significance: Regulation of methionine import at the protein level may minimize wasteful consumption of ATP when adequate intracellular supplies are available.
Keywords: ABC Transporter, Amino Acid Transport, ATPase, Enzyme Mechanism, Methionine
Abstract
The MetNI methionine importer of Escherichia coli, an ATP binding cassette (ABC) transporter, uses the energy of ATP binding and hydrolysis to catalyze the high affinity uptake of d- and l-methionine. Early in vivo studies showed that the uptake of external methionine is repressed by the level of the internal methionine pool, a phenomenon termed transinhibition. Our understanding of the MetNI mechanism has thus far been limited to a series of crystal structures in an inward-facing conformation. To understand the molecular mechanism of transinhibition, we studied the kinetics of ATP hydrolysis using detergent-solubilized MetNI. We find that transinhibition is due to noncompetitive inhibition by l-methionine, much like a negative feedback loop. Thermodynamic analyses revealed two allosteric methionine binding sites per transporter. This quantitative analysis of transinhibition, the first to our knowledge for a structurally defined transporter, builds upon the previously proposed structurally based model for regulation. This mechanism of regulation at the transporter activity level could be applicable to not only ABC transporters but other types of membrane transporters as well.
Introduction
Early in vivo studies by Kadner and colleagues (1, 2) established that Escherichia coli utilizes a high affinity transport system that mediates the energy-dependent uptake of d- and l-methionine into cells. These elegant studies further demonstrated that uptake of external methionine is repressed by the level of the internal methionine pool, a phenomenon called transinhibition (3). As a result, the cell is able to regulate uptake of extracellular methionine to maintain an appropriate intracellular concentration, thereby minimizing the wasteful consumption of energy.
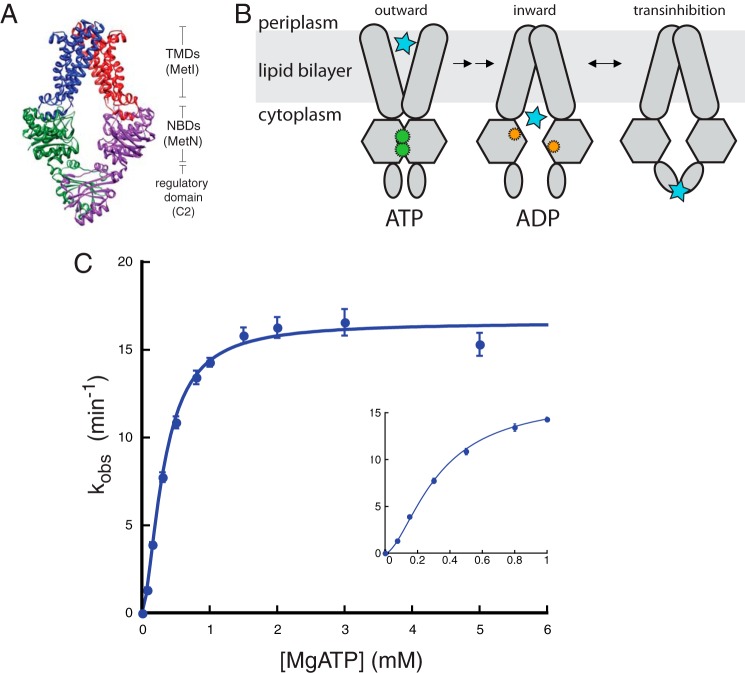
The high affinity import system of E. coli was subsequently identified as MetNI, a member of the ATP binding cassette (ABC)2 family of transporters (4–6). The MetNI importer, as with all ABC transporters, consists of two nucleotide binding domains (NBDs) and two transmembrane domains (TMDs) (7). ATP binding and hydrolysis occur at the interface between the highly conserved NBDs, where ATP is sandwiched between the Walker A motif of one monomer and the ABC motif (LSGGQ) on the other (8–10). Based on the TMD fold observed in the crystal structure (see Fig. 1A), MetNI was categorized as a Type I importer, with other structurally characterized members including the ModBC molybdate transporter and the MalFGK2 maltose transporter (11–14). For this class of importers, the alternating access transport mechanism proposes that ATP binding at the NBD interface induces the closure, or “dimerization” of the two domains, thereby promoting an outward-facing conformation of the TMDs (15). The internal cavity of the transporter is thus open to receive substrate from the cognate periplasmic binding protein. Subsequent ATP hydrolysis releases NBD dimerization and converts the TMDs to an inward-facing conformation where substrate can be released to the cytoplasm (see Fig. 1B).
FIGURE 1.
Binding of ATP to MetNI is cooperative. A, representation of MetNI structure and domains within the cell membrane (11). B, alternating access model for transport with the addition of the transinhibition model. C, ATPase activity of MetNI in the absence of inhibitors. Inset, observed rate constant at low concentrations of MgATP. Reaction conditions consisted of 60 mm Tris, pH 7.5, 5 mm TAPS, pH 8.5, 0.055% DDM, 55 mm NaCl, 200 μm 2-amino-6-mercapto-7-methylpurine riboside, 0.1 units of purine nucleoside phosphorylase per 100 μl, and equimolar amounts of MgCl2 and ATP at 33 °C. Equations for fitting are described under “Experimental Procedures.” Error bars represent S.E. from at least three independent experiments.
In addition to this common domain architecture, the MetNI NBDs are fused to two carboxyl-terminal domains, termed C2, which are believed to be crucial for transinhibition (11, 12). The MetNI C2 domains are distinct from those found on the ModBC and MalFGK2 transporters, although all are implicated in regulatory roles of transport (13, 16). The high-resolution crystal structure of MetNI shows the NBDs widely separated due to dimerization of their C2 domains (see Fig. 1A). Selenomethionine has been shown to bind the C2 domains and inhibit ATPase activity. Based on these findings, it has been proposed that ligand-dependent association of the C2 domains prevents dimerization of the NBDs. Because the two ATP binding pockets are created at this dimerization interface, physical separation of the NBDs will prevent ATP binding and hydrolysis, thereby inhibiting transport (see Fig. 1B) (11).
MetNI provides a rare opportunity to develop detailed kinetic and thermodynamic analyses to study the mechanism of a structurally characterized ABC transporter. As a starting point, we examine the kinetics of ATP hydrolysis to elucidate the mechanism of MetNI transinhibition. To focus on the inhibitory effects of methionine, we employed a basic experimental system consisting of detergent-solubilized MetNI in the absence of MetQ, the substrate-containing cognate periplasmic binding protein. Although MetQ will clearly influence the structure and activity of MetNI (see Ref. 17 for an informative discussion of the maltose transport system), it will also complicate an analysis of the role of methionine because the stimulating effect of MetQ on the ATPase activity is opposite of the allosteric effect of methionine. As a consequence, we have not included MetQ in these studies. Additionally, the basal ATPase activity of detergent-solubilized ABC transporters typically increases and the coupling efficiency decreases, relative to the behavior in a phospholipid bilayer, as again exemplified by the maltose transporter (see Ref. 18). Although ATPase activity uncoupled to transport is not the physiological reaction of ABC transporters, comparisons of ABC subunits in isolation and in transporters are suggestive of a common mechanism for ATP hydrolysis (19). Consequently, meaningful kinetic interpretations can be derived from the effects of methionine on the relative ATPase activity of MetNI in our simplified system. We find that transinhibition is due to noncompetitive inhibition by l-methionine and that there are two allosteric methionine binding sites per transporter. These findings are consistent with the structurally based model for transinhibition and could be extended to understanding the regulation of the broader ABC family.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The wild-type His-tagged MetNI expression plasmid (11) was transformed into BL21-Gold (λDE3) cells (Novagen). Cell growth was carried out at 37 °C in Terrific Broth containing 1% glucose and 100 μg/ml ampicillin. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to 1 mm for 1 h. Cells were harvested by centrifugation and stored at −80 °C.
Frozen cell pellets were homogenized in purification buffer (50 mm TAPS, pH 8.5, 250 mm NaCl, 0.05% n-dodecyl-β-d-maltopyranoside (DDM, Anatrace)) supplemented with DNase, lysozyme, and protease inhibitor tablets (Roche Diagnostics GmbH). To extract MetNI protein, DDM was added to 1.0% final concentration. Cells were stirred at 4 °C for 20 min and then passed through a Microfluidizer three times. The cell lysate was stirred for 20 min and then spun for 20 min at 16,000 rpm in a JLA 17 rotor. The supernatant was recovered, and imidazole was added to 25 mm final concentration. The supernatant was then flowed over three tandem 5-ml HisTrap FF columns (GE Healthcare) and washed with 2 column volumes of purification buffer containing 25 mm imidazole and 5 column volumes of purification buffer containing 75 mm imidazole. MetNI protein was eluted with purification buffer containing 350 mm imidazole. Eluate was then buffer-exchanged into purification buffer without imidazole using a HiPrep 26/10 desalting column (GE Healthcare). The protein was then sized over a Superdex 200 column (GE Healthcare) and concentrated to ∼60 μm in an Amicon Ultra 100-kDa molecular mass cutoff centrifugal filter (Millipore) prewashed in purification buffer. Purified protein was flash-frozen in LN2 and stored at −80 °C.
ATPase Assays
The amount of inorganic phosphate generated in solution was measured in real time using the Enzchek phosphate assay kit (Molecular Probes). Each 100-μl reaction contained 60 mm Tris, pH 7.5, 5 mm TAPS, pH 8.5, 0.055% DDM, 55 mm NaCl, 200 μm 2-amino-6-mercapto-7-methylpurine riboside, 0.1 units of purine nucleoside phosphorylase, equimolar amounts of MgCl2 and ATP and/or ADP, and l-methionine as indicated. Data were collected on an Infinite M200 microplate reader (Tecan Group) at 33 °C. Reactions were incubated for 2 min and then initiated by automatic injection of MetNI (supplemental Fig. S1). For measurement of KI(ADP), 3 μm MetNI final concentration was injected into reactions containing 100 μm MgATP. For all other measurements, reactions contained 410 nm MetNI. Initial rates were obtained by calculating the linear portion of the change in absorbance at 360 nm as a function of time using Magellan software. Error bars represent S.E. from at least three independent experiments.
All data were fit using KaleidaGraph (Synergy Software). For determination of KI values, the initial rate was plotted against inhibitor concentration and fit to the following equation
 |
where [I] is the inhibitor concentration, KI is the inhibition constant, and nI is the Hill coefficient for the inhibitor.
For determination of Km(ATP), Vmax, and the Hill coefficient nATP, the initial rate was plotted against ATP concentration and fit to the following equation
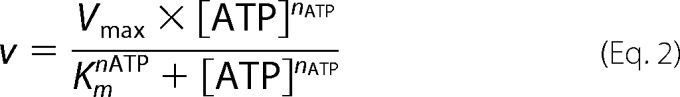 |
Isothermal Titration Calorimetry
MetNI was further concentrated to 350–450 μm in an Amicon Ultra 100-kDa molecular mass cutoff centrifugal filter (Millipore) prewashed in purification buffer. Samples were then dialyzed overnight in purification buffer in 100-kDa molecular mass cutoff Slide-a-Lyzer MINI dialysis devices (Thermo Scientific). The next day, samples were recovered and cleared at 95,000 rpm for 10 min in a TLA100 rotor. l-Methionine (BioUltra, Sigma) was reconstituted in the purification buffer used for dialysis. Binding data were collected on a MicroCal iTC-200 calorimeter at 25 °C. 300–450 μm MetNI was titrated with 8–10 mm l-methionine. For each experiment, an initial injection of 0.4 μl was followed by 19 injections of 2 μl each. The sample cell was allowed to equilibrate for 180 s between titrations. Data were processed using Origin v7.0 (OriginLab). Error bars represent S.E. from at least three independent experiments.
RESULTS
To establish the kinetic mechanism of transinhibition, we measured the ATPase activity of detergent-solubilized, wild-type MetNI using a continuous spectrophotometric coupled enzymatic assay (see “Experimental Procedures”). To define the basic parameters of the assay, MetNI was automatically injected into reactions containing varying concentrations of MgATP. The initial rates were plotted and fit to a sigmoidal curve with Km = 330 ± 20 μm and a Hill coefficient of 1.7 ± 0.1, indicating positive cooperativity between the two ATP binding pockets (Fig. 1C and supplemental Fig. S1). Cooperative binding of ATP is a common feature of several ABC transporters, including the vitamin B12, maltose, and histidine transporters (20–23).
We then verified the competitive nature of nucleotide inhibition of MetNI ATPase activity. Because canonical nucleotide analogs for ATP, including AMPPNP, AMPPCP, and ATPγS, did not completely inhibit ATPase activity in the concentration range tested (data not shown), we measured ATPase inhibition using ADP. Under subsaturating ATP concentrations, ADP inhibited MetNI ATPase activity with a KI(ADP) of 41 ± 2 μm and a Hill coefficient of 1.3 ± 0.1, indicating low cooperativity for the binding of ADP (Fig. 2A and Table 1). Next, the ATP concentration was varied under constant ADP concentrations both above and below the measured KI(ADP). In the presence of ADP, the apparent Vmax remained unchanged, whereas the apparent Km increased with increasing inhibitor concentration (Fig. 2B and Table 2). As expected, these experiments confirmed that ADP is indeed a competitive inhibitor.
FIGURE 2.
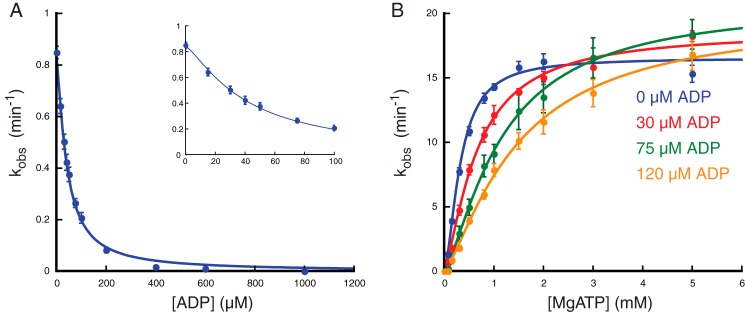
ADP is a competitive inhibitor of MetNI ATPase activity. A, inhibition of MetNI ATPase activity as a function of ADP concentration. Inset, observed rate constant at low concentrations of MgATP. B, ATPase activity as a function of ATP concentration in the presence of different concentrations of ADP. Reaction conditions were identical with those in Fig. 1. Error bars represent S.E. from at least three independent experiments.
TABLE 1.
Inhibition constants
| Inhibitor | KI | Hill coefficient |
|---|---|---|
| μm | ||
| ADP | 41 ± 2 | 1.3 ± 0.1 |
| l-Met | 41 ± 1 | 1.4 ± 0.1 |
TABLE 2.
Kinetic parameters from the ATPase reaction
| Inhibitor | Vmaxapp | Kmapp |
|---|---|---|
| min−1 | μm | |
| None | 17 ± 1 | 330 ± 20 |
| 30 μm ADP | 19 ± 1 | 660 ± 40 |
| 75 μm ADP | 21 ± 1 | 1170 ± 80 |
| 120 μm ADP | 20 ± 1 | 1490 ± 140 |
| 25 μm l-Met | 13 ± 1 | 309 ± 19 |
| 50 μm l-Met | 7 ± 1 | 325 ± 15 |
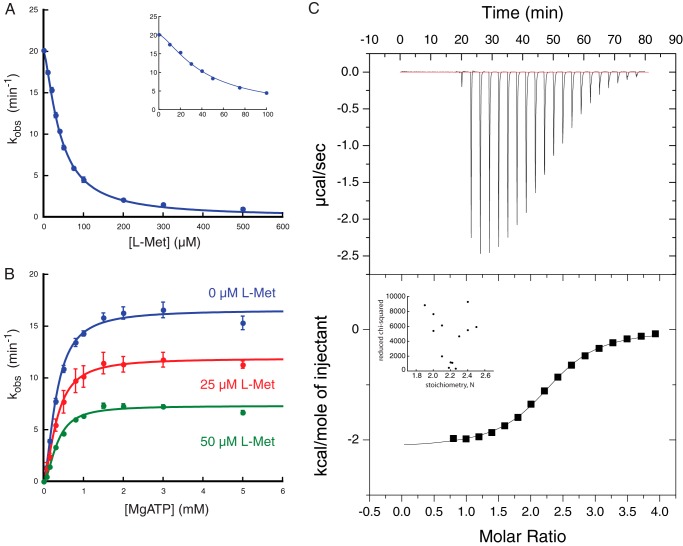
We next tested l-methionine inhibition of MetNI ATPase activity. Inhibition experiments under saturating ATP concentrations were fit to a KI(l-Met) of 41 ± 1 μm and a Hill coefficient of 1.4 ± 0.1, indicating that there is more than one methionine binding site per MetNI transporter and low cooperativity between the sites (Fig. 3A and Table 1). Furthermore, with increasing l-methionine concentrations, the apparent Vmax significantly decreased, whereas the apparent Km remained constant (Fig. 3B and Table 2). These data suggest that l-methionine is a noncompetitive inhibitor of MetNI ATPase activity. To further support this model, we conducted isothermal titration calorimetry experiments using l-methionine as the ligand (Fig. 3C). l-Methionine bound to MetNI at 2.2 ± 0.1 sites, with a KD(l-Met) of 33 ± 4 μm. Although the error estimates provided by the software analysis are artificially low (see reduced χ-squared values for fixed values of stoichiometry, Fig. 3C, inset), the measured value for the KD(l-Met) is in the same order of magnitude of the KI(l-Met) for the ATPase reaction (41 ± 1 μm).
FIGURE 3.
l-Methionine is a noncompetitive inhibitor. A, inhibition of MetNI ATPase activity as a function of l-Met concentration. Inset, observed rate constant at low concentrations of MgATP. B, ATPase activity as a function of ATP concentration in the presence of different concentrations of l-Met. Reaction conditions for ATPase assays were identical with those in Fig. 1. Error bars represent S.E. from at least three independent experiments. C, top, representative ITC thermogram profile for association of l-Met ligand with MetNI reactant. Bottom, binding curve from plot of heats from each injection as a function of the ratio of l-Met to MetNI, c = 14. Buffer conditions for ITC experiments consisted of 50 mm TAPS, pH 8.5, 250 mm NaCl, 0.05% n-dodecyl-β-d-maltopyranoside at 25 °C. Binding of l-methionine was exothermic with ΔH = −2140 ± 40 cal mol−1. Inset: reduced χ-squared value for ITC data fit as function of stoichiometry, N.
DISCUSSION
We have demonstrated that l-methionine can bind to MetNI at allosteric sites, resulting in the noncompetitive inhibition of ATP hydrolysis. This direct modulation of MetNI activity is consistent with the model based on the crystallographically observed ATPase-inactive state of MetNI. Our data suggest that the transinhibition phenomenon observed by Kadner (3) can be attributed to the regulation of methionine import at the protein activity level. The de novo biosynthesis of methionine in E. coli is costly, requiring 7 ATPs and 8 NADPHs per molecule of methionine (24). If extracellular methionine is available, uptake of the exogenous methionine at an estimated cost of ∼2 ATPs per methionine molecule would be energetically preferable to de novo biosynthesis. The homeostatic intracellular concentration of methionine is estimated to be 150 μm (25), maintained by the balance between the rate of appearance of methionine via the MetNI import system and/or the methionine biosynthesis pathway and the rate of disappearance via protein synthesis and other processes. Given the KI for methionine inhibition measured in this study (∼40 μm), uptake should occur under conditions when the intracellular levels of methionine are well below 150 μm, with the rate of transport decreasing as the interior concentration increases.
Several aspects of our study are worth detailing. We have kinetically analyzed the influence of methionine on the ATPase activity of detergent-solubilized MetNI in the absence of the MetQ binding protein and in the absence of phospholipid. For the kinetic analysis, we have used the simplest models for noncompetitive and competitive inhibition to treat the effects of methionine and ADP, respectively, on ATPase activity. These models do not include binding interactions between ligands. We have explored more complex kinetic models (see the supplemental text), which suggest that little interaction occurs in the binding of these ligands. Given the correlation of parameters intrinsic to these binding models (26), it is not possible to confidently assess the magnitudes of these effects. The MetQ binding protein was not utilized in these studies to avoid the opposing effects of methionine on ATPase activity due to transport (requiring methionine-bound MetQ) and to transinhibition (requiring methionine acting as an allosteric ligand). We have focused on detergent-solubilized MetNI to directly characterize the influence of methionine on the ATPase activity of the form of MetNI that has been crystallographically characterized. Although the physiological function of ABC transporters requires membranes, detergent-solubilized transporters can capture important functional properties. The most extensive mechanistic studies of ABC importers have been conducted for the MalFGK2 transporter; although the detailed properties differ, the cooperativity of ATP hydrolysis can be observed for both detergent-solubilized and proteoliposome-reconstituted ABC transporters (22). Furthermore, complexes between the maltose transporter and binding protein can be trapped using ATP + vanadate with either membrane-bound or detergent-solubilized transporter (27). Unquestionably, the structural studies on detergent-solubilized maltose transporter from the Chen group (14, 17) and on detergent-solubilized vitamin B12 transporter from the Locher group (20, 28) have provided an invaluable framework for the interpretation of mechanistic studies of transport in proteoliposomes.
ABC transporters have adopted a wide range of mechanisms to regulate their activity. The molybdate/tungstate transporter ModBC shares a similar architecture with MetNI and also demonstrates transinhibitory behavior at higher concentrations of substrate (13). The maltose transporter, in contrast, is regulated by an additional protein that responds to nutrient availability. Unphosphorylated EIIAGlc binds MalFGK2 when preferred sources of carbon, such as glucose, are available (29). Two EIIAGlc molecules bind the NBDs and promote the inward-facing conformation reminiscent of the MetNI inhibited state (16). Interestingly, it has been demonstrated that the binding of EIIAGlc does not affect the binding of ATP but rather inhibits ATP hydrolysis by MalK (30). Quantitative modeling of our MetNI data suggests that methionine may similarly inhibit ATP hydrolysis but not ATP binding (see the supplemental text).
Although the above examples highlight inhibition as the result of a stabilized inward-facing conformation, ABC transporters are also regulated at the level of transcription, glycosylation, and direct phosphorylation. For example, the osmoregulatory transporter OpuA is regulated by the transcription factor OpuR in response to osmotic stress (31). CFTR (cystic fibrosis transmembrane conductance regulator), an ABC-type chloride channel whose malfunction is linked to cystic fibrosis, is gated by PKA-mediated phosphorylation of its cytoplasmic regulatory domain (32–35). Lastly, it is hypothesized that glycosylation may help stabilize the rod photoreceptor ABC transporter (ABCR) against degradation while in the endoplasmic reticulum (36).
The regulation of ABC transporters, whether through accessory proteins, regulatory domains, or posttranslational modifications, is critical to balance the transport of metabolites for optimal growth. The MetNI transporter, through transinhibition, is effectively able to shut off import when adequate intracellular methionine is available for crucial cellular processes. A possible mechanism for this phenomenon envisioned nearly 40 years ago by Kadner (3) on the basis of transport studies in E. coli cells (“ … internal methionine binds to the carrier on the internal face of the membrane. This methionine-carrier complex might be unable, or at least less able, to undergo the energy-dependent change necessary for active transport … ”) was prescient, as the present studies have established using purified components. Significantly, the underlying mechanism (ligand binding to regulatory domains inhibits NBD dimerization, thereby preventing ATP hydrolysis and effectively “turning off the engine”) exemplifies a broadly relevant mechanism for regulating the activity of ABC transporters at the protein level.
Supplementary Material
Acknowledgments
We thank Eric Johnson and Chris Vercollone for early initial studies, Kana Takematsu for discussion regarding modeling, and Jeff Lai for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM45162 (to D. C. R.).

This article contains supplemental Figs. S1–S3, Table S1, and Modeling of MetNI Inhibition.
- ABC
- ATP binding cassette
- NBD
- nucleotide binding domain
- TMD
- transmembrane domain
- C2
- carboxy-terminal domains
- DDM
- n-dodecyl-β-d-maltopyranoside
- TAPS
- 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate
- EIIAGlc
- glucose-specific EIIA.
REFERENCES
- 1. Kadner R. J. (1974) Transport systems for l-methionine in Escherichia coli. J. Bacteriol. 117, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kadner R. J., Watson W. J. (1974) Methionine transport in Escherichia coli: physiological and genetic evidence for two uptake systems. J. Bacteriol. 119, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kadner R. J. (1975) Regulation of methionine transport activity in Escherichia coli. J. Bacteriol. 122, 110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Merlin C., Gardiner G., Durand S., Masters M. (2002) The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184, 5513–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gál J., Szvetnik A., Schnell R., Kálmán M. (2002) The metD d-methionine transporter locus of Escherichia coli is an ABC transporter gene cluster. J. Bacteriol. 184, 4930–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Z., Feige J. N., Chang A. B., Anderson I. J., Brodianski V. M., Vitreschak A. G., Gelfand M. S., Saier M. H., Jr. (2003) A transporter of Escherichia coli specific for l- and d-methionine is the prototype for a new family within the ABC superfamily. Arch. Microbiol. 180, 88–100 [DOI] [PubMed] [Google Scholar]
- 7. Rees D. C., Johnson E., Lewinson O. (2009) ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dawson R. J., Locher K. P. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 9. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 10. Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadaba N. S., Kaiser J. T., Johnson E., Lee A., Rees D. C. (2008) The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson E., Nguyen P. T., Yeates T. O., Rees D. C. (2012) Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition. Protein Sci. 21, 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerber S., Comellas-Bigler M., Goetz B. A., Locher K. P. (2008) Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321, 246–250 [DOI] [PubMed] [Google Scholar]
- 14. Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J. (2007) Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450, 515–521 [DOI] [PubMed] [Google Scholar]
- 15. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 16. Chen S., Oldham M. L., Davidson A. L., Chen J. (2013) Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 499, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oldham M. L., Chen J. (2011) Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 18. Bao H., Dalal K., Wang V., Rouiller I., Duong F. (2013) The maltose ABC transporter: action of membrane lipids on the transporter stability, coupling and ATPase activity. Biochim. Biophys. Acta 1828, 1723–1730 [DOI] [PubMed] [Google Scholar]
- 19. Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korkhov V. M., Mireku S. A., Locher K. P. (2012) Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature 490, 367–372 [DOI] [PubMed] [Google Scholar]
- 21. Tal N., Ovcharenko E., Lewinson O. (2013) A single intact ATPase site of the ABC transporter BtuCD drives 5% transport activity yet supports full in vivo vitamin B12 utilization. Proc. Natl. Acad. Sci. U.S.A. 110, 5434–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davidson A. L., Laghaeian S. S., Mannering D. E. (1996) The maltose transport system of Escherichia coli displays positive cooperativity in ATP hydrolysis. J. Biol. Chem. 271, 4858–4863 [PubMed] [Google Scholar]
- 23. Liu C. E., Liu P. Q., Ames G. F. (1997) Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J. Biol. Chem. 272, 21883–21891 [DOI] [PubMed] [Google Scholar]
- 24. Ingraham J. L., Maaløe O., Neidhardt F. C. (1983) Growth of the Bacterial Cell, pp. 129–132, Sinauer Associates, Sunderland, MA [Google Scholar]
- 25. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hines K. E., Middendorf T. R., Aldrich R. W. (2014) Determination of parameter identifiability in nonlinear biophysical models: a Bayesian approach. J. Gen. Physiol. 143, 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J., Sharma S., Quiocho F. A., Davidson A. L. (2001) Trapping the transition state of an ATP-binding cassette transporter: evidence for a concerted mechanism of maltose transport. Proc. Natl. Acad. Sci. U.S.A. 98, 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korkhov V. M., Mireku S. A., Veprintsev D. B., Locher K. P. (2014) Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F. Nat. Struct. Mol. Biol. 21, 1097–1099 [DOI] [PubMed] [Google Scholar]
- 29. Deutscher J., Francke C., Postma P. W. (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bao H., Duong F. (2013) Phosphatidylglycerol directs binding and inhibitory action of EIIAGlc protein on the maltose transporter. J. Biol. Chem. 288, 23666–23674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romeo Y., Obis D., Bouvier J., Guillot A., Fourçans A., Bouvier I., Gutierrez C., Mistou M. Y. (2003) Osmoregulation in Lactococcus lactis: BusR, a transcriptional repressor of the glycine betaine uptake system BusA. Mol. Microbiol. 47, 1135–1147 [DOI] [PubMed] [Google Scholar]
- 32. Baldursson O., Ostedgaard L. S., Rokhlina T., Cotten J. F., Welsh M. J. (2001) Cystic fibrosis transmembrane conductance regulator Cl− channels with R domain deletions and translocations show phosphorylation-dependent and -independent activity. J. Biol. Chem. 276, 1904–1910 [DOI] [PubMed] [Google Scholar]
- 33. Chappe V., Irvine T., Liao J., Evagelidis A., Hanrahan J. W. (2005) Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J. 24, 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Csanády L., Chan K. W., Seto-Young D., Kopsco D. C., Nairn A. C., Gadsby D. C. (2000) Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J. Gen. Physiol. 116, 477–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rich D. P., Berger H. A., Cheng S. H., Travis S. M., Saxena M., Smith A. E., Welsh M. J. (1993) Regulation of the cystic fibrosis transmembrane conductance regulator Cl− channel by negative charge in the R domain. J. Biol. Chem. 268, 20259–20267 [PubMed] [Google Scholar]
- 36. Bungert S., Molday L. L., Molday R. S. (2001) Membrane topology of the ATP binding cassette transporter ABCR and its relationship to ABC1 and related ABCA transporters: identification of N-linked glycosylation sites. J. Biol. Chem. 276, 23539–23546 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.