Background: PB2cap is critical for the initiation of influenza virus transcription.
Results: FluB PB2cap binds to GDP and m7GDP utilizing unique structural features, which is corroborated by data from ITC.
Conclusion: FluB PB2cap has a unique cap recognition mechanism compared with FluA PB2cap.
Significance: We characterize the cap recognition mechanism of FluB PB2cap, consequently providing insight into inhibitor design targeting FluB PB2cap.
Keywords: Crystal Structure, Influenza Virus, Nucleoside/Nucleotide Analogue, Protein Structure, RNA Polymerase, GDP, PB2, Cap Binding, m7GDP
Abstract
The influenza RNA-dependent RNA polymerase is a core enzyme required for both transcription and replication of the virus RNA genome, making it a potential drug target for the influenza virus. To detect the feature of cap-dependent transcription of influenza B virus (FluB) polymerase, we determined the crystal structures of the wild-type FluB polymerase PB2 subunit cap-binding domain (PB2cap) with bound GDP and the mutant FluB Q325F PB2cap with bound m7GDP or GDP. These structures revealed that, distinct from influenza A virus (FluA) PB2cap, the guanine and ribose moieties of substrates invert in FluB PB2caps. Moreover, we characterized the substrate specificity and affinity of the PB2caps using isothermal titration calorimetry. FluB PB2cap has a weaker affinity for m7GDP than FluA PB2cap. Unlike FluA PB2cap that has a preference for m7GDP in comparison with GDP, FluB PB2cap shows an analogous affinity for both substrates. Replacement of FluB PB2 Glu325 by Phe, the corresponding residue of FluA PB2, increased the binding affinity of FluB PB2cap for m7GDP to a level approximate to that of FluA PB2cap and caused a significant higher affinity to GDP. This study indicated that FluB PB2cap has a unique cap recognition mechanism compared with FluA PB2cap, providing molecular insight into inhibitor design targeting FluB PB2cap.
Introduction
The influenza virus is subdivided into three different categories: influenza A virus (FluA),2 influenza B virus (FluB), and influenza C virus (FluC). FluA is well studied as it frequently causes both pandemic and seasonal influenza. FluB mainly infects humans as a cause of seasonal influenza and is also detected in seals (1, 2). As an important pathogen of influenza-associated hospitalizations, FluB infection causes influenza and even mortality in humans, predominantly in children (3–5). The report for influenza-associated pediatric mortality showed that FluB, as 26% of circulating influenza viruses, caused 38% of all influenza-associated pediatric deaths in the United States during 2010–2011 (6). The latest report from the World Health Organization FluNet showed that FluB is prevalent as 24.8% of the seasonal epidemics each year and co-circulates with certain subtypes of FluA, such as FluA (H3N2) (29.5%) and FluA (H1N1) pdm09 (16.5%) viruses. In contrast, FluC has a distant evolutionary relationship with FluA and FluB and causes rare but still severe flu and sometimes local epidemics (7).
FluB is a negative-sense RNA virus, and its eight viral genome RNA segments (vRNA) encode 11 proteins. PA, PB1, and PB2 subunits form the influenza RNA-dependent RNA polymerase for both transcription and replication of vRNA. In the host nucleus, the polymerase transcribes vRNA by the “cap-snatching” mechanism (8). In transcription, the cap-binding domain of PB2 (PB2cap) binds to the 5′ cap structures of host pre-mRNAs (9), and then the cap, together with 10–13 nucleotides downstream of the cap, is cleaved off by the N-terminal endonuclease domain of PA (10, 11). Subsequently, the generated 5′-capped RNA fragment serves as the primer to synthesize viral mRNA by the PB1 subunit. As a milestone contribution, the newest work from Cusack and co-workers (12, 13) clarified the atomic level assembly mode of the whole polymerase, including PA, PB1, and PB2 subunits and the viral RNA promoter.
The N7-methyl group of guanine in the cap structure of eukaryotic mRNA is essential for FluA to prime transcription (14). However, FluB polymerase recognizes not only the N7-methyl group of guanine (m7G-capped RNAs) but also the unmethylated GpppG-RNAs efficiently (15). Moreover, FluB polymerase is demonstrated to have a weaker cap binding activity than FluA polymerase (15). A study recently proved that VX-787, which is an inhibitor targeting FluA PB2cap, is active against FluA but not against FluB (16). Taken together, these data suggest that FluB functions differently from FluA during the cap binding process.
The structures of the cap recognition mechanism for FluA PB2cap have been extensively studied. The structure of FluA PB2cap (amino acids 318–483) was first determined by Stephen Cusack and co-workers (9), and the cap-binding pocket complex with the cap analog m7GTP was characterized. Furthermore, the conservative characteristics of the cap-binding pocket were evaluated (17), and the unliganded structure was solved (17, 18). Recently, inhibitors targeting the PB2 cap-binding pocket were reported (19, 20). These efforts provided clear atomic information for FluA PB2cap and indicated that the conserved cap-binding pocket is indeed a potential drug target. However, because several key amino acids in the cap-binding pocket are different from those of FluA, how FluB PB2cap functions in the cap binding mechanism and whether the cap-binding pocket of FluB is also a suitable drug target still remain to be resolved. Most recently, the structure of the whole FluB polymerase without cap binding for PB2 subunit was reported (12). The substrate binding information is still necessary for understanding the cap recognition mechanism of FluB PB2cap.
In this study, we first report the crystal structure of FluB Q325F PB2cap with bound m7GDP. Compared with FluA PB2cap, the structure reveals that the guanine and ribose moieties of m7GDP invert, resulting in the N7-methyl group and hydroxyl group of guanine facing outward of the binding pocket. We then solved the crystal structures of wild-type FluB PB2cap and FluB Q325F PB2cap with bound GDP. Here GDP shows the same cap binding pattern as m7GDP, and the side chain of Gln325 in FluB PB2cap-GDP shows conformational flexibility. Moreover, we characterize the cap binding affinity and specificity of these FluB PB2 caps and compare them with the FluA PB2cap by isothermal titration calorimetry (ITC). Our results indicate that the FluB polymerase possesses a novel cap recognition mechanism.
MATERIALS AND METHODS
Cloning, Protein Expression, and Purification
The cDNA encoding amino acids 320–484 of FluB polymerase PB2 subunit was obtained from influenza B/Jiangxi/BV/2006.3 The Q325F, W359H, and Y434H mutations (amino acids 320–484) were constructed using a QuikChangeTM site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. FluB PB2cap (amino acids 318–484) that contains two more wild-type residues Gly318-Leu319 at the N terminus was cloned and is designated as FluB PB2cap in the following text. The FluA PB2cap (amino acids 318–483) gene from influenza A/Puerto Rico/8/34 (H1N1) was cloned as described previously (17, 22). All the cDNAs were digested with NdeI and XhoI and inserted into the pET28a vector with a His6 tag at the N terminus. The recombinant FluB PB2cap plasmids were overexpressed in Escherichia coli strain BL21(DE3), and FluA PB2cap was expressed in E. coli strain Rosetta(DE3). Cells were grown at 37 °C until an A600 of 0.8–1.0 was reached, and then the protein expressions were induced with 0.3 mm isopropyl d-1-thiogalactopyranoside (Sigma) at 18 °C overnight. Cells were then harvested in a binding buffer (50 mm Tris, pH 8.0, 500 mm NaCl) and lysed by sonication. The cell lysate was centrifuged twice at 38,900 × g at 4 °C for 30 min each. The supernatant was filtered with a 0.22-μm membrane and purified by metal affinity chromatography using a HiTrap chelating HP column (GE Healthcare). The FluB PB2cap and FluB Q325F PB2cap proteins were subjected to thrombin digestion at 4 °C overnight, whereas FluA PB2cap was digested at 22 °C for 1 h, and then proteins were applied to a nickel affinity column to remove the His tag and undigested protein followed by purification using Superdex 75 gel filtration chromatography (GE Healthcare). The FluB Q325F PB2cap was further purified by ion exchange chromatography using a Q column (GE Healthcare). The protein was then dialyzed against buffer containing 10 mm Tris, pH 8.0, 200 mm NaCl for crystal screening or buffer containing 10 mm HEPES, pH 7.4, 150 mm NaCl for isothermal titration calorimetry.
Crystallization
10 mg ml−1 FluB Q325F PB2cap proteins were incubated with 5 mm m7GTP (Sigma) and 5 mm GDP (Sigma) separately on ice for 1 h. The crystals used for data collection were obtained via the sitting drop vapor diffusion method at 20 °C. The FluB Q325F PB2cap with m7GTP grew crystal clusters from 0.2 m ammonium acetate, 0.1 m Tris, pH 8.5, 25% (w/v) PEG 3350 after approximately 2 weeks, and a single crystal was extracted for data collection. The crystal of FluB Q325F PB2cap with GDP grew from 0.15 m dl-malic acid, pH 7.0, 20% (w/v) PEG 3350 within 2–4 days. 10–12 mg ml−1 FluB PB2cap (amino acids 318–484) was incubated with 5 mm m7GTP, 5 mm, and 10 mm m7GDP (Sigma), and 5 mm GDP separately for crystal screening purposes, and only the crystal of FluB PB2capwith GDP was obtained from 0.2 m sodium formate, 20% (w/v) PEG 3350 within 2 days.
Data Collection and Structure Determination
All of the x-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility (China) beamline BL17U. The corresponding reservoir solutions with a gradient of 5 and 10% (v/v) glycerol as a cryoprotectant for crystals of FluB Q325F PB2cap-m7GDP and FluB Q325F PB2cap-GDP and with 5% (v/v) glycerol for FluB PB2cap-GDP were used. After soaking, the crystals were flash cooled in liquid nitrogen and maintained at 100 K in cooled nitrogen gas for data collection. All of the data were processed with HKL 2000. The FluB Q325F PB2cap-m7GDP crystal structure was solved by molecular replacement using Molrep in the CCP4 suite (23) with the H3N2 PB2cap-m7GTP structure (Protein Data Bank code 4EQK) used as a search model. The 424-loop was rebuilt manually in Coot (24). The FluB Q325F PB2cap-GDP and FluB PB2cap-GDP were solved with the FluB Q325F PB2cap-m7GDP structure used as a search model. All the initial structures were refined by REFMAC5 (25) and built in Coot (24). The structure model of FluB Q325F PB2cap-GDP was further refined with the geometry restraints by the refinement program in PHENIX (26). The final models were assessed with the program PROCHECK (27). The final statistics for data collection and structure refinement are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
Mol/Asym, molecules per asymmetric unit; r.m.s., root mean square.
| Protein Data Bank code | 4OR4 | 4OR6 | 4Q46 |
| Protein | FluB Q325F PB2cap | FluB Q325F PB2cap | FluB PB2cap |
| Ligand | m7GDP | GDP | GDP |
| Data collection | |||
| Space group | P 212121 | P 212121 | P 1211 |
| Cell dimensions | |||
| a, b, c (Å) | 42.94, 90.70, 95.77 | 42.97, 90.15, 95.71 | 43.07, 88.54 52.15 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 105.18, 90 |
| Mol/Asym | 2 | 2 | 2 |
| Wavelength (Å) | 0.9793 | 1.0000 | 0.9792 |
| Resolution (Å) | 50-2.20 (2.24-2.20) | 50-2.30 (2.30-2.36) | 50-1.80 (1.83-1.80)a |
| Rmerge (%)b | 16.0 (76.9) | 21.2 (83.0) | 7.7 (21.7) |
| I/σ | 22.6 (4.6) | 10.9 (3.8) | 19.9 (8.5) |
| Completeness (%) | 99.9 (100) | 99.9 (99.9) | 94.6 (98.2) |
| Redundancy | 7.8 (7.9) | 9.3 (9.3) | 5.5 (5.3) |
| Refinement | |||
| Resolution (Å) | 42.35-2.21 | 30.13-2.29 | 24.57-1.80 |
| No. reflections | 19,227 | 17,022 | 31,459 |
| Rwork/Rfree (%)c | 17.6/24.2 | 18.5/25.0 | 17.0/23.0 |
| Average B-factors (Å2) | 26.84 | 25.22 | 18.0 |
| Protein (chain A, B) | 27.02, 25.88 | 25.54, 24.37 | 16.51,17.77 |
| Ligand (chain A, B) | 21.87, 24.53 | 20.59, 22.85 | 13.79, 14.01 |
| Water | 34.91 | 30.94 | 30.91 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.022 | 0.002 | 0.025 |
| Bond angles (°) | 2.197 | 0.614 | 2.170 |
| Ramachandran plot (%) | |||
| Favored region | 97.5 | 96.9 | 98.5 |
| Allowed region | 2.5 | 3.1 | 1.5 |
| Outlier region | 0 | 0 | 0 |
a Numbers in parentheses are for highest resolution shell.
bRmerge = Σ|Ii − I|/Σ|Ii|.
c R = Σh|Fo(h) − Fc(h)|/Σ|hFo(h)|. Rfree is calculated using 5% of the data excluded from the refinement.
Isothermal Titration Calorimetry
ITC was carried out at 20 °C with a MicroCal iTC200 (MicroCal Inc.). All proteins were desalted into a buffer containing 10 mm HEPES, pH 7.4, 150 mm NaCl with either m7GDP or GDP dissolved in the same buffer. For FluA PB2cap and FluB PB2cap, 240–280 μm protein (in the cell) and 2.5 mm m7GDP or GDP (in the syringe) were used for titration. For FluB Q325F PB2cap, 85 μm protein (in the cell) and 1.2 mm m7GDP or GDP (in the syringe) were used. The first injection of 0.5 μl was followed by 34 injections of 1 μl with a 110-s spacer. All of the data were fitted to a single binding site model using Origin 7.0 program analysis.
RESULTS
The Crystal Structures of FluB Q325F PB2cap with Bound m7GDP and GDP
Unlike FluA PB2cap, FluB PB2cap readily precipitated out of solution at room temperature. During protein purification efforts, we noticed that the addition of m7GTP improved the stability of FluB PB2cap, which is similar to FluA PB2cap. We obtained crystals of m7GTP with FluA PB2cap (17) but not m7GTP with FluB PB2cap. Comparison of the residues of the cap-binding pocket between FluA and FluB revealed that the key difference(s) in the m7GTP-binding site (cap-binding pocket) of PB2cap might result in the weak stability of FluB PB2cap. Based on the sequence alignment (Fig. 1), Gln325, Trp359, and Tyr434 are the three key residues in the cap-binding pocket of FluB PB2cap that are different from FluA, and we thus mutated these residues to the corresponding residues of FluA PB2 (Q325F, W359H, and Y434H) and screened the three single mutations. FluB W359H PB2cap was insoluble, and FluB Y434H PB2cap exhibited the same instability as the wild type. However, FluB Q325F PB2cap showed greater stability than the wild type and was used for further crystallographic studies.
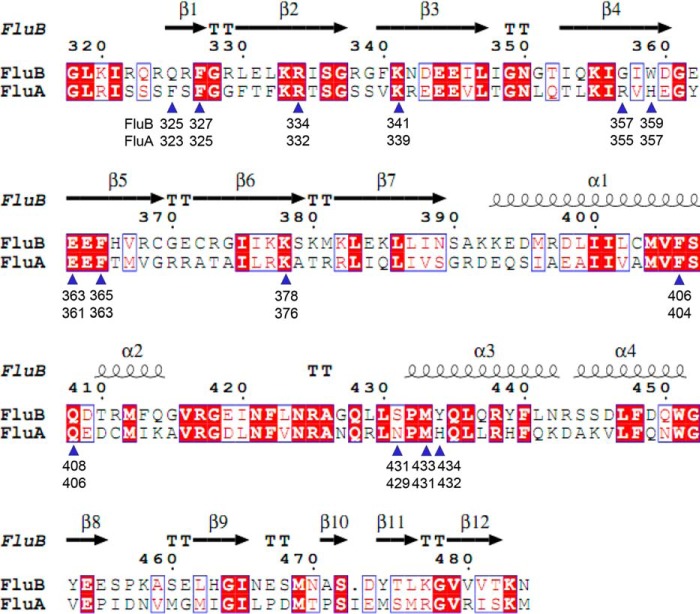
FIGURE 1.
Structure-based amino acid sequence alignments of FluB and FluA PB2caps. Amino acid sequence alignments of the PB2caps, including FluA (influenza A/Puerto Rico/8/34 (H1N1)) and FluB (influenza B/Jiangxi/BV/2006), were analyzed by ESPript (37). The secondary structure and amino acid number of FluB PB2cap-GDP are shown at the top. There are 14 residues associated with substrate binding in FluB or FluA PB2caps that are marked with blue triangles, and the respective amino acid number is noted for FluB and FluA PB2caps.
We first solved the crystal structure of FluB Q325F PB2cap-m7GTP at 2.21-Å resolution, and it was refined with an R-factor of 0.176 (Rfree = 0.242) (Fig. 2A and Table 1). In the crystal structure, there was clear electron density for the substrate without the γ-phosphate observed (Fig. 3B), and we therefore named this structure as FluB Q325F PB2cap-m7GDP.
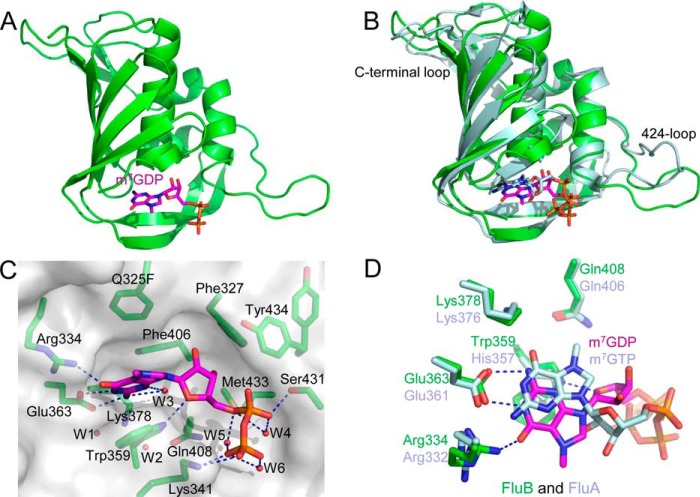
FIGURE 2.
Crystal structure of the FluB Q325F PB2cap with m7GDP. A, ribbon diagram of the structure of FluB Q325F PB2cap-m7GDP. The overall structure is in green with m7GDP colored purple. B, structural superposition of FluB Q325F PB2cap-m7GDP (colored as in A) with FluA PB2cap-m7GTP (pale cyan; Protein Data Bank code 4EQK). C, close-up view of the cap-binding pocket of FluB Q325F PB2cap-m7GDP. Side chains are shown together with a translucent surface, and hydrogen bonds are indicated by dotted lines. Water molecules W1–W6 are presented as red spheres. D, comparison of the cap-binding pocket between FluB Q325F PB2cap-m7GDP and FluA PB2cap-m7GTP (colored as in B). The structure models in all figures were prepared with PyMOL (21).
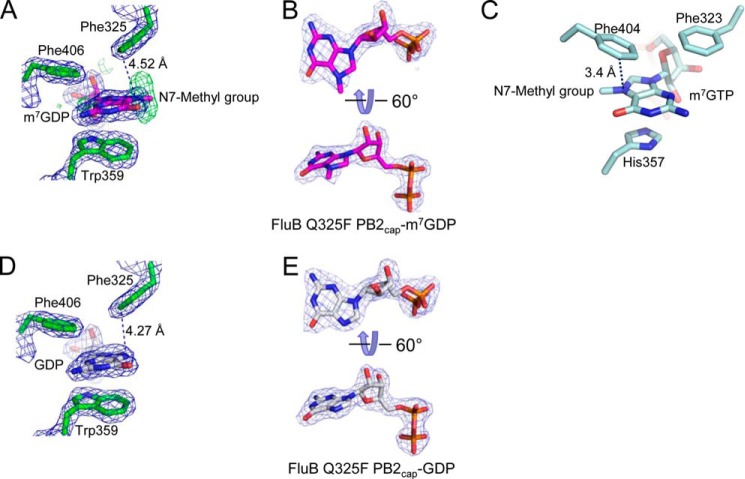
FIGURE 3.
Comparison of the cap-binding pockets of FluB Q325F PB2cap with m7GDP and GDP, and FluA PB2cap-m7GTP. A and B, electron density of the cap-binding pocket of molecule A of FluB Q325F PB2-m7GDP (purple) with the vertical Fo − Fc map (green) of the N7-methyl group in A and the 2Fo − Fc map (blue) of single m7GDP in B. C, the cap-binding pocket of FluA PB2cap-m7GTP. Phe323, His357, and Phe404 are shown to bind m7GTP. D and E, electron density of the cap-binding pocket of molecule A of FluB Q325F PB2-GDP (white) in D with the electron density of single GDP in E. The dotted lines in A, C, and D point to the shortest distance between Phe325 and N7 of m7GDP/GDP/m7GTP. The 2Fo − Fc (blue) and Fo − Fc (green; in A) maps contoured at 1.4 and 2.5 σ, respectively, were generated by the FFT program in CCP4 (23) and drawn by PyMOL (21).
The structure of FluB Q325F PB2cap-m7GDP shares a similar conformation with FluA PB2cap-m7GTP (Protein Data Bank code 4EQK) with a 1.31-Å root mean square deviation mainly in the 424-loop and C-terminal loop (Fig. 2B). Surprisingly, FluB shows a distinct binding pattern to the substrate in the cap-binding pocket (Fig. 2, C and D). Compared with FluA PB2cap-m7GTP, the guanine moiety of m7GDP in FluB Q325F PB2cap flips 180° around the long axis of the base, resulting in the N7-methyl group and hydroxyl group facing outward of the binding pocket and leaving room for a water molecule (W3) inside of the pocket. The broadened Fo − Fc map (Fig. 3A, green map) suggests oscillation of the N7-methyl group, which may result because it points out into the solvent. In this case, Lys378 and Gln408 lose their interaction to the guanine moiety, whereas Arg334 forms a new 2.59-Å hydrogen bond with the hydroxyl group. Glu363 keeps conserved hydrogen bonds with the N1 and N2 of the guanine moiety. Trp359 sandwiches the methylated guanine moiety instead of His357 in FluA PB2cap. Phe406 and Phe325 (the Q325F mutation) form the same upper cover as FluA PB2cap for stacking interactions to the guanine moiety (Fig. 3, A and C). Phe325 has a 4.52-Å distance to the N7 of the guanine moiety (Fig. 3A) compared with the 3.4-Å distance between Phe404 and the N7-methyl group in FluA PB2cap (Fig. 3C). The ribose moiety is also inverted in the structure, and the ribose hydroxyl groups face inward toward the pocket and are close to Phe327 and Met433. The O4 of the ribose moiety forms a 2.85-Å hydrogen bond with Trp359. Although the guanine and ribose moieties invert, the diphosphate moiety of m7GDP shows a similar trend as that of m7GTP in FluA PB2cap. The diphosphate moiety of m7GDP forms hydrogen bonds with Ser431 (α-phosphate, 2.78 Å) and Lys341 (β-phosphate, 2.69 and 3.18 Å) with three well ordered water molecules internally placed (W4, W5, and W6). Tyr434 loses its interaction with the substrate, whereas in FluA PB2cap, His432 binds to the α-phosphate of the m7GTP at the corresponding position. As found in the FluA PB2caps structure (9, 17), there are also two water molecules (W1 and W2) that are buried among residues Glu363, Lys378, and Gln408.
The FluB polymerase recognizes both the m7G-capped RNAs and the unmethylated GpppG-RNAs efficiently (15). The structure of FluB Q325F PB2cap-m7GDP shows the N7-methyl group pointing out toward the solvent. To test whether the unmethylated GpppG-RNA binds in the same orientation, we determined the crystal structure of GDP-bound FluB Q325F PB2cap. The crystal structure of FluB Q325F PB2cap-GDP was obtained within 2–4 days after co-crystallization with 5 mm GDP. The 2.29-Å FluB Q325F PB2cap-GDP structure was refined with an R-factor of 0.185 (Rfree = 0.250) (Table 1). In the cap-binding pocket here, GDP shows the same conformation as m7GDP in FluB Q325F PB2cap-m7GDP (Fig. 3, D and E).
Gln325 Is Flexible in the Cap-binding Pocket of Wild-type FluB PB2cap
The structure of the wild-type FluB PB2cap with its substrate is essential for understanding how the wild-type residue Gln325 interacts with substrate. As mentioned above, we failed to crystallize FluB PB2cap with m7GTP even though the addition of m7GTP improved the stability of FluB PB2cap. Based on the successful crystallization of FluB Q325F PB2cap with m7GDP and GDP, we further screened the crystals of wild-type FluB PB2cap with m7GDP and GDP. We obtained crystals and solved the structure of FluB PB2cap (amino acids 318–484) bound with GDP but not with the methylated substrate m7GDP.
The structure of FluB PB2cap-GDP was solved to 1.8-Å resolution and refined with an R-factor of 0.170 (Rfree = 0.230) (Fig. 4, A and B, and Table 1). The GDP in the structure (Fig. 4, C and D) shows the same orientation as the GDP and m7GDP in FluB Q325F PB2cap (Fig. 3, D and A). There are two molecules in the crystallographic asymmetric unit. In molecule B, the side chain of Gln325 shows a 3.96-Å distance to GDP (Fig. 4D), and in molecule A, the side chain of Gln325 is disordered with weak electron density (Fig. 4C). These observations suggest that Gln325 is flexible and may contribute weakly to cap binding of FluB PB2cap.
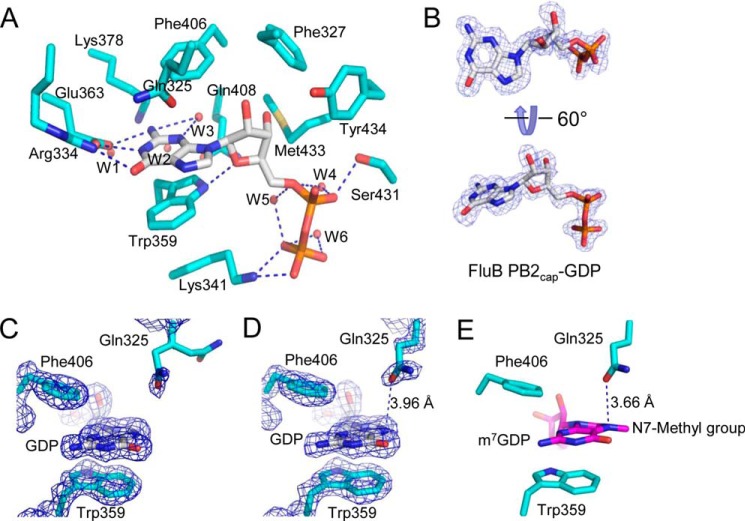
FIGURE 4.
Crystal structure of the cap-binding pocket of FluB PB2cap-GDP. A, close-up view of the cap-binding pocket of FluB PB2cap-GDP (FluB PB2cap is colored cyan, and GDP is colored white). B, electron density of GDP in molecule A of FluB PB2cap-GDP. C and D, electron density of the cap-binding pockets of molecule A (C) and molecule B (D) of FluB PB2cap-GDP. E, proposed structural model of FluB PB2 binding m7GDP based on the structures shown in Figs. 3, A and C, and 4D. The dotted lines in D and E point to the shortest distance between Gln325 and N7 of GDP/m7GDP. The 2Fo − Fc (blue) map contoured at 1.4 σ was generated by the FFT program in CCP4 (23) and drawn by PyMOL (21).
Structural analysis revealed that the N7-methyl group is oscillating in FluB Q325F PB2cap-m7GDP (Fig. 3A) and that Gln325 in the wild-type FluB PB2cap is more flexible than Phe325 of the mutant (Fig. 4C). These data may explain why we could not obtain the crystal of FluB PB2cap-m7GDP. Based on the crystal structures solved in this investigation, we propose a model for wild-type FluB PB2cap binding to m7GDP (Fig. 4E). In the model, m7GDP keeps the same orientation as m7GDP in the crystal structure of FluB Q325F PB2cap-m7GDP (Fig. 3A) and is consistent with GDP in FluB Q325F PB2cap-GDP (Fig. 3D) and FluB PB2cap-GDP (Fig. 4, C and D). The orientation of Gln325 is the same as that of Gln325 in FluB PB2cap-GDP (Fig. 4D) and is similar to that of Phe325 in FluB Q325F PB2cap-m7GDP (Fig. 3A) and FluB Q325F PB2cap-GDP (Fig. 3D). All three available crystal structures and the model presented here show that the guanine and ribose moieties of substrates invert in FluB PB2caps compared with those of FluA PB2cap.
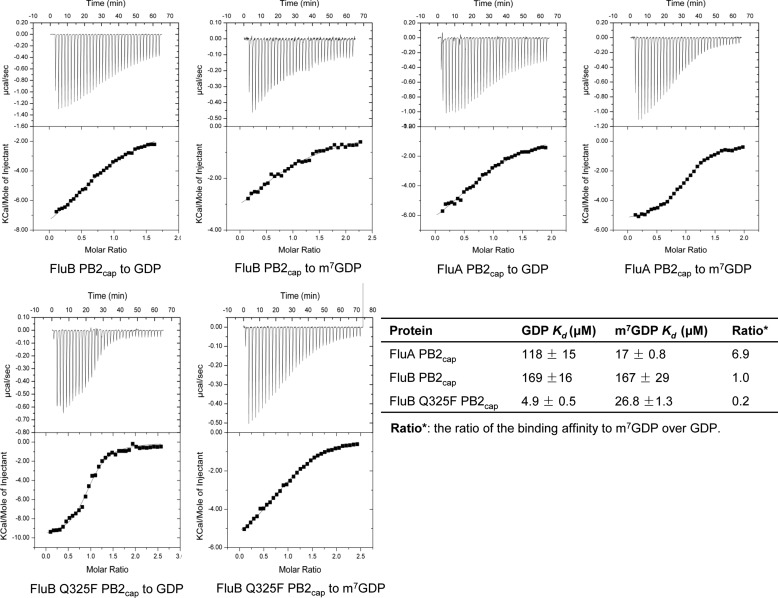
FluB and FluA PB2caps Show Different Substrate Specificity and Affinity
To investigate the cap binding affinity resulting from the structural differences between FluB and FluA PB2caps, we examined the affinity of m7GDP and GDP to FluA, FluB, and FluB Q325F PB2caps using ITC, respectively (Fig. 5). ITC was carried out at 20 °C in a buffer containing 10 mm HEPES, pH 7.4, 150 mm NaCl. Consistent with previous reports (9), FluA PB2cap was favored a methylated substrate with a dissociation equilibrium constant (Kd) of 17 μm for m7GDP and 118 μm for GDP. In contrast, FluB PB2cap showed an approximated binding to m7GDP (167 μm) and GDP (169 μm) with about one-tenth of the affinity of FluA PB2cap to m7GDP and slightly weaker affinity to GDP than FluA PB2cap. Notably, the Q325F mutation significantly improves the affinity of FluB PB2cap to both m7GDP (26.8 μm; 6.2-fold) and GDP (4.9 μm; 34.5-fold) and shows great specificity for GDP versus m7GDP. Compared with FluA PB2cap, FluB Q325F PB2cap shows a similar affinity to m7GDP and tighter binding characteristics to GDP. These results indicate that FluB PB2cap possesses different substrate specificity and affinity compared with those of FluA PB2cap.
FIGURE 5.
Comparison of the substrate specificity and affinity between FluA, FluB and FluB Q325F PB2caps by ITC. The binding of FluA PB2cap, FluB PB2cap, and FluB Q325F PB2cap to m7GDP and GDP was analyzed by ITC. The raw data are depicted in the top panels with the integrated data in the bottom panels. All data are shown with continuous lines fitting to a one-site binding model. The dissociation equilibrium constants (Kd) are listed in the table. The standard errors of affinity represent data fitted to binding isotherms in the lower panel.
DISCUSSION
Influenza B virus has a devastating mortality rate in humans, especially in children (3, 4, 6), which is an essential characteristic of seasonal influenza. Structural information for FluB PB2cap, a core executor for the transcription initiation of influenza polymerase, had previously been unknown. The structure of the whole FluB polymerase was reported recently; however, it still lacks substrate binding information for cap snatching (12). Whether FluB PB2cap functions with the same cap binding mechanism as FluA is unclear. Conversely, the neuraminidase structures of FluA and FluB viruses show a common drug-binding pocket (28); therefore, FluB can be treated with the same clinical drugs, such as oseltamivir (Tamiflu®) and zanamivir (Relenza®), as FluA. As a potential drug target, whether FluB PB2cap and FluA PB2cap can bind to the same inhibitors is currently unknown. Herein, we have determined the structures of substrate-binding FluB PB2cap and FluB Q325F PB2cap and characterized their substrate specificity and affinity.
Our results suggest that, unlike FluA PB2cap, FluB PB2cap has a novel cap recognition feature. In both structures of FluB wild-type and Q325F PB2caps, the guanine and ribose moieties of m7GDP (GDP) invert around the long axis of the base compared with FluA PB2cap, leaving the N7-methyl group and hydroxyl group of the guanine moiety facing outward and the ribose hydroxyl group facing toward the cap-binding pocket (Fig. 2). According to the structure and sequence alignments between FluA and FluB Q325F PB2caps (Figs. 1 and 2D), this inversion may be caused by the orientation of Trp359 in FluB PB2caps, whereas a His357 occupies the same position of FluA PB2cap.
Lys378 and Gln408 of FluB PB2cap lose their interactions with m7GDP (GDP) due to the inversion of the guanine moiety, consistent with the report that these two residues contribute less to cap binding (15). Conversely, Arg334 of FluB PB2cap is a newly identified residue utilized for the recognition of the outward facing hydroxyl group of guanine moiety that functions similarly to Lys376 in FluA PB2cap (at the corresponding position of Lys378 of FluB PB2cap; Figs. 1 and 2D). Conservatively, Glu363 of FluB PB2cap retains recognition of N1 and N2 of guanine moiety as in FluA (Figs. 1 and 2D). In conclusion, the available structural information for PB2caps indicates that both FluA and FluB need an acidic glutamate and an alternative basic residue for the recognition of the guanine moiety in the cap-snatching mechanism (Fig. 2D).
The combination of Trp359, Phe406, and Gln325 is used to clamp the guanine moiety and is associated with the weak cap binding of FluB PB2cap to m7GDP. In contrast, the mutation Q325F can recover the affinity of FluB PB2cap to m7GDP (Fig. 5). The weak cap binding affinity of methylated substrate to FluB PB2cap could further explain why the RNA elongation and cap binding activities of the FluB polymerase are lower than those of the FluA polymerase (15). However, it is unclear whether some other factors contribute to the unexpectedly low affinity of the FluB PB2cap to substrates under physiological conditions.
The inversion of the guanine moiety also allows FluB PB2cap to accept GDP well. The ITC results revealed that FluB PB2cap has an equal affinity for GDP and m7GDP (Fig. 5). The side chain of Gln325 shows conformational flexibility in molecule A of FluB PB2cap-GDP (Fig. 4C). These observations suggest that the flexibility of Gln325 may contribute to the weak recognition of the N7-methyl and unmethylated substrates.
In contrast, both FluB Q325F PB2cap and FluA PB2cap could distinguish m7GDP from GDP (Fig. 5) because Phe325 in FluB Q325F PB2cap and Phe404 in FluA PB2cap are used for rigid packing to the N7-methyl group (Fig. 3, A and C). In a comparison of FluA PB2 with FluB PB2, FluB PB2 could recognize GpppG for cap primer generation, although it fails in elongation (15). It has been reported that the transcript from the non-methylated cap primer could not be translated by host ribosomes (29–31). Whether the recognition of non-methylated cap primer by FluB has a physiological role is unclear; therefore further investigation is needed.
Cap-binding proteins in humans, such as the heterodimeric nuclear cap-binding complex (32) and eIF4E (33), show a higher affinity to methylated substrates and significant discrimination against non-methylation (34–36). Therefore, the cap recognition of FluB PB2cap for both m7GDP and GDP with a low affinity provides a good opportunity for rational inhibitor design. First, the binding cavity allows FluB PB2cap to accommodate both GDP and m7GDP derivatives. Second, a modification of N3 of the guanine moiety, such as the addition of a hydroxyl to N3 (Protein Data Bank code 3G9L), may recover its interaction with Lys378 and Gln408. FluA PB2cap may also utilize this modification, which has the potential to interact with Arg332 of FluA PB2cap (Fig. 2D). Third, N7 of the guanine moiety could be modified to interact with Gln325. Finally, modification of the ribose hydroxyl groups facing inward toward the pocket is likely to build a stronger interaction to Tyr434 or Phe327. In summary, our data have provided the structural and biochemical basis for cap recognition of FluB PB2cap, which in turn helps in understanding how FluB PB2cap executes the cap binding process and provides further insight into rational inhibitor design targeting FluB PB2cap.
Acknowledgments
We thank Dr. Kun Qin for providing the cDNA of influenza virus. We also thank the staff at the Shanghai Synchrotron Radiation Facility beamline BL17U and the Beijing Synchrotron Radiation Facility beamline 3W1A for assistance in data collection and Drs. Tao Jiang, Sheng Ye, and Yun Zhu at the Institute of Biophysics, Chinese Academy of Sciences; Zhaoyang Ye at Peking University; and Zengqiang Gao at the Beijing Synchrotron Radiation Facility for assistance with crystal testing and data collection. We also thank Dr. Lorenzo Finci and Linglong Qu (Peking University) for careful editing of the manuscript.
This work was supported by Beijing Natural Science Foundation Grant 5152012, National Science Foundation of China Grants 31470754 and 31170709, and Doctoral Fund Grant 20130001130003 of the Ministry of Education of China.
The atomic coordinates and structure factors (codes 4OR4, 4OR6, and 4Q46) have been deposited in the Protein Data Bank (http://wwpdb.org/).
K. Qin, unpublished sequence.
- FluA
- influenza A virus
- FluB
- influenza B virus
- FluC
- influenza C virus
- vRNA
- viral genome RNA segments
- PB2cap
- cap-binding domain of PB2
- ITC
- isothermal titration calorimetry
- W1–W6
- water molecules 1–6.
REFERENCES
- 1. Osterhaus A. D., Rimmelzwaan G. F., Martina B. E., Bestebroer T. M., Fouchier R. A. (2000) Influenza B virus in seals. Science 288, 1051–1053 [DOI] [PubMed] [Google Scholar]
- 2. Bodewes R., Morick D., de Mutsert G., Osinga N., Bestebroer T., van der Vliet S., Smits S. L., Kuiken T., Rimmelzwaan G. F., Fouchier R. A., Osterhaus A. D. (2013) Recurring influenza B virus infections in seals. Emerg. Infect. Dis. 19, 511–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCullers J. A., Hayden F. G. (2012) Fatal influenza B infections: time to reexamine influenza research priorities. J. Infect. Dis. 205, 870–872 [DOI] [PubMed] [Google Scholar]
- 4. Paddock C. D., Liu L., Denison A. M., Bartlett J. H., Holman R. C., Deleon-Carnes M., Emery S. L., Drew C. P., Shieh W. J., Uyeki T. M., Zaki S. R. (2012) Myocardial injury and bacterial pneumonia contribute to the pathogenesis of fatal influenza B virus infection. J. Infect. Dis. 205, 895–905 [DOI] [PubMed] [Google Scholar]
- 5. Thompson W. W., Shay D. K., Weintraub E., Brammer L., Bridges C. B., Cox N. J., Fukuda K. (2004) Influenza-associated hospitalizations in the United States. JAMA 292, 1333–1340 [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention (CDC) (2011) Influenza-associated pediatric deaths—United States, September 2010-August 2011. MMWR. Morb. Mortal. Wkly. Rep. 60, 1233–1238 [PubMed] [Google Scholar]
- 7. Roy Mukherjee T., Mukherjee A., Mullick S., Chawla-Sarkar M. (2013) Full genome analysis and characterization of influenza C virus identified in Eastern India. Infect. Genet. Evol. 16, 419–425 [DOI] [PubMed] [Google Scholar]
- 8. Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 [DOI] [PubMed] [Google Scholar]
- 9. Guilligay D., Tarendeau F., Resa-Infante P., Coloma R., Crepin T., Sehr P., Lewis J., Ruigrok R. W., Ortin J., Hart D. J., Cusack S. (2008) The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15, 500–506 [DOI] [PubMed] [Google Scholar]
- 10. Yuan P., Bartlam M., Lou Z., Chen S., Zhou J., He X., Lv Z., Ge R., Li X., Deng T., Fodor E., Rao Z., Liu Y. (2009) Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458, 909–913 [DOI] [PubMed] [Google Scholar]
- 11. Dias A., Bouvier D., Crépin T., McCarthy A. A., Hart D. J., Baudin F., Cusack S., Ruigrok R. W. (2009) The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 [DOI] [PubMed] [Google Scholar]
- 12. Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crépin T., Hart D., Lunardi T., Nanao M., Ruigrok R. W., Cusack S. (2014) Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366 [DOI] [PubMed] [Google Scholar]
- 13. Pflug A., Guilligay D., Reich S., Cusack S. (2014) Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–360 [DOI] [PubMed] [Google Scholar]
- 14. Bouloy M., Plotch S. J., Krug R. M. (1980) Both the 7-methyl and the 2′-O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 77, 3952–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wakai C., Iwama M., Mizumoto K., Nagata K. (2011) Recognition of cap structure by influenza B virus RNA polymerase is less dependent on the methyl residue than recognition by influenza A virus polymerase. J. Virol. 85, 7504–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byrn R. A., Jones S. M., Bennett H. B., Bral C., Clark M. P., Jacobs M. D., Kwong A. D., Ledeboer M. W., Leeman J. R., McNeil C. F., Murcko M. A., Nezami A., Perola E., Rijnbrand R., Saxena K., Tsai A. W., Zhou Y., Charifson P. S. (2015) Preclinical activity of VX-787, a first in class, orally bioavailable inhibitor of the influenza virus polymerase PB2 subunit. Antimicrob. Agents Chemother. 59, 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y., Qin K., Meng G., Zhang J., Zhou J., Zhao G., Luo M., Zheng X. (2013) Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus. J. Biol. Chem. 288, 11013–11023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsurumura T., Qiu H., Yoshida T., Tsumori Y., Hatakeyama D., Kuzuhara T., Tsuge H. (2013) Conformational polymorphism of m7GTP in crystal structure of the PB2 middle domain from human influenza A virus. PLoS One 8, e82020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark M. P., Ledeboer M. W., Davies I., Byrn R. A., Jones S. M., Perola E., Tsai A., Jacobs M., Nti-Addae K., Bandarage U. K., Boyd M. J., Bethiel R. S., Court J. J., Deng H., Duffy J. P., Dorsch W. A., Farmer L. J., Gao H., Gu W., Jackson K., Jacobs D. H., Kennedy J. M., Ledford B., Liang J., Maltais F., Murcko M., Wang T., Wannamaker M. W., Bennett H. B., Leeman J. R., McNeil C., Taylor W. P., Memmott C., Jiang M., Rijnbrand R., Bral C., Germann U., Nezami A., Zhang Y., Salituro F. G., Bennani Y. L., Charifson P. S. (2014) Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 57, 6668–6678 [DOI] [PubMed] [Google Scholar]
- 20. Pautus S., Sehr P., Lewis J., Fortuné A., Wolkerstorfer A., Szolar O., Guilligay D., Lunardi T., Décout J. L., Cusack S. (2013) New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J. Med. Chem. 56, 8915–8930 [DOI] [PubMed] [Google Scholar]
- 21. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 22. Liu Y., Meng G., Luo M., Zheng X. (2013) Crystallization and x-ray crystallographic analysis of the cap-binding domain of influenza A virus H1N1 polymerase subunit PB2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Stereochemical quality of protein structure coordinates. Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 28. Taylor N. R., Cleasby A., Singh O., Skarzynski T., Wonacott A. J., Smith P. W., Sollis S. L., Howes P. D., Cherry P. C., Bethell R., Colman P., Varghese J. (1998) Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J. Med. Chem. 41, 798–807 [DOI] [PubMed] [Google Scholar]
- 29. Graff J. R., Zimmer S. G. (2003) Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Metastasis 20, 265–273 [DOI] [PubMed] [Google Scholar]
- 30. von der Haar T., Gross J. D., Wagner G., McCarthy J. E. (2004) The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11, 503–511 [DOI] [PubMed] [Google Scholar]
- 31. Culjkovic B., Topisirovic I., Borden K. L. (2007) Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 6, 65–69 [DOI] [PubMed] [Google Scholar]
- 32. Mazza C., Segref A., Mattaj I. W., Cusack S. (2002) Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21, 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89, 951–961 [DOI] [PubMed] [Google Scholar]
- 34. Kinkelin K., Veith K., Grünwald M., Bono F. (2012) Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA 18, 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosettani P., Knapp S., Vismara M. G., Rusconi L., Cameron A. D. (2007) Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J. Mol. Biol. 368, 691–705 [DOI] [PubMed] [Google Scholar]
- 36. Osborne M. J., Volpon L., Kornblatt J. A., Culjkovic-Kraljacic B., Baguet A., Borden K. L. (2013) eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc. Natl. Acad. Sci. U.S.A. 110, 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]