Background: Matriptase, the activator of PDGF-D, can abolish its function.
Results: The R340R341GR343A motif within the growth factor domain is the matriptase cleavage site through which matriptase destroys PDGF-D activity and binding to the extracellular matrix (ECM).
Conclusion: Matriptase has dual roles in regulating PDGF-D activity and affinity for the ECM.
Significance: Our study provides molecular insight into the functional interplay between matriptase and PDGF-D.
Keywords: Fibroblast, Growth Factor, Prostate Cancer, Receptor Tyrosine Kinase, Serine Protease
Abstract
The oncogenic roles of PDGF-D and its proteolytic activator, matriptase, have been strongly implicated in human prostate cancer. Latent full-length PDGF-D (FL-D) consists of a CUB domain, a growth factor domain (GFD), and the hinge region in between. Matriptase processes the FL-D dimer into a GFD dimer (GFD-D) in a stepwise manner, involving generation of a hemidimer (HD), an intermediate product containing one FL-D subunit and one GFD subunit. Although the HD is a pro-growth factor that can be processed into the GFD-D by matriptase, the HD can also act as a dominant-negative ligand that prevents PDGF-B-mediated β-PDGF receptor activation in fibroblasts. The active GFD-D can be further cleaved into a smaller and yet inactive form if matriptase-mediated proteolysis persists. Through mutagenesis and functional analyses, we found that the R340R341GR343A (P4–P1/P1′) motif within the GFD is the matriptase cleavage site through which matriptase can deactivate PDGF-D. Comparative sequence analysis based on the published crystal structure of PDGF-B predicted that the matriptase cleavage site R340R341GR343A is within loop III of the GFD, a critical structural element for its binding with the β-PDGF receptor. Interestingly, we also found that matriptase processing regulates the deposition of PDGF-D dimer species into the extracellular matrix (ECM) with increased binding from the FL-D dimer, to the HD, and to the GFD-D. Furthermore, we provide evidence that R340R341GR343A within the GFD is critical for PDGF-D deposition and binding to the ECM. In this study, we report a structural element crucial for the biological function and ECM deposition of PDGF-D and provide molecular insight into the dynamic functional interplay between the serine protease matriptase and PDGF-D.
Introduction
PDGF family members are primary mitogens of cells with mesenchymal origin. They play important roles in many physiological and pathological conditions (1, 2). There are four PDGF ligands (PDGF-A, PDGF-B, PDGF-C, and PDGF-D) that exert their biological functions through binding to their cognate receptors, α-PDGFR2 and β-PDGFR. PDGF-A and PDGF-B undergo intracellular proteolytic processing by furin before they are secreted as readily active growth factor dimers (3–5). In contrast, PDGF-C and PDGF-D are secreted as latent growth factor dimers with an N-terminal CUB domain, a C-terminal growth factor domain (GFD), and the hinge region in between (6–8). Proteolytic removal of the CUB domain is required for the GFD of PDGF-C and PDGF-D to activate the α-PDGFR and β-PDGFR, respectively (8, 9). Previously, we showed that serine proteases, such as urokinase plasminogen activator and matriptase, can remove the CUB domain from the latent full-length PDGF-D (FL-D) dimer in a stepwise manner, producing a hemidimer (HD) containing one FL-D monomer and one GFD monomer (GFD-M) and subsequently a biologically active GFD dimer (GFD-D) (10, 11). At present, the biological function of the HD is largely unknown. Matriptase can further cleave the active PDGF-D GFD-D into a smaller and yet inactive dimer, thus revealing dual roles of matriptase in activation and deactivation of PDGF-D, referred to as biphasic proteolytic processing (11). We previously reported that YR247GR249S (P4–P1/P1′) within the PDGF-D hinge region is the cleavage site for urokinase plasminogen activator or matriptase to remove the inhibitory CUB domain and release the active GFD-D (11). However, the cleavage site within the PDGF-D GFD, where matriptase processes the active GFD-D into an inactive form, has not been identified. In this study, we identified the matriptase cleavage site within the PDGF-D GFD and also characterized the biological activity of the HD.
The binding and retention within the extracellular matrix (ECM) are important determinants for the functions of PDGF ligands, as demonstrated by PDGF-B in vascular maturation and stabilization (12–15). Clustered basic or positively charged amino acid residues in the C terminus of PDGF-B, referred to as a retention motif (16), mediate the storage of PDGF-B within the ECM of endothelial cells. Endothelial PDGF-B retention was proven to be indispensable for proper alignment of pericytes to the microvessel wall. Deleting the retention motif of PDGF-B by gene targeting in mice impairs the recruitment of pericytes to the microvascular wall, giving rise to abnormal vasculature in the kidney and retina (14, 17, 18). At present, the association of PDGF-D dimer species with the ECM has not yet been investigated. Unlike PDGF-B, the C-terminal retention motif for ECM binding is absent in PDGF-D (7, 8); thus, we and others previously speculated that if PDGF-D is sequestered in the ECM, the CUB domain, known to play a role in protein-protein interactions (19), is likely to mediate PDGF-D interactions with the ECM (20). In this study, we investigated the association of PDGF-D dimer species with the ECM and the molecular basis for their interactions. We report the unexpected findings that the removal of the CUB domain by matriptase increases the binding of the PDGF-D dimer to the ECM and that further matriptase-mediated cleavage within the GFD reduces its association with the ECM.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human HeLa S3 cells (ATCC CCL-2.2) were purchased from American Type Culture Collection and grown in suspension in minimum essential medium with Spinner's modification (Quality Biologicals, Inc., Gaithersburg, MD) supplemented with 5% horse serum and antibiotics. Control and PDGF-D expression vector-transfected human prostate carcinoma LNCaP cell lines were maintained in RPMI 1640 medium supplemented with 5% FBS as described previously (21). The green monkey kidney cell line CV-1 and COS-1 cells were cultured in DMEM supplemented with heat-inactivated 10% FBS (HyClone, Logan, UT), 2 mm glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin (Gibco). The mouse fibroblast cell line NIH3T3 was cultured in DMEM/nutrient mixture F-12 with 10% bovine serum. Rat cerebral microvascular endothelial cells, also referred to as resistant vessel endothelial cells, were a kind gift from Dr. C. A. Diglio (Wayne State University, Detroit, MI) (22). The human glioblastoma cell line U87 was kind gift from Dr. R. Fridman (Wayne State University). All culture media were purchased from Invitrogen. Human mesenchymal stem cells (hMSCs) were purchased from Lonza (Walkersville, MD) and cultured in MesenPRO RS medium (Life Technologies, Inc.).
Anti-PDGF-D Antibodies
Polyclonal antibody against the PDGF-D GFD was raised in rabbits using synthetic peptides (N′-RKSKVDLDRLNDDAKRYS-C′) representing amino acids 254–271 in the GFD of PDGF-D as an antigen. The resulting antibodies were affinity-purified (Zymed Laboratories Inc.) and are referred to as anti-PDGF-D antibody (21). The additional anti-PDGF-D antibody used in this study was purchased from R&D Systems (catalog no. AF1159).
Recombinant PDGF-D (rPDGF-D) Proteins
Human rPDGF-Dprotein with a C-terminal histidine tag was produced by infecting HeLa S3 cells with a vaccinia expression vector for PDGF-D and recombinant vaccinia virus vTF7-3 in the presence of aprotinin (5 μg/ml) as described previously (21). rPDGF-D-His proteins in the conditioned medium (CM) were collected through a HisTrap HP column (GE Healthcare) and eluted with a sodium chloride gradient from 0.5 to 2 m in the presence of 200 mm imidazole (Sigma-Aldrich). The resulting eluates were tested for the presence of PDGF-D dimer species by immunoblot analysis. Fractions containing FL-D or the HD exclusively were pooled together and used as purified recombinant FL-D or HD, respectively. The concentration and purity of recombinant FL-D and HD proteins were assessed by silver staining and compared with known concentrations of BSA. A silver staining kit (SilverSNAP) and BSA standard were purchased from Pierce.
Cloning and Production of Mutant PDGF-D Proteins
WT FL-D cDNA without a histidine tag in the pCR2.1-TOPO cloning vector (Invitrogen) was used as a template to perform point mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. Primers used to generate the corresponding site-directed mutants are listed in Table 1. The fidelity of the mutants was verified by DNA sequence analysis in both directions (Elim Biopharmaceuticals, Hayward, CA). These cDNA inserts encoding either WT or mutant FL-D proteins were then cloned into the pTF7 vaccinia virus expression plasmid.
TABLE 1.
Primers used in this study
| Mutant | Sense primer (5′–3′) | Antisense primer (5′–3′) |
|---|---|---|
| R340A | CCT GGC CAC ATC AAG GCG AGG GGT AGA GCT AAG ACC ATG GC | GC CAT GGT CTT AGC TCT ACC CCT CGC CTT GAT GTG GCC AGG |
| R341A | CCT GGC CAC ATC AAG AGG GCG GGT AGA GCT AAG ACC ATG GC | GC CAT GGT CTT AGC TCT ACC CGC CCT CTT GAT GTG GCC AGG |
| R343A | CCT GGC CAC ATC AAG AGG AGG GGT GCA GCT AAG ACC ATG GC | GC CAT GGT CTT AGC TGC ACC CCT CCT CTT GAT GTG GCC AGG |
| K345A | GGG GTA GAG CTG CGA CCA TGG CTC TAG TTG ACA T | ATG TCA ACT AGA GCC ATG GTC GCA GCT CTA CCC C |
| R340A/R341A | CCT GGC CAC ATC AAG GCG GCG GGT AGA GCT AAG ACC ATG GC | GC CAT GGT CTT AGC TCT ACC CGC CGC CTT GAT GTG GCC AGG |
| R340A/R341A/R343A | CCT GGC CAC ATC AAG GCG GCG GGT GCA GCT AAG ACC ATG GC | GC CAT GGT CTT AGC TGC ACC CGC CGC CTT GAT GTG GCC AGG |
To produce WT or mutant PDGF-D proteins, the CV-1 cells were sequentially infected with vTF7-3 virus and then transfected with pTF7 expression plasmids encoding WT or mutant PDGF-D proteins as described previously (21). CM containing rPDGF-D was collected after 18–20 h of infection/transfection. Cell debris was pelleted by centrifugation at 2000 rpm for 5 min, and the supernatant was concentrated using Millipore Amicon centrifugal filter concentrators with a cutoff molecular mass of 10 kDa (EMD Millipore, Billerica, MA).
Generation of N-terminal Deletion Mutants of PDGF-D
N-terminal deletion mutants of PDGF-D lacking the CUB domain and different lengths of the hinge region were cloned into the pSecTag vector (Life Technologies, Inc.). These expression vectors were transiently transfected into the indicated cell lines using Lipofectamine 2000 (Life Technologies, Inc.) for 2 days according to the manufacturer's instruction. CM containing rPDGF-D proteins was collected after removing cell debris by centrifugation.
Generation of Cleavage-resistant HD (CR-HD) Protein
The CR-HD was generated by infecting COS-1 cells with vTF7-3 virus, followed by transient cotransfection with a cleavage-resistant FL-D (CR-FL-D) construct (R247A/R249A) (10) in the pTF7 vaccinia virus expression vector and the N-terminal CUB domain-deleted GFD without a C-terminal histidine tag (GD2; see Fig. 4) in the pSecTag vector for 2 days. The resulting PDGF-D dimer proteins in CM were run through a HisTrap nickel column. CR-FL-D and CR-HD dimers were trapped in the column through their histidine tags. CR-FL-D and CR-HD dimers were separated by different concentrations of imidazole in the elution buffer. The CR-HD-containing fractions were pooled together.
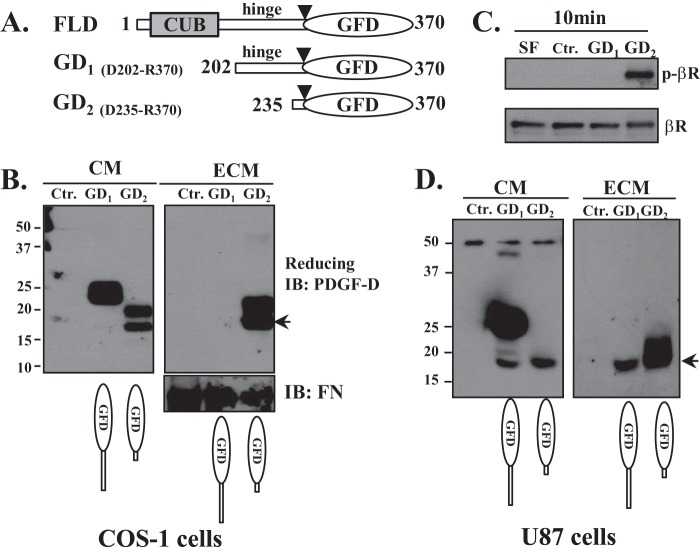
FIGURE 4.
Inhibitory role of the CUB domain and hinge region against PDGF-D activity and deposition into the ECM. A, diagrams depicting expression vectors encoding the FL-D and GFD with different lengths of the hinge region (GD1 and GD2). COS-1 (B) and U87 (D) cells were transiently transfected with GD1 and GD2 expression vectors for 48 h. Equal volumes of CM and the ECM were analyzed by immunoblot (IB) analysis under reducing conditions using anti-PDGF-D antibody. The 18-kDa GFD is indicated by arrows. FN, fibronectin. C, serum-starved NIH3T3 cells were treated with CM collected from GD1- or GD2-expressing COS-1 cells for 10 min, and β-PDGFR activation was assessed by immunoblot analysis using anti-phospho-β-PDGFR antibody. The same membrane was stripped and reprobed with anti-β-PDGFR antibody. SF, serum-free medium; Ctr., control.
β-PDGFR Activation Assay
Serum-starved NIH3T3 cells or hMSCs were treated with rPDGF-D proteins as indicated. Cell lysates were collected in radioimmune precipitation assay buffer as described previously (21). Activated β-PDGFR was detected by immunoblot analysis using anti-phospho-β-PDGFR (phospho-Tyr-751) antibody (Cell Signaling Technology) following the manufacturer's instruction. After stripping, the same blot was reprobed with anti-β-PDGFR antibody (sc-432, Santa Cruz Biotechnology).
Detection of HD Interactions with the β-PDGFR
Serum-starved hMSCs were treated with or without the CR-HD on ice for 1 h. Cells were then washed twice with ice-cold PBS, and the cell lysates were collected in radioimmune precipitation assay buffer. Equal amounts of proteins were incubated with nickel-nitrilotriacetic acid (Ni-NTA) magnetic beads or immunoprecipitated with mixed anti-PDGF-D antibodies (2 μg of our custom-made anti-PDGF-D antibody and 1 μg of anti-PDGF-D antibody from R&D Systems). The eluates from Ni-NTA beads and the PDGF-D immunocomplex were subjected to immunoblot analysis under reducing conditions using anti-β-PDGFR antibody. The IgG heavy and light chains served as loading controls.
Preparation of Acellular ECM-coated Dishes and Extraction of ECM Proteins
Cells were plated at a subconfluent density. The next day, confluent cells were washed with PBS and cultured with serum-free medium for 2 days. Serum-free medium was removed, and the remaining cells were stripped off with 50 mm EDTA (pH 8.0) in PBS for 20 s. After extensive washes with PBS, the absence of cells in the culture dish was confirmed by microscopy. This is referred to as the “acellular ECM-coated dish” used for the protein binding assay. For extraction of ECM proteins, proteins in acellular ECM-coated dishes were collected in 2% SDS lysis buffer as described (23–25). Equal volumes of ECM extracts were subjected to immunoblot analyses, and fibronectin was used as a loading control (24).
Binding of rPDGF-D Dimers to the Acellular ECM
WT or mutant rPDGF-D dimer proteins in serum-free medium were incubated with acellular ECM-coated plates prepared from the indicated cell lines. At the indicated time points, non-binding proteins were collected in the supernatant. The acellular ECM and binding proteins were washed twice with cold PBS and extracted with 2% SDS lysis buffer.
RESULTS
HD Has a Dual Function as a Pro-growth Factor and a Dominant-negative Ligand for the β-PDGFR
We previously determined that PDGF-D is a substrate of matriptase by incubating rPDGF-D in CM with the purified serine protease domain of matriptase, followed by immunoblot analysis using anti-PDGF-D antibody (21). In this study, using purified recombinant FL-D protein, we confirmed the matriptase-mediated proteolytic processing of PDGF-D in a two-step manner. Consistent with our previous report, the HD (∼58 kDa), which consists of a FL-D monomer and a GFD-M, was detected as an intermediate product under nonreducing conditions before the FL-D dimer was fully processed into GFD-D (data not shown).
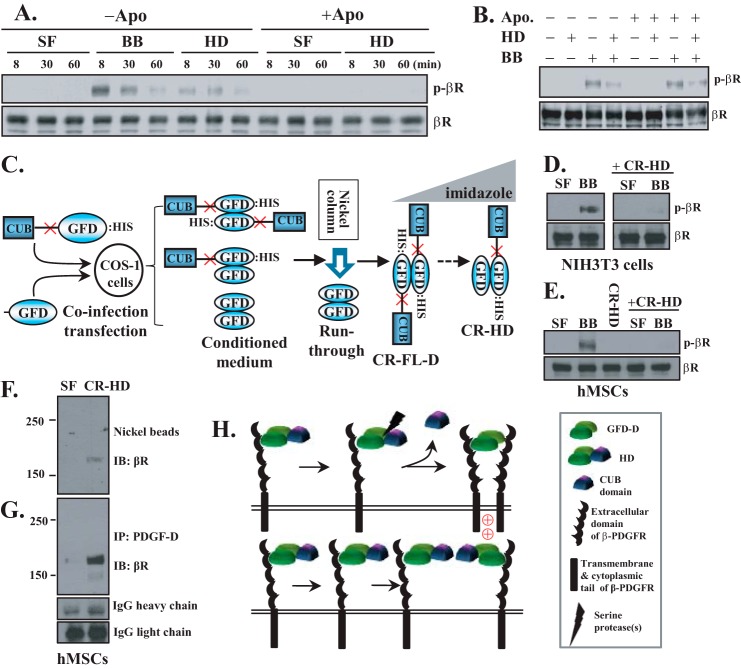
Although the FL-D dimer and GFD-D are well known as inactive and active forms of growth factor, respectively, the biological function of the HD is largely unknown. To address this, we purified recombinant HD and examined its ability to activate the β-PDGFR in NIH3T3 cells. As a positive control, a comparable molar concentration of PDGF-B was also applied under the same conditions. As expected, activation of the β-PDGFR was readily detected in NIH3T3 cells upon treatment with PDGF-B for 8 min and sharply decreased after 30 min (Fig. 1A). In contrast, the HD showed weak and delayed activity, with a peak reached at 30 min after treatment. The delayed activity of the HD suggests that further proteolytic cleavage of the HD into the GFD-D at the pericellular milieu of NIH3T3 cells may be required for the activation of the β-PDGFR. To test this hypothesis, serum-starved NIH3T3 cells were pretreated with the serine protease inhibitor aprotinin (10 μg/ml) for 30 min, followed by treatment with the HD in the presence of aprotinin. As shown in Fig. 1A, in the presence of aprotinin, the HD was unable to activate the β-PDGFR in NIH3T3 cells, indicating that the HD is not an active ligand for the β-PDGFR. It is thought that the CUB domain masks the β-PDGFR binding site in the GFD (8). Because the HD consists of a FL-D monomer and a proteolytically processed GFD-M (11, 26), we tested the possibility that the HD binds to one β-PDGFR subunit through its processed GFD subunit but fails to induce receptor dimerization and activation, thereby acting as a dominant-negative ligand. To test this hypothesis, serum-starved NIH3T3 cells were incubated with the HD in the presence of aprotinin for 30 min on ice, followed by treatment with PDGF-B for 10 min at 37 °C. As shown in Fig. 1B, PDGF-B activation of the β-PDGFR was inhibited when NIH3T3 cells were preincubated with the HD. These results show conflicting dual functions of the HD as both a pro-growth factor and a potentially dominant-negative ligand for the β-PDGFR.
FIGURE 1.
Dual roles of the HD as a precursor of active growth factor and a dominant-negative inhibitor of the β-PDGFR. A, serum-starved NIH3T3 cells were incubated with serum-free medium (SF), PDGF-B (BB; 0.2 nm), or the HD (0.14 nm) in the presence or absence of aprotinin (Apo; 10 μg/ml). At the indicated time points, activation of the β-PDGFR was detected by immunoblot analysis using anti-phospho-β-PDGFR (p-βR; phospho-Tyr-751) antibody. The same membrane was reprobed with anti-β-PDGFR (βR) antibody as a loading control. B, serum-starved NIH3T3 cells were preincubated with the HD (0.7 nm) in the presence or absence of aprotinin (10 μg/ml) for 30 min on ice, followed by treatment with PDGF-B (1 nm) at 37 °C for 10 min. The β-PDGFR activation assay was performed as described for A. C, diagram depicting the strategy used to generate the CR-HD as described under “Experimental Procedures.” D and E, serum-starved NIH3T3 or hMSCs cells, as indicated, were treated with 1 nm CR-HD or 1 nm PDGF-B with or without pretreatment with 2 nm CR-HD on ice for 30 min. The β-PDGFR activation assay was performed as described for A. F and G, serum-starved hMSCs were treated with serum-free medium or 3 nm recombinant CR-HD containing a histidine tag on ice for 1 h. PDGF-D-interacting proteins were isolated using Ni-NTA magnetic beads (F) or anti-PDGF-D antibody (G), followed by immunoblot (IB) analysis using anti-β-PDGFR antibody. The IgG heavy and light chains were used as loading controls (G). IP, immunoprecipitation. H, working model depicting the dual functions of the HD as a precursor (upper panel) of the active GFD-D and a dominant-negative inhibitor (lower panel) of β-PDGFR activation.
Because aprotinin inhibits matriptase-mediated proteolytic processing effectively but not completely, the dominant-negative nature of the HD was further confirmed using CR-HD proteins. As depicted in Fig. 1C, the CR-HD was generated by co-infection-transfection with a GFD expression vector and a CR-FL-D expression vector in which the matriptase cleavage site in the hinge region was mutated (see “Experimental Procedures”). Pretreatment of cells with the CR-HD effectively blocked PDGF-B-induced β-PDGFR activation in NIH3T3 cells (Fig. 1D), as well as in hMSCs (Fig. 1E). Using the CR-HD, we also confirmed that the HD was unable to activate the β-PDGFR in these cells (Fig. 1E). Next, we examined whether the HD indeed binds to the β-PDGFR. To this end, serum-starved hMSCs were treated with the CR-HD on ice for 1 h, and then the PDGF-D-interacting protein complexes were pulled down using either Ni-NTA beads or anti-PDGF-D antibodies. As shown in Fig. 1 (F and G), the β-PDGFR was readily detected as a PDGF-D-interacting protein by both methods. Taken together, these results suggest that although the HD is a precursor of the active GFD-D (Fig. 1H, upper panel), the HD interacts with one β-PDGFR subunit but fails to induce β-PDGFR dimerization in the absence of proteolytic processing and thereby can act as a dominant-negative ligand in cells of mesenchymal origin (Fig. 1H, lower panel).
Matriptase Deactivates PDGF-D through Cleavage at R340R341GR343A (P4–P1/P1′) within the GFD
As we reported previously (11), the 18-kDa GFD can be further processed into an even smaller fragment with a molecular mass of ∼15 kDa as matriptase-mediated proteolysis persists. The β-PDGFR activation assay showed that the 15-kDa GFD is an inactive form, demonstrating the biphasic nature of matriptase regulation of PDGF-D activity. Because our custom-made antibody (raised against the N terminus of the GFD) recognizes both the 18- and 15-kDa GFD fragments, we anticipated that the matriptase cleavage site resides close to the C terminus of the GFD. Sequence analysis identified a putative matriptase cleavage site (R340RGRAK345) within the GFD close to its C terminus (Fig. 2A). On the basis of the proposed matriptase consensus cleavage sequences with Lys/Arg (P1), a small amino acid (P2), and basic and non-basic (or vice versa) amino acids (P3 and P4) (27), we hypothesized that arginine at position 343 or lysine at position 345 of PDGF-D would be the P1 site for matriptase cleavage within the GFD (P4–P1/P1′: R340RGR343/A or G342RAK345/T). Because arginine or lysine is the preferred amino acid for P1 and P3, a single substitution of arginine or lysine with alanine was made by site-directed mutagenesis. A Lys-to-Ala substitution at position 345 (G342RAA345) failed to block matriptase processing of the 18-kDa GFD into a 15-kDa fragment. Instead, this single substitution made the PDGF-D mutant even more susceptible to matriptase cleavage, as shown by increases in the generation of the 15-kDa GFD fragment compared with the WT PDGF-D protein (Fig. 2B). This result indicates that G342RAK345T (P4–P1/P1′) is unlikely the matriptase cleavage motif within the GFD.
FIGURE 2.
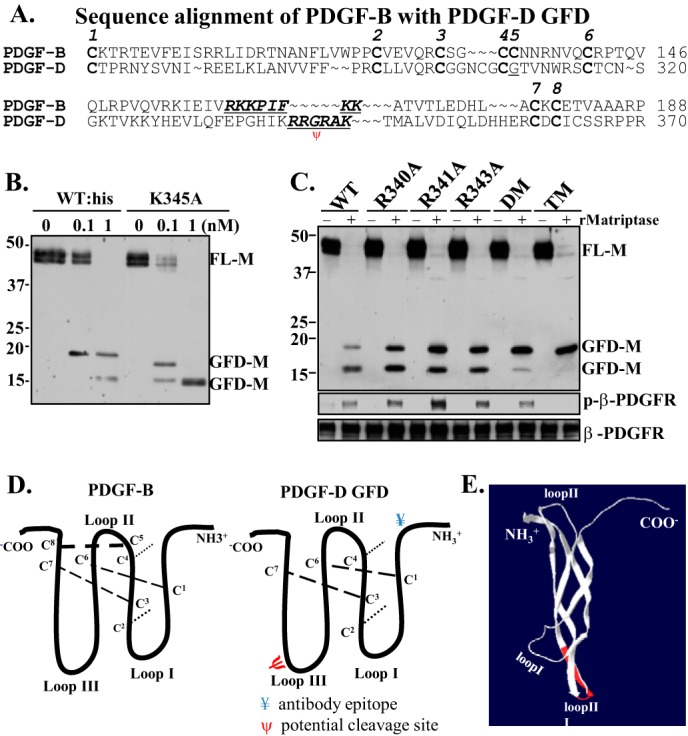
Identification of the matriptase cleavage site (R340RGRAK345, P4–P1/P1′P2′) for deactivation of PDGF-D. A, amino acid sequence alignment of PDGF-B and the PDGF-D GFD. B, WT recombinant FL-D and its mutant K345A were incubated with the indicated concentrations of recombinant matriptase at 37 °C for 1 h. The resultant samples were analyzed by immunoblot analysis under reducing conditions. FL-M, FL-D monomer. C, WT, single-mutant, double-mutant (DM; R340A/R341A), and triple-mutant (TM; R340A/R341A/R343A) PDGF-D were incubated with 1 nm recombinant matriptase (rMatriptase) at 37 °C for 1 h. The resultant samples were analyzed by immunoblot analysis under reducing conditions (upper panel). The corresponding samples were applied to serum-starved NIH3T3 cells for 10 min. Activation of the β-PDGFR was analyzed as described in Fig. 1A. D, diagram depicting the well established three-loop structure of PDGF-B (left panel) and a deduced PDGF-D GFD (right panel). The locations of the epitope for antibody raised against the PDGF-D GFD (¥) and the putative matriptase cleavage site (ψ) are indicated. E, a three-dimensional ribbon model of PDGF-B (Protein Data Bank ID 3MJG) was visualized in Swiss-PdbViewer. The R160KKPIFKK167 motif in PDGF-B is shown in red on the ribbon model, which is mapped at the loop III region of PDGF-B.
Next, we examined whether R340R341GR343A (P4–P1/P1′) is the preferential cleavage site for matriptase. When each of these arginine residues was mutated to alanine, matriptase cleaved the 18-kDa GFD into a 15-kDa fragment less efficiently (Fig. 2C), suggesting that each of these arginine residues contributes to matriptase cleavage of the GFD. Consistent with accumulation of the 18-kDa GFD, these GFD mutants resulted in increased β-PDGFR phosphorylation in NIH3T3 cells (Fig. 2C, lower panels). To further test the putative matriptase cleavage site, we generated multiple Arg-to-Ala substitutions. The double-alanine mutation (A340A341GR343) and especially the triple-alanine mutation (A340A341GA343) effectively reduced matriptase cleavage of the 18-kDa GFD into the 15-kDa fragment (Fig. 2C). Taken together, these results indicate that the R340RGR343A (P4–P1/P1′) motif is the matriptase cleavage site within the GFD of PDGF-D. However, it should be noted that the 18-kDa GFD with double- or triple-alanine mutations displayed little to no biological activity, as determined by the β-PDGFR activation assay (Fig. 2C, lower panels).
To address the molecular mechanism by which matriptase abolishes the biological activity of the PDGF-D GFD and to establish the structure-function relationship, key structural elements conserved among the PDGF family members were considered. Generally, there are eight conserved cysteine residues in PDGFs committed to the formation of the three-loop structure via three intrachain disulfide bonds and two interchain disulfide bonds, as depicted in Fig. 2D. The dimeric three-loop structure is known to be critical in maintaining the ability of PDGF ligands to bind and activate their receptors (28). Alignment of the amino acid sequences of PDGF-B and the PDGF-D GFD suggests that the matriptase cleavage site R340R341GR343A is within loop III of PDGF-D, corresponding to R160KKPIFKK in PDGF-B (Fig. 2D). As shown using the Swiss-PdbViewer, these amino acids mapped to loop III of PDGF-B (Fig. 2E, marked in red) in agreement with a previous report (29). Importantly, these positively charged amino acids in loop III of PDGF-B (R160KK162) were shown to be critical for its binding to the β-PDGFR (29). Consistently, when the positively charged arginine residues in PDGF-D (R340R341GR343), corresponding to the R160KK162 motif in PDGF-B according to our model, were mutated to non-charged alanine residues (A340A341GR343 or A340A341GA343), PDGF-D failed to activate the β-PDGFR. Thus, we propose that the matriptase cleavage site is within a critical motif of PDGF-D, possibly the β-PDGFR-binding site.
Matriptase Regulates the ECM Binding Properties of PDGF-D and Inhibitory Roles of the CUB Domain and Hinge Region in PDGF-D Interactions with the ECM
The association with the ECM is a key mechanism by which VEGF/PDGF family members establish their tissue gradient, through which they spatially regulate morphogenesis, wound healing, and angiogenesis in vivo (2). For instance, PDGF-B bound within the ECM of endothelial tip cells plays a critical role in the regulation of angiogenesis by recruiting pericytes that express PDGFRs to nascent vasculature in vivo (14). Previous studies showed that the binding of PDGF-B within the ECM is mainly regulated by basic amino acids clustered at its C terminus, referred to as the retention motif (16). Unlike PDGF-B, a cluster of basic amino acids is absent in the C terminus of PDGF-D (7). Because the CUB domain is known to mediate protein-protein interactions, we and others (7, 21) previously speculated that FL-D dimers containing two CUB domains are likely to be stored in the ECM as a latent growth factor. We further speculated that with physiological or pathological stimuli, the serine protease-mediated removal of the CUB domain might result in the release the GFD-Ds from the ECM. In this study, we asked whether PDGF-D interacts with the ECM and, if so, whether PDGF-D dimer species exhibit differential binding to the ECM. To this end, we examined the spatial distribution of PDGF-D dimer species in CM versus the acellular ECM of LNCaP cells engineered to express PDGF-D. As shown in Fig. 3, the GFD-D and HD were detected mostly in the acellular ECM, whereas the FL-D dimer was detected in CM, suggesting that the differential distribution of PDGF-D dimer species in the extracellular milieu is tightly associated with proteolytic processing for the removal of the N-terminal CUB domain, which contradicts our previous speculation.
FIGURE 3.
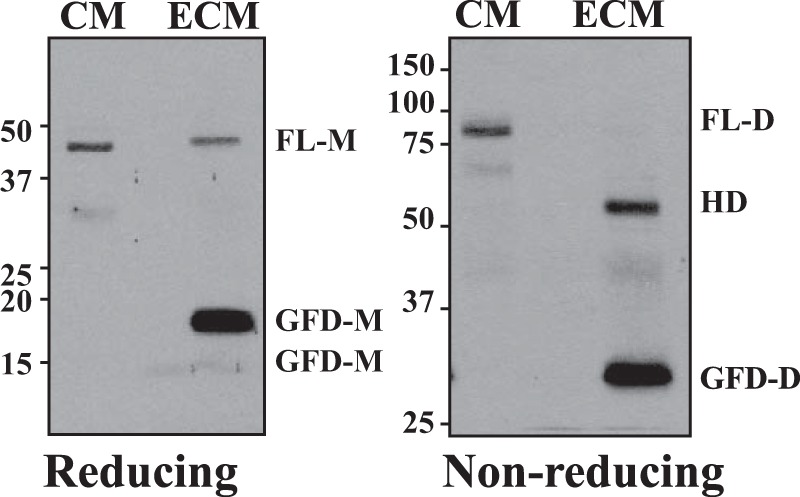
Differential distribution of PDGF-D dimer species in CM and the ECM. Shown are the results from immunoblot analysis of the PDGF-D dimer species in CM and ECM samples collected from PDGF-D-overexpressing LNCaP cells as described under “Experimental Procedures” under reducing (left panel) and nonreducing (right panel) conditions. FL-M, FL-D monomer.
To determine whether removal of the CUB domain is sufficient for GFD interactions with the ECM or whether the hinge region also participates in preventing PDGF-D from interacting with the ECM, we constructed PDGF-D GFD expression plasmids GD1 and GD2 containing different lengths of the hinge region (Fig. 4A). GD1 (Asp202–Arg370) contained most of the hinge region with an observed molecular mass of 25 kDa under reducing conditions, whereas GD2 (Arg235–Arg370) had a shorter hinge region with an observed molecular mass of 20 kDa. When COS-1 cells were transiently transfected, GD1 was present almost exclusively in CM, whereas GD2 was more readily detected in the ECM fraction (Fig. 4B). As shown in Fig. 4D, these results were reproduced using U87 human glioblastoma cells, which are well known to produce abundant ECM (30, 31). The GD2 protein was susceptible to proteolytic cleavage, as it was cleaved to the 18-kDa GFD (Fig. 4, B and D, arrows). As shown by the β-PDGFR activation assay, GD2 (but not GD1) was biologically active (Fig. 4C). These data demonstrate that removal of the CUB domain and hinge region of PDGF-D is required for growth factor activity and deposition of PDGF-D into the ECM.
Positively Charged Amino Acid Residues (R340R341GR343) in the GFD Mediate PDGF-D GFD Interactions with the ECM
The above results demonstrate that the R340R341GR343 motif in the GFD is critical for PDGF-D ligand activity, as determined by the β-PDGFR activation assay (Fig. 2). Here, we asked whether these positively charged amino acids contribute to deposition of PDGF-D into the ECM. To address this, WT or triple-alanine mutant (A340A341GA343) PDGF-D expression plasmid was transiently transfected into LNCaP cells. CM and the acellular ECM were collected at the indicated time points and analyzed for the distribution of PDGF-D by immunoblot assay under reducing conditions. As shown in Fig. 5, whereas both WT and triple-alanine mutant PDGF-D proteins were secreted into CM and underwent proteolytic cleavage into the GFD, only the GFD of WT PDGF-D was detected in the acellular ECM (Fig. 5B). These results indicate that the positively charged arginine residues in the R340R341GR343A motif of the GFD are crucial for interactions between PDGF-D and the ECM.
FIGURE 5.
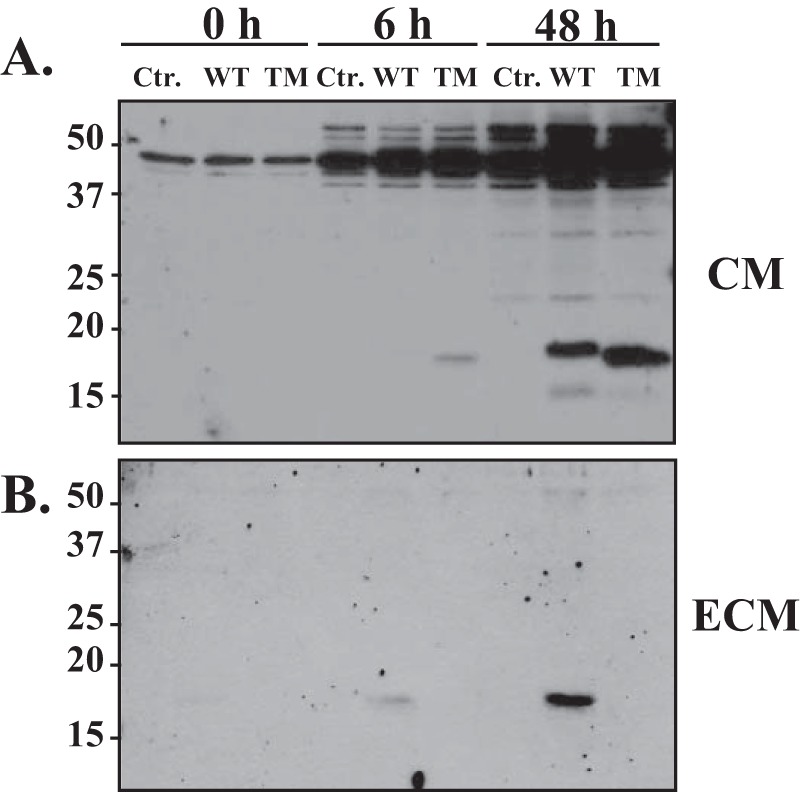
Critical role of positively charged amino acids in the R340R341GR343A motif in the deposition of PDGF-D into the ECM. LNCaP cells were transiently transfected with control (Ctr.), WT PDGF-D, or triple-alanine mutant (TM) PDGF-D expression vectors. CM and the ECM were collected at the indicated time points under serum-free conditions. PDGF-D dimer species in CM (A) and the ECM (B) were analyzed by immunoblot analysis under reducing conditions.
To further test the significance of the arginine residues in the R340R341GR343A motif for interactions of PDGF-D with the ECM, we directly tested the ECM binding of WT or triple-alanine mutant rPDGF-D dimer species. As shown in Fig. 6A, WT rPDGF-D dimers effectively interacted with the acellular ECM prepared from U87 cells (23, 24). WT GFD-D was found exclusively in the ECM. In contrast, triple-alanine mutant PDGF-D dimer species were barely detected in the ECM even after 60 min of incubation with acellular ECM-coated dishes (Fig. 6B), demonstrating the significance of the R340R341GR343A motif in mediating PDGF-D deposition into the ECM.
FIGURE 6.
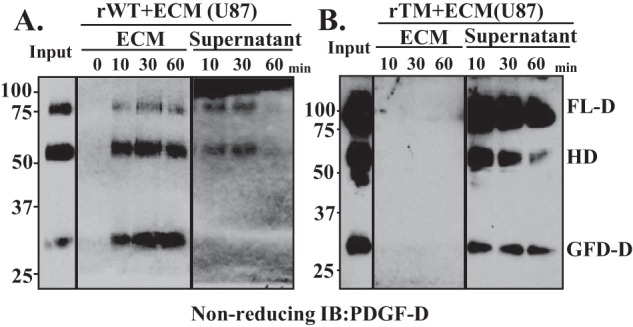
Differential binding of rPDGF-D dimer species to the acellular ECM. WT (rWT; A) or triple-alanine mutant (rTM; B) rPDGF-D proteins was incubated with the acellular ECM prepared from U87 cells for the indicated time periods. PDGF-D dimer species bound to the ECM or remaining in the supernatant were detected by immunoblot (IB) analyses using anti-PDGF-D antibody under nonreducing conditions.
DISCUSSION
We previously identified matriptase as a PDGF-D-inducible gene (11, 21) and further demonstrated that PDGF-D-specific signaling results in activation and shedding of matriptase in prostate epithelial cells (32). We also identified matriptase as a proteolytic activator of PDGF-D by removing the N-terminal inhibitory CUB domain from latent PDGF-D proteins (11), demonstrating a positive-feedback signaling loop between the serine protease matriptase and PDGF-D signaling. Interestingly, the results from this study demonstrate that matriptase is not merely an activator of PDGF-D but is also a deactivator. In addition, we demonstrated that matriptase-mediated proteolytic processing is coupled with the regulation of extracellular spatial distribution of PDGF-D dimers. Matriptase activates PDGF-D in a two-step manner involving generation of the HD, which consists of one FL-D monomer and one active GFD-M. We showed that generation of the HD is not an intermediate byproduct when recombinant FL-D proteins are incubated with recombinant matriptase in a test tube, as evidenced by existence of the HD in the ECM prepared from the in vitro culture of PDGF-D-expressing cells. Importantly, our results suggest the functional significance of the HD in the regulation of PDGFR signaling, as well as spatial distribution of extracellular PDGF-D. Activation of receptor tyrosine kinases, including the PDGFR, is initiated by receptor dimerization, which is driven entirely by ligand binding (33). The PDGF monomer has a three-loop structure and forms a dimer in a head-to-tail manner. As a result, in the PDGF dimer, loop II of one subunit resides close to loops I and III of the other subunit, forming two binding sites for its receptor. Studies by others and us (11, 20, 26) demonstrated that the CUB domains in the PDGF-D dimer are not connected by disulfide linkages, thus individually providing steric hindrance against each receptor binding site. The GFD-M in the HD, free from CUB domain hindrance, can interact with one β-PDGFR subunit, whereas the remaining CUB domain in the FL-D monomer prevents the HD from inducing β-PDGFR dimerization and subsequent activation of classic β-PDGFR signal transduction. Thus, the HD on the cell surface at saturating concentrations can function as a dominant-negative isoform for β-PDGFR signaling, as depicted in Fig. 1H. However, the HD can further undergo serine protease-mediated pericellular processing for the release of the remaining CUB domain, resulting in the generation of the GFD-D, revealing the HD as a precursor of an active growth factor.
In addition to the role in the regulation of PDGF-D activity, matriptase-mediated proteolytic processing also regulates PDGF-D interactions with the ECM. The removal of the CUB domain and hinge region results in the binding of PDGF-D to the ECM. PDGF-D dimers stored within the ECM may be released with physiological or pathological stimuli and brought into the pericellular milieu for interactions with the β-PDGFR. At present, the molecular nature of PDGF-D-interacting proteins in the ECM or molecular actions underlying the release of PDGF-D from the ECM is unknown. It is likely that the levels/activity of serine proteases and PDGF-D-interacting proteins within the ECM and on the cell surface will co-regulate extracellular spatial distribution and PDGF-D activity in the regulation of PDGFR signaling.
Recently, we discovered a novel function of PDGF-D in activation of osteoclastogenesis critical for prostate cancer cell growth in the bone microenvironment. Although both PDGF-B and PDGF-D are potent activators of the β-PDGFR, only PDGF-D induces osteoclastic differentiation (34). Importantly, we found that the PDGF-D HD is responsible for inducing unique signal transduction pathways leading to osteoclast differentiation.3 In fibroblasts, as shown in this study, the HD is biologically inactive, whereas it has the potential to act as both an inhibitor and activator of the β-PDGFR (Fig. 1). These results suggest that HD-specific cell signaling occurs in a cell type-specific manner possibly involving the CUB domain. The HD may induce heterodimerization of cell-surface receptors with the β-PDGFR for the formation of novel signaling complexes in some cell types. The identification of HD-specific signaling complexes in osteoclasts will shed light on molecular mechanisms underlying the novel biological functions of PDGF-D.
The matriptase-activated GFD of PDGF-D with a molecular mass of ∼18 kDa is vulnerable to further proteolytic processing, resulting in generation of an ∼15-kDa fragment with loss of growth factor activity. This study identified the matriptase cleavage motif within the PDGF-D GFD to be R340R341AR343. Structural delineation of the PDGF-D GFD with PDGF-B predicted that the matriptase cleavage motif is within the GFD receptor-binding site. Interestingly, the same residues are critical for PDGF-D interactions with the ECM. The binding of PDGF-D dimer species increased as the FL-D dimer was processed into the HD and further into the GFD-D. Consistent with our finding that the positively charged amino acids in the R340R341AR343 motif are critical for both growth factor activity and the ability to interact with the ECM, the HD containing one GFD liberated from the CUB domain-mediated steric hindrance showed higher binding to the ECM compared with the FL-D dimer, but lower binding compared with the GFD-D, in which the R340R341AR343 motif in both subunits is unmasked. Although the processed GFD-D can readily undergo serine protease-mediated deactivation (generation of the 15-kDa fragment) and further degradation in solution, the HD and 18-kDa GFD appear to be more stable in the ECM, as evidenced by accumulation of the GFD in the acellular ECM (Fig. 6). Interestingly, the 15-kDa GFD fragment was barely detected in the ECM, supporting our finding that the R340R341AR343 motif is critical for PDGF-D binding to the ECM. We surmise that matriptase-processed PDGF-D binding to the ECM not only contributes to PDGF-D deposition in the ECM, but also protects against further proteolytic degradation.
To our knowledge, this study has provided the first evidence of serine protease-regulated PDGF-D deposition into the ECM. This may have profound biological implications. Differential distribution of PDGF-D dimer species (GD-D > HD > FL-D dimer) may play a role in forming the growth factor gradient in vivo. The latent FL-D dimer may circulate in the body and travel further within the tissue until it undergoes serine protease-mediated proteolysis in the extracellular milieu of target cells. This idea is supported by the observation that PDGF-D is highly produced by the adrenal gland and in experimental nephritis, where plasma levels of PDGF-D (but not PDGF-B) are drastically increased, with PDGF-D apparently acting as an endocrine growth factor (7, 8, 35, 36). Unlike PDGF-B, which is secreted as an active growth factor and contains the C-terminal retention motif, PDGF-D activity and extracellular spatial distribution are regulated by the serine protease matriptase, as demonstrated in this study. These results provide important information for a better understanding of the differential functions of PDGF-B and PDGF-D despite both being agonists of the β-PDGFR.
Acknowledgments
We thank Xuwen Liu and Carolyn V. Ustach for technical support and Abdo J. Najy for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA123362 from NCI (to H.-R. C. K.).
W. Huang and H.-R. C. Kim, manuscript in preparation.
- PDGFR
- PDGF receptor
- GFD
- growth factor domain
- FL-D
- full-length PDGF-D
- HD
- hemidimer
- GFD-M
- GFD monomer
- GFD-D
- GFD dimer
- ECM
- extracellular matrix
- hMSC
- human mesenchymal stem cell
- rPDGF-D
- recombinant PDGF-D
- CM
- conditioned medium
- CR-HD
- cleavage-resistant HD
- CR-FL-D
- cleavage-resistant FL-D
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Yu J., Ustach C., Kim H.-R. C. (2003) Platelet-derived growth factor signaling and human cancer. J. Biochem. Mol. Biol. 36, 49–59 [DOI] [PubMed] [Google Scholar]
- 2. Andrae J., Gallini R., Betsholtz C. (2008) Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegfried G., Basak A., Prichett-Pejic W., Scamuffa N., Ma L., Benjannet S., Veinot J. P., Calvo F., Seidah N., Khatib A. M. (2005) Regulation of the stepwise proteolytic cleavage and secretion of PDGF-B by the proprotein convertases. Oncogene 24, 6925–6935 [DOI] [PubMed] [Google Scholar]
- 4. Siegfried G., Khatib A. M., Benjannet S., Chrétien M., Seidah N. G. (2003) The proteolytic processing of pro-platelet-derived growth factor-A at RRKR86 by members of the proprotein convertase family is functionally correlated to platelet-derived growth factor-A-induced functions and tumorigenicity. Cancer Res. 63, 1458–1463 [PubMed] [Google Scholar]
- 5. Heldin C.-H., Westermark B. (1999) Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283–1316 [DOI] [PubMed] [Google Scholar]
- 6. Li X., Pontén A., Aase K., Karlsson L., Abramsson A., Uutela M., Bäckström G., Hellström M., Boström H., Li H., Soriano P., Betsholtz C., Heldin C.-H., Alitalo K., Ostman A., Eriksson U. (2000) PDGF-C is a new protease-activated ligand for the PDGF α-receptor. Nat. Cell Biol. 2, 302–309 [DOI] [PubMed] [Google Scholar]
- 7. LaRochelle W. J., Jeffers M., McDonald W. F., Chillakuru R. A., Giese N. A., Lokker N. A., Sullivan C., Boldog F. L., Yang M., Vernet C., Burgess C. E., Fernandes E., Deegler L. L., Rittman B., Shimkets J., Shimkets R. A., Rothberg J. M., Lichenstein H. S. (2001) PDGF-D, a new protease-activated growth factor. Nat. Cell Biol. 3, 517–521 [DOI] [PubMed] [Google Scholar]
- 8. Bergsten E., Uutela M., Li X., Pietras K., Ostman A., Heldin C.-H., Alitalo K., Eriksson U. (2001) PDGF-D is a specific, protease-activated ligand for the PDGF [beta]-receptor. Nat. Cell Biol. 3, 512–516 [DOI] [PubMed] [Google Scholar]
- 9. Gilbertson D. G., Duff M. E., West J. W., Kelly J. D., Sheppard P. O., Hofstrand P. D., Gao Z., Shoemaker K., Bukowski T. R., Moore M., Feldhaus A. L., Humes J. M., Palmer T. E., Hart C. E. (2001) Platelet-derived growth factor C (PDGF-C), a novel growth factor that binds to PDGF α and β receptor. J. Biol. Chem. 276, 27406–27414 [DOI] [PubMed] [Google Scholar]
- 10. Ustach C. V., Kim H.-R. C. (2005) Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Mol. Cell. Biol. 25, 6279–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ustach C. V., Huang W., Conley-LaComb M. K., Lin C. Y., Che M., Abrams J., Kim H.-R. C. (2010) A novel signaling axis of matriptase/PDGF-D/ss-PDGFR in human prostate cancer. Cancer Res. 70, 9631–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enge M., Bjarnegård M., Gerhardt H., Gustafsson E., Kalén M., Asker N., Hammes H. P., Shani M., Fässler R., Betsholtz C. (2002) Endothelium-specific platelet-derived growth factor-B ablation mimics diabetic retinopathy. EMBO J. 21, 4307–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindahl P., Johansson B. R., Levéen P., Betsholtz C. (1997) Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245 [DOI] [PubMed] [Google Scholar]
- 14. Lindblom P., Gerhardt H., Liebner S., Abramsson A., Enge M., Hellstrom M., Backstrom G., Fredriksson S., Landegren U., Nyström H. C., Bergström G., Dejana E., Ostman A., Lindahl P., Betsholtz C. (2003) Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes Dev. 17, 1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bjarnegård M., Enge M., Norlin J., Gustafsdottir S., Fredriksson S., Abramsson A., Takemoto M., Gustafsson E., Fässler R., Betsholtz C. (2004) Endothelium-specific ablation of PDGFB leads to pericyte loss and glomerular, cardiac and placental abnormalities. Development 131, 1847–1857 [DOI] [PubMed] [Google Scholar]
- 16. Ostman A., Andersson M., Betsholtz C., Westermark B., Heldin C.-H. (1991) Identification of a cell retention signal in the B-chain of platelet-derived growth factor and in the long splice version of the A-chain. Cell Regul. 2, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nyström H. C., Lindblom P., Wickman A., Andersson I., Norlin J., Fäldt J., Lindahl P., Skøtt O., Bjarnegård M., Fitzgerald S. M., Caidahl K., Gan L. M., Betsholtz C., Bergström G. (2006) Platelet-derived growth factor B retention is essential for development of normal structure and function of conduit vessels and capillaries. Cardiovasc Res. 71, 557–565 [DOI] [PubMed] [Google Scholar]
- 18. Abramsson A., Lindblom P., Betsholtz C. (2003) Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J. Clin. Invest. 112, 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bork P., Beckmann G. (1993) The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 231, 539–545 [DOI] [PubMed] [Google Scholar]
- 20. Reigstad L. J., Varhaug J. E., Lillehaug J. R. (2005) Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family. FEBS J. 272, 5723–5741 [DOI] [PubMed] [Google Scholar]
- 21. Ustach C. V., Taube M. E., Hurst N. J., Jr., Bhagat S., Bonfil R. D., Cher M. L., Schuger L., Kim H.-R. C. (2004) A potential oncogenic activity of platelet-derived growth factor D in prostate cancer progression. Cancer Res. 64, 1722–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diglio C. A., Wolfe D. E., Meyers P. (1983) Transformation of rat cerebral endothelial cells by Rous sarcoma virus. J. Cell Biol. 97, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Z., Jing H., Tao Z., Choi H., Aboulfatova K., Moake J., Li R., Dong J. F. (2009) Effects of naturally occurring mutations in CUB-1 domain on synthesis, stability, and activity of ADAMTS-13. Thromb. Res. 124, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goetz J. G., Minguet S., Navarro-Lérida I., Lazcano J. J., Samaniego R., Calvo E., Tello M., Osteso-Ibáñez T., Pellinen T., Echarri A., Cerezo A., Klein-Szanto A. J., Garcia R., Keely P. J., Sánchez-Mateos P., Cukierman E., Del Pozo M. A. (2011) Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell 146, 148–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu Y. C., Liou Y. M. (2011) The anti-cancer effects of (−)-epigallocatechin-3-gallate on the signaling pathways associated with membrane receptors in MCF-7 cells. J. Cell. Physiol. 226, 2721–2730 [DOI] [PubMed] [Google Scholar]
- 26. Li X., Eriksson U. (2003) Novel PDGF family members: PDGF-C and PDGF-D. Cytokine Growth Factor Rev. 14, 91–98 [DOI] [PubMed] [Google Scholar]
- 27. Takeuchi T., Harris J. L., Huang W., Yan K. W., Coughlin S. R., Craik C. S. (2000) Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J. Biol. Chem. 275, 26333–26342 [DOI] [PubMed] [Google Scholar]
- 28. Oefner C., D'Arcy A., Winkler F. K., Eggimann B., Hosang M. (1992) Crystal structure of human platelet-derived growth factor BB. EMBO J. 11, 3921–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schilling D., Reid I. J., IV, Hujer A., Morgan D., Demoll E., Bummer P., Fenstermaker R. A., Kaetzel D. M. (1998) Loop III region of platelet-derived growth factor (PDGF) B-chain mediates binding to PDGF receptors and heparin. Biochem. J. 333, 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziu M., Schmidt N. O., Cargioli T. G., Aboody K. S., Black P. M., Carroll R. S. (2006) Glioma-produced extracellular matrix influences brain tumor tropism of human neural stem cells. J. Neurooncol. 79, 125–133 [DOI] [PubMed] [Google Scholar]
- 31. Wang C., Tong X., Yang F. (2014) Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Mol. Pharm. 11, 2115–2125 [DOI] [PubMed] [Google Scholar]
- 32. Najy A. J., Won J. J., Movilla L. S., Kim H.-R. C. (2012) Differential tumorigenic potential and matriptase activation between PDGF B versus PDGF D in prostate cancer. Mol. Cancer Res. 10, 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang Y., Yuzawa S., Schlessinger J. (2008) Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang W., Fridman Y., Bonfil R. D., Ustach C. V., Conley-LaComb M. K., Wiesner C., Saliganan A., Cher M. L., Kim H.-R. C. (2012) A novel function for platelet-derived growth factor D: induction of osteoclastic differentiation for intraosseous tumor growth. Oncogene 31, 4527–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Floege J., Eitner F., Van Roeyen C., Ostendorf T. (2003) PDGF-D and renal disease: yet another one of those growth factors? J. Am. Soc. Nephrol. 14, 2690–2691 [DOI] [PubMed] [Google Scholar]
- 36. Ostendorf T., van Roeyen C. R., Peterson J. D., Kunter U., Eitner F., Hamad A. J., Chan G., Jia X. C., Macaluso J., Gazit-Bornstein G., Keyt B. A., Lichenstein H. S., LaRochelle W. J., Floege J. (2003) A fully human monoclonal antibody (CR002) identifies PDGF-D as a novel mediator of mesangioproliferative glomerulonephritis. J. Am. Soc. Nephrol. 14, 2237–2247 [DOI] [PubMed] [Google Scholar]