Summary
The relationship between commensal microbes and their hosts has been studied for many years. Commensal microorganisms are known to have a significant role in regulating the physiology of their hosts and preventing pathogenic infections while the hosts’ immune system is important in determining the composition of the microbiota. More recently, specific effects of the intestinal microbiota on the local and distal immune systems have been uncovered with important consequences for health and disease, and alterations in intestinal microbial composition has been associated with various disease states. Here, we will review the current understanding of the microbiota/immune system crosstalk, highlight the clinical consequences of changes in the microbiota and consider how to harness this symbiotic relationship to improve public health.
Introduction
Humans exist as metaorganisms consisting of host cells and symbiotic microbes. The microbes approximate 100 trillion cells, outnumbering host cells by a factor of 10, with greater than 10-fold more microbial than eukaryotic genes being expressed [1]. This complex community of microbes includes fungi [2], viruses [3] and bacteria [4], which play a fundamental role in modulating the physiology of their host. For instance, the microbiota are important for host metabolism such as conjugation and de-conjugation of bile acids, and production of short chain fatty acids, essential vitamins and amino acids [5]. In addition to complementing the host’s metabolism, commensal bacteria can control the virulence of pathogens through competition for similar energy sources and production of anti-microbial molecules [6].
Importantly, the microbiota are also essential for the development of the mucosal immune system [7]. As early as 1963, it was documented that germ-free (GF) mice responded more weakly to bacterial antigens than conventional mice, in part because of less-developed secondary lymphoid organs (SLO) [8]. Although more recent studies have shown that mesenteric lymph nodes (mLNs) and Peyer’s patches (PPs) in mice start to develop during embryogenesis, which is believed to be sterile, further formation of lymphoid tissue can be induced by commensal bacteria [9]. Moreover, maturation of intestinal cryptopatches and isolated lymphoid follicles (ILFs), both considered gut-associated lymphoid tissue, is dependent on the microbiota after birth [7].
The immune system provides a formidable and flexible network to maintain host homeostasis in response to different challenges from the environment. The acquisition of an adaptive immune system during evolution coincides with the emergence of microbial colonization [4], suggesting that the immune system co-evolved with the microbial community to attain a well-balanced symbiotic relationship. Immune system cells can respond to microbes following their recognition, by pattern-recognition receptors (PRRs), of microbial-associated molecular patterns (MAMPs) expressed on microbes but not on host cells [10]. PRRs are expressed on the cell surface as well as intra-cellularly, and include among others Toll-like receptors (TLRs), which can signal in a manner dependent on the adaptors MyD88 and/or TRIF and activate NF-κB or interferon-response factors, inflammasome-dependent receptors that drive production of IL-1β and IL-18, and cytosolic sensors that induce interferon response factors. Microbial recognition can have distinct consequences depending on the cell-type and particular microbe involved, the location of the interaction in the body, and the additional contextual signals that may accompany a local or systemic invasion versus a barrier surface-protected interaction.
Limiting Intestinal Immune Exposure to the Microbiota
The intestine is constantly facing challenges from food antigens, commensals and pathogens and has to respond quickly and precisely to mount appropriate responses. According to Hooper et al., intestinal homeostasis can be achieved if the microbiota is stratified and compartmentalized [11], with stratification referring to limiting contact between luminal antigens and the immune system, and compartmentalization to localizing the immune response to the intestine to prevents systemic immunity.
Mucus and goblet cells
The first line of defense in this barrier is the mucus layer produced by goblet cells. Mucus consists of heavily glycosylated mucins, which can be subdivided into secreted gel-forming and non-gel-forming, and cell-surface mucins, with gel-forming mucins, especially MUC2, as the major constituents [12]. Due to the anatomy and absorptive requirements of the intestine, the mucus layer in the small intestine differs from that in the colon, going from discontinuous in the small intestine to continuous in the colon [13]. Furthermore, the mucus in the colon can be subdivided into an outer and an inner layer, where the inner layer is denser and devoid of bacteria [14]. It is likely that this distribution of mucus is an evolutionary product of symbiosis, as the number of bacteria in the colon is higher than in the small intestine. Besides providing physical segregation, mucus also traps on its outer layer antimicrobial peptides (AMPs) and secretory immunoglobulin A (sIgA) that are at the first line of our immune defenses and help shape the microbial community. Interestingly, intestinal mucus has been shown to promote oral tolerance [15]. The porosity of the mucus layer in the small intestine allows antigen-MUC2 complexes to be uptaken by DCs and the glycans associated with MUC2 have been found to deliver tolerogenic signals and induce anti-inflammatory properties in DCs.
Signals from the microbiota appear necessary for the normal function of goblet cells, as goblet cell MyD88-dependent sensing of the luminal microbiota was necessary to reduce the formation of goblet cell-associated passage formation, conduits known to increase luminal antigen delivery to the lamina propria [16]. Microbial signals can also regulate mucus secretion by signaling via the inflammasome NLRP6, to induce autophagy and mucus production by goblet cells [17]. Underscoring the importance of microbiota signals, the function of goblet cells was impaired when the microbial community was perturbed during gastrointestinal infection or antibiotic treatment [18,19].
AMPs and IgA
AMPs and IgA, which are produced by Paneth cells and IgA+ B cells, respectively, adhere to the outer mucus layer and, in addition to mucus, maintain microbial segregation in the intestinal lumen. AMPs include defensins and C-type lectins, and they are essential to contain the microbiota and minimize bacterial contact with epithelial cells. AMPs’ secretion by Paneth cells is regulated by IL-22 produced by innate lymphoid cells (ILCs) [20], the secretion of which is modulated by the microbiota [21]. Consistent with the microbiota regulating AMP secretion, expression by Paneth cells of RegIIIγ, a C-type lectin that preferentially binds to gram-positive bacteria, was decreased in GF mice [22]. Signaling by the microbiota to Paneth cells is dependent on MyD88, as deleting MyD88 from the non-hematopoietic compartment of conventional mice (which includes Paneth cells) led to a significant reduction in RegIIIγ production and failure to fight Listeria. monocytogenes infection [23].
Like production of mucus by goblet cells, secretion of AMPs appears dependent on autophagy, as mutations in the autophagy gene ATG16L1 were shown to affect the normal function of Paneth cells in mice and humans [24].
Similarly to AMPs, B cell-produced IgA plays an important role in maintaining the microbiota-host relationship by neutralizing bacteria, reducing intestinal pro-inflammatory signals and balancing expression of bacterial epitopes [25,26].
Intestinal Epithelial Cells
Intestinal epithelial cells (IECs), an umbrella term that includes classical epithelial cells, M cells that sample luminal antigens, goblet cells and Paneth cells, serve as a major barrier to separate the intestinal luminal content from the mucosal immune system and are responsible for various functions, including the production of mucus and AMPs and the facilitation of secretory IgA transcytosis into the gut lumen. More than just a physical barrier, IECs signal in response to luminal microbial products to maintain homeostasis. For instance, IEC-intrinsic MyD88-dependent autophagy contributes to intestinal homeostasis upon bacterial invasion [27], and inflammasome activation is essential to control enteric bacterial colonization [28–30]. In addition to responding to gut microbiota, IECs were recently found to respond to systemic microbial products to sustain host-commensal symbiosis during severe illness [31]. Systemic LPS resulted in α(1,2)-fucosylation of IECs and fucosylated proteins shed into the intestinal lumen were shown to be metabolized by commensals and reduce expression of bacterial virulence genes thus improving tolerance to mild intestinal pathogens that could perhaps outgrow during sepsis-mediated anorexia.
Finally, IECs also interact with the underlying local host immune system to achieve mucosal integrity [32–34]. In conclusion, the intestinal barrier has evolved to adapt to the luminal and systemic environment to maintain intestinal homeostasis.
Local Intestinal Immune Responses to the Microbiota
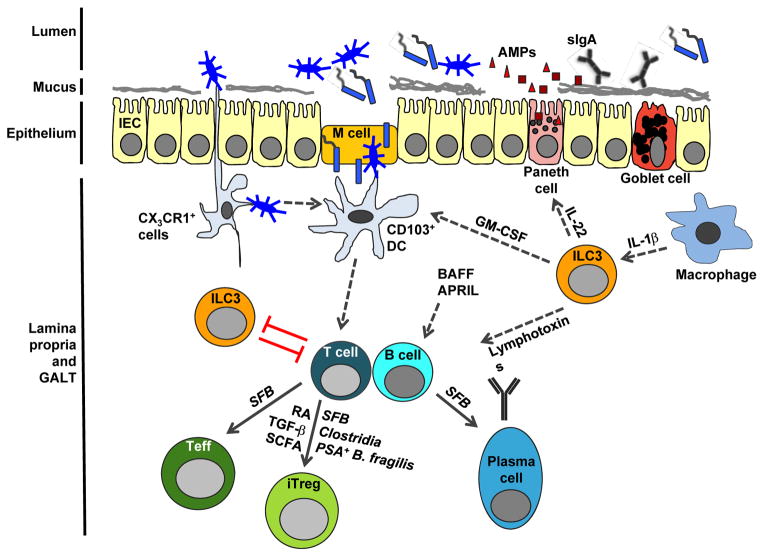
Host immune cells reside in the lamina propria of the intestine, below the epithelial layer. Despite the physical barrier between the lamina propria and luminal commensals, the host’s immune system is constantly exposed to commensal microbes and their products. To avoid systemic inflammation, the mucosal immune system has evolved to localize its response. For instance, intestinal dendritic cells (DCs) activated by commensals are restricted to mLNs and specifically induce the production of IgA in B cells [35]. The crosstalk between resident microbiota and immune cells of the lamina propria is key to maintaining homeostasis in the intestine. Figure 1 summarizes some of the interactions discussed in this review.
Figure 1. A model of the interplay between the intestinal microbiota and the mucosal immune system.
Intestinal bacteria are segregated to the intestinal lumen by a physical barrier that includes goblet cells and Paneth cells which produce mucus and anti-microbial peptides (AMPs), respectively. The AMPs, secretory IgA (sIgA), mucus layer and IEC epidermal layer help control bacterial translocation and shape the microbial composition. Dendritic cells (DCs) constantly sample luminal contents to keep the mucosal immune system at its basal poised state and induce a balance of effector and regulatory cells. CX3CR1+ DCs are shown to sample luminal antigens and transfer them to CD103+ DCs that drive downstream adaptive immune responses, such as activation of T and B cells. With the help of various bacteria or microbial components and metabolic products, activated T cells differentiate into effector (Teff) and regulatory T (Treg) cells. B cells receive signals from various cell types and differentiate into IgA-secreting plasma cells, whose product, IgA, translocates across the epithelial barrier. RORγt+ ILC3s that can respond to intestinal macrophages support the function of Paneth cells, CD103+ DCs and B cells, whereas there can be a reciprocal negative regulation between ILC3s and T cells.
Mononuclear Phagocytes
Mononuclear phagocytes (MNPs) in the intestine are a diverse population that includes DCs and macrophages [36]. They specialize in uptake of luminal antigens to promote downstream adaptive mucosal immune responses. However, the identity of the intestinal antigen-presenting cell that activates the adaptive mucosal immune system remains controversial. In 2001, it was shown that DCs can sample luminal content by expressing their own tight junction proteins and extending dendrites through these tight junctions into the lumen [37]. When finer phenotypic characterization of intestinal MNPs became available, it was suggested that CX3CR1+ DCs are responsible for forming trans-epithelial dendrites and sampling antigen, leading to clearance of infectious bacteria [38]. This idea was further supported by Diehl et al., who reported that signals from the microbiota through MyD88 in CX3CR1+ cells restricted trafficking of antigen to mLNs [39]. On the other hand, CD103+ DCs were also suggested to be the antigen sampling cells that migrate to mLNs [40]. Evidence now suggests that division of labor exists between MNPs and that both subsets play a role. Indeed, it was recently demonstrated that CX3CR1+ cells uptake antigen and transfer it to CD103+ DCs [41] in a gap junction protein-dependent manner [42].
With the unique environment of the intestine where commensals and potentially pathobionts are in close proximity to the mucosal immune system, it is important for MNPs not only to activate pro-inflammatory adaptive responses, but also regulatory ones. One of the critical roles of activated intestinal DCs is the generation of induced regulatory T cells (iTregs), in a TGFβ- and retinoic acid-dependent process [43] and in a manner that involves TRAF6 in DCs [44]. Recently, Mortha et al demonstrated that a cross-talk between GMCSF-producing ILC3s and IL-1β-producing macrophages was necessary to regulate the number and function of CD103+ DCs and that this cross-talk was important for the generation of iTregs [45].
T Regulatory Cells
A few phyla of bacteria have been specifically implicated in mucosal tolerance via iTreg induction. Bacteroides fragilis has been shown to enhance Treg function via the symbiosis factor polysaccharide A (PSA), as PSA+ but not PSA-deficient B. fragilis-monocolonized animals had the ability to suppress Th17 inflammatory responses [46,47]. In addition, GF mice infected with the pathogenic bacterium Helicobacter hepaticus developed colitis, but when co-colonized with B. fragilis, PSA-induced IL-10 secretion by iTregs and allowed for suppression of colitis [48]. PSA-induced TLR2 signaling on CD4+ T cells was shown to be necessary for iTreg differentiation and increased IL-10 secretion [47]. These results show that B. fragilis can mediate gut regulatory responses via a bacterium-associated molecule.
The Clostridium genus of commensals has also been shown to play a role in prevention of inflammatory bowel disease (IBD), particularly clusters IV and XIVa [49]. Mice colonized with mixtures of commensal clostridial strains have been shown to have reduced symptoms of colitis [50,51]. Whereas B. fragilis appears responsible for enhanced Treg activity, Clostridia have been shown to induce iTreg differentiation and frequency [50,51]. When commensal Clostridia strains were isolated and identified and GF or SPF animals were colonized with the mixture of strains, they developed decreased Th17 responses and an increased frequency of IL-10-producing Tregs [51]. Several mechanisms have been implicated in the ability of Clostridia strains to enhance iTreg differentiation, including induction of TGFβ in IELs [50], and production of the bacterial fermentation products, short-chain fatty acids (SCFAs) [52]. The effects of SCFAs, butyrate in particular, on iTreg differentiation was demonstrated both in vitro and in vivo when butyrate was added to T cell tissue cultures [53], or mice received oral butyrate [53,54]. The major mechanism of action of butyrate is thought to be its function as a histone deacetylase inhibitor (HDAC). Butyrate has been shown to increase acetylation at the FoxP3 promoter and CNS elements, and its function as a promoter of Treg induction suggests that its activity as an HDAC inhibitor allows for a greater accessibility of FoxP3 and other regulatory transcription factors [53,54].
Tregs not only receive signals from non-specific receptors that respond to microbial molecular patterns or microbial metabolic products, but also from direct TCR recognition of microbial antigens [55]. This, in the context of anti-inflammatory signals such as TGF-β, retinoic acid and SCFA may allow differentiation of conventional T cells into iTregs to provide tolerance to commensal microbiota and prevent immunopathology such as inflammatory bowel disease [54,56].
Mechanisms antagonistic to Tregs appear in place to ensure the mucosal immune system is not over-suppressed at the time of infection. DNA from the microbial community was found to be suppressive for Treg conversion, instead facilitating the differentiation of IFNγ- and IL-17-producing effector T cells to fight against fungal infection [57]. While this interaction was TLR9-dependent, the origin of the DNA remains unclear. Nevertheless, the microbiota contribute to the homeostasis of the Treg population.
T Effector Cells
The microbiota can also directly affect the differentiation of effector T cells. Segmented filamentous bacteria (SFB), non-culturable, gram-positive bacteria that are closely related to the genus Clostridium and tightly adhering to intestinal epithelial surfaces [58], are sufficient to promote the full development of the T helper cell response, including IFNγ-, IL-17- and IL10- producing CD4 T cells [59]. Th17 cells are particularly known to have a prominent role in intestinal immunity [60]. In the intestine, where TGF-β is highly abundant, naïve T cells differentiate into Th17 cells in the presence of the pro-inflammatory cytokine IL-6 [61]. Colonization with SFB in mice previously devoid of it resulted in IL-17 and IL-22 expression by CD4+ T cells in the lamina propria of the small intestine and promoted antimicrobial defense. The mechanisms by which SFB induce Th17 responses are still being investigated. Recently, a study suggested that intestinal DCs can present SFB peptides directly to the TCR of T cells and induce SFB-specific Th17cells in an MHC class II-dependent manner [62]. Interestingly, naïve T cells that bore a TCR transgene specific for SFB had a strong tendency to differentiate into Th17 cells even in the presence of the strong Th1 inducer L. monocytogenes [63]. Thus, differentiation of T cells in the intestine may depend on direct TCR-dependent microbial antigen recognition and/or the balance of pro- and anti-inflammatory molecules present at any given time providing equilibrium between effector T cells and Tregs. Importantly, intestinal homeostasis can be broken and tolerance to commensals be lost if the barrier is breached during intestinal infection. Toxoplasma gondii infection was shown to result in active immunization and development of T cell memory against commensals [64].
B Cells
SFB also potently induce IgA production [65]. Intestinal IgA can be produced at multiple sites, such as Peyer’s patches, isolated lymphoid follicles, lamina propria and mesenteric lymph nodes. A new study has shown that SFB can facilitate postnatal development of isolated lymphoid follicles with germinal centers, which are critical for IgA induction [66].
The mechanism of intestinal IgA induction is not completely understood. In general, IgA can be induced by B cells following CD40L-CD40 interactions with activated T cells, but can also occur in a T cell-independent manner, whereby B cell-activating factor (BAFF) and a proliferation-inducing ligand (APRIL) secreted by IECs and DCs promote IgA induction [67]. For T cell-dependent induction of IgA, both Th17 [68] and Tregs [69] cells are capable of being reprogrammed into T follicular helper (Tfh) cells to activate B cells to produce IgA. Additionally, soluble lymphotoxin from RORγt+ ILCs was shown to facilitate T cell-dependent IgA production [70]. In contrast, membrane lymphotoxin was important for T cell-independent IgA induction in the intestine [70].
Whereas it is known that the microbiota have a role in intestinal B cell activation and IgA production [35,65], how the microbiota contribute to the intestinal B cell repertoire remains ambiguous. Vossenvämper et al. showed that human transitional B cells responded to intestinal bacteria in vitro [71], suggesting that B cells may be able to acquire a repertoire that recognizes commensals. Indeed, the microbiota drive early B cell development in the intestine, where receptor recombination and editing processes change the immunoglobulin repertoire. Colonization of GF mice increased Igλ-expression in intestinal B cells, an indication of receptor editing [72]. Furthermore, production of TNFα and inducible nitric oxide synthase by IgA+ plasma cells was dependent on signals from the microbiota [73]. Activation of immature B cells in gut-associated lymphoid tissue may also be important to remove autoreactive B cells and protect against autoimmunity [71]. Indeed, B cells in patients with systemic lupus erythematosus exhibited a defect in expression of the gut-homing β7 integrin receptor [71].
Innate Lymphoid Cells
In recent years, ILCs were found to be crucial for organogenesis, wound-healing, antimicrobial and anti-helminth defenses [74]. Among three groups of ILCs, RORγt+ group 3 ILCs (ILC3s) play an important role in maintaining intestinal homeostasis. Specifically, these RORγt+ NKp46+ cells secrete IL-22, which is critical for the production of the AMPs RegIIIβ and RegIIIγ. Moreover, commensal-driven IL-22 production by ILC3s was also critical for mucus production [20] and for defense against Citrobacter rodentium infection [21].
ILC3s can also act on innate immune cells to maintain homeostasis. As mentioned before, RORγt+ ILC3s, responding to microbiota-driven IL-1β production by macrophages, facilitate DC-mediated Treg induction by providing GM-CSF for DC differentiation [45]. Finally there seems to be an inhibitory crosstalk between some ILCs and CD4+ T cells. Indeed, Hepworth et al [75] showed that RORγt+ ILCs express MHC class II and are able to process and present antigen to CD4+ T cells to inhibit T cell proliferation, as disrupting this interaction resulted in commensal-driven spontaneous intestinal inflammation. Reciprocally, a recent study showed that CD4+ T cells in turn limited ILCs both in terms of their numbers and ability to produce IL-22 and AMPs, which were all elevated in mice lacking T cells or MHC class II[76].
The mechanism by which the microbiota regulate ILC3s is starting to be elucidated and implicates signals through the aryl hydrocarbon receptor (AhR). Qiu et al [77] showed that AhR signals are required for the postnatal maturation of the ILC compartment and that lacking AhR in ILCs reduced their production of IL-22. Moreover, ILC3 signaling through AhR was suggested to be responsible for the inhibition of T cell-mediated intestinal inflammation [78]. Diet-derived ligands could drive AhR activation and induce the production of IL-22 [79]. In addition, indole-3-aldehyde, a tryptophan metabolite produced by Lactobacillus reuteri, could induce IL-22 production in NKp46+ cells and confer protection against fungal infection [80].
The Case of the Liver
The liver is generally considered to be devoid of microbial colonization. However, it receives blood from the portal vein, which drains the intestine, such that it is likely constantly exposed to intestinal microbial products. TLRs are known to be expressed by different immune cells in the liver [81], and it may be evolutionarily beneficial for the liver to develop unique local strategies to prevent continuous inflammation.
A recent study showed that the liver serves as a vascular firewall to capture and clear microbes during intestinal pathology [82]. Patients with nonalcoholic fatty liver diseases had increased systemic immune responses specific for commensal bacteria, implicating the normal liver in the prevention of these responses. In support of this idea, Lunz et al. showed that commensal bacteria increase the activation threshold of hepatic DCs through the IL-6-STAT3 axis in mice, thus diminishing possible downstream adaptive immune activation [83]. Reducing signals from the microbiota by antibiotic treatment resulted in increased functional capacity of hepatic DCs, though the pathological consequences of this remain to be explored. Additionally, the numbers of Kupffer cells, a type of liver-resident macrophage, are directly influenced by the microbiota in a manner dependent on intracellular adhesion molecule-1 on liver sinusoidal endothelium [84]. This suggests that the microbiota may play a role in the seeding and maturation of tissue resident macrophage precursors.
The specific microbial candidates that affect hepatic immune function are largely unknown. In a recent clinical study, where metagenomic data on gut microbiota were collected from healthy individuals and patients with liver cirrhosis, differences in microbial species were observed between the two cohorts [85]. However, it remains to be determined whether the disease state of the patients was a cause or a consequence of the altered microbiota.
Systemic Effects of Intestinal Microbiota
Arthritis
In addition to the local influence of the intestinal microbiota, some studies support a systemic role for gut commensals [86]. For instance, in a genetic mouse model of spontaneous arthritis, GF mice displayed a reduction in autoantibodies, autoantibody-secreting cells and Th17 cells, which was associated with an attenuation of arthritis [87]. Reconstitution with SFB restored the pathological state of these mice. It was suggested that SFB promote autoimmunity by altering the activation threshold of T cells, which makes them prone to activation by endogenous ligands [88]. SFB may not colonize humans, but intestinal Prevotella copri (P. copri) was associated with susceptibility to arthritis in patients [89] and increased the risk for chemically-induced colitis in mice, suggesting that other pro-inflammatory species may drive distal clinical diseases.
Metabolic Diseases
The intestinal microbiota have been associated with the occurrence of diabetes. In non-obese diabetes (NOD) mice, deficiency in MyD88 protected against the development of type 1 diabetes in conventional but not GF mice, suggesting that the microbiota in MyD88-deficient mice may inhibit autoimmunity [90]. Within the microbial community, SFB was shown to confer protection against insulitis in females but not males NOD mice [91]. In addition to this bacterial lineage affecting males and females differently, 2 studies suggest that species differentially represented in males versus females may be responsible for the higher prevalence of type 1 diabetes in female mice [92,93]. Indeed, castration in male mice narrowed the differences in the microbiota, suggesting hormonal shaping of microbial constituents [92]or hormonal-dependent responsiveness to microbial products [93].
The gut microbiota also affects the amount of energy harvested from the diet and consequentially can play a role in obesity [94]. A recent study in mice showed that lymphotoxin contributed to obesity by controlling the composition of the microbiota, perhaps in part by driving the expansion of SFB[95]. Conversely to lymphotoxin signaling, innate sensing through the NLRP6 and NLRP3 inflammasomes protected hosts from non-alcoholic fatty liver disease and obesity via reducing influx of microbial products to the liver and modulating microbial composition [96]. Accordingly, genetic defects in the inflammasome resulted in hepatic inflammation and obesity. These results suggest that a crosstalk between microbiota and immune system is essential to maintain a balanced metabolic state.
Immune Defense
In healthy individuals, the microbiota may constantly calibrate and arm the immune system to be ready to fight potential infections. For example, the microbiota has been shown to confer protection against sepsis induced by systemic infection with Escherichia coli. Liu et al [97] demonstrated that antibiotic-induced dysbiosis resulted in reduced production of IL-17 and granulocyte-colony stimulating factor (G-CSF), and lethality from E coli. K1-mediated sepsis. Additionally, in a model of lung influenza infection, antibiotic-treated mice exhibited a reduction in influenza-specific CD4+ and CD8+ T cells, resulting in increased pulmonary viral titers [98]. This was associated with decreased migration of DCs after viral infection, an IL-1β- and inflammasome-dependent event, suggesting that the intestinal microbiota may participate in priming the distal immune system. Indeed, MNPs in GF mice were less capable of producing type I IFNs [99] and type II IFNs [100], which resulted in defects in antiviral immunity, implicating microbiota signals in making the distal immune system competent. Whether dysbiosis from antibiotic treatment also reduces this distal innate immunity in patients remains to be determined.
In immunocompromised patients, intestinal dysbiosis may occur as a result of antibiotic therapy and perhaps of reduced immune function, such that opportunistic pathogenic bacteria may rise and possibly translocate and provoke systemic infections [101]. Correction of microbial imbalances may help prevent outgrowth of pathogens and infectious complications.
Allergy
The hygiene hypothesis suggests that a shift in commensal flora over the past century may be responsible for the higher prevalence of allergic and autoimmune diseases, with the idea that modern humans may be missing protective microbes [102]. In support of microbes that can suppress allergic responses, the local lung microbiota has been shown to facilitate tolerance to allergens [103]. In addition, it has been suggested that the intestinal microbiota can also reduce airway allergic responses by producing short-chain fatty acids that can drive generation of lung-seeding DC precursors less capable of inducing Th2 differentiation, a cell type implicated in allergic responses [104].
Similarly, a new study reported that Clostridial groups could confer protection against food allergy in an IL-22 dependent manner [105]. In this study, GF or antibiotic-treated mice displayed elevated immune responses against an allergen and reconstituting GF mice with normal microbiota or a consortium of Clostridia restored protective immune responses. These studies suggest that the intestinal microbiota is important to prevent the development of allergic diseases.
Neurological Diseases
Recent research has also revealed a relationship between the intestinal microbiota and the central nervous system [106]. Presence of the microbiota exacerbated spontaneous experimental autoimmune encephalomyelitis (EAE), correlating with increased migration of autoreactive B cells to draining lymph nodes [107], greater proliferation of myelin-specific T cells and a decreased Treg population [108]. Interestingly, a recent study showed that the intestinal microbiota could also modulate psychological disorders that accompany neurodevelopmental defects, as treatment with B. fragilis mitigated defects in neurological behavior [109]. In particular, PSA from B. fragilis provided protection against demyelination in EAE via TLR2-mediated CD39 signaling in CD4+ T cells [110], reinforcing the idea that bacterial components from the intestinal microbiota have distal consequences though it is not known whether it is such components or the local effector cells they modulate that travel to distal regions to affect disease.
Antitumor Response
There is an increasing interest in the role of the microbiota on anti-tumor immune responses, including in the context of lymphoma [111], colorectal [112] and pancreatic cancer [113]. In 2 recent studies involving transplantable tumors, GF or antibiotic-treated mice were found to be refractory to CpG-oligonucleotide immunotherapy and platinum [114] and cyclophosphamide [115] chemotherapies. Iida and colleagues correlated the increased tumor growth with a reduction in TNFα and iNOS production in the myeloid cell compartment, whereas Viaud et al. found that cyclophosphamide promoted translocation of gram positive bacteria, driving Th17 and memory Th1 responses, a phenomenon impaired in GF and antibiotic-treated mice. These studies imply that a normal microbiota is necessary for responsiveness to anti-tumor therapies.
Transplantation
Little is known about the role of the intestinal microbiota on transplant outcomes. Lactobacilliales have been linked with better transplant survival and less severe graft-versus-host disease (GvHD) in clinical small bowel [116] and bone marrow [117] transplantation, respectively. Protection from GvHD-associated intestinal inflammation was recapitulated following Lactobacilliales reconstitution in mice [117], suggesting a causal role for this bacterial family in the protection against GvHD. As transplanted organs differ in their microbial colonization, an intriguing question in the field is whether organ colonization plays a role in the shorter half-life of colonized versus sterile allografts.
The studies described in this section have addressed the potential distal effects of the gut microbiota. However, one must keep in mind that GF mice lack microbial colonization on all barrier surfaces and that some oral antibiotics can be resorbed and target extra-intestinal flora. Thus, it is not certain in some cases whether effects observed are due to distal effects of the intestinal microbiota and/or to a more local action of the microbiota colonizing an adjacent barrier surface. Future studies will require experimental strategies to distinguish between the distal effects of intestinal microbiota and the local effects of microbiota residing at other barrier surfaces.
Microbiota-derived Therapeutics
In various mouse models discussed in this review, reconstitution of GF or dysbiotic animals with normal, or specific intestinal microbiota has been shown to rescue defects caused by disrupted microbiota. In the clinic, an important recent study has demonstrated the therapeutic potential of transferring healthy microbiota via fecal transplantation to cure refractory Clostridium difficile infection [118]. Thus, several applications of probiotic therapies can be cautiously considered as therapeutic options.
First, transfer of fecal material from healthy donors may be a therapeutic option to correct patients’ dysbiosis in various pathological conditions such as IBD or GvHD. However, the microbiota likely contains both pro- and anti-inflammatory species such that the net effect of a blind transfer of whole intestinal communities may be hard to predict. Moreover, the definition of a healthy donor would need to be articulated. These caveats may explain the variable results to date of fecal transplantation in IBD[119].
Second, more targeted transfers of particular bacterial species with known immune consequences are attractive to consider. However, it is important to keep in mind that, in mice, SFB is protective for autoimmune diabetes but detrimental for autoimmune arthritis [87] such that much more would need to be known about the genetic susceptibilities of patients, and of their microbial communities, before being able to anticipate beneficial or harmful effects of adding discrete bacterial members. In addition, a better understanding not only of the members of the microbiota but also of the functional properties of the genes they express may be needed as the latter seem to determine physiological states [120]. Additional factors such as diet may also be required to acquire the full potential of transferred microbiota and attain a desirable physiological state [121].
Third, microbiota-derived metabolites, such as SCFAs [52,104] and tryptophan catabolites [80,122] may be able to be used to specifically target particular compartments of the immune system.
Fourth, a better understanding of specific species that may drive disease in particular genetic backgrounds could prompt the development of highly specific narrow spectrum antibiotics. These may be less disruptive to the overall microbiota and thus less likely to result in outgrowth of pathogenic opportunistic bacteria.
Fifth, altering the diet, with its short- and long-term consequences on the microbiota [123–126] may have an important impact on maintenance of a poised but controlled immune system and therefore on health.
Concluding Remarks
The microbiota co-exist with the host in a unique symbiotic relationship. The complex crosstalk between the microbiota and the immune system is critical not only to maintain intestinal homeostasis, but also to prevent systemic pathology. An increasing number of microbial candidates or products are being discovered as a potential cause for certain physiological and pathologic states. Future studies will require a combination of metagenomic, metabolomic and gnotobiotic experiments to further advance the understanding of this intricate and complex relationship.
Acknowledgments
This work was supported by NIAID grant AI097113 to MLA.
Abbreviations
- AhR
aryl hydrocarbon receptor
- AMP
antimicrobial peptides
- APRIL
a proliferation-inducing ligand
- BAFF
B cell-activating factor
- DC
dendritic cell
- EAE
experimental autoimmune encephalomyelitis
- GALT
gut-associated lymphoid tissue
- GvHD
graft-versus-host disease
- HDAC
histone deacetylase
- IBD
inflammatory bowel disease
- IEC
intestinal epithelial cell
- ILC
innate lymphoid cell
- ILF
isolated lymphoid follicles
- iTreg
induced regulatory T cell
- mLN
mesenteric lymph node
- MNP
mononuclear phagocyte
- NOD
non-obese diabetic
- PP
Peyer’s Patches
- PSA
polysaccharide A
- SCFA
short-chain fatty acid
- sIgA
immunoglobulin A
- SLO
secondary lymphoid organs
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nature Publishing Group. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Micro. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nature Publishing Group. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randall TD, Mebius RE. The development and function of mucosal lymphoid tissues: a balancing act with micro-organisms. 2014;7:455–466. doi: 10.1038/mi.2014.11. [DOI] [PubMed] [Google Scholar]
- 8.HOROWITZ RE, BAUER H, PARONETTO F, ABRAMS GD, WATKINS KC, POPPER H. THE RESPONSE OF THE LYMPHATIC TISSUE TO BACTERIAL ANTIGEN. STUDIES IN GERMFREE MICE. The American Journal of Pathology. 1964;44:747–761. [PMC free article] [PubMed] [Google Scholar]
- 9.Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 10.Medzhitov R. Decoding the Patterns of Self and Nonself by the Innate Immune System. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunology. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nature Publishing Group. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 14.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USa. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, et al. Mucus Enhances Gut Homeostasis and Oral Tolerance by Delivering Immunoregulatory Signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunology. 2014:1–13. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang J-P, Brown EM, et al. NLRP6 Inflammasome Orchestrates the Colonic Host- Microbial Interface by Regulating Goblet Cell Mucus Secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergstrom KSB, Guttman JA, Rumi M, Ma C, Bouzari S, Khan MA, et al. Modulation of Intestinal Goblet Cell Function during Infection by an Attaching and Effacing Bacterial Pathogen. Infection and Immunity. 2008;76:796–811. doi: 10.1128/IAI.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, et al. Antibiotic Treatment Alters the Colonic Mucus Layer and Predisposes the Host to Exacerbated Citrobacter rodentium-Induced Colitis. Infection and Immunity. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL- 22. Nature Publishing Group. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 21.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial Flora Drives Interleukin 22 Production in Intestinal NKp46+ Cells that Provide Innate Mucosal Immune Defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Cash HL. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII and protect mice against intestinal Listeria monocytogenes infection. Journal of Experimental Medicine. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host and Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Boullier S, Tanguy M, Kadaoui KA, Caubet C, Sansonetti P, Corthesy B, et al. Secretory IgA-Mediated Neutralization of Shigella flexneri Prevents Intestinal Tissue Destruction by Down-Regulating Inflammatory Circuits. The Journal of Immunology. 2009;183:5879–5885. doi: 10.4049/jimmunol.0901838. [DOI] [PubMed] [Google Scholar]
- 27.Benjamin JL, Sumpter R, Jr, Levine B, Hooper LV. Intestinal Epithelial Autophagy Is Essential for Host Defense against Invasive Bacteria. Cell Host and Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song-Zhao GX, Srinivasan N, Pott J, Baban D, Frankel G, Maloy KJ. Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. 2013;7:763–774. doi: 10.1038/mi.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordlander S, Pott J, Maloy KJ. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. 2013;7:775–785. doi: 10.1038/mi.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knodler LA, Crowley SM, Sham HP, Yang H, Wrande M, Ma C, et al. Noncanonical Inflammasome Activation of Caspase-4/Caspase-11 Mediates Epithelial Defenses against Enteric Bacterial Pathogens. Cell Host and Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, et al. Rapid fucosylation of intestinal epithelium sustains hostcommensal symbiosis in sickness. Nature. 2014:1–16. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 33.Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO, et al. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature. 2014:1–19. doi: 10.1038/nature13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rescigno M. Dendritic cell-epithelial cell crosstalk in the gut. Immunol Rev. 2014;260:118–128. doi: 10.1111/imr.12181. [DOI] [PubMed] [Google Scholar]
- 35.Macpherson AJ. Induction of Protective IgA by Intestinal Dendritic Cells Carrying Commensal Bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 36.Cerovic V, Bain CC, Mowat AM, Milling SWF. Intestinal macrophages and dendritic cells: what’s the difference? Trends in Immunology. 2014;35:270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 38.Niess JH. CX3CR1-Mediated Dendritic Cell Access to the Intestinal Lumen and Bacterial Clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 39.Diehl GE, Longman RS, Zhang J-X, Breart B, Galan C, Cuesta A, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. Journal of Experimental Medicine. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossini V, Zhurina D, Radulovic K, Manta C, Walther P, Riedel CU, et al. CX3CR1+ cells facilitate the activation of CD4 T cells in the colonic lamina propria during antigen-driven colitis. Mucosal Immunology. 2014;7:533–548. doi: 10.1038/mi.2013.70. [DOI] [PubMed] [Google Scholar]
- 42.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1+ Macrophages to CD103+ Dendritic Cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Coombes JL, Siddiqui KRR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF- and retinoic acid dependent mechanism. Journal of Experimental Medicine. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J, et al. Dendritic Cell Expression of the Signaling Molecule TRAF6 Is Critical for Gut Microbiota-Dependent Immune Tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343:1249288–1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USa. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 49.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USa. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013:1–7. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 52.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013:1–7. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 54.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013:1–6. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nutsch KM, Hsieh C-S. T cell tolerance and immunity to commensal bacteria. Current Opinion in Immunology. 2012;24:385–391. doi: 10.1016/j.coi.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, et al. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 60.Blaschitz C, Raffatellu M. Th17 Cytokines and the Gut Mucosal Barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, et al. Reciprocal TH17 and Regulatory T Cell Differentiation Mediated by Retinoic Acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 62.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented Filamentous Bacteria Antigens Presented by Intestinal Dendritic Cells Drive Mucosal Th17 Cell Differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute Gastrointestinal Infection Induces Long-Lived Microbiota-Specific T Cell Responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infection and Immunity. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lécuyer E, Rakotobe S, Lengliné-Garnier H, Lebreton C, Picard M, Juste C, et al. Segmented Filamentous Bacterium Uses Secondary and Tertiary Lymphoid Tissues to Induce Gut IgA and Specific T Helper 17 Cell Responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 67.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 68.Hirota K, Turner J-E, Villa M, Duarte JH, Demengeot J, Steinmetz OM, et al. Plasticity of TH17 cells in Peyer’s patches is responsible for the induction of T cell–dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 70.Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, Seleznik GM, et al. Nonredundant Function of Soluble LT 3 Produced by Innate Lymphoid Cells in Intestinal Homeostasis. Science. 2013;342:1243–1246. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- 71.Vossenkamper A, Blair PA, Safinia N, Fraser LD, Das L, Sanders TJ, et al. A role for gut-associated lymphoid tissue in shaping the human B cell repertoire. Journal of Experimental Medicine. 2013;210:1665–1674. doi: 10.1084/jem.20122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, Rubino S, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells — how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 75.Hepworth MR, Monticelli LA, Fung TC, Ziegler CGK, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korn LL, Thomas HL, Hubbeling HG, Spencer SP, Sinha R, Simkins HM, et al. Conventional CD4+ T cells regulate IL-22-producing intestinal innate lymphoid cells. Mucosal Immunology. 2014;7:1045–1057. doi: 10.1038/mi.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qiu J, Heller JJ, Guo X, Chen Z-ME, Fish K, Fu Y-X, et al. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu J, Guo X, Chen Z-ME, He L, Sonnenberg GF, Artis D, et al. Group 3 Innate Lymphoid Cells Inhibit T-Cell-Mediated Intestinal Inflammation through Aryl Hydrocarbon Receptor Signaling and Regulation of Microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Veldhoen M, Brucklacher-Waldert V. Dietary influences on intestinal immunity. Nat Rev Immunol. 2012;12:696–708. doi: 10.1038/nri3299. [DOI] [PubMed] [Google Scholar]
- 80.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 81.Kanai T. Role of Toll-like receptors in immune activation and tolerance in the liver. 2014:1–8. doi: 10.3389/fimmu.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balmer ML, Slack E, de Gottardi A, Lawson MAE, Hapfelmeier S, Miele L, et al. The Liver May Act as a Firewall Mediating Mutualism Between the Host and Its Gut Commensal Microbiota. Science Translational Medicine. 2014;6:237ra66–237ra66. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- 83.Lunz JG, III, Specht SM, Murase N, Isse K, Demetris AJ. Gut-derived commensal bacterial products inhibit liver dendritic cell maturation by stimulating hepatic interleukin-6/signal transducer and activator of transcription 3 activity. Hepatology. 2007;46:1946–1959. doi: 10.1002/hep.21906. [DOI] [PubMed] [Google Scholar]
- 84.Corbitt N, Kimura S, Isse K, Specht S, Chedwick L, Rosborough BR, et al. Gut Bacteria Drive Kupffer Cell Expansion via MAMP-Mediated ICAM-1 Induction on Sinusoidal Endothelium and Influence Preservation- Reperfusion Injury after Orthotopic Liver Transplantation. The American Journal of Pathology. 2013;182:180–191. doi: 10.1016/j.ajpath.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014:1–19. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 86.Chervonsky AV. Influence of microbial environment on autoimmunity. Nature Publishing Group. 2009;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 87.Wu H-J, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut- Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chappert P, Bouladoux N, Naik S, Schwartz RH. Specific Gut Commensal Flora Locally Alters T Cell Tuning to Endogenous Ligands. Immunity. 2013;38:1198–1210. doi: 10.1016/j.immuni.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202–e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kriegel MA, Sefik E, Hill JA, Wu H-J, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USa. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender Bias in Autoimmunity Is Influenced by Microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 94.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 95.Upadhyay V, Poroyko V, Kim T-J, Devkota S, Fu S, Liu D, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nature Publishing Group. 2012:1–8. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2013;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nature Medicine. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USa. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 100.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Current Opinion in Infectious Diseases. 2013;26:332–337. doi: 10.1097/QCO.0b013e3283630dd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blaser MJ. Missing Microbes: How the Overuse of Antibiotics Is Fueling Our Modern Plagues. 1. Henry Holt and Co; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gollwitzer ES, Saglani S, Trompette AEL, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nature Medicine. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 104.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Medicine. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 105.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain, Behaviour, and Immunity. 2014:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 108.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell. 2013:1–13. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, Telesford KM, Ochoa-Repáraz J, Haque-Begum S, Christy M, Kasper EJ, et al. An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signalling. Nature Communications. 2014;5:4432. doi: 10.1038/ncomms5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamamoto ML, Schiestl RH. Intestinal microbiome and lymphoma development. Cancer J. 2014;20:190–194. doi: 10.1097/PPO.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer J. 2014;20:225–231. doi: 10.1097/PPO.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zambirinis CP, Pushalkar S, Saxena D, Miller G. Pancreatic cancer, inflammation, and microbiome. Cancer J. 2014;20:195–202. doi: 10.1097/PPO.0000000000000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Viaud S, re RDE, Boneca IG, Lepage P, Langella P, Chamaillard M, et al. Gut microbiome and anticancer immune response: really hot Sh*t! Cell Death and Differentiation. 2014:1–16. doi: 10.1038/cdd.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oh PL, Martínez I, Sun Y, Walter J, Peterson DA, Mercer DF. Characterization of the Ileal Microbiota in Rejecting and Nonrejecting Recipients of Small Bowel Transplants. American Journal of Transplantation. 2011;12:753–762. doi: 10.1111/j.1600-6143.2011.03860.x. [DOI] [PubMed] [Google Scholar]
- 117.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. Journal of Experimental Medicine. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 119.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: A systematic review and meta-analysis. Journal of Crohn’s and Colitis. 2014:1–13. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. The Journal of Immunology. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 124.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]