Abstract
Sequencing studies have identified many recurrent coding mutations in human cancer genes; however, highly recurrent mutations involving regulatory regions have rarely been observed. Here we describe two independent mutations within the TERT core promoter that, when taken together, were observed in 71% (50 of 70) of melanomas and generate de novo consensus ETS binding motifs. Reporter assays showed that these mutations increase transcriptional activity from the TERT promoter by 2–4-fold. Examination of cancer cell lines derived from diverse tumor types revealed the same mutations in 16% (24 of 150) of cases, with preliminary evidence of elevated frequency in bladder and hepatocellular cancer cells. Thus, somatic mutations in regulatory regions of the genome may represent an important tumorigenic mechanism.
Systematic characterization of human cancer genomes has led to the discovery of a wide range of mutated genes that contribute to tumor development and progression. Most of the somatic mutations in tumors reside within the protein-coding regions of genes or at splice junctions. To determine whether tumor genomes harbor recurrent mutations outside of protein-coding regions, we systematically queried noncoding somatic mutations using published whole-genome sequencing data.
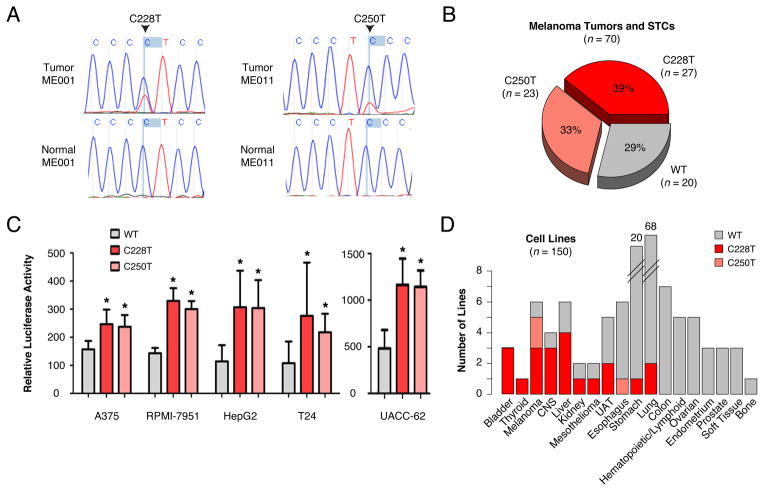
Analysis of whole-genome sequencing data from malignant melanomas (1, 2) revealed two somatic telomerase reverse transcriptase (TERT) gene promoter mutations in 17 of 19 (89%) cases examined. The average sequence coverage at the TERT promoter locus was 30-fold in normal samples and 60-fold in tumor samples (fig. S1A). Each of these promoter mutations resulted in a cytidine-to-thymidine transition at a dipyrimidine motif indicative of ultraviolet (UV) light–induced damage (chr5, 1,295,228 C>T and 1,295,250 C>T; hereafter termed C228T and C250T, respectively), and both mutations localized within 100 base pairs (bp) of the TERT transcriptional start site (TSS) (mean allelic fraction, 0.32; range, 0.07 to 0.55) (table S1). We validated these mutations by means of polymerase chain reaction and Sanger sequencing tumor/normal sample pairs from both the discovery set (Fig. 1A and fig. S1, B and C) and an extension set of 51 additional melanoma tumor/normal sample pairs. Within this extension set, 33 tumors (65%) harbored one of the mutations. Moreover, the mutations were mutually exclusive in both the discovery and extension sets (P = 5.4 × 10−7, Fisher’s one-sided exact test). Two tumors with a C228T transition also contained an adjacent C>T transition (at position chr5, 1,295,229), which is indicative of a dinucleotide CC>TT transition. Together, these TERT promoter mutations were observed in 50 of 70 (71%; 95% confidence interval: 59 to 82%, Clopper-Pearson method) melanomas examined (Fig. 1B and table S1).
Fig. 1. Identification of TERT promoter mutations in melanoma and cancer cell lines.
(A) Sequence chromatograms of matched tumor and normal DNA representing somatic mutations chr 5: 1,295,228 C>T (C228T) and chr 5: 1,295,250 C>T (C250T) in the TERT promoter locus.
(B) Pie chart of C228T and C250T somatic mutation status in 70 surveyed melanoma tumors and short-term cultures. Sum of percentages is greater than 100% due to rounding.
(C) Luciferase reporter assays for transcriptional activity from the TERT core promoter (−200 to +73) with either the C228T or C250T mutation compared to wild-type promoter in A375, RPMI-7951, UACC-62, T24 or HepG2 cell lines. The results depicted are the average of at least 3 independent experiments. Values are mean ± s.d. * P < 0.05.
(D) Bar plot of 150 cancer cell lines of the Cancer Cell Line Encyclopedia (3) depicting TERT promoter mutation status. Individual bars represent the total number of cell lines of a given tumor type interrogated for C228T and C250T mutations, with mutation status indicated by colors defined in the legend.
Both C228T and C250T generated an identical 11-bp nucleotide stretch (5′-CCCCTTCCGGG-3′) containing a consensus binding site for E-twenty-six (ETS) transcription factors (GGAA, reverse complement) within the TERT promoter region. Because ETS transcription factors may become activated through dysregulation of mitogen-activated protein kinase (MAP kinase) signaling, we hypothesized that these promoter mutations might augment gene expression. To test this hypothesis, we used a reporter assay system in which the relevant portion of the mutant or wild-type TERT core promoter was cloned upstream of the firefly luciferase gene (2). Here, we tested both a core promoter fragment (−132 to +5 relative to the TSS) and the full core promoter (−200 to +73). In comparison to the wild-type TERT promoter, both mutations conferred approximately two- to fourfold increased transcriptional activity in five distinct cell line contexts (Fig. 1C and fig. S1D). Thus, each mutation was capable of augmenting transcriptional activity from the TERT promoter.
To investigate whether similar TERT promoter mutations occur in other cancer types, we examined sequencing data from this locus in 150 cell lines from the Cancer Cell Line Encyclopedia (CCLE) (3). Overall, 24 CCLE lines (16%) contained either C228T or C250T (mean allelic fraction, 0.61; range, 0.17 to 1.00) (table S1). An increased frequency in melanoma was again noted (five of six lines tested), with additional evidence suggesting possible heightened prevalence (>25%; one-sided 95% confidence interval) in bladder (three of three lines) and hepatocellular cancer cell lines (four of six lines) (Fig. 1D).
Several lines of evidence support the hypothesis that these promoter mutations may function as driver events that contribute to oncogenesis through TERT dysregulation and undergo positive selection, at least in human melanoma. First, the TERT promoter mutations showed a combined frequency that exceeded those of BRAF and NRAS mutations, which activate known melanoma driver oncogenes (4, 5). In an analysis restricted to somatic mutations present at an allelic fraction of 0.2 or greater [to reduce artifacts of mutation calling (1)], the four most recurrent melanoma nucleotide substitutions included BRAF [chr7, 140,453,136 A>T (V600E)], NRAS [chr1, 115,256,529 T>C (Q61R)], and the TERT core promoter mutations C228T and C250T. Second, although highly recurrent, C228T and C250T occurred in a wholly mutually exclusive fashion. This suggests the possibility that the mutations might be functionally redundant. Third, the absence of other recurrent somatic mutations in the 3 kb upstream of the TERT transcription start site in the queried melanomas (1) coupled with the absence of the described TERT promoter mutations in 24 lung adenocarcinomas with comparably high somatic mutation rates (6) reduces the possibility that these recurrent TERT promoter mutations are solely due to an increased background mutation rate at this locus. Although the role of telomerase in tumorigenesis is well established, details regarding its dysregulation in cancer cells remain incompletely understood, particularly in melanoma (7). The TERT promoter mutations identified here may link telomerase gene regulation and tumorigenic activation in this malignancy. The high prevalence of C228T and C250T suggests that these TERT promoter mutations may comprise early genetic events in the genesis of melanoma and other cancer types. Although TERT expression alone is not sufficient to bypass oncogene-induced senescence, genomic TERT activation may potentiate mechanisms by which melanocytes achieve immortalization in the setting of oncogenic mutations (8). These results therefore suggest that renewed efforts to develop clinically effective telomerase inhibitors may be warranted.
At the same time, promoter mutations likely represent only one potential mechanism of TERT reactivation in a subset of human cancers. Indeed, recurrent chromosomal copy gains spanning the TERT locus have been described previously for several cancers, including melanoma (9, 10).
Highly recurrent somatic mutations within a cancer gene promoter region have not previously been described. Similarly, the de novo mutational generation of transcription factor binding motifs in tumor genomes was heretofore unknown, although an ETS transcription factor binding motif was previously associated with a single-nucleotide polymorphism insertion at the MMP-1 locus (11). Together, these findings raise the possibility that recurrent somatic mutations involving regulatory regions, in addition to coding sequences, may represent important driver events in cancer.
Supplementary Material
(A) Representative screenshot of TERT promoter mutations chr 5: 1,295,228 C>T (C228T) and chr 5: 1,295,250 C>T (C250T) from Integrative Genomics Viewer. Average depth of coverage in the 19 melanoma tumor-normal pairs with whole genome sequence coverage at the relevant loci was 58x in the tumor and 30x in the normal at chr 5: 1,295,228 and 61x in the tumor and 30x in the normal at chr 5: 1,295,250, with minimum base quality score of 30 and minimum read mapping quality of 60.
(B) Additional sequence chromatograms of matched tumor and normal DNA representing somatic mutations C228T and C250T in the TERT promoter locus.
(C) Subcloning of TERT core promoter mutations C228T and C250T. Sequence chromatograms depict the reverse complement G>A transition.
(D) Luciferase reporter assays for transcriptional activity from a portion of the TERT core promoter (−132 to +5) with either the C228T or C250T mutation compared to wild-type promoter in A375, T24 or HepG2 cell lines. The results depicted are the average of at least 3 independent experiments. Values are mean ± s.d. * P < 0.01.
Melanoma samples and cell lines are listed with indicated TERT promoter mutation. Two samples had a dinucleotide mutation CC>TT causing C228T and C229T, were counted as C228T samples, and also generated a consensus ETS motif.
One Sentence Summary.
This report describes the identification of two recurrent mutations in the TERT promoter in human melanoma that are also observed in other cancer types.
Acknowledgments
We thank S.N. Wagner, D. Schadendorf, J.A. Wargo and D.S.B. Hoon for contribution of samples; K. Cibulskis for assistance with whole genome sequence data analysis; and E. Nickerson for coordination of validation/extension studies. This work was supported by NIH grant T32 CA009172 (F.W.H.), the Mittelman Family Fellowship (F.W.H.), NIGMS T32GM07753 (E.H.), the American Cancer Society (M.J.X.), NIH New Innovator Award DP2OD002750 (L.A.G.), NCI R33CA126674 (L.A.G.), the Novartis Institutes for Biomedical Research (G.K., L.A.G.), the Melanoma Research Alliance (L.A.G.), and the Starr Cancer Consortium (L.C., L.A.G.). L.A.G. is on the Scientific Advisory Board of Millennium Pharmaceuticals; is a paid consultant for and holds equity in Foundation Medicine, a company developing cancer genome-based diagnostic tests; and is a paid consultant for and sponsored research recipient from Novartis. The BAM files for the CCLE WGS data corresponding to the TERT locus can be accessed at ftp://tertguest:broad@ftp.broadinstitute.org/.
Footnotes
References and Notes
- 1.Berger M, et al. Nature. 2012;485:502. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Materials and methods are available as supplementary materials on Science Online.
- 3.Barretina J, et al. Nature. 2012;483:603. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodis E, et al. Cell. 2012;150:251. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krauthammer M, et al. Nature Genetics. 2012;44:1006. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imielinski M, et al. Cell. 2012;250:1107. [Google Scholar]
- 7.Bennett DC. Pigment Cell Melanoma Res. 2008;21:27. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- 8.Michaloglou C. Nature. 2005;436:720. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 9.Zhang A, et al. Cancer Res. 2000;22:6320. [Google Scholar]
- 10.Pirker C, et al. Melanoma Research. 2003;13:483. doi: 10.1097/00008390-200310000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Rutter JL, et al. Cancer Res. 1998;58:5321. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative screenshot of TERT promoter mutations chr 5: 1,295,228 C>T (C228T) and chr 5: 1,295,250 C>T (C250T) from Integrative Genomics Viewer. Average depth of coverage in the 19 melanoma tumor-normal pairs with whole genome sequence coverage at the relevant loci was 58x in the tumor and 30x in the normal at chr 5: 1,295,228 and 61x in the tumor and 30x in the normal at chr 5: 1,295,250, with minimum base quality score of 30 and minimum read mapping quality of 60.
(B) Additional sequence chromatograms of matched tumor and normal DNA representing somatic mutations C228T and C250T in the TERT promoter locus.
(C) Subcloning of TERT core promoter mutations C228T and C250T. Sequence chromatograms depict the reverse complement G>A transition.
(D) Luciferase reporter assays for transcriptional activity from a portion of the TERT core promoter (−132 to +5) with either the C228T or C250T mutation compared to wild-type promoter in A375, T24 or HepG2 cell lines. The results depicted are the average of at least 3 independent experiments. Values are mean ± s.d. * P < 0.01.
Melanoma samples and cell lines are listed with indicated TERT promoter mutation. Two samples had a dinucleotide mutation CC>TT causing C228T and C229T, were counted as C228T samples, and also generated a consensus ETS motif.