Abstract
Nipple aspirate fluid (NAF) is a noninvasively obtained biofluid from the duct openings of the breast. NAF components are constantly secreted, metabolized, and reabsorbed by the epithelial lining of the lactiferous ducts of the breast. NAF has been studied as a potential breast tissue surrogate for the discovery of novel breast cancer risk, early detection, and treatment response biomarkers. We report the first unsupervised metabolite characterization of nipple aspirate fluid using NMR and GC-MS using convenience samples previously collected from four premenopausal and four postmenopausal women. A total of 38 metabolites were identified using the two analytical techniques, including amino acids, organic acids, fatty acids, and carbohydrates. Analytical reproducibility of metabolites in NAF by GC-MS was high across different extraction and analysis days. Overall, 31 metabolites had a coefficient of variation below 20%. By GC-MS, there were eight metabolites unique to NAF, 19 unique to plasma, and 24 shared metabolites. Correlative analysis of shared metabolites between matched NAF and plasma samples from pre- and postmenopausal women shows almost no correlations, with the exception being lactic acid, which was significantly negatively correlated (R2 = 0.57; P = 0.03). These results suggest that NAF is metabolically distinct from plasma and that the application of metabolomic strategies may be useful for future studies investigating breast cancer risk and intervention response biomarkers.
Keywords: metabolomics, metabonomics, nipple aspirate fluid, breast cancer, GC-MS, NMR
INTRODUCTION
With the exception of genetic testing in high risk women and mammography-based detection of benign proliferative diseases (i.e., atypical ductal hyperplasia, lobular carcinoma in situ, and ductal carcinoma in situ), there are currently no minimally invasive biological markers in routine use for risk assessment or early detection of breast cancer in average risk individuals.1 Nipple aspirate fluid (NAF) is a biological fluid produced by the breast ductal epithelium that can be collected from the breast nipple by gentle aspiration in 36–98% of healthy, nonlactating women.2–4 The breast is first massaged before NAF is aspirated from the nipple using a hand-held suction cup. Microcapillary tubes are used to collect the fluid, which is then diluted with phosphate buffered saline (PBS) to ease handling. The actual volume of undiluted NAF obtained appears to vary between women. Maskarinec et al. reported a mean NAF volume of ~33 μL, though 15% of the women in the study were able to produce >90 μL.3 A more invasive procedure called ductal lavage has also been described, involving the use of a microcatheter to cannulate ductal orifices on the nipple.5 Although ductal lavage and ductoscopy contain higher cellularity, the volume of NAF collected is not improved using these procedures. These procedures are painful for participants and so not attractive for prevention. Further, they need to be performed by someone with a medical degree (physician or nurse), whereas NAF can be collected by any trained clinical study personnel.6
NAF contains cells and extracellular fluid produced within the breast duct. NAF composition includes high concentrations of proteins and metabolites produced, secreted, and reabsorbed predominantly by the epithelial lining of the ductal/alveolar system.7,8 Our group, as well as others, has characterized drug, protein, and hormone levels in NAF under the premise that NAF, as a biological fluid produced in the breast duct, more closely reflects the products of breast tissue metabolism and exposures than does the study of plasma or serum factors.9–12 Cytological evaluation of the epithelial cells in NAF is very specific for breast cancer detection; however, it has not proven to be sufficiently sensitive to be clinically useful.9 Arguably though, with the improved understanding of the molecular heterogeneity of breast cancer and advances in more sensitive “omics” technologies, the study of NAF is re-emerging as a viable biological compartment for breast tissue biomarker discovery under minimally invasive conditions.
NAF has a high protein content (range 1–200 mg/mL).13 As such, there have been efforts to characterize NAF-related proteins.14 Since the introduction of mass spectrometry (MS)-based proteomics, there have been a few reports on the protein composition of NAF and potential use for biomarker discovery. Using surface-enhanced laser desorption ionization (SELDI) time-of-flight (TOF) MS, Pawlik et al. described 17 “protein” peaks (identity unknown) that were overexpressed in the cancer-bearing breast of patients (N = 23) compared to those of healthy matched volunteers (N = 5) (p < 0.0005).15 The same group identified 39 unique proteins that were differentially expressed in the tumor-bearing side compared to the contralateral unaffected breast using isotope-coded affinity tag tandem mass spectrometry (ICAT-MS).16 Sauter et al. found seven candidate protein masses in NAF using SELDI-TOF-MS that were predictive of breast cancer in a prospective clinical trial.17 While promising, these findings have not been replicated and proteomic profiling of NAF has failed to advance any candidate breast cancer biomarkers into clinical practice.
There is now considerable interest in metabolomic approaches applied to plasma and serum for the discovery of risk and response biomarkers for applications in breast cancer;18–20 however, to our knowledge, there are no reports describing the NAF metabolome. Here we describe the analysis of NAF collected from healthy women participating in an early phase clinical trial21 using nuclear magnetic resonance (NMR) and gas chromatography mass spectrometry (GC-MS). Our results demonstrate the feasibility of obtaining metabolic profiles in NAF by GC-MS and NMR. We describe some of the challenges of working with this highly proteinaceous biofluid and demonstrate that, similar to protein studies, the metabolic profile of NAF is distinct from that of matched plasma samples.
RESULTS AND DISCUSSION
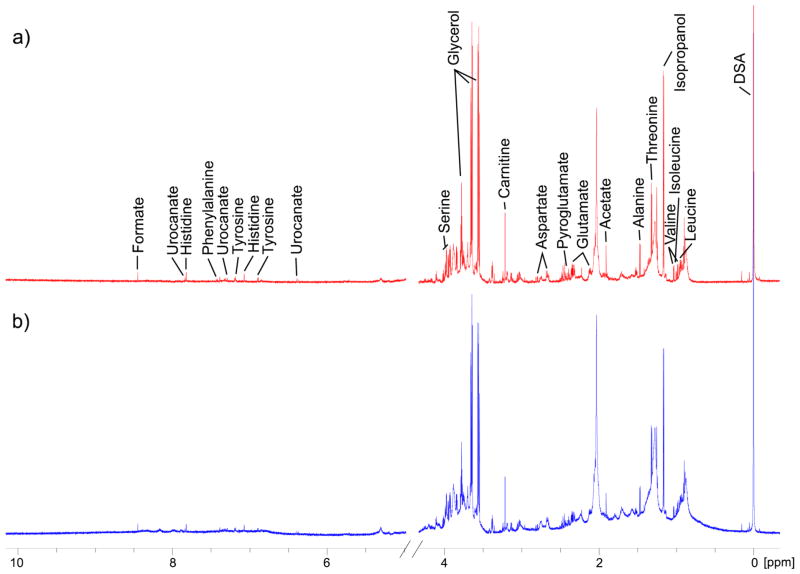
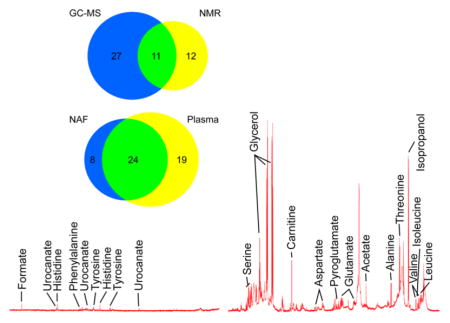
In order to avoid diluting the NAF sample further, small volume NMR microtubes were used and spectra were obtained on an 800 MHz NMR spectrometer with a cryoprobe to increase sensitivity. The 1H NMR spectra of NAF, annotated for assigned metabolites, are shown in Figure 1. Large broad peaks from lipoproteins/glycoproteins were clearly present (δ 0.8, 1.2, and 2.1 ppm) in the 1D “Noesy-presat” experiment, but a CPMG experiment (see Materials and Methods) improved the baseline, aiding metabolite identification.22 Currently, we have identified 24 metabolites from the pooled NAF sample using a Chenomx library, and their concentrations relative to the internal standard DSS are reported in Table 1. Glycerol (glycerin) was the most abundant metabolite as measured by NMR, likely from the high levels contained in the lotion used for breast massage. Other identified metabolites largely consisted of amino acids, organic acids, and carbohydrates. Apart from the unusually high levels of glycerol, the metabolite coverage was similar to that typically observed for human plasma.22 Isopropyl alcohol was present at fairly high concentrations, but this is likely to be contamination from the alcohol swab performed prior to NAF collection. Clearly, for future metabolite profiling studies, care should be taken to limit the sources of contamination from the NAF collection procedure as much as possible.
Figure 1.
1H NMR spectrum (800 MHz) of a pooled NAF sample showing assigned metabolites. (a) Carr–Purcell–Meiboom–Gill (CPMG) experiment; (b) 1D Noesy-presat experiment.
Table 1.
Metabolite Concentrations from the 1H NMR Analysis of a Pooled NAF Samplea
| metabolite | concentration (mM) |
|---|---|
| acetate | 0.77 |
| alanine | 1.66 |
| arabinitol | 4.61 |
| aspartic acid | 1.20 |
| carnitine | 0.66 |
| formate | 0.98 |
| glutamic acid | 3.74 |
| glutamine | 1.55 |
| glycerol | 20.52 |
| glycine | 1.98 |
| histidine | 0.42 |
| isoleucine | 0.57 |
| isopropyl alcohol | 3.71 |
| lactic acid | 3.78 |
| leucine | 0.65 |
| phenylalanine | 0.20 |
| pyroglutamic acid | 0.92 |
| serine | 4.42 |
| threonine | 2.23 |
| tryptophan | 0.05 |
| tyrosine | 0.29 |
| urocanate | 0.000489 |
| valine | 0.000884 |
| cis-aconitate | 0.000179 |
The concentrations have been adjusted to reflect actual metabolite concentrations in NAF before dilution with phosphate-buffered saline.
The high protein content of NAF combined with the low sensitivity of NMR and overlapping broad macromolecule peaks presented a challenge for NMR metabolite analysis. Further, the low sample volume of NAF limited the detectability of many metabolites that we typically identify in plasma and other biofluids by NMR where larger sample volumes are available. Therefore, it was determined that mass spectrometry might be a more suitable analytical platform for NAF metabolite measurements.
Using GC-MS, we next investigated the intra- and interday variation of both the extraction and the analytical protocols. From the pooled NAF sample, 12 aliquots were prepared for separate metabolite extraction as follows: 6 on one day and 6 on a following day. These samples were then split and analyzed by GC-MS on separate days. Specifically, for a given run day, 3 samples from extraction day 1 and 3 samples from extraction day 2 were included.
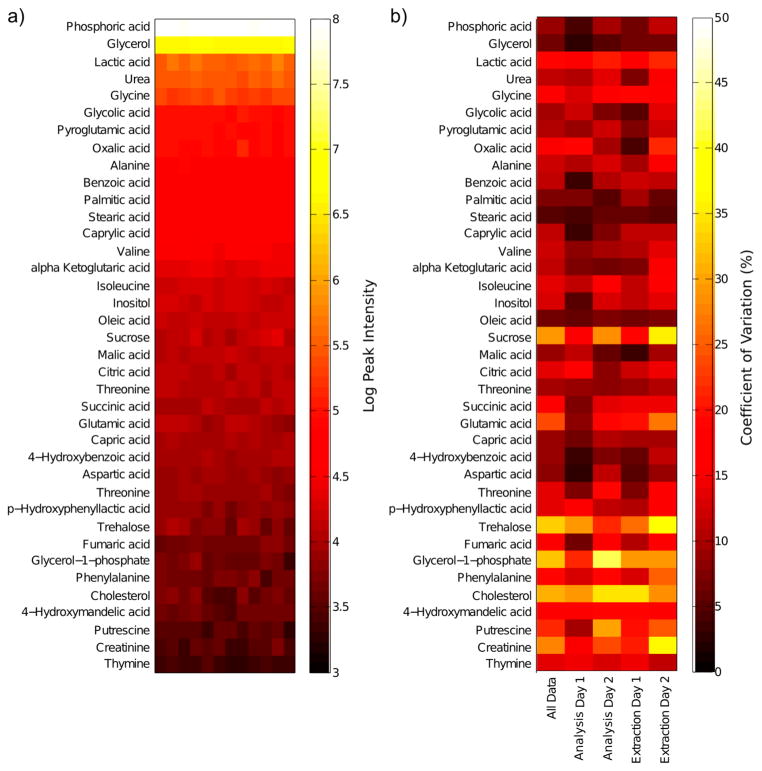
Currently, we have identified 38 metabolites from the GC-MS analysis of NAF using the commercial Fiehn library. Peak intensities for the metabolites that are currently assigned are shown in Figure 2a, sorted by their mean peak intensity to give an indication of their levels in NAF. The peak intensities for a large number of metabolites were highly reproducible for metabolite extraction and GC-MS analysis, for both within and between days (Figure 2b). A total of 31 metabolites had coefficients of variation (CV) below 20%, with lower intensity peaks showing greater variation.
Figure 2.
GC-MS data for the analysis of a pooled NAF sample. Multiple aliquots from the same sample were analyzed to determine intra- and interday variation of the extraction and the GC-MS analysis protocols. (a) GC-MS metabolite peak intensity data for all aliquots (N = 12); (b) coefficient of variation of GC-MS metabolite peaks for different analysis and extraction days (all data, N = 12; analysis day 1, N = 6; analysis day 2, N = 6; extraction day 1, N = 6; extraction day 2, N = 6).
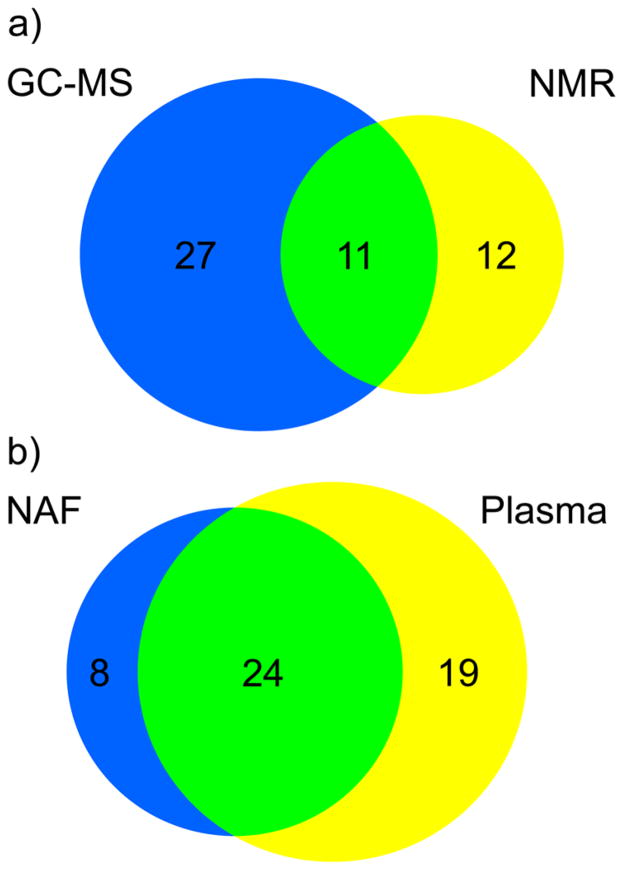
A wide range of metabolites were identified by the GC-MS analysis of NAF. Glycerol was again detected with high abundance, second only to phosphoric acid. Identified metabolites included amino acids, organic acids, fatty acids, and carbohydrates. These classes were also observed in the NMR spectra of NAF, although only 11 common metabolites were identified by both analytical approaches (Figure 3a). Due to increased sensitivity, our results suggest that GC-MS analysis is perhaps better suited as a first approach given the low sample volumes of NAF obtained from subjects, although additional information is obtained from NMR.
Figure 3.
(a) Comparison of the metabolites identified in the pooled NAF sample by NMR (yellow) and GC-MS (blue); 11 metabolites were identified by both platforms (green). (b) Comparison of the metabolites identified from the GC-MS analysis of individual NAF (blue) and matched plasma samples (yellow); 24 metabolites were identified in both samples (green).
These GC-MS protocols were applied to the analysis of a series of NAF samples that included paired plasma samples, from pre-and postmenopausal women (N = 8). A total of 32 metabolites were identified in the NAF samples and 43 metabolites in the plasma samples (Figure 3b). There were eight metabolites unique to NAF: 4-hydroxybenzoic acid, benzoic acid, caprylic acid, glycolic acid, lactose, oxalic acid, porphine, and a disaccharide (trehalose/maltose). There was no evidence of correlation between the relative concentrations of the 24 metabolites identified in NAF and its paired plasma sample (Table 2) with the exception that lactic acid was significantly inversely correlated (R2 = 0.57, P value = 0.03). The lack of direct correlation of metabolite concentration between the two biological fluids highlights the fact that they are metabolically distinct and capable of providing different biological information.
Table 2.
Correlations of Metabolite Peak Intensities Measured from NAF Samples with Matched Plasma Samples from Pre- and Postmenopausal Women
| Pearson’s r | r2 | P value | |
|---|---|---|---|
| lactic acid | −0.75 | 0.57 | 0.03 |
| alanine | 0.37 | 0.13 | 0.37 |
| valine | −0.33 | 0.11 | 0.43 |
| urea | 0.06 | 0.00 | 0.89 |
| glycerol | −0.45 | 0.20 | 0.26 |
| phosphoric acid | 0.15 | 0.02 | 0.73 |
| isoleucine | 0.27 | 0.08 | 0.51 |
| threonine | 0.18 | 0.03 | 0.68 |
| glycine | −0.28 | 0.08 | 0.51 |
| succinic acid | −0.05 | 0.00 | 0.90 |
| fumaric acid | −0.36 | 0.13 | 0.39 |
| capric acid | 0.60 | 0.36 | 0.12 |
| aspartic acid | −0.20 | 0.04 | 0.63 |
| pyroglutamic acid | 0.49 | 0.24 | 0.22 |
| clutamic acid | −0.10 | 0.01 | 0.81 |
| phenylalanine | −0.01 | 0.00 | 0.97 |
| citric acid | 0.03 | 0.00 | 0.94 |
| myristic acid | 0.16 | 0.03 | 0.70 |
| palmitic acid | −0.02 | 0.00 | 0.95 |
| inositol | 0.08 | 0.01 | 0.85 |
| oleic acid | 0.22 | 0.05 | 0.60 |
| stearic acid | 0.14 | 0.02 | 0.74 |
| sucrose | −0.24 | 0.06 | 0.57 |
| cholesterol | 0.41 | 0.17 | 0.31 |
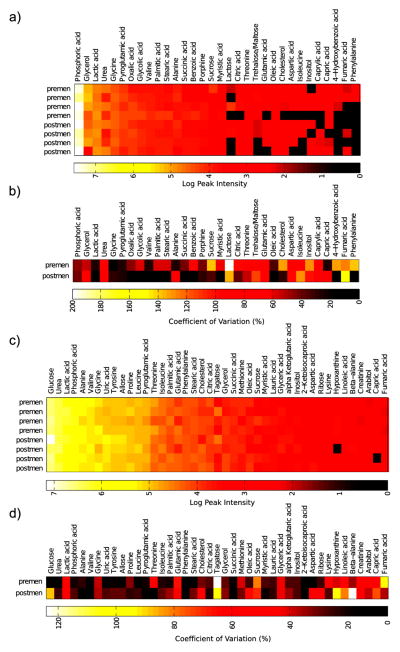
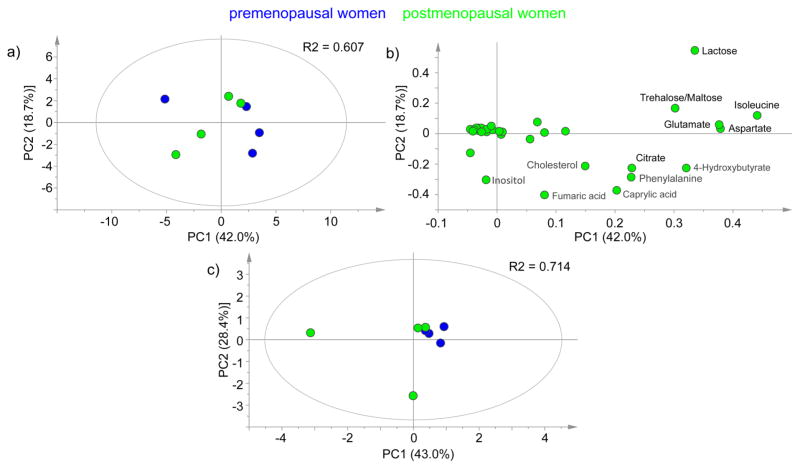
Peak intensities and coefficient of variation within the two sample groups for both NAF and plasma samples are shown in Figure 4. NAF metabolite levels were highly variable, and not all metabolites were detectable in all samples. The premenopausal NAF samples showed extensive variation with a median metabolite peak intensity CV of 62% compared to 38% for the postmenopausal NAF samples, whereas median CV values for the plasma metabolite data were 27 and 33% for the pre- and postmenopausal samples, respectively. A principal components analysis of the NAF and plasma data did not show a clear separation of the pre- and postmenopausal samples (Figure 5).
Figure 4.
GC-MS analysis of matched NAF and plasma samples from pre- and postmenopausal women. (a) Metabolite peak intensity data from NAF samples; (b) coefficient of variation of NAF metabolite peak intensities within each of the sample groups; (c) metabolite peak intensity data from matched plasma samples; (d) coefficient of variation of plasma metabolite peak intensities within each of the sample groups.
Figure 5.
Principal component analysis of GC-MS measured metabolites from pre- and postmenopausal women. (a) NAF metabolite data for all samples (N = 8); (b) loadings of the PCA described in (a); (c) plasma metabolite data for all samples (N = 8). All data are log transformed and mean centered.
Two of the NAF samples (1 premenopausal and 1 post-menopausal) were observed to be unusually clear in appearance, and exclusion of these samples improved the median metabolite CV values to 45 and 22% for the pre- and postmenopausal NAF samples, respectively. This suggests that turbidity/translucency could be a quality control criterion. While our sample size is too small to relate metabolites to clinical characteristics, NAF color has been previously related to both cancer risk and metabolite content. In a cohort of 521 women, red/brown NAF was highly correlated with the presence of breast cancer (P < 0.001).23 Petrakis et al. found that “dark” NAF compared to “light” NAF had significantly higher concentrations of cholesterol, cholesterol epoxides, lipid peroxides, and estrogens.24 Notably, given the sample size, we did not correct for time in menstrual cycle among the premenopausal samples. That might also explain the extensive between-individual variations observed. Future work will need to establish the within-individual variation in the NAF metabolite profile.
CONCLUSION
To the best of our knowledge, this is the first metabolite profiling study of NAF. We have applied a protocol for the analysis of NAF by GC-MS involving metabolite extraction by organic solvents, followed by derivatization by methoxyamination and silylation, which was highly reproducible for a large number of identified metabolites on different run days. This study demonstrates that, even restricted by the low volume of NAF samples, reliable metabolite measurements can be made, supporting future metabolic studies of NAF samples. Because NAF is a noninvasively obtained breast biofluid, further characterization of its metabolite composition has potential to have a high impact on breast cancer prevention, diagnosis, and treatment.
MATERIALS AND METHODS
Samples
Matched NAF and plasma were convenience samples previously collected from a phase I clinical trial with healthy premenopausal (N = 4) and postmenopausal (N = 4) women age 18–65 who were able to produce NAF.21 The study was approved by the University of Arizona Human Subjects Committee, and written consent was obtained from all participants. Prior to NAF collection, the breast was washed with St. Ives Apricot Scrub (containing water, Juglans regia (walnut) shell powder, glyceryl stearate, glycerin) and water. The breast was then wiped with Kendall Curity Alcohol Prep Pads (70% isopropyl alcohol). Suave lavender vanilla lotion (containing water, glycerin, stearic acid, mineral oil, glycol stearate) was used to massage the breast. A medela breast pump and a capillary tube were used to collect the NAF, which was then diluted 1:10 in PBS, and immediately placed in the −80 °C until analysis. The volume of diluted NAF available for analysis was approximately 20 μL. A small number NAF samples that did not have matched plasma samples were pooled together in order to assess the reproducibility of the analytical methods.
NMR Analysis
A pooled NAF sample (45 μL) that had been previously diluted 1:10 in PBS was prepared for NMR with the addition of D2O (5 μL) containing 5 mM DSS as an internal standard. After centrifugation, the solution was transferred to a 1.7 mm NMR microtube (Norell) and inserted into a 5 mm NMR microtube holder. Spectra were acquired on a Bruker Avance II NMR spectrometer (Bruker BioSpin, Rheinstetten, Germany), operating at a 1H frequency of 800.32 MHz and a temperature of 300 K, using a z-gradient triple-channel inverse cryoprobe (TXI, 1H/13C/15N). One-dimensional “Noesy-presat” and Carr–Purcell–Meiboom–Gill (CPMG) spectra were acquired following the procedure described by Beckonert et al.22 In brief, each spectrum was recorded with presaturation of the water resonance during the relaxation delay, 1024 transients (scans), a spectral width of 16 kHz, and a total acquisition time of 2.04 s. For the 1D NOESY experiment, presaturation was applied during the mixing time of 100 ms. The T2 delay for the CPMG experiment was 64 ms. Spectra were processed in iNMR 3.4 (Nucleomatica, Molfetta, Italy). Fourier transform of the free-induction decay was applied with a line broadening of 0.5 Hz, with zero filling to give 64k frequency domain data points. Spectra were manually phased, and automated first-order baseline correction was applied. Metabolites were assigned and quantified using the Chenomx NMR Suite 5.1 (Chenomx, Inc., Edmonton, Alberta, Canada) relative to 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS).
Metabolite Extraction
Metabolites were extracted from NAF or plasma using a dual-phase aqueous methanol/chloroform method. Prediluted NAF (20 μL) or plasma (50 μL) was added to 2:1 chloroform/methanol (300 μL) on ice and vortexed. H2O (300 μL) chilled on ice was then added to the samples, which were vortexed and centrifuged at 13 000 rpm for 5 min. The upper aqueous fraction and lower organic fraction were transferred to separate silanized GC-MS vials. The extraction was repeated, and the aqueous and organic fractions were pooled with the corresponding vials.
To assess reproducibility of the sample preparation and analytical method, a pooled NAF sample was prepared. Metabolites from prediluted NAF samples (6 × 20 μL) were extracted with the above aqueous methanol/chloroform method, and this was repeated on a different day. Three NAF samples from extraction day 1 and three samples from extraction day 2 were prepared for GC-MS analysis by the following derivitization procedures and injected onto the GC-MS. This was repeated for the remaining samples on a following day after running unrelated sample sets.
GC-MS Analysis
The aqueous fraction was derivatized for GC-MS by a two-step methoximation/silylation derivatization procedure.25 U–13C-glucose (20 μL, 1 mM), 13C-serine (20 μL, 1 mM), and myristic acid d27 (10 μL, 1.5 mg/mL) were added as internal standards before drying under reduced pressure. Dried samples were first methoximated with a solution of 20 mg/mL methoxyamine hydrochloride in anhydrous pyridine (20 μL) and incubated at 30 °C for 90 min. Samples were then silylated by adding 80 μL of MSTFA (with 1% TMCS) (Thermo) and incubating at 37 °C for 30 min. Following derivatization, 2-fluorobiphenyl in anhydrous pyridine (10 μL, 1 mM) was added to the samples as an injection standard, and the samples were transferred to deactivated glass vial inserts.
GC-MS analysis was performed on an Agilent 7890 GC equipped with a 30 m DB-5MS capillary column with a 10 m Duraguard column connected to an Agilent 5975 MSD operating under electron impact (EI) ionization (Agilent Technologies UK Ltd.). Samples were injected with an Agilent 7693 autosampler injector into deactivated splitless liners according to the method of Fiehn et al.,25 using helium as the carrier gas. Metabolites were assigned using the Fiehn library,25 with the deconvolution program AMDIS,26 and the MATLAB program GAVIN, developed in-house, was used to integrate metabolite peak areas for all samples.27 Data were normalized by the probabilistic quotient normalization method described by Dieterle et al.28 Multivariate analysis was performed using SIMCA 13.0 (Umetrics). Data for principal components analysis (PCA) was log scaled and mean centered.
Acknowledgments
The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London and United States Department of Defense Grant No. BC061529. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Nowsheen S, et al. Molecular markers for cancer prognosis and treatment: Have we struck gold? Cancer Lett. 2012;327(1–2):142–152. doi: 10.1016/j.canlet.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Higgins SA, et al. Patterns of reduced nipple aspirate fluid production and ductal lavage cellularity in women at high risk for breast cancer. Breast Cancer Res. 2005;7(6):R1017–R1022. doi: 10.1186/bcr1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maskarinec G, et al. The volume of nipple aspirate fluid is not affected by 6 months of treatment with soy foods in premenopausal women. J Nutr. 2011;141(4):626–630. doi: 10.3945/jn.110.133769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannello F, et al. Intracrinology of breast microenvironment: hormonal status in nipple aspirate fluid and its relationship to breast cancer. Expert Rev Endocrinol Metab. 2009;4(5):493–505. doi: 10.1586/eem.09.28. [DOI] [PubMed] [Google Scholar]
- 5.Dooley WC, et al. Ductal lavage for detection of cellular atypia in women at high risk for breast cancer. J Natl Cancer Inst. 2001;93(21):1624–1632. doi: 10.1093/jnci/93.21.1624. [DOI] [PubMed] [Google Scholar]
- 6.Sauter ER, et al. Nipple aspirate fluid and ductoscopy to detect breast cancer. Diagn Cytopathol. 2009;38(4):244–251. doi: 10.1002/dc.21177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malatesta M, et al. Biochemical and ultrastructural features of human milk and nipple aspirate fluids. J Clin Lab Anal. 2000;14(6):330–335. doi: 10.1002/1098-2825(20001212)14:6<330::AID-JCLA14>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrakis NL. Physiologic, biochemical, and cytologic aspects of nipple aspirate fluid. Breast Cancer Res Treat. 1986;8(1):7–19. doi: 10.1007/BF01805919. [DOI] [PubMed] [Google Scholar]
- 9.Sauter ER, et al. Nipple aspirate fluid: a promising non-invasive method to identify cellular markers of breast cancer risk. Br J Cancer. 1997;76(4):494–501. doi: 10.1038/bjc.1997.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrakis NL, et al. Gross cystic disease fluid protein in nipple aspirates of breast fluid of Asian and non-Asian women. Cancer Epidemiol Biomarkers Prev. 1993;2(6):573–579. [PubMed] [Google Scholar]
- 11.Thompson PA, et al. Sulindac and sulindac metabolites in nipple aspirate fluid and effect on drug targets in a phase I trial. Cancer Prev Res. 2010;3(1):101–107. doi: 10.1158/1940-6207.CAPR-09-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JA, et al. Expression of epidermal growth factor, transforming growth factor-beta 1 and adiponectin in nipple aspirate fluid and plasma of pre and post-menopausal women. Biomarker Res. 2013;1:18. doi: 10.1186/2050-7771-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varnum SM, et al. Proteomic characterization of nipple aspirate fluid: identification of potential biomarkers of breast cancer. Breast Cancer Res Treat. 2003;80(1):87–97. doi: 10.1023/A:1024479106887. [DOI] [PubMed] [Google Scholar]
- 14.Mannello F, et al. Protein profile analysis of the breast microenvironment to differentiate healthy women from breast cancer patients. Expert Rev Proteomics. 2009;6(1):43–60. doi: 10.1586/14789450.6.1.43. [DOI] [PubMed] [Google Scholar]
- 15.Pawlik TM, et al. Significant differences in nipple aspirate fluid protein expression between healthy women and those with breast cancer demonstrated by time-of-flight mass spectrometry. Breast Cancer Res Treat. 2005;89(2):149–157. doi: 10.1007/s10549-004-1710-4. [DOI] [PubMed] [Google Scholar]
- 16.Pawlik TM, et al. Proteomic analysis of nipple aspirate fluid from women with early-stage breast cancer using isotope-coded affinity tags and tandem mass spectrometry reveals differential expression of vitamin D binding protein. BMC Cancer. 2006;6:68. doi: 10.1186/1471-2407-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauter ER, et al. Proteomic analysis of nipple aspirate fluid using SELDI-TOF-MS. Int J Cancer. 2004;114(5):791–796. doi: 10.1002/ijc.20742. [DOI] [PubMed] [Google Scholar]
- 18.Keun H, et al. Serum molecular signatures of weight change during early breast cancer chemotherapy. Clin Cancer Res. 2009;15(21):6716. doi: 10.1158/1078-0432.CCR-09-1452. [DOI] [PubMed] [Google Scholar]
- 19.Stebbing J, et al. A metabolic phenotyping approach to understanding relationships between metabolic syndrome and breast tumour responses to chemotherapy. Ann Oncol. 2012;23(4):860–866. doi: 10.1093/annonc/mdr347. [DOI] [PubMed] [Google Scholar]
- 20.Denkert C, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4(4):37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller TP, et al. Safety and feasibility of topical application of limonene as a massage oil to the breast. J Cancer Ther. 2012;3:749–54. doi: 10.4236/jct.2012.325094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2(11):2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 23.Sauter ER, et al. Nipple aspirate fluid color is associated with breast cancer. Cancer Detect Prev. 2006;30(4):322–328. doi: 10.1016/j.cdp.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Petrakis NL, et al. Coloration of breast fluid related to concentration of cholesterol, cholesterol epoxides, estrogen, and lipid peroxides. Am J Clin Pathol. 1988;89(1):117–120. doi: 10.1093/ajcp/89.1.117. [DOI] [PubMed] [Google Scholar]
- 25.Kind T, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein SE. An integrated method for spectrum extraction and compound identification from GC/MS Data. J Am Soc Mass Spectrom. 1999;10:770–781. [Google Scholar]
- 27.Behrends V, et al. A software complement to AMDIS for processing GC-MS metabolomic data. Anal Biochem. 2011;415(2):206–208. doi: 10.1016/j.ab.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Dieterle F, et al. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]