Abstract
The microbiota represents the complex collections of microbial communities that colonize a host. In health, the microbiota is essential for metabolism, protection against pathogens and maturation of the immune system. In return, the immune system determines the composition of the microbiota. Altered microbial composition (dysbiosis) has been correlated with a number of diseases in humans. The tight reciprocal immune/microbial interactions complicate determining whether dysbiosis is a cause and/or a consequence of immune dysregulation and disease initiation or progression. However, a number of studies in germ-free and antibiotic-treated animal models support causal roles for intestinal bacteria in disease susceptibility. The role of the microbiota in transplant recipients is only starting to be investigated and its study is further complicated by putative contributions of both recipient and donor microbiota. Moreover, both flora may be affected directly or indirectly by immunosuppressive drugs and anti-microbial prophylaxis taken by transplant patients, as well as by inflammatory processes secondary to ischemia/reperfusion and allorecognition, and the underlying cause of end-organ failure. Whether the ensuing dysbiosis affects alloresponses and whether therapies aimed at correcting dysbiosis should be considered in transplant patients constitutes an exciting new field of research.
Introduction
The microbiota (Table I) is the collective term for the complex communities of microorganisms comprising bacteria, viruses, parasites and fungi that inhabit the body surfaces exposed to the outside world. These include the skin, the oropharynx-gastro-intestinal tract, the genito-urinary tract, and the airways [microbial density decreasing from upper to lower airways (1)], with the greatest concentration of microbes present in the distal part of the intestine. Most of the published work to date focuses on the bacterial communities of the intestine, although colonization of all surfaces and presence of non-bacterial microbes also most likely profoundly impact the host. Intestinal bacteria have a mutualistic relationship with their host as they profit from the habitat that provides them with nutrients for their growth, but in turn contribute to the fitness of their host. Indeed, they play an emerging role in host energy balance, including colonic metabolism and fermentation of complex carbohydrates in dietary fibers to generate short chain fatty acids important for providing energy to colonic epithelial cells and to serve as substrates for gluconeogenesis and lipogenesis (2). Other metabolic roles include conversion, de-convolution and reabsorption of bile acids, production of vitamin K, and increased absorption of amino acids. In addition, the microbiota drive the maturation of the host immune system, being important for the normal architecture of secondary lymphoid organs, the generation of IgA-secreting B cells or the differentiation of induced regulatory T cells (iTregs). Finally, the microbiota also help prevent pathogenic infections by filling intestinal niches and competing for nutrients with pathobionts (disease-causing microbes).
Table I.
Glossary of terms.
| Term | Definition |
|---|---|
| Microbiota | The complex of microorganisms (bacteria, parasites, viruses, fungi) that colonize a host or a habitat |
| Microbiome |
|
| Metagenome | All the genetic material contained in a sample |
| Dysbiosis | Alteration in the normal composition of the microbiota; can be associated with disease |
| Commensalism | Relationship between 2 organisms in which 1 is helped and 1 is not affected |
| Mutualism | Relationship between 2 organisms in which both benefit |
| Parasitism | One organism benefits at the expense of another |
| Symbiosis | Broader category encompassing relationships that are commensal, mutualistic or parasitic |
| Pathobiont | Disease-causing microorganism |
| Taxonomy | Microbial classification into ordered taxa (categories) |
| Taxon (plural taxa) | A grouping of microbes defined by the degree of genetic identity (often >75% for phylum, >80% for class, >85% for order, >90% for family, >95% for genus and >97% for species) |
It is thought that the commensal flora of a tissue play a major influence on local immunity. Intriguingly, gut commensals are also thought to control distal immune responses, thus modulating diseases of distant tissues in conditions such as rheumatoid arthritis, obesity, multiple sclerosis, and autism. Thus, the intestinal microbiota of the host may have extra-intestinal effects influencing alloimmune responses to any transplanted organ, whereas it is tempting to speculate that the local microbiota in colonized organs such as the lung (containing colonized airways), the intestine, or composite grafts containing skin may influence how the immune system responds to those organs following their transplantation. A number of studies are characterizing the commensal flora of transplant patients hypothesizing that microbial composition, which may be affected by diet, immunosuppressive and anti-microbial drugs, underlying disease and inflammatory responses, impacts transplant outcomes. This review will summarize results from the Human Microbiome Project, focus on the reciprocal effects between the immune system and the commensal flora as have been defined in mouse models, review current studies on the microbiota in animal and human transplantation and speculate on how the microbiota may affect alloresponses and transplant fate.
Tools for exploring the relationship between microbiota and immune system
Many insights into the importance of the microbiota in health and disease have come from analyses of germ-free mice that are kept in sterile micro-isolators to limit microbial exposure. These mice can also be used as recipients of single or defined bacterial species, a technique known as gnotobiotics, a term of Greek origin meaning known life. In parallel with germ-free mice, conventional mice treated with cocktails of broad-spectrum antibiotics to reduce bacterial diversity and/or load are often used.
New sequencing tools such as next generation sequencing (NGS) have greatly facilitated the identification of commensal communities, many of which cannot be cultured in the laboratory. NGS can target gene segments whose polymorphisms determine the taxonomic rank (Table I) of bacterial communities (3). In particular, the gene encoding the 16S ribosomal RNA subunit contains 9 hypervariable regions whose sequences differ between bacterial species. Sequencing 1 to 3 of these hypervariable areas is utilized to define bacterial communities at higher taxonomic ranks, a useful approach to compare communities over time or across treatments, but that does not have sufficient resolution to identify which bacterial strains are actually present. A complementary technique is whole-genome sequencing of DNA extracted from microbial communities in bulk to catalog all the metagenome, the genetic material present in a sample, with deep sequencing required to identify genes from minor microbial constituents that may still play important functional roles. These approaches get at finer specification of the colonizing species, though still depend on aligning the DNA reads with reference genome sequences. Because not all bacterial genomes have been sequenced, many shotgun reads remain anonymous. Definitive identification of the strains present in a given habitat requires whole genome sequencing of individual single-cell sorted bacteria.
Finally, because genomic sequencing does not necessarily distinguish between live and dead bacteria and expressed versus silent genes, these techniques can be coupled with transcriptomics to detect transcribed genes and metabolomics to measure metabolic products indicative of gene functionality.
Distribution and composition of the microbiota
Recently, the Human Microbiome Project defined the variability and similarity of the gut microbiota in a population of healthy North American adults (4). While there was a wide variation among individuals in the composition of bacterial genera present in a spontaneously passed stool sample, the composition of each individual’s fecal bacterial metagenome was surprisingly uniform from person to person. It is therefore inferred that the metabolic and catabolic function of the microbiota is similar in healthy adults (although the actual metabolome or global transcriptome of the bacterial microbiota was not measured to test whether microbiota protein expression or protein function was similarly uniform). In this healthy population, the prevalence of known gut pathobionts was very low (5), and while there were significant associations of specific body site microbiota with ethnicity, there were modest to low associations with gender, age and body mass index.
Another recent study of the healthy human gut microbiota divided individuals into three major enterotypes of bacterial composition enriched for certain bacteria and metabolic features, (Bacteroides and biotin synthesis, Prevotella and thiamine synthesis, Ruminococcus and heme synthesis), but the importance and validity of these described subgroups is unclear (6, 7). Thus, among healthy adults, the gut microbiota represented in a stool sample can have wide diversity from person to person but is expected to have more uniform functions across hosts.
Some of the important questions about the gut microbiota, particularly in light of the variation seen from person to person, include how these populations establish themselves and how stable they are once established. Studies of human infants show that the earliest detectable gut bacterial microbiota reflect commensal flora of the birth canal and later, if breastfed, those associated with human breast milk consumption (along with a metagenome enriched for milk-digestion enzymes) (8–11). Changes are seen following the introduction of solid food or formula, with an adult-like microbiota becoming established by one year of age and relatively stable for the next two years, illustrated by an in depth study of the developing gut microbiota of a single infant (8). Once the adult gut microbiota is established, it appears to be relatively stable over long periods, with 60% of the original bacterial microbes still detected during 5 years of follow-up (12, 13). The stability of the gut microbiota is also supported by the observation that microbial communities are more similar between adult twin siblings (mono- and dizygotic) and between adult twins and their mother than between the twin and an unrelated person (14).
Association between dysbiosis and disease
The intestinal microbiota has now been associated with several human disease states including obesity, diabetes/metabolic syndrome, and inflammation. The idea that gut microbes are not mere commensals but a community of microorganisms co-evolving with its host as symbionts and pathobionts, has led to the hypothesis that changes in the composition and function of the microbiota are associated with disease. The challenge of course is to determine whether any such change is cause or effect. Supporting the importance of specific commensals early in life, children born by C-section that are initially colonized by maternal skin commensals have some increased susceptibility to developing allergies and asthma compared to those that are colonized by the vaginal flora (15), despite rapid diversification of the microbial communities after birth. Similarly, mice treated with Vancomycin at birth but not in adulthood developed more severe disease in a model of allergic asthma (16). Later in life, there are several examples where dysbiosis is associated with disease states. The primary example of this is found in inflammatory bowel disease, particularly Crohn’s disease where lower proportions of Firmicutes and relative lack of Faecalibacterium have been confirmed in multiple studies (17–21). Whether the dysbiosis results from the inflammatory milieu or underlying host genetic factors, and/or whether the dysbiosis drives the inflammation is unclear. In other diseases, data from murine models show that obesity can be prevented in genetically obesity-predisposed mouse strains using germ-free derivation or after gut-sterilizing antibiotic treatment, and furthermore that obesity can be induced in lean mice by fecal transplantation from obese mouse donors and even gnotobiotic colonization (22). In humans, the overall data are conflicting as to whether a decreased ratio of Bacteroidetes:Firmicutes is always present, but it seems that genes involved in carbohydrate metabolism are typically increased in obese persons’ fecal microbiota metagenome (14, 23). That gut microbiota from obese persons can induce excess weight gain in germ-free mice and that the metabolic syndrome can be improved after fecal transplant in some patients also supports a causal role for the microbiota in the pathophysiology of these conditions (24, 25).
Despite little to no association of microbiota variation with gender in humans, murine experiments suggest that sex differences may be consequential. Using the non-obese diabetic (Nod) mouse model of spontaneous autoimmune diabetes that has a higher prevalence in females, 2 recent studies suggest that gender differences reflected in altered microbiota may be responsible for differential autoimmune susceptibility. One study found that androgen production dictated distinct intestinal colonization as male castration eliminated gender differences in microbial composition (26), whereas the other suggested that particular microbiota could promote elevated testosterone levels that were in turn protective if androgen receptor was present (27). Other associations of gut dysbiosis have been proposed in diabetes, steatohepatitis, atherosclerosis, multiple sclerosis and autism (28–30), although definitive proof that human disease is caused by dysbiosis of the gut or other colonized surfaces is lacking. Nevertheless, this causality has been established in several experimental models as described below.
The intestinal immune system maintains bacterial homeostasis and prevents dysbiosis
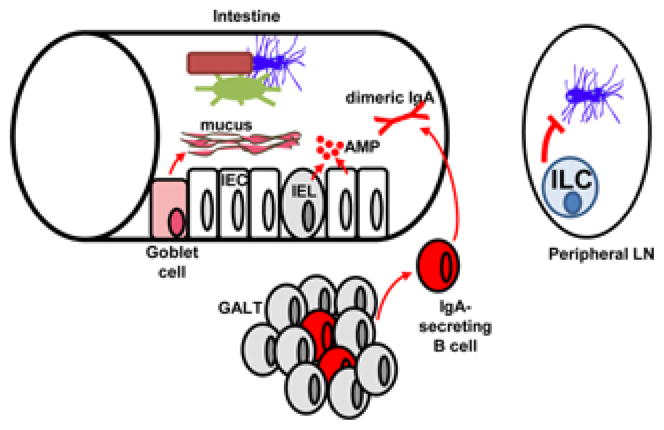
To prevent dysbiosis and disease, epithelial, mucosal and immune cells at barrier surfaces all play critical roles in maintaining bacterial homeostasis and containing the microbiota via production of mucus, anti-microbial peptides or luminal immunoglobulins (Figure 1). Although each barrier surface is organized in its own specialized mode (31), we will focus here on the intestine for which more data are available. The intestinal tract comprises many immune cells, some of which are organized into lymphoid structures, including mesenteric lymph nodes (MLNs), Peyer’s patches (PPs), and isolated lymphoid follicles (ILFs) while others reside intercalated between intestinal epithelial cells (IECs) like intraepithelial lymphocytes (IELs), or underneath the epithelium such as lamina propria immune cells. The important role of these immune structures in preventing dysbiosis is exemplified by the fact that mice that lack ILFs have a profoundly altered microbiota with over-representation of Gram-negative bacteria (32).
Figure 1. The intestinal epithelium and immune system control the local microbiota.
The immune system, whose maturation is helped by the microbiota, in turn contains it and prevents outgrowth of pathogenic species via production of mucus that physically separates the microbiota from the host, antimicrobial peptides (AMP) that create a sterility gradient and secretory IgA that neutralizes biologically active antigens. In peripheral lymph nodes, innate lymphoid cells (ILC) help contain translocated bacteria. IEL, intra-epithelial lymphocyte; IEC, intestinal epithelial cell; GALT, gut-associated lymphoid tissue.
Presence of secretory IgA (sIgA) is important to contain the microbiota. Breast milk is rich in maternal sIgA that travels to the intestine of breastfed infants and whose specificities have been shaped by the maternal microbiota. Intestinal B cells then take over to produce sIgA. Perhaps because IgA is poor at fixing complement, sIgA appears to promote a tolerogenic environment in which intestinal bacteria and the developing immune system coexist and shape each other (33). If the affinity of IgA for the microbiota is impaired, dysbiosis ensues, as observed in mice deficient in programmed cell death-1 (PD-1) (34), an inhibitory receptor expressed on activated T cells. These mice developed altered T follicular helper (Tfh) cells, a subset of T cells essential for helping antibody production by B cells in secondary lymphoid structures and PPs. In addition to their role as producers of sIgA, B cells also help to contain the microbiota during intestinal injury, as bacteria disseminated systemically in mice subjected to DSS-induced colonic damage when their B cells were selectively deficient in MyD88, an adaptor downstream of most Toll-like receptors (TLRs) (35).
Examples of how global immune dysfunctions can result in dysbiosis that then modulates disease susceptibility include deficiencies in MyD88, whose absence protected Nod mice from autoimmune diabetes (36), mutations in the inflammasome where dysbiosis predisposed to colitis (37) and non-alcoholic steato-hepatitis (38), colitis in T-bet−/− x Rag2−/− mice (39), and protection from diet-induced obesity in mice deficient in lymphotoxin (40). In each case, disease protection or exacerbation could be transferred to wildtype mice by co-housing or fecal transplant. However, all of these results need to be reconciled with the observation that the composition of the microbiota is more affected by physical familial transmission to the progeny than by the presence of immune mutations, at least for TLR mutations (41).
Recent data indicate that some bacteria do normally translocate outside of the intestine and reach distal immune structures in the host. These bacteria appear to be contained by innate lymphoid cells (ILCs), a cytokine receptor common γ chain-dependent population that segregates into cytokine profiles similar to T helper (Th) cells, with ILC1s producing IFNγ, ILC2s secreting IL-5 and IL-13 and ILC3s making IL-17A and IL22 (42). Indeed, depletion of ILCs has been shown to result in peripheral dissemination of commensal bacteria of the Alcaligenes species out of lymphoid tissues and in systemic inflammation, which was prevented by administration of IL-22 (43). It seems that ILC3s express MHC class II and present microbial antigens to CD4+ T cells to limit their response to the microbiota, as selective deletion of MHC class II in ILC3s resulted in CD4-mediated dysregulated inflammation to commensal bacteria (44). Thus, in addition to helping maintain intestinal homeostasis, ILCs have critical roles in tolerance to microbiota that has reached lymphoid tissues.
Together, these examples indicate the ability of immune cells to shape the composition of the intestinal and translocated microbiota and conversely how genetic immunodeficiencies can result in dysbiosis which in turn modulates disease susceptibility. These data may be relevant to the transplant patient where immunosuppressive medications on top of host genetic backgrounds and anti-microbial prophylaxis may predispose to dysbiosis. Whether such dysbiosis can in turn modulate the alloresponse remains at this point a matter of speculation.
Effects of the microbiota on the local immune system
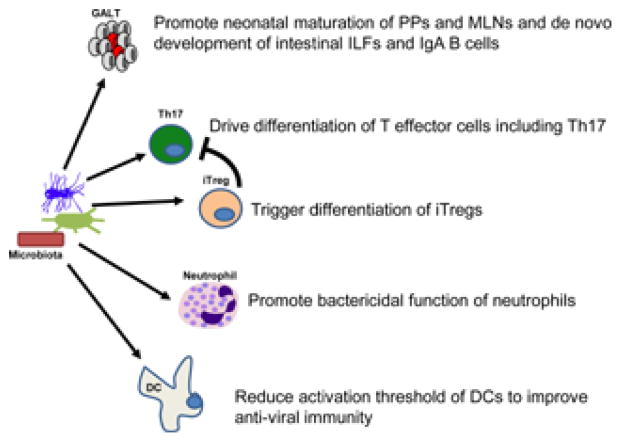
In parallel with the immune system shaping microbial composition and preventing dysbiosis, the microbiota has profound converse effects on the immune system (Figure 2). This is in keeping with the hygiene hypothesis that correlates the higher prevalence of autoimmunity and allergies in economically advanced societies over the last century with reduced exposure to protective microbes that still exist in developing countries and may enable immunoregulatory networks (45, 46). The loss of such protective strains may be a consequence of increased cleanliness, wider use of antibiotics or consumption of meat from antibiotic-raised animals for instance.
Figure 2. The microbiota promotes maturation and differentiation of the immune system.
The microbiota carries many essential functions to ensure that optimal immune responses can be generated, including helping development and maturation of lymphoid structures, differentiation of T cell subsets and potentiation of the function of innate immune cells. GALT, gut-associated lymphoid tissue; PP, Peyer’s patches; mLN, mesenteric lymph node; IEL, intestinal epithelial cell; iTreg, induced T regulatory cell; DC, dendritic cell.
To communicate with the immune system, commensal microbes, similarly to pathogens, express microbial-associated molecular patterns (MAMPs) that can be sensed by specialized receptors on a variety of cells, including immune cells, epithelial and endothelial cells. These pattern-recognition receptors (PRRs) comprise surface or endosomal membrane-associated TLRs and C-type lectin receptors (CLRs), as well as cytosolic NOD-like receptors (NLRs), RIG-I receptors (RIRs) and DNA sensors (47). PRR signaling depends on intracellular adaptors, of which the most studied has been MyD88, a molecule downstream of all TLRs except TLR3. PRR signaling in cells exposed to MAMPs results in upregulation of major histocompatibility complex (MHC) and costimulatory molecules in particular in antigen-presenting cells (APCs), but also some endothelial and epithelial cells (48), which can enhance antigen presentation to T cells. PRR signaling in APCs also results in the production of cytokines whose profile depends on the type of PRR engaged including type I interferons (IFN), tumor necrosis factor (TNF), interleukin (IL)-1 and IL-6. These cytokines in turn can direct the differentiation of effector T cells, and activate and attract innate immune cells which themselves exert effector functions. Interestingly, the expression levels of TLRs, at least on intestinal epithelial cells (IECs), have been shown to vary with the circadian clock, such that the continuous presence of MAMPs may be transformed into pulsed circadian signals (49). Notably, despite engaging the same cellular machinery, symbionts and pathobionts trigger different outcomes for the microbes (symbiosis versus clearance) and for the host (immune homeostasis versus inflammation), although it is not always clear how these different fates come to be.
The initial sensing of the microbiota after birth is thought to be carried out by epithelial cells at barrier interfaces which then instruct the behavior of the immune system. Signals from the microbiota to IECs are crucial to induce post-partum maturation of PPs and MLNs and de novo development of ILF, which are in turn essential to maintain host/microbiota homeostasis. Indeed germ-free mice have reduced size and cellularity of mucosal lymphoid structures and secondary lymphoid organs in general and lack ILCs, CD8αβ IELs and secretory IgA (sIgA) (50). In particular, peptidoglycan, a product of Gram-negative bacteria, has been shown to activate the pattern-recognition receptor NOD1 in IECs to induce factors that recruit B cells to ILFs where they express sIgA specific for microbial antigens (32).
Of particular relevance to transplantation, as T cells orchestrate transplant rejection, several studies indicate that specific bacterial species in the intestine can direct T cell differentiation towards distinct effector subsets with deleterious or protective effects on local inflammatory diseases. Colonization of mice with Segmented Filamentous Bacterium (SFB), a Clostridia-related Gram-positive species that attaches tightly to the intestinal epithelium of the terminal ileum, resulted in accumulation of Th17 and to a lesser extent of Th1 cells in the lamina propria (51, 52) and resistance to infection by Citrobacter rodentium (52), a mouse model of enteritis. The particular mechanism by which SFB promotes Th17 responses is not yet clear. This effect is not unique to SFB, as Lactobacillus johnsonii, for example has been shown to drive Th17 responses in rats (53).
Conversely, several bacterial species have been shown to promote expansion or differentiation of Foxp3-expressing regulatory T cells (Tregs). Intestinal colonization of germ-free mice with a set of defined benign commensals termed altered Schaedler flora resulted in expansion of colonic Tregs and resistance to chemically-induced colitis (54). Similarly, colonization of germ-free mice with a cocktail of 46 strains of the Clostridium genus belonging mainly to clusters IV and XIVa resulted in expansion of lamina propria Tregs (55). This correlated with an increase in local TGF-β, a cytokine that can help conversion of conventional CD4+ T cells into induced Tregs. The human microbiota appears to contain species of the Clostridium genus that may have similar Treg-inducing effects (56). Clostridia are not the only bacteria that can promote regulation. Polysaccharide A (PSA) from Bacteroides fragilis was shown to be sufficient to correct the Th1/Th2 imbalances present at birth (57), protect mice from experimental colitis induced by Helicobacter hepaticus (58), drive conversion of CD4+ T cells into iTregs in the intestine (59) and promote IL-10 production by engaging TLR-2 on Tregs (27). B. fragilis-mediated expansion of Tregs in the intestine in turn repressed Th17 responses against it, thus enabling its establishment as a resident commensal while benefiting the host as the immunoregulatory networks it helped drive could protect from immune pathology or autoimmunity. Effects of the intestinal flora on Tregs can also be indirect via DCs, as depletion of TRAF6 selectively in DCs prevented them from producing IL-2, a homeostatic factor for Tregs (60), and resulted in a Th2-driven eosinophilic colitis reminiscent of animals who lacked the capacity to generate iTregs (61).
The particular metabolites produced by bacteria that can drive increased numbers of Tregs in the lamina propria are starting to be elucidated (62). The short chain fatty acid butyrate, and to a lesser extent propionate but not acetate, produced by bacteria upon fermentation of complex polysaccharides were shown to drive iTreg differentiation by acting directly on Tregs and enhancing histone H3 acetylation and therefore transcriptional accessibility of the FoxP3 promoter (63, 64). These short chain fatty acids can leave the intestinal lumen and be detected in the plasma, such that they may also affect distal Tregs (63). Short chain fatty acids also bind the G-protein-coupled receptor 43, which was necessary for the normal resolution of colitis, arthritis and asthma (65). Importantly, treatment with butyrate reduced disease severity in a model of colitis, demonstrating the potential therapeutic impact of bacterial metabolites. In addition to providing metabolites that can facilitate iTreg differentiation, the intestinal microbiota is also a source of antigens such that subsets of intestinal Tregs bear a T cell receptor (TCR) specific for the indigenous intestinal flora (66). Whether these Tregs are mostly iTregs or thymic Tregs remains controversial (67) but these data support a role for microbiota-reactive Tregs in the maintenance of tolerance to the intestinal flora.
Thus, specific strains of bacteria can help generate effector T cells with particular cytokine profiles while others promote Treg expansion or differentiation. Whereas at least some intestinal Tregs directly recognize bacterial antigens as discussed above, whether the effect of the microbiota on Teffectors is in part due to TCR recognition of microbial peptides or is purely an adjuvant effect of microbial molecular patterns costimulating T cell activation and differentiation independently of TCR recognition of the microbes remains to be determined. Understanding this relationship is critical to determine how microbial effects on T cell immunity are relevant to alloimmunity and transplantation, be they an adjuvant effect (68, 69) or a microbe-specific effect that mediates cross-reactivity with alloantigen (70).
Systemic immune effects of the microbiota
As mentioned previously, the microbiota not only helps mature local lymphoid structures, but is also necessary for the normal architecture of distal secondary lymphoid organs. Whereas the architecture of the bone marrow and thymus, the 2 primary lymphoid structures home to immune precursors, appears normal in germ-free mice, spleens and lymph nodes are smaller with reduced number and size of germinal centers (71). In addition, the basal production of IgA, IgM and IgG appears diminished suggesting that the microbiota affects systemic B cell maturation and isotype switching although some of these effects may be due to microbial influences on T cells that then fail to provide optimal help to B cells.
Indeed, systemic and not only local T cells are influenced by the intestinal microbiota. For instance, SFB that can drive Th17 differentiation in the lamina propria, could also promote systemic Th17 responses in a mouse model of rheumatoid arthritis (72). Whereas germ-free mice had reduced disease severity, intestinal colonization with SFB restored arthritis and antibiotic treatment of conventionally housed mice to eliminate SFB reduced arthritis severity. Similar results were reported in a mouse model of multiple sclerosis (73). In addition to driving Th17 responses, SFB could also render the TCR of Th1 cells more responsive to antigen, in a manner dependent on intestinal inflammatory cytokines. This resulted in increased systemic responses and disease severity in a model of arthritis (74). Although these results cannot be extrapolated to humans directly because SFB may not colonize the human colon, recent data have associated human intestinal colonization with Prevotella copri with rheumatoid arthritis (75), raising the possibility that particular gut bacteria in humans may affect systemic immunity, although whether joint-specific immune responses were modified by P. copri remains to be shown.
Increases in systemic Treg numbers can also be observed after colonization of germ-free mice with the Treg-inducing Clostridia mixture (55). Systemic Tregs in turn can travel to the colonic lamina propria to limit local inflammation if they express GPR15, whose expression is modulated by the microbiota and TGF-β(76). It remains to be established if distant effects of the microbiota are due to T cells differentiated in the intestine that recirculate systemically or to translocation of microbes or microbial products outside of the intestine that drive cellular differentiation distally, and, as mentioned before whether the effects are dependent on TCR recognition of microbial peptides or on an adjuvant effect of microbial products. The recent report of bacterial outgrowth in lymph nodes from mice depleted of ILCs demonstrates that bacterial translocation does normally occur (43).
Although the microbiota does not appear to affect the numbers of most innate immune cells, it does affect their function. For instance, neutrophils from the bone marrow of germ-free mice or conventional mice treated with antibiotics displayed reduced killing of pathogens (77). In this case, translocation of intestinal peptidoglycan and signaling via NOD1 on neutrophils resulted in enhanced killing activity. Importantly for immune responses, splenic dendritic cells (DCs), a crucial cell type for presentation of antigen (and alloantigen) to T cells and therefore priming of the adaptive immune response, had impaired function in germ-free mice, showing reduced production of type I IFN and IL-15 upon activation. This defect in turn impaired NK cell function and antiviral immunity (78). Thus, microbes or microbial products appear to travel systemically to prime DCs at sites distal from the gut and poise these cells for anti-viral immunity (79). Consequently, germ-free mice or mice treated with antibiotics showed reduced immune responses to pathogens when compared with conventional mice. Whether the microbiota can similarly enable DCs to better present alloantigen remains to be determined.
Another interesting example of the effect of microbiota on immunity is that of autoimmune diabetes. Whereas MyD88-deficient Nod mice were protected from the disease, germ-free mice developed accelerated disease with equal penetrance in males and females (36). This suggests that microbiota in MyD88-deficient mice and to some extent in wildtype mice may inhibit anti-β-cell immunity and these protective effects are lost in the absence of commensals. SFB in particular has been associated with protection from autoimmune diabetes in mice (80). Thus, the same bacterial strain can have protective (diabetes) or detrimental (arthritis) effects on autoimmune diseases experienced by different mouse strains, prompting caution for implementing probiotic therapies in patients with different genetic backgrounds that may confer distinct disease susceptibility.
Of particular interest for transplantation, acute intestinal infections that breach barrier integrity and allow direct exposure of the immune system to commensals result in long-lived anti-commensal memory T cells (81). Because anti-microbial T cells can cross-react with alloantigens and memory T cells are resistant to immunosuppression (82), a combination of these events could provide a mechanism by which patients become allosensitized through heterologous immunity (Figure 3).
Figure 3. Speculative model for how the microbiota may affect alloresponses.
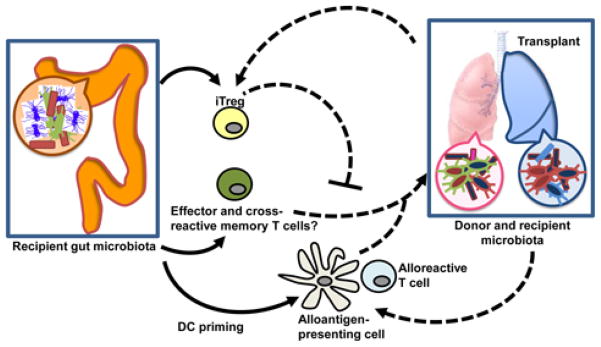
Although it is not known if the microbiota can affect alloresponses, parallels drawn from other fields suggest mechanisms by which the microbiota could both inhibit and promote anti-transplant immunity. For instance, intestinal bacteria and microbial products can promote the differentiation of local and possibly distal regulatory and effector T cells. These T cells may cross-react with alloantigen and therefore inhibit or promote alloresponses, respectively. The microbiota has also been reported to prime antigen-presenting cells (APC) such that alloantigen could potentially be presented more efficiently to alloreactive T cells. In addition, when transplanting colonized organs such as composite grafts, intestine or lung (airways contain resident microbiota although bacterial density decreases with diminishing size of the airways), the microbiota from the donor organ may control the organ’s own local immunity and perhaps also distal immunity and thus further influence regulatory and/or effector alloresponses. Dashed lines represent speculative effects that need to be investigated.
Other microbiota
Little is known about the effects of commensals that reside outside of the gut. Clearly the skin, airways and genito-urinary tracts also harbor great microbial diversity as well as site-specific immune networks that play important roles in maintaining barrier function and local immune homeostasis. Recently, the skin, but not the gut, microbiota was shown to be important for tuning local T cells and enabling immune responses to a cutaneous parasite (83). Whether the site-specific donor or host microbiota can modulate alloresponses against colonized transplanted organs such as the lung remains to be determined (Figure 3).
Although the effect of commensal non-bacterial microbes on the immune system and immune diseases has been less studied, some examples hint at their importance. Resident fungi live both as branching filamentous hypha and as individual cells on the different surfaces of the host (84, 85). In a mouse model of arthritis resulting from a mutation in the T cell signaling enzyme ZAP-70, arthritis did not develop in germ-free mice despite the presence of arthritogenic T cells, unless the fungal β-glucan that can bind Dectin-1 on DCs was introduced (86). Conversely dectin-1-deficient mice were more susceptible to chemically-induced colitis (87). Together, these results suggest that commensal fungi can prevent disease in normal hosts, but also be the trigger to autoimmunity is susceptible animals.
Metagenomics of the intestinal, cutaneous and respiratory microbiota have identified many viral genes in addition to the bacterial ones. The ‘virobiota’ in vertebrates is comprised of viruses that replicate in bacterial cells (bacteriophages) and viruses that replicate in eukaryotic cells. The intestinal virobiota also contain plant viruses that are probably associated with ingested vegetables (88). Stable viral phages may confer metabolic benefits to their hosts. However, it is unclear if eukaryotic viruses sampled during metagenomic analyses represent innocuous/beneficial residents stably associated with healthy host tissue versus acute, chronic or latent viral infections. Analysis of the virome at 3 time points from fecal samples of human female monozygotic twins and their mothers revealed remarkable interpersonal variation compared to greater similarity of the bacteriome from related individuals; however, the virome displayed very low intra-personal variation over time (89). Whether phages can influence immune responses during their associations with a host remains to be determined, though it is clear that they can engage intracellular PRRs if they end up in eukaryotic cells during an immune response to the bacteria that carry them (90). Interestingly, the transmission of mouse mammary tumor virus (MMTV) from mother to newborn suckling pups was recently shown to depend on the presence of bacteria (91). MMTV bound bacterial LPS which triggered IL-10 production by the host to inhibit antiviral responses and thus tolerate the establishment of the virus in the murine progeny. Similarly, intestinal commensal bacteria may help support infection by enteric viruses, as antibiotic-treated mice were less susceptible to poliovirus and reovirus infection (90). Poliovirus bound bacterial LPS which promoted viral infection of host cells. Thus, viruses appear to exploit intestinal bacteria for their replication and transmission. Importantly, endogenous retroviruses appear to be activated and become infectious in immunodeficient mice in the presence, but not absence of bacterial microbiota (92), prompting caution for the interpretation of results in mice with various genetic immunodeficiencies which may be in part due to virus reactivation. Whether viral MAMPs can modulate the alloresponse or affect the signals transmitted by their bacterial host to the immune system remains to be determined.
The microbiota and experimental transplantation
Little is known about the influence of the microbiota on alloresponses, but a suggestion that the microbiota may have a direct effect on alloresponses comes from experiments in mice lacking MyD88. In these mice, survival of skin grafts bearing minor mismatches is prolonged when the donor similarly lacks MyD88 such that TLR signals cannot be transmitted (93). However, because TLRs can be engaged not only by MAMPs but also by DAMPs induced during ischemia/reperfusion injury and the latter are known to augment alloimmunity (94), it is not possible to distinguish whether prolonged graft survival is due to diminished DAMP or PAMP signaling and more direct experiments analyzing the effects of the microbiota on alloimmunity are required in experimental settings. Studies in experimental or clinical transplantation so far have been mostly limited to observing the changes in microbial composition after transplantation and trying to correlate alterations in microbial communities with transplant fate. One exception is the attack of the immune system on the intestine during graft versus host disease (GVHD) following bone marrow transplantation (BMT), as it was shown to cause dysbiosis while the dysbiosis itself was also reported to modulate the severity of the GVHD in the only causal link to date demonstrating that the microbiota can affect alloimmunity. GVHD was associated with loss of Paneth cell function and a reduction in α-defensins, resulting in expansion of pathogenic bacteria such as E. coli (95). Moreover, the onset of intestinal GVHD was correlated both in mice and humans with a considerable loss of diversity in resident bacteria, with a shift towards Lactobacillales, and loss of Clostridiales (96). Importantly, in mice, reduction of Lactobacillales aggravated GVHD, whereas the introduction of Lactobacillus reduced GVHD, indicating an impact of this bacterial family on alloimmunity. In contrast to the importance of the recipient’s microbiota, the microbiota of the donor did not appear to influence the severity of murine GVHD after allogeneic BMT (97).
In solid organ transplantation, recent mouse studies highlight the importance of the type of organ transplanted in terms of its response to the host microbiota. In the case of liver, Kupfer cells seem to respond to MAMPs that reach the portal circulation. Indeed, intestinal bacteria can shed flagellin, a TLR-5 agonist, that upregulates ICAM-1 on hepatic sinusoidal endothelium resulting in activation of liver-derived Kupfer cells. These MAMPs could increase Kupfer cell proliferation, suppress phagocytic activity, and increase MHC class II expression, resulting in enhanced ischemia/reperfusion injury as assessed by transaminase expression in a mouse model of orthotopic liver transplantation (98). This was in part ameliorated by antibiotic treatment with down-regulated MAMP expression. Thus portal microbial products can shape the outcome of liver allografts, and these novel findings have potential implications on the outcome of both adaptive and innate responses following transplantation.
The microbiota and clinical transplantation
There are limited data to date in regards to the effects of microbiota on clinical transplant outcomes. Lymphocyte depletional therapy with alemtuzumab may perturb intestinal epithelial lymphocytes, thereby affecting the microbiota inadvertently (99). The recognition of the impact of immunosuppression on the microbiota and the routine utilization of anti-bacterial prophylaxis post-transplantation has led to immense interest by the clinical transplant community in assessing whether the microbiota plays a role in transplant outcomes. In kidney transplant recipients, a comprehensive analysis of saliva, stool, blood, and urine before and 1 and 6 months after transplantation demonstrated substantial changes in the flora of recipients (100). Moreover, there was significant inter-individual variation, suggesting that there may not be a specific microbiota signature that can mark outcomes such as renal function or rejection, although the studied population was relatively small, and transplant outcomes such as rejection were uncommon in the study period. The authors suggested that patients must be followed individually and longitudinally in order to reveal microbial changes possibly associated with pathologic conditions. This observation is in keeping with the notion seen in several other studies described below that intra-individual changes in the microbiota composition may be more important than inter-individual differences. Thus, in addition to longitudinal follow up of microbial changes within each individual, it may be important to complement these studies with profiles of the fecal metagenome, transcriptome, proteome and metabolome and how they are altered after transplantation, as microbial gene expression and function may be more conserved between individuals than the actual microbial sources of those genes and functions. Analysis of population-level effects of the microbiota during disease may help understand disease pathogenesis and help develop novel therapeutics.
A number of studies have investigated the impact of liver transplantation on the intestinal microbiota. In a recent analysis of stool flora in 12 liver transplant recipients, changes in the microbiota were assessed and correlated to post-transplant infections. A loss of microbial diversity was associated with infection; the authors suggested that the shift to pathogenic strains of bacteria due to the use of prophylactic antibiotics may be contributing to post-transplant complications (101). In a larger study, Wu et al. demonstrated marked changes in the gut microbiota post transplantation with an increase in Enterobacteriaceae and Enterococcus, and reduction in Eubacteria, Bifidobacterium, and Lactobacillus species (102). These changes however resolved over time such that by 6 months, at times when bacterial prophylaxis ends and immunosuppression is reduced, there was near complete restoration of pre-transplant patterns, with the exception of Enterococcus, as detected by quantitative PCR.
In contrast to other organs, dysbiosis was associated with rejection of small bowel allografts (103). Mercer and colleagues studied 10 transplant recipients and analyzed gut flora from ileal effluents. Compared to non-rejection samples, the phylum Firmicutes was significantly reduced while Proteobacteria were increased; moreover, Firmicutes abundance was correlated significantly with rejection supporting the notion that monitoring gut microbiota may provide an alternative diagnostic marker for small bowel rejection, which is associated with substantial morbidity and graft loss.
In 21 orthotopic lung transplant recipients, bronchoalveolar lavage and oropharyngeal washes were monitored in the first 15 months post-transplantation, and compared to findings in normal volunteers enrolled in a surveillance project. BAL from lung transplant recipients displayed a higher bacterial burden but loss of microbial diversity compared to controls (104). Candida and Aspergillus dominated the fungal species identified. However, due to the small number of subjects, it was not feasible to correlate these findings with specific transplant outcomes. In contrast, analysis of 4 recipients over three time periods demonstrated more bacterial diversity although significantly different in composition than in normal healthy individuals, with enrichment in the family Burkholderiaceae (105). In an attempt to correlate the microbiota with bronchiolitis obliterans, the leading cause of late allograft loss, 57 recipients, 8 with bronchiolitic obliterans syndrome (BOS), underwent analysis of bronchial washings (106). There were differences in microbial content in patients with cystic fibrosis compared with other transplant recipients with Pseudomonas dominating in cystic fibrosis patients and Streptococcus and Veilonella most abundant in other transplant patients and controls. Loss of pre-transplant flora was associated with BOS development. Further, return to pre-transplant microbiota predominance was protective. Thus, avoiding de novo acquisition of microbes, particularly in the same genera, could reduce the risk of BOS development.
Thus, these clinical studies reveal a loss of bacterial diversity after solid organ transplantation, but the contribution of peri-transplant therapies to these changes, the consequences of these bacterial changes on the alloresponse and whether the presence of particular microbial signatures will have some potential as biomarkers of graft fate all require additional extensive research.
Therapies and the microbiota
The stability of the microbiota can be challenged by antibiotics and dietary consumption. Clearly antibiotics can abruptly and dramatically alter the composition of the gut microbiota after 3–4 days with a return to similar pre-treatment states in weeks to months even after a second dose of ciprofloxacin (107). However, there are also examples of long-term effects of antibiotic treatment where the gut microbiota has a vastly different composition at 2 to 4 years post-treatment, with changes that appear to be individual-specific (108, 109). By comparison, the effects of diet seem to reflect a more contemporaneous relationship with the microbiota. Studies of short-term diet changes such as animal-based (meat, eggs, cheese) versus plant-based (grain, legume, fruits, vegetables) showed the former diet to more readily change the microbial composition toward bacteria associated with amino acid metabolism, then revert to baseline within 3 days of stopping the diet (110). High fat/low fiber versus low fat/high fiber also induced rapid changes in different taxa within individuals (111). The long-term effects of dietary composition have been inferred from studies of different geographic populations with fairly stable but vastly different diets (Florence, Italy versus Boulpon, Burkina Faso) and the same geographic population with different self-reported diets. Results from both studies showed a correlation between plant-based diets and either enrichment for Prevotella species among the gut microbiota or presence of previously described Prevotella enterotype (111, 112).
Recent data show that chemotherapies can also alter the microbiota, as may be expected of drugs that target cell replication. For instance, cyclophosphamide was shown to allow translocation of intestinal bacteria to secondary lymphoid organs resulting in Th17 differentiation and anti-tumor immune effects that did not occur in germ-free mice (113). Immune responses elicited by other chemotherapies were also shown to depend on the microbiota (114). How individual immunosuppressive drugs affect the microbiota and whether commensals are necessary for the efficacy of drugs taken by transplant patients remains to be investigated.
Indeed, particular bacterial species can metabolize specific drugs as recently shown with some strains of Eggerthella lenta. These strains could metabolize digoxin into inactive hydrodigoxin but their metabolic activity was inhibited by proteins from the diet, resulting in toxic rather than therapeutic drug levels (115). The impact of the microbiota on immunosuppressive drug metabolism remains to be investigated but could theoretically play a role in individual drug requirements.
Along with the interest in defining the dysbiosis associated with disease states, there has been an equal interest in developing approaches to correcting the dysbiosis. These strategies include use of antibiotics, probiotics (specific bacterial species), prebiotics (nondigestible nutrients that promote the growth of beneficial microbes), synbiotics (combinations of probiotic and the specific prebiotic that enhances its growth), postbiotics (bacterial metabolic products such as short chain fatty acids to promote immune regulation) and even fecal microbiota transplantation (38, 63, 64, 116). The latter has been used successfully to treat refractory C. difficile colitis in humans (117).
Conclusions and future directions
Thus, the gut microbiota while highly individual in composition (but highly similar in representation of functional microbial genes), established early in life and showing a capacity for resilience, can change quickly with environmental exposures perhaps sometimes irrevocably. The change in microbial composition may mean more than the change in the global representation of microbial genes, however, as microbial changes can have a profound impact on the immune system and on blood levels of oral drugs. Specific disease states are associated with gut dysbiosis but in many cases it is still unclear whether this is a cause or an effect and the application of therapy targeting the gut microbiota is very early in development. Transplant patients present a special group when considering the consequences of dysbiosis as they have been subjected to severe metabolic alterations (azotemia, hepatic insufficiency, chronic hypoxemia), extended antibiotic exposures, and finally immunomodulation which all affect the gut microbiota. More research is needed to establish how the microbiota changes during their pre-existing disease and after transplantation. Direct experiments are required to test whether commensal microbes modulate the quality and strength of the alloimmune response to solid organs, and if so by what mechanisms. Because tissues such as intestine, lung and composite grafts are colonized locally, experimental studies investigating the effect on alloresponses of donor versus recipient microbiota at the local transplant site versus in the host’s intestine which may also modulate distal immunity are also needed. If the microbiota does indeed modulate alloresponses, transplantation immunology will join other disciplines in having to decide whether and how to focus on effects of microbial gene products, versus microbial species (many genes and gene products), versus microbial communities (many organisms and many genes) on the immune system and on pharmaceuticals. Progress in bacterial culturing techniques to be able to grow presently unculturable species, more extensive genomic cataloging of the microbial commensals that have not yet been sequenced and high throughput integration of microbiome data with metabolomics and proteomics may better enable these types of studies in the future. Use of germ-free animals reconstituted not only with a human immune system but also with human microbiota are also emerging as test tubes of host-microbial interactions and may prove useful in transplantation studies. Finally, the virome, in addition and in synergy to the bacteriome, is likely to profoundly affect immune and alloresponses, but the study of its effects is in its infancy. The microbiota provides a unique opportunity for immune intervention. As different microbial communities can promote effector and regulatory networks, the hope is that therapies could harness the effects of protective microbes for the induction of transplantation tolerance.
References
- 1.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 3.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Structure function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome biology. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS computational biology. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 10.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome biology and evolution. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 16.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 18.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, et al. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflammatory bowel diseases. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- 20.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol. 2013;11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson F, Tremaroli V, Nielsen J, Backhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62:3341–3349. doi: 10.2337/db13-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridaura V. Gut microbiota from twins discordant for obseity modulate metabolism in mice. Science. 2013;341:1241214,1–10. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain, behavior, and immunity. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–55. doi: 10.1093/carcin/bgt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ussher JR, Lopaschuk GD, Arduini A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis. 2013;231:456–461. doi: 10.1016/j.atherosclerosis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25:370–377. doi: 10.1016/j.smim.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 33.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 35.Kirkland D, Benson A, Mirpuri J, Pifer R, Hou B, DeFranco AL, et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Upadhyay V, Poroyko V, Kim TJ, Devkota S, Fu S, Liu D, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 43.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol. 2012;42:5–15. doi: 10.1007/s12016-011-8285-8. [DOI] [PubMed] [Google Scholar]
- 46.Brown EM, Arrieta MC, Finlay BB. A fresh look at the hygiene hypothesis: How intestinal microbial exposure drives immune effector responses in atopic disease. Semin Immunol. 2013;25:378–387. doi: 10.1016/j.smim.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Kawai T, Akira S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Alegre ML, Leemans J, Le Moine A, Florquin S, De Wilde V, Chong A, et al. The Multiple Facets of Toll-Like Receptors in Transplantation Biology. Transplantation. 2008;86:1–9. doi: 10.1097/TP.0b013e31817c11e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 50.Brandtzaeg P. Secretory IgA: Designed for Anti-Microbial Defense. Front Immunol. 2013;4:222,1–17. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 52.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6. 2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 54.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 55.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 57.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 59.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J, et al. Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota-dependent immune tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Deroos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 65.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alegre ML, Chen L, Wang T, Ahmed E, Wang CR, Chong A. Antagonistic effect of toll-like receptor signaling and bacterial infections on transplantation tolerance. Transplantation. 2009;87(9 Suppl):S77–79. doi: 10.1097/TP.0b013e3181a2b90f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porrett PM, Yuan X, LaRosa DF, Walsh PT, Yang J, Gao W, et al. Mechanisms underlying blockade of allograft acceptance by TLR ligands. J Immunol. 2008;181:1692–1699. doi: 10.4049/jimmunol.181.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gras S, Kjer-Nielsen L, Chen Z, Rossjohn J, McCluskey J. The structural bases of direct T-cell allorecognition: implications for T-cell-mediated transplant rejection. Immunol Cell Biol. 2011;89:388–395. doi: 10.1038/icb.2010.150. [DOI] [PubMed] [Google Scholar]
- 71.Kuhn KA, Stappenbeck TS. Peripheral education of the immune system by the colonic microbiota. Semin Immunol. 2013;25:364–369. doi: 10.1016/j.smim.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chappert P, Bouladoux N, Naik S, Schwartz RH. Specific gut commensal flora locally alters T cell tuning to endogenous ligands. Immunity. 2013;38:1198–1210. doi: 10.1016/j.immuni.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–1459. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 79.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adams AB, Pearson TC, Larsen CP. Heterologous immunity: an overlooked barrier to tolerance. Immunol Rev. 2003;196:147–160. doi: 10.1046/j.1600-065x.2003.00082.x. [DOI] [PubMed] [Google Scholar]
- 83.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, et al. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. 2005;201:949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal Microbial Diversity during Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goldstein DR, Tesar BM, Akira S, Lakkis FG. Critical role of the Toll-like receptor signal adaptor protein MyD88 in acute allograft rejection. J Clin Invest. 2003;111:1571–1578. doi: 10.1172/JCI17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen H, Song Y, Colangelo CM, Wu T, Bruce C, Scabia G, et al. Haptoglobin activates innate immunity to enhance acute transplant rejection in mice. J Clin Invest. 2012;122:383–387. doi: 10.1172/JCI58344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eriguchi Y, Takashima S, Oka H, Shimoji S, Nakamura K, Uryu H, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 96.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tawara I, Liu C, Tamaki H, Toubai T, Sun Y, Evers R, et al. Influence of donor microbiota on the severity of experimental graft-versus-host-disease. Biol Blood Marrow Transplant. 2013;19:164–168. doi: 10.1016/j.bbmt.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Corbitt N, Kimura S, Isse K, Specht S, Chedwick L, Rosborough BR, et al. Gut bacteria drive Kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. The American journal of pathology. 2013;182:180–191. doi: 10.1016/j.ajpath.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li QR, Wang CY, Tang C, He Q, Li N, Li JS. Reciprocal interaction between intestinal microbiota and mucosal lymphocyte in cynomolgus monkeys after alemtuzumab treatment. Am J Transplant. 2013;13:899–910. doi: 10.1111/ajt.12148. [DOI] [PubMed] [Google Scholar]
- 100.Fricke WF, Maddox C, Song Y, Bromberg JS. Human Microbiota Characterization in the Course of Renal Transplantation. Am J Transplant. 2014;14:416–427. doi: 10.1111/ajt.12588. [DOI] [PubMed] [Google Scholar]
- 101.Lu H, He J, Wu Z, Xu W, Zhang H, Ye P, et al. Assessment of microbiome variation during the perioperative period in liver transplant patients: a retrospective analysis. Microb Ecol. 2013;65:781–791. doi: 10.1007/s00248-013-0211-6. [DOI] [PubMed] [Google Scholar]
- 102.Wu ZW, Ling ZX, Lu HF, Zuo J, Sheng JF, Zheng SS, et al. Changes of gut bacteria and immune parameters in liver transplant recipients. Hepatobiliary Pancreat Dis Int. 2012;11:40–50. doi: 10.1016/s1499-3872(11)60124-0. [DOI] [PubMed] [Google Scholar]
- 103.Oh PL, Martinez I, Sun Y, Walter J, Peterson DA, Mercer DF. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am J Transplant. 2012;12:753–762. doi: 10.1111/j.1600-6143.2011.03860.x. [DOI] [PubMed] [Google Scholar]
- 104.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS Microbiol Lett. 2013;339(1):57–65. doi: 10.1111/1574-6968.12053. Epub 12012 Dec 12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Willner DL, Hugenholtz P, Yerkovich ST, Tan ME, Daly JN, Lachner N, et al. Reestablishment of Recipient-associated Microbiota in the Lung Allograft Is Linked to Reduced Risk of Bronchiolitis Obliterans Syndrome. Am J Respir Crit Care Med. 2013;187:640–647. doi: 10.1164/rccm.201209-1680OC. [DOI] [PubMed] [Google Scholar]
- 107.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PloS one. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 110.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]