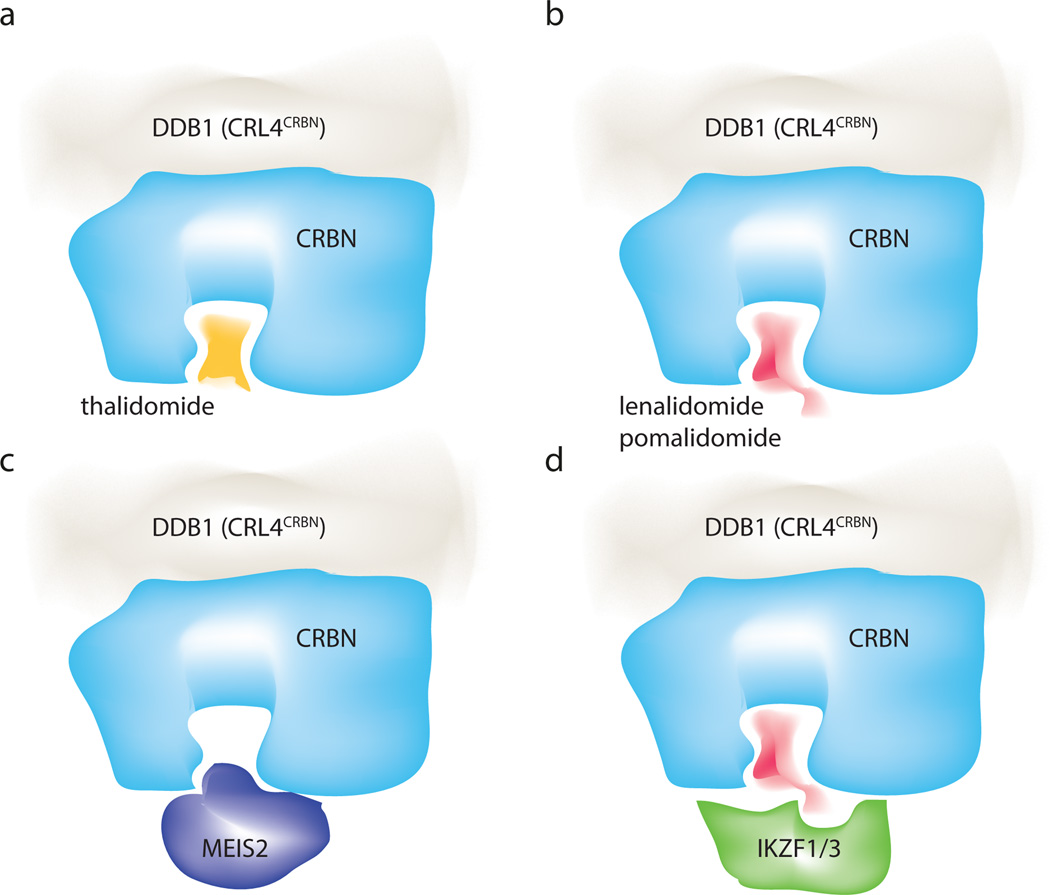
Abstract
In the 1950s the drug thalidomide administered as a sedative to pregnant women led to the birth of thousands of children with multiple defects. Despite its teratogenicity, thalidomide and its derivatives lenalidomide and pomalidomide (together known as Immunomodulatory Drugs: IMiDs) recently emerged as effective treatments for multiple myeloma and 5q-dysplasia. IMiDs target the CUL4-RBX1-DDB1-CRBN (CRL4CRBN) E3 ubiquitin ligase and promote the ubiquitination of Ikaros/Aiolos transcription factors by CRL4CRBN. Here we present the crystal structure of the DDB1-CRBN complex bound to thalidomide, lenalidomide and pomalidomide. The structure establishes CRBN as a CRL4CRBN substrate receptor, which enantioselectively binds IMiDs. Through an unbiased screen we identify the homeobox transcription factor MEIS2 as an endogenous substrate of CRL4CRBN. Our studies suggest that IMiDs block endogenous substrates (MEIS2) from binding to CRL4CRBN when recruiting Ikaros/Aiolos for degradation. This dual activity implies that small molecules can principally modulate a ligase to up- or down-regulate the ubiquitination of proteins.
Thalidomide (α-(N-phthalimido)glutarimide) was introduced to the market in 1954 by Chemie Grünenthal. Initially promoted as a sedative with anti-emetic properties1,2, it became popular in the treatment of ‘morning sickness’3. In 1961, thalidomide taken in the first trimester of pregnancy was implicated in frequent limb deformities in infants4,5. Between 8,000 and 12,000 affected children were born before the drug was banned. Interest in thalidomide revived in 1965 when it was shown to have immunomodulatory and anti-inflammatory properties in erythema nodosum leprosum (ENL), an inflammatory complication of leprosy6. In 1994, thalidomide was found to inhibit fibroblast growth factor (bFGF)-induced formation of new blood vessels7. These findings prompted clinical trials exploring thalidomide use in anti-angiogenic cancer therapy. The efficacy of thalidomide and its derivatives lenalidomide and pomalidomide (collectively known as IMiDs) has since been demonstrated in several blood cancers8 : newly diagnosed multiple myeloma (thalidomide)9, refractory multiple myeloma (lenalidomide/pomalidomide) and 5q-deletion-associated myelodysplastic syndrome (lenalidomide).
The target of thalidomide, cereblon (CRBN), is a ubiquitously expressed protein that is part of the cullin-4 E3 ubiquitin ligase complex, CUL4-RBX1-DDB1 (CRL4)10. Mutations in CRBN are associated with autosomal recessive non-syndromic mental retardation (MR)11. In myeloma cells, the anti-proliferative activities of IMiDs are linked to CRBN expression12,13, making IMiDs the first clinically approved E3 ubiquitin ligase inhibitors with specificity for the CRL4CRBN ligase12. The IMiD anti-proliferative and immunomodulatory effects have recently been linked to drug-induced ubiquitination and degradation of Ikaros (IKZF1) and Aiolos (IKZF3) transcription factors by CRL4CRBN14 –16. Accordingly, loss of CRBN is a common determinant of drug resistance in myeloma cells12. How IMiD binding affects the CRL4CRBN ligase at the molecular level remains unclear. We set out to examine the role of CRBN within the CUL4-RBX1-DDB1-CRBN (CRL4CRBN) E3-ligase complex, characterising the effect of IMiD binding on ligase activity.
Structure of DDB1-CRBN bound to IMiDs
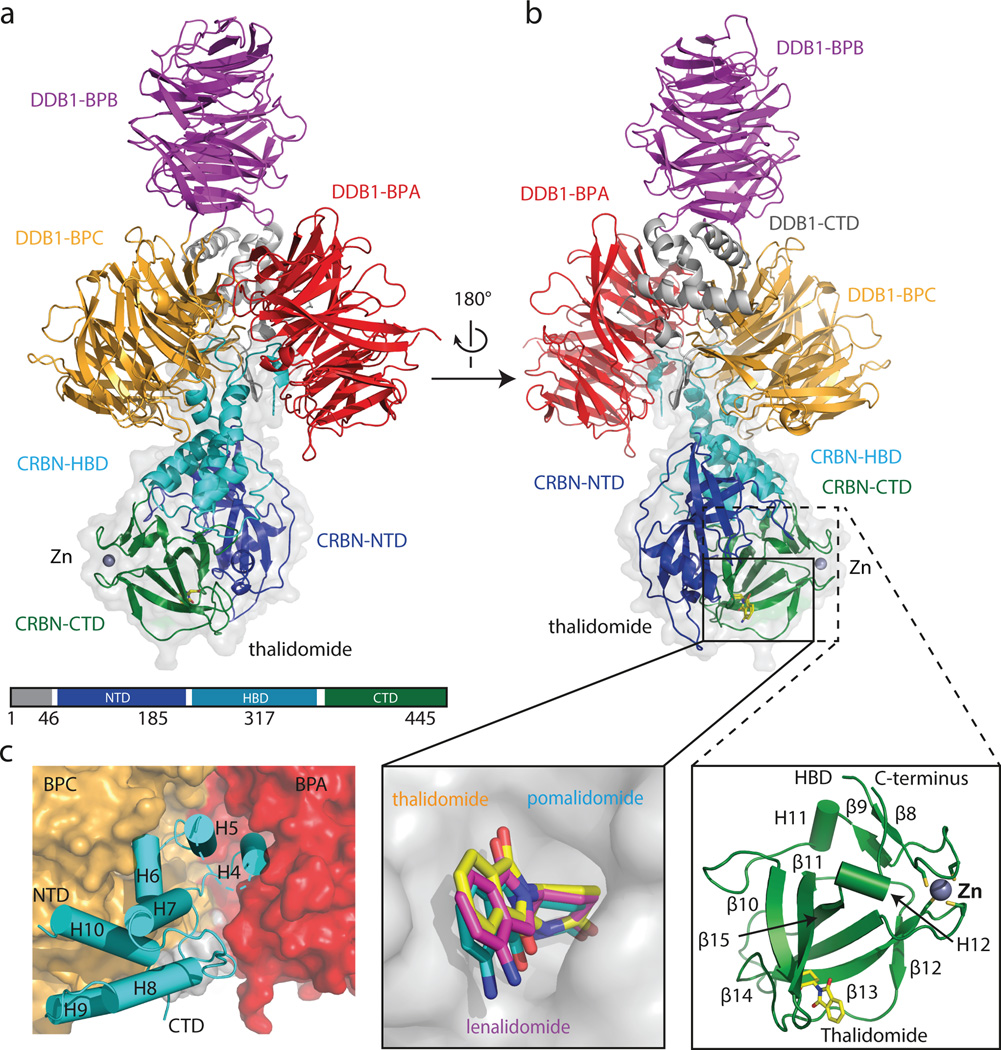
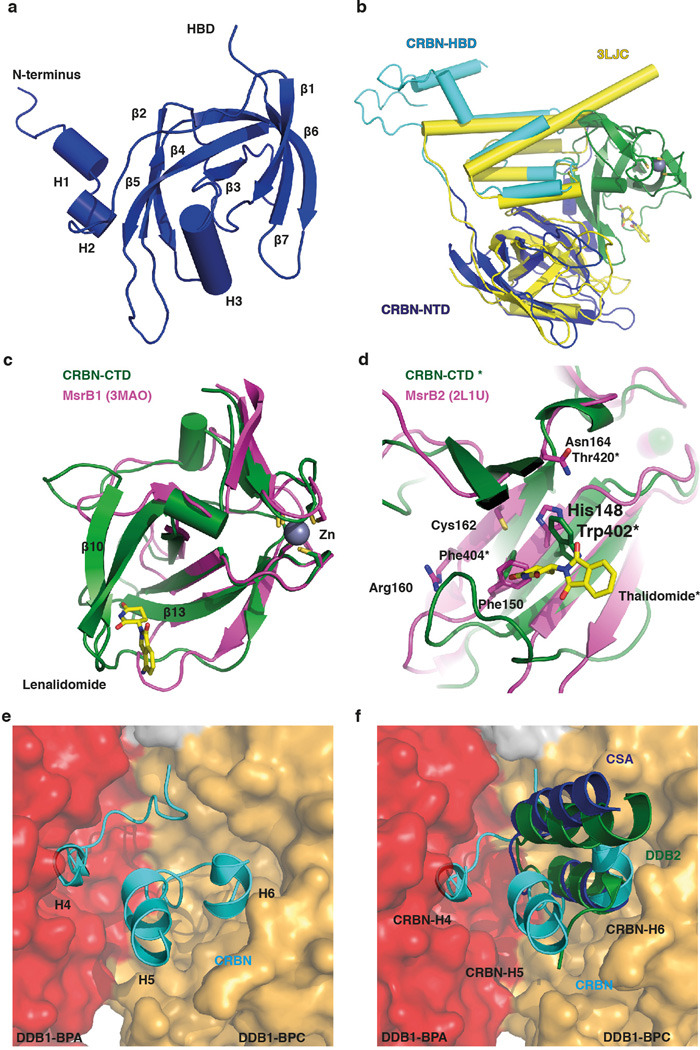
We crystallized a chimeric complex of human DDB1 (DDB1) and chicken CRBN (ggCRBN) bound to thalidomide (refined to 3.0 Å), lenalidomide (3.0 Å) and pomalidomide (3.5 Å) (Fig. 1a, b and Extended Data Table 1). The high level of sequence conservation between human and chicken CRBN (Extended Data Fig. 1a, b) allows structural insights to be inferred directly from chicken to human CRBN. All subsequent biochemical and cell-biological experiments were performed with human full-length proteins. ggCRBN consists of three sub-domains (Extended Data Fig. 2a–f): a seven-stranded β-sheet located in the N-terminal domain (NTD, residues 1–185) (Extended Data Fig. 2a), a 7-α-helical bundle domain (HBD, residues 186–317) involved in DDB1 binding (Fig. 1c), and a C-terminal domain composed of 8 β-sheets (CTD, residues 318–445) (Fig. 1b). DDB1 comprises three seven-bladed WD40 β-propellers arranged in a triangular fashion (BPA, BPB and BPC)17 with ggCRBN attaching to a cavity between the BPA and BPC propellers (Fig. 1c). The molecular basis of the HBD-mediated attachment of ggCRBN to DDB1 defines a novel class of DDB1 binders and differs in detail from previous DDB1 attachment modules17 –20 (Extended Data Fig. 2e, f).
Figure 1. Overall structure of the DDB1-CRBN complex.
(a) Cartoon representation of the hsDDB1-ggCRBN-thaliomide structure: DDB1 highlighting domains BPA (red), BPB (magenta), BPC (orange) and DDB1-CTD (grey); ggCRBN highlighting domains NTD (blue), HBD (cyan) and CTD (green). The Zn2+-ion is drawn as a grey sphere. (b) As in (a) with the thalidomide shown as yellow sticks. A close-up showing that all IMiDs occupy a common binding site on CRBN, and a close-up of the overall ggCRBN-CTD architecture. (c) ggCRBN-HBD helices and their interactions with DDB1.
The ggCRBN N-terminal region (residues 46–317) including the NTD and HBD resembles the N-terminal domain of bacterial Lon proteases (PDB: 3LJC - RMSD of 2.7 Å over 178 residues aligned) (Extended Data Fig. 2b). The CTD harbours the thalidomide-binding pocket and contains a conserved Zn2+-binding site situated approximately 18 Å from the compound (Fig. 1a, b). The Zn2+ ion is coordinated through conserved cysteine residues 325, 328, 393 and 396. The ggCRBN-CTD shares structural similarity with the pseudouridine synthase and archaeosine transglycosylase (PUA) fold family21 involved in the binding of diverse sets of ligands (Extended Data Fig. 2c, d).
IMiD binding to CRBN
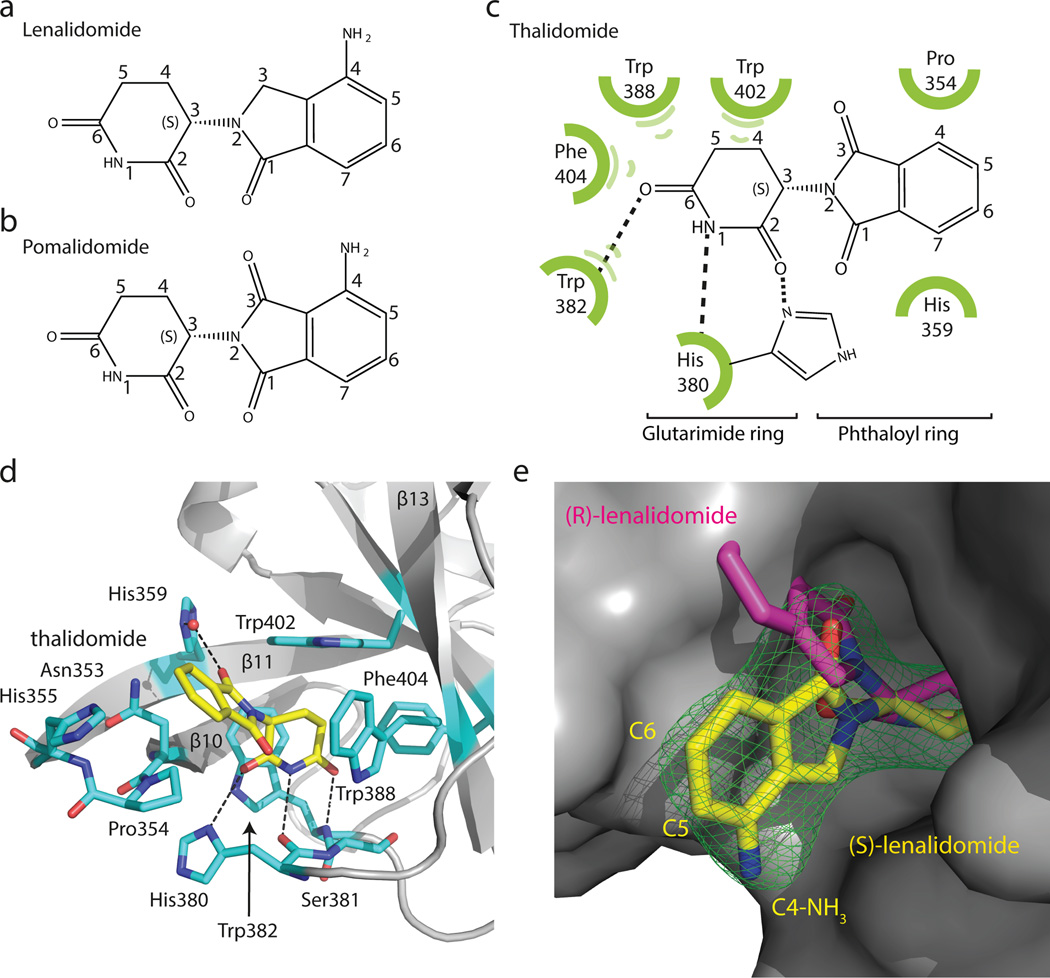
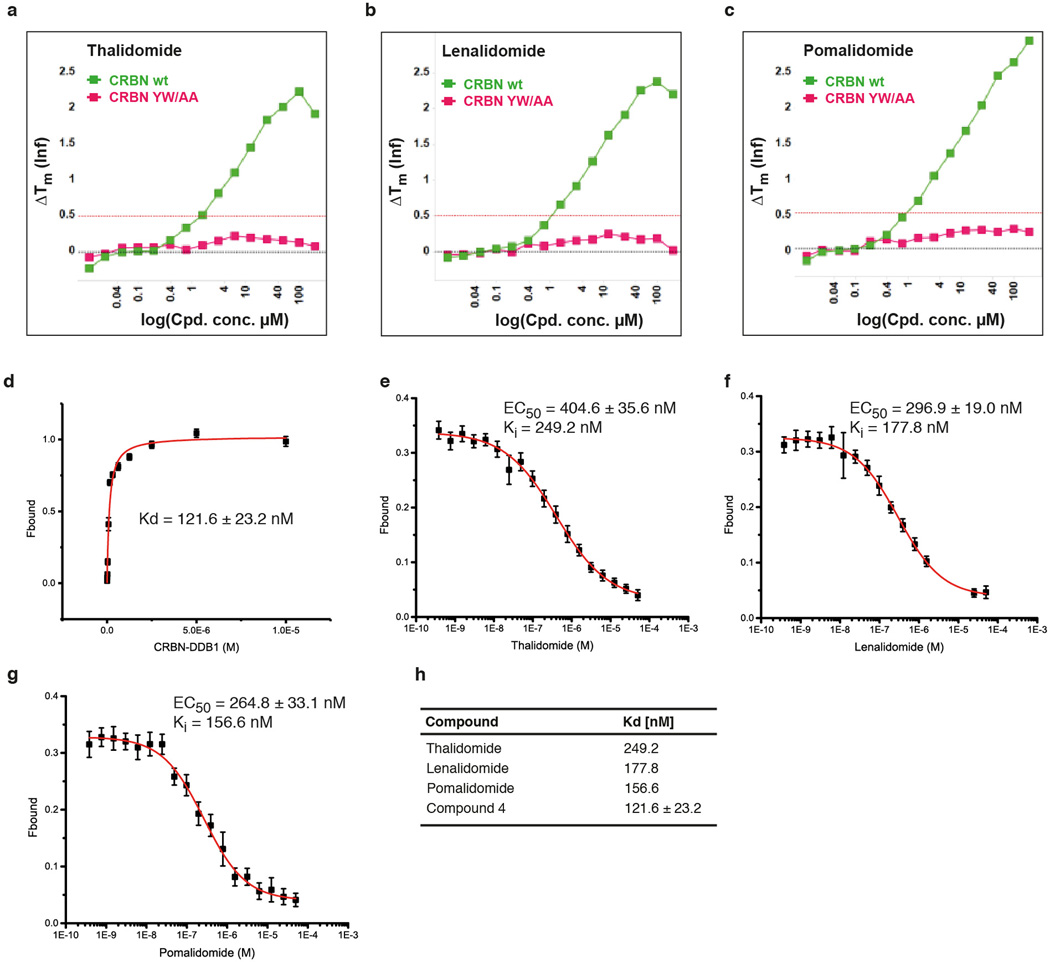
Thalidomide, lenalidomide and pomalidomide (Fig. 2a–c and Extended Data Fig. 3a–i) bind a pocket on the ggCRBN-CTD (Fig. 1b) situated in a surface groove that is highly conserved across CRBN orthologues (Extended Data Fig. 1b). The three ligands superimpose with very little deviation in the α-(isoindolinone-2-yl) glutarimide moiety, which contributes the majority of interactions between the receptor and the compounds and represents the main pharmacophore22. The glutarimide group is held in a buried cavity between ggCRBN sheets β10 and β13(Fig. 2d). Glutarimide carbonyls (C2, C6) and the intervening amide (N1) are in hydrogen-bonding distance to ggCRBN residues His380 and Trp382, respectively (Fig. 2c, d). A delocalised lone pair connects the glutarimide nitrogen with the two glutarimide carbonyls (C2-N1-C6) and is coplanar with Trp382. The opposing aliphatic face of the glutarimide ring (C3, C4 and C5) is in tight Van-der-Waals contact with a hydrophobic pocket lined by Trp382, Trp388, Trp402 and Phe404. In vitro, mutations of Tyr386 (affecting the integrity of the binding pocket) and Trp388 (directly involved in compound binding) to alanine ablate binding of all three IMiDs to CRBN (Extended Data Fig. 4a–c)10. Mutations of the equivalent residues render CRL4CRBN insensitive to the presence of thalidomide and lenalidomide in vivo10,12. In addition to near identical binding modes, we found that thalidomide, lenalidomide and pomalidomide have similar affinities for CRBN with dissociation constants of ~250 nM, ~178 nM and ~157 nM, respectively (Extended Data Fig. 4d–h).
Figure 2. Thalidomide binding to CRBN.
(a) Chemical structure of lenalidomide and (b) pomalidomide. (c) Sketch of thalidomide and its interactions with ggCRBN. (d) IMiDs are anchored through the glutarimide functionality hydrogen-bonding to ggCRBN His380 and Trp382 and the aliphatic face of the glutarimide engulfed in a hydrophobic cage. (e) Surface representation of ggCRBN (grey) and (S)-lenalidomide shown as yellow sticks, together with its positive mFo-DFc electron density (σ=3.5) shown in green. The fit of the (S)- and (R)-enantiomers is indicated.
Thalidomide and lenalidomide/pomalidomide differ in the C4 phthalimide aniline functionality (Fig. 2a–c), which is found in a solvent-exposed position. The common carbonyl at the phthaloyl C1-position contributes a water mediated hydrogen bond to His359, which anchors the phthaloyl ring-system together with stacking interactions provided by the aliphatic face of Pro354 (Fig. 2d). The phthalimide C5 and C6 positions are fully solvent exposed. The overall shape of the buried IMiD binding pocket, favours binding of (S)-thalidomide over the (R)-enantiomer (Fig. 2e), which is in agreement within vivo experiments12.
CRBN functions as a DCAF for the CRL4CRBN ligase
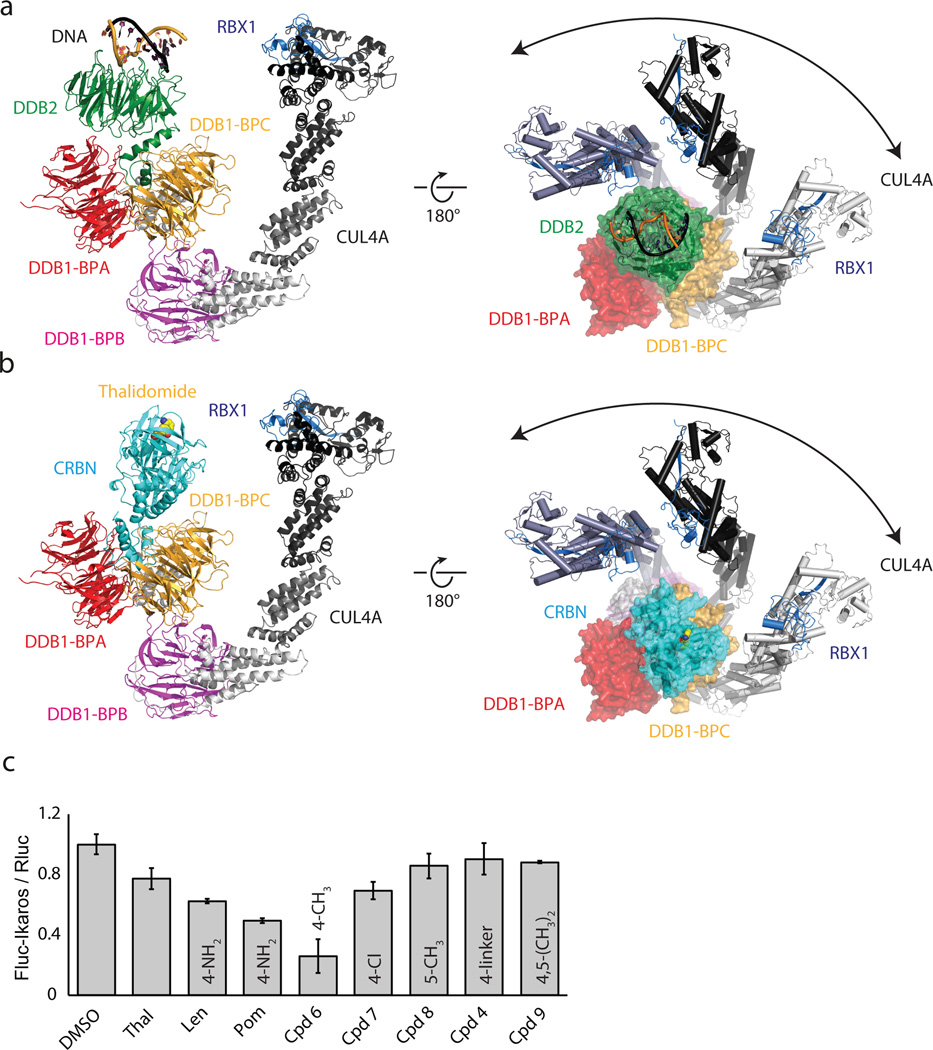
Within the CRL4 ligase family, DDB1 functions as the adaptor connecting the substrate receptor to the CUL4 ligase17,19,23. More than a dozen substrate receptors, including CRBN, have been identified (designated DCAFs: DDB1 CUL4 associated factor). ggCRBN, despite lacking the canonical DCAF WD40 fold, resembles a substrate receptor in dimensions and position on DDB1 (Extended Data Fig. 5a, b). The thalidomide binding site is situated where substrates generally bind to WD40 DCAFs (see e.g. DDB2-engaging DNA Fig. 3a and Extended Data Fig. 5a, b). Equivalent residues in the structurally related PUA domain-containing proteins are directly engaged in ligand binding (Extended Data Fig. 5c–e). The PUA domain of CRBN has striking structural similarity to a member of the methionine sulfoxide reductase (Extended Data Fig. 2c, d). In ggCRBN, the now defunct active centre of the reductase interacts with the IMiDs. Truncation of the CRBN C-terminus bordering the conserved thalidomide-binding pocket has been found in non-syndromic mental retardation (see Extended Data Fig. 6a–c for analysis of CRBN mutations).
Figure 3. CRBN serves as a substrate receptor to the CRL4CRBN ligase.
(a) Architecture of the CRL4DDB2 complex bound to DNA (pdb:4A0K). (b) Model of the CRL4CRBN ligase bound to thalidomide. (c) Firefly-luciferase (Fluc) to renilla-luciferase (Rluc) ratios (Fluc/Rluc) of IKZF1-reporter-plasmid transfected HEK293T cells following incubation with the indicated compounds. Data shown are the means ± SEM (n=4).
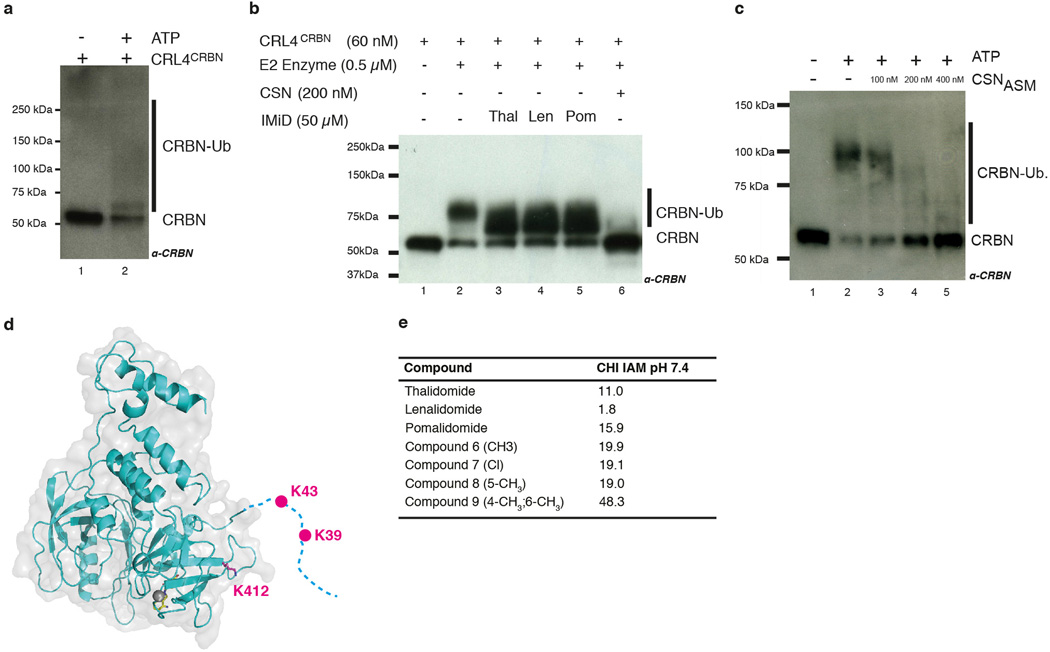
Within CUL4-RBX1-DDB1-DCAF complexes, the cullin was found to freely rotate up to 150° around DDB1 (Fig. 3a, b)24,25. CUL4 mobility is undirected and driven exclusively by Brownian motion. Given the strictly modular architecture of the CRL4 family23,24,25, the structure of the CRL4CRBN ligase can be predicted with high confidence (Fig. 3b). Rotation of the CUL4 ligase arm around DDB1 and CRBN establishes a ubiquitination zone with dimensions of up to 340Å × 110Å × 30Å (Fig. 3b), with the centre of rotation near the thalidomide-binding site. The CRL4 ligase is promiscuous, targeting lysines that cross this ubiquitination zone. Accordingly, we observe in vitro that CRBN is autoubiquitinated (Extended Data Fig. 7a–d) at the unstructured N-terminal tail (residues K39 and K43, see also Extended Data Fig. 7d). We find that CRBN autoubiquitination persisted in the presence of IMiDs in vitro (Extended Data Fig. 7b) and is subject to inhibition by the Cop9 signalosome (CSN) (Extended Data Fig. 7b, c).
Pomalidomide and lenalidomide effectively target the Ikaros transcription factors IKFZ1 and 3 for degradation by CRL4CRBN. Thalidomide, on the other hand, is here less efficient14 –16 (Fig. 3c). All three IMiDs have comparable affinities for CRBN (Extended Data Fig. 4e–h) and similar predicted membrane permeabilities (Extended Data Fig. 7e). The major structural difference between CRBN-bound lenalidomide/pomalidomide and thalidomide lies in the presence of the solvent-exposed C4aniline functionality in the former. Therefore, different functional groups at the phthaloyl C4, C5 and C6 positions were tested for their ability to degrade Ikaros in a dual luciferase reporter assay14 (see Methods). Small groups attached to C4 (-NH3; -CH3 and to some extent -Cl) promote Ikaros degradation (Fig. 3c). On the other hand, larger substituents at the C4, or C5 and C6 positions were less effective in Ikaros degradation. These modifications are not expected to affect CRBN binding, as even a large substituent at the C4 position had no adverse effect on affinity (Extended Data Fig. 4d). Assuming comparable cellular concentrations27,28,29, this would imply that solvent-exposed bulky groups at C4 (and methyl groups at C5 and C6) interfere with Ikaros binding. The aniline and methyl substituents at C4, on the other hand, appear to contribute to Ikaros degradation, likely through direct interactions.
MEIS2 is an endogenous CRL4CRBN ligase substrate
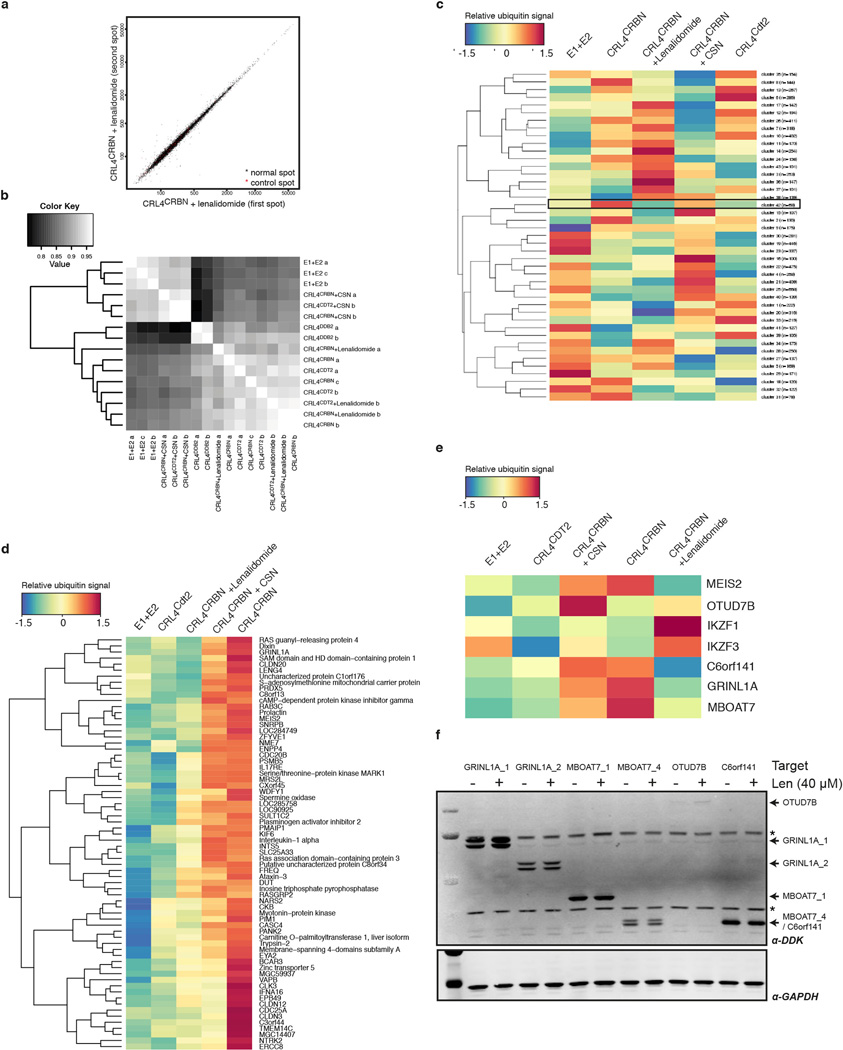
Next, we set out to examine the effects of IMiDs on endogenous CRBN substrates, which have so far remained elusive. An unbiased biochemical screen was established to identify proteins ubiquitinated by CRL4CRBN. Human protein microarrays (~9,000 proteins) were used for on-chip ubiquitination by CRL4ACRBN in the presence of E1 (Uba1), E2 (UbcH5a), ubiquitin (biotin-ubiquitin) and ATP (Extended Data Fig. 8a–e). We reasoned that a substrate would be subject to ubiquitination by CRL4CRBN but not by a control ligase (CRL4Cdt2), that a substrate would overcome inhibition of the CRL4CRBN ligase by CSN24, and that ubiquitination of such substrates should be inhibited by lenalidomide. Following cluster analysis (43 clusters), we identified one cluster that best matched the expected ubiquitination profile (see Methods for details). In an orthogonal screen, the top five candidate genes (GRINL1A, MBOAT7, OTUD7B, C6orf141 and MEIS2) where overexpressed in HEK293T cells and assessed for lenalidomide induced changes insteady-state levels (Extended Data Fig. 8f). Of these, we focused on MEIS2, a transcription factor implicated in different aspects of human development30 –32, which we found was stabilised upon lenalidomide treatment (see below).
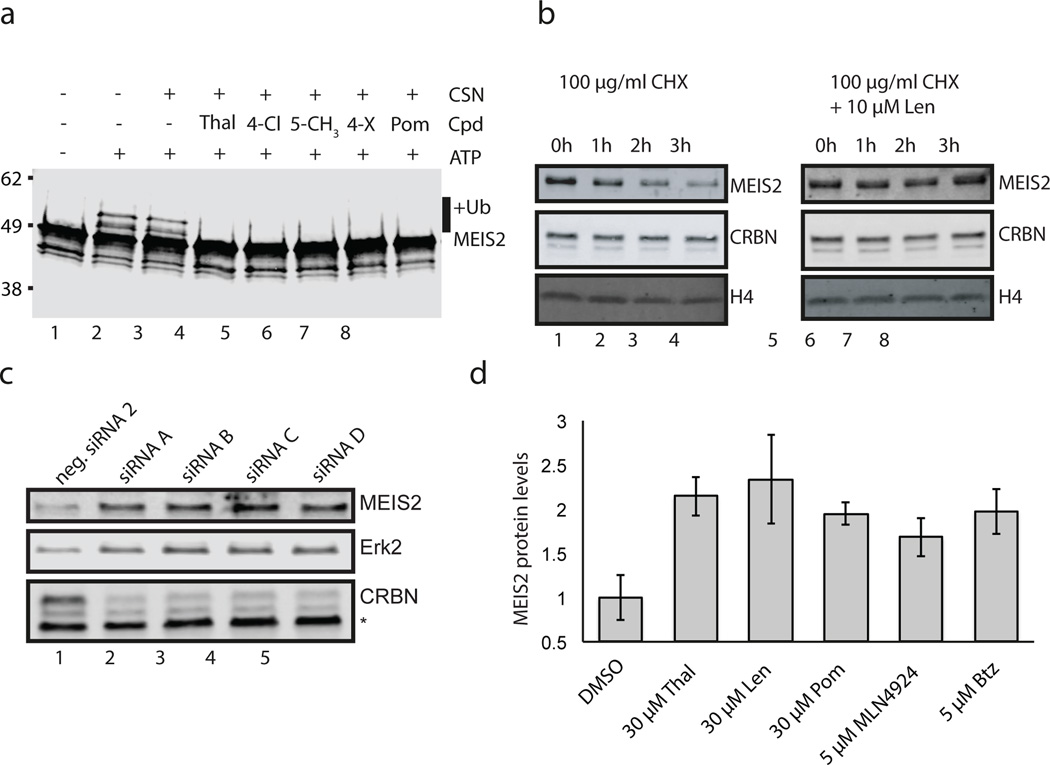
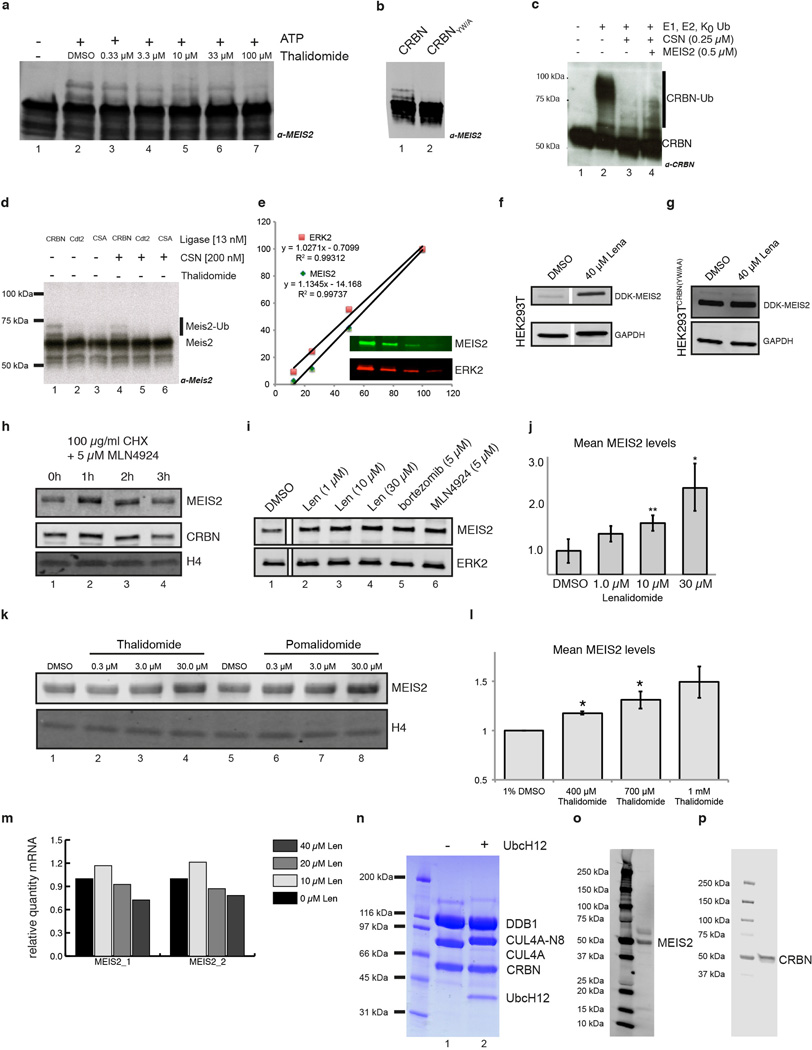
To recapitulate the on-chip results in solution, insect cell derived MEIS2 was used to establish a fully recombinant CRL4CRBN ubiquitination system. MEIS2 expressed from insect cells was isolated in a hyper-phosphorylated form. Dephosphorylation was found to improve protein behaviour, resulting in increasedMEIS2 ubiquitinationusing lysine-free (K0) ubiquitin (Fig. 4a compare lanes 1 and 2). MEIS2 ubiquitination was also observed using phosphorylated MEIS2 (data not shown). MEIS2 ubiquitination by CRL4CRBN was subject to inhibition by different IMiDs (Fig. 4a, lanes 4–8 and Extended Data Fig. 9a), irrespective of the chemical substituent at the C4, C5 and C6 positions. A CRL4CRBN mutant carrying the Y384A/W386A substitutions (CRBNYW/AA) that rendered the receptor unable to bind IMiDs (Extended Data Fig. 3a–c) also impaired MEIS2 ubiquitination (Extended Data Fig. 9b). In accordance with our on-chip results, we find that MEIS2 is able to relieve the inhibition of CRL4CRBN by CSN (Extended Data Fig. 9c) and is not ubiquitinated by the control ligases CRL4CSA and CRL4Cdt2 (Extended Data Fig. 9d). This data suggests that, within the CRL4CRBN complex, CRBN targets MEIS2 for ubiquitination in vitro with the aid of its IMiD binding site.
Figure 4. MEIS2 is an endogenous substrate of the CRL4CRBN ligase.
(a) CRL4CRBN ubiquitinates MEIS2 in vitro (lane 2), a reaction inhibited by thalidomide (lane 4), pomalidomide (lane 8), compound 7 (lane 5), compound 8 (lane 6), and compound 4 (lane 7). K0 ubiquitin was used to obtain a defined band-shift detected by anti-MEIS2 immunoblotting. (b) SK-N-DZ cells were pretreated with 10 µM lenalidomide or DMSO before addition of 100 µg/ml CHX for the indicated times. MEIS2 and CRBN levels were detected using anti-MEIS2 and anti-CRBN immunoblots. Histone H4 served as a loading control. (c) Four siRNA constructs (labelled A–D) targeting CRBN and a negative control were transfected into M059J cells and the levels of endogenous CRBN and MEIS2 monitored. An asterisk indicates a non-specific band and Erk2 served as a loading control. (d) Treatment of M059J cells for 4 h with the indicated amounts of IMiDs, MLN4924, or bortezomib (Btz) resulted in an increase in steady-state MEIS2 levels. Data shown are the means ± SEM (IMiDs: n=3; MLN4924 and Btz: n=2).
We next sought to test the effect of lenalidomide on MEIS2 half-life in cells by performing cycloheximide (CHX) chase experiments. RNA-seq. data from the cancer genome atlas (TCGA) identified the neuroblastoma cell line SK-N-DZ as having high levels of endogenous MEIS2 and CRBN transcripts. Following treatment with 100 µg/ml CHX, we found MEIS2 protein levels to be largely depleted after 3 h (Fig. 4b, lanes 1–4). Addition of 10 µM lenalidomide stabilised MEIS2 protein levels under these conditions (Fig. 4b, lanes 5–8). As SK-N-DZ cells were recalcitrant to transfection, we subsequently employed M059J cells, similarly characterized by high levels of MEIS2 mRNA (TCGA), to examine the effects of CRBN depletion by RNAi on MEIS2 abundance. For all experiments, endogenous MEIS2 levels were monitored by quantitative western blot using infrared detection (see Extended Data Fig. 9e–p). Four siRNAs were used to transfect M059J or HEK293T cells, resulting in efficient CRBN knockdown (Fig. 4c) and increased MEIS2 steady-state levels in M059J (Fig. 4c) and HEK293T (data not shown), implicating CRBN in MEIS2 turnover. To test whether lenalidomide treatment results in increased steady-state levels of MEIS2, M059J cells were treated with lenalidomide or a DMSO control. Following lenalidomide treatment endogenous MEIS2 was elevated up to 2.5-fold (Fig. 4d and Extended Data Fig. 9i–k). A similar level of steady-state MEIS2 stabilisation was observed with MLN4924 (Extended Data Fig. 9i, lane 6), a Nedd8-activating enzyme inhibitor, and with the proteasome inhibitor bortezomib (Extended Data Fig. 9i, lane 5). Increases in MEIS2 protein levels following lenalidomide exposure were not due to mRNA changes, as determined by quantitative RT-PCR (Extended Data Fig. 9m). When examining different IMiD derivatives, we found thalidomide, lenalidomide and pomalidomide to stabilise steady-state MEIS2 levels to a similar extent (Fig. 4d and Extended Data Fig. 9i, k). The effect of thalidomide on MEIS2 levels was also observed in vivo, where whole zebrafish embryos 24 h post-fertilisation showed a 1.5-fold increase in MEIS2 levels, comparable to that observed in cell lines (Extended Data Fig. 9l).
Thalidomide is an agonist and antagonist
We find in vitro and in vivo that IMiD binding is mutually exclusive with the recruitment of MEIS2 (Fig. 4a, 5a–c and Extended Data Fig. 9b). Accordingly, a stable cell line overexpressing an epitope-tagged MEIS2 together with a CRBNYW/AA mutant (Extended Data Fig. 9f, g) exhibited no changes in MEIS2 levels following addition of 40 µM lenalidomide. MEIS2 thus is an endogenous, IMiD-sensitive, target of CRBN (see Supplementary Discussion for a putative role of MEIS2 in IMiD mediated teratogenicity). Most CRL ligases studied so far target multiple substrates33,34. Given that IMiDs occupy the canonical ligand interface of the CRBN PUA domain, we also expect other endogenous substrates to be pre-empted from CRBN binding by thalidomide and its derivatives. On the other hand, IMiD-dependent targeting of Ikaros transcription factors (Fig. 5d) is facilitated by specific solvent-exposed functionalities that are not involved in CRBN binding. We propose a model where the interaction surface used for Ikaros binding comprises CRBN (in its IMiD-bound form) and the C4, C5 and C6 phthaloyl positions of the IMiD. While structural studies on CRBN-IMiD-Ikaros complexes are required to shed insights on the detailed binding mode, conceptually this mechanism bears striking similarities to auxin-induced degradation of Aux/IAA by the TIR1 ligase35 and to cyclosporine/FK506-induced cyclophilin/FKBP12 binding to calcineurin36,37. The CRL4 architecture supports ubiquitination in its vicinity, a property exploited by viral proteins in recruiting cellular targets for degradation by CRL4s17. As small molecules are apparently able to mimic this behaviour, it will be important to explore whether synthetic small molecules can promote the degradation of other substrates, which are not typically targeted by a specific ubiquitin ligase.
Figure 5. Molecular basis of IMiD function.
(a) Thalidomide binds the CRBN receptor at the canonical substrate binding site. (b) The potent anti-myeloma derivatives lenalidomide and pomalidomide occupy the same site, while having different solvent exposed moieties. (c) Binding of the endogenous substrate MEIS2 and the IMiDs to this site is mutually exclusive. (d) We propose a direct interaction of the C4 amine of lenalidomide/pomalidomide with the Ikaros transcription factor recruitment. Binding of Ikaros likely also involves surrounding residues of the CRBN receptor.
Our structure/function analysis indicates that IMiD-mediated Ikaros degradation interferes at the same time with the recruitment of endogenous substrates (such as MEIS2) to CRL4CRBN. Depending on the cell type and the proteins expressed, administration of thalidomide and its derivatives will thus simultaneously affect the levels of two groups of proteins: upregulating the endogenous substrates, while decreasing the neo-substrates. Thalidomide and its IMiD derivatives give rise to pleiotropic clinical manifestations ranging from anti-cancer and immunomodulatory properties to pronounced teratogenicity. Both loss of ubiquitination, as seen for MEIS2, as well as a gain of function seen for the Ikaros family and even complex synthetic combinations of these opposing changes need to be considered as underlying causes for these diverse clinical effects.
Extended Data
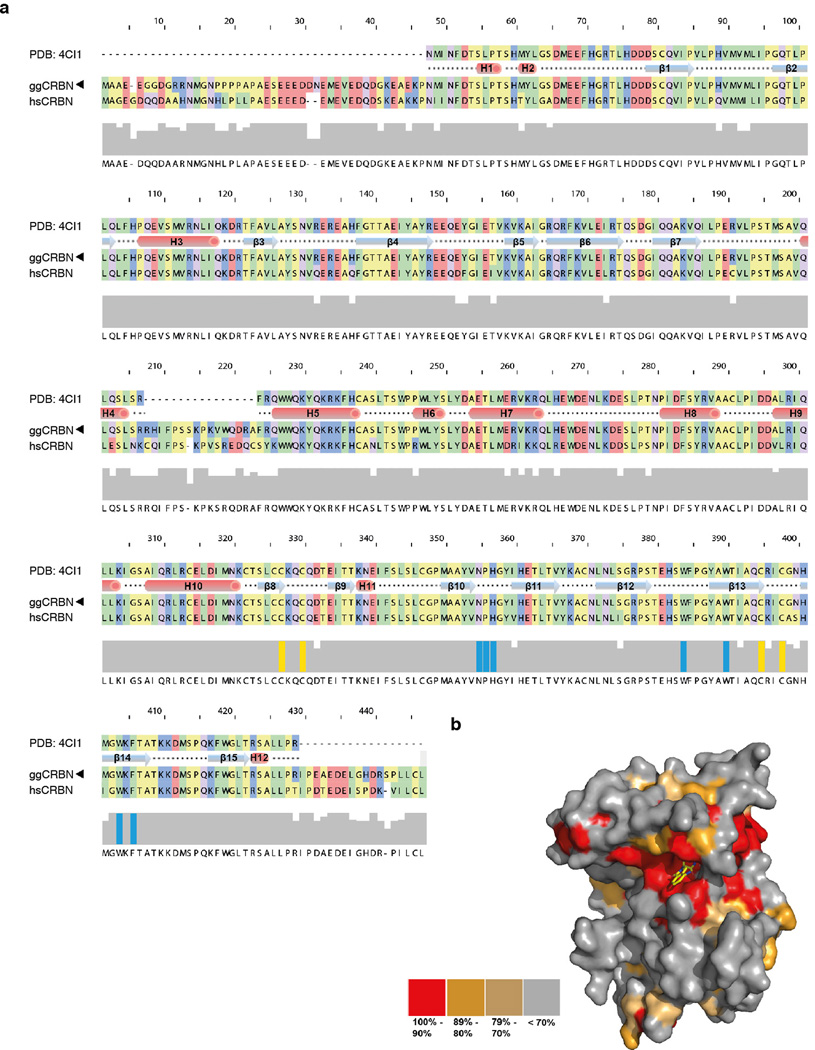
Extended Data Figure 1. Sequence conservation of CRBN.
(a) Alignment of ggCRBN and hsCRBN; the Zn coordinating domain and residues involved in compound binding are highlighted in yellow and cyan, respectively. CRBN protein secondary structure is shown at the top. (b) Surface representation of CRBN orthologues highlighting evolutionary conserved residues (sequences used for the alignment include human, gibbon, rhesus, gorilla, giant panda, marmoset, cat, wolf, horse, ferret, greater galapagos, elephant, naked mole rat, orang utan, guinea pig, sheep, chinese hamster, mouse, rat, vampire bat, cow, opossum, anoli carol, chicken, zebra finch, turkey, xenopus, nile tilapia, pufferfish, fugo, and zebrafish). Residues that cluster around the compound binding pocket are highly conserved38.
Extended Data Figure 2. Structure of the DDB1-CRBN complex bound to thalidomide and derivatives.
(a) Close-up view and topology of the ggCRBN N-terminal domain (NTD). (b) Overlay of the ggCRBN-NTD (dark-blue) and the helical bundle domain (HBD) (light-blue) with the Lon-protease domain coloured yellow (PDB: 3LJC - RMSD of 2.7 Å over 178 residues aligned). How Lon-proteases recognise their substrates is unclear at present. As both the NTD and the HBD are present in the Lon-fold, it is conceivable that CRBN originated from a fusion of a Lon-protease with a PUA fold-containing enzyme (see below). In the course of divergent evolution, the helical element of the Lon-fold appeared to be utilised for DDB1 binding. (c) The ggCRBN C-terminal domain (CTD: residues aa318-aa427) harbours the thalidomide, lenalidomide, or pomalidomide binding pocket and displays structural similarity to pseudouridine synthase and archaeosine transglycosylase (PUA) folds. The alignment with the methionine sulfoxide reductase subfamily, the closest structural ggCRBN orthologue (PDB: 3MAO - RMSD of 2.0 Å over 79 residues) is shown. The PUA domain fold family frequently carries a conserved metal coordination site, which in ggCRBN appears to bind Zn2+ and is coordinated by Cys325, 328, 393 and 396. (d) Residues involved in the catalytic activity in the MsrB2 reductase (Trp103, His148, Phe150, Arg160, Cys162 and Arg162). The catalytic cysteine (Cys 162) residue is not conserved in ggCRBN, which makes it unlikely that ggCRBN acts as an MsrB2-like reductase. Interestingly, however, residues equivalent to Trp402 and Phe404 involved in ggCRBN thalidomide binding are also involved in substrate binding and catalysis in MsrB2. The conserved binding patch surrounding lenalidomide may thus act as a substrate binding interface, with thalidomide possibly blocking binding of a hitherto unknown protein substrate, or a specific post-translation modification. (e) The ggCRBN interaction with DDB1 is mediated through the HBD motif utilizing ggCRBN α-helices H5: 224–233, H6: 244–249, and H4: 200–204, a 310 helix. ggCRBN helices H5 and H6 together with the intervening loop segment form extensive interactions with the DDB1-BPC propeller, with additional contributions from residues ggCRBN 190–195. ggCRBN H4 and the preceding loop (197–205) form interactions with DDB1-BPA. ggCRBN binding to BPA occurs through a H1 310 helix located at the central cavity of the WD40 propeller, the narrow side of the WD40-propeller cone where ligands typically bind. (f) ggCRBN binding to DDB1 differs from that previously seen in DCAF-DDB1 complexes, where the majority of interactions outside the WD40 propeller domain originated from a consecutive helix-loop-helix (HLH) motif that predominantly contacts DDB1-BPC (golden-brown surface). Extensive interactions between a DCAF and DDB1-BPA domain (red surface) have not been observed previously, in either the DDB1 co-crystal structures with DDB2 (pdb: 3EI3; green), CSA (pdb: 4A11; dark blue) or other HLH motifs to which DDB1 binds. The novel DDB1 binding mode found in CRBN precluded prior sequence-based identification and suggested considerable plasticity of DCAF binding to DDB1.
Extended Data Figure 3. Thalidomide derivatives used in this study.
(a) Skeletal formula of thalidomide, (b) lenalidomide, and (c) pomalidomide. (d) A thalidomide derivative containing a flexible pendant amine linker at the C4 position (4-OCH C(O)N(CH2)4NH2) of the phthalimide ring system has been synthesised as described in the Material and Methods to derive (e) Cy5-labeled thalidomide. Thalidomide analogues have been synthesised, including the substitution of the C4 amino group with (f) -CH3 or (g) –Cl, (h) a –CH3 at the C5 position and (i) a –CH3 at the C4 and C6 positions.
Extended Data Figure 4. Detailed analysis of CRBN-compound interactions.
(a) Thermal denaturation assays of thalidomide binding to hsDDB1-hsCRBN wild type (CRBN wt) and to an hsDDB1-hsCRBN mutant harbouring mutations Tyr386Ala and Trp388Ala (CRBNYW/AA). While the mutant showed no sign of binding to thalidomide, wild type CRBN caused a shift in the thermal denaturation curve indicative of compound binding. (b) As in (a), but using lenalidomide or (c) pomalidomide. (d) Increasing amounts of hsDDB1-hsCRBN were mixed with 20 nM Cy5-coupled thalidomide (compound 5). Protein-compound interactions were quantified using fluorescence polarisation of the Cy5 dye. Curve fitting to a model assuming one binding site resulted in a KD of 121.6 ± 23.2 nM. Data shown are the means ± SD (n=3).(e) Competitive titration of thalidomide to hsDDB1-hsCRBN (50 nM) and Cy5-thalidomide (20 nM); EC50 values were used to calculate a Ki50 (equivalent to the KD of thalidomide to hsDDB1-hsCRBN) of 249.20 nM. Similar titrations were carried out for (f) lenalidomide and (g) pomalidomide resulting in Ki values of 177.80 nM and 156.60 nM, respectively. For all competitive titrations (e–g) data shown are the means ± SD (n=3).(h) Affinities of thalidomide analogues for the CRBN receptor.
Extended Data Figure 5. Details of CRBN as a substrate receptor to the CRL4CRBN ubiquitin ligase complex.
(a)) Architecture of CRL4DCAF complexes: DDB1-DDB2 (pdb: 3EI2), DDB1-CSA (pdb: 4A11) and the DDB1-CRBN complex are shown side-by-side. (b) Overlay of the structure of DDB1-CRBN, with CRBN shown in cyan, and that of DDB1-DDB2 (pdb:3EI2) with DDB2 shown in green. The DDB2 footprint on DDB1 fully overlaps with that of CRBN, rendering binding to DDB1 mutually exclusive. Mutual exclusivity of DDB2 and CRBN binding has been noted previously10. As other WD40-DCAFs attach to DDB1 through similar HLH helical-binding motifs (see also Extended Data Fig. 2f)39, we expected CRBN to sterically exclude the majority of if not all DCAFs from binding DDB1. (c) PUA-fold (CRBN-CTD in green) aligns with other PUA complexes, such as the MSS4-Rab8 complex (pdb:2FU5; rmsd of 3.47 Å over 107 residues). The small helix from Rab8 occupies a binding grove on the PUA-domain similar to thalidomide/lenalidomide/pomalidomide binding. (d) As in (c) but with the PUA surface coloured according to conservation within the CRBN orthologues (see Extended Data Fig. 1). The lenalidomide-binding groove on which Rab8 impinges is conserved. (e) PUA domain proteins are also involved in RNA binding, as shown by the superposition of CRBN and Rig1 (pdb:3OG8). While thalidomide somewhat resembles a nucleotide (with the phthoyl-group chemically related to a purine), we note that the binding mode observed for the compound is likely not compatible with RNA or DNA binding to CRBN, due to severe steric clashes. However, the PUA-domain itself appears adaptable for a broad range of ligands. It is not clear at present whether CRBN recognises a distinct post-translational modification on a protein that somewhat resembles thalidomide.
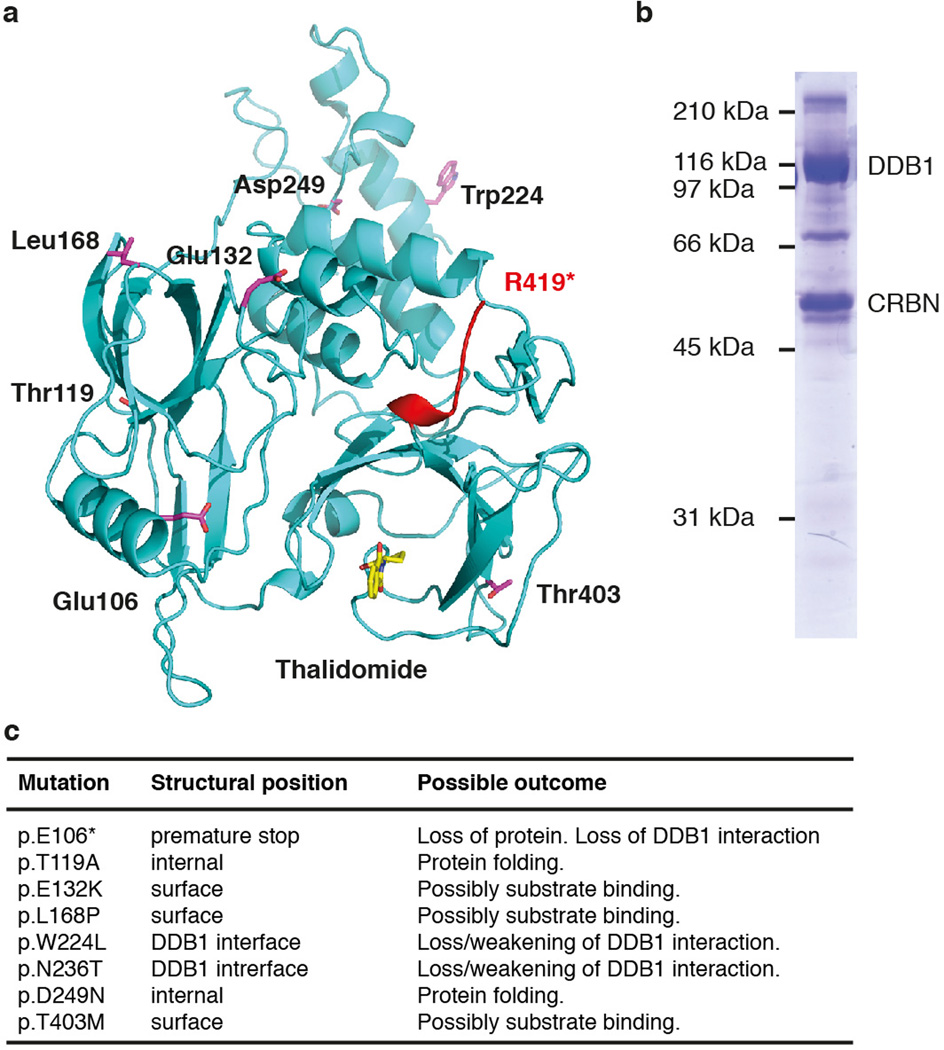
Extended Data Figure 6. Mutations in CRBN are found in mental retardation and cancer.
(a) Mutations of CRBN associated with mental retardation (indicated in red) and CRBN mutations found in cancer (shown as sticks in magenta). CRBN was first identified as a gene mutated in a mild form of mental retardation11. The identified nonsense mutation causes a premature stop codon after residue Arg419, resulting in a truncated form of CRBN. When tested in cells, the truncated CRBN bound DDB1 but exhibited a higher rate of autoubiquitination40. (b) In analogy, we found in vitro that a recombinant ggCRBN construct [1 to 426] apparently remained properly folded, with no obvious association of heat-shock factors. This construct was also capable of complex formation with DDB1. Two not mutually exclusive structural rationales can be proposed for the detrimental effect of the truncation seen in non-syndromic mental retardation: (i) the conserved C-terminus may be involved in substrate binding; in addition, the absence of the C-terminus could also give rise to temporary unfolding that impinges on the ubiquitination zone and, thus, renders the CRBN receptor subject to CRL4CRBN ubiquitination. (ii) The removal of the CRBN C-terminal tail could also delete the binding site for a potential de-ubiquitination enzyme that frequently appears bound at the tails of CRL4 substrate receptors, thus counteracting receptor autoubiquitination; DDB2 is complexed to USP2441 and CSA in conjunction with USP742. (c) The COSMIC database43 reports a number of somatic mutations in the CRBN gene in various tumours: one nonsense mutation (p.E106*) and seven missense substitutions have been reported. The structure now provides a molecular rationale for the reported mutations. The premature stop codon after E106 likely leads to an unfolded protein or at least abolishes the interaction with DDB1, resulting in a phenotype similar to a total loss of CRBN expression. W224L and N236T are surface-exposed residues involved in DDB1 binding, likely weakening this interaction. The T119A and D249N mutations affect the core of the protein and likely impair correct folding. Mutations E132K, L168P and T403M are surface mutations and could be involved in substrate binding.
Extended Data Figure 7. Thalidomide and derivatives modulate in vitro ubiquitination of MEIS2.
(a)In vitro autoubiquitination assays were performed with neddylated CRL4CRBN, E1 (Uba1), E2 (UbcH5A), and ubiquitin. CRBN ubiquitination was detected by anti-CRBN immunoblotting. The use of ubiquitin resulted in a high molecular smear, similar to that observed for CRBN autoubiquitination in cells10. (b) This contrasts with the use of K0 ubiquitin, which gave rise to a defined banding pattern. The amount of non-ubiquitinated CRBN remained largely constant as judged by anti-CRBN western blotting, with no gross overall inhibition of autoubiquitination observed following addition of thal(idomide) (lane 3), len(alidomide) (lane 4) and pom(alidomide) (lane 5). Autoubiquitination was inhibited by addition of CSN (lane 6).(c) Autoubiquitination of CRBN within the CRL4CRBN ligase was monitored in the presence of ATP, E1 (Uba1), and E2 (UbcH5a) (lane 2). We found that CRBN autoubiquitination was repressed in the presence of CSN in a dose-dependent manner (lanes 3–5). (d) Mass spectrometry-based identification of ubiquitin modification on CRBN using K0 ubiquitin. One phosphorylation site (S25) as well as three ubiquitination sites were detected in recombinant CRBN. The location of the ubiquitin sites on the ggCRBN structure is indicated: K39 and K43 are located on the unstructured CRBN N-terminal tail, which is not ordered in the structure and is indicated as a dashed line. The K412 site is also indicated in magenta and is found within 10–15Å of the presumed radius of gyration of disordered residues K39/K43. (e) Immobilised artificial membrane chromatography experiments predict similar cell permeability properties for the compounds used in this study.
Extended Data Figure 8. Details of protein array CRL4CRBN ligase substrate profiling.
(a) Assessment of intra-array reproducibility: comparison of fluorescence intensities of the two replicate spots of each protein for an array probed with E1, E2, CRL4CRBN and lenalidomide. (b) Assessment of between-array reproducibility: pairwise correlation plot showing Pearson correlation coefficients between spot intensities for all pairs of arrays, reordered according to similarity. (c) Heatmap of cluster averages (mean across all genes in a cluster). Cluster 42 contains MEIS2. (d) Heatmap of individual genes in cluster 42, which contains MEIS2. (e) Heatmap of individual genes of interest, including MEIS2 and the Ikaros transcription factors IKZF1/3. (f) The top 5 candidate genes were transiently overexpressed in HEK293T cells and assessed for stabilisation upon lenalidomide treatment (40 µM for 4 h).
Extended Data Figure 9. Thalidomide and derivatives modulate cellular abundance of MEIS2.
(a) MEIS2 ubiquitination by CRL4CRBN in vitro was inhibited by thalidomide in a dose-dependent manner.(b) A CRL4CRBN complex harbouring a CRBN variant carrying Y386A and W388A substitutions was found severely impaired in its ability to ubiquitinate MEIS2 in vitro. (c) CRL4CRBN autoubiquitination (lane 2) was inhibited by CSN (lane 3). The inhibition of CRBN autoubiquitination was partially overcome by addition of MEIS2 (lane 4). (d) E3 ligase specificity control showing that MEIS2 is ubiquitinated by CRL4CRBN (lane 1) but not by CRL4CDT2 (lane 2) or CRL4CSA (lane 3). Specificity was further improved in the presence of CSN, where MEIS2 ubiquitination by CRL4CRBN did not change (compare lanes 1 and 4), while MEIS2 ubiquitination by CRL4CDT2 (lane 5) and CRL4CSA (lane 6) was further suppressed.(e) All western blots were carried out in a quantitative fashion with infrared detection on a LiCor Odyssey reader. A representative example of a linearity control included on every blot is shown in the figure. (f) Overexpressed DDK tagged MEIS2 was stabilised upon treatment with 40 µM lenalidomide in HEK293T cells, but not (g) in HEK293T cells that stably overexpress a CRBNYW/AA mutant protein. We found that overexpressing wild type CRBN but not CRBNYW/AA mutant protein resulted in a severe growth defect of HEK293T cells (data not shown).(h) similar to what was observed with lenalidomide (Fig. 4b), MLN4924 was found to stabilise MEIS2 protein levels in a cycloheximide chase experiment. (i) Endogenous MEIS2 levels in M059J cells (DMSO control, lane 1). Anti-ERK2 immunoblot was used as a loading control. MEIS2 levels higher than the DMSO control were seen following addition of 1 – 30 µM lenalidomide (lanes 2–4), 5 µM bortezomib, and 5 µM MLN4924 following a 4h incubation. (j) Average increase of MEIS2 protein level following treatment with the indicated amounts of lenalidomide for 4 h. Data are shown as means ± SEM (n=3);* P<0.05; ** P<0.01 (two-tailed, unpaired Student’s t-test). (k) Thalidomide and pomalidomide show an increase in MEIS2 protein levels similar to that observed for lenalidomide. An anti-H4 immunoblot was used as a loading control. (l) Zebrafish embryos were treated with the indicated amounts of thalidomide 2 h post fertilisation and subjected to western blot analysis after a 22h incubation. An up to 1.5-fold increase in MEIS2 protein levels was observed in whole embryo lysate. Data shown as means ± SEM (n=3); * P<0.05 (two-tailed, unpaired Student’s t-test).(m) Lenalidomide treatment does not influence accumulation of MEIS2 mRNA. HEK293T cells treated with the indicated amounts of lenalidomide for 12 h were subjected to quantitative RT-PCR to assess levels of MEIS2 mRNA. mRNA levels were normalised to GAPDH mRNA. The MEIS2 mRNA level remained stable or even decreased, which does not explain the increase in MEIS2 protein levels observed. (n) SDS-PAGE analysis of CRL4CRBN (lane 1) and neddylated CRL4CRBN (lane 2). (o) Immunoblots with anti-MEIS2 (Abnova) and (p) anti-CRBN (Novus) antibodies for endogenous proteins from SK-N-DZ cells display no significant unspecific signal.
Extended Data Table 1.
Data collection and refinement statistics.
| hsDDB1-ggCRBN Thalidomide |
hsDDB1-ggCRBN Lenalidomide |
hsDDB1-ggCRBN Pomalidomide |
|
|---|---|---|---|
| Data collection | P3221 | P3221 | P3221 |
| Cell dimensions | |||
| a, b, c (Å) | 172.2, 172.2, 140.1 | 172.1, 172.1, 139.8 | 172.1, 172.1, 137.9 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 30.0-3.0 (3.1-3.0)* | 30.0-3.0 (3.2-3.0) | 30.0-3.5 (3.6-3.5) |
| Rmerge (%) | 11.9 (137.4) | 11.1 (124.4) | 14.1 (131.7) |
| I/σI | 15.77 (1.22) | 16.21 (1.66) | 7.05 (0.91) |
| Completeness (%) | 100.0 (99.9) | 99.9 (99.9) | 84.4 (83.4) |
| Redundancy | 7.9 (5.6) | 7.0 (7.0) | 7.0 (7.4) |
| CC(1/2) | 0.99 (0.40) | 0.99 (0.53) | 0.99 (0.29) |
| Refinement | |||
| Resolution (Å) | 30.0-3.0 | 30.0-3.0 | 30.0-3.5 |
| No. reflections | 49199 | 50542 | 25108 |
| Rwork / Rfree | 19.7 / 23.3 | 19.3 / 23.4 | 21.2 / 23.5 |
| No. atoms | |||
| Protein | 11405 | 11411 | 11389 |
| Ligand/ion | 20 | 20 | 21 |
| Water | 35 | 8 | 0 |
| B-factors | |||
| Protein | 87.8 | 95.4 | 139.3 |
| Ligand/ion | 67.2 | 54.3 | 93.5 |
| Water | 56.3 | 60.5 | n.a. |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.008 | 0.009 | 0.008 |
| Bond angles (°) | 1.11 | 1.18 | 1.16 |
Highest resolution shell is shown in parentheses.
Two crystals were combined for this dataset.
Supplementary Material
Acknowledgements
This work was supported by the Novartis Research Foundation and grants to N.H.T. from the European Research Council (ERC-2010-StG 260481-MoBa-CS), and to J.W.H. from the NIH (AG011085). J.R.L. was supported by a Damon Runyon Postdoctoral Fellowship DRG 2061-10. We acknowledge John Reilly (Novartis) for performing IAM experiments. We thank John Tallarico, Jeff Porter, William Sellers, Sylvain Cottens and Martin Renatus for help and comments. Daniel Hess, Ragna Sack and Jan Seebacher (FMI protein analysis facility) for the mass spectrometry analysis, and Jeremy Keusch and Heinz Gut (FMI protein structure facility) for support. We thank William Kaelin for kindly providing the IKZF1 reporter plasmid (pCMV-IRES-Renilla Luciferase-IRES-Gateway-Firefly Luciferase). Part of this work was performed at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions E.S.F., N.H.T., J.L.J and W.C.F initiated the project. E.S.F. and K.B. conducted the protein purification and crystallisation. S.C. pre-screened protein complexes by EM. E.S.F collected data, processed and refined x-ray data. E.S.F. and N.H.T. analysed the structures. E.S.F. performed in vitro experiments and, with the help of U.H., developed and performed TR-FRET and FP assays. E.S.F. performed protein array experiments, M.B.S. and E.S.F. analysed the data. E.S.F., K.B., J.R.L., H.Y., M.H., J.W.H. and N.H.T. conceived and performed the cell biological characterisation. R.B.T. and R.E.J.B. conceived and conducted chemical syntheses. J.N. and M.S. performed proteomics. V.A. and J.O. did the DSF experiments. F.S. and M.S. did the zebrafish experiments. E.S.F. and N.H.T. wrote the manuscript. All authors assisted in editing the manuscript.
Author Information The following structural coordinates have been reported and were submitted to the Protein Data Bank under accession numbers: hsDDB1-ggCRBN-thalidomide (pdb:4CI1), hsDDB1-ggCRBN-lenalidomide (pdb:4CI2), hsDDB1-ggCRBN-pomalidomide (pdb:4CI3). Human protein microarray data sets generated for this study are available from GEO (http://www.ncbi.nlm.nih.gov/geo) under accession GSE57554.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nature reviews Cancer. 2004;4:314–322. doi: 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- 2.Shortt J, Hsu AK, Johnstone RW. Thalidomide-analogue biology: immunological, molecular and epigenetic targets in cancer therapy. Oncogene. 2013 doi: 10.1038/onc.2012.599. [DOI] [PubMed] [Google Scholar]
- 3.Melchert M, List A. The thalidomide saga. The international journal of biochemistry & cell biology. 2007;39:1489–1499. doi: 10.1016/j.biocel.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 4.McBride WG. Thalidomide and congenital abnormalities. The Journal of American Medical Association. 1961;2 [Google Scholar]
- 5.Lenz W. Thalidomide and congenital abnormalities. The Journal of American Medical Association. 1962 [Google Scholar]
- 6.Sheskin J. Thalidomide in the treatment of lepra reactions. Clinical pharmacology and therapeutics. 1965;6:303–306. doi: 10.1002/cpt196563303. [DOI] [PubMed] [Google Scholar]
- 7.D'Amato RJ, Loughnan MS, Flynn E, Folkman J. Thalidomide is an inhibitor of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4082–4085. doi: 10.1073/pnas.91.9.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan B, Lentzsch S. The application and biology of immunomodulatory drugs (IMiDs) in cancer. Pharmacol Ther. 2012;136:56–68. doi: 10.1016/j.pharmthera.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Singhal S, et al. Antitumor activity of thalidomide in refractory multiple myeloma. The New England journal of medicine. 1999;341:1565–1571. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science (New York, NY) 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 11.Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Girona A, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2012 doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu YX, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) British journal of haematology. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in Complex with a Paramyxovirus V Protein: Viral Hijack of a Propeller Cluster in Ubiquitin Ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 18.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scrima A, et al. Detecting UV-lesions in the genome: The modular CRL4 ubiquitin ligase does it best! FEBS letters. 2011;585:2818–2825. doi: 10.1016/j.febslet.2011.04.064. [DOI] [PubMed] [Google Scholar]
- 21.Hur S, Stroud RM, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J Biol Chem. 2006;281:38969–38973. doi: 10.1074/jbc.R600034200. [DOI] [PubMed] [Google Scholar]
- 22.Ruchelman AL, et al. Isosteric analogs of lenalidomide and pomalidomide: Synthesis and biological activity. Bioorganic & medicinal chemistry letters. 2012;23(1):360–365. doi: 10.1016/j.bmcl.2012.10.071. [DOI] [PubMed] [Google Scholar]
- 23.Jin J, Arias E, Chen J, Harper J, Walter J. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Molecular cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Angers S, et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 25.Fischer ES, et al. The Molecular Basis of CRL4DDB2/CSA Ubiquitin Ligase Architecture, Targeting, and Activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmeister CC, et al. Phase I trial of lenalidomide and CCI-779 in patients with relapsed multiple myeloma: evidence for lenalidomide-CCI-779 interaction via P-glycoprotein. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3427–3434. doi: 10.1200/JCO.2010.32.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S, et al. Transport of thalidomide by the human intestinal caco-2 monolayers. European journal of drug metabolism and pharmacokinetics. 2005;30:49–61. doi: 10.1007/BF03226408. [DOI] [PubMed] [Google Scholar]
- 29.Roche S, et al. Development, validation and application of a sensitive LC-MS/MS method for the quantification of thalidomide in human serum, cells and cell culture medium. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;902:16–26. doi: 10.1016/j.jchromb.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Crowley MA, et al. Further evidence for the possible role of MEIS2 in the development of cleft palate and cardiac septum. Am J Med Genet A. 2010;152A:1326–1327. doi: 10.1002/ajmg.a.33375. [DOI] [PubMed] [Google Scholar]
- 31.Capdevila J, Tsukui T, Rodríquez Esteban C, Zappavigna V, Izpisúa Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Molecular cell. 1999;4:839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- 32.Paige SL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 36.Huai Q, et al. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc Natl Acad Sci U S A. 2002;99:12037–12042. doi: 10.1073/pnas.192206699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kissinger CR, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 38.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic acids research. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu G, Jiang X, Jaffrey SR. A mental retardation-linked nonsense mutation in cereblon is rescued by proteasome inhibition. J Biol Chem. 2013 doi: 10.1074/jbc.M113.472092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Lubin A, Chen H, Sun Z, Gong F. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell cycle (Georgetown, Tex) 2012;11 doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 43.Forbes SAS, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2010;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.