Abstract
Stable and monodisperse phenylboronic acid-functionalized nanoparticles (PBA-NPs) were fabricated using 3-((acrylamido)methyl)phenylboronic acid homopolymer (PBAH) via solvent displacement technique. The effect of operating parameters, including stirring time, initial polymer concentration and the proportion of methanol on the self-assembly process were systematically investigated. The diameters of the PBA-NPs were increased as increasing the initial PBAH concentration and the proportion of methanol. Likewise, there was a linear dependence between the size of self-assembled nanoparticles and the polymer concentration. Moreover, the dissipative particle dynamics (DPD) simulation technique was used to investigate the mechanism of self-assembly behavior of PBAH, which indicated that the interior of PBA-NPs was hydrophobic and compact, and the boronic acid groups were displayed on both the outermost and interior of PBA-NPs. The resulting PBA-NPs could successfully encapsulate emodin through PBA-diol interaction and the encapsulation efficiency (EE%) and drug loading content (DLC%) of drug-loaded PBA-NPs were 78% and 2.1%, respectively. Owing to the acid-labile feature of the boronate linkage, a reduction in environmental pH from pH 7.4 to 5.0 could trigger the disassociation of the boronate ester bonds, which could accelerate the drug release from PBA-Emodin-NPs. Besides, PBA-Emodin-NPs showed a much higher cytotoxicity to HepG2 cells (cancer cells) than that to MC-3T3-E1 cells (normal cells). These results imply that PBA-NPs would be a promising scaffold for the delivery of polyphenolic drugs.
Introduction
Polyphenols are natural plant secondary metabolites found in fruits, vegetables and wines etc.1 They have been shown to possess beneficial biological properties through their strong antioxidative activity. However, the utilization of some polyphenols including resveratrol and emodin et al., as a bioactive agent in pharmaceuticals, is currently limited due to its poor water solubility, high chemical instability and low oral bioavailability.2 Encapsulation of these drugs can be used to improve its water-dispersibility, chemical stability, and bioavailability. Delivery systems are available to encapsulate, protect and release drugs, and highlight their potential applications within clinical therapeutics. Among those various carriers, polymeric nanoparticles, which have simple preparation, high drug loading capacity, high stability and easy surface modification, have been widely applied for hydrophobic drug delivery.3–11 Moreover, the nanoparticles in the size range from 50–400 nm can escape from the leaky vasculature around the tumor and accumulate in the tumor.12
Phenylboronic acid (PBA) polymers as the polymeric nanoparticles carriers have attracted significant research interests because of their reversible reactions with cis-diol compounds such as polyphenols, carbohydrates and glycans in aqueous systems.13–15 Li and Yang et al. have developed PBA-functionalized nanoparticles self-assembled by amphiphilic copolymers, such as LAMA-r-AAPBA copolymer and PEG-g-PBA copolymer, as the glucose-responsive nanocarriers for self-regulated insulin release.16,17 In another example, a complex micelles prepared by two amphiphilic copolymers using a facile cross-linking strategy based on PBA-diol interaction, have been developed for protein delivery, which display superior physiological stability compared with their non-cross-linked counterparts.18 In addition, benefiting from the interaction between PBA groups and glycoproteins on the cell membrane, the PBA-functionalized micelles fabricated by Pluronic-PMCC-BA copolymer, could recognize special cells with good affinity.9 Therefore, we presume that the polymer compose of PBA groups could be a good resultant material for polyphenolic drug delivery through PBA-diol interaction.
In this work, we reported on the development of PBA-functionalized nanoparticles using 3-((acrylamido)methyl)phenylboronic acid homopolymer (PBAH) via solvent displacement method. The PBAH was synthesized by a reversible addition-fragmentation chain-transfer (RAFT) polymerization reaction with PBA as pendant group in each monomer unit. The size of these PBA-functionalized nanoparticles could be readily tuned by changing the polymer concentration and the ratio of the organic phase to aqueous phase. Based on the PBA-diol interaction, the resulting PBA-NPs could successfully encapsulate emodin, a polyphenolic drug, and the drug release profile from nanoparticles follows a pH-dependent manner. To the best of our knowledge, this is the first time reports the fabrication of polyphenolic drug delivery by PBA homopolymer through PBA-diol interaction directly. Additionally, the PBA groups located on both the outermost and interior of PBA-NPs, which was benefited to encapsulate polyphenolic drugs and interact with sugar compounds on the cell surface at the same time. We believe our study will pave the way for new application of PBA polymers in targeting polyphenolic drug delivery to the cancer cells.
Materials and methods
Materials
3-(aminomethyl)phenylboronic acid hydrochloride was purchased from Frontier Scientific, Inc. Sodium borohydride and acryloyl chloride were purchased from Aladdin Industrial Inc. Emodin (95%) was purchased from J&K Scientific Ltd. Dubelcoo’s Modified Eagle’s Medium (DMEM) with no glucose was purchased from Genom Pharmaceutical Co., Ltd. 4',6-diamidino-2-phenylindole (DAPI) was obtained from Sigma-Aldrich. All reagents were purchased from Beijing Chemical Works without any further purification. Ultrapure water was obtained from UNIQUE-R20 system (18.2 MΩ cm−1). Carbon-coated copper grids were purchased from Beijing Xinxing Braim Technology Co., Ltd.
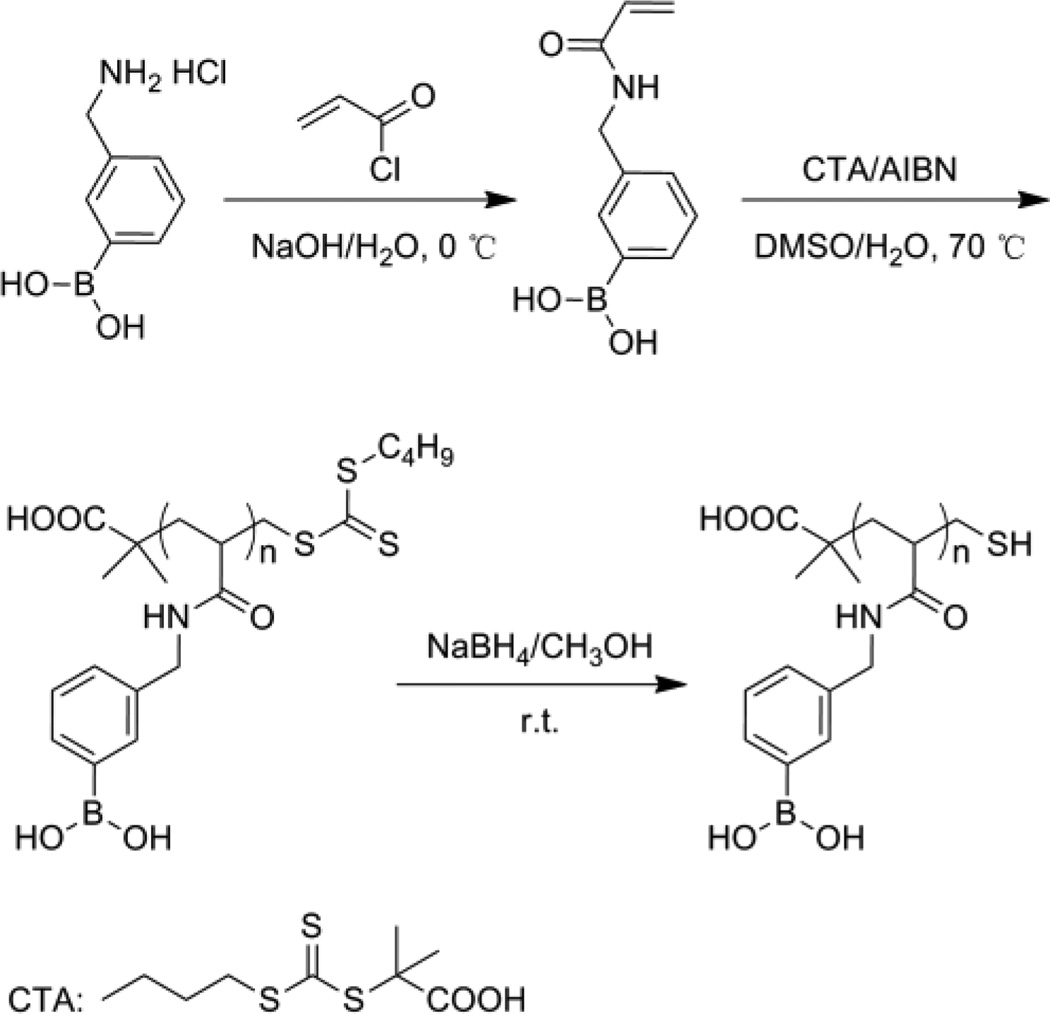
Synthesis of PBA homopolymer
3-((acrylamido)methyl)phenylboronic acid homopolymer was abbreviated as PBAH in this paper. The general procedure for synthesis of PBAH was shown in Scheme 1. 1H NMR and 13C NMR spectra of monomer were obtained on a Bruker AVANCE DRX 400 MHz spectrometer using DMSO-d6 as the solvent. 1H NMR spectrum of PBAH used the CD3OD as the solvent.
Scheme 1.
Synthesis of poly(3-((acrylamido)methyl)phenylboronic acid)
Preparation of monomer
Monomer was synthesized following the literature published procedure.19 Briefly, 3-(aminomethyl)phenylboronic acid hydrochloride (1.0 g, 5.34 mmol) was dissolved in NaOH solution (2.0 M, 10.7 mL) at 0 °C. Subsequently, cold acryloyl chloride (2.2 mL, 26.7 mmol) was added dropwise under stirring. Then HCl solution (1.0 M) was slowly added to the reaction mixture until the pH reached 1.0. The precipitate was collected by filtration and washed with cold water to afford the final product as white solid. 1H NMR (400 MHz, DMSO-d6, ppm, Fig. S1A): δ 8.55 (t, 1H, NHCO), 8.00 (s, 2H, BOH), 7.68 (m, 2H, benzene-H) 7.30 (m, 2H, benzene-H), 6.28 (m, 1H, CH2CHCO), 6.11 (m, 1H, CH2CHCO), 5.60(m, 1H,CH2CHCO), 4.35 (d, 2H, CH2NH); 13C NMR (75 MHz, DMSO-d6, ppm, Fig S1B): δ 164.5, 138.0, 133.3, 132.6, 131.8, 129.2, 127.4, 125.3, 42.4.
Polymerization of monomer
Monomer was polymerized according to the reference20 by RAFT at 70 °C with the chain transfer agent (CTA) and AIBN as the initiator in DMSO/water = 95/5 (v/v). The molar ratio of monomer, CTA and AIBN was 100:2:2. The reaction mixture was pumped with ultrapure nitrogen for 2 h at room temperature to exclude oxygen. Then the reaction was initiated at 70 °C and allowed to proceed for 24 h under nitrogen. The solvent was removed under reduced pressure. The crude solid was dissolved in methanol, precipitated in ether, washed with acetone and dried under vacuum to give light-yellow powder.
Reduction
The polymerized product was dissolved in methanol. Then 50 equiv molar excess of NaBH4 was added slowly under vigorously stirring. The reaction was allowed to proceed for 12 h at room temperature. Then the resulting mixture was dialyzed against ultrapure water for 72 h (cut-off Mw 1000 Da dialysis bag). The water was removed under vacuum to yield white powder. 1 H NMR (400MHz, CD3OD, ppm, Fig. S2): δ 7.27–7.61 (m, 4H, benzene-H), 4.30 (m, 2H, CH2NH), 2.35 (m, 1H, COCH), 1.75 (m, 2H, CH2).
Preparation of PBA-functionalized nanoparticles
The formation of PBA-functionalized nanoparticles (PBA-NPs) was simply carried out by adding PBAH (2.0 mg mL−1, in methanol) dropwise to deionized water within 30 sec, the solution was further stirred for 30 min at room temperature. The mixed solution showed a light blue color instantly indicating the formation of nanoparticles. Several operating parameters such as stirring time, polymer concentration, and methanol quantity were screened to test their impacts on the self-assembly process of PBAH. However, the diameter distributions of PBA-NPs were similar before and after dialysis. Therefore, the PBA-NPs solution was not dialyzed before TEM or DLS tests.
The hydrodynamic diameter and the polydispersity of self-assembled nanoparticles were determined by dynamic light scattering (DLS) measurement with a Zetasizer Nano-ZS from Malvern Instruments (Worcestershire, UK). The morphology of the nanoparticles was measured by transmission electron microscopy (TEM) performed on a JEOL JEM-1011 electron microscope operating at an acceleration voltage of 100 kV.
Drug loading
The drug-loaded nanoparticles were readily obtained by mixing 0.1 mg PBAH and 0.1 mg emodin in DMSO first. Then the mixture was added dropwise to deionized water under stirring 30 min as previously described. Final volume of the PBA-Emodin solution was 1.0 mL (DMSO/H2O = 3:7, v/v). Then the mixture was transferred to a dialysis cut-off bag (MWCO 3500 Da) and dialyzed against 2000 mL of deionized water for 24 h. The UV absorbance at 430 nm was measured to calculate the drug concentration with UV-vis spectrophotometer (Beijing Purkinje General Instrument Co., Ltd, Model TU-1901). The drug encapsulation efficiency (EE) and drug loading content (DLC) were calculated according to the following equations:
In vitro drug release
The release of emodin from the PBA-Emodin-NPs was examined using a dialysis method against a 10 mM potassium phosphate buffer (KPS) at pH 5.0 and 7.4, respectively. Due to the poor solubility of emodin, the released emodin may precipitate in the dialysis bag during the drug release process. Therefore, at different time points, 100 µL of samples were withdrawn from the dialysis tube and were centrifuged at 2,000 rpm for 1 min. The supernatant was collected for UV/Vis spectrophotometer analysis to measure the absorbance at 430 nm. The amounts of released emodin were determined according to the standard curves pre-generated for emodin. After each analysis, the supernatant collected was reintroduced into dialysis bag to maintain the total volume of particle solution.
Cell viability assay and fluorescent stain
In vitro cytotoxicity assay
HepG2 and MC-3T3-E1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose containing 10% fetal bovine serum (FBS), 100 units mL−1 penicillin and 100 µg mL−1 streptomycin at 37 °C in a 5% CO2 humidified atmosphere. Cells were seeded in 96-well flat-bottomed plates and cultured for 24 h. After that the medium was replaced by the PBA-NPs, PBA-Emodin-NPs and free emodin diluted in DMEM glucose-free medium at various emodin concentrations of 0.1, 1, 5, 10, 12, 15 and 20 µg mL−1. After incubation for 24 h, 20 µL of MTT solution in PBS (5 mg mL−1) was added to each well and the plates were incubated for another 4 h at 37 °C. Then the medium containing MTT was replaced with DMSO to dissolve the MTT formazan crystals. Finally, the plates were shaken for 10 min before the UV measurement (Tecan infinite 200, Tecan Group Ltd.) at 490 nm. The cell viability was calculated based on the following equation:
Cell fluorescent staining
Coverslips were sterilized with 75% ethanol, and washed with PBS buffer before putting into 24-well culture plates. HepG2 and MC-3T3-E1 cells were seeded at a density of 3×104 cells well−1 and incubated at 37 °C with 5% CO2 for 24 h. Then the cells were treated with PBA-Emodin-NPs in DMEM glucose-free medium at 37 °C with 5% CO2 for 24 h. After incubation, cells were washed twice with PBS. Subsequently, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with PBS again. Later, cell nucleus was stained with DAPI (1.25 µg mL−1) for 10 min followed by washing with PBS. The fluorescence images were obtained using confocal laser scanning microscopy (CLSM, LSM 700, Carl Zeiss Microscopy). In the process of taking the images, all the camera parameters (laser intensity, image brightness, exposure, magnification, etc.) are fixed.
Simulation details
Dissipative particle dynamics (DPD) simulation technique21 was chosen to study the self-assembled structure of PBA-NPs. The groups of PBAH molecules are represented by different beads by introducing bead spring type model. The conservative interaction strength in DPD between the different types of beads (between the same type beads αij = 25) can be chosen according to the linear relation with Flory-Huggins parameters χ:
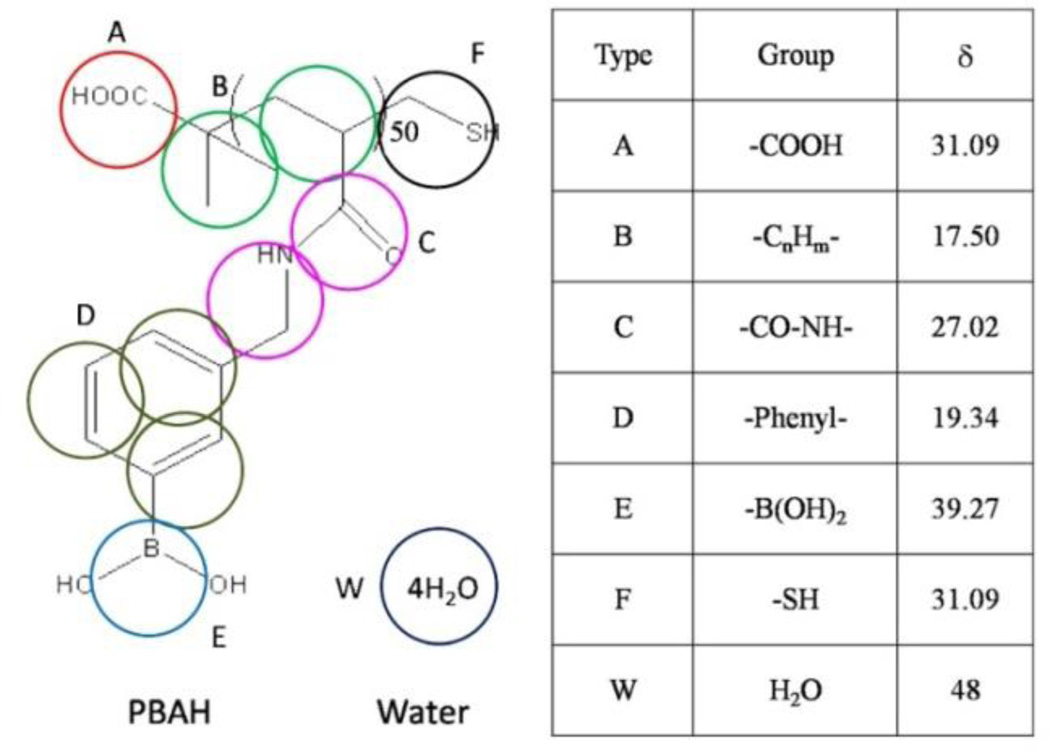
The Flory-Huggins χ-parameters of groups can be calculated by the Hansen solubility parameters according to:22
where Vr is the reference volume. The coarse-graining scheme and the solubility parameters of ingredients are listed in Fig. 1. The coarse-graining scheme of PBAH is shown in the left picture and the Hansen solubility parameters of PBAH groups with unit of MPa1/2 are given in the right table. The simulation system with the initial configuration of randomly distributed PBAH in aqueous solution has been studied. The 50 PBAH chains (one chain of 50 monomer units and represented by 353 beads) are simulated in a box of size 505050. The number of beads which represent water molecules is fixed by keeping the total reduced number density to be 3. The simulation works are finished by software GALAMOST with the acceleration of GPU.23
Fig. 1.
The coarse-graining scheme and the solubility parameters of PBAH.
Results and discussion
The self-assembly of PBAH
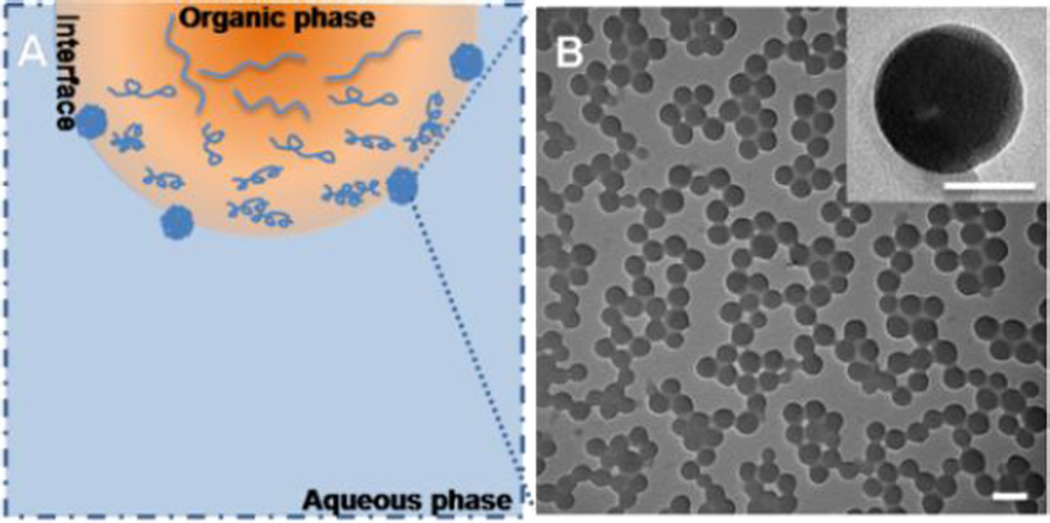
In this work, the PBA-functionalized nanoparticles were developed using PBAH via solvent displacement. The solvent displacement method was developed by Fessi et al, which system consists of the polymer, the polymer solvent and the non-solvent of the polymer.24 The organic solvent must be able to dissolve polymer, also be miscible with non-solvent and be easily to remove. Therefore, methanol and deionized water were chosen respectively as polymer good-solvent and non-solvent on account of solubility of PBAH. The procedure parameters such as stirring time, initial polymer concentration and the solvent composition were screened to study their effects on the self-assembly of PBAH. The PBA-NPs were prepared by dropwise addition of polymer solution (2 mg mL−1, in methanol) within 30 sec into deionized water under stirring, and the volume ratio of methanol and deionized water was 1:9. During the addition process, methanol diffused from organic phase into water and carried the polymers to the interface, which led to the nucleation, growth and formation of nanoparticles (Fig. 2A). This process was governed by the Marangoni effect, wherein the movement from organic phase to aqueous phase at the interface was driven by longitudinal variations of interfacial tension.24
Fig. 2.
(A) Schematic illustration of the formation process of the PBA-NPs. (B) TEM analysis of PBA-NPs. The inset shows an individual particle. The scale bar is 100 nm and the scale bar of inset is 50 nm.
The micelle-like PBA-NPs were observed by TEM with the diameter of 81±11 nm (Fig. 2B) and the average hydrodynamic diameter (Dh) determined by DLS was 84±3 nm and the polydispersity index (PDI) was 0.092, indicating a good homogeneity of PBA-NPs (Fig. S3A). Additionally, the surface charge of the PBA-NPs was measured. The zeta potential was −30.2±2.5 mV, suggesting that most of carboxyl groups were on the surface of the nanoparticles because the aggregation of the hydrophobic chains forced the ionic end groups to stay on the periphery.25 Besides, as shown in Fig S3B, the sizes of the PBA-NPs had no significantly changes upon incubating in a 20 mM dithiothreitol (DTT) solution after 24 h, which indicated that the disulfide bonds were not the main driving force to maintain the structure of PBA-NPs.26 These results suggested that stable PBA-NPs could be prepared by this facile and surfactant-free method.
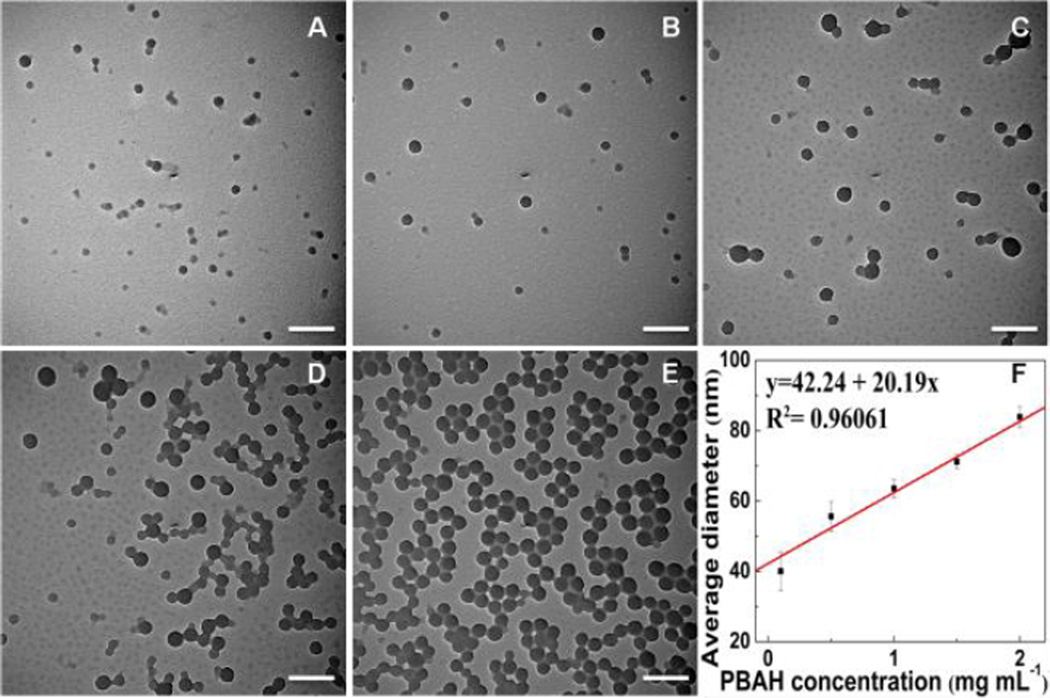
The formation of PBA-NPs is a fast process, as shown in Fig. S4. The self-assembled nanoparticles with an average diameter of 32±7 nm spontaneously formed within 30 sec. The diameter of nanoparticles reached 46±11 nm upon stirring for 30 min and thereafter remained constant. Therefore, in all the following experiments, 30 min was chosen as the fixed reaction time. As shown in Fig. 3, the diameter of the PBA-NPs increased from 40±2 nm to 84±3 nm when the initial PBAH concentration was increased from 0.1 to 2.0 mg mL−1. The particle size showed a linear dependence on the initial polymer concentration. At high initial polymer concentration, more polymers were brought to the interface as the methanol diffusing into the aqueous solution, which could lead to more polymer aggregation and the formation of larger NPs. Additionally, a higher initial polymer concentration contributed to a more viscous organic phase, resulting in a higher mass transfer resistance, thus the diffusion of polymer-solvent phase into the external aqueous phase was reduced and bigger nanoparticles formed.27
Fig. 3.
TEM images of PBA-NPs with increasing initial PBAH concentration: (A) 0.1 mg mL−1, (B) 0.5 mg mL−1, (C) 1 mg mL−1, (D) 1.5 mg mL−1, and (E) 2 mg mL−1. (F) The correlation between the average diameter (Dh) and the initial PBAH concentration. Dh was measured with DLS. The volume ratio of methanol and water was 1:9. The scale bars are 200 nm.
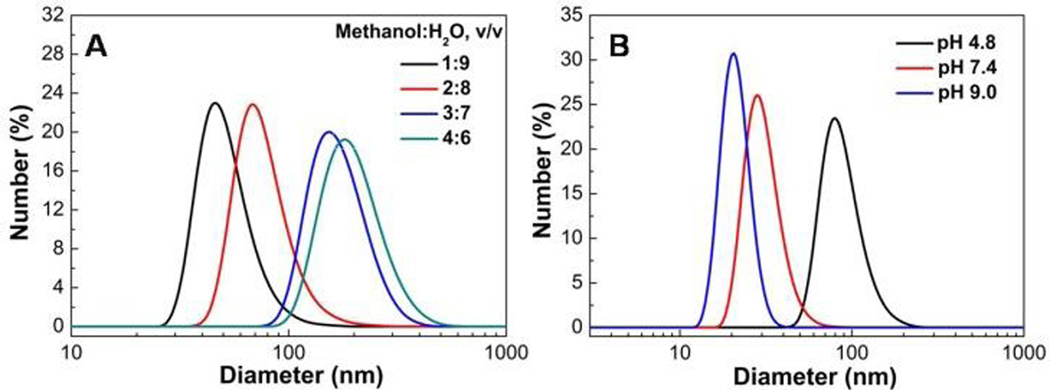
As shown in Fig. 4A and Fig. S5, the Dh of PBA-NPs increased from 64±3 nm to 197±6 nm as the ratio of methanol and water was changed from 1:9 to 4:6. This is attributed to the fact that increasing the proportion of methanol can improve the polymer solubility in the methanol-water mixture, resulted in a lower nucleation rate and thus increased final mean particle size. When the ratio of methanol and water was increased to 5:5, the solubility of PBAH was enhanced greatly, the suspension became unstable, and regular nanoparticles could not be obtained (Fig. S5E). At the same time, the Dh of PBA-NPs decreased with the increase of solution pH (Fig. 4B). The pKa of PBAH was ca. 7.7 examined by UV absorption changes (Fig. S6), and increasing the solution pH will lead to a rising proportion of tetrahedral boronate ion.14 Therefore, it would increase the hydrophilicity of the homopolymer and decrease the size of PBA-NPs.10
Fig. 4.
The diameter distributions of PBA-NPs assembled in different solution conditions. (A) PBA-NPs were formed at various volume ratios of methanol and water. The initial PBAH concentration was 1.0 mg mL−1. (B) PBA-NPs were formed at various pH of aqueous solution (0.01 M KPS). The initial PBAH concentration was 0.5 mg mL−1 and the volume ratio of methanol and buffer was 1:9.
Drug loading
PBA are known to bind with compounds containing 1,2 or 1,3 diol moieties with high affinity through reversible ester formation,14,28,29 and many drugs contain a polyphenol skeleton. The pharmaceutical efficacy of polyphenols often suffers from their sub-optimal solubility and instability under an oxidative environment. Emodin, i.e. 3-methyl-1,6,8-trihydroxyanthranquinone, as a typical member of polyphenols, can be isolated from herb rhubarb and giant knotweed rhizome. It has been shown that emodin exhibits potent anticancer, anti-inflammatory and anti-bacterial properties.30–33 Especially, emodin shows a potential excellent cytotoxic agent against a variety of malignant human cancers, because it can inhibit cancer cell growth and regulate genes related to cell apoptosis, cell proliferation, tumor cell invasion and metastasis.34,35 However, the application of emodin-based therapeutics is hampered by its poor pharmacokinetics.36 In past years, various delivery systems, including liposome, solid lipid nanoparticles (SLN), silver nanoparticles and polyvinylpyrrolidone blended nanofibrous have been developed to enhance emodin availability.37–40 Using emodin as a model drug, we test the feasibility of using PBA-NPs as a carrier to deliver polyphenolic drugs based on the formation of boronate ester bonds between PBA groups and diols.
First, Alizarin Red S (ARS), a cis-diol-containing fluorescent dye, was used as a model compound to examine the binding between diols and PBA-NPs. Considering on the solubility of drugs, the polymer solvent was changed from methanol to DMSO to form PBA-NPs loaded with cargoes. Before adding polymer solution to deionized water, ARS was firstly mixed with PBAH in DMSO. Then, the procedure for the preparation of PBA-ARS-NPs was the same as used for PBA-NPs (DMSO/H2O = 3:7, v/v) except the mixture was further dialyzed (MWCO 3500 Da) against 2000 mL of deionized water for 24 h to remove the free ARS. As shown in Fig. S7, the fluorescent intensity of the PBA-ARS-NPs was significantly higher than the free ARS solution at the same concentration, indicating the formation of boronate ester bonds between ARS and PBA groups in the PBA-ARS-NPs.14
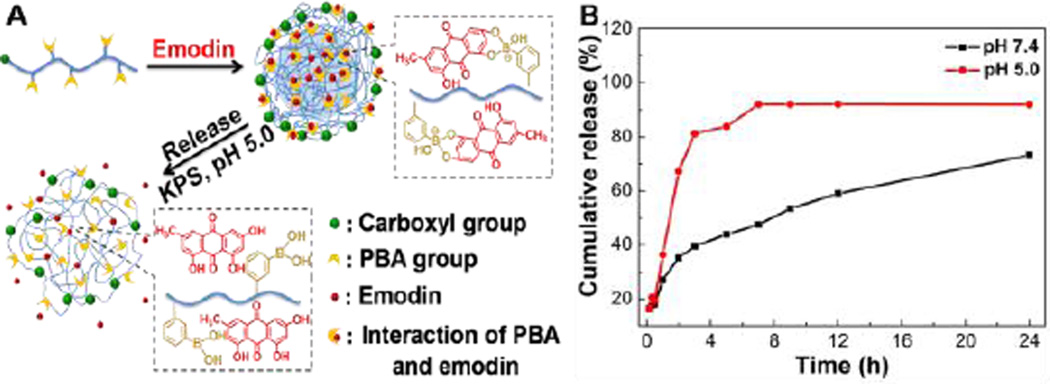
Similarly, emodin could be entrapped in the PBA-NPs by adding PBAH and emodin mixture to deionized water to form a transparent PBA-Emodin solution (Fig. 5A), which was very stable even upon dialysis for 24 h and centrifugation at 5,000 rpm for 5 min. As comparison, pure emodin would precipitate out at the same concentration (Fig. 5B). Under TEM, the PBA-Emodin-NPs appeared as well-defined spherical particles (Fig. 5C). The encapsulation efficiency (EE%) and drug loading content (DLC) of PBA-Emodin-NPs fabricated using this method were 78%, and 2.1%, respectively. Meanwhile, the diameter of PBA-Emodin-NPs was decreased to 117 nm, which was smaller than that of drug-free nanoparticles (Table 1). The reason of the diameter decreased might be attributed to the increase of hydrophilicity of PBAH, because of the formation of hydrophilic boronate ester bonds in the presence of emodin. The above results suggested that emodin could be successfully loaded into PBA-NPs and its solubility in aqueous solution was significantly enhanced.
Fig. 5.
(A–B) Digital camera images of PBA-Emodin and Emodin solution: (A) before dialysis and centrifugation, emodin solution shows strong turbidity; (B) after dialysis and centrifugation, the PBA-Emodin remains the same transparent solution while emodin precipitates out affording a colorless solution. (C) The morphology of PBA-Emodin nanoparticles. The scale bar is 200 nm.
Table 1.
The properties of PBA-NPs and PBA-Emodin-NPs
| Initial mass of Emodin (µg/mL) |
Average diameter (nm) |
EE (%) | DLC (%) |
|---|---|---|---|
| 0 | 165±7 | / | / |
| 100a | 117±6 | 78.0 | 2.1 |
| 100b | 162±7 | 19.0 | 0.5 |
: adding the mixture of emodin and PBAH to deionized water
: adding the emodin to the preformed PBA-NPs solution
In addition, the other method was used to fabricate PBA-Emodin-NPs by adding the emodin (in DMSO) to the preformed PBA-NPs solution stirred for 30 min at room temperature. After the dialysis and centrifugation, the EE% and the DLC% of this PBA-Emodin-NPs were only 19% and 0.5%, respectively (Table 1), mainly due to the adsorption of drugs on the PBA-NPs surface. This indicated that little of PBA groups located on the outermost of the nanoparticles and the high inside density inhibited emodin penetrating into the inside of the performed particles
Simulation structure of self-assembled PBA-NPs
Dissipative particle dynamics (DPD) simulation technique was chosen to investigate the self-assembly mechanism of PBAH and the structure of PBA-NPs. For simplicity, the chains of PBAH were directly blended into water in the initial simulation.
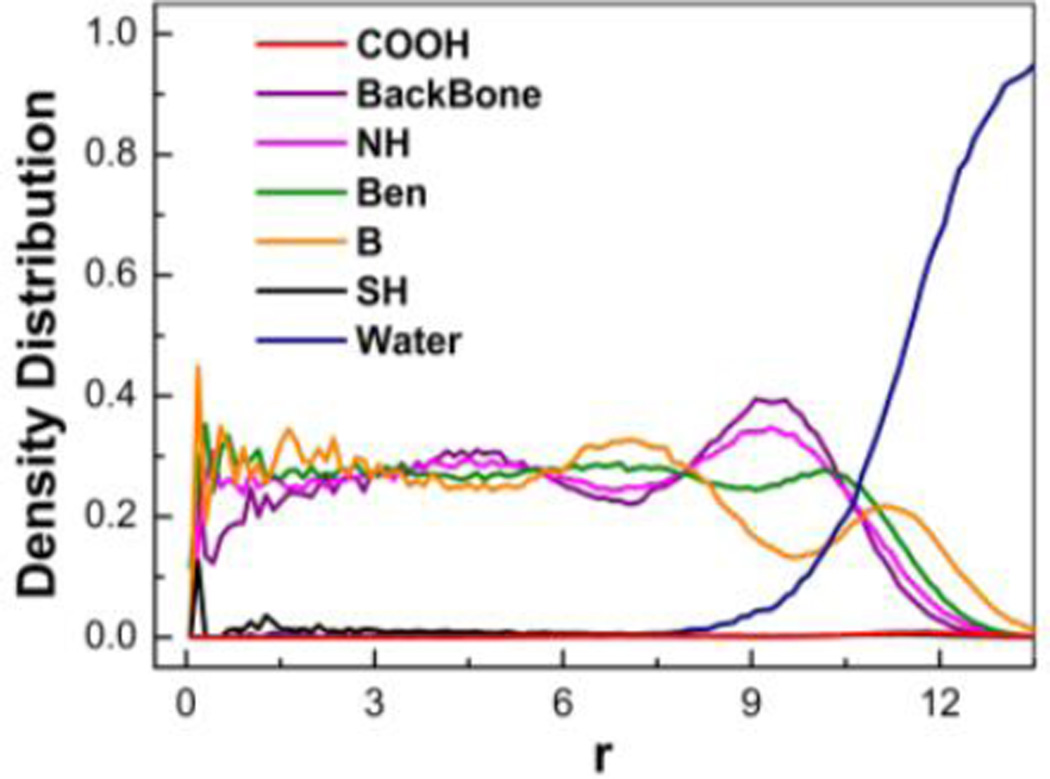
As shown in Fig. 6, the boronic acid groups (shown in blue color) exist on both the outermost and interior of PBA-NPs and the hydrocarbon groups (shown in green color) as the hydrophobic ingredient mostly accumulate in interior. For a more accurate description of the group distribution, we calculated the radial density of each ingredient of PBA-NPs. As shown in Fig. 7, the density distributions of different groups keep uniform in the range from 0 to 9, which indicates that the chains of PBAH was closely packed in the interior of the particles. This correlates well with the TEM results, where electron dense cores were observed (Fig. 2B). The high polymer density at the interior would limit the drug diffusion, which could be the reason why the lower EE% and DLC% of PBA-Emodin-NPs formed by adding emodin to performed PBA-NPs was observed (Table 1). Meanwhile, the boronic acid groups become the predominate group near the surface. Therefore, together with the carboxyl groups, boronic acid groups contributed as the hydrophilic part to ensure the PBA-NPs well-dispersed in water.
Fig. 6.
The snapshots of self-assembled PBA-NPs: (A) the outside snapshot of PBA-NPs; (B) the sectional snapshot of PBA-NPs.
Fig. 7.
The radial density of ingredients of PBA-NPs assembled in solution. r indicates the radius of the PBA-NPs.
In vitro drug release
The PBA-Emodin-NPs release behaviors were investigated at pH 7.4 and pH 5.0, respectively. As shown in Fig. 8B, the emodin was slowly released and less than 70% of the drug leaked out across 24 h period at pH 7.4. Conversely, the emodin release was greatly promoted and approached to 90% after 7 h at pH 5.0, indicating that the drug release profile from nanoparticles follows a pH-dependent manner. This pH-sensitive characteristic of PBA-NPs loaded with drug relied on the ability of PBA group to form robust covalent bonding with emodin under neutral conditions, and reversible cleavability of boronate ester bonds in an acidic environment.16,28 Decreasing the environmental pH results in a quick disassociation of the boronate ester bonds and an acceleration of the drug release (Fig. 8A).41 The short residence inside lysosomes decreases the risk of drug deactivation. Therefore, the acid-accelerated drug release is beneficial for enhanced drug efficacy inside the cells in consideration of the substantial pH decline occurring in the lysosomal compartment.42,43
Fig. 8.
(A) Schematic illustration of the drug loading and drug delivery triggered by acidic pH. (B) Drug release profiles of PBA-Emodin-NPs at various pH in vitro.
Meanwhile, the stability of PBA-Emodin-NPs at pH 7.4 and pH 5.0 was studied by DLS. Fig. S8 showed that the diameter of PBA-Emodin-NPs did not change much after 24 h at pH 7.4, while the diameter of PBA-Emodin-NPs increased drastically after 12 h incubation at pH 5.0, which suggested that the PBA-Emodin-NPs were swelled and aggregated during the drug release under an acidic environment. This observation agreed fairly well with the pH-dependent drug release of the PBA-Emodin-NPs.
In vitro cytotoxicity assay
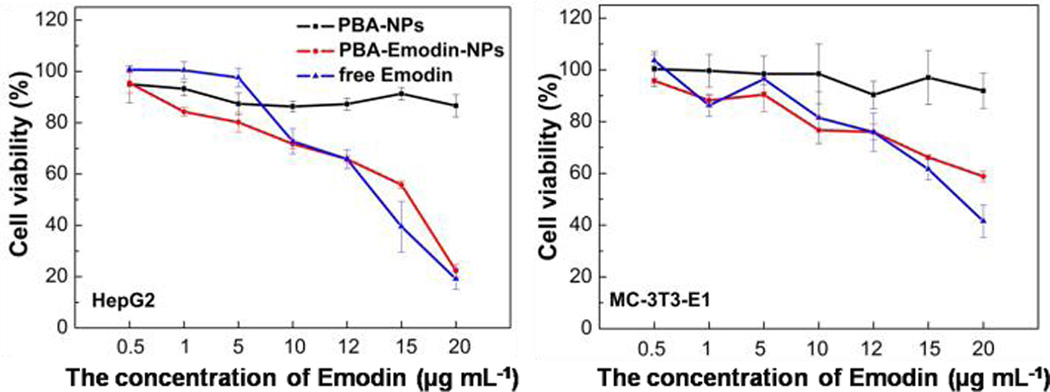
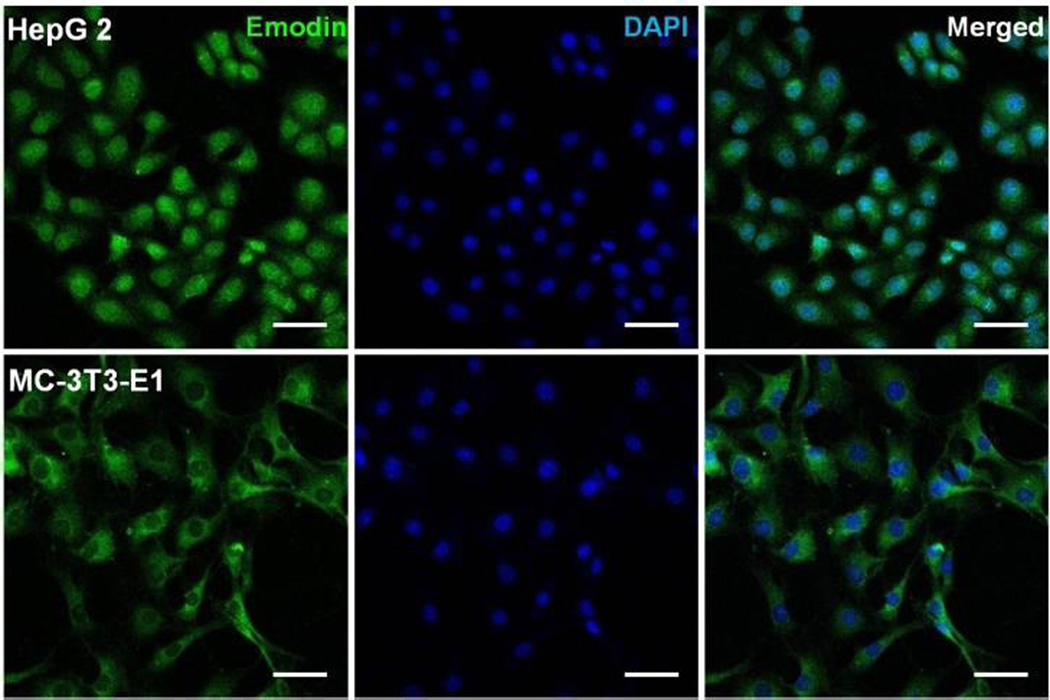
Herein, in vitro cytotoxicity profiles of PBA-NPs and PBA-Emodin-NPs to HepG2 cells and MC-3T3-E1 cells were evaluated by MTT assay. Considering the interaction between the PBA group and glucose, glucose-free medium was used in the MTT assay. As shown in Fig. 9, PBA-NPs showed negligible cytotoxicity in both cell lines, indicating good cell biocompatibility of these nanoparticles. In comparison, the loading of emodin can drastically increase the cytotoxicity. PBA-Emodin-NPs showed much higher cytotoxicity to HepG2 cells, a cancer cell line, than to MC-3T3-E1 cells, a normal cell line. The anti-cancer effect of emodin and its derivatives is due to the DNA intercalation.44 According to recent research, emodin can inhibit pulmonary metastasis of breast cancer through reducing STAT6 phosphorylation and C/EBPβ expression.35 Therefore, the confocal laser scanning microscopy was used to observe the fluorescence of emodin in the nucleus of cells after 24 h incubation. As shown in Fig. 10, the fluorescent intensity of emodin in the nucleus of HepG2 cells is higher than that in MC-3T3-E1 cells, which may contribute to the higher cytotoxicity of PBA-Emodin-NPs to HepG2 cells. Therefore, PBA-NPs will be a promising biomaterial for targeted polyphenolic drug delivery to the cancer cells. Based on our previous researches,45 we will use the mouse model to investigate in vivo effects of drug-loaded PBA-NPs focusing on examining the contribution of emodin’s multiple biological activities in the future research.
Fig. 9.
Relative cell viabilities at 24 h in HepG2 cells and MC-3T3-E1 cells in the presence of free emodin, PBA-NPs, and PBA-Emodin-NPs measured by MTT assays.
Fig. 10.
Confocal microscopy images of HepG2, and MC-3T3-E1 cell lines incubated with PBA-Emodin-NPs for 24 h. The original concentration of emodin is 5 µg mL−1. The images show the emodin fluorescence (green), and nuclei (blue, stained by DAPI). The scale bars are 50 µm.
Conclusions
In summary, an efficient procedure was developed for the preparation of uniform PBA-functionalized nanoparticles using PBAH via solvent displacement. The diameters of the PBA-NPs could be easily tuned by adjusting the initial PBAH concentration and the proportion of methanol. Moreover, the dissipative particle dynamics simulation technique was successfully used to simulate the self-assemble behavior of PBAH. Through a reversible interaction between boronic acid and diols, PBA-NPs can be as a carrier used to load polyphenolic drugs. Therefore, the facile synthetic protocol and low cytotoxicity of PBA-NPs make them promising candidates for controlled polyphenolic drug delivery.
Supplementary Material
Acknowledgement
The authors would like to thank the financial support from the Chinese Academy of Sciences, and the National Natural Science Foundation of China (Programs 21429401, 21374119). D.F. acknowledge the financial support from NIH grants (NIH R21AT006767 and R01HL116626). Y.L also acknowledge the financial support from Project supported by State key laboratory of precision measuring technology and instruments (Tianjin University).
Footnotes
Electronic Supplementary Information (ESI) available: [The procedure of PBA-ARS nanoparticles preparation, 1H NMR and 13C NMR spectra of monomer, 1H NMR spectrum of PBAH, Size distribution profile of PBA-NPs determined by DLS. The time course DLS analysis of the diameter distributions of PBA-NPs in 20 mM DTT solution, proportions of methanol and water, TEM image of PBA-ARS-NPs, UV absorbance spectra for PBAH, fluorescent spectra of PBA-ARS-NPs solution and ARS solution, and the time course DLS analysis of the diameter distributions of PBA-Emodin-NPs at various pH.]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Bhullar K, Rupasinghe H. Food Chem. 2015;168:595–605. doi: 10.1016/j.foodchem.2014.07.103. [DOI] [PubMed] [Google Scholar]
- 2.Davidov-Pardo G, McClements D. Trends Food Sci. Technol. 2014;38:88–103. [Google Scholar]
- 3.Zhang Z, Ma R, Shi L. Acc. Chem. Res. 2014;47:1426–1437. doi: 10.1021/ar5000264. [DOI] [PubMed] [Google Scholar]
- 4.Rao J, Geckeler K. Prog. Polym. Sci, 2011;36:887–913. [Google Scholar]
- 5.Dube N, Shu J, Dong H, Seo J, Ingham E, Kheirolomoom A, Chen P, Forsayeth J, Bankiewicz K, Ferrara K, Xu T. Biomacromolecules. 2013;14:3697–3705. doi: 10.1021/bm4010518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luk B, Zhang L. ACS Appl. Mater. Interfaces. 2014 doi: 10.1021/am5036225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng C, Jiang Y, Cheng R, Meng F, Zhong Z. Nano Today. 2012;7:467–480. [Google Scholar]
- 8.Scarano W, Duong H, Lu H, De Souza P, Stenzel M. Biomacromolecules. 2013;14:962–975. doi: 10.1021/bm400121q. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Zhang Z, Su X, Cai M, Zhuo R, Zhong Z. Biomaterials. 2013;34:10296–10304. doi: 10.1016/j.biomaterials.2013.09.042. [DOI] [PubMed] [Google Scholar]
- 10.Savariar E, Aathimanikandan S, Thayumanavan S. J. Am. Chem. Soc. 2006;128:16224–16230. doi: 10.1021/ja065213o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu S, Vutukuri D, Thayumanavan S. J. Am. Chem. Soc. 2005;127:16794–16795. doi: 10.1021/ja056042a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haag R, Kratz F. Angew. Chem. Int. Ed. 2006;45:1198–1215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- 13.Narla S, Pinnamaneni P, Nie H, Li Y, Sun X. Biochem. Biophys. Res. Commun. 2014;443:562–567. doi: 10.1016/j.bbrc.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Springsteen G, Wang B. Tetrahedron. 2002;58:5291–5300. [Google Scholar]
- 15.Wang Y, Chalagalla S, Li T, Sun X, Zhao W, Wang P, Zeng X. Biosens. Bioelectron. 2010;26:996–1001. doi: 10.1016/j.bios.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Xiao W, Xiao K, Berti L, Luo J, Tseng H, Fung G, Lam K. Angew. Chem. Int. Ed. 2012;51:2864–2869. doi: 10.1002/anie.201107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Ma R, Liu G, Li Z, Sun X, An Y, Shi L. J. Controlled Release. 2013;172:e103. [Google Scholar]
- 18.Ren J, Zhang Y, Zhang J, Gao H, Liu G, Ma R, An Y, Kong D, Shi L. Biomacromolecules. 2013;14:3434–3443. doi: 10.1021/bm4007387. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Davis E, Anderson J, Lin Q, Wang Q. Biomacromolecules. 2009;10:113–118. doi: 10.1021/bm8009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy D, Cambre N, Sumerlin B. Chem. Commun. 2009;16:2106–2108. doi: 10.1039/b900374f. [DOI] [PubMed] [Google Scholar]
- 21.Hoogerbrugge P, Koelman J. Europhys. Lett. 1992;19:155–160. [Google Scholar]
- 22.Maiti A, McGrother S. J. Chem. Phys. 2004;120:1594–1601. doi: 10.1063/1.1630294. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Liu H, Li Z, Qian H, Milano G, Lu Z. J. Comput. Chem. 2013;34:2197–2211. doi: 10.1002/jcc.23365. [DOI] [PubMed] [Google Scholar]
- 24.Fessi H, Puisieux F, Devissaguet J, Ammoury N, Benita S. Int. J. Pharm. 1989;55:R1–R4. doi: 10.1002/jps.2600790902. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Hu T, Liang H, Jiang M, Wu C. Macromolecules. 2000;33:8640–8643. [Google Scholar]
- 26.Lee S, Lee H, In I, Park S. Eur. Polym. J. 2014;57:1–10. [Google Scholar]
- 27.Galindo-Rodriguez S, Allemann E, Fessi H, Doelker E. Pharmaceutical Research. 2004;21:1428–1439. doi: 10.1023/b:pham.0000036917.75634.be. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, Springsteen G, Deeter S, Wang B. Tetrahedron. 2004;60:11205–11209. [Google Scholar]
- 29.Shinkai S, Tsukagoshi K, Ishikawa Y, Kunitake T. J. Chem. Soc., Chem. Commun. 1991;15:1039–1041. [Google Scholar]
- 30.Wang H. J. Chung. Curr. Microbiol. 1997;35:262–266. doi: 10.1007/s002849900250. [DOI] [PubMed] [Google Scholar]
- 31.Chang C, Lin C, Yang J, Namba T, Hattori M. Am. J. Chin. Med. 1996;24:139–142. doi: 10.1142/S0192415X96000189. [DOI] [PubMed] [Google Scholar]
- 32.Gao Q, Wang F, Guo S, Li J, Zhu B, Cheng J, Jin Y, Li B, Wang H, Shi S, Gao Q, Zhang Z, Cao W, Tian Y. Ultrasound in Medicine and Biology. 2011;37:1478–1485. doi: 10.1016/j.ultrasmedbio.2011.05.846. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Chan R, Yu P, Chan S. Pharm. Biol. 2013;51:1175–1181. doi: 10.3109/13880209.2013.782322. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Lin S, Liu D, Wang Z. Oncol. Rep. 2013;30:2555–2562. doi: 10.3892/or.2013.2741. [DOI] [PubMed] [Google Scholar]
- 35.Jia X, Yu F, Wang J, Iwanowycz S, Saaoud F, Wang Y, Hu J, Wang Q, Fan D. Breast Cancer Res. Treat. 2014;148:291–302. doi: 10.1007/s10549-014-3164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Duan J, Guo T, Xie W, Yan S, Li B, Zhou Y, Chen Y. J. Ethnopharmacol. 2009;124:522–529. doi: 10.1016/j.jep.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Wang T, Yin X, Lu Y, Shan W, Xiong S. Int. J. Nanomed. 2012;7:2325–2337. doi: 10.2147/IJN.S31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gobin A, Rhea R, Newman R, Mathur A. Int. J. Nanomed. 2006;1:81–87. doi: 10.2147/nano.2006.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai X, Nie W, Wang Y, Shen Y, Li Y, Gan S. J. Mater. Sci.: Mater. Med. 2012;23:2709–2716. doi: 10.1007/s10856-012-4728-x. [DOI] [PubMed] [Google Scholar]
- 40.Singh D, Rawat M, Semalty A, Semalty M. J. Therm. Anal. Calorim. 2012;108:289–298. [Google Scholar]
- 41.Evans D, Searles D, Mittag E. Phys. Rev. E. 2001;63:051105. doi: 10.1103/PhysRevE.63.051105. [DOI] [PubMed] [Google Scholar]
- 42.Onaca O, Enea R, Hughes D, Meier W. Macromol. Biosci. 2009;9:129–139. doi: 10.1002/mabi.200800248. [DOI] [PubMed] [Google Scholar]
- 43.Meng F, Zhong Z, Feijen J. Biomacromolecules. 2009;10:197–209. doi: 10.1021/bm801127d. [DOI] [PubMed] [Google Scholar]
- 44.Narender T, Sukanya P, Sharma K, Bathula S. Phytomedicine. 2013;20:890–896. doi: 10.1016/j.phymed.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Jia X, Iwanowycz S, Wang J, Saaoud F, Yu F, Wang Y, Hu J, Chatterjee S, Wang Q, Fan D. Exp. Biol. Med. 2014;239:1025–1035. doi: 10.1177/1535370214530247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.