Abstract
Nanopores have been used to detect molecules, to sequence DNA or to investigate chemical reactions at the single-molecule level. Because they approach the absolute limit of sensor miniaturisation, nanopores are amenable to parallelisation and could be used in single-cell measurements. Here we show that single enzymes can be functionally and reversibly trapped inside the confined space of a ClyA nanopore. Remarkably, the binding of ligands to the internalised proteins is mirrored by specific changes to the nanopore conductance. Conveniently, the manipulation of the charge of the protein allowed increasing of the residence time of the protein inside the nanopore. Nanopores with internalised protein adaptors can be used to study proteins in real-time or can be incorporated into inexpensive portable devices for the detection of analytes with high selectivity.
Introduction
Over the past two decades nanopore analysis has emerged as a promising analytical tool for single-molecule analysis1-4. Nanopore technology allows the investigation of native molecules with high sampling bandwidth without the need for labelling, chemical modifications or surface immobilisation. Further, the ionic current output signal can be easily interfaced with miniaturised and portable electronic devices. For instance, arrays of nanopores integrated into a MinION™ sequencer have been recently used for the profiling of genomic DNA5-7. Furthermore, biological nanopores have been reconstituted into bilayers formed on glass nanopipettes8 and on glass tips for scanning ion-conductance microscopy9. Therefore, nanopore-functionalized nanopipettes that can detect and quantify metabolites are promising platforms for measurement in single cells.
Previous studies showed that small molecules binding to cyclodextrin10 and cyclic peptide11 adaptors or cucurbiturils carriers12 could be detected by ionic current recordings using the α-hemolysin (αHL) nanopore. However, such guest adaptors and carriers do not bind selectively to host molecules, making the identification of analytes in a complex mixture of compounds a real challenge. By contrast, proteins have evolved to identify their ligands with high specificity in a sea of very similar chemical species. Therefore, nanopores equipped with protein adaptors would be ideal elements for integration into nanopore-based sensing devices for the analysis of complex biological samples. However, building such hybrid devices is challenging. Proteins are too large to be incorporated into the αHL and other biological nanopores13-15 and mostly translocate through solid-state nanopores too fast to be properly sampled 16. Further, it is not known if the environment of the nanopore lumen is compatible with enzymatic functions, as experiments with solid-state nanopores revealed that proteins might be stretched by the electrical field17 and unfolded under applied potentials higher than +200 mV18.
Recently, we showed that folded proteins enter the lumen of Type I ClyA-AS (C87A/L99Q/E103G/F166Y/I203V/C285S/K294R/H307Y), a dodecameric19 engineered version of ClyA from Salmonella Typhi selected for its favourable properties in planar lipid bilayers20. Conveniently, ClyA also assembles into higher oligomeric forms (Type II and Type III ClyA)20 that are large enough to accommodate, for example, protein-DNA complexes 21. Notably, we showed that electroosmotic and electrophoretic forces allowed trapping proteins such as thrombin inside the ~240 nm3 cavity of Type I ClyA nanopores for tens of minutes20,22, suggesting that protein adaptors might be paired to ClyA nanopores without the use of covalent chemistry or other immobilisation techniques. In this work we report that the binding of analytes to two model proteins incorporated inside a ClyA nanopore is reflected by changes in the nanopore conductance, indicating that proteins immobilised inside the nanopore remain functional. Moreover, electrical readouts of nanopore-confined proteins will have applications in the fabrication of sensor arrays for the discovery of new therapeutics or the detection of biomarker analytes in biological samples.
Results and discussion
As a first model protein we selected E. coli AlkB demethylase (Mw=25 kDa), a globular protein that is expected to pass the cis entry of ClyA, but is too large to traverse the trans exit of the nanopore (Figure 1a,b). In complex with iron ions (AlkB-Fe++) AlkB co-oxidises methylated DNA and its cofactor 2-oxoglutarate (2-OG), producing succinate (SUC), carbon dioxide and formaldehyde23,24. 2-oxoglutarate is an important metabolite that influences aging and age-related diseases25, and is a biomarker for non-alcoholic fatty liver disease26, heart failure27 and cardiorenal syndrome 27. The level of succinate in urine is a biomarker for kidney damage 28.
Figure 1.
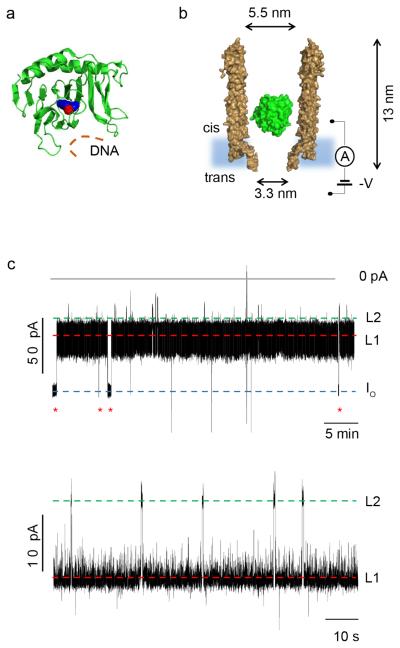
Internalization of AlkB-Fe++ into ClyA-AS. a, Cartoon representation of E. coli AlkB (green) containing a metal ion (Co2+, red sphere) and binding to the cofactor (2-OG, blue spheres). The DNA binding site is depicted by an orange line. PDB_ID 3KHB. b, Representation of a single AlkB-Fe++ enzyme (green) confined in a ClyA-AS nanopore (brown, shown as cross-section) embedded in a planar lipid bilayer (light blue) under a negative applied potential. The dimensions of the pore consider the Van der Waals radii of the atoms. c, Top: Typical current blockades provoked by AlkB-Fe++ molecules (~4 nM, cis) entering a ClyA-AS nanopore at −60 mV. The open pore current (IO) is represented by a blue dashed line, while Level 1 and Level 2 are shown by red and green dashed lines, respectively. The red asterisks represent the restoration of IO upon the exiting of AlkB-Fe++ from the pore. Bottom: Detail of a single AlkB-Fe++ blockade, showing Level 1 (red) and Level 2 (green) current levels. The current traces were collected by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz. An additional Bessel 8-pole filter with 50 Hz cut-off was digitally applied to the trace shown in c, bottom. All recordings were carried out in 150 mM NaCl, 15 mM Tris HCl pH 8.0, at 28°C, and the AlkB was added to the cis compartment.
Individual AlkB-Fe++ molecules were studied using Type I ClyA-AS (ClyA-AS hereafter). In 150 mM NaCl, 15 mM Tris HCl and pH 8.0 ClyA-AS formed nanopores with a steady open pore conductance (IO = −1.7±0.1 nSi, average±SD, N=38, −60 mV, 28°C) under a wide range of applied potentials. Here and hereafter N indicates the number of independent single nanopore experiments, np the number of individual protein blockades and nl the total number of ligand binding events analysed. The addition of AlkB-Fe++ (~4 nM) to the cis side of ClyA-AS provoked current blockades (IB), quoted here as residual currents calculated as a percentage of the open pore current (IRES%), due to the electroosmotic confinement of AlkB-Fe++ between the wider cis entrance and the narrower trans exit of the protein nanopore (Figure 1b)20,22. Conveniently, AlkB-Fe++ remained trapped inside the nanopore for several minutes (Figure 1c). The signal induced by AlkB-Fe++ fluctuated between two distinctive current levels, L1 (IRES% = 52.6±2.0%, np=15, N=7) and L2 (IRES% = 39.0±1.0%, np=15, N=7, Figure 1c), possibly due to two residence sites for the protein within the lumen of the ClyA-AS nanopore22.
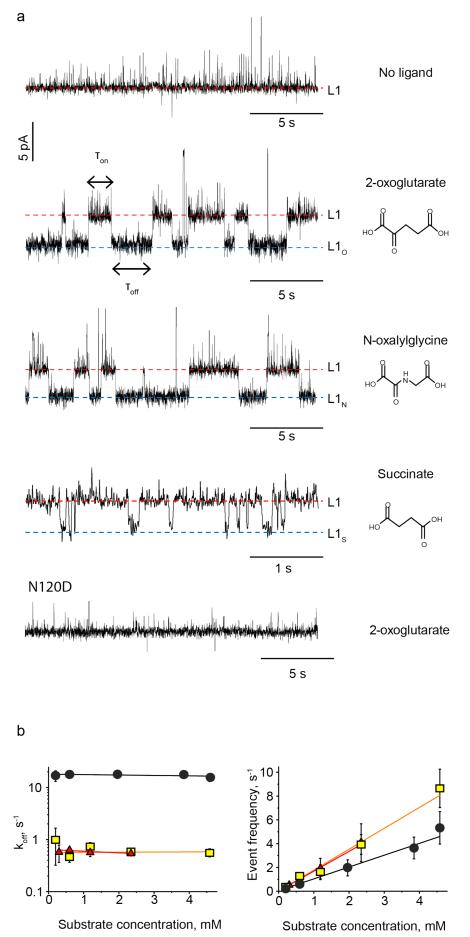
At −60 mV the addition to the cis reservoir of the cofactor (2-OG), an isosteric inhibitor (N-oxalylglycine, N-OG) or the processed cofactor (SUC) induced reversible current enhancements within the AlkB-Fe++ blockades (ΔIRES%= +4.7±1.3%, +4.9±1.0 and +4.6±1.3, respectively, np>15, nl > 75, N>4, Figure 2a, Figure S1,2 and Table S1) that showed a mean duration (τoff) of 1.7±0.5 s, 1.8±0.4 s and 61±11 ms, respectively (nl>4500, N>8, and np>80). The current enhancements were also observed from the current level L2 (Figure S2). We hypothesised that such current events reflected the conformational changes occurring during the transition from the open conformation of the apo-enzyme to the closed state of the ligand-bound form of AlkB-Fe++ (Figure 2a)29,30. To confirm this hypothesis we tested an AlkB mutant where the asparagine at position 120, which has been reported to be involved in the binding of 2-OG to AlkB29, was substituted by aspartate (N120D). The addition of 7.2 mM of 2-OG did not induce current transitions to the N120D-AlkB-Fe++ blockades (N=4, Figure 2a), suggesting that the affinity of this AlkB mutant for 2-OG is strongly reduced. As expected for a protein-ligand association process the dissociation rate constants (koff, Table S2), measured from the inverse of the dwell times of the ligand-binding events (1/τoff), did not depend on the concentration of the ligand, while the frequencies of the ligand-induced events (f = 1/τon) increased linearly with the concentration of the three ligands, from which slopes the association rate constants (kon) could be calculated (Figure 2b, Table S2).
Figure 2.
Binding of ligands to AlkB-Fe++ confined inside ClyA-AS. a, Typical ligand-induced blockades to individual AlkB-Fe++ enzymes confined inside ClyA-AS at −60 mV. The ligand used is shown on the right of the trace. The bound Level 1 current levels (L1O, L1N, L1S) are represented by the blue dashed lines. The substrate concentration was 0.6 mM for 2-OG, 0.6 mM for N-OG and 2 mM for SUC binding to wild type AlkB-Fe++, and 7.2 mM for 2-OG binding to N120D-AlkB-Fe++. b, Left: Dissociation rate constants (koff) as a function of the ligand concentration at −60 mV. Right: Event frequency (1/τon) as a function of the ligand concentration at −60 mV. 2-OG is shown in yellow squares, SUC in black circles and N-OG in red triangles. All current traces were collected by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz. An additional Bessel 8-pole filter with 50 Hz cut-off was digitally applied to the current traces. All recordings were carried out in 150 mM NaCl, 15 mM Tris HCl pH 8.0, at 28°C, and the ligands were added to the cis compartment. Errors are given as standard deviations.
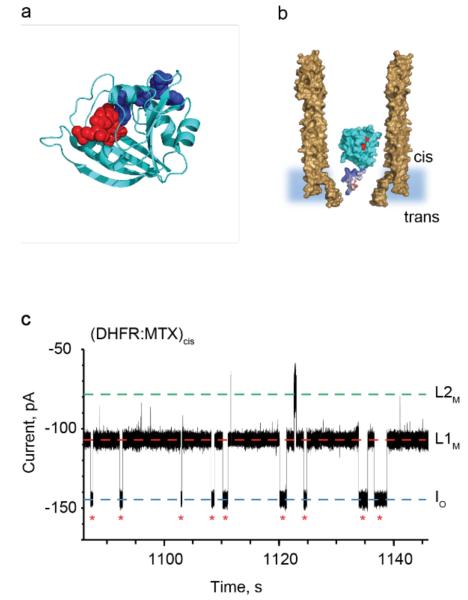
E. coli dihydrofolate reductase (DHFR, Mw=19 kDa) was selected as a second model protein adaptor (Figure 3a,b). During the DHFR catalytic cycle dihydrofolate is reduced to tetrahydrofolate and the cofactor NADPH is oxidised to NADP+. Tetrahydrofolate is a cofactor in many metabolic reactions, thus inhibitors of DHFR such as methotrexate (MTX) are antibiotic and anticancer agents. The ratio of the NADP+ and NADPH intracellular concentrations is used to monitor the oxidative stress in cells32. We found that apo-DHFR, which is smaller than AlkB, dwelled inside ClyA-AS only for a few milliseconds. Upon the addition of MTX to the cis solution the frequency and the dwell time of the protein blockades decreased, while the residual current increased (supporting results). The blockades were then abolished by the subsequent addition of NADPH to the same side (Figure S3). Since both the inhibitor and the cofactor are negatively charged, these results suggested that the additional negative charges increased the electrophoretic/electrostatic drag force opposing DHFR entry and residence inside the nanopore. In order to increase the residence time of the protein, we engineered DHFR by introducing a polypeptide tag containing four additional positive charges at the C-terminus of the protein (DHFRtag, supporting results, Figure S4). In complex with MTX, DHFRtag, added to the cis compartment, induced current blockades with a mean dwell of 3.1±1.4 s (N=5, np=230, Figure 3c) that was three orders of magnitude longer than DHFRtag or DHFR:MTX blockades mean dwell times. A possible explanation to this result is that, tuned by the additional positive charges, the binary DHFRtag:MTX complex is at a potential minimum inside the nanopore where the electroosmotic, electrophoretic and electrostatic forces are balanced. The dissociation of MTX from the binary complex was slower than the residence time of the complex inside the nanopore and could not be observed by ionic current recordings. As shown before with apo-AlkB-Fe++, DHFRtag:MTX blockades showed a main current level L1 (L1M, IRES% = 74.7±0.5%, np =25, N=5) that rarely visited a second current level L2 (L2M, IRES%, = 53.5±0.9%, np =25, N=5, Figure 3c).
Figure 3.
DHFR as a protein adaptor. a, Cartoon representation of E. coli DHFR (cyan) with bound methotrexate (MTX, red spheres) and NADPH (blue spheres), PDB_ID 1RH3. b, Representation of a single DHFRtag enzyme (cyan) in complex with MTX (red) confined in a ClyA-AS nanopore (brown, shown as cross-section) embedded in a planar lipid bilayer (light blue) under a negative applied potential. The positively charged polypeptide tag added at the C-terminus of DHFR is shown in blue. c, Typical current blockades provoked by the capture of DHFRtag:MTX complexes (20 nM DHFRtag, 400 nM MTX, cis) by the ClyA-AS nanopore at −90 mV. The open pore current (IO) is represented by a blue dashed line, while L1M and L2M are shown by red and green dashed lines, respectively. Red asterisks represent restoration of IO upon the exiting of DHFRtag:MTX from the pore. The current traces were collected in 150 mM NaCl, 15 mM Tris HCl pH 7.5, at 28°C by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz.
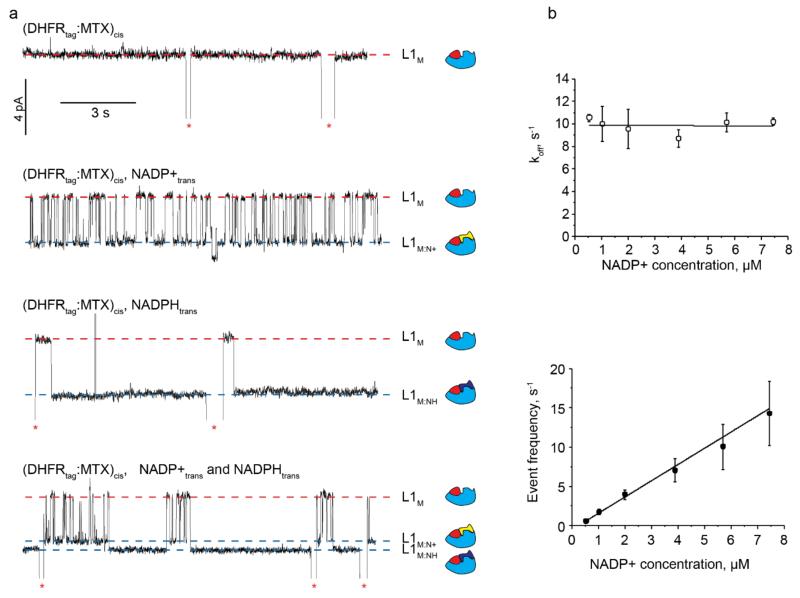
At −90 mV the addition of the oxidised cofactor NADP+ to the trans compartment of ClyA-AS produced reversible current enhancements to the DHFRtag:MTX complex blockades formed in the cis solution (L1M:N+, ΔIRES%= +2.3±0.5%, np =15 blockades, nl > 225, N=3; and τM:N+ = 102±11 ms, nl =19,000, N=9 np > 800, Figure 4a, Table S3, Figure S5). Association and dissociation rate constants could be measured from titration experiments (Figure 4b, Table S4). NADPH added to the trans compartment also induced additional current enhancements to the binary complex blockades (Figure 4a). Remarkably, the current events induced by NADPH showed a slightly higher residual current (L1M:NH, ΔIRES%= +2.7±0.7%, np =15 nl=15, N=4 Table S3) than the NADP+ blockades (ΔIRES%= +2.3±0.5%) and had a dwell time longer than the residence time of the ternary complex inside the nanopore (Figure 4a). As a consequence, despite the minute difference between NADPH and NADP+ (a hydride ion), the binding of the two ligands to DHFRtag:MTX could be clearly differentiated (Figure 4a).
Figure 4.
Ligands binding to DHFRtag. a, Ligand-induced current enhancements to individual DHFRtag:MTX blockades at −90 mV. NADP+ and NADPH are added to the trans compartment after addition of 20 nM DHFRtag and 400 nM MTX to the cis compartment. From top to bottom: no ligand; 5.7 μM of NADP+; 0.7 μM of NADPH; 7.4 μM of NADP+ together with 0.7 μM of NADPH. Free and bound Level 1 are shown by red and blue dashed lines, respectively. Red asterisks represent restoration of IO upon the exit of DHFRtag:MTX from the pore. On the right of the current traces is the schematic representation of the interaction of DHFRtag (cyan) with MTX (red), NADP+ (yellow) or NADPH (blue). b, Top: Dissociation rate constants (koff) as a function of the NADP+ concentration added to the trans compartment at −90 mV. Bottom: Event frequency (1/τon) as a function of the NADP+ concentration added to the trans compartment at −90 mV. Errors are shown as standard deviations. All current traces were collected by applying a Bessel-low pass filter with a 2 kHz cut-off and sampled at 10 kHz. An additional Bessel 8-pole filter with 50 Hz cut-off was digitally applied to the traces shown in a. All recordings were carried out in 150 mM NaCl, 15 mM Tris HCl pH 7.5, at 28°C.
Although the bulk kinetic constants for the binding of NADP+ and NADPH to MTX:DHFR could not be retrieved from the literature, the equilibrium dissociation constant for the binding of 2-OG to AlkB-Mn++ was recently measured by an intrinsic tryptophan fluorescence quenching assay (KDbulk= 4.1±0.6 10−6 M at 24 °C)31. By comparison, the equilibrium dissociation constant of 2-OG for AlkB-Fe++ inside the nanopore measured from the ratio of the association and dissociation constants (KDpore =koff/kon) was about two orders of magnitude higher than the bulk value (KDpore=3.7±1.9 10−4 M, −60 mV, 28°C). This effect is likely to be related to the confinement of AlkB-Fe++ inside the nanopore and to the effect of the applied potential. ClyA nanopores have a negatively charged interior and are, therefore, cation selective.33 Thus, under negative applied potentials (trans) the diffusion of the negatively charged ligands added to the cis solution through the nanopore is likely to be opposed. This is probably to be further accentuated by the unfavourable electrostatic interaction between the ligands and the wall of the nanopore lumen. This complication might be overcome by using nanopores with an internal charge with an opposite sign to that of the ligand to detect.
Conclusions
The results presented here indicate that the binding of analytes to proteins trapped inside ClyA can be monitored by specific changes in the nanopore conductance, suggesting that proteins confined inside nanopores remain functional. Proteins with suitable size and shape, such as AlkB, are sterically trapped between the wider cis entrance and the narrower trans exit of the pore. Smaller proteins, such as DHFR, that escape ClyA too quickly to allow the sampling of ligand binding kinetics, can be engineered with genetically encoded extensions to increase their residence time inside the nanopore. Our approach should also be applicable to larger protein adaptors, which could be internalised into larger nanopores such as higher oligomeric forms of ClyA 20, Phi29 34, pneumolysin 35 or solid-state nanopores. Since most biologically active molecules have a protein target, nanopores with an internal protein adaptor are promising systems for integration in miniaturised low-cost electronic devices for medical, forensics or environmental monitoring or for single-cell analysis.
Supplementary Material
ACKNOWLEDGMENT
We thank the European Research Council (European Commission’s Seventh Framework Programme, project n° 260884) for funding. AB is funded by a Ph.D. grant from the Agency for Innovation by Science and Technology (IWT) Flanders.
Footnotes
ASSOCIATED CONTENT
Additional text, Materials and methods, Supporting Tables, Supporting Figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- (1).Howorka S, Siwy Z. Chemical Society Reviews. 2009;38:2360. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- (2).Bayley H. Clinical chemistry. 2015;61:25. doi: 10.1373/clinchem.2014.223016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Luchian T, Shin SH, Bayley H. Angewandte Chemie-International Edition. 2003;42:3766. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- (4).Bezrukov SM, Vodyanoy I, Parsegian VA. Nature. 1994;370:279. doi: 10.1038/370279a0. [DOI] [PubMed] [Google Scholar]
- (5).Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O’Grady J. Nature biotechnology. 2014 doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- (6).Mikheyev AS, Tin MM. Molecular ecology resources. 2014;14:1097. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- (7).Quick J, Quinlan AR, Loman NJ. GigaScience. 2014;3:22. doi: 10.1186/2047-217X-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).White RJ, Ervin EN, Yang T, Chen X, Daniel S, Cremer PS, White HS. J Am Chem Soc. 2007;129:11766. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- (9).Zhou Y, Bright LK, Shi W, Aspinwall CA, Baker LA. Langmuir. 2014;30:15351. doi: 10.1021/la504097f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- (11).Sanchez-Quesada J, Ghadiri MR, Bayley H, Braha O. Journal of the American Chemical Society. 2000;122:11757. [Google Scholar]
- (12).Braha O, Webb J, Gu LQ, Kim K, Bayley H. Chemphyschem. 2005;6:889. doi: 10.1002/cphc.200400595. [DOI] [PubMed] [Google Scholar]
- (13).Jung Y, Cheley S, Braha O, Bayley H. Biochemistry. 2005;44:8919. doi: 10.1021/bi0473713. [DOI] [PubMed] [Google Scholar]
- (14).Movileanu L, Howorka S, Braha O, Bayley H. Nature Biotechnology. 2000;18:1091. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- (15).Fahie M, Chisholm C, Chen M. ACS nano. 2015;9:1089. doi: 10.1021/nn506606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C. Nano Letters. 2013;13:658. doi: 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Oukhaled A, Cressiot B, Bacri L, Pastoriza-Gallego M, Betton JM, Bourhis E, Jede R, Gierak J, Auvray L, Pelta J. ACS nano. 2011;5:3628. doi: 10.1021/nn1034795. [DOI] [PubMed] [Google Scholar]
- (18).Freedman KJ, Haq SR, Edel JB, Jemth P, Kim MJ. Sci Rep. 2013;3:1638. doi: 10.1038/srep01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. Nature. 2009;459:726. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- (20).Soskine M, Biesemans A, De Maeyer M, Maglia G. J Am Chem Soc. 2013;135:13456. doi: 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Van Meervelt V, Soskine M, Maglia G. ACS nano. 2014;8:12826. doi: 10.1021/nn506077e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. Nano Letters. 2012;12:4895. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Aravind L, Koonin EV. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- (25).Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, Hu E, Whelan SA, Wang JX, Jung G, Solis GM, Fazlollahi F, Kaweeteerawat C, Quach A, Nili M, Krall AS, Godwin HA, Chang HR, Faull KF, Guo F, Jiang M, Trauger SA, Saghatelian A, Braas D, Christofk HR, Clarke CF, Teitell MA, Petrascheck M, Reue K, Jung ME, Frand AR, Huang J. Nature. 2014;510:397. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rodriguez-Gallego E, Guirro M, Riera-Borrull M, Hernandez-Aguilera A, Marine-Casado R, Fernandez-Arroyo S, Beltran-Debon R, Sabench F, Hernandez M, Del Castillo D, Menendez JA, Camps J, Ras R, Arola L, Joven J. International journal of obesity. 2014 doi: 10.1038/ijo.2014.53. [DOI] [PubMed] [Google Scholar]
- (27).Nikolaidou T, Mamas M, Oceandy D, Neyses L. Heart. 2010;96:e14. [Google Scholar]
- (28).Peti-Peterdi J. Google Patents. 2014
- (29).Bleijlevens B, Shivarattan T, Flashman E, Yang Y, Simpson PJ, Koivisto P, Sedgwick B, Schofield CJ, Matthews SJ. EMBO reports. 2008;9:872. doi: 10.1038/embor.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bleijlevens B, Shivarattan T, van den Boom KS, de Haan A, van der Zwan G, Simpson PJ, Matthews SJ. Biochemistry. 2012;51:3334. doi: 10.1021/bi201699e. [DOI] [PubMed] [Google Scholar]
- (31).Ergel B, Gill ML, Brown L, Yu B, Palmer AG, 3rd, Hunt JF. The Journal of biological chemistry. 2014;289:29584. doi: 10.1074/jbc.M114.575647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ogasawara Y, Funakoshi M, Ishii K. Biological & pharmaceutical bulletin. 2009;32:1819. doi: 10.1248/bpb.32.1819. [DOI] [PubMed] [Google Scholar]
- (33).Ludwig A, Bauer S, Benz R, Bergmann B, Goebel W. Molecular Microbiology. 1999;31:557. doi: 10.1046/j.1365-2958.1999.01196.x. [DOI] [PubMed] [Google Scholar]
- (34).Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Nature nanotechnology. 2009;4:765. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gilbert RJ, Jimenez JL, Chen S, Tickle IJ, Rossjohn J, Parker M, Andrew PW, Saibil HR. Cell. 1999;97:647. doi: 10.1016/s0092-8674(00)80775-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.