Reducing isozyme competition between host and transgenic acyltransferases increases the accumulation of ricinoleic and α-eleostearic acid in seed triacylglycerol of Arabidopsis.
Abstract
One goal of green chemistry is the production of industrially useful fatty acids (FAs) in crop plants. We focus on hydroxy fatty acids (HFAs) and conjugated polyenoic FAs (α-eleostearic acids [ESAs]) using Arabidopsis (Arabidopsis thaliana) as a model. These FAs are found naturally in seed oils of castor (Ricinus communis) and tung tree (Vernicia fordii), respectively, and used for the production of lubricants, nylon, and paints. Transgenic oils typically contain less target FA than that produced in the source species. We hypothesized that competition between endogenous and transgenic isozymes for substrates limits accumulation of unique FAs in Arabidopsis seeds. This hypothesis was tested by introducing a mutation in Arabidopsis diacylglycerol acyltransferase1 (AtDGAT1) in a line expressing castor FA hydroxylase and acyl-Coenzyme A:RcDGAT2 in its seeds. This led to a 17% increase in the proportion of HFA in seed oil. Expression of castor phospholipid:diacylglycerol acyltransferase 1A in this line increased the proportion of HFA by an additional 12%. To determine if our observations are more widely applicable, we investigated if isozyme competition influenced production of ESA. Expression of tung tree FA conjugase/desaturase in Arabidopsis produced approximately 7.5% ESA in seed lipids. Coexpression of VfDGAT2 increased ESA levels to approximately 11%. Overexpression of VfDGAT2 combined with suppression of AtDGAT1 increased ESA accumulation to 14% to 15%. Our results indicate that isozyme competition is a limiting factor in the engineering of unusual FAs in heterologous plant systems and that reduction of competition through mutation and RNA suppression may be a useful component of seed metabolic engineering strategies.
Production of vegetable oils in the form of triacylglycerols (TAGs) is of great importance for human nutrition and as a source of chemicals for industry. The vegetable oils produced in our food crops are composed of five major fatty acids (FAs; Bates et al., 2013). Other than these common FAs, numerous uncommon FAs are produced in nature, such as hydroxy fatty acids (HFAs), conjugated FAs, epoxy FAs, and short-chain FAs, that are or could be used for industrial purposes (Badami and Patil, 1980). However, the species producing these uncommon FAs are often not suitable for large-scale industrialized agriculture (Voelker and Kinney, 2001; Dyer et al., 2008). To solve this problem, attempts have been made to produce these uncommon FAs in seeds of crop plants. This has been a long-standing goal in the field of lipid research and seemed initially quite straightforward (Voelker and Kinney, 2001; Napier, 2007; Napier and Graham, 2010; Carlsson et al., 2011; Bates and Browse, 2012; Bates et al., 2013; Vanhercke et al., 2013). The approach taken was to identify the enzyme responsible for synthesis of the desired FA and express the corresponding gene in seeds of high-yielding crop plants. Unfortunately, in general, only low levels of the desired FAs were produced compared with levels in the native plant (Broun and Somerville, 1997; Cahoon et al., 2006). One reason for this discrepancy is that enzymes of TAG synthesis often lack proper substrate specificity and selectivity, leading to poor utilization of substrates containing these unusual FAs (Knutzon et al., 1999; Burgal et al., 2008; Li et al., 2010; Kim et al., 2011; van Erp et al., 2011).
In this article, we focus on the engineering of HFAs (including ricinoleic acid) and α-eleostearic acid (ESA) in heterologous plant systems. HFAs are used in many industrial applications, including the production of nylon, plastics, and lubricants, and they are produced at high levels in the seeds of castor (Ricinus communis). However, castor is not suitable for industrialized agriculture and produces the toxic protein ricin as well as other proteins that can cause allergenic reactions in humans. Currently, most of the cultivation of castor for the production of HFAs occurs in China, India, and Brazil. ESA is used in industrial applications, such as inks, coatings, and resins, and produced in the seeds of tung tree (Vernicia fordii; formerly Aleurites fordii; Sonntag, 1979). Tung tree also has problematic agronomic characteristics and can only be grown in limited areas of the United States that are prone to damage from hurricanes. To create cheap and reliable sources of these FAs, their synthesis has been studied, and attempts have been made to produce them in heterologous plant systems.
The gene responsible for the synthesis of HFA in castor is FATTY ACID HYDROXYLASE12 (RcFAH12). This enzyme hydroxylates the ∆12 position of oleic acid esterified to the stereospecifically numbered2 (sn-2) position of phosphatidylcholine (PC) and is a homolog of FATTY ACID DESATURASE2 (FAD2; Van de Loo et al., 1995). Tung tree Fatty Acid Conjugase/Desaturase (FADX; Dyer et al., 2002) is responsible for the synthesis of ESA, and also, it is an FAD2 homolog. It converts PC-bound linoleoyl groups to eleostearate (18:3Δ9cis, 11trans, and 13trans; Dyer et al., 2002). After synthesis on PC, these FAs can be incorporated into TAG by several different metabolic routes and enzymes (Fig. 1). Only the routes and enzymes relevant to this article will be discussed here (a more exhaustive description is in van Erp et al., 2011). The modified FAs can be hydrolyzed from the sn-2 position of PC by phospholipid:diacylglycerol acyltransferase (PDAT), which then esterifies these FAs to the sn-3 position of diacylglycerol (DAG) to generate TAG. The lysophosphatidylcholine generated by this reaction can be used by acyl-CoA:lysophosphatidylcholine acyltransferase to regenerate PC. PC can be converted into DAG by phospholipase C or phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT). Phospholipase C is involved in the removal of the choline head group of PC to generate DAG with the same FA composition as PC. PDCT exchanges the choline head group between PC and DAG. The DAG formed by these enzymes can subsequently be used by PDAT or acyl-CoA:diacylglycerol acyltransferase (DGAT) to generate TAG. A reverse activity of lysophosphatidylcholine acyltransferase can hydrolyze the modified FAs from the sn-2 position of PC to generate acyl-CoA and lysophosphatidylcholine. These acyl-CoAs can subsequently be used by the acyltransferase enzymes of the Kennedy pathway (acyl-CoA:glycerol-3-P acyltransferase, acyl-CoA:lysophosphatidic acid acyltransferase, and DGAT) to generate TAG-containing modified FAs.
Figure 1.
Overview of TAG biosynthesis in transgenic Arabidopsis seeds overexpressing FAH12 or FADX. These enzymes modify 18:1-PC and 18:2-PC, respectively, to generate HFA-PC or ESA-PC, respectively (m indicates FAs with these modifications). The modified FAs (as well as 18:2 and 18:3) are subsequently incorporated into TAG by the various enzymes involved in the synthesis of this storage lipid. GPAT, Acyl-CoA:glycerol-3-P acyltransferase; G3P, glycerol-3-P; LPA, lysophosphatidic acid; LPAT, acyl-CoA:lysophosphatidic acid acyltransferase; LPC, lysophosphatidylcholine; LPCAT, acyl-CoA:lysophosphatidylcholine acyltransferase; PA, phosphatidic acid; PAP, phosphatidic acid phosphatase; PLC, phospholipase C.
Expression of an RcFAH12 complementary DNA (cDNA) in Arabidopsis (Arabidopsis thaliana) under control of several seed-specific promoters led to accumulation of HFA to only 17% of total seed FAs (Broun and Somerville, 1997; Smith et al., 2000, 2003; Lu et al., 2006) compared with approximately 90% in native castor seed oil (Badami and Patil, 1980). Engineering attempts to overcome this problem have focused on coexpression of castor acyltransferases, such as RcDGAT2 (Burgal et al., 2008), RcPDAT1A (Kim et al., 2011; van Erp et al., 2011), or the castor electron transfer system in Arabidopsis seeds expressing RcFAH12 (Wayne et al., 2013). Coexpression of RcDGAT2 and RcPDAT1A led to significant increases in HFA levels from 17% to 26% to 28% of total seed FAs, whereas coexpression of the electron transfer system did not result in an increase. Production of ESA in Arabidopsis has met similar challenges. Expression of the FADX genes from either tung tree or bitter gourd (Momordica charantia; another species containing high levels of ESA in seeds) in an fad3 fatty acid elongase1 (fae1) Arabidopsis line resulted in 7% to 13% ESA (Cahoon et al., 2006). Two types of DGAT from tung tree showed significantly different affinities toward ESA-containing substrates when expressed in yeast (Saccharomyces cerevisiae)-fed tung oil FAs (Shockey et al., 2006), but the effects of these and other tung tree TAG metabolic enzymes in planta have not previously been reported. The experiments reported here represent a valuable opportunity to investigate possible commonalities between metabolic engineering strategies for production of various types of value-added oils.
Subsequent research into the underlying causes of suboptimal production of HFA revealed that RcFAH12 expression in Arabidopsis seeds caused metabolic perturbations, leading to the poor accumulation of HFA in TAG and a reduction in total FA content of seeds (Dauk et al., 2007; Bates and Browse, 2011; van Erp et al., 2011). Overexpression of castor PDAT and DGAT restored FA content in RcFAH12 transgenic seeds to nearly wild-type levels, while simultaneously increasing HFA content of the seeds (van Erp et al., 2011). Bates et al., 2014 elucidated the mechanism behind this decrease in FA content of seeds expressing RcFAH12 by showing that RcFAH12 expression resulted in feedback inhibition of FA synthesis. These results indicate that efficient transfer of HFA from their site of synthesis on PC into TAG is essential for the engineering of high levels of HFA in heterologous plant systems.
In this article, we investigated whether it is possible to improve the flux of HFA out of PC and into TAG by reducing substrate competition between endogenous Arabidopsis isozymes and the transgenic counterparts from castor and tung tree. We focused on substrate competition between AtDGAT1 and/or AtPDAT1 and either RcDGAT2 and RcPDAT1A from castor or VfDGAT2 from tung tree. To increase synthesis of HFA, we attempted to replace AtDGAT1 and AtPDAT1 with RcDGAT2 and RcPDAT1A by introducing a null mutation in AtDGAT1 and suppressing the expression of AtPDAT1 with artificial microRNAs (amiRNAs). Introduction of an Atdgat1 null mutation in a plant line expressing RcFAH12 and RcDGAT2 resulted in a significant increase in HFA levels. Subsequent overexpression of RcPDAT1A resulted in a further increase in HFA levels. In the case of ESA, an established homozygous FADX line (producing approximately 8% ESA in total seed lipids) was retransformed with either VfDGAT2 alone or VfDGAT2 together with a seed-specific AtDGAT1 RNA interference (RNAi) construct. Analysis of seed lipids from homozygous double-transgenic versus parental single-transgenic FADX lines showed higher levels of ESA when tung tree DGAT2 was present, which were enhanced further when the AtDGAT1 RNAi was present. The similarities between these two data sets suggest that endogenous competition is a limitation common to many such engineering strategies.
RESULTS
Eliminating AtDGAT1 Increases HFA Accumulation in an FAH12 RcDGAT2 Transgenic Line
AtDGAT1 and AtPDAT1 catalyze the final step in the synthesis of TAG in Arabidopsis seeds (Zhang et al., 2009). AtDGAT1 and AtPDAT1 preferentially incorporate common FAs into TAG, whereas RcDGAT2 and RcPDAT1A prefer HFA (Burgal et al., 2008; van Erp et al., 2011). We hypothesize that substrate competition for common FAs versus unusual FAs occurs between AtPDAT1/AtDGAT1 and the transgenic acyltransferases and that this competition limits the accumulation of unusual FAs in our FAH12 RcDGAT2 transgenic lines.
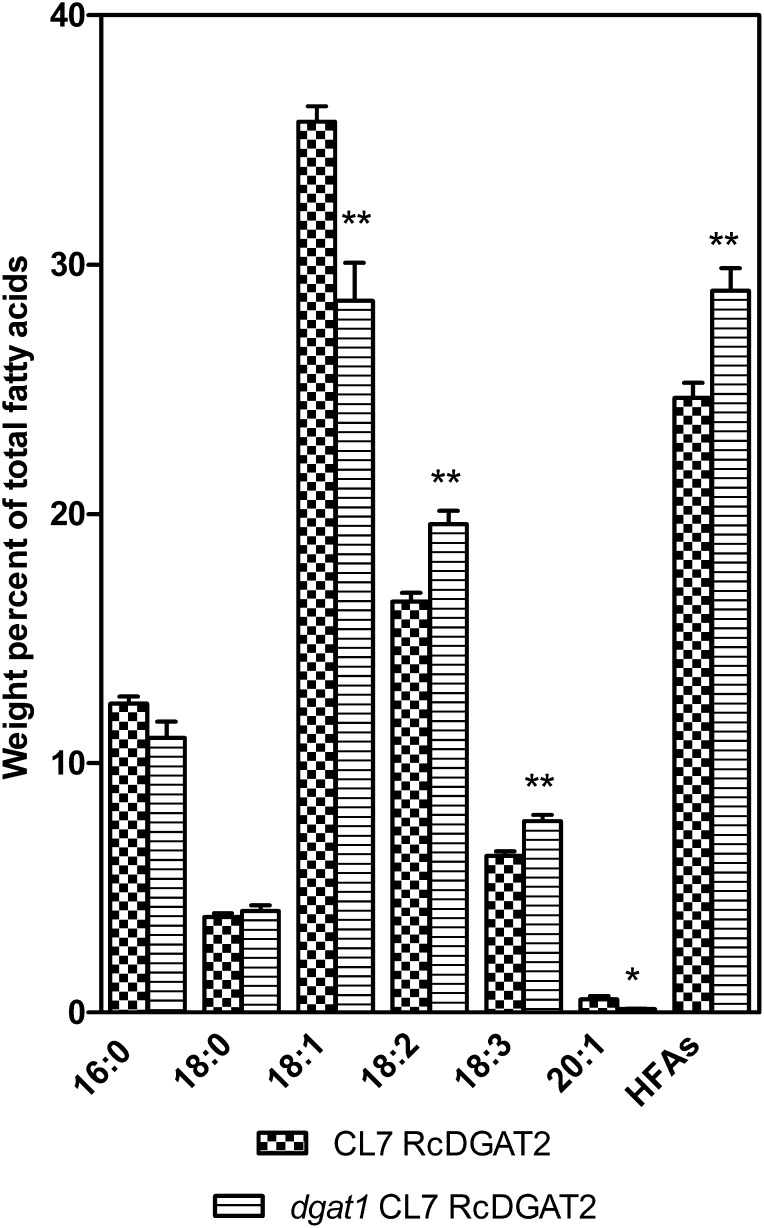
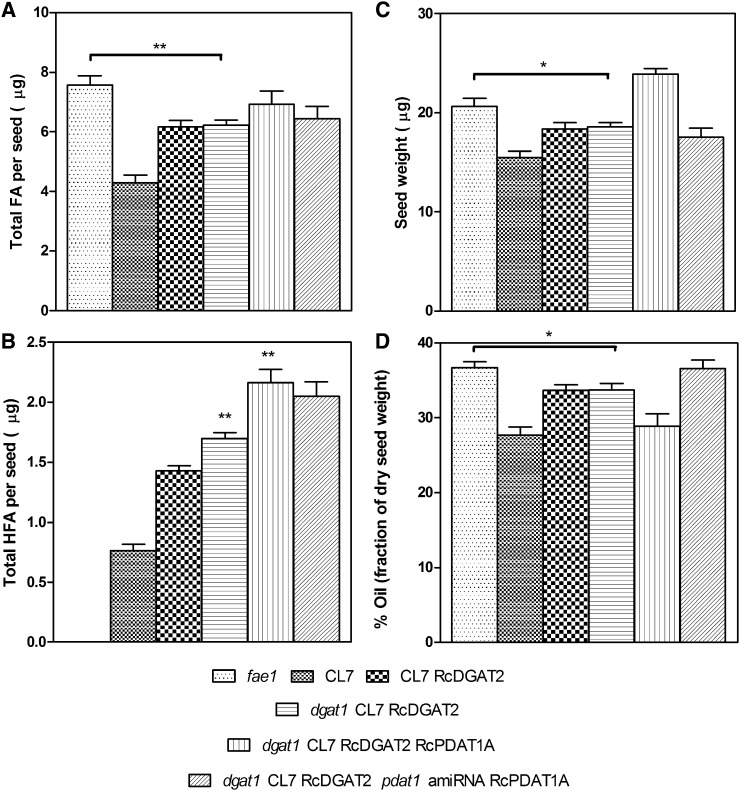
To test this proposal, the Arabidopsis dgat1-2 mutant (Routaboul et al., 1999) was crossed with the Chaofu Lu7 (CL7) RcDGAT2 line 544 #5 (Burgal et al., 2008) followed by selfing of F1 plants. The CL7 line expresses RcFAH12 in seeds of the fae1 mutant (Kunst et al., 1992; Lu et al., 2006). Segregation of the four loci (FAH12, RcDGAT2, fae1, and dgat1) in an F2 population of 460 plants was monitored by PCR assays and gas chromatography of seed FA compositions. An F2 plant determined to be heterozygous for dgat1 and homozygous for fae1, RcFAH12, and RcDGAT2 was grown to maturity. A population of F3 progeny of this plant (n = 60) was grown, and sequencing of DGAT1 was used to identify nine wild-type and nine dgat1-2 segregants. Analysis of seed samples from these plants shows a 17% increase in the proportion of HFA from an average of 24.7% ± 0.61% in the DGAT1 wild-type plants to an average of 29.0% ± 0.9% in the dgat1 CL7 RcDGAT2 line (Fig. 2; Supplemental Fig. S1), and the amount of HFA per seed increased by 23% (Fig. 3). Introducing the dgat1 mutation also leads to a reduction in 18:1 and increases in 18:2 and 18:3 (Fig. 2). These changes are likely the result of the known increases in FAD2, FAD3, and PDAT1 expression that occur in dgat1-mutant seed (Xu et al., 2012). Mutations at the dgat1 locus cause a 45% reduction in seed FA content (Katavic et al., 1995; Routaboul et al., 1999; Zou et al., 1999). However, introducing a dgat1 mutation in the CL7 RcDGAT2 background did not lead to any additional reduction in total FA/seed or percentage of oil relative to CL7 RcDGAT2 controls (Fig. 3, A and D), indicating that the castor DGAT2 can maintain rates of oil synthesis in a dgat1 PDAT1 genetic background. Nevertheless, relative to the fae1 parental line, total FA/seed, seed weight, and percentage of oil are reduced by 14%, 10%, and 8%, respectively (Fig. 3). Results from previous studies have indicated that fae1 is comparable with the wild type in all of these parameters (Kunst et al., 1992; van Erp et al., 2011).
Figure 2.
Seed FA composition of CL7 RcDGAT2 and dgat1 CL7 RcDGAT2 lines. The data represent the average of nine individual plants ± se. Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
Figure 3.
Results of extended analysis of mature seeds from the transgenic lines. A, Total FAs per seed. B, HFA per seed. C, Seed weight. D, Percentage of oil content. The data represent the average of five to 27 individual plants ± se. Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
Expression of RcPDAT1A in the dgat1 CL7 RcDGAT2 Background Further Increases HFA Levels
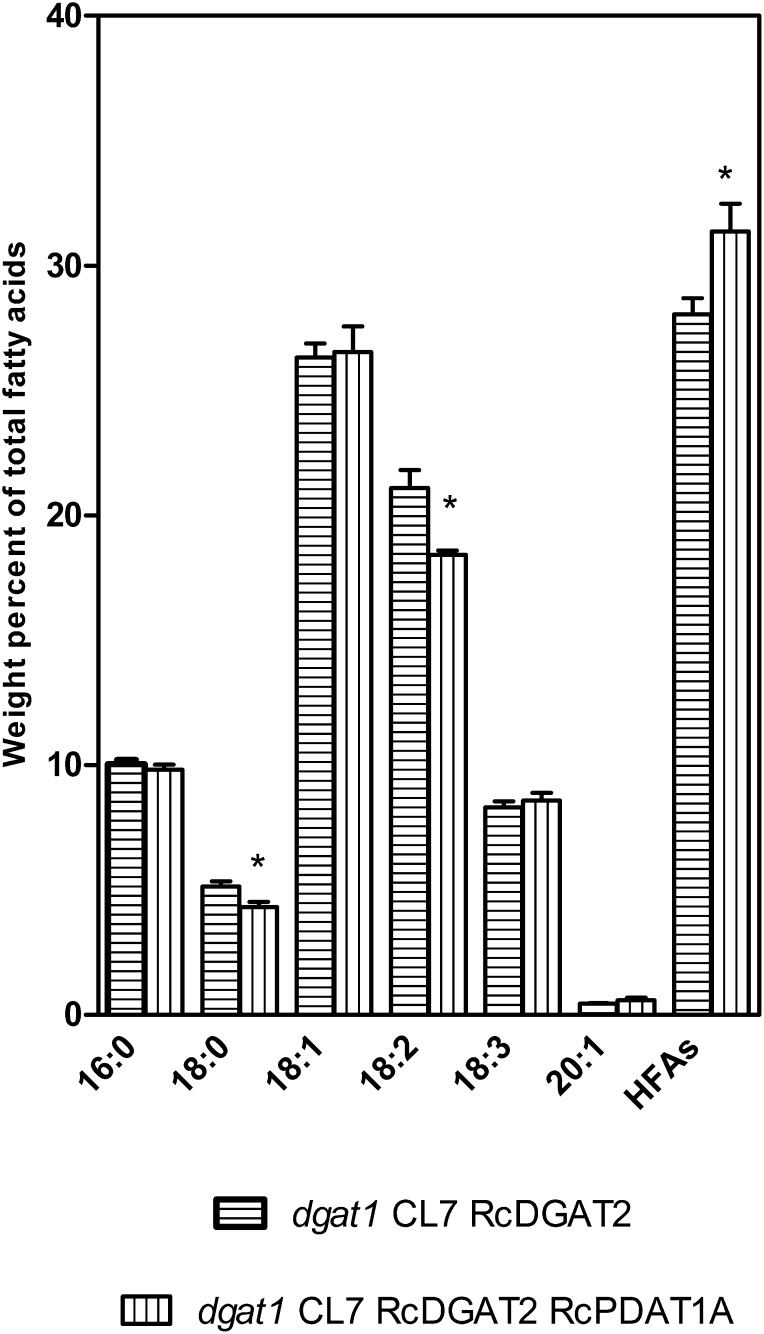
Expression of RcPDAT1A in the dgat1 CL7 RcDGAT2 background could shift the balance in substrate competition between castor and Arabidopsis enzymes further in favor of incorporation of HFA into TAG. To test this hypothesis, RcPDAT1A was transformed in the dgat1 CL7 RcDGAT2 background using Discosoma spp. red fluorescent protein (DsRed) as a selection marker (Stuitje et al., 2003). Thirty-nine lines with independent transgene insertion sites were generated. From these, four lines with high levels of HFA segregating 1:3 for brown:red seeds, indicating a single functional insertion allele, were selected. To determine if the increase in HFA levels is caused by the presence of RcPDAT1A, a T2 population of 40 plants segregating for RcPDAT1A was planted. The four tested lines gave similar results, and Figure 4 shows data for one of these lines. HFA levels in T3 seeds increased from an average of 28.0% ± 0.72% in the dgat1 CL7 RcDGAT2 plants to an average of 31.4% ± 1.12% in the dgat1 CL7 RcDGAT2 RcPDAT1A plants. There were no significant changes in the amount of total FA per seed (Fig. 3A), but there was a 23% increase in the amount of HFA per seed (Fig. 3B) in the dgat1 CL7 RcDGAT2 RcPDAT1A plants compared with the dgat1 CL7 RcDGAT2 segregants. These increases are somewhat larger than observed previously (van Erp et al., 2011) when RcPDAT1A was expressed in the CL7 RcDGAT2 background (26.7% HFA). These improvements likely arise from the introduction of the dgat1 mutation, which shifts the balance in substrate competition further in favor of incorporation of HFA.
Figure 4.
Seed FA composition of dgat1 CL7 RcDGAT2 and dgat1 CL7 RcDGAT2 RcPDAT1A lines. The data represent the average of five to 11 individual plants ± se. Two-tailed Student’s t test. *, P < 0.05.
Reduced Expression of AtPDAT1 in the dgat1 CL7 RcDGAT2 RcPDAT1A Background Does Not Further Increase HFA Levels
In the dgat1 CL7 RcDGAT2 RcPDAT1A line, substrate competition between AtPDAT1 and RcPDAT1A/RcDGAT2 can still occur. The dgat1-1 pdat1 double mutant of Arabidopsis is not viable (Zhang et al., 2009); therefore, to determine if reducing AtPDAT1 activity in the dgat1 CL7 RcDGAT2 RcPDAT1A line might further increase HFA accumulation, we used an amiRNA approach. A 21-mer sequence targeting the 3′-untranslated region (UTR) of the AtPDAT1 mRNA was designed and used to replace the stem loops in the micro-RNA 319a (MIR319a) precursor (Palatnik et al., 2003; Ossowski et al., 2008). The pdat1-amiRNA was cloned in a multigene vector behind the glycinin seed-specific promoter, and RcPDAT1A was cloned behind the oleosin promoter in the same vector, which expresses the DsRed selectable marker. The resulting pdat1-amiRNA/RcPDAT1A construct was transformed into the dgat1 CL7 RcDGAT2 line. Most of the transgenic DsRed T1 seeds were wrinkled and had a low FA content. This resembles the observations made when AtPDAT1 was suppressed in the dgat1 background (Zhang et al., 2009). Germination of the T1 seeds was reduced to less than 50%, but 45 T1 plants were grown successfully. T2 seeds from these plants were analyzed, and lines with high levels of HFA showing Mendelian segregation patterns, indicating a single functional insertion allele, were selected for additional analysis. Two segregating populations of lines with the highest HFA levels were planted (50 plants of each line), and the HFA levels in T3 seeds were determined. Figure 5 shows the data for one line. The HFA level in the dgat1 CL7 RcDGAT2 pdat1-amiRNA RcPDAT1A segregants (32.2% ± 0.53% HFA) was not statistically different from that of the dgat1 CL7 RcDGAT2 RcPDAT1A line (31.4% ± 1.12% HFA). There were also no significant changes in the micrograms of FA and HFA per seed (Fig. 3, A and B). A possible explanation for this result is that T1 seeds with the strongest suppression of AtPDAT1 expression were inviable and did not germinate. Consistent with this possibility, quantitative reverse transcription (RT)-PCR results indicated that AtPDAT1 transcript levels in dgat1 CL7 RcDGAT2 pdat1-amiRNA RcPDAT1A plants were more than 40% of those measured in the dgat1 CL7 RcDGAT2 RcPDAT1A controls. These experiments indicate that RcPDAT1A and RcDGAT2 cannot fully replace the function of AtPDAT1 and AtDGAT1 during seed development. Numerical data for the seed FA compositions of four different transgene combinations investigated are included in Supplemental Table S1.
Figure 5.
Seed FA composition of dgat1 CL7 RcDGAT2 and dgat1 CL7 RcDGAT2 pdat1-amiRNA RcPDAT1A lines. The data represent the average of 11 individual plants ± se. Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
Analysis of TAG Composition and Regiochemistry
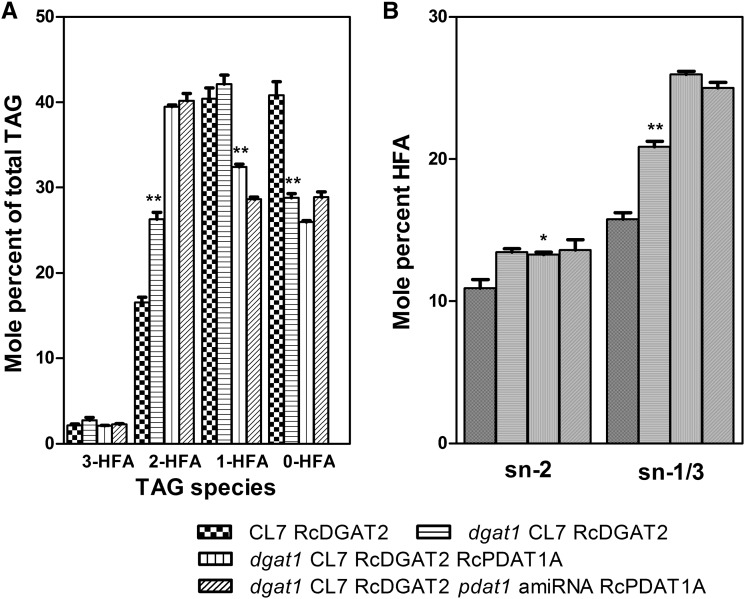
To determine the biochemical basis for the changes in HFA levels observed in our transgenic lines, the composition of seed TAG was investigated. TAG was extracted from the seeds of the transgenic plant lines and separated by thin-layer chromatography into molecular species with zero, one, two, or three HFAs. Quantitative FA analysis by gas chromatography was used to determine the relative amounts of four molecular species. Introduction of the dgat1 null mutation in the CL7 RcDGAT2 background increased the level of 2-HFA-TAG from 16.6% ± 0.60% to 26.3% ± 0.85% of total seed TAGs (a 59% increase), caused no change in 1-HFA-TAG, and decreased the amount of 0-HFA-TAG from 40.8% ± 1.62% to 28.8% ± 0.51% (a 29% decrease; Fig. 6A). This change in TAG species composition is most likely caused by a difference in substrate preference between AtDGAT1 and RcDGAT2. Burgal et al., 2008 showed that RcDGAT2 preferentially uses HFA-DAG as a substrate compared with 18:1- and 18:2-DAG. The decrease in 0-HFA-TAG is probably caused by acylation of 0-HFA-DAG with an HFA at the sn-3 position by RcDGAT2. The fact that this does not result in an increase in 1-HFA-TAG levels might be caused by acylation of sn-2 HFA-DAG with an HFA at the sn-3 position to produce 2-HFA-TAG (Fig. 6A).
Figure 6.
Analysis of TAG compositions of mature seeds from the transgenic lines. A, TAG molecular species; 0-HFA to 3-HFA represent molecular species with zero to three HFAs, respectively. B, Percentage of HFAs at the sn-2 and sn-1/3 positions of TAG. The data represent the average of three replicates ± se. Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
Expression of RcPDAT1A in the dgat1 CL7 RcDGAT2 background further increased 2-HFA-TAG from 26.3% ± 0.85% to 39.5% ± 0.21% (a 50% increase), decreased 1-HFA-TAG levels from 42.1% ± 1.05% to 32.4% ± 0.33% (a 30% decrease), and caused a small (10%) decrease from 28.8% ± 0.51% to 26.0% ± 0.14% in the 0-HFA-TAG levels (Fig. 6A). A possible explanation for these observations could be that RcPDAT1A might preferentially acylate HFA-DAG over normal DAG. The decrease in 1-HFA-TAG is probably caused by RcPDAT1A transferring HFA from the sn-2 position of PC to HFA-DAG. Consistent with this interpretation, regiochemical analysis of TAG by incubation with Rhizomucor miehei lipase (van Erp et al., 2011) indicated that increases in HFA seen in our lines was predominantly found at the sn-1/3 position of the TAG molecule (Fig. 6B).
Suppression of AtPDAT1 in the dgat1 CL7 RcDGAT2 RcPDAT1A background did not lead to any significant additional change in TAG species composition, except that 1-HFA-TAG slightly decreased and 0-HFA-TAG slightly increased (Fig. 6A). This is consistent with the low level of suppression of expression of AtPDAT1 in the viable pdat1-amiRNA lines described above.
Substrate Competition between AtDGAT1 and VfDGAT2 Limits the Accumulation of ESA
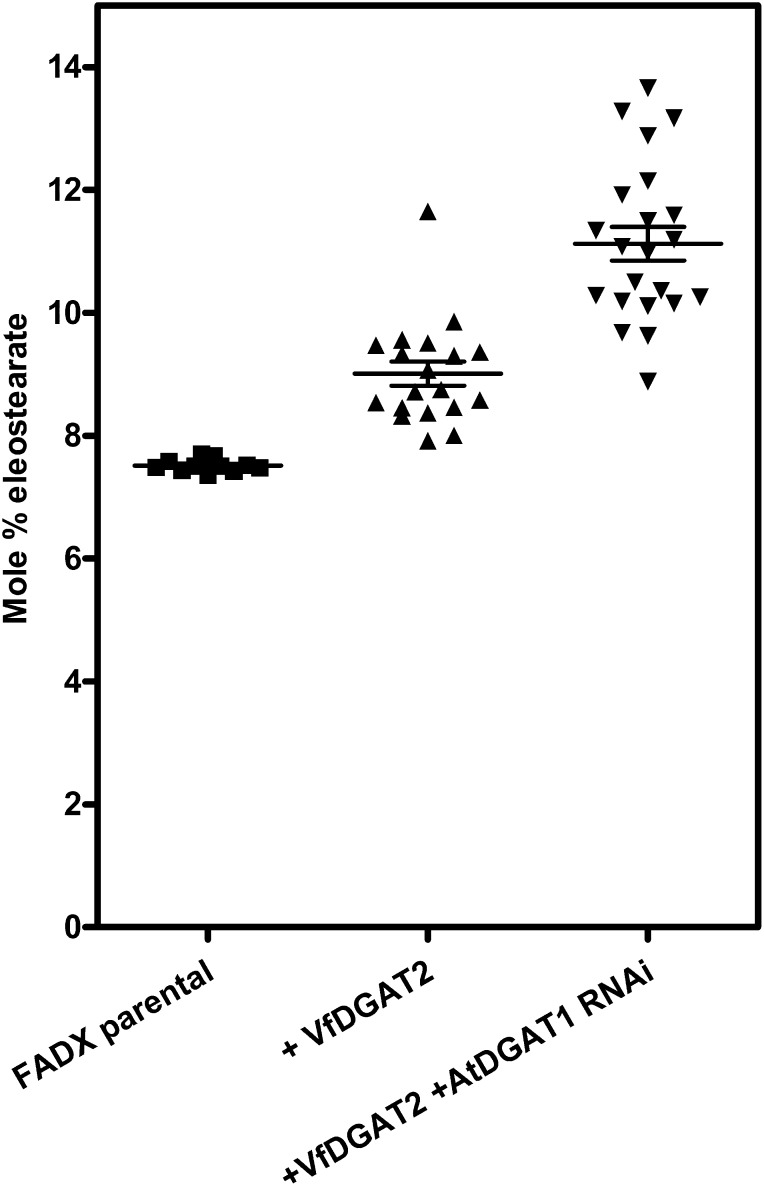
To determine if substrate competition between endogenous acyltransferases and introduced transgenic enzymes limits the accumulation of other unusual FAs, we investigated the synthesis of ESA in Arabidopsis seeds. To examine how ESA accumulation was affected by competition between endogenous and transgenic enzymes, the fad3 fae1 double mutant of Arabidopsis (Smith et al., 2003) was first transformed with a construct containing the tung tree FA conjugase FADX (Dyer et al., 2002) driven by the strong seed-specific phaseolin promoter (Slightom et al., 1983). Using this line as a starting point, we explored the ability of VfDGAT2 to increase ESA content in Arabidopsis seeds when coexpressed with tung tree FADX. A DGAT2 was chosen for this role based on previous studies of VfDGAT2 in yeast (Shockey et al., 2006), which showed that this enzyme has a strong preference for ESA-containing substrates, and similar findings regarding the substrate selectivities of other related DGAT2 enzymes (Burgal et al., 2008; Li et al., 2010). T5 FADX plants grown from homozygous transgenic T4 plants producing 7.5% ± 0.1% ESA were retransformed with VfDGAT2 transgene under control of the Arabidopsis 2S seed storage protein3 (At2S-3) promoter (Guerche et al., 1990) either alone or combined with a seed-specific AtDGAT1 RNAi cassette driven by the soybean (Glycine max) β-conglycinin α′-subunit promoter (Doyle et al., 1986). Multiple transgenic T1 seeds were selected by observation of fluorescence from the DsRed marker included in the binary constructs. Segregating seed samples were harvested from mature T1 plants, and their lipids were analyzed by gas chromatography. Nineteen independent transgenic events yielded an average of 9.01% ESA at this stage, representing a 20% increase relative to the parental FADX plants. Twenty-two transgenic lines coexpressing the combination of VfDGAT2 and AtDGAT1 RNAi averaged 11.1% ± 1.29% ESA (a 48% increase relative to the parental lines; Fig. 7).
Figure 7.
Distribution of ESA levels in transgenic plants expressing tung tree FADX alone or with tung tree DGAT2 or tung tree DGAT2 + AtDGAT1 RNAi. A parental FADX line was retransformed with VfDGAT2 or VfDGAT2 + AtDGAT1 RNAi. Seed pools from segregating T1 plants representing independent transgenic events were analyzed by gas chromatography; the corresponding ESA levels are shown. The data are plotted as a scatterplot, with the mean and se indicated. Differences between the three data sets are statistically significant (two-tailed Student’s t test; P < 0.001).
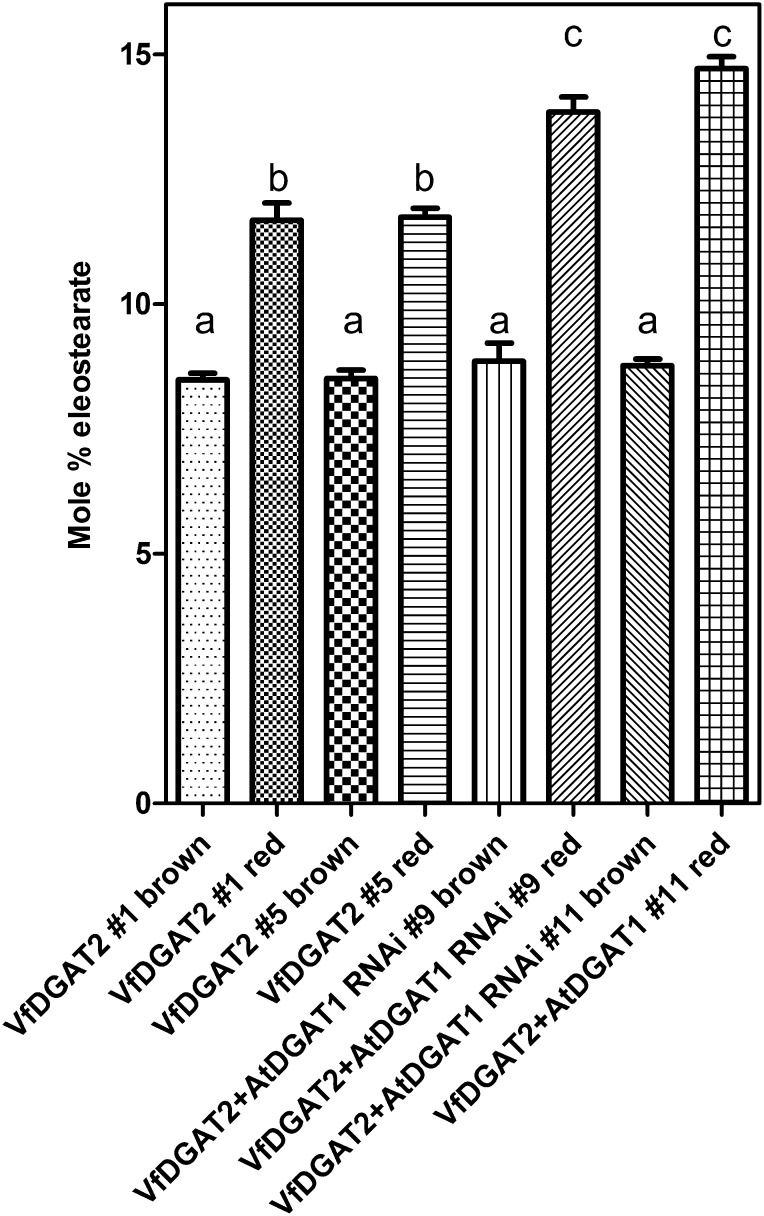
To ascertain the full degree of change affected by expression of VfDGAT2 overexpression and AtDGAT1 silencing, two T2 lines for each of the double transformants were chosen for analysis in the T3 generation. Seeds were harvested from between 18 and 32 progeny from each of the chosen T2 lines. Several homozygous samples (consisting of 100% red seeds) and FADX parental revertants (displaying uniform brown seeds devoid of fluorescence) from each line were analyzed. All four sets of brown FADX parental revertant seeds averaged between 8.49% and 8.85% ESA. Individual FADX plants homozygous for VfDGAT2 averaged 11.6% ± 0.71% and 11.7% ± 0.62% ESA, respectively (Fig. 8), including individual plants with seeds that contained as much as 12.47% and 12.64% ESA, respectively. As in the T2 sample distributions, FADX plants coexpressing both VfDGAT2 and AtDGAT1 RNAi produced higher levels of the unique FA in terms of both plants with the highest seed ESA (14.9% and 16.1%, respectively) and average ESA across all homozygous plants (13.8% ± 0.69% and 14.7% ± 0.85%, respectively; Fig. 8). Complete FA analyses of seeds of these eight lines are included in Supplemental Table S2.
Figure 8.
Determination of ESA levels in homozygous transgenic plants coexpressing FADX and VfDGAT2 with or without AtDGAT1I RNAi. Seeds from two high-performing lines from each of the transgenic genotypes were sown on soil and grown to maturity. Seed samples from homozygous transgenic (red) and nontransgenic (brown) plants were collected separately and analyzed by gas chromatography. Mean ESA levels ± se are shown. Lowercase letters indicate different statistically significant groups (Student’s t test; P < 0.01).
To provide additional data on the seed oil composition of three different transgene combinations, we analyzed additional seed batches of line #5 red (homozygous VfFADX VfDGAT2) and line #9 red (homozygous VfFADX VfDGAT2 AtDGAT1 RNAi) along with brown segregants (homozygous VfFADX) as controls. The data obtained confirmed the increases in percentage of ESA in the oil conferred by expression of VfDGAT2 and VfDGAT2 + AtDGAT1 RNAi (Supplemental Fig. S2A). These increases were associated with increases in ESA per milligram of seed weight (Supplemental Fig. S2B), whereas there was no significant change in the total FA per milligram of seed (Supplemental Fig. S2C).
DISCUSSION
AtDGAT1 and AtPDAT1 are the two enzymes that are responsible for the acylation of the sn-3 position of DAG in seeds and some other Arabidopsis tissues. When expression of AtPDAT1 was suppressed in the atdgat1 background, most of the pollen was not viable, likely because of a severe reduction in pollen storage lipid, and the reduced number of seeds that did develop had very low oil content (Zhang et al., 2009). These results indicate that, in Arabidopsis, DGAT1 and PDAT1 have an essential, redundant role and that other enzymes, such as AtDGAT2 and AtDGAT3, do not contribute significantly to acylation of the sn-3 position of DAG in Arabidopsis seed lipids. AtDGAT1 has the quantitatively more significant role, which was indicated by the reduction in oil content in the atdgat1 background (Katavic et al., 1995; Routaboul et al., 1999; Zou et al., 1999). By contrast, characterization of an atpdat1 null mutant showed no reduction in seed oil content (Mhaske et al., 2005).
The goal of our investigation was to determine if substrate competition between transgenic castor or tung tree enzymes and their endogenous Arabidopsis counterparts is a limiting factor for the accumulation of HFA or ESA in our transgenic plant lines. This possibility arises, because the endogenous Arabidopsis enzymes are known or proposed to favor substrates containing FA normally found in Arabidopsis seed TAG, whereas the castor and tung tree enzymes have been shown to have specificity for HFA- or ESA-containing substrates, respectively (Shockey et al., 2006; Burgal et al., 2008). We chose to focus on potential competition between AtDGAT1 and either RcDGAT2 or VfDGAT2, because these enzymes are known to play major roles in TAG synthesis in the respective plant species (Ståhl et al., 2004; Kroon et al., 2006; Shockey et al., 2006; Burgal et al., 2008; Zhang et al., 2009; van Erp et al., 2011). To test if substrate competition between AtDGAT1 and RcDGAT2 is limiting for accumulation of HFA, the atdgat1-2 mutation was crossed into the CL7 RcDGAT2 line. Homozygous dgat1 CL7 RcDGAT2 plants had significantly higher HFA levels than the CL7 RcDGAT2 segregants (Fig. 2). Subsequent analyses of the derived dgat1 and wild-type DGAT1 lines indicated that there was no reduction in seed oil content associated with the loss of the DGAT1 enzyme (Fig. 3). These results indicate that eliminating competition from the endogenous DGAT enzyme results in a 20% increase in HFA accumulation in the seed oil compared with the parental CL7 RcDGAT2 line.
To determine if substrate competition is a more general problem for the engineering of unusual FAs, seed-specific overexpression of tung tree DGAT2 with or without accompanying silencing of AtDGAT1 expression by RNAi was also performed in an established homozygous transgenic line producing ESA. Analysis of T2 and T3 lines showed that VfDGAT2 expression resulted in expected increases in ESA levels compared with the parental lines. However, an additional 22% increase in ESA was observed when AtDGAT1 expression was reduced by RNAi. As with the results for castor DGAT2, the tung tree DGAT2 data clearly indicate that AtDGAT1 supports incorporation of normal FA into TAG and that AtDGAT1 and transgenic VfDGAT2 compete to mobilize different DAG and acyl-CoA substrates into TAG.
Arabidopsis does not use the classic Kennedy pathway for the synthesis of TAG but instead, uses PC as an intermediate in TAG synthesis (Fig. 1). It is PC that is the substrate for FA desaturation and modification enzymes (including RcFAH12 and VfFADX). Thus, DAG and acyl-CoA derived from PC will contain HFA or ESA, and these, in turn, become available for TAG synthesis through DGAT and PDAT activities (Bates and Browse, 2011). In transgenic Arabidopsis seeds expressing RcFAH12, this pathway creates a metabolic bottleneck for the incorporation of HFA into HFA-TAG. In these plants, 1-HFA-DAG is synthesized but not efficiently converted into PC. Radiolabeling experiments indicate that 1-HFA-TAG is converted into 2-HFA-TAG and degraded (Bates and Browse, 2011), and the metabolic bottleneck also results in feedback inhibition of FA synthesis and reduced seed oil content (Bates et al., 2014). Both RcDGAT2 and RcPDAT1A expressions can increase incorporation of HFA into TAG and partially alleviate the reduction of seed oil in RcFAH transgenic lines (van Erp et al., 2011; Bates et al., 2014). Consistent with these previous findings, our dgat1 CL7 RcDGAT2 RcPDAT1A transgenics showed a significant additional increase in seed HFA content (Figs. 3B and 4). These plants also had a very substantial increase in the proportion of 2-HFA-TAG species relative to the dgat1 CL7 RcDGAT2 parental line (Fig. 6). However, the elimination of AtDGAT1 and the coexpression of RcDGAT2 and RcPDAT1A did not significantly change the proportion of 3-HFA-TAG.
Our attempts to reduce competition from the endogenous AtPDAT1 isozyme were evidently unsuccessful. The pdat1-amiRNA/RcPDAT1A lines that we were able to recover had seed characteristics that are indistinguishable from the RcPDAT1A lines lacking the pdat1-amiRNA construct (Figs. 4 and 5) and contained levels of AtPDAT1 mRNA that were at least 40% of the levels in parental control lines. Many of the dgat1 CL7 RcDGAT2 pdat1-amiRNA RcPDAT1A T1 seeds were shrunken and did not germinate on either soil or agar medium supplemented with 1% (w/v) Suc. It is possible that these inviable seeds included transgenics with strong suppression of AtPDAT1 expression.
Our results indicate that substrate competition between endogenous and transgenic acyltransferases may be a general problem in the engineering of unusual FAs in heterologous plant systems. The greater than 20% increases in HFA and ESA that we observed indicate that reducing enzyme competition can provide unique avenues for metabolic engineering. Hopefully this strategy will bring us a step closer to the engineering of crop plants with high levels of unusual FAs for industrial or health purposes.
Although our results characterize examples of isozyme competition that lead to reduced HFA or ESA accumulation, it is also possible for endogenous and transgenic isozymes to act synergistically. The Reduced Oleate Desaturation1 (ROD1) gene encodes PDCT that interconverts PC and DAG (Lu et al., 2009). PDCT is required for efficient HFA incorporation into TAG in transgenic Arabidopsis expressing the castor hydroxylase, because the proportion of HFA in seed oil is reduced by >50% in rod1 mutants compared with ROD1 RcFAH12 controls (Hu et al., 2012). In additional experiments, Hu et al. (2012) showed that RcROD1 expression could compensate for the loss of the Arabidopsis enzyme when transformed into rod1 mutants expressing RcFAH12 or could lead to increased HFA when expressed in an RcFAH12 transgenic line that is the wild type at the ROD1 locus. Importantly, expression of RcROD1 also provided an increase in HFA (from 24.7% to 28.5% of seed FA) when expressed in the CL7 RcDGAT2 background used in our experiments (Hu et al., 2012). These results point to the potential for obtaining further increases in HFA accumulation in seeds through the production of higher order multitransgenic lines.
MATERIALS AND METHODS
Plant Growth Conditions and Transformation
The Arabidopsis (Arabidopsis thaliana) lines and growth conditions were similar to those described in van Erp et al. (2011) unless otherwise mentioned. Plant transformation was achieved using the floral dip method (Clough and Bent, 1998). All lines are in the Columbia-0 wild-type background, except that dgat1-2 is in the Wassilewskija background.
For experiments comparing genotypes, plants of the different lines were randomly distributed across the pots and trays that were used. Plants were grown in environmentally controlled chambers under continuous fluorescent illumination of 120 to 150 µmol quanta m−2 s−1 with 70% relative humidity at 22°C.
Genotyping of Plants and RT-PCR Analysis
For genotyping of AtDGAT1, plant material was ground (30 Hz for 30 s; Tissuelyser II; QIAGEN) followed by extraction of genomic DNA using a QIAcube (QIAGEN). A portion of AtDGAT1 was amplified with gene-specific primers (Supplemental Table S3) flanking both sides of the point mutation, and the amplified fragment was gel purified with a QIAcube. Sequencing was performed (Eurofins MWG Operon) to determine which plants were the wild type, heterozygous, or homozygous for the point mutation in AtDGAT1. To prepare RNA, young siliques were harvested approximately 8 to 12 d after flowering from plants, flash frozen in liquid nitrogen, and stored at −80°C. Developing seeds were scraped from each silique on a petri dish on dry ice and collected in 1.5-mL Eppendorf tubes kept in liquid nitrogen. For RT-PCR analysis, total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Samples were treated with RNase-Free DNase (Qiagen) using the on-column DNase digestion method according to the manufacturer’s protocol. cDNA was synthesized using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen/Life Technologies). To confirm that the transgenic plants were expressing their respective transgenes, RT-PCR was performed using primers listed in Supplemental Table S3. This confirmed that the transgenic lines expressed the genes of interest.
Design and Cloning of amiRNA against AtPDAT1
To suppress expression of AtPDAT1 in our transgenic plant lines, a 21-mer sequence targeting AtPDAT1 was designed (WMD3 Web MicroRNA Designer, Arabidopsis cDNA [The Arabidopsis Information Resource 9]; minimum no. of included targets, one; accepted off targets, zero; S. Ossowski, J. Fitz, R. Schwab, M. Riester, and D. Weigel, personal communication). To prevent suppression of expression of other Arabidopsis genes in our plant lines, a Target Search was performed against Arabidopsis cDNAs with the 21-mer sequences (The Arabidopsis Information Resource 9_cdna_20090619; no. of mismatches, five), and no targets were found. To prevent suppression of the castor (Ricinus communis) genes in our transgenic plant lines (RcFAH12, RcDGAT2, and RcPDAT1A), a Target Search was performed against castor mRNAs with the 21-mer sequences (castor PUT v163a [Plant Genome Database]; no. of mismatches, five), and no targets were found. The best 21 mer with no other targets in Arabidopsis and castor was selected for additional analysis (5′-TTGCGGGTTATACGTAGTGTA-3′; hybridization energy = −40.49 kcal mol−1). This 21 mer targets the AtPDAT1 mRNA in the 3′-UTR.
Cloning of the pdat1-amiRNA and RcPDAT1A in a Multigene Vector
The 21-mer sequence was used to replace the stem loops in the MIR319a precursor (Ossowski et al., 2008). Primer sequences used for cloning are described in Supplemental Table S1. The pdat1-amiRNA construct was cloned in the RS3GSeed DsRed vector in between the glycinin promoter and the glycinin 3′-UTR. The pdat1-amiRNA construct was digested with EcoRI and XbaI. The RS3GSeed DsRed vector was digested with EcoRI and XbaI and dephosphorylated with calf intestinal phosphatase (New England Biolabs). The EcoRI/XbaI pdat1-amiRNA fragment was ligated into the linearized vector using the Quick Ligation Kit (NEB). RcPDAT1A was cloned in the pdat1-amiRNA vector behind the oleosin promoter and followed by the oleosin 3′-UTR. The RcPDAT1A cDNA was PCR amplified with primers containing NotI sites (Supplemental Table S1) and digested with NotI HF (New England Biolabs). The PCR product was cloned in the NotI side of the calf intestinal phosphatase-treated pKMS2 vector behind the oleosin promoter. The oleosin promoter RcPDAT1A oleosin 3′-UTR construct was cut out of the pKMS2 vector using AscI. This construct was ligated into the AscI site of the pdat1-amiRNA vector to generate the pdat1-amiRNA RcPDAT1A vector.
Construction of Tung Tree Gene Vectors and Expression in Plants
The open reading frames (ORFs) for tung tree (Vernicia fordii) FADX and tung tree DGAT2 as well as the AtDGAT1 RNAi construct were assembled in various components of a flexible set of cloning vectors and plant binary plasmids before transformation into Agrobacterium tumefaciens. The cDNA for tung tree FADX (Dyer et al., 2002) was PCR amplified using a forward primer containing an NotI site and a reverse primer containing an SacII site adjacent to the stop codon. This product was digested with NotI and SacII and ligated into plasmid pK8 that had been similarly treated. pK8 contains the strong seed-specific promoter from the Phaseolus vulgaris gene (Slightom et al., 1983) and the cauliflower mosaic virus 35S transcriptional terminator; both are flanked on their respective distal ends by AscI sites. The resulting plasmid was named pB190. The AscI cassette from pB190 was transferred to the AscI site of the plant binary plasmid pB9, which carries a kanamycin resistance gene for bacterial selection and a gene for basta (glufosinate ammonium) herbicide resistance for selection in plants. The resulting VfFADX binary plasmid, designated pE181, was transformed into Agrobacterium spp. strain GV3101; kanamycin- and gentamycin-resistant colonies were cultured in liquid media and used to transform the fad3 fae1 double-mutant line of Arabidopsis (Smith et al., 2003) by floral dip.
A seed-specific shuttle plasmid containing an N-terminal myelocytomatosis viral oncogene homolog (myc) epitope-tagged tung tree DGAT2 was generated by PCR amplification of the native DGAT2 ORF with a forward primer in which the initiator Met codon has been replaced by a KasI site and a reverse primer containing an SacII site adjacent to the stop codon. After KasI/SacII digestion, this product was ligated into similarly digested pB50, a shuttle plasmid containing the Arabidopsis 2S-3 promoter (Guerche et al., 1990), the soybean (Glycine max) glycinin G1 subunit transcriptional terminator (Sims and Goldberg, 1989), and a multiple cloning site containing sites that allow for production of N-terminal myc epitope fusions. The AscI fragment (representing the promoter-gene-terminator cassette) from this plasmid, pB240, was transferred into the corresponding site of the plant binary vector pB110 to form plasmid E278. In turn, pE278 was modified by MluI digestion and ligation of the AscI fragment of pJ6, which contains the soybean β-conglycinin promoter (Sato et al., 2004) driving the expression of an RNAi hairpin for AtDGAT1. The RNAi portion of this plasmid contains an intron from the 5′-UTR of AtFAD2 flanked by a 592-bp region of the AtDGAT1 ORF (base pairs 303–894) cloned in inverted orientations. The binary plasmid carrying both the tung tree DGAT2 overexpression cassette and the AtDGAT1 RNAi cassette is called pE290. The sequences of all primers used to generate these plasmid constructs are included in Supplemental Table S1.
ESA production stabilized at approximately 8% in the T4 generation of E181 plants. T5 plants from this line were retransformed with Agrobacterium spp. bearing either pE278 or pE290. Red T1 seeds were sown on soil and grown to maturity followed by seed lipid extraction and analysis for determination of ESA content. Two lines, one representing the highest T2 18:3Δ9cis, 11trans, and 13trans producer and one representing a level near the average of the T2 population, were selected for each double transgenic and carried forward to the T3 generation. Seed samples from T3 plants producing either uniformly red or uniformly brown seeds were analyzed by gas chromatography.
FA Analysis by Gas Chromatography
Analysis of HFA was performed as described in van Erp et al., 2011. Lipids from Arabidopsis seed containing ESA were extracted as follows. Approximately 30 mg of seeds and three to four 2.3-mm chrome steel beads (BioSpec Products, Inc.) were added into a 2-mL Eppendorf tube with 500 µL of hexane followed by 5 min of agitation on a Bead Beater. The extract was centrifuged (13,000g for 2 min) to remove debris, and a portion (300 µL) of the extract was transferred to a 13- × 100-mm glass Corning culture tube. Hexane (700 µL) and sodium methoxide in methanol (400 µL) were added followed by 10 min of incubation at room temperature with intermittent shaking. The reactions were quenched by the addition of 2 mL of hexane and 2 mL of saturated NaCl solution, and the phases were separated by centrifugation in a tabletop swinging bucket centrifuge. Two milliliters of upper layer was transferred to vials, capped, and analyzed immediately or stored at −20° until needed. Gas chromatography was conducted as described in Shockey et al. (2011). Whenever possible, all samples containing ESA were prepared using amber-colored glassware to reduce exposure of ESA to light. Regiochemical analysis of TAG was performed as described (van Erp et al., 2011).
Determination of Seed FA Content
Seed FA content was determined according to the protocol in Li et al. (2006), except that 200 µL of toluene was added to each sample, butylated hydroxytoluene was omitted, and 20 seeds were used for each measurement. Seed weights were determined by counting seed numbers in samples of 1 to 2 mg of seeds.
Lipid Extraction and Characterization of TAG Species
Lipid extraction and characterization of TAG species were performed as described in van Erp et al. (2011).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of RcDGAT2 transcript in the parental CL7 RcDGAT2 line and the dgat1 CL7 RcDGAT2 line obtained by crossing.
Supplemental Figure S2. Extended analysis of lines accumulating ESA.
Supplemental Table S1. FA compositions of samples of T3 seeds of transgenic lines accumulating HFAs characterized in this study.
Supplemental Table S2. FA compositions of samples of T3 seeds of transgenic lines accumulating ESA characterized in this study.
Supplemental Table S3. Primers used.
Supplementary Material
Acknowledgments
We thank all of the members of the laboratory of J.B. who contributed to this article, the greenhouse staff for help with growing plants, Dr. Jim Wallis (Institute of Biological Chemistry, Washington State University) for discussions and help with formatting the figures, and Catherine Mason (Southern Regional Research Center, U.S. Department of Agriculture-Agricultural Research Service) for technical assistance with seed lipid extraction and gas chromatography analysis.
Glossary
- cDNA
complementary DNA
- DAG
diacylglycerol
- ESA
α-eleostearic acid
- FA
fatty acid
- HFA
hydroxy fatty acid
- ORF
open reading frame
- PC
phosphatidylcholine
- PDCT
phosphatidylcholine:diacylglycerol cholinephosphotransferase
- RT
reverse transcription
- TAG
triacylglycerol
- UTR
untranslated region
Footnotes
This work was supported by the U.S. National Science Foundation Plant Genome Research Program (grant nos. DBI–0701919 and IOS–1339385) and the Agricultural Research Center at Washington State University.
Articles can be viewed without a subscription.
References
- Badami RC, Patil KB (1980) Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res 19: 119–153 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2012) The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci 3: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Johnson SR, Cao X, Li J, Nam JW, Jaworski JG, Ohlrogge JB, Browse J (2014) Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc Natl Acad Sci USA 111: 1204–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Stymne S, Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol 16: 358–364 [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67: 1166–1176 [DOI] [PubMed] [Google Scholar]
- Carlsson AS, Yilmaz JL, Green AG, Stymne S, Hofvander P (2011) Replacing fossil oil with fresh oil - with what and for what? Eur J Lipid Sci Technol 113: 812–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dauk M, Lam P, Kunst L, Smith MA (2007) A FAD2 homologue from Lesquerella lindheimeri has predominantly fatty acid hydroxylase activity. Plant Sci 173: 43–49 [Google Scholar]
- Doyle JJ, Schuler MA, Godette WD, Zenger V, Beachy RN, Slightom JL (1986) The glycosylated seed storage proteins of Glycine max and Phaseolus vulgaris. Structural homologies of genes and proteins. J Biol Chem 261: 9228–9238 [PubMed] [Google Scholar]
- Dyer JM, Chapital DC, Kuan JC, Mullen RT, Turner C, McKeon TA, Pepperman AB (2002) Molecular analysis of a bifunctional fatty acid conjugase/desaturase from tung: implications for the evolution of plant fatty acid diversity. Plant Physiol 130: 2027–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS (2008) High-value oils from plants. Plant J 54: 640–655 [DOI] [PubMed] [Google Scholar]
- Guerche P, Tire C, De Sa FG, De Clercq A, Van Montagu M, Krebbers E (1990) Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2: 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ren Z, Lu C (2012) The phosphatidylcholine diacylglycerol cholinephosphotransferase is required for efficient hydroxy fatty acid accumulation in transgenic Arabidopsis. Plant Physiol 158: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L (1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Lee KR, Go YS, Jung JH, Suh MC, Kim JB (2011) Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol 52: 983–993 [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Hayes TR, Wyrick A, Xiong H, Maelor Davies H, Voelker TA (1999) Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiol 120: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon JT, Wei W, Simon WJ, Slabas AR (2006) Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67: 2541–2549 [DOI] [PubMed] [Google Scholar]
- Kunst L, Taylor DC, Underhill EW (1992) Fatty-acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem 30: 425–434 [Google Scholar]
- Li R, Yu K, Hatanaka T, Hildebrand DF (2010) Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J 8: 184–195 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG, Browse J (2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45: 847–856 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaske V, Beldjilali K, Ohlrogge J, Pollard M (2005) Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol Biochem 43: 413–417 [DOI] [PubMed] [Google Scholar]
- Napier JA. (2007) The production of unusual fatty acids in transgenic plants. Annu Rev Plant Biol 58: 295–319 [DOI] [PubMed] [Google Scholar]
- Napier JA, Graham IA (2010) Tailoring plant lipid composition: designer oilseeds come of age. Curr Opin Plant Biol 13: 330–337 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem 37: 831–840 [DOI] [PubMed] [Google Scholar]
- Sato S, Xing AQ, Ye XG, Schweiger B, Kinney A, Graef G, Clemente T (2004) Production of gamma-linolenic acid and stearidonic acid in seeds of marker-free transgenic soybean. Crop Sci 44: 646–652 [Google Scholar]
- Shockey J, Chapital D, Gidda S, Mason C, Davis G, Klasson KT, Cao H, Mullen R, Dyer J (2011) Expression of a lipid-inducible, self-regulating form of Yarrowia lipolytica lipase LIP2 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 92: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims TL, Goldberg RB (1989) The glycinin Gy1 gene from soybean. Nucleic Acids Res 17: 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom JL, Sun SM, Hall TC (1983) Complete nucleotide sequence of a French bean storage protein gene: phaseolin. Proc Natl Acad Sci USA 80: 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Moon H, Kunst L (2000) Production of hydroxy fatty acids in the seeds of Arabidopsis thaliana. Biochem Soc Trans 28: 947–950 [PubMed] [Google Scholar]
- Smith MA, Moon H, Chowrira G, Kunst L (2003) Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217: 507–516 [DOI] [PubMed] [Google Scholar]
- Sonntag NOV. (1979) Composition and characteristics of individual fats and oils. InSwern D, ed, Bailey's Industrial Oil and Fat Products. John Wiley & Sons, New York, pp 289–477 [Google Scholar]
- Ståhl U, Carlsson AS, Lenman M, Dahlqvist A, Huang B, Banas W, Banas A, Stymne S (2004) Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol 135: 1324–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap JP, Kneppers TJ (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1: 301–309 [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92: 6743–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Bates PD, Burgal J, Shockey J, Browse J (2011) Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol 155: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, Wood CC, Stymne S, Singh SP, Green AG (2013) Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnol J 11: 197–210 [DOI] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52: 335–361 [DOI] [PubMed] [Google Scholar]
- Wayne LL, Wallis JG, Kumar R, Markham JE, Browse J (2013) Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell 25: 3052–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, Taylor DC (2012) Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19: 645–653 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.