Grape berry skin shows a specific induction of the resveratrol synthetic pathway by UV-C.
Abstract
Grape (Vitis vinifera) accumulates various polyphenolic compounds, which protect against environmental stresses, including ultraviolet-C (UV-C) light and pathogens. In this study, we looked at the transcriptome and metabolome in grape berry skin after UV-C irradiation, which demonstrated the effectiveness of omics approaches to clarify important traits of grape. We performed transcriptome analysis using a genome-wide microarray, which revealed 238 genes up-regulated more than 5-fold by UV-C light. Enrichment analysis of Gene Ontology terms showed that genes encoding stilbene synthase, a key enzyme for resveratrol synthesis, were enriched in the up-regulated genes. We performed metabolome analysis using liquid chromatography-quadrupole time-of-flight mass spectrometry, and 2,012 metabolite peaks, including unidentified peaks, were detected. Principal component analysis using the peaks showed that only one metabolite peak, identified as resveratrol, was highly induced by UV-C light. We updated the metabolic pathway map of grape in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and in the KaPPA-View 4 KEGG system, then projected the transcriptome and metabolome data on a metabolic pathway map. The map showed specific induction of the resveratrol synthetic pathway by UV-C light. Our results showed that multiomics is a powerful tool to elucidate the accumulation mechanisms of secondary metabolites, and updated systems, such as KEGG and KaPPA-View 4 KEGG for grape, can support such studies.
Grape (Vitis vinifera) accumulates many polyphenolic compounds, such as anthocyanin, catechin, and resveratrol, in the berry skin. These compounds are important not only for resistance to abiotic and biotic stresses but also for berry qualities such as color, astringency, and consumer health benefits. Resveratrol is a polyphenolic compound categorized as a stilbenoid, and it, as well as its dimer, ε-viniferin, act as phytoalexins (Langcake and Pryce, 1977). Resveratrol has been identified as a key compound associated with the French paradox (Renaud and de Lorgeril, 1992). Resveratrol has also been reported to prevent heart disease and Alzheimer’s disease and to have anticancer effects (Jang et al., 1997; Marambaud et al., 2005). Recently, some clinical trials have been presented showing the evidence of favorable effects of resveratrol not only on mouse but also on human health (Vang et al., 2011).
The accumulation of polyphenolic compounds occurs during berry maturation, but it can also be induced by external stimuli, such as light, plant hormones, and pathogen infection. The amounts of resveratrol and its analogs are increased by pathogen infection in leaves and berries (Aziz et al., 2003; Pezet et al., 2003). Resveratrol accumulation in mature grape berries is also induced by UV-C irradiation (Adrian et al., 2000; Versari et al., 2001; Takayanagi et al., 2004; Nishikawa et al., 2011).
Recent EST analyses (Quackenbush et al., 2001) and whole-genome sequencing of grape (Jaillon et al., 2007; Velasco et al., 2007) have allowed a more comprehensive approach to the study of its secondary metabolites. However, most transcriptome analyses have used EST-based microarrays, and this information is limited, because not all genes are represented on these microarrays. Profiling of metabolites has also been targeted, and only a few selected compounds have been analyzed. Few analyses in grape have used genome-wide microarrays, and even fewer studies have combined different omes, such as combining transcriptome analysis and metabolome analysis (Zamboni et al., 2010).
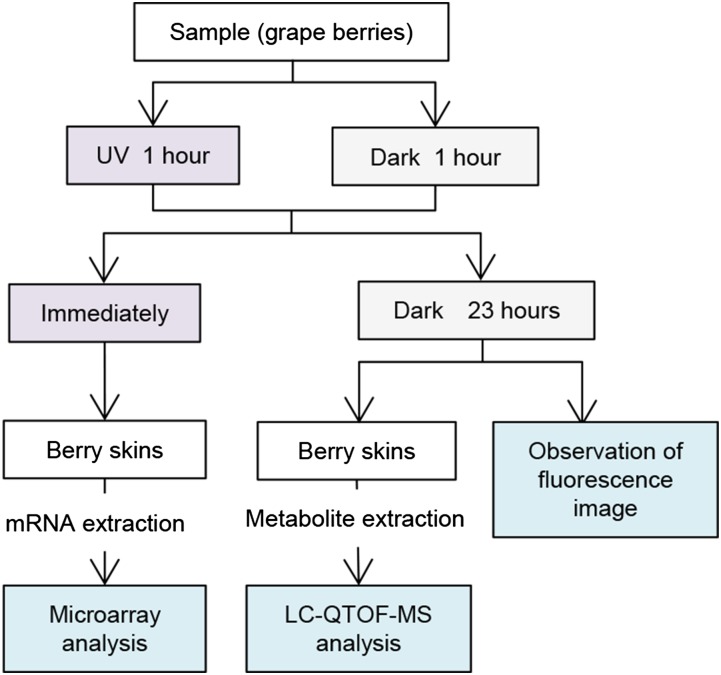
In this study, we performed multiomics analysis, combining transcriptome and metabolome analyses to study metabolic changes in grape berry skin after UV-C irradiation (Fig. 1). A genome-wide microarray, but not an EST-based microarray, was used to determine the expression of whole grape genes, and the obtained data underwent Gene Ontology (GO) enrichment analysis. Nontargeted metabolome analysis in grape berry skin was performed by liquid chromatography-quadrupole time-of-flight mass spectrometry (LC-QTOF-MS), allowing the detection of more than 2,000 metabolite peaks. Principal component analysis (PCA) revealed the induction of a single metabolite by UV-C irradiation. Data from the transcriptome and metabolome analyses were projected on a metabolic map using the KaPPA-View 4 Kyoto Encyclopedia of Genes and Genomes (KEGG) system (http://kpv.kazusa.or.jp/kpv4-kegg-1402/; Sakurai et al., 2011) after modification to apply the whole grape gene expression. The map demonstrated the induction of the stilbene synthetic pathway by UV-C irradiation.
Figure 1.
Overview of the experimental design in this study.
RESULTS
Annotation of Grape Genes
The grape genome has been sequenced by two groups, the Istituto Agrario San Michele all'Adige Research Center (Velasco et al., 2007) and the French-Italian Public Consortium for Grapevine Genome Characterization (Jaillon et al., 2007). The latter group first released a set of gene predictions of the grape genome called version 0 (V0; Jaillon et al., 2007), which is available on the GENOSCOPE Web site (http://www.genoscope.cns.fr/spip/). Later gene predictions of the grape genome, called version 1 (V1), have been made available online independently on the Centro di Ricerca Interdipartimentale per le Biotecnologie Innovative (CRIBI) Web site (http://genomes.cribi.unipd.it/grape/) by the CRIBI Biotechnology Centre, which includes members of the latter group (Grimplet et al., 2012). The NimbleGen microarray 090818 Vitis exp HX12 (Roche NimbleGen), which was used in this study, was designed based on the V1 gene predictions. To obtain more information about grape gene functions, we searched best-hit homologs of each grape gene in the Arabidopsis (Arabidopsis thaliana) gene database (The Arabidopsis Information Resource version 10; http://www.arabidopsis.org) and in the UniprotKB database (http://www.uniprot.org). Out of the 29,549 genes on the grape microarray, 28,801 could be assigned to an Arabidopsis gene, while 28,322 genes had hits in UniprotKB. Some genes did not have a hit to any Arabidopsis genes but did to genes in UniprotKB. Among the grape genes, 1,847 could be assigned to grape genes in UniprotKB. As a result, 20,302 grape genes were tagged with GO slim terms (http://www.geneontology.org), which could be assigned based on the results of a BLAST search against the National Center for Biotechnology Information (NCBI) nonredundant protein database using the Blast2GO algorithm (Conesa and Götz, 2008; see “Materials and Methods”). In addition, 13,651 grape genes were tagged with network names in VitisNet (Grimplet et al., 2009; http://www.sdstate.edu/ps/research/vitis/pathways.cfm), which are similar to GO terms but specific for grape (Supplemental Data Set S1).
Transcriptome Analysis
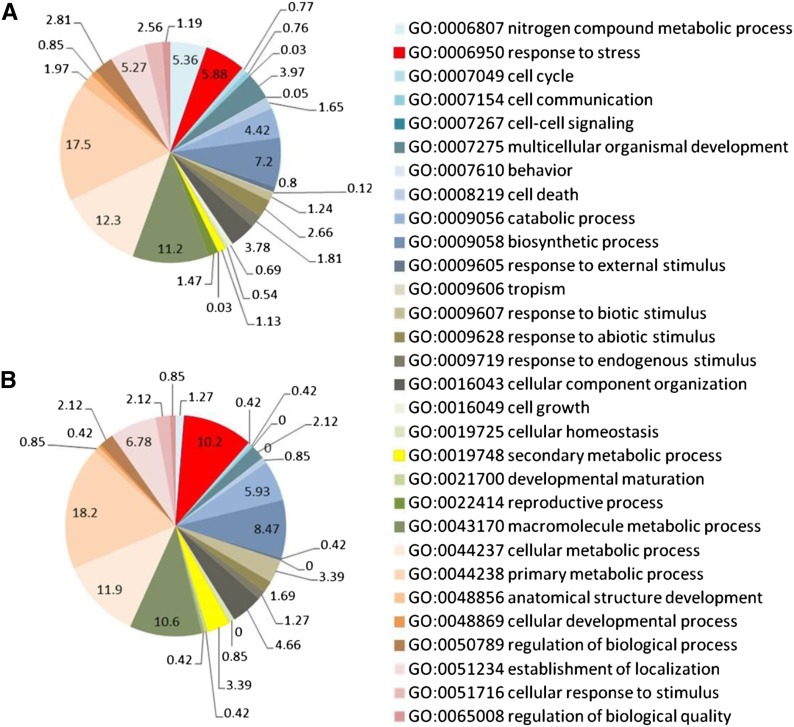
The transcriptome of grape berry skin after UV-C irradiation was analyzed using a microarray covering the whole grape genes. Following UV-C irradiation, 238 genes were more than 5-fold up-regulated, while 24 genes were more than 0.2-fold down-regulated (Supplemental Data Sets S2 and S3; Supplemental Fig. S1). Among the down-regulated genes, cytochrome P450 (VIT_01s0137g00410, VIT_07s0031g01680, and VIT_02s0025g03320), Wax2 (VIT_09s0018g01340 and VIT_09s0018g01360), and pleiotropic drug resistance 4 (VIT_11s0016g04540) were found (Supplemental Data Set S3). The 238 up-regulated genes (Supplemental Data Set S2) were investigated for GO functional categories using Blast2GO. Based on a comparison between the rates of occurrence of GO slim terms in all grape genes and those in the genes up-regulated by UV-C irradiation, response to stress (GO:0006950) increased from 5.88% to 10.2% and secondary metabolic process (GO:0019748) increased from 1.13% to 3.39% in the up-regulated genes (Fig. 2). Response to stress included genes related to resistance to pathogens, such as bacteria-induced peroxidase (Chassot et al., 2007), calmodulin-binding protein (Wang et al., 2011; Wan et al., 2012), and stilbene synthase (STS; Delaunois et al., 2009), while secondary metabolic process included genes related to secondary metabolite synthesis, such as laccase14-like (Turlapati et al., 2011) and Phe ammonia lyase (Huang et al., 2010).
Figure 2.
Distribution of grape genes based on GO slim classification. A, Functional classifications for all microarray genes (n = 29,549). B, Functional classifications for up-regulated genes (n = 238) by UV-C irradiation. For illustration purposes only, Biological Process level 3 ontology terms were selected. Values indicate the percentage of each GO category in total.
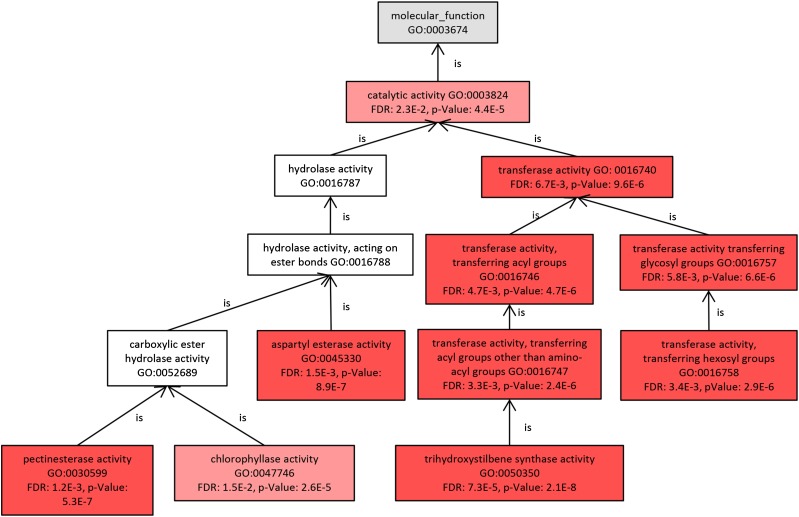
GO enrichment analysis revealed that two GO terms in Biological Process and 11 GO terms in Molecular Function were enriched in the genes up-regulated by UV-C irradiation (Table I). Cell wall modification (GO:0042545) and lipid glycosylation (GO:0030259) were the GO terms in Biological Process. As shown in Figure 3, among the GO terms found in Molecular Function were pectinesterase activity (GO:0030599), chlorophyllase activity (GO:0047746), aspartyl esterase activity (GO:0045330), trihydroxystilbene synthase activity (GO:0050350), and transferase activity, transferring hexosyl groups (GO:0016758). Also found were pectinesterase activity (GO:0030599) and aspartyl esterase activity (GO:0045330), which includes pectinesterase2-like (Louvet et al., 2006; Table II). The category transferase activity, transferring hexosyl groups (GO:0016758) includes UDP-glycosyltransferase, related to the formation of cell wall and cell plate (Eudes et al., 2008), while trihydroxystilbene synthase activity includes STS (Table II). STS is a key enzyme of resveratrol synthesis from coumaroyl-CoA and malonyl-CoA (Delaunois et al., 2009). As described above, the annotated genes of the GO slim term response to stress, including STS, were enriched remarkably after UV-C irradiation, and this category includes STS. Our transcriptome analysis showed that UV-C irradiation strongly and conspicuously induces STS gene expression.
Table I. List of enrichment GO terms.
Thirteen GO terms were picked out from the enrichment analysis graph by Blast2GO. Categories are as follows: F, Molecular Function; and P, Biological Process.
| GO Identifier | Term | Category | False Discovery Rate | P |
|---|---|---|---|---|
| GO:0052689 | Carboxylic ester hydrolase activity | F | 1.E-05 | 2.13E-09 |
| GO:0050350 | Trihydroxystilbene synthase activity | F | 7.E-05 | 2.10E-08 |
| GO:0030599 | Pectinesterase activity | F | 1.E-03 | 5.25E-07 |
| GO:0045330 | Aspartyl esterase activity | F | 2.E-03 | 8.89E-07 |
| GO:0016747 | Transferase activity, transferring acyl groups other than amino-acyl groups | F | 3.E-03 | 2.35E-06 |
| GO:0016758 | Transferase activity, transferring hexosyl groups | F | 3.E-03 | 2.90E-06 |
| GO:0016746 | Transferase activity, transferring acyl groups | F | 5.E-03 | 4.71E-06 |
| GO:0016757 | Transferase activity, transferring glycosyl groups | F | 6.E-03 | 6.62E-06 |
| GO:0042545 | Cell wall modification | P | 6.E-03 | 8.19E-06 |
| GO:0016740 | Transferase activity | F | 7.E-03 | 9.57E-06 |
| GO:0030259 | Lipid glycosylation | P | 2.E-02 | 2.45E-05 |
| GO:0047746 | Chlorophyllase activity | F | 2.E-02 | 2.62E-05 |
| GO:0003824 | Catalytic activity | F | 2.E-02 | 4.38E-05 |
Figure 3.
GO hierarchy and enrichment statistics for the Molecular Function ontology. Some GO terms within the Molecular Function ontology were significantly enriched by UV-C irradiation. Different shades of pink indicate activity levels according to GO term analysis. Significance was calculated by false discovery rate (FDR).
Table II. List of enrichment GO terms and gene annotations for the Molecular Function ontology.
Five GO terms were picked out as terms of the lowest level of the hierarchy in the enrichment graph (Fig. 3).
| GO | Gene Identifier | Fold Change (UV Light/Dark) | Annotation of Blast2GO (NCBI, BLASTX) |
|---|---|---|---|
| GO:0030599, pectinesterase activity | VIT_06s0009g02630 | 14.9 | Pectinesterase2-like |
| VIT_06s0009g02590 | 39.5 | Pectinesterase2-like | |
| VIT_03s0038g03570 | 5.6 | l-Ascorbate oxidase homolog | |
| VIT_02s0154g00600 | 8.6 | Probable pectinesterase68-like | |
| VIT_06s0009g02570 | 9.8 | Pectinesterase2-like | |
| VIT_06s0009g02600 | 18.6 | Pectinesterase2-like | |
| VIT_06s0009g02560 | 6.0 | Pectinesterase2-like | |
| VIT_07s0005g00720 | 7.5 | Probable pectinesterase pectinesterase inhibitor41-like | |
| GO:0047746, chlorophyllase activity | VIT_07s0151g00210 | 12.9 | Chlorophyllase1 |
| VIT_07s0151g00130 | 10.2 | Chlorophyllase1 | |
| VIT_07s0151g00270 | 10.2 | Chlorophyllase2 | |
| GO:0045330, aspartyl esterase activity | VIT_06s0009g02630 | 14.9 | Pectinesterase2-like |
| VIT_06s0009g02590 | 39.5 | Pectinesterase2-like | |
| VIT_02s0154g00600 | 8.6 | Probable pectinesterase68-like | |
| VIT_06s0009g02570 | 9.8 | Pectinesterase2-like | |
| VIT_06s0009g02600 | 18.6 | Pectinesterase2-like | |
| VIT_06s0009g02560 | 6.0 | Pectinesterase2-like | |
| VIT_07s0005g00720 | 7.5 | Probable pectinesterase pectinesterase inhibitor41-like | |
| GO:0050350, trihydroxystilbene synthase activity | VIT_16s0100g01000 | 6.5 | Stilbene synthase4-like |
| VIT_16s0100g01140 | 6.2 | Stilbene synthase1 | |
| VIT_16s0100g01130 | 6.0 | Resveratrol synthase | |
| VIT_10s0042g00890 | 5.4 | Stilbene synthase1 | |
| VIT_10s0042g00870 | 6.1 | Stilbene synthase | |
| VIT_16s0100g00770 | 9.9 | Stilbene synthase | |
| VIT_16s0100g01190 | 9.3 | Resveratrol synthase | |
| GO:0016758, transferase activity, transferring hexosyl groups | VIT_05s0062g00310 | 5.6 | UDP-glycosyltransferase75d1-like |
| VIT_05s0062g00270 | 8.0 | UDP-glycosyltransferase75d1-like | |
| VIT_05s0062g00700 | 8.0 | UDP-glycosyltransferase75d1-like | |
| VIT_05s0062g00660 | 5.6 | UDP-glycosyltransferase75d1-like | |
| VIT_05s0062g00300 | 7.8 | UDP-glycosyltransferase75d1-like | |
| VIT_03s0017g01990 | 7.6 | UDP-Glc flavonoid 3-O-glucosyltransferase6-like | |
| VIT_17s0000g08100 | 5.7 | UDP-glycosyltransferase-like protein | |
| VIT_06s0004g07250 | 6.0 | UDP-glycosyltransferase87a1-like | |
| VIT_06s0004g07240 | 6.1 | UDP-glycosyltransferase87a1-like | |
| VIT_12s0034g00030 | 5.2 | UDP-Glc flavonoid 3-O-glucosyltransferase6-like | |
| VIT_04s0023g01120 | 6.1 | Cazy family gt8 | |
| VIT_18s0041g00740 | 5.3 | UDP-glycosyltransferase88a1-like | |
| VIT_15s0021g02060 | 17.8 | Hydroquinone glucosyltransferase | |
| VIT_03s0017g02000 | 5.8 | UDP-Glc flavonoid 3-O-glucosyltransferase6 | |
| VIT_01s0011g03850 | 6.3 | Protein | |
| VIT_06s0004g01670 | 8.2 | UDP-glycosyltransferase92a1-like | |
| VIT_05s0062g00340 | 6.2 | UDP-glycosyltransferase75d1-like | |
| VIT_17s0000g04760 | 6.5 | UDP-glycosyltransferase89b1-like | |
| VIT_02s0025g01860 | 5.1 | Cellulose synthase-like protein g3 |
Metabolome Analysis
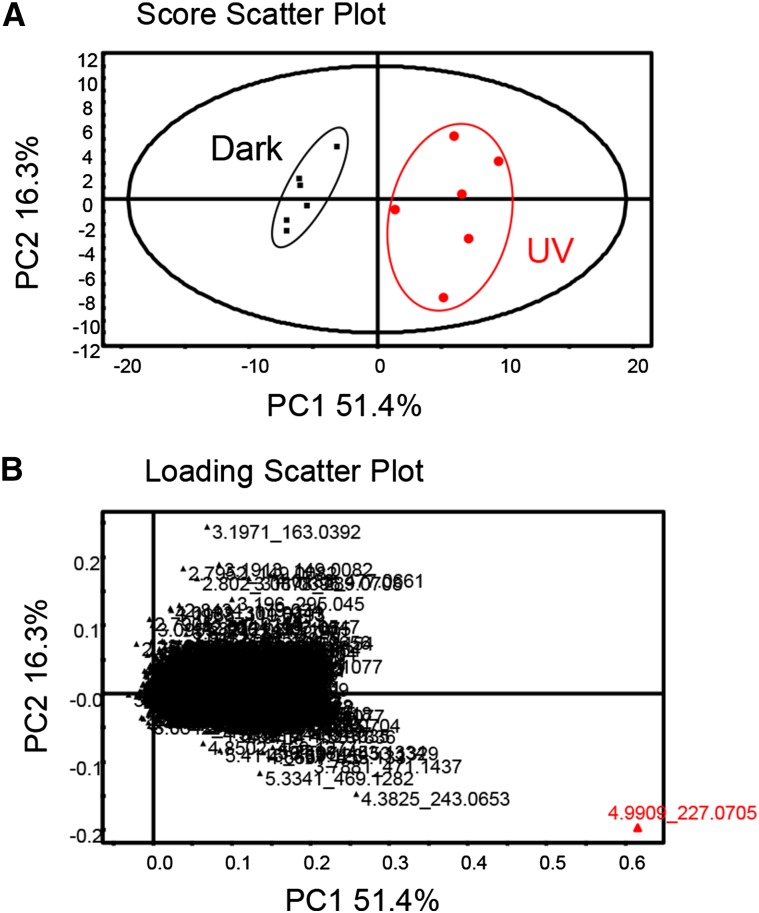
Next, we analyzed the metabolome, focusing on secondary metabolites, using LC-QTOF-MS. In positive ion mode, 1,197 metabolite peaks were detected, and in negative ion mode, 2,012 peaks were detected, including unidentified peaks. The data from negative ion mode were normalized before PCA. A score scatterplot from PCA showed two groups of data, control (dark) and UV-C irradiation (Fig. 4). Interestingly, a loading scatterplot showed only one metabolite related to UV-C irradiation (Fig. 4), and the metabolite was identified as resveratrol based on chemical reference standards. Its intensity was high, meaning that resveratrol accumulation was strongly induced by UV-C irradiation. The same results were obtained when data from the positive ion mode were used.
Figure 4.
PCA of the grape berry skin metabolome. A, Score scatterplot of a PCA model on the berry skin metabolome in darkness (squares) and after UV-C light treatment (circles; n = 6 for each treatment). B, Loading scatterplot from the PCA analysis of berry skin in darkness and after UV-C light treatment (n = 2,012 metabolite peaks). Each annotation showed the retention time and mass-to-charge ratio (m/z).
We attempted to identify and quantify major phenolic compounds and their intermediates in the phenylpropanoid pathway with reference to 24 standard chemical compounds (Supplemental Table S1). As a result, 12 metabolite peaks were identified in positive ion mode and 17 in negative ion mode. The concentrations of 10 metabolites identified in negative ion mode were high enough to calculate, while those of seven metabolites were not (Table III).
Table III. List of metabolites identified by one-point calibration in darkness and after treatment with UV light.
| Category | KEGG Identifier | Compound | Intensity | UV Light-Dark Ratio | Concentrationb | ||
|---|---|---|---|---|---|---|---|
| Dark | UV Light | Dark | UV Light | ||||
| μg g−1 fresh wt | |||||||
| C00079 | l-Phe | 0.152 | 0.103 | 0.68 | 61.8 | 41.9 | |
| Resveratrol | C03582 | Trans-resveratrol, 3,4′,5-trihydroxystilbene | 0.091 | 32.430 | 355.18 | 9.8 | 3,492.3 |
| –c | Cis-resveratrol | 0.051a | 0.088 | 1.72 | – | 6.3 | |
| C10275 | Piceid | 0.051a | 0.171 | 3.35 | – | 45.3 | |
| C10289 | (−)-ε-Viniferin, ε-viniferin | 0.389 | 2.273 | 5.84 | 79.2 | 462.3 | |
| Anthocyanin | C12138 | Delphinidin 3-glucoside (mirtillin) | 0.051a | 0.052a | 1.01 | – | – |
| C12140 | Malvidin 3-glucoside (oenin) | 0.051a | 0.052a | 1.01 | – | – | |
| C12139 | Petunidin 3-glucoside | 0.051a | 0.052a | 1.01 | – | – | |
| C08604 | Cyanidin 3-glucoside | 0.051a | 0.052a | 1.01 | – | – | |
| C12141 | Peonidin 3-O-glucoside | 0.051a | 0.052a | 1.01 | – | – | |
| Proanthocyanidin | C06562 | (+)-Catechin | 21.313 | 22.850 | 1.07 | 3,771.2 | 4,043.3 |
| C09727 | (−)-Epicatechin | 0.635 | 0.598 | 0.94 | 114.6 | 108.0 | |
| C12136 | l-Epigallocatechin | 0.061 | 0.062 | 1.01 | 17.9 | 18.0 | |
| –c | (−)-Epicatechin 3-O-gallate | 0.051a | 0.052a | 1.01 | – | – | |
| Flavonol | C00389 | Quercetin | 0.051a | 0.073 | 1.42 | – | 8.2 |
| C10107 | Myricetin | 0.051a | 0.052a | 1.01 | – | – | |
| C05623 | Quercetin-3-O-glucoside (hirsutrin) | 0.946 | 1.162 | 1.23 | 227.6 | 279.4 | |
Low intensities (less than or equal to 0.052) were detected at background noise levels.
The concentrations of the compound of low intensity are indicated by a dash.
It has not been registered in KEGG.
The intensity of catechin was high in both control and UV-C-irradiated samples. Its concentrations were 3,771 and 4,043 µg g−1 fresh weight in control samples and UV-C-irradiated samples, respectively (Table III). Catechin was the most abundant metabolite among those identified, but the difference in catechin concentration between the two sets of samples was only minor. Some reports in the literature have reported high accumulation of proanthocyanidins, including catechin, in young berries before veraison (Bogs et al., 2005; Fujita et al., 2007). Our results agree with these reports.
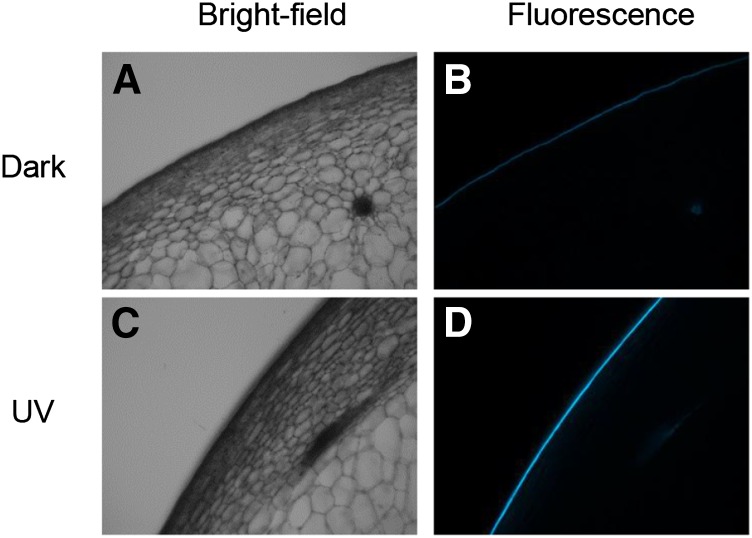
On the other hand, the intensity of trans-resveratrol was low in control samples but very high in UV-C-irradiated samples. The trans-resveratrol concentration in UV-C-irradiated samples was 3,492 µg g−1 fresh weight, 355 times higher than that in controls (Table III). In addition, we observed much stronger fluorescence, perhaps due to resveratrol (Del Nero and de Melo, 2002), in the grape berry skin of UV-C-irradiated samples than in control samples (Fig. 5).
Figure 5.
Accumulation of fluorescent substances at the berry skin. Sections of UV-C-irradiated (A and B) and control (C and D) grape berry are shown. A and C, Bright-field images. B and D, Fluorescence images.
Cis-resveratrol is a minor compound compared with trans-resveratrol in grape berry skin (De Nisco et al., 2013). In our metabolome analysis, measurable cis-resveratrol was detected in UV-C-irradiated samples, although its concentration was 6.3 µg g−1 fresh weight, far lower than the trans-resveratrol concentration (Table III). The resveratrol analogs viniferin and piceid were induced by UV-C irradiation at detectable levels (Table III). These minor analogs are thought to be synthesized by the modification of trans-resveratrol.
Transcriptome and metabolome analyses in this study found clear induction of STS gene expression and the accumulation of resveratrol.
Update of the Metabolic Pathway Map of Grape in KEGG and the KaPPA-View 4 KEGG System
The transcriptome and metabolome data were integrated and applied to a metabolic pathway map using the KaPPA-View 4 KEGG system (Sakurai et al., 2011).
GENESCOPE released the V0 grape genome data in 2007 (Jaillon et al., 2007), and the V0 data were registered as reference sequences in NCBI. Recently, the data set was updated from V0 to V1 by CRIBI (Grimplet et al., 2012). However, the reference sequences of the grape genome in the NCBI remain V0. The grape metabolic pathway map in the KaPPA-View 4 KEGG system was constructed based on the grape pathway map in KEGG (http://www.genome.jp/kegg; Kanehisa 2000). The KEGG system collects gene catalogs from the NCBI as well as another draft genome dataset, DGENES (http://www.genome.jp/kegg/catalog/org_list1.html).
On the other hand, the NimbleGen microarray was designed based on the V1 grape genome data (http://genomes.cribi.unipd.it/grape/). Thus, NimbleGen microarray data cannot be applied to the KaPPA-View 4 KEGG system. Therefore, we updated the KaPPA-View 4 KEGG system for this study. We added the V1 coding sequence (CDS) to the DGENES data set (T number T10027) with an automatic annotation server, KAAS (Moriya et al., 2007). Then we applied the new pathway map based on the DGENES data of the V1 CDS to the KaPPA-View 4 KEGG system. As a result, 5,440 genes of the V1 CDS are now reflected on the pathway maps. The updated system is available on the KaPPA-View 4 KEGG Web page (http://kpv.kazusa.or.jp/kpv4-kegg-1402/).
Integration of Transcriptome and Metabolome Data on the Metabolic Pathway Map
Our experimental values from transcriptome and metabolome analysis were included at appropriate scales (see “Materials and Methods”) on the metabolic pathway map. Using the updated KaPPA-View 4 KEGG system, we produced an updated map (Fig. 6). It includes levels of 73 genes and 17 compounds and is based on four maps (Supplemental Fig. S2).
Figure 6.
Modulation of secondary metabolism after UV-C irradiation. The schematic representation was determined by transcriptome and metabolome data integration using the updated KaPPA-View 4 KEGG. Fold change (FC) of each gene expression after log10 transformation is divided into 10 levels and depicted using the following colors: red, up-regulated by UV-C light; blue, down-regulated by UV-C light; gray, and not detected. Fold change of the contents of each metabolite is depicted using the following colors: red, up-regulated by UV-C light; and white, not identified. PAL, Phe ammonia-lyase; 4CL, 4-coumarate:CoA ligase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; ROMT, resveratrol O-methyltransferase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol-4-reductase; LDOX, leucoanthocyanidin dioxygenase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; BZ1, anthocyanidin 3-O-glucosyltransferase; 3MaT1, anthocyanin 3-O-glucoside-6′′-O-malonyltransferase; GT1, anthocyanidin 5,3-O-glucosyltransferase; UGAT, cyanidin-3-O-glucoside 2′′-O-glucuronosyltransferase.
Phenolic compounds, such as flavonol, proanthocyanidin, anthocyanin, and resveratrol, have a common synthetic pathway from Phe to coumaroyl-CoA, and these pathways branch after coumaroyl-CoA. Coumaroyl-CoA is the substrate of both STS and chalcone synthase. Chalcone synthase mediates the reaction from coumaroyl-CoA to naringenin chalcone; naringenin chalcone is converted to naringenin by chalcone isomerase, and this reaction pathway feeds into the proanthocyanidin, flavonol, and anthocyanin synthetic pathways. Figure 6 shows that not only STS but also genes involved in the pathway from Phe to coumaroyl-CoA (i.e. Phe ammonia-lyase, 4-coumarate:CoA ligase, and cinnamate 4-hydroxylase) were up-regulated by UV-C irradiation. On the other hand, expression of the genes involved in the other pathway branches for phenolic compounds, such as those encoding chalcone synthase, chalcone isomerase, and flavonoid 3′,5′-hydroxylase, were not changed by UV-C irradiation. Figure 6 also shows that UV-C irradiation induced only the accumulation of resveratrol and its analogs, not any other phenolic compounds. This map reveals dynamic and specific metabolic changes in grape berry skin due to UV-C irradiation.
DISCUSSION
To elucidate the effects of UV-C irradiation on grape berry skin, we performed transcriptome analysis using a genome-wide microarray and nontargeted metabolome analysis. The advantages of this study compared with previous studies are the comprehensiveness of ome analyses and the data integration of two ome analyses on a metabolic pathway map using the updated KaPPa-View 4 KEGG system. In this study, to apply the transcriptome data of the whole grape genes, we added V1 CDS to the KEGG database and then updated the KaPPa-View 4 KEGG system. By these updates, the KEGG database and KaPPa-View 4 KEGG system become more powerful tools for grape research.
Omics-Based Studies in Grape
Before the release of the grape genome sequence data by Jaillon et al. (2007) and Velasco et al. (2007), a few omics-based studies had been reported in grape, such as the transcriptome analyses of pathogen resistance in leaves (Fung et al., 2008; Polesani et al., 2010) and the transcriptome and proteome analyses focused on grape berry development (Deluc et al., 2007; Negri et al., 2008; Lücker et al., 2009). Particularly, the accumulation of secondary metabolites is an active topic in omics-based research in grape (Ali et al., 2011). Although a certain number of publications have reported the results of transcriptome analyses in grape, most used EST-based microarrays and their data did not cover the whole grape genes. After the update of the grape genome data by Grimplet et al. (2012), transcriptome analysis using a microarray covering the whole grape genes became popular (Pastore et al., 2011, 2013; Fasoli et al., 2012; Gambino et al., 2012; Lijavetzky et al., 2012; Young et al., 2012; Dal Santo et al., 2013; Carbonell-Bejerano et al., 2014a, 2014b; Dai et al., 2014; Rienth et al., 2014a, 2014b). Recently, RNA sequencing has also become popular in the transcriptome analysis of grape (Zenoni et al., 2010; Fasoli et al., 2012; Perazzolli et al., 2012; Sweetman et al., 2012; Venturini et al., 2013; Chitwood et al., 2014; Li et al., 2014; Vitulo et al., 2014; Xu et al., 2014).
On the other hand, metabolome analyses of various plants have also been reported (Saito and Matsuda, 2010). However, only a few studies reporting the grape metabolome detected more than 1,000 metabolite peaks (Zamboni et al., 2010; Sternad Lemut et al., 2013; Marti et al., 2014). Other studies detected far fewer metabolite peaks and discussed only a small number of metabolites (Dai et al., 2013).
With the availability of mass spectrometry and microarrays covering the whole grape genes, it has become possible to perform comprehensive multiomics studies. In this context, there have been a few studies involving integrated multiomics analyses in grape (Zamboni et al., 2010; Agudelo-Romero et al., 2013). We analyzed the expression of 29,549 genes and detected 2,012 metabolite peaks corresponding to metabolites. In addition, we combined these data using a metabolic pathway map. Thus, this study is one of the most comprehensive multiomics studies in grape research. The identification of genes responsible for key enzymes and the compounds produced by them from the detailed pathway map is the first step toward a better understanding of the metabolic changes in grape berry skin during the accumulation of polyphenolic components.
Omics Studies in Grape Berry Skin
It should be noted that, in this study, we focused on grape berry skin. Grape berry skin accumulates various secondary metabolites at higher concentrations than other tissues such as leaves and berry flesh (Kader, 2002; Steyn, 2009). These secondary metabolites determine the important traits of grape berries. Some researchers have used omics approaches to study secondary metabolite accumulation in grape berry skin, such as changes in the transcriptome before and after ripening (Waters et al., 2005), tissue-specific transcriptome profiles (Grimplet et al., 2007), transcriptome changes after abscisic acid (ABA) treatment at the veraison stage (Koyama et al., 2010), and comparison of transcriptomes in berry flesh with that in berry skin (Ali et al., 2011). Recently, transcriptome analyses of grape berry with or without the usage of solar UV light was reported by Carbonell-Bejerano et al. (2014a). They observed both flavonol and stilbenoid accumulation and the induction of their synthetic genes (i.e. phenylpropanoid and stilbenoid synthetic genes with solar UV light condition). Although we irradiated grape berries with UV-C light in this study, interestingly, we observed only specific stilbenoid accumulation and its biosynthetic gene induction and no flavonol accumulation and its synthetic gene induction (Fig. 3; Table II).
Transcription Factors Associated with Secondary Metabolite Accumulation
Carbonell-Bejerano et al. (2014a) reported that some transcription factors (TFs) such as MYB and basic helix-loop-helix were up-regulated by solar UV radiation in grape berry skin. In contrast, these TFs were not induced in our experiment (Supplemental Data Set S1). Pontin et al. (2010) reported that some TFs, including ethylene response factor, WRKY, and NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF1-2], and cup-shaped cotyledon [CUC2]), were up-regulated by UV-B irradiation in grape leaves. Interestingly, ethylene response factors (VIT_07s0005g03220, VIT_15s0021g01630, VIT_05s0049g00490, VIT_07s0005g03260, VIT_07s0005g03230, and VIT_17s0000g00200), WRKYs (VIT_04s0069g00970, VIT_14s0068g01770, and VIT_04s0008g05750), and NACs (VIT_06s0004g00020) were up-regulated in our experiment (Supplemental Data Set S2). Carbonell-Bejerano et al. (2014a) applied natural solar UV light to berries, whereas in our study and the study by Pontin et al. (2010), artificial UV-B or UV-C light was applied. Differences in the quality and quantity of UV light may cause differences in TF expression, and the strong artificial UV light in our study and the study by Pontin et al. (2010) may act as a stress and also induce stress-related TFs.
Höll et al. (2013) identified the R2R3-MYB TFs (i.e. MYB14 and MYB15) as TFs implicated in the regulation of stilbene biosynthesis, given that they are coexpressed with STS genes. However, the expression of neither MYB14 (VIT_07s0005g03340) nor MYB15 (VIT_05s0049g01020) was induced by UV irradiation in other studies (Pontin et al., 2010; Carbonell-Bejerano et al., 2014a) or in our study (Supplemental Data Set S1). This difference may be due to the use of different cultivars, tissues, and experimental conditions, which suggests that various TFs are responsible for the regulation of stilbene biosynthesis by different stimuli.
Different Effects of UV-B and UV-C Light on the Accumulation of Phenolic Compounds
Plants accumulate phenolic compounds to prevent UV light damage (Caldwell et al., 1983). However, in our experiment, the levels of none of the studied phenolic compounds, including anthocyanin and flavonol, increased after UV-C irradiation (Table III), although strong resveratrol accumulation was observed (Fig. 4).
Different accumulation patterns of phenolic compounds in grapes in response to UV-B and UV-C light have been observed. Anthocyanin and flavanol contents in grape berry skin were at the same level during the conditions of the presence and absence of solar UV light, although flavonol content was higher during the presence of solar UV light (Carbonell-Bejerano et al., 2014a). Martínez-Lüscher et al. (2014) showed that UV-B irradiation increased flavonol accumulation in grape berry skin but did not modify anthocyanin content. Berli et al. (2010) reported that anthocyanin accumulation in grape leaves was not induced by UV-B irradiation alone but by a combination of UV-B irradiation and ABA treatment. These results show that anthocyanin accumulation is not induced by UV-B alone and that other factors, including ABA, are needed for its induction. ABA is known as a trigger for the ripening of grape berry and induces anthocyanin accumulation (Coombe and Hale, 1973; Koyama et al., 2010). Therefore, ABA must be an important cofactor for UV-B light-dependent anthocyanin accumulation in grape.
Stilbene biosynthesis is induced not only by UV-C light but also by pathogen infection, elicitors, and methyl jasmonate (Adrian et al., 2000; Versari et al., 2001; Aziz et al., 2003; Pezet et al., 2003; Takayanagi et al., 2004; Bru et al., 2006; Nishikawa et al., 2011; Belchí-Navarro et al., 2012). UV-C light induced pathogenesis-related proteins (Colas et al., 2012), antioxidant defense (Xie et al., 2012), a DNA repair mechanism (Molinier et al., 2004), programmed cell death (He et al., 2008), and phenolic compound accumulation, including stilbenenoid accumulation (Douillet-Breuil et al., 1999; Adrian et al., 2000; Versari et al., 2001; Pezet et al., 2003; Takayanagi et al., 2004; Nishikawa et al., 2011). As mentioned above, UV-C light may act as a stress and induce many stress responses, including stilbenoid accumulation.
Our research provided an overview of grape berry skin metabolism and identified a key factor involved in the response to UV-C irradiation. Based on the transcriptional analysis, genes from the whole genome were tagged with GO terms and used for enrichment analysis. As a result, 238 genes were identified as up-regulated genes (Supplemental Data Set S2), and we found enrichment of trihydroxystilbene synthase activity (GO:0050350), which includes STS, a key enzyme of resveratrol synthesis (Table II). In the 238 up-regulated genes by UV-C irradiation, seven STS genes were found (Table II). On the other hand, using an EST-based microarray, Pontin et al. (2010) found four STS genes as more than 5-fold up-regulated in grape leaves by UV-B irradiation. Only one gene, VIT_10s0042g00870 (GSVIVP00031875001), was overlapped between two experiments, and six STS genes were found newly in this study. Thirty-three STS genes, including those six genes, were not included in the EST-based microarray, although the genome-wide microarray includes all 39 STS genes of grape.
In the nontargeted metabolome analysis, the profiles of metabolite peaks were analyzed by PCA to detect fundamental differences between treatments. Surprisingly, in the 2,012 detected metabolite peaks, only resveratrol was identified as a compound strongly responsive to UV-C irradiation (Fig. 4). These data clarified that one of the metabolic changes causing resveratrol to accumulate in berry skin was the uniquely up-regulated expression of the genes for STS. In addition, our updated metabolic map showed the UV-C light-induced metabolic change in polyphenolic compounds visually (Fig. 6). Because we modified the KaPPA-View 4 KEGG system to include V1 grape genome data, this system has become a more powerful tool in grape omics research. The NimbleGen microarray data directory can be uploaded into the KaPPA-View 4 KEGG system to easily project transcriptome data on a metabolic pathway map.
CONCLUSION
In this study, we focused on the UV-C light-inducible accumulation of phenolic compounds in grape berry skin and showed the specific induction of the resveratrol synthetic pathway by a microarray covering the whole grape genes and LC-QTOF-MS analysis. The development of analytical tools (i.e. the KEGG database and the KaPPA-View 4 KEGG system) and integration of the two thorough comprehensive omics data using the analytical tools could demonstrate metabolic change much more clearly than previous studies. This study is an example demonstrating the effectiveness of omics approaches to the analysis of specific traits of grape.
MATERIALS AND METHODS
Plant Materials and Treatments
Berry clusters of grape (Vitis vinifera ‘Pinot Noir’) before veraison stage were harvested at the vineyard of the Azumi Apple Corporation in August 2011. Berry clusters were irradiated using a UV-C lamp (253.7 nm; GL-15; Toshiba) from a distance of 50 cm (0.25 μW cm−2) for 1 h. Dark-treated control samples were covered with a box and placed next to the samples undergoing UV-C irradiation. For transcriptome analysis, berry skins were collected immediately after irradiation. For metabolite extraction, after irradiation, berry clusters were kept for 23 h in the dark at room temperature, and then the berry skins were harvested (Fig. 1). All samples were frozen immediately in liquid nitrogen and stored at −80°C. Three biological replicates were performed for each treatment. Each biological replicate comprised a pool of berries from four clusters collected from four different plants randomly.
RNA Extraction and Microarray Analysis
Total RNA was extracted from berry skins according to the hot borate method (Wan and Wilkins, 1994). The RNA obtained was further purified using an RNeasy Plant Mini Kit (Qiagen). RNA quality and quantity were determined using a spectrophotometer and a Bioanalyzer 2100 instrument (Agilent). The NimbleGen microarray 090818 Vitis exp HX12 (Roche NimbleGen), representing 29,549 predicted genes on the basis of the 12X grape V1 gene prediction (http://genomes.cribi.unipd.it/grape/), was used for microarray analysis according to the manufacturer’s specifications. Microarray data were processed using the Subio platform (Subio Inc.). The signal intensities of the four probes for each gene were averaged. Averaged raw data were then analyzed using the Subio platform. After the removal of noise signals with intensity less than 3,000, the triplicate data were standardized by a global normalization method with log formation and centering. When the fold change (UV-C light versus dark) was greater than 5 or less than 0.2, the gene was considered significantly up-regulated or down-regulated, respectively, by UV-C light. All microarray expression data are available in the Gene Expression Omnibus under the series entry GSE59436 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59436).
Gene Annotation and GO Analyses
The best-hit homolog of each grape gene (V1 coding sequences) in CRIBI (http://genomes.cribi.unipd.it/grape/) was found by searching the Arabidopsis CDS (The Arabidopsis Information Resource 10; http://arabidopsis.org) or whole organism amino acid sequences in UniprotKB (http://www.uniprot.org) using the BLAST within the NCBI (http://www.ncbi.nlm.nih.gov). VitisNet identifiers and the network name of VitisNet (http://www.sdstate.edu/ps/research/vitis/pathways.cfm) were also searched. GO annotations, classification, and enrichment analysis were performed using Blast2GO software (http://www.blast2go.org) based on sequence similarity. For annotation, the default configuration settings were used (BLASTX against the NCBI nonredundant protein database; E-value filter of 1.0E-3, 20 BLAST hits per sequence to sequence description tool, and annotation cutoff of 55). GO enrichment analysis was performed directly using the default settings. The GO slim goslim_plant.obo annotation was added to the data set to obtain plant-related GO terms.
Metabolome Analysis
Fresh samples were extracted with 5 μL of 80% (v/v) methanol containing 2.5 mm lidocaine and 10-camphor sulfonic acid per mg fresh weight using a mixer mill with zirconia beads for 7 min at 18 Hz and 4°C. After centrifugation for 10 min, the supernatant was filtered using an HLB μElution plate (Waters). The extracts (1 μL) were analyzed using LC-QTOF-MS (Waters Acquity UPLC system liquid chromatograph and Waters Xevo G2 quadrupole time-of-flight mass spectrometer). Analytical conditions were as follows: liquid chromatograph column, Acquity bridged ethyl hybrid C18 (1.7 μm, 2.1 × 100 mm; Waters); solvent system, solvent A (0.1% [v/v] formic acid in water) and solvent B (acetonitrile including 0.1% formic acid); gradient program, 99.5% A/0.5% B at 0 min, 99.5% A/0.5% B at 0.1 min, 20% A/80% B at 10 min, 0.5% A/99.5% B at 10.1 min, 0.5% A/99.5% B at 12 min, 99.5% A/0.5% B at 12.1 min, and 99.5% A/0.5% B at 15 min; flow rate, 0.3 mL min−1 at 0 min, 0.3 mL min−1 at 10 min, 0.4 mL min−1 at 10.1 min, 0.4 mL min−1 at 14.4 min, and 0.3 mL min−1 at 14.5 min; column temperature, 40°C; MS capillary voltage, +3 keV; cone voltage, 25 V; source temperature, 120°C; desolvation temperature, 450°C; cone gas flow, 50 L h−1; desolvation gas flow, 800 L h−1; collision energy, 6 V; mass range, m/z 100 to 1,500; scan duration, 0.1 s; interscan delay, 0.014 s; data acquisition, centroid mode; polarity, positive/negative; Lockspray (Leu enkephalin) scan duration, 1 s; and interscan delay, 0.1 s. The data matrix was aligned using MassLynx version 4.1 software (Waters). After alignment, deisotoping, and cutoff of the low-intensity peaks (less than 500 counts), the intensity values of the remaining peaks were divided by those of 10-camphor sulfonic acid ([M − H]−, m/z 231.06910) for normalization. The processed data were used for PCA using the SIMCA-P 11.5 program (Umetrics).
Chemicals
Standard compounds were purchased from Wako Pure Chemical Industries (http://www.wako-chem.co.jp/), Sigma-Aldrich (http://www.sigmaaldrich.com/), ChromaDex (https://chromadex.com/default.aspx), Cayman Chemical (https://www.caymanchem.com/app/template/Home.vm), Enzo Life Sciences (http://www.enzolifesciences.com/), Polyphenols Laboratories (http://www.polyphenols.no/), Extrasynthese (http://www.extrasynthese.com/), Nacalai Tesque (http://www.nacalai.co.jp/), and Tokyo Chemical Industry (http://www.tcichemicals.com/ja/jp/). A list is shown in Supplemental Table S1.
Fluorescence Microscopy
UV-C light-treated and control samples were prepared into 100-μm thin sections by a vibrating-type microtome (VT1000 S; Leica). Sections were placed on a glass slide, covered by a coverslip, and observed immediately. The sections were observed with a BZ-9000 fluorescence microscope (Keyence) using a mercury-vapor lamp and filter setting for 4′,6-diamidino-2-phenylindole (OP-66834 BZ filter; Keyence).
Gene Expression and Metabolite Profiling
The pathway map from Phe to phenolic compounds, such as resveratrol, flavonol, proanthocyanidin, and anthocyanin, was originally based on the KEGG pathway database (http://www.genome.jp/kegg; Kanehisa 2000). The KaPPA-View 4 KEGG system (Sakurai et al., 2011) was used for the representation of transcriptome and metabolome data on pathway maps. The experimental values were input on a log scale for the gene intensity and on a linear scale for the metabolite intensity in negative ion mode.
The data reported in this article have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE59436.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Volcano plot of microarray data.
Supplemental Figure S2. The grape metabolic pathway maps from KaPPA View 4 KEGG.
Supplemental Table S1. Chemical compounds for identification of metabolites.
Supplemental Data Set S1. Transcriptome data after UV-C irradiation.
Supplemental Data Set S2. List of 238 up-regulated genes by UV-C irradiation.
Supplemental Data Set S3. List of 24 down-regulated genes by UV-C irradiation.
Supplementary Material
Acknowledgments
We thank Hiroya Saito and Chiharu Uchikata (Azumi Apple Corporation) for supplying grape berries, Yutaka Nishikawa and Kenji Wada (Mie Prefecture Agricultural Research Institute) and Dr. Takafumi Tezuka (Nagoya University) for helpful advice on UV-C light treatment of grape berries, Dr. Mikio Nakazono (Nagoya University) for help with preparing grape skin sections, Tetsuya Mori (RIKEN Center for Sustainable Resource Science) for technical assistance, and Dr. Stefan Reuscher (Nagoya University) for proofreading the article.
Glossary
- GO
Gene Ontology
- LC-QTOF-MS
liquid chromatography-quadrupole time-of-flight mass spectrometry
- PCA
principal component analysis
- V0
version 0
- V1
version 1
- NCBI
National Center for Biotechnology Information
- STS
stilbene synthase
- ABA
abscisic acid
- TF
transcription factor
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the Programme for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry from the Bio-oriented Technology Research Advancement Institution and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
References
- Adrian M, Jeandet P, Douillet-Breuil AC, Tesson L, Bessis R (2000) Stilbene content of mature Vitis vinifera berries in response to UV-C elicitation. J Agric Food Chem 48: 6103–6105 [DOI] [PubMed] [Google Scholar]
- Agudelo-Romero P, Erban A, Sousa L, Pais MS, Kopka J, Fortes AM (2013) Search for transcriptional and metabolic markers of grape pre-ripening and ripening and insights into specific aroma development in three Portuguese cultivars. PLoS ONE 8: e60422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MB, Howard S, Chen S, Wang Y, Yu O, Kovacs LG, Qiu W (2011) Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Poinssot B, Daire X, Adrian M, Bézier A, Lambert B, Joubert JM, Pugin A (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol Plant Microbe Interact 16: 1118–1128 [DOI] [PubMed] [Google Scholar]
- Belchí-Navarro S, Almagro L, Lijavetzky D, Bru R, Pedreño MA (2012) Enhanced extracellular production of trans-resveratrol in Vitis vinifera suspension cultured cells by using cyclodextrins and methyljasmonate. Plant Cell Rep 31: 81–89 [DOI] [PubMed] [Google Scholar]
- Berli FJ, Moreno D, Piccoli P, Hespanhol-Viana L, Silva MF, Bressan-Smith R, Cavagnaro JB, Bottini R (2010) Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ 33: 1–10 [DOI] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru R, Sellés S, Casado-Vela J, Belchí-Navarro S, Pedreño MA (2006) Modified cyclodextrins are chemically defined glucan inducers of defense responses in grapevine cell cultures. J Agric Food Chem 54: 65–71 [DOI] [PubMed] [Google Scholar]
- Caldwell MM, Robberecht R, Flint SD (1983) Internal filters: prospects for UV-acclimation in higher plants. Physiol Plant 58: 445–450 [Google Scholar]
- Carbonell-Bejerano P, Diago MP, Martínez-Abaigar J, Martínez-Zapater JM, Tardáguila J, Núñez-Olivera E (2014a) Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant Biol 14: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Rodríguez V, Royo C, Hernáiz S, Moro-González LC, Torres-Viñals M, Martínez-Zapater JM (2014b) Circadian oscillatory transcriptional programs in grapevine ripening fruits. BMC Plant Biol 14: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C, Nawrath C, Métraux JP (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49: 972–980 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Ranjan A, Martinez CC, Headland LR, Thiem T, Kumar R, Covington MF, Hatcher T, Naylor DT, Zimmerman S, et al. (2014) A modern ampelography: a genetic basis for leaf shape and venation patterning in grape. Plant Physiol 164: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas S, Afoufa-Bastien D, Jacquens L, Clément C, Baillieul F, Mazeyrat-Gourbeyre F, Monti-Dedieu L (2012) Expression and in situ localization of two major PR proteins of grapevine berries during development and after UV-C exposition. PLoS ONE 7: e43681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe BG, Hale CR (1973) The hormone content of ripening grape berries and the effects of growth substance treatments. Plant Physiol 51: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZW, Léon C, Feil R, Lunn JE, Delrot S, Gomès E (2013) Metabolic profiling reveals coordinated switches in primary carbohydrate metabolism in grape berry (Vitis vinifera L.), a non-climacteric fleshy fruit. J Exp Bot 64: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZW, Meddar M, Renaud C, Merlin I, Hilbert G, Delrot S, Gomès E (2014) Long-term in vitro culture of grape berries and its application to assess the effects of sugar supply on anthocyanin accumulation. J Exp Bot 65: 4665–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, Guzzo F, Delledonne M, Pezzotti M (2013) The plasticity of the grapevine berry transcriptome. Genome Biol 14: r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B, Cordelier S, Conreux A, Clément C, Jeandet P (2009) Molecular engineering of resveratrol in plants. Plant Biotechnol J 7: 2–12 [DOI] [PubMed] [Google Scholar]
- Del Nero J, de Melo CP (2002) Quantum chemistry calculation of resveratrol and related stilbenes. Opt Mater 21: 455–460 [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nisco M, Manfra M, Bolognese A, Sofo A, Scopa A, Tenore GC, Pagano F, Milite C, Russo MT (2013) Nutraceutical properties and polyphenolic profile of berry skin and wine of Vitis vinifera L. (cv. Aglianico). Food Chem 140: 623–629 [DOI] [PubMed] [Google Scholar]
- Douillet-Breuil AC, Jeandet P, Adrian M, Bessis R (1999) Changes in the phytoalexin content of various Vitis spp. in response to ultraviolet C elicitation. J Agric Food Chem 47: 4456–4461 [DOI] [PubMed] [Google Scholar]
- Eudes A, Bozzo GG, Waller JC, Naponelli V, Lim EK, Bowles DJ, Gregory JF III, Hanson AD (2008) Metabolism of the folate precursor p-aminobenzoate in plants: glucose ester formation and vacuolar storage. J Biol Chem 283: 15451–15459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, et al. (2012) The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24: 3489–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A, Soma N, Goto-Yamamoto N, Mizuno A, Kiso K, Hashizume K (2007) Effect of shading on proanthocyanidin biosynthesis in the grape berry. J Jpn Soc Hortic Sci 76: 112–119 [Google Scholar]
- Fung RW, Gonzalo M, Fekete C, Kovacs LG, He Y, Marsh E, McIntyre LM, Schachtman DP, Qiu W (2008) Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 146: 236–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino G, Cuozzo D, Fasoli M, Pagliarani C, Vitali M, Boccacci P, Pezzotti M, Mannini F (2012) Co-evolution between grapevine rupestris stem pitting-associated virus and Vitis vinifera L. leads to decreased defence responses and increased transcription of genes related to photosynthesis. J Exp Bot 63: 5919–5933 [DOI] [PubMed] [Google Scholar]
- Grimplet J, Cramer GR, Dickerson JA, Mathiason K, Van Hemert J, Fennell AY (2009) VitisNet: “omics” integration through grapevine molecular networks. PLoS ONE 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Van Hemert J, Carbonell-Bejerano P, Díaz-Riquelme J, Dickerson J, Fennell A, Pezzotti M, Martínez-Zapater JM (2012) Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res Notes 5: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2008) Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25: 4135–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. (1997) Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275: 218–220 [DOI] [PubMed] [Google Scholar]
- Kader AA. (2002) Fruits in the global market. In Knee M, ed, Fruit Quality and Its Biological Basis. Sheffield Academic Press, Sheffield, UK, pp 1–16 [Google Scholar]
- Kanehisa M. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama K, Sadamatsu K, Goto-Yamamoto N (2010) Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Funct Integr Genomics 10: 367–381 [DOI] [PubMed] [Google Scholar]
- Langcake P, Pryce RJ (1977) A new class of phytoalexins from grapevines. Experientia 33: 151–152 [DOI] [PubMed] [Google Scholar]
- Li Q, He F, Zhu BQ, Liu B, Sun RZ, Duan CQ, Reeves MJ, Wang J (2014) Comparison of distinct transcriptional expression patterns of flavonoid biosynthesis in Cabernet Sauvignon grapes from east and west China. Plant Physiol Biochem 84: 45–56 [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, Fenoll J, Hellín P, Oliveros JC, Martínez-Zapater JM (2012) Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PLoS ONE 7: e39547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guénin S, Roger D, Gillet F, Guerineau F, Pelloux J (2006) Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 224: 782–791 [DOI] [PubMed] [Google Scholar]
- Lücker J, Laszczak M, Smith D, Lund ST (2009) Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genomics 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Zhao H, Davies P (2005) Resveratrol promotes clearance of Alzheimer’s disease amyloid-beta peptides. J Biol Chem 280: 37377–37382 [DOI] [PubMed] [Google Scholar]
- Marti G, Schnee S, Andrey Y, Simoes-Pires C, Carrupt PA, Wolfender JL, Gindro K (2014) Study of leaf metabolome modifications induced by UV-C radiations in representative Vitis, Cissus and Cannabis species by LC-MS based metabolomics and antioxidant assays. Molecules 19: 14004–14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Lüscher J, Torres N, Hilbert G, Richard T, Sánchez-Díaz M, Delrot S, Aguirreolea J, Pascual I, Gomès E (2014) Ultraviolet-B radiation modifies the quantitative and qualitative profile of flavonoids and amino acids in grape berries. Phytochemistry 102: 106–114 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ramos C, Fritsch O, Hohn B (2004) CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri AS, Prinsi B, Rossoni M, Failla O, Scienza A, Cocucci M, Espen L (2008) Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa Y, Tominori S, Wada K, Kondo H (2011) Effect of cultivation practices on resveratrol content in grape berry skins. Hortic Res 10: 249–253 (in Japanese) [Google Scholar]
- Pastore C, Zenoni S, Fasoli M, Pezzotti M, Tornielli GB, Filippetti I (2013) Selective defoliation affects plant growth, fruit transcriptional ripening program and flavonoid metabolism in grapevine. BMC Plant Biol 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Zenoni S, Tornielli GB, Allegro G, Dal Santo S, Valentini G, Intrieri C, Pezzotti M, Filippetti I (2011) Increasing the source/sink ratio in Vitis vinifera (cv Sangiovese) induces extensive transcriptome reprogramming and modifies berry ripening. BMC Genomics 12: 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M, Moretto M, Fontana P, Ferrarini A, Velasco R, Moser C, Delledonne M, Pertot I (2012) Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics 13: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet R, Perret C, Jean-Denis JB, Tabacchi R, Gindro K, Viret O (2003) δ-viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. J Agric Food Chem 51: 5488–5492 [DOI] [PubMed] [Google Scholar]
- Polesani M, Bortesi L, Ferrarini A, Zamboni A, Fasoli M, Zadra C, Lovato A, Pezzotti M, Delledonne M, Polverari A (2010) General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics 11: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontin MA, Piccoli PN, Francisco R, Bottini R, Martinez-Zapater JM, Lijavetzky D (2010) Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol 10: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, de Lorgeril M (1992) Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet 339: 1523–1526 [DOI] [PubMed] [Google Scholar]
- Rienth M, Torregrosa L, Kelly MT, Luchaire N, Pellegrino A, Grimplet J, Romieu C (2014a) Is transcriptomic regulation of berry development more important at night than during the day? PLoS ONE 9: e88844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienth M, Torregrosa L, Luchaire N, Chatbanyong R, Lecourieux D, Kelly MT, Romieu C (2014b) Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol 14: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Ogata Y, Sano R, Ohno T, Sugiyama K, Hiruta A, Yamazaki K, Yano K, Aoki K, et al. (2011) KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res 39: D677–D684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternad Lemut M, Sivilotti P, Franceschi P, Wehrens R, Vrhovsek U (2013) Use of metabolic profiling to study grape skin polyphenol behavior as a result of canopy microclimate manipulation in a ‘Pinot Noir’ vineyard. J Agric Food Chem 61: 8976–8986 [DOI] [PubMed] [Google Scholar]
- Steyn WJ. (2009) Prevalence and functions of anthocyanins in fruits. In Winefield C, Davies K, Gould K, eds, Anthocyanins. Springer, New York, pp 86–105 [Google Scholar]
- Sweetman C, Wong DC, Ford CM, Drew DP (2012) Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genomics 13: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi T, Okuda T, Mine Y, Yokotsuka K (2004) Induction of resveratrol biosynthesis in skins of three grape cultivars by ultraviolet irradiation. J Jpn Soc Hortic Sci 73: 193–199 [Google Scholar]
- Turlapati PV, Kim KW, Davin LB, Lewis NG (2011) The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233: 439–470 [DOI] [PubMed] [Google Scholar]
- Vang O, Ahmad N, Baile CA, Baur JA, Brown K, Csiszar A, Das DK, Delmas D, Gottfried C, Lin HY, et al. (2011) What is new for an old molecule? Systematic review and recommendations on the use of resveratrol. PLoS One 6: e19881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al. (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L, Ferrarini A, Zenoni S, Tornielli GB, Fasoli M, Dal Santo S, Minio A, Buson G, Tononi P, Zago ED, et al. (2013) De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C (2001) Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J Agric Food Chem 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D’Angelo M, Zimbello R, Corso M, Vannozzi A, Bonghi C, et al. (2014) A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol 14: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Wan D, Li R, Zou B, Zhang X, Cong J, Wang R, Xia Y, Li G (2012) Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep 31: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen V, Katagiri F, Glazebrook J (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Waters DLE, Holton TA, Ablett EM, Lee LS, Henry RJ (2005) cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct Integr Genomics 5: 40–58 [DOI] [PubMed] [Google Scholar]
- Xie Y, Xu D, Cui W, Shen W (2012) Mutation of Arabidopsis HY1 causes UV-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defence. J Exp Bot 63: 3869–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Li R, Zhang N, Ma F, Jiao Y, Wang Z (2014) Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol Biol 86: 527–541 [DOI] [PubMed] [Google Scholar]
- Young PR, Lashbrooke JG, Alexandersson E, Jacobson D, Moser C, Velasco R, Vivier MA (2012) The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genomics 13: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S, Ferrarini A, Tononi P, Toffali K, Desiderio A, Lilley KS, et al. (2010) Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152: 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.