Three R2R3-MYB transcription factors regulate floral pigmentation in Phalaenopsis spp. for diverse floral color patterns.
Abstract
Orchidaceae are well known for their fascinating floral morphologic features, specialized pollination, and distinctive ecological strategies. With their long-lasting flowers of various colors and pigmentation patterning, Phalaenopsis spp. have become important ornamental plants worldwide. In this study, we identified three R2R3-MYB transcription factors PeMYB2, PeMYB11, and PeMYB12. Their expression profiles were concomitant with red color formation in Phalaenopsis spp. flowers. Transient assay of overexpression of three PeMYBs verified that PeMYB2 resulted in anthocyanin accumulation, and these PeMYBs could activate the expression of three downstream structural genes Phalaenopsis spp. Flavanone 3-hydroxylase5, Phalaenopsis spp. Dihydroflavonol 4-reductase1, and Phalaenopsis spp. Anthocyanidin synthase3. In addition, these three PeMYBs participated in the distinct pigmentation patterning in a single flower, which was revealed by virus-induced gene silencing. In the sepals/petals, silencing of PeMYB2, PeMYB11, and PeMYB12 resulted in the loss of the full-red pigmentation, red spots, and venation patterns, respectively. Moreover, different pigmentation patterning was regulated by PeMYBs in the sepals/petals and lip. PeMYB11 was responsive to the red spots in the callus of the lip, and PeMYB12 participated in the full pigmentation in the central lobe of the lip. The differential pigmentation patterning was validated by RNA in situ hybridization. Additional assessment was performed in six Phalaenopsis spp. cultivars with different color patterns. The combined expression of these three PeMYBs in different ratios leads to a wealth of complicated floral pigmentation patterning in Phalaenopsis spp.
Phalaenopsis spp., one genus of Orchidaceae, have become very popular worldwide for their long-lived flowers with various colors and pigmentation patterns. In addition to the two lateral petals, the third petal is modified into a labellum or lip to attract pollinators. Their flower colors range from black to purple, red, yellow, and white, and differential coloration generates various pigmentation patterns, such as spots, irregular blotches, stripes overlaying veins (venation pattern), or combinations of these (Supplemental Fig. S1). Pigmentation patterning may increase successful pollination by both increasing the frequency of pollinator visits and providing color guides for the location of rewards, pollen, and nectar or preferred landing platforms of the flower (Medel et al., 2003; Heuschen et al., 2005; Lunau et al., 2006; Ushimaru et al., 2007). Of note, both the color and the pigmentation patterning usually differ between the sepals/petals and the lip of a Phalaenopsis spp. flower. This observation indicates a distinct regulatory mechanism within a single Phalaenopsis spp. flower for the accumulation of anthocyanins, the water-soluble pigments occurring in almost all plants that are responsible for most of the orange, red, purple, and blue colors of flowers. With all of the above-mentioned characteristics, Phalaenopsis spp. with natural variations in flower colors would be an excellent model plant for studying the molecular mechanism regulating floral pigmentation patterning.
The biosynthetic pathway for anthocyanins is one of the most extensively studied plant secondary metabolisms (Grotewold, 2006). Many regulatory genes involved in the anthocyanin biosynthetic pathway have been cloned and characterized from a wide variety of plants. R2R3-MYB, basic helix-loop-helix (bHLH) transcription factors, and WD40 repeat (WDR) proteins are the three major families of regulatory proteins for anthocyanin biosynthesis (Koes et al., 2005; Feller et al., 2011; Hichri et al., 2011; Petroni and Tonelli, 2011).
The R2R3-MYB transcription factors play a major role in determining the spatial and temporal patterning of anthocyanins in Antirrhinum spp. and Petunia spp. (Schwinn et al., 2006; Albert et al., 2011; Davies et al., 2012). Irregular pigmentation patterning, such as color flecks and sectors, was studied in morning glory (Pharbitis purpurea) and other plants and found to be linked to the activated transposon insertion in anthocyanin biosynthetic genes (Inagaki et al., 1994; Iida et al., 1999; Itoh et al., 2002). Other patterns, such as in Petunia spp. Red Star and Petunia spp. picotee, flowers are the result of reduced RNA expression of Chalcone synthase (CHS), possibly because of short-interfering RNA degradation (Koseki et al., 2005; Saito et al., 2006; Griesbach et al., 2007). The spot pattern is associated with the differential expression of Dihydroflavonol 4-reductase2 (Dfr2) in the red-purple spot in Clarkia gracilis (Martins et al., 2013) or light-induced Lilium hybrid MYB6 (LhMYB6) in the red spots compared with the pink background regulated by LhMYB12 in the Asiatic hybrid lily (Lilium spp.) ‘Montreux’ (Yamagishi et al., 2010). Splatter-type spots in the Asiatic hybrid lily ‘Latvia’ are regulated by a unique allele of LhMYB12-Lat (Yamagishi et al., 2014). Moreover, the anthocyanin phenotypes of leaf blotching, calyx spotting, and corolla banding were varied within the populations of Mimulus guttatus and controlled by Petal Lobe Anthocyanin1 locus, which contains three tandem repeats of R2R3-MYB genes (Lowry et al., 2012). However, the genetic control of petal spots has not been well studied and may differ among species.
In Orchidaceae, Phalaenopsis schilleriana DFR (PsDFR) and PsMYB express in the purple flowers of P. schilleriana and spots of Phalaenopsis spp. ‘Ever-spring Fairy’ petals but not in white-flower Phalaenopsis amabilis (Ma et al., 2009). In Oncidium spp. ‘Gower Ramsey,’ the expression of Oncidium gower Chalcone isomerase (OgCHI), OgDFR, and OgMYB1 confers a lip crest with a mosaic red pigmentation in flowers (Chiou and Yeh, 2008).
Virus-induced gene silencing (VIGS) is a reverse genetics approach used for functional analysis of genes in plants, especially those with long lifecycles and few genetic resources, such as Phalaenopsis spp. A Cymbidium mosaic virus (CymMV)-based VIGS vector and its modified vector have been established (Lu et al., 2007, 2012) and were used to determine the effect of Phalaenopsis spp. TF15 (PhaTF15) on disease resistance (Lu et al., 2012), Phalaenopsis equestris UDP glucose: flavonoid 3-O-glucosyltransferase (PeUFGT3) on anthocyanin biosynthesis (Chen et al., 2011), P. equestris MADS5 (PeMADS5), PeMADS6, P. equestris SEPALLATA1 (PeSEP1), PeSEP2, PeSEP3, and PeSEP4 on floral morphogenesis (Hsieh et al., 2013a; Pan et al., 2014), and high-throughput silencing of 126 transcription factors for identifying genes involved in flower development of Phalaenopsis spp. (Hsieh et al., 2013b).
In this study, we identified three R2R3-MYB transcription factors (PeMYB2, PeMYB11, and PeMYB12) in Phalaenopsis spp. and analyzed their expression profiles. We used transient assay of gene overexpression to verify that these three PeMYBs can activate the expression of the downstream structural genes and result in anthocyanin accumulation. Moreover, these three PeMYBs individually participated in regulating the distinct pigmentation patterning in Phalaenopsis spp., which was revealed by VIGS and RNA in situ hybridization. The expression of the three PeMYBs and their mutual interaction were confirmed in various Phalaenopsis spp. cultivars.
RESULTS
Phylogenetic Relationships of R2R3-MYB Transcription Factors Isolated from Phalaenopsis spp.
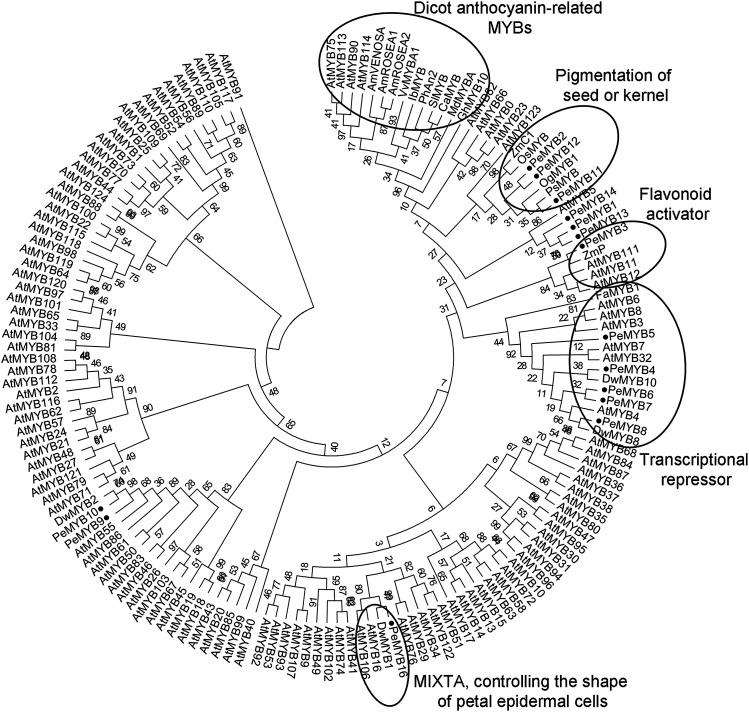
To study the MYB transcription factors that regulate anthocyanin biosynthesis in Phalaenopsis spp., we identified 16 R2R3-MYB transcription factors (PeMYB1–PeMYB16), including 14 PeMYBs isolated from OrchidBase (Fu et al., 2011; Tsai et al., 2013) and 2 PeMYBs amplified from P. equestris with degenerated primers. Phylogenetic analysis showed PeMYB2, PeMYB11, and PeMYB12 in the same clade as Zea mays C1 (ZmC1), which controls the pigmentation of seeds or kernels (Paz-Ares et al., 1987; Lepiniec et al., 2006), and the same large cluster with dicot anthocyanin-promoting MYBs, including Petunia hybrida Anthocyanin2 (PhAN2) and Antirrhinum majus VENOSA (AmVENOSA), AmROSEA1 (AmROS1), and AmROS2 (Fig. 1). PeMYB11 was most similar to purple-flower orchid PsMYB (Ma et al., 2009) and Oncidium spp. OgMYB1 in regulating red color formation (Chiou and Yeh, 2008; Fig. 1). PeMYB1, PeMYB13, and PeMYB14 were grouped together, and PeMYB3 was in another clade with Z. mays P (ZmP; Fig. 1). Otherwise, PeMYB4, PeMYB5, PeMYB6, PeMYB7, and PeMYB8 were grouped with AtMYB4 (Jin et al., 2000), Fragaria ananassa MYB1 (FaMYB1; Aharoni et al., 2001), and other MYBs that are repressors of flavonoid biosynthesis (Fig. 1).
Figure 1.
Phylogenetic tree inferred from the amino acid sequences of the R2R3 region of PeMYBs with anthocyanin-related MYBs and all Arabidopsis (Arabidopsis thaliana) R2R3-MYBs. This phylogenetic tree was constructed by the Maximum Likelihood method with 1,000 bootstrapping data sets. Black circles indicate 15 PeMYBs identified in this study. MIXTA is an R2R3-MYB gene in A. majus and controls the development of conical cell shape in petal epidermal cells (Baumann et al., 2007).
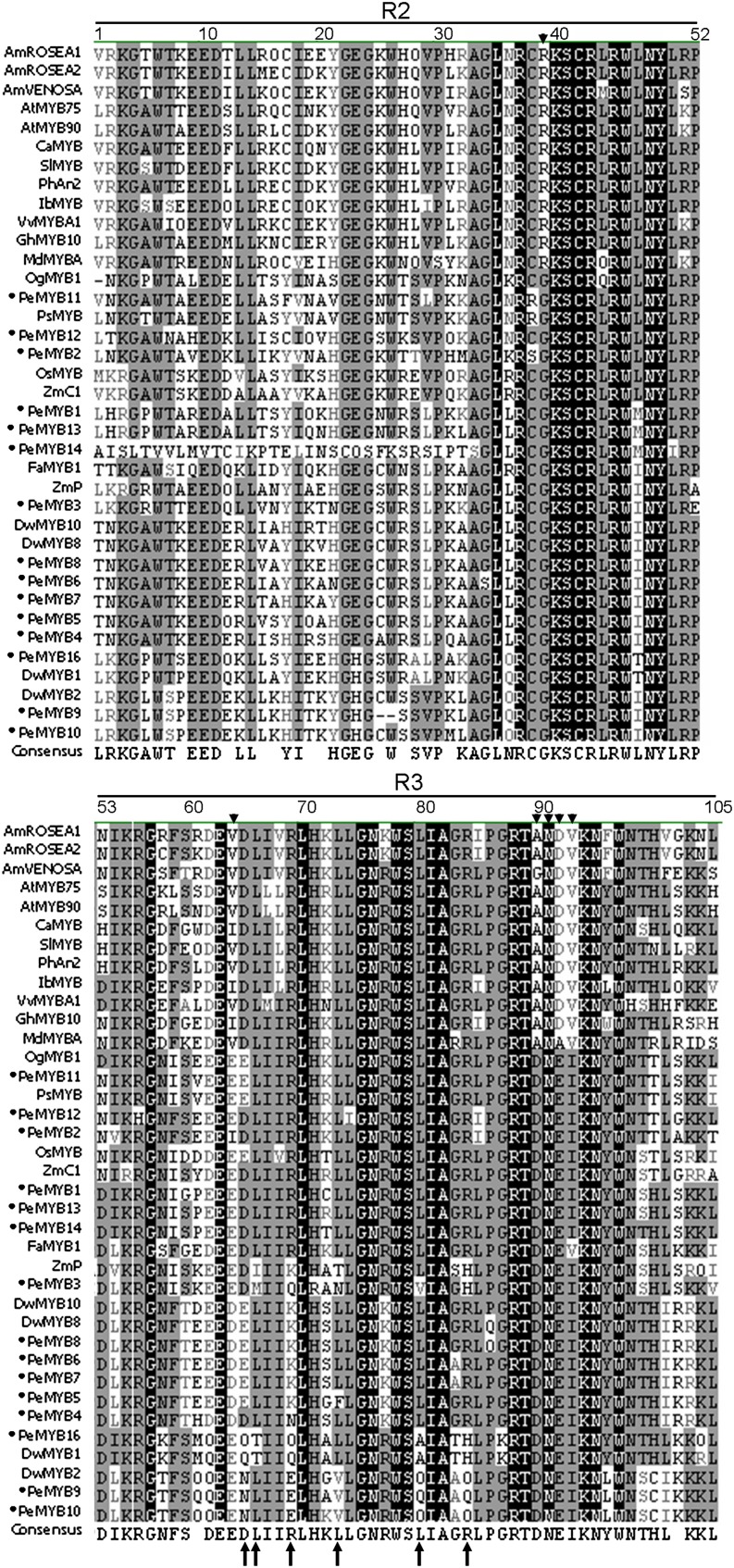
Multiple alignment of the MYB-R2R3 region showed that the sequences of PeMYB2, PeMYB11, and PeMYB12 contained a conserved [D/E]Lx2[R/K]x3Lx6Lx3R motif at positions 65 to 84 required for interaction with R/B-like bHLH proteins (Zimmermann et al., 2004; Fig. 2, arrows). However, they lack the three conserved amino acid residues (R39, V64, and A90), and a convenient identifier, ANDV, at positions 90 to 93 of dicot anthocyanin-promoting MYBs (Lin-Wang et al., 2010; Fig. 2, arrowheads).
Figure 2.
Multiple alignment of the amino acid sequences in the R2R3 region of PeMYBs with anthocyanin-promoting MYBs. Arrowheads indicate the conserved R39, V64, A90, and ANDV at positions 90 to 93 for dicot anthocyanin-promoting MYBs. Arrows indicate specific residues that contribute to a motif ([D/E]Lx2[R/K]x3Lx6Lx3R) interacting with a bHLH transcription factor in Arabidopsis (Zimmermann et al., 2004). Black circles indicate 15 PeMYBs isolated in this study.
Expression of PeMYB2, PeMYB11, and PeMYB12 Was Concomitant with Red Color Formation in Flowers
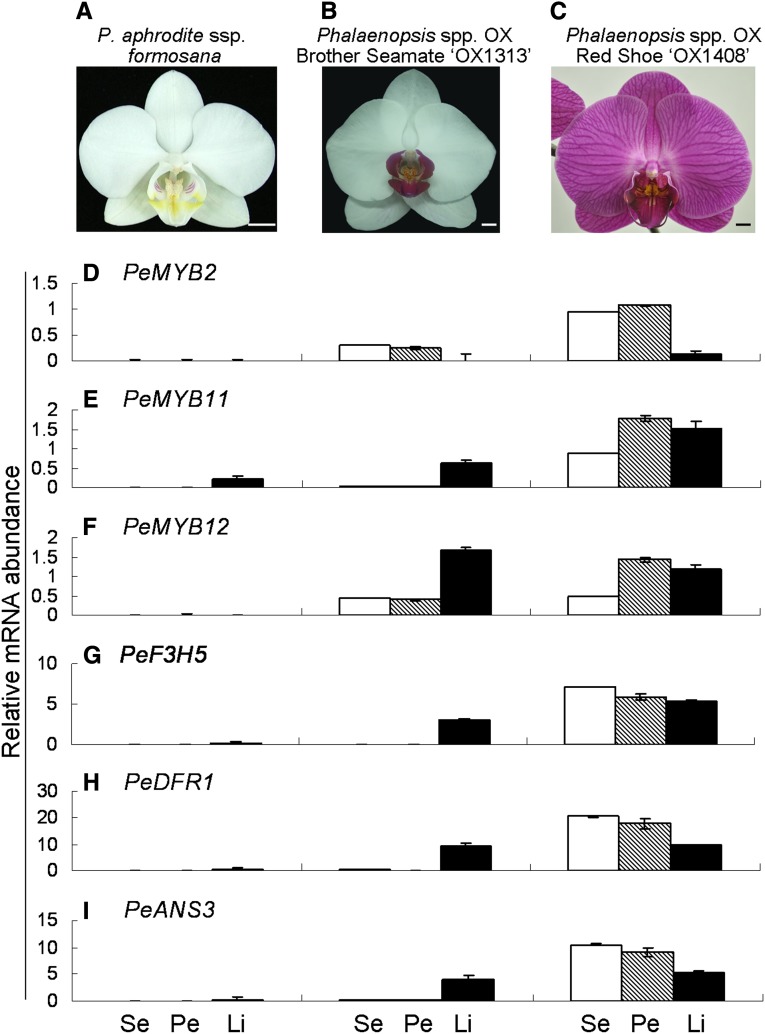
The expression profiles of the PeMYBs were first assessed in Phalaenopsis aphrodite ssp. formosana with white sepals/petals and a yellow lip (Fig. 3A) and Phalaenopsis spp. OX Brother Seamate ‘OX1313’ with white sepals/petals and a red lip (Fig. 3B) by reverse transcription (RT)-PCR (Supplemental Fig. S2F). In P. aphrodite ssp. formosana, transcripts of PeMYB2 and PeMYB11 were detected in the sepals and lip, respectively (Supplemental Fig. S2F), concomitant with the pink color formation on the abaxial surface of the sepals (Supplemental Fig. S2C) and the red spots on the callus and lateral lobes of the lip (Supplemental Fig. S2B), respectively. In Phalaenopsis spp. OX Brother Seamate ‘OX1313,’ the expression of PeMYB11 and PeMYB12 was detected in the red lip (Supplemental Fig. S2F), but PeMYB2 expressed in the sepals and petals, which was concomitant with the pink color on the abaxial surfaces of both sepals and petals (Supplemental Fig. S2, D and E). In contrast, PeMYB3, PeMYB4, PeMYB5, PeMYB7, PeMYB8, and PeMYB16 expressed in all three floral organs at different levels (Supplemental Fig. S2F), with little or no expression of PeMYB1, PeMYB6, PeMYB9, PeMYB10, and PeMYB13 to PeMYB15 in these floral organs (Supplemental Fig. S2F).
Figure 3.
qRT-PCR analysis of expression profiles of the structural and regulatory genes of flower color in P. aphrodite ssp. formosana (A), Phalaenopsis spp. OX Brother Seamate ‘OX1313’ (B), and Phalaenopsis spp. OX Red Shoe ‘OX1408’ (C). The expression profiles of the regulatory genes PeMYB2 (D), PeMYB11 (E), and PeMYB12 (F) and structural genes PeF3H5 (G), PeDFR1 (H), and PeANS3 (I) were analyzed in sepals (Ses; white bars), petals (Pes; hatched bars), and lips (Lis; black bars). Data are means ± sem from three technological replicates and three biological samples independently and normalized to those of PeAct4. Bars = 1 cm.
The expression profiles of PeMYB2, PeMYB11, and PeMYB12 were further assessed in P. aphrodite ssp. formosana and Phalaenopsis spp. OX Brother Seamate ‘OX1313’ and compared with the red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408’ by quantitative real-time reverse transcription (qRT) -PCR (Fig. 3). Little or no expression of PeMYB2 and PeMYB12 was detected in any of the floral organs of the white-flower P. aphrodite ssp. formosana, with PeMYB11 slightly expressed in the lip (Fig. 3, A, D, and E), which was associated with the red spots in the callus and lateral lobes of the lip (Supplemental Fig. S2B). For white-flower/red-lip Phalaenopsis spp. OX Brother Seamate ‘OX1313,’ the expression of PeMYB12 was higher than that of both PeMYB2 and PeMYB11 in the lip (Fig. 3, D and E), and low-level expression of PeMYB2 and PeMYB12 in the sepals and petals was associated with the pink color on abaxial surfaces (Fig. 3, D and E; Supplemental Fig. S2, D and E). PeMYB2, PeMYB11, and PeMYB12 were highly expressed in the floral organs of the red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408,’ except that PeMYB2 showed lower expression in the lip (Fig. 3, C–E). Thus, the differential expression of PeMYB2, PeMYB11, and PeMYB12 was concomitant with the red color formation in these three Phalaenopsis spp. with different flower colors, which indicates that they are involved in the regulation of the anthocyanin biosynthesis in Phalaenopsis spp.
Expression of PebHLH1, PebHLH2, PebHLH3, and PeWDR1 Was Not Concomitant with the Red Color Formation
R2R3-MYB transcription factors are known to form complexes with bHLH and WDR factors to regulate anthocyanin biosynthesis. We identified three bHLH transcription factors and one WDR protein from OrchidBase. PebHLH1 was grouped with PhAN1 (Spelt et al., 2000), and PebHLH2 and PebHLH3 were categorized with Z. mays Leaf color (ZmLc; Ludwig et al., 1989) and A. majus DELILA (Goodrich et al., 1992; Supplemental Fig. S3). Real-time RT-PCR results showed that, in white-flower P. aphrodite ssp. formosana, high expression of PebHLH3 was only detected in the lip, whereas PebHLH1 and PebHLH2 expressed slightly in the sepals, petals, and lip (Supplemental Fig. S4). For white-flower/red-lip Phalaenopsis spp. OX Brother Seamate ‘OX1313,’ no differential expression of PebHLH2 and PebHLH3 was detected in these three floral organs, whereas PebHLH1 expressed higher in sepal than petal and lip (Supplemental Fig. S4). In the red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408,’ PebHLH3 was highly expressed in all three floral organs, whereas PebHLH2 expressed similarly in these three floral organs (Supplemental Fig. S4). The expression pattern of PebHLH1 was similar in Phalaenopsis spp. OX Brother Seamate ‘OX1313’ and Phalaenopsis spp. OX Red Shoe ‘OX1408’ (Supplemental Fig. S4). PeWDR1 was chosen for its 87% identity to Arabidopsis AtAN11, which is responsible for anthocyanin accumulation in Arabidopsis (de Vetten et al., 1997). PeWDR1 expressed in all three floral organs of P. aphrodite ssp. formosana and Phalaenopsis spp. OX Brother Seamate ‘OX1313’ (Supplemental Fig. S2F). Together, these results showed that the expression of PebHLH1, PebHLH2, PebHLH3, and PeWDR1 was not concomitant with the red color formation in these three cultivars, which suggests that bHLH transcription factors are not the limiting regulators for the anthocyanin accumulation in Phalaenopsis spp.
Transient Assay of Overexpression of PeMYB2-, PeMYB11-, and PeMYB12-Induced Expression of Their Downstream Genes
To identify the putative downstream genes that may be regulated by these PeMYBs, we identified the structural genes involved in the anthocyanin biosynthetic pathway in OrchidBase. Flavanone 3-hydroxylase5 (PeF3H5), PeDFR1, and Anthocyanidin synthase3 (PeANS3) expressed only in the red lip of Phalaenopsis spp. OX Brother Seamate ‘OX1313’ (Supplemental Fig. S2, A and G). In contrast, we detected no differential expression of PeCHS, PeCHI, and other candidates among various colors of flower organs (Supplemental Fig. S2, A and G). Moreover, PeF3H5, PeDFR1, and PeANS3 were highly expressed in the red lip of Phalaenopsis spp. OX Brother Seamate ‘OX1313’ and all floral organs of Phalaenopsis spp. OX Red Shoe ‘OX1408’ by qRT-PCR assay (Fig. 3, G–I).
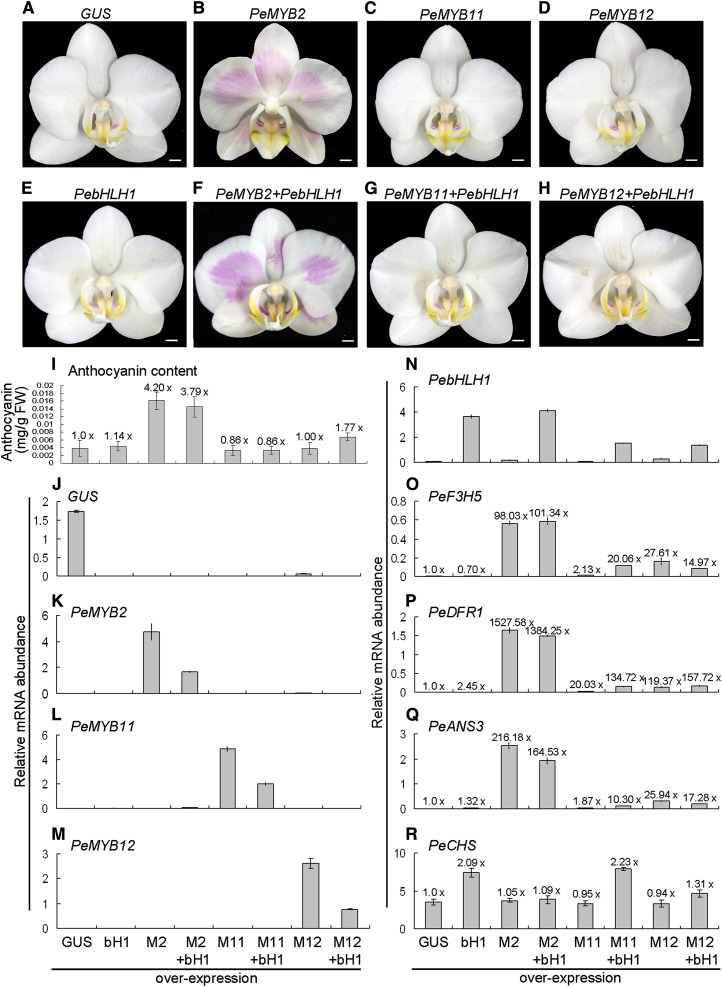
We wondered whether these PeMYBs can activate the expression of the downstream genes PeF3H5, PeDFR1, and PeANS3 and thus, accumulate anthocyanin in Phalaenopsis spp. To test this, Agrobacterium tumefaciens containing various PeMYBs was infiltrated into white sepals/petals of P. aphrodite ssp. formosana. Overexpression of PeMYB2 with or without the addition of PebHLH1 resulted in red pigmentation (Fig. 4, B and F). However, overexpression of PebHLH1, PeMYB11, and PeMYB12 per se did not result in any red color, which was similar to the negative control of GUS overexpression (Fig. 4). We found a 3.79- or 4.20-fold increase in anthocyanin content in PeMYB2-overexpressed flowers with or without the addition of PebHLH1, respectively, compared with the negative control (Fig. 4I). However, we detected low anthocyanin contents in both PeMYB11- and PeMYB12-overexpressed flowers with or without the addition of PebHLH1. PeMYB12 conferred a 1.77-fold increase in anthocyanin content in the presence of PebHLH1 (Fig. 4I), and therefore, PeMYB12 also activated anthocyanin accumulation but to a much lower extent than PeMYB2.
Figure 4.
Anthocyanin production and qRT-PCR analysis of expression profile analysis in the flowers in transient assay of overexpression of GUS (A), PeMYB2 (B), PeMYB11 (C), PeMYB12 (D), and PebHLH1 (E) with or without the addition of PebHLH1 (F–H) in P. aphrodite ssp. formosana by A. tumefaciens infiltration. I to R, Anthocyanin contents and expression profiles in flowers with overexpression of GUS, PebHLH1 (bH1), PeMYB2 (M2), PeMYB2 + PebHLH1 (M2 + bH1), PeMYB11 (M11), PeMYB11 + PebHLH1 (M11 + bH1), PeMYB12 (M12), and PeMYB12 + PebHLH1 (M12 + bH1). The anthocyanin content and expression level detected in the negative control of overexpressed GUS were set to 1×, and those in the other overexpression flowers were calculated as fold change to those of the negative control as shown above each bar. This result was for three technological repeats, and the experiment was performed three times independently. FW, Fresh weight. Bars = 0.5 cm.
GUS, PebHLH1, and PeMYBs were successfully expressed in individual ectopic overexpression plants (Fig. 4, J–N). PebHLH1 expressed highly in plants with overexpression of both PebHLH1 per se and PeMYB2 + PebHLH1 (Fig. 4N). The expression of PeF3H5, PeDFR1, and PeANS3 was up-regulated highly in plants with overexpression of PeMYB2 and PeMYB2 + PebHLH1 for a 98.03- to 1,527-fold increase in expression compared with the overexpressed GUS flower (Fig. 4, O–Q). Therefore, the red color formation was greatly up-regulated by PeMYB2. Interestingly, the expression of these three structural genes was also activated by PeMYB11 with the addition of PebHLH1 and PeMYB12 with or without the addition of PebHLH1, with a 10.30- to 157.72-fold increase in expression (Fig. 4, O–Q). In contrast, we detected little or no up-regulated expression of PeF3H5, PeDFR1, and PeANS3 with PebHLH1 overexpressed alone (Fig. 4, O–Q). PeCHS was constitutively expressed and showed only a 2.23-fold increase in expression with the addition of PebHLH1 to PeMYB11; therefore, PeCHS was not regulated by these transgenes (Fig. 4R). All of these results confirmed that PeMYB2 mainly regulated red pigmentation in Phalaenopsis spp. flower, and all of these PeMYBs could activate the expression of the late anthocyanin biosynthetic genes in Phalaenopsis spp. but to different levels.
Distinct Pigmentation Patterning in the Sepals/Petals within a Single Flower Regulated by Three PeMYBs
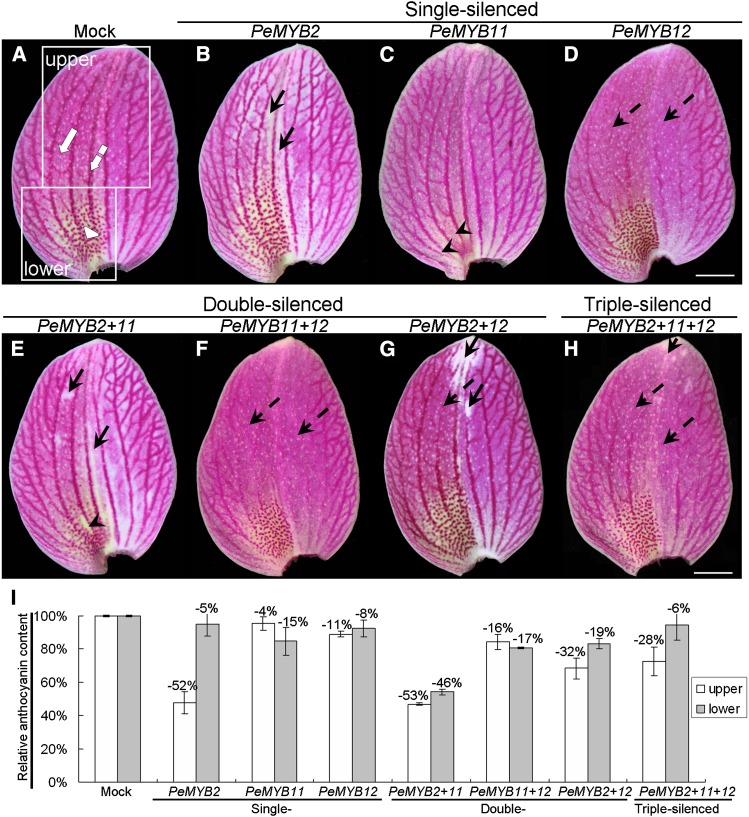
To further validate the in planta roles of three PeMYBs in regulating floral pigmentation patterning in Phalaenopsis spp., we used VIGS strategy with a CymMV-based vector (Lu et al., 2012) in the red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408.’ This plant was chosen, because it has various pigmentation patterning in the sepals/petals of flowers, including the full-red pigment (Fig. 5A, white arrow), venation pattern (Fig. 5A, white dashed arrow), and red spots (Fig. 5A, white arrowhead; Supplemental Fig. S5A). Intriguingly, distinct phenotypic changes on the floral pigmentation patterning resulted from silencing each individual PeMYB. PeMYB2-silenced flowers showed overall reduced pigmentation in the entire sepals but with unaffected venation pattern (Fig. 5B, black arrows). The red spots of PeMYB11-silenced sepals were bleached (Fig. 5C, arrowheads). PeMYB12-silenced sepals showed reduced venation pattern, with no effect on full-red and spot patterns (Fig. 5D, black dashed arrows). Double-silenced PeMYB2+11 flowers showed a similar phenotype as PeMYB2-silenced flowers, with reduced pigment in the full-red sepals, and as PeMYB11-silenced sepals but with a slight change in red spots (Fig. 5, B, C, and E). The sepals of PeMYB11+12-silenced flowers showed almost the same phenotype as that of PeMYB12-silenced flowers, with reduced venation pattern (Fig. 5, D and F). PeMYB2+12-silenced sepals contained bleached regions with loss of full-red and venation patterns (Fig. 5G). Because of the DNA replication competition among three viral vectors, the triple-silenced PeMYB2+11+12 sepals showed weaker phenotype changes than single- or double-silenced sepals, with a reduced venation pattern but unaffected full-red pigmentation and red spots (Fig. 5H). Thus, PeMYB2, PeMYB11, and PeMYB12 may participate in distinct pigmentation patterning in the sepals of a single flower: PeMYB2 for full-red pigmentation, PeMYB11 for red-spot formation, and PeMYB12 for the venation pattern.
Figure 5.
The sepal phenotypes of Phalaenopsis spp. OX Red Shoe ‘OX1408’ by VIGS. The adaxial surface of sepals in mock (A); single-silenced PeMYB2 (B), PeMYB11 (C), and PeMYB12 (D); double-silenced PeMYB2+11 (E), PeMYB11+12 (F), and PeMYB2+12 (G); and triple-silenced PeMYB2+11+12 (H) plants. White arrow, white arrowhead, and white dashed arrow indicate full-red, red-spot, and venation patterns, respectively (A). Black arrows indicate the reduced pigmentation in whole sepals (B, E, G, and H). Black arrowheads show the absent red spots (C and E). Black dashed arrows indicate the reduced venation pigmentation (D and F–H). White boxes in A indicate the regions used to analyze anthocyanin contents of the upper (white bars) and lower (gray bars) parts of sepals (I). The anthocyanin content detected in the mock-treated sepals was set to 100%, and that in the silenced sepals was calculated as the percentage relative to that of the mock-treated sepals. Data are means ± sem from the first blooming flower of three plants, and they were repeated two times for VIGS experiments. Bars = 1 cm.
The ratios of the affected flowers to the total blossomed flowers were calculated from two independent VIGS experiments (Table I). The silencing efficiencies of single-silenced PeMYB2, PeMYB11, or PeMYB12 were high: 63.4% to 94.7% in the sepals for each individual pigmentation pattern (Table I). Double and triple silencing of PeMYB2 with PeMYB11 or PeMYB12 resulted in silencing efficiencies of 22.9% to 90.9% with reduced full-red pigmentation (Table I). Cosilencing of PeMYB11 with PeMYB2 showed high silencing efficiency, with bleached red spots only in experiment 1 of PeMYB2+11, although the silencing efficiencies of double and triple silencing of PeMYB11 with PeMYB2 or PeMYB12 were very low (Table I). Moreover, the silencing efficiencies in the reduced venation pattern were always high: 68.8% to 87.5% in the double and triple silencing of PeMYB12 with PeMYB2 or PeMYB11 (Table I).
Table I. Ratios of features of plant organs affected by PeMYB2, PeMYB11, and PeMYB12 silencing in Phalaenopsis spp. OX Red Shoe ‘OX1408’.
| Treatments/ Replicationsa | Flowers with Color Changed |
||||||
|---|---|---|---|---|---|---|---|
| Sepal |
Petal |
Lip | |||||
| Full Red | Spot | Venation | Full Red | Spot | Venation | ||
| PeMYB2 | |||||||
| Experiment 1 | 25/34b (73.5)c | —d | — | 21/34 (61.8) | — | — | — |
| Experiment 2 | 36/38 (94.7) | — | — | 24/38 (63.2) | — | — | — |
| PeMYB11 | |||||||
| Experiment 1 | — | 33/38 (86.8) | — | — | 0e/38 (0) | — | — |
| Experiment 2 | — | 25/33 (75.8) | — | — | 0/33 (0) | — | — |
| PeMYB12 | |||||||
| Experiment 1 | — | — | 26/41 (63.4) | — | — | 25/41 (61.0) | 31/41 (75.6) |
| Experiment 2 | — | — | 30/39 (76.9) | — | — | 28/39 (71.8) | 30/39 (76.9) |
| PeMYB2+11 | |||||||
| Experiment 1 | 30/33 (90.9) | 30/33 (90.9) | — | 26/33 (78.8) | 0/33 (0) | — | — |
| Experiment 2 | 20/38 (52.6) | 1/38 (2.6) | — | 15/38 (39.5) | 0/38 (0) | — | — |
| PeMYB11+12 | |||||||
| Experiment 1 | — | 1/38 (2.6) | 28/38 (73.7) | — | 0/38 (0) | 30/38 (78.9) | 26/38 (68.4) |
| Experiment 2 | — | 0/32 (0) | 22/32 (68.8) | — | 0/32 (0) | 17/32 (53.1) | 12/32 (37.5) |
| PeMYB2+12 | |||||||
| Experiment 1 | 35/39 (89.7) | — | 30/39 (76.9) | 21/39 (53.8) | — | 25/39 (64.1) | 27/39 (69.2) |
| Experiment 2 | 30/40 (75.0) | — | 35/40 (87.5) | 15/40 (37.5) | — | 29/40 (72.5) | 27/40 (67.5) |
| PeMYB2+11+12 | |||||||
| Experiment 1 | 18/36 (50.0) | 0/36 (0) | 28/36 (77.8) | 15/36 (41.7) | 0/36 (0) | 27/36 (75.0) | 30/36 (83.3) |
| Experiment 2 | 8/35 (22.9) | 0/35 (0) | 25/35 (71.4) | 0/35 (0) | 0/35 (0) | 24/35 (68.6) | 28/35 (80.0) |
Replications of experiment per treatment.
Number of affected flowers to total number of blossomed flowers in five treated plants.
The ratio of affected flowers to total flowers (%).
The dashes indicate that the features should not be changed by this treatment.
The red spots in petal are not as obvious as in sepal.
Anthocyanin content was measured from the upper and lower parts of the mock-treated and PeMYB-silenced sepals. The upper part showed the full-red color, with venation patterns at the peripherial region, and the lower part showed red spots at the base in addition to the full-red and venation patterns (Fig. 5A, boxes labeled upper and lower). PeMYB2-silenced sepals contained reduced anthocyanin content in the upper part, with a 52% decrease compared with that of the mock-treated sepals (Fig. 5I), which reflects PeMYB2’s role in full-red pigmentation in the upper part of sepals. The sepals of PeMYB11-silenced plants with no red spots showed a 15% decrease in anthocyanin content in the lower part, which was associated with the lack of red spots. PeMYB12-silenced sepals contained slightly reduced anthocyanin content in the upper and lower parts, with 11% and 8% decrease, respectively; therefore, the venation pattern extended to the lower part of sepals. Double-silenced PeMYB2+11 sepals showed highly reduced anthocyanin content in both upper and lower parts, with 53% and 46% decreases, respectively, in content. The double-silenced PeMYB11+12 and PeMYB2+12 and triple-silenced PeMYB2+11+12 sepals showed reduced content from 6% to 32% in the upper and lower parts of sepals. Together, these results suggest that the absent pigmentation patterns in VIGS sepals resulted from the decreased anthocyanin content.
The petals of Phalaenopsis spp. have a similar micromorphological structure as the sepals (Hsieh et al., 2013a, 2013b; Pan et al., 2014). Similarly, the VIGS phenotypes of petals resembled those of sepals for changed pigmentation patterning on silencing of these PeMYBs, although much weaker (Supplemental Fig. S5).
Distinct Pigmentation Patterning in the Lip Regulated by PeMYB11 and PeMYB12
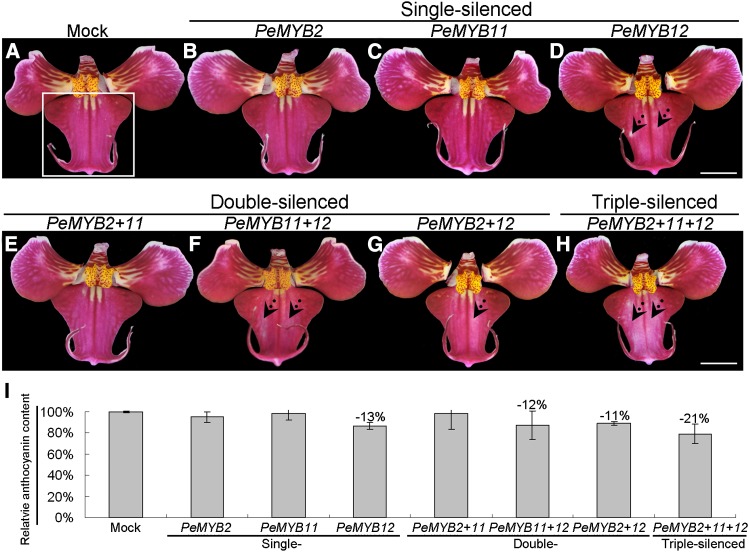
Although the Phalaenopsis spp. lip is a modified petal, its micromorphological structure is distinct from that of petals (Hsieh et al., 2013b; Pan et al., 2014). In the VIGS experiment, only PeMYB12-silenced lips showed reduced red color in the central lobe compared with the mock-treated and single-silenced PeMYB2 and PeMYB11 flowers (Fig. 6, A–D). These results suggest that PeMYB12 was the major regulator of lip pigmentation, especially for the central lobe. The lips of double-silenced PeMYB11+12 and PeMYB2+12 and triple-silenced PeMYB2+11+12 flowers showed bleached areas in the central lobe, whereas the double-silenced PeMYB2+11 flowers did not (Fig. 6, E–H). Interestingly, the bleached spots were most apparent in the triple-silenced PeMYB2+11+12 flowers (Fig. 6H). We compared the anthocyanin content of the central lobes of lips in mock-treated (Fig. 6A, white box) and silenced plants. The lips of silenced flowers showed slightly decreased anthocyanin content (11%–21% reduction) compared with mock-treated flowers (Fig. 6I). However, because the two wings in the central lobes were not affected, they may level off the bleaching effect on anthocyanin content in these silenced plants.
Figure 6.
The lip phenotypes of Phalaenopsis spp. OX Red Shoe ‘OX1408’ by VIGS. The lips of mock (A); single-silenced PeMYB2 (B), PeMYB11 (C), and PeMYB12 (D); double-silenced PeMYB2+11 (E), PeMYB11+12 (F), and PeMYB2+12 (G); and triple-silenced PeMYB2+11+12 (H) plants. Dotted arrows indicate the reduced pigmentation region in the central lobe of the lip (D and F–H). The white box in A indicates the central lobes used to analyze anthocyanin content in I. The anthocyanin content detected in the mock-treated lip was set to 100%, and that in the silenced lips was calculated as the percentage relative to that of the mock-treated lip. Data are means ± sem from the first blooming flower of three plants, and they were repeated two times for VIGS experiments. Bars = 1 cm.
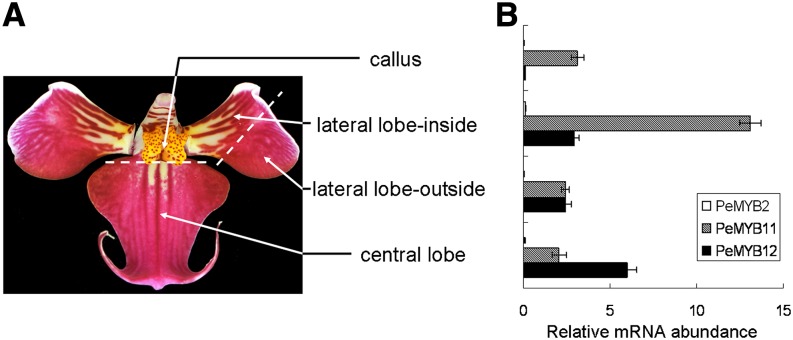
We further analyzed whether these PeMYBs also regulated the distinct pigmentation patterning in the lip. For this, the lip of Phalaenopsis spp. OX Red Shoe ‘OX1408’ was analyzed by dissecting it into four parts, including red spots on the callus, red stripes on the lateral lobe inside, and red pigmentation on the lateral lobe outside and central lobe (Fig. 7A). The expression of PeMYB11 was high in the callus and lateral lobe inside (Fig. 7B). This result was consistent with only PeMYB11 expression in the red spots in the callus and lateral lobe of the lip in P. aphrodite ssp. formosana (Fig. 3; Supplemental Fig. S2B); the red stripes on the lateral lobe inside of lip of Phalaenopsis spp. OX Red Shoe ‘OX1408’ may suggest the fusion of red spots. In contrast, the expression of PeMYB12 was high in the central lobe (Fig. 7B). Both PeMYB11 and PeMYB12 may participate in the distinct pigmentation patterning in the lip: PeMYB11 for the red spots in the callus and PeMYB12 for the red pigmentation in the central lobe of the lip.
Figure 7.
Expression profiles of PeMYB11 and PeMYB12 in the four parts of a dissected lip of Phalaenopsis spp. OX Red Shoe ‘OX1408’: callus, lateral lobe outside, lateral lobe inside, and central lobe (A). B, qRT-PCR analysis of expression profiles of PeMYB2 (white bars), PeMYB11 (hatched bars), and PeMYB12 (black bars). Data are means ± sem from three technological replicates and three biological samples independently and normalized to that of PeAct4.
Cytological Characterization of Various Pigmentation Patterns in the Sepals of Phalaenopsis spp. OX Red Shoe ‘OX1408’
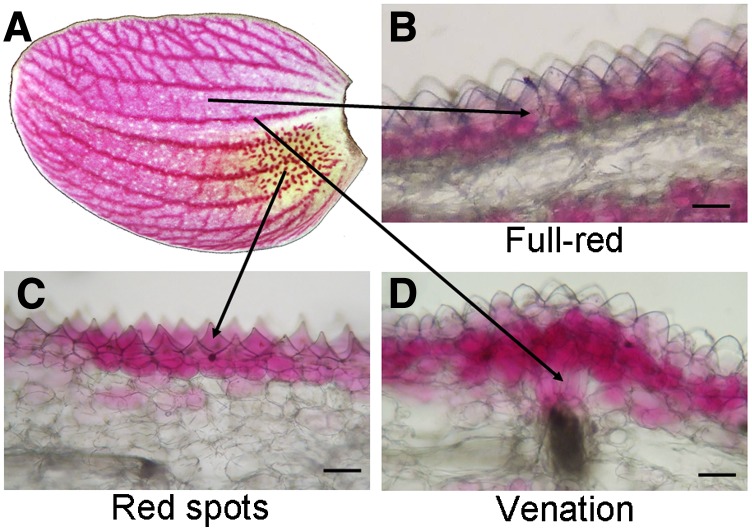
To study the distribution of anthocyanin production in cells with three pigmentation patterns, we analyzed cross sections of the sepal from Phalaenopsis spp. OX Red Shoe ‘OX1408’ (Fig. 8A). The full-red pigmentation showed anthocyanin concentrated in the outer layer of subepidermal cells (Fig. 8B). The red spots showed anthocyanin accumulated in epidermal cells (Fig. 8C). The venation pattern revealed anthocyanin content in the region from subepidermal cells to the xylem (Fig. 8D). The various pigmentation patterns regulated by PeMYB2, PeMYB11, and PeMYB12 in full-red, red-spot, and venation patterns, respectively, may result from their expression inducing anthocyanin accumulation in different cell layers of floral organs.
Figure 8.
Cross section of the sepal of Phalaenopsis spp. OX Red Shoe ‘OX1408’ showing the distribution of anthocyanin production in full-red, red-spot, and venation patterns. The sepal phenotype (A) and the locations in cross sections of full-red (B), red-spot (C), and venation (D) patterns. Bars = 0.1 mm.
RNA in Situ Hybridization Reveals the Distinct Expression Profiles of PeMYBs Associated with Their Pigmentation Patterns
To test whether the expression of these three PeMYBs is associated with their distinct pigmentation patterns, we used RNA in situ hybridization. We assessed the expression profiles of all three PeMYBs in various floral stages and three floral organs—sepals, petals, and lip—of the red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408’ (Supplemental Fig. S6, A–C). These genes showed distinct expression patterns in sepals/petals and lip. In sepals/petals, the expression of PeMYB2 was high at stages 1 and 2 (Supplemental Fig. S6, D and E), and PeMYB11 and PeMYB12 were highly expressed at stage 4 (Supplemental Fig. S6, D and E). In the lip, the expression of PeMYB11 and PeMYB12 was high at stage 4 (Supplemental Fig. S6F), which agreed with the previous findings. PeMYB2 was not expressed in the lip at any stages (Supplemental Fig. S6F). Therefore, we used stage 3 floral buds of Phalaenopsis spp. OX Red Shoe ‘OX1408’ for RNA in situ hybridization to further analyze the expression profiles of three PeMYBs and PebHLH1.
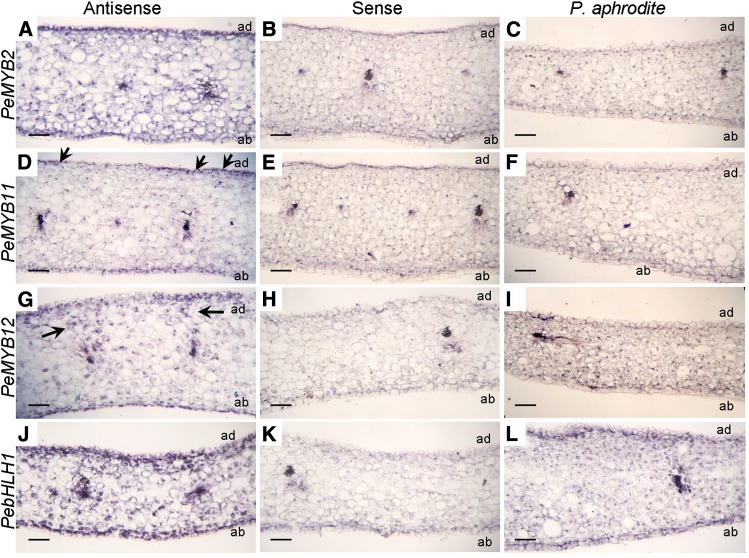
The expression of PeMYB2 was detected in the subepidermal cells of both adaxial and abaxial surfaces of sepals (Fig. 9A). PeMYB11 was detected in the cell clusters of the adaxial surface (Fig. 9D, black arrows), which reflected its red-spot pattern. PeMYB12 was expressed in the range of cells between the xylem and epidermal cells (Fig. 9G, black arrows). Therefore, PeMYB2, PeMYB11, and PeMYB12 showed differential expression profiles for pigmentation patterns on in situ hybridization. Moreover, the expression of PebHLH1 was detected in subepidermal cells (Fig. 9J). As a negative control, we used hybridization with sense probes (Fig. 9, B, E, H, and K). As another negative control, the white-flower P. aphrodite ssp. formosana showed lack of expression of PeMYB2, PeMYB11, and PeMYB12 but high expression of PebHLH1 (Supplemental Fig. S2). RNA in situ hybridization revealed transcripts of PebHLH1 in subepidermal cells but not PeMYBs (Fig. 9, C, F, I, and L).
Figure 9.
RNA in situ hybridization analysis of PeMYB2, PeMYB11, PeMYB12, and PebHLH1 transcripts in the stage 3 sepals of Phalaenopsis spp. Transverse sections of the sepals of Phalaenopsis spp. OX Red Shoe ‘OX1408’ were hybridized with antisense or sense RNA probes of PeMYB2 (A and B), PeMYB11 (D and E), PeMYB12 (G and H), and PebHLH1 (J and K). Transverse sections of the sepals of P. aphrodite ssp. formosana were hybridized with antisense probes of PeMYB2 (C), PeMYB11 (F), PeMYB12 (I), and PebHLH1 (L). Black arrows indicate the detected signals. ab, Abaxial surface; ad, adaxial surface. Bars = 0.1 mm.
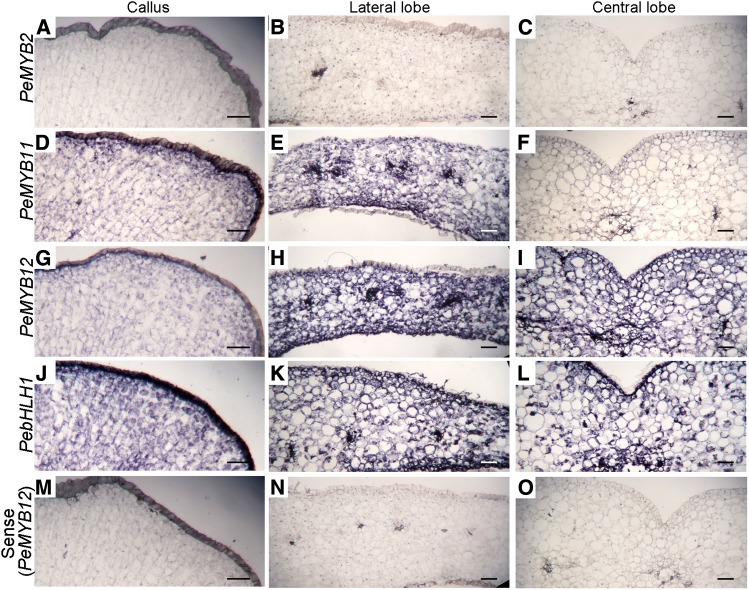
In addition, we used RNA in situ hybridization to examine the expression of PeMYBs and PebHLH1 in the lip of Phalaenopsis spp. OX Red Shoe ‘OX1408.’ PeMYB11 was expressed in the lateral lobe and callus but not the central lobe (Fig. 10, D–F). We detected the expression of PeMYB12 in the lateral and central lobes (Fig. 10, G–I). PebHLH1 (Fig. 10, J–L) but not PeMYB2 (Fig. 10, A–C) was detected in the lip. As a negative control, hybridization with the sense probe of PeMYB12 in the lateral and central lobes and callus revealed no signals (Fig. 10, M–O). All of these results confirmed that PeMYB11 and PeMYB12 may participate in the distinct pigmentation patterning in the lip.
Figure 10.
RNA in situ hybridization analysis of PeMYB2, PeMYB11, PeMYB12, and PebHLH1 transcripts in the stage 3 lip of Phalaenopsis spp. OX Red Shoe ‘OX1408.’ Transverse sections of the lips were hybridized with antisense RNA probes of PeMYB2 (A–C), PeMYB11 (D–F), PeMYB12 (G–I), and PebHLH1 (J–L) and sense probes of PeMYB12 (M–O) in the callus and lateral and central lobes. Bars = 0.1 mm.
Confirmation of Distinct Expression of PeMYBs in Various Cultivars with Different Color Patterns
We further verified the distinct pigmentation patterning in the sepals/petals regulated by three PeMYBs by analyzing the expression profiles in the sepals/petals of six Phalaenopsis spp. cultivars with different color patterns. The flowers of Phalaenopsis spp. Luchia Pink ‘KHM1078’ and Phalaenopsis spp. OX Firebird ‘OX1527’ contain full-red pigmentation with different intensities (Fig. 11, A and B). The expression of PeMYB2 was high in Phalaenopsis spp. Luchia Pink ‘KHM1078’ and Phalaenopsis spp. OX Firebird ‘OX1527’ (Fig. 11, C and D) associated with their full-red pigmentation. Phalaenopsis spp. I-Hsin White Tiger ‘KHM2215’ and Phalaenopsis spp. I-Hsin Sesame ‘OX1224’ contain red spots (Fig. 11, E and F). We detected a combination of moderate to high expression of PeMYB2 and PeMYB11 in Phalaenopsis spp. I-Hsin White Tiger ‘KHM2215’ and Phalaenopsis spp. I-Hsin Sesame ‘OX1224’ (Fig. 11, G and H), which indicates the role of PeMYB11 in red-spot formation. Phalaenopsis spp. I-Hsin Spot Leopard ‘KHM1499’ and Phalaenopsis spp. Leopard Prince ‘KHM2267’ contain the venation pattern with red spots on the base of the sepals (Fig. 11, I and J). We detected a combination of PeMYB11 and PeMYB12, with higher expression of PeMYB12, associated with their major venation pattern and some red-spot formation at the base of sepals (Fig. 11, K and L). Overall, the expression profiles of PeMYB2, PeMYB11, and PeMYB12 were concomitant with distinct floral pigmentation patterns in these six cultivars (Fig. 11). These results suggest that the combinational expression of these three PeMYBs contributes a wealth of natural variation in floral color patterns of Phalaenopsis spp. with different pigmentation intensities and patterning.
Figure 11.
The expression profiles of PeMYB2, PeMYB11, and PeMYB12 in the sepals and petals of six Phalaenopsis spp. cultivars with one of three pigmentation patterns. Expression of PeMYB2 (M2; white bars), PeMYB11 (M11; hatched bars), and PeMYB12 (M12; black bars) was detected in Phalaenopsis spp. Luchia Pink ‘KHM1078’ (A and C), Phalaenopsis spp. OX Firebird ‘OX1527’ (B and D), P. I-Hsin White Tiger ‘KHM2215’ (E and G), Phalaenopsis spp. I-Hsin Sesame ‘OX1224’ (F and H), Phalaenopsis spp. I-Hsin Spot Leopard ‘KHM1499’ (I and K), and Phalaenopsis spp. Leopard Prince ‘KHM2267’ (J and L). Data are means ± sem from three technological replicates and three biological samples independently and normalized to that of PeAct4. Bars = 1 cm.
DISCUSSION
Phalaenopsis spp. are popular ornamental plants worldwide for their long-lived flowers with various flower colors and pigmentation patterning that may be linked to their high interactions with their pollinators or artificial selection. However, the molecular mechanism of anthocyanin biosynthesis and pigmentation patterning for Phalaenopsis spp. flowers has not been well studied. Our results suggest that PeMYB2, PeMYB11, and PeMYB12 activate the expression of anthocyanin biosynthetic genes PeF3H5, PeDFR1, and PeANS3 and result in the red pigmentation in Phalaenopsis spp. flowers. Interestingly, PeMYB2, PeMYB11, and PeMYB12 participated in the regulation of distinct floral pigmentation patterning, which contributed to the three differential patterns present in a single flower. In addition, different pigmentation patterns were regulated by these PeMYBs between the sepals/petals and lip, which suggests two sets of regulatory mechanisms related to the morphologic features of Phalaenopsis spp. flowers. The combined expression of different PeMYBs resulted in a wealth of various floral pigmentation patterning in Phalaenopsis spp.
Distinct Pigmentation Patterning Revealed by VIGS and RNA in Situ Hybridization Assays
The abundant Phalaenopsis spp. species and cultivars provide good materials for studying the regulatory mechanism of floral pigmentation patterning and other flower features (Hsiao et al., 2011). The VIGS approach with a modified CymMV-based vector was useful for screening the in planta functions of most genes for functional genomics in Phalaenopsis spp. (Hsieh et al., 2013b), which is of benefit for breeding these plants with long lifecycles. In this study, we used VIGS to screen the in planta functions of PeMYBs and verified their roles in the distinct pigmentation patterning in Phalaenopsis spp. flowers by using RNA in situ hybridization assays. Although VIGS data were not perfect for revealing the real gene functions, they provided a quick approach for gene functions in nonmodel plants. However, four small defects were present for VIGS experiments in this study as well as previous studies (Hsieh et al., 2013b). First, the changed phenotype was most severe in the first four blooming flowers and became weakened with each consecutive flower. The temporal expression of genes was during floral development, and the first four blooming flowers were at the inflorescence stage, which is more sensitive to loss of function of PeMYBs. A similar observation was reported previously (Hsieh et al., 2013b). Second, the changes appeared only as a mosaic phenotype; therefore, only the regions along the central veins of the sepals became bleached, but the other regions were rarely affected, which suggests the route of viral spread through vascular bundles. Third, the sepals and petals of flowers were similar in morphologic aspects, but the phenotype changes were more severe in sepals than petals, despite similar gene expression in these two floral organs. Previously, silencing of a B-class MADS-box gene, PeMADS6, caused sepals with reduced size and leaf-like green areas along the central veins, whereas petals showed only reduced cell size (Hsieh et al., 2013a). Fourth, compared with single-silenced plants, double- and triple-silenced plants showed reduced phenotypic changes. This finding is because of the competition of viral replication as previously reported (Hsieh et al., 2013a, 2013b; Pan et al., 2014). Overall, despite the fact that the patchy nature of VIGS is not easy to overcome, it is still a useful approach to study the regulation of floral pigmentation patterning, especially for plants with complicated color patterns and long lifecycles, such as Phalaenopsis spp.
Distinct Pigmentation Patterning Was Regulated by R2R3-MYB Transcription Factors in Antirrhinum spp., Petunia spp., and Phalaenopsis spp.
R2R3-MYB transcription factors play a major role in determining the pattern and intensity of floral pigmentation in Antirrhinum spp. and Petunia spp. (Schwinn et al., 2006; Albert et al., 2011). Ros1 contributes to the strong full-red corolla pigmentation in A. majus (Schwinn et al., 2006). With loss of the Ros1 function, Ros2 affects the weak pigmentation in the inner epidermis of the corolla lobes, and Venosa results in the epidermal-specific venation pattern (Schwinn et al., 2006; Shang et al., 2011). Venosa transcript is detected in a wedge of cells between the vein and the adaxial epidermis (Shang et al., 2011). Delila, a bHLH factor, expresses in an epidermal-specific pattern in the corolla (Goodrich et al., 1992; Jackson, 1992), whereas the WDR protein is constitutively expressed and may move to neighboring cells (Walker et al., 1999; Bouyer et al., 2008). Thus, the epidermal-specific venation pattern in Antirrhinum spp. is determined by the overlap of the expression domains of the MYB and bHLH factors involved in anthocyanin production (Shang et al., 2011).
In Petunia spp., AN2 controls strong, full-petal limb pigmentation, and AN4 regulates pigments accumulated in the flower tube and anther (Quattrocchio et al., 1998, 1999). In Petunia spp. lines without AN2 and AN4 function, light-regulated pigmentation on the abaxial petal surface of flower buds (bud-blush pattern) is regulated by PURPLE HAZE (PHZ), and the venation pattern in the flower tube is controlled by DEEP PURPLE (DPL; Albert et al., 2011). All AN2, AN4, PHZ, and DPL share common bHLH (AN1) and WDR (AN11) factors (Albert et al., 2011).
In this study, we showed that three R2R3-MYB transcription factors (PeMYB2, PeMYB11, and PeMYB12) participated in the distinct pigmentation patterns in Phalaenopsis spp. flowers. These three PeMYBs seemed to interact with endogenous, common bHLH and WDR factors, because the expression of PebHLH1, PebHLH2, PebHLH3, and PeWDR1 was either not concomitant with the red-color formation or not specific to distinct pigmentation patterns.
In addition, for both Antirrhinum spp. and Petunia spp., Ros1 and AN2 regulate the strong, full-red flower color, and the other pigmentation patterns only appeared when Ros1 or AN2 is loss of function. In contrast, in Phalaenopsis spp., these pigmentation patterns regulated by three PeMYBs were concomitantly present in one single flower of Phalaenopsis spp. OX Red Shoe ‘OX1408.’ The expression of PeMYB2, PeMYB11, and PeMYB12 was majorly detected in accordance with their distinct pigmentation patterns as revealed by in situ hybridization results. It is possible that these three PeMYBs not only regulate the main patterns but also, partially participate in the regulation of the other patterns. This possibility may explain why the boundary between each pigmentation pattern was difficult to define, such as the full-red and venation pigmentation or the venation pattern extending to the red spots at the central region of the sepals. Furthermore, the expression profiles of PeMYBs overlapped, such as both PeMYB2 and PeMYB12 expressed in the subepidermal cells above the vein. Therefore, the ratio of the expression of PeMYB2, PeMYB11, and PeMYB12 may contribute to various pigmentation patterns appearing in one single flower.
MYB Transcription Factors Interacted with bHLH Factors for Their Functions
In Orchidaceae, PeMYB2, PeMYB11, and PeMYB12 from P. equestris, PsMYB from purple-flower P. schilleriana, and OgMYB1 from Oncidium spp. ‘Gower Ramsey’ all harbor the [D/E]Lx2[R/K]x3Lx6Lx3R motif for interacting with bHLH factors. Transient overexpression of both PsMYB and OgMYB1 independently induces anthocyanin accumulation after particle bombardment, but PsMYB needs a bHLH transcription factor, ZmLc, from Z. mays (Ma et al., 2009) but not OgMYB1 (Chiou and Yeh, 2008). Here, overexpression of PeMYB2 and PeMYB12 per se activated the expression of downstream genes either with or without the addition of PebHLH1, whereas PeMYB11 required the addition of PebHLH1 to activate downstream gene expression (Fig. 4). Because other R2R3-MYBs with this motif participate in more physiological functions, such as Arabidopsis Transparent Testa2 (Baudry et al., 2004) and Arabidopsis MYB-like2 (Sawa, 2002) for proanthocyandin biosynthesis and trichrome development, respectively, the presence of this motif suggests that these MYBs require a bHLH partner but not is indicative of the candidate MYBs within the anthocyanin-promoting clade (Lin-Wang et al., 2010). Therefore, we think that the bHLH and WDR are required for anthocyanin biosynthesis, and these three PeMYBs may exert their functions with endogenous WDR and bHLH. In the case of PeMYB2 and PeMYB12, they may perform their functions by interacting with an endogenous bHLH transcription factor (PebHLH1, PebHLH2, or PebHLH3), because these three PebHLHs were expressed at low level in all floral organs of P. aphrodite ssp. formosana (Supplemental Fig. S4). In contrast, PeMYB11 may prefer interacting with PebHLH1 because of the fact that overexpression of PebHLH1 was required for the function of PeMYB11.
The Distinct Pigmentation Patterning Regulated by PeMYB2, PeMYB11, and PeMYB12 Might Be Concomitant with the Morphological Features in Phalaenopsis spp.
PeMYB2 showed the remarkable high transactivational activity for anthocyanin accumulation within transient expression assay, but interestingly, the transcript of PeMYB2 was detected only in the sepals/petals but not lip (Fig. 3), which suggests that the regulatory strategy for anthocyanin production is coordinated with their distinct morphologic features in a single flower. Previous study showed that overexpression of a B-class Torenia fournieri GLOBOSA revealed altered cell shapes on the surfaces of sepals to a petal-like shape, and the sepals accumulated anthocyanin similar to petals of wild-type torenia (Sasaki et al., 2010). In Phalaenopsis spp., the flowers of a down-regulated B-class PISTILLTA-like PeMADS6 showed discolored areas in sepals, petals, and lip (Hsieh et al., 2013a, 2013b). The link between the expression of MADS-box genes and the accumulation of anthocyanin has been reported in Arabidopsis, bilberry (Vaccinium myrtillus), and sweet potato (Ipomoea batatas; Nesi et al., 2002; Debeaujon et al., 2003; Lalusin et al., 2006; Jaakola et al., 2010). Previously, we have shown that PeMADS2, a B-class MADS-box gene, is strongly expressed in the sepals and petals (Tsai et al., 2004). Whether there is any correlation between PeMADS2 and PeMYB2 requires further study.
PeMYB11 participated in the regulation of the red spots in sepals/petals and lip of Phalaenopsis spp. However, red spots were absent only in the sepals/petals of single-silenced PeMYB11 but not double- and triple-silenced PeMYB11 or the lip of PeMYB11-silenced flowers. The red spots were located in the basal region of the sepals/petals and lip, and therefore, the red-spot pattern may be difficult to change during flower development. Previous study reported that the double-silenced flower of C-class PeMADS1 and B-class PeMADS6 showed only the margins of sepals and petals as curly and discolored (Hsieh et al., 2013b), which suggests that the margins of flowers are easier to affect by gene silencing than the basal region. Therefore, only the single-silenced PeMYB11 sepal showed the absence of red spots but not the double- and triple-silenced sepals with decreasing silencing efficiency. Moreover, because of the location and special structure of the callus of the lip, the red spots were not affected in all VIGS flowers.
PeMYB12 participated in the regulation of different pigmentation patterns in various floral organs in a single flower: the venation pattern in the sepals/petals (Figs. 5D and 9G) but the full pigmentation pattern in the central lobe of the lip (Figs. 6D, 7, and 10I). The results of the venation pattern caused by Venosa and DPL in Antirrhinum spp. and Petunia spp., respectively, suggest a common mechanism for venation pigmentation in a wide range of plant species. Several Phalaenopsis spp. cultivars verified the role of PeMYB12 in the venation pattern (Fig. 11, I–L), and confirmation is needed for whether all Phalaenopsis spp. cultivars with the venation pattern also contained the full pigmentation in the central lobe of lip. This differential regulatory mechanism may result from the distinct floral organs causing a diverse morphology between the sepals/petals and lip.
PeMYB2, PeMYB11, and PeMYB12 for Molecular Breeding of Cultivars with Various Color Patterns
We examined the pigmentation patterning of full-red, red-spot, and venation patterns regulated by PeMYB2, PeMYB11, and PeMYB12, respectively, in six Phalaenopsis spp. cultivars. Our findings suggest their involvement in regulating the pigmentation patterning of a single Phalaenopsis spp. flower and that they are required but not sufficient for deciphering the pigmentation pattering in Phalaenopsis spp. flowers. Thus, PeMYB2, PeMYB11, and PeMYB12 could be molecular markers for predicting various pigmentation patterning and choosing suitable parents for molecular breeding in Phalaenopsis spp. A wealth of various Phalaenopsis spp. cultivars facilitates the study of floral pigmentation patterning, which is difficult to investigate in many other plant species.
PeMYB2 Showed High Transactivational Activity for Anthocyanin Biosynthesis
Although PeMYB2, PeMYB11, and PeMYB12 were grouped together in the same clade in the phylogenetic analysis, PeMYB2 showed high transactivational activity for anthocyanin biosynthesis, whereas PeMYB11 and PeMYB12 contained fewer activities. The amino acid sequence of PeMYB2 showed about 80% identity and 41.9% to 43.0% identity to the MYB-R2R3 regions and the full-length complementary DNA (cDNA) of PeMYB11 and PeMYB12, respectively (Supplemental Table S1). Compared with PsMYB and OgMYB1, PeMYB11 was most similar to PsMYB, with 95.2% identity and 85.4% identity to the MYB-R2R3 region and the full-length cDNA, respectively (Supplemental Table S1). However, transient overexpression of PsMYB can induce anthocyanin accumulation after particle bombardment, but PeMYB11 cannot. Therefore, the sequence homologies between PeMYB2, PeMYB11, PeMYB12, PsMYB, and OgMYB1 did not directly reflect their transactivational activity for activating the anthocyanin biosynthetic pathway. Alternatively, it is possible that repressors may be present in P. aphrodite ssp. formosana and negatively regulate the transactivational activities of PeMYB11 and PeMYB12, such as R3-MYB and R2R3-MYB repressors (Albert et al., 2014).
Other PeMYBs May Also Participate in Anthocyanin Accumulation in Phalaenopsis spp.
We isolated 16 R2R3-MYB transcription factors by screening OrchidBase, an Orchidaceae transcriptome database, and characterized three PeMYBs (PeMYB2, PeMYB11, and PeMYB12) in this study. For the other PeMYBs, PeMYB1, PeMYB13, and PeMYB14 were grouped together and categorized into the same large clade with PeMYB2, PeMYB11, PeMYB12, and dicot anthocyanin-related MYBs (Fig. 1). However, we detected no expression of PeMYB1, PeMYB13, and PeMYB14 in P. aphrodite ssp. formosana and Phalaenopsis spp. OX Brother Seamate ‘OX1313’ (Supplemental Fig. S2F). These three genes may express and regulate anthocyanin content in other Phalaenopsis spp. cultivars. In addition, PeMYB4, PeMYB5, PeMYB6, PeMYB7, and PeMYB8 were grouped with AtMYB4 (Jin et al., 2000), FaMYB1 (Aharoni et al., 2001), and other MYBs that are repressors of flavonoid biosynthesis and shared the Ethylene Responsive Factor-associated amphiphilic repression repressive motif (conserved PDLNLELSIS; Aharoni et al., 2001) in their C terminus. PeMYB4, PeMYB5, PeMYB6, PeMYB7, and PeMYB8 may exert a negative regulation on anthocyanin biosynthesis and interact with PeMYB2, PeMYB11, and PeMYB12.
CONCLUSION
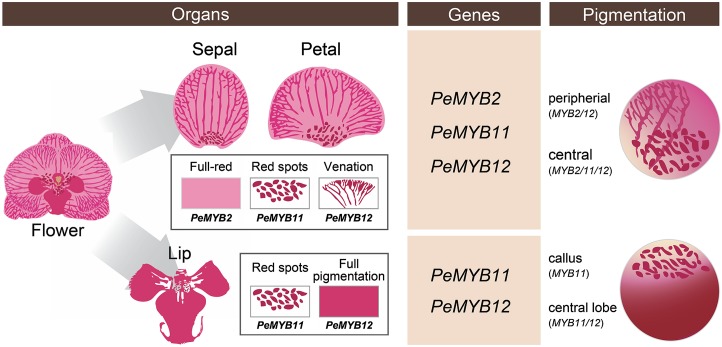
We propose a model of three R2R3-MYB transcription factors (PeMYB2, PeMYB11, and PeMYB12) involved in floral anthocyanin pigmentation patterning in Phalaenopsis spp. (Fig. 12). In the sepals/petals, PeMYB2, PeMYB11, and PeMYB12 regulate full-red pigmentation, red spots, and venation pattern, respectively. Moreover, the regulation of pigmentation patterning by PeMYBs differed in sepals/petals and the lip. PeMYB11 controls the red spots in the callus, and PeMYB12 is the major factor for pigmentation in the central lobe of the lip. We detected the combined expression of two or three PeMYBs resulting in various pigmentation patterns in a single flower. This study will benefit understanding of the genetic basis of the regulatory mechanism for flower color and pigmentation patterning in Phalaenopsis spp. and molecular breeding of Phalaenopsis spp. cultivars with various pigmentation patterns.
Figure 12.
Proposed model for PeMYB2, PeMYB11, and PeMYB12 involved in anthocyanin pigmentation patterning in Phalaenopsis spp. flowers. In the sepals/petals, PeMYB2, PeMYB11, and PeMYB12 regulate full-red pigmentation, red spots, and venation pattern, respectively. Moreover, PeMYB11 controls the red spots in the callus, and PeMYB12 regulates full pigmentation in the central lobe of the lip. The combined expression of two or three PeMYBs results in various pigmentation patterns in Phalaenopsis spp.
MATERIALS AND METHODS
Plant Materials
Phalaenopsis aphrodite ssp. formosana with white sepals/petals and a yellow lip and Phalaenopsis spp. OX Brother Seamate ‘OX1313’ with white sepals/petals and a red lip were used for identifying anthocyanin-related genes. The former was also used for transient gene overexpression analysis, because the white sepals/petals were beneficial for detecting anthocyanin accumulation. A third plant, red-flower Phalaenopsis spp. OX Red Shoe ‘OX1408’ containing various floral pigmentation patterning, was used for VIGS analysis. In addition, six Phalaenopsis spp. cultivars were used for verifying the regulatory roles of PeMYBs on the pigmentation patterning, and they included Phalaenopsis spp. Luchia Pink ‘KHM1078,’ Phalaenopsis spp. OX Firebird ‘OX1527,’ Phalaenopsis spp. I-Hsin White Tiger ‘KHM2215,’ Phalaenopsis spp. I-Hsin Sesame ‘OX1224,’ Phalaenopsis spp. I-Hsin Spot Leopard ‘KHM1499,’ and Phalaenopsis spp. Leopard Prince ‘KHM2267’ (Fig. 9).
All plants were purchased from Taiwan Sugar Corp. and OX Orchid Farm and grown in the greenhouse at National Cheng Kung University under natural light and controlled temperature from 23°C to 27°C.
Identification and Phylogenetic Analysis of PeMYBs
The sequences of 125 R2R3-MYB transcription factors from Arabidopsis (Arabidopsis thaliana) and the anthocyanin-promoting MYBs from other plant species were used to screen the homologous genes in OrchidBase, a collection of transcriptomic sequences from Phalaenopsis spp. cDNA libraries (Fu et al., 2011; Tsai et al., 2013), by using a BLASTX algorithm. Moreover, the degenerated primers designed within the conserved R2R3-MYB domain were also used to amplify MYB transcription factors from the cDNA of floral buds of P. equestris. The R2R3 domain of MYB transcription factors was first aligned by using the default settings in ClustalW and MUSCLE implemented in MEGA version 5.2 (Tamura et al., 2011) and used to construct the phylogenetic tree with the Maximum Likelihood method. We used 1,000 bootstrapping data sets to estimate the confidence of each tree clade. Sequences of R2R3-MYBs from other plants were obtained from GenBank (http://www.ncbi.nlm.nih.gov/), and the accession numbers are as follows: Antirrhinum majus AmROS1 (ABB83826), AmROS2 (ABB83827), and AmVENOSA (ABB83828); apple (Malus domestica) MdMYBA (BAF80582); Capsicum annuum CaMYB (CAE75745); Dendrobium spp. DwMYB1 (AAO49410), DwMYB2 (AAO49411), DwMYB8 (AAO49417), and DwMYB10 (AAO49419); Fragaria ananassa FaMYB1 (AAK84064); Gerbera hybrid GhMYB10 (CAD87010); sweet potato (Ipomoea batatas) IbMYB (BAF45115); Oncidium spp. Gower Ramsey OgMYB1 (ABS58501); Oryza sativa OsMYB (CAA75509); Petunia hybrida PhAN2 (AAF66727); P. schilleriana PsMYB (ACH95792); Solanum lycopersicum SlMYB (AAQ55181); Vitis vinifera VvMYBA1 (BAD18977); and Zea mays ZmC1 (AAA33482) and ZmP (AAA19821). The complete R2R3 region for PeMYB15 could not be amplified by using RACE; therefore, it was excluded from the analysis.
Isolation of Plant RNA and RT to cDNA
For RNA extraction, various floral organs, including sepals, petals, and lip, in 1- to 1.5-cm floral buds were collected. Floral organs of Phalaenopsis spp. OX Red Shoe ‘OX1408’ were divided into five stages from floral buds to blooming stage (stage 1, 0–0.5 cm; stage 2, 0.5–1 cm; stage 3, 1–2 cm; stage 4, 2–3 cm; and stage 5, blooming flower; Supplemental Fig. S5, A–C). Each sample was immersed in liquid nitrogen and stored at −80°C. Total RNA was extracted by the guanidium thiocyanate method (O’Neill et al., 1993), treated with RNase-Free DNase I (New England Biolabs) to remove residual DNA, and reverse transcribed to cDNA by use of SuperScript III (Invitrogen).
5′- and 3′-RACE
The full-length cDNA was obtained by extending the 5′ and 3′ ends of cDNA with use of the SMART RACE cDNA Amplification Kit (Clontech). The full-length sequences were obtained by two rounds of PCR amplifications with primary PCR with a universal primer and the first gene-specific primer. Nested PCR involved the nested universal primer and the second gene-specific primer. The primers are in Supplemental Table S2. The PCR products were cloned into pGEM-T Easy Vector (Promega); 10 to 12 colonies were randomly selected for sequencing.
RT-PCR and qRT-PCR
Primer pairs for each gene within the gene-specific regions were designed and are in Supplemental Table S2. The PCR protocol was initial denaturation at 94°C for 5 min, 30 cycles of amplification (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min), and extension at 72°C for 7 min. PeActin9 of P. equestris was used as an internal control (Pan et al., 2011). The amplified products were separated on 1% (w/v) agarose gel.
For qRT-PCR, the cDNA template was mixed with 2× SYBR Green PCR Master Mix (Applied Biosystems) in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Each sample was analyzed in triplicate. Reactions involved incubation at 95°C for 10 min and thermocycling for 40 cycles (95°C for 15 s and 60°C for 1 min). After amplification, melting curve analysis was used to verify amplicon specificity and primer dimer formation. The housekeeping gene PeActin4 (AY134752) was used for normalization (Chen et al., 2005). Data are means ± sem calculated from three technological replicates and three biological samples independently.
Transient Assay of Overexpression of PeMYBs by Agrobacterium tumefaciens Infiltration
For transient assay, the binary vector pCAMBIA1304 was digested with XbaI and NheI, filled in with the Klenow fragment (New England Biolabs), and then ligated to produce the p1304NhXb vector with a reduced vector size. GUS, PebHLH1, PeMYB2, PeMYB11, and PeMYB12 were amplified, digested with XhoI, and ligated to p1304NhXb to replace the gene hygromycin phosphotransferase to produce the overexpression vectors of these genes driven by the duplicated cauliflower mosaic virus 35S promoter. The recombinant vectors were transformed into A. tumefaciens EHA105 by electroproration. Vector-containing A. tumefaciens was cultured overnight at 28°C. After centrifugation, bacterial cell pellets were resuspended by adding 500 µL of Murishige and Skoog medium containing 100 µm acetosyringone to an optical density at 600 nm of 1 and allowed to stand at room temperature for 0.5 h without shaking before infiltration. The suspensions were injected into the basal regions of the sepals/petals of flowers of P. aphrodite ssp. formosana. The A. tumefaciens-infiltrated plants were incubated at 25°C in an incubator with a 10-h-light/14-h-dark photoperiod for 5 d. After being photographed, the flowers were stored for RNA extraction. The transient assay was performed for five plants in each experiment and repeated for three experiments independently.
VIGS of PeMYBs
VIGS of PeMYB2, PeMYB11, and PeMYB12 was performed in Phalaenopsis spp. OX Red Shoe ‘OX1408’ containing various floral pigmentation patterning as described (Hsieh et al., 2013a, 2013b). The sequences located downstream from the MYB-R2R3 region to the stop codon were selected, with 435-, 328-, and 311-bp fragments for PeMYB2, PeMYB11, and PeMYB12, respectively. The primers are in Supplemental Table S2. In addition to single silencing, we combined two or three vectors to perform the double silencing of PeMYB2+11, PeMYB2+12, and PeMYB11+12 and triple silencing of PeMYB2+11+12. Mock-treated plants were injected with an empty plasmid of Cymbidium mosaic virus with a Gateway system vector as a negative control to confirm that any flower color changes were not caused by the viral infection. For each VIGS experiment, we used a B-class MADS-box gene (PeMADS6) as a positive control because of its silencing phenotypes, such as leaf-like characteristics and inability to fully expand (Hsieh et al., 2013a). Each treatment involved five plants and was repeated for two VIGS experiments independently.
Determination of Anthocyanin Content
Anthocyanin content was quantified as previously described (Hsieh et al., 2013a). Samples were collected from the first blooming flower of six plants that showed VIGS phenotypes for each treatment and ground in liquid nitrogen. The ground powder was extracted with methanol containing 5% (v/v) trifluoroacetic acid at 4°C for 20 h and centrifuged at 10,000g for 20 min at 4°C. The absorbance of supernatants was measured for anthocyanin content with use of a spectrophotometer at 530 nm (Agilent 8453; Agilent Technologies). The anthocyanin content was calculated as the average of three measurements from each flower, with pure anthocyanin extracted from Phalaenopsis spp. used as an internal standard (provided by Ping-Chung Kuo, Department of Biotechnology, National Formosa University, Yunlin, Taiwan). Relative anthocyanin content was calculated as the percentage of the anthocyanin content from silenced plants to that from mock-treated plant. Data were means ± sem from the first blooming flower of three plants and repeated for two VIGS experiments.
RNA in Situ Hybridization
RNA in situ hybridization was performed as previously described (Pan et al., 2011). The stage 3 floral organs of Phalaenopsis spp. OX Red Shoe ‘OX1408’ were fixed in fixation buffer (4% [w/v] paraformaldehyde and 0.5% [v/v] glutaraldehyde) at 4°C for 16 to 24 h. The floral organs were dehydrated through a graded ethanol series, embedded in Histoplast, and sectioned at 10 mm with use of a rotary microtome (MICROM, HM 310; Walldorf). Tissue sections were deparaffinized with xylene, rehydrated through an ethanol series, and pretreated with proteinase K (1 µg mL−1) in 1× PBS at 37°C for 30 min. Prehybridization and hybridization followed previous protocols with more stringent wash conditions: two times of 1× SSC at 45°C for 20 min and two times of 0.5× SSC at 42°C for 15 min (Tsai et al., 2005). DNA substrates containing a partial C-terminal region and the 30-untranslated region were used for riboprobe synthesis. PCR products amplified with primers are in Supplemental Table S2. Each sequence of these PCR products showed no significant similarity found against the other two PeMYBs by using BLAST-2 sequences. The resulting PCR fragments were used as templates for synthesis of antisense and sense riboprobes with digoxigenin (DIG)-labeled UTP-DIG (Roche Applied Science) and the T7/SP6 Riboprobe In Vitro Transcription System (Promega) following the manufacturer’s instructions. The RNA in situ hybridization was performed for three hybridized samples and repeated for three experiments independently.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: PeF3H5 (KF769464), PeDFR1 (KF769462), PeANS3 (KF769463), PeMYB1 to PeMYB16 (KF769466–KF769481), and PebHLH1 (KF769482).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Examples of Phalaenopsis spp. cultivars with various pigmentation patterns.
Supplemental Figure S2. Expression profiles of the structural and regulatory genes of flower color in P. aphrodite ssp. formosana and Phalaenopsis spp. OX Brother Seamate ‘OX1313.’
Supplemental Figure S3. Phylogenetic analysis of PebHLH1, PebHLH2, and PebHLH3 with anthocyanin-related bHLH transcription factors.
Supplemental Figure S4. qRT-PCR analysis of expression profiles of the bHLH transcription factors PebHLH1, PebHLH2, and PebHLH3 in P. aphrodite ssp. formosana, Phalaenopsis spp. OX Brother Seamate ‘OX1313,’ and Phalaenopsis spp. OX Red Shoe ‘OX1408.’
Supplemental Figure S5. The petal phenotypes of plants with VIGS.
Supplemental Figure S6. qRT-PCR analysis of spatial and temporal expression of three PeMYBs in Phalaenopsis spp. OX Red Shoe ‘OX1408.’
Supplemental Table S1. Protein sequence identities between PeMYB2, PeMYB11, PeMYB12, PsMYB, and OgMYB1.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Chi-Kuang Wen (National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China), Drs. Tuan-Hua David Ho and Na-Sheng Lin (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan), and Drs. Kai-Wun Yeh and Keqiang Wu (Institute of Plant Biology, National Taiwan University, Taiwan) for helpful discussion of this article; Dr. Zhao-Jun Pan (Department of Life Science, National Taiwan University) for the art work in Fig. 12; and Dr. Ming-Hsien Hsieh (Tainan District Agricultural Research and Extension Station, Council of Agriculture, Taiwan) for assistance with VIGS experiments.
Glossary
- cDNA
complementary DNA
- CymMV
Cymbidium mosaic virus
- qRT
quantitative real-time reverse transcription
- RT
reverse transcription
- VIGS
virus-induced gene silencing
Footnotes
This work was supported by the Ministry of Science and Technology, Taiwan (grant no. NSC 103–2321–B–006–002).
Articles can be viewed without a subscription.
References
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65: 771–784 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hülskamp M (2008) Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Hsu CY, Cheng HY, Chang H, Chen HH, Ger MJ (2011) Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep 30: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Chen YH, Tsai YJ, Huang JZ, Chen FC (2005) Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Res 15: 639–657 [DOI] [PubMed] [Google Scholar]
- Chiou CY, Yeh KW (2008) Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol Biol 66: 379–388 [DOI] [PubMed] [Google Scholar]
- Davies KM, Albert NW, Schwinn KE (2012) From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct Plant Biol 39: 619–638 [DOI] [PubMed] [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11: 1422–1434 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, Tsai WC, Chen HH (2011) OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol 52: 238–243 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964 [DOI] [PubMed] [Google Scholar]
- Griesbach RJ, Beck RM, Hammond J (2007) Gene expression in the star mutation of Petunia x hybrida Vilm. J Am Soc Hortic Sci 132: 680–690 [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Heuschen B, Gumbert A, Lunau K (2005) A generalised mimicry system involving angiosperm flower colour, pollen and bumblebees’ innate colour preferences. Plant Syst Evol 252: 121–137 [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Hsiao YY, Pan ZJ, Hsu CC, Yang YP, Hsu YC, Chuang YC, Shih HH, Chen WH, Tsai WC, Chen HH (2011) Research on orchid biology and biotechnology. Plant Cell Physiol 52: 1467–1486 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Lu HC, Pan ZJ, Yeh HH, Wang SS, Chen WH, Chen HH (2013a) Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci 201-202: 25–41 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Pan ZJ, Lai PH, Lu HC, Yeh HH, Hsu CC, Wu WL, Chung MC, Wang SS, Chen WH, et al. (2013b) Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J Exp Bot 64: 3869–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Hoshino A, Johzuka-Hisatomi Y, Habu Y, Inagaki Y (1999) Floricultural traits and transposable elements in the Japanese and common morning glories. Ann N Y Acad Sci 870: 265–274 [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S (1994) Isolation of a Suppressor-Mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. Plant Cell 6: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Higeta D, Suzuki A, Yoshida H, Ozeki Y (2002) Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation (Dianthus caryophyllus). Plant Cell Physiol 43: 578–585 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al. (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Roberts K, Martin C (1992) Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinum majus. Plant J 2: 425–434 [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A (2005) The star-type color pattern in Petunia hybrida ‘red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol 46: 1879–1883 [DOI] [PubMed] [Google Scholar]
- Lalusin AG, Nishita K, Kim SH, Ohta M, Fujimura T (2006) A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Mol Genet Genomics 275: 44–54 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Sheng CC, Lasky JR, Willis JH (2012) Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am J Bot 99: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang DC, Yeh HH (2007) Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol 143: 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HC, Hsieh MH, Chen CE, Chen HH, Wang HI, Yeh HH (2012) A high-throughput virus-induced gene-silencing vector for screening transcription factors in virus-induced plant defense response in orchid. Mol Plant Microbe Interact 25: 738–746 [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86: 7092–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunau K, Fieselmann G, Heuschen B, van de Loo A (2006) Visual targeting of components of floral colour patterns in flower-naïve bumblebees (Bombus terrestris; Apidae). Naturwissenschaften 93: 325–328 [DOI] [PubMed] [Google Scholar]
- Ma H, Pooler M, Griesbach R (2009) Anthocyanin regulatory/structural gene expression in Phalaenopsis. J Am Soc Hortic Sci 134: 88–96 [Google Scholar]
- Martins TR, Berg JJ, Blinka S, Rausher MD, Baum DA (2013) Precise spatio-temporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytol 197: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medel R, Botto-Mahan C, Kalin-Arroyo M (2003) Pollinator-mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology 84: 1721–1732 [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L (2002) The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill SD, Nadeau JA, Zhang XS, Bui AQ, Halevy AH (1993) Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell 5: 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZJ, Chen YY, Du JS, Chen YY, Chung MC, Tsai WC, Wang CN, Chen HH (2014) Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol 202: 1024–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, Hu JM, Chen HH (2011) The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol 52: 1515–1531 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6: 3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181: 219–229 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Saito R, Fukuta N, Ohmiya A, Itoh Y, Ozeki Y, Kuchitsu K, Nakayama M (2006) Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci 170: 828–834 [Google Scholar]
- Sasaki K, Aida R, Yamaguchi H, Shikata M, Niki T, Nishijima T, Ohtsubo N (2010) Functional divergence within class B MADS-box genes TfGLO and TfDEF in Torenia fournieri Lind. Mol Genet Genomics 284: 399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S. (2002) Overexpression of the AtmybL2 gene represses trichome development in Arabidopsis. DNA Res 9: 31–34 [DOI] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin CR, Davies KM (2011) The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol 189: 602–615 [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JN, Koes R (2000) anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Fu CH, Hsiao YY, Huang YM, Chen LJ, Wang M, Liu ZJ, Chen HH (2013) OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol 54: e7. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Kuoh CS, Chuang MH, Chen WH, Chen HH (2004) Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol 45: 831–844 [DOI] [PubMed] [Google Scholar]
- Tsai WC, Lee PF, Chen HI, Hsiao YY, Wei WJ, Pan ZJ, Chuang MH, Kuoh CS, Chen WH, Chen HH (2005) PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol 46: 1125–1139 [DOI] [PubMed] [Google Scholar]
- Ushimaru A, Watanabe T, Nakata K (2007) Colored floral organs influence pollinator behavior and pollen transfer in Commelina communis (Commelinaceae). Am J Bot 94: 249–258 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010) Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol 51: 463–474 [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Toda S, Tasaki K (2014) The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol 201: 1009–1020 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.