A mutation in a laccase gene alters lignin deposition and cell wall recalcitrance to saccharification.
Abstract
The oxidation of monolignols is a required step for lignin polymerization and deposition in cell walls. In dicots, both peroxidases and laccases are known to participate in this process. Here, we provide evidence that laccases are also involved in the lignification of Brachypodium distachyon, a model plant for temperate grasses. Transcript quantification data as well as in situ and immunolocalization experiments demonstrated that at least two laccases (LACCASE5 and LACCASE6) are present in lignifying tissues. A mutant with a misspliced LACCASE5 messenger RNA was identified in a targeting-induced local lesion in genome mutant collection. This mutant shows 10% decreased Klason lignin content and modification of the syringyl-to-guaiacyl units ratio. The amount of ferulic acid units ester linked to the mutant cell walls is increased by 40% when compared with control plants, while the amount of ferulic acid units ether linked to lignins is decreased. In addition, the mutant shows a higher saccharification efficiency. These results provide clear evidence that laccases are required for B. distachyon lignification and are promising targets to alleviate the recalcitrance of grass lignocelluloses.
Lignins are cell wall phenolic heteropolymers, primarily made from p-coumaryl, coniferyl, and sinapyl alcohols, referred to as monolignols. These monolignols are synthesized in the cytoplasm from the phenylpropanoid pathway and then transported to the cell walls, where they are oxidatively polymerized into p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units. This oxidative polymerization step is driven by hydrogen peroxide-dependent peroxidases and/or oxygen-dependent laccases (Vanholme et al., 2010). While the involvement of peroxidases in lignification has been indicated for many years, the occurrence of lignin-specific laccases has been unambiguously established more recently in Arabidopsis (Arabidopsis thaliana), the genetic model plant. While the transfer DNA mutants of AtLAC4 (for LACCASE4) and AtLAC17 showed moderate alterations in lignification, the double mutant lac4 lac17 displayed more severe effects, together with poorly lignified interfascicular fibers and collapsed vessels in stems, when grown in continuous light (Berthet et al., 2011). More recently, AtLAC4 and AtLAC17 were shown to direct lignification in secondary cell walls and during protoxylem tracheary element development (Schuetz et al., 2014).
In this study, we aim to establish whether laccases are also involved in the lignification of grass cell walls. Indeed, to our knowledge, no mutants or transgenics affected in laccase activity with a convincing impact on lignification have been reported in grasses, despite a report of a laccase cloned from sugarcane (Saccharum officinarum), SofLAC, shown to partially complement Atlac17 (Cesarino et al., 2013). To this aim, we used the Brachypodium distachyon mutant collection and the targeting-induced local lesion in genome platform established previously (Dalmais et al., 2013) to isolate laccase mutants. By doing so, we demonstrated that the BdLAC5 gene is involved in the lignification of B. distachyon culms and that laccases might be good candidates to target for cell wall engineering.
RESULTS AND DISCUSSION
BdLAC5 and BdLAC6 Proteins Are Closely Related to Lignin-Specific AtLAC17 and AtLAC4
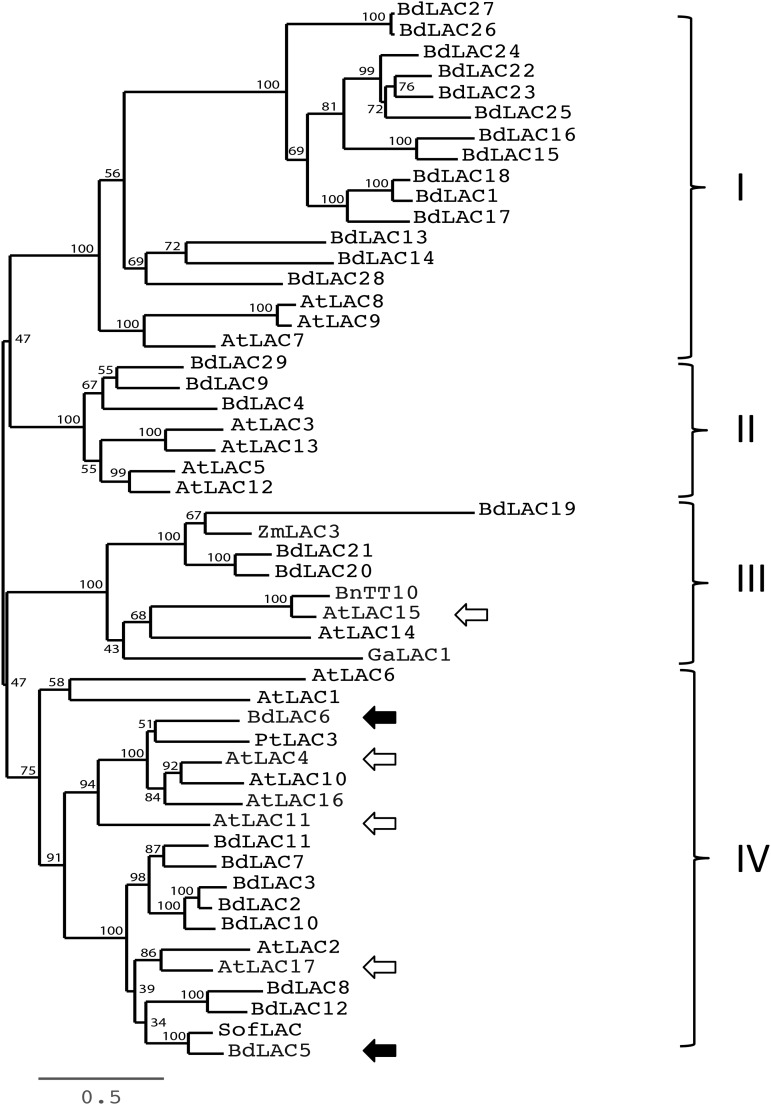
In order to identify the laccases of B. distachyon in silico, we BLASTed the amino acid sequences of Arabidopsis AtLAC4 or AtLAC17 (http://mips.helmholtz-muenchen.de/plant/brachypodium/) against proteins from the B. distachyon genome. This search allowed the identification of 29 nonredundant laccase genes named BdLAC1 to BdLAC29, according to their chromosome localization (Supplemental Table S1). A phylogenetic tree was constructed with the deduced 29 B. distachyon laccase proteins together with the 17 Arabidopsis laccases (Fig. 1). In addition, we incorporated in this tree some other laccases reported to catalyze the oxidative polymerization of various phenolics, such as SofLAC from sugarcane (Cesarino et al., 2013), ZmLAC3 from maize (Zea mays; Caparrós-Ruiz et al., 2006), PtLAC3 from Populus trichocarpa (Ranocha et al., 2002), GaLAC1 from Gossypium arboretum (Wang et al., 2008), and Brassica napus Transparent Testa10 (BnTT10; Zhang et al., 2013; Supplemental Table S2). The resulting phylogeny comprised four large clades (Fig. 1). Clades I and II gathered 14 and three B. distachyon laccases, respectively, and some Arabidopsis proteins of unknown functions. Clade III comprised three B. distachyon laccases with AtLAC15 and BnTT10 laccase proteins involved in proanthocyanidin polymerization (Pourcel et al., 2005; Zhang et al., 2013). This result suggests that other members of clade III might also catalyze the oxidation of flavonoids. The other members clustered in clade IV with the Arabidopsis lignin-specific laccases AtLAC17 and AtLAC4 (Fig. 1). More importantly, only one protein, BdLAC6 (Bradi1g74320), tightly gathered in a subcluster with AtLAC4 (66%/83% identity/similarity), AtLAC11 (Zhao et al., 2013), and PtLAC3 (Ranocha et al., 2002), also was reported to be involved in lignification. BdLAC5 (Bradi1g66720), BdLAC8 (Bradi2g23370), and BdLAC12 (Bradi2g54740) gathered together in a subcluster including AtLAC17. BdLAC5, which shares 64% identity and 77% similarity with AtLAC17, is also the closest ortholog of SofLAC, a protein able to partially restore the lignin level of the lignin-deficient Atlac17 mutant (Cesarino et al., 2013). Based on this analysis, the BdLAC5 and BdLAC6 genes were chosen as the best lignin-specific candidates for further investigation.
Figure 1.
Phylogenetic analysis of laccase proteins. Twenty-nine laccases from B. distachyon and 17 laccases from Arabidopsis were used to build the tree. Other proteins putatively involved in the polymerization of phenolics were also included: SofLAC from sugarcane, ZmLAC3 from maize, PtLAC3 from P. trichocarpa, GaLAC1 from G. arboretum, and BnTT10-1 from B. napus. Branch length is proportional to the number of substitutions per site and represents evolutionary distance as indicated by the scale bar. Names and protein sequences are available in Supplemental Tables S1 and S2. White arrows show Arabidopsis laccases for which mutants have already been published. Black arrows show BdLAC5 and BdLAC6.
BdLAC5 and BdLAC6 Are Mainly Expressed in Lignified Tissues
The expression and coexpression of the selected BdLAC5 and BdLAC6 genes were studied in different organs and at different development stages using the PlaNet tools (http://aranet.mpimp-golm.mpg.de/; Supplemental Fig. S1). Both genes were highly expressed in lignified organs (internode, node, and peduncle) and poorly expressed in organs with low lignin levels (e.g. leaf) or in nonlignified tissue (endosperm). This coexpression study (Supplemental Table S3) revealed that both laccases are coexpressed with genes involved in monolignol biosynthesis, such as O-methyltransferase (Bradi3g16530; Bragg et al., 2012; Dalmais et al., 2013), cinnamoyl-CoA reductase (Bradi3g36887), or Phe ammonia lyase (Bradi3g49250 and Bradi3g49260). They are also coexpressed with three cellulose synthase genes specific to the secondary cell wall, CESA4 (Bradi3g28350), CESA7 (Bradi4g30540), and CESA8 (Bradi2g49912; Handakumbura et al., 2013).
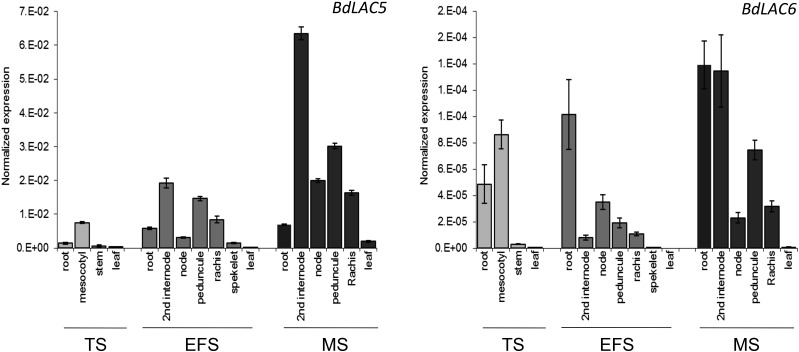
The expression levels of BdLAC5 and BdLAC6 were analyzed by quantitative reverse transcription real-time (RT)-PCR on RNA isolated from different organs at three developmental stages (see “Materials and Methods”), the tillering stage (TS), early flowering stage (EFS), and maturity stage (MS). BdLAC5 and BdLAC6 transcripts were detected in all tested organs and stages, but with considerable variation (Fig. 2). BdLAC5 is expressed mainly in the stem, internode or peduncle, and floral rachis, whereas BdLAC6 is expressed mainly in the root or mesocotyl at earlier stages and also in the internode at MS. Both BdLAC5 and BdLAC6 transcript levels were found to be very low in poorly lignified organs, such as young leaves and young spikelets.
Figure 2.
Expression patterns of BdLAC5 and BdLAC6 mRNA at different stages of development in wild-type plants (accession Bd21-3). BdLAC5 and BdLAC6 transcript levels were quantified with quantitative RT-PCR. The relative transcript levels of BdLAC5 and BdLAC6 were normalized with the housekeeping gene BdUBI4. The data represent means ± sd from biological replicates (n = 3).
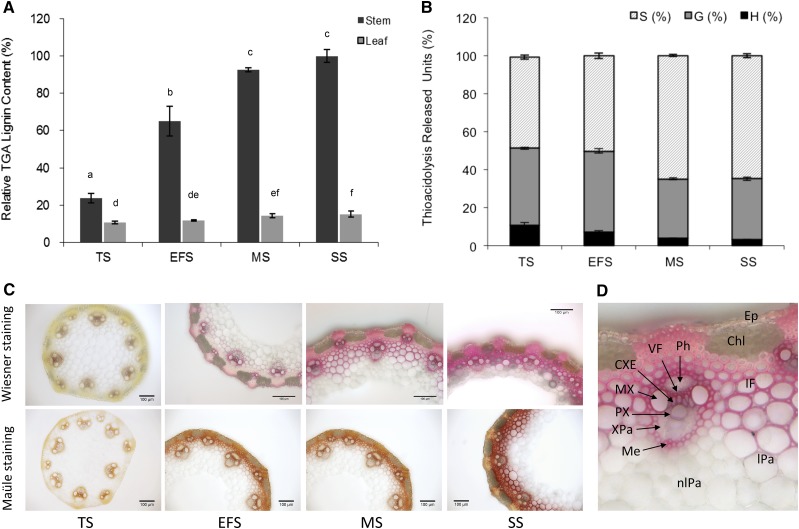
In parallel with this transcript study, the kinetic monitoring of B. distachyon lignification was performed on stem and leaf samples at the same stages in addition to the senescence stage (SS). Lignin content and the structure of extractive-free material were estimated using thioglycolic acid (TGA) and thioacidolysis methods (Fig. 3, A and B). In agreement with Terashima et al. (1993), the stem lignin level as well as the percentage of S lignin units was found to increase with maturity, whereas H thioacidolysis monomers were recovered in higher frequency at early developmental stages (Fig. 3, A and B). Histochemical Wiesner and Maüle stainings of B. distachyon stems (Fig. 3C) were consistent with these chemical analyses. At TS, using Wiesner staining, lignins could be detected mainly in vessels (metaxylem and protoxylem), mestome, and intrafascicular fibers of vascular bundles. It is only at the EFS and later stages that interfascicular fibers were substantially lignified, with enrichment of S units as revealed by its bright red color with the Maüle reagent. By contrast, and whatever the stage, xylem elements remained orange with Maüle staining, which suggests the occurrence of G-rich lignins. In summary, the lignification of vascular bundles occurs early, with deposition of G-rich lignins, while interfascicular fibers lignify gradually, with a distribution of G-lignin and S-lignin units.
Figure 3.
Lignin content and histochemical detection of lignified tissues in wild-type plants at different stages. A, Relative TGA lignin content in stems and leaves. Data are shown as percentages of the highest value (stem at SS). Lowercase letters represent significant differences. B, Thioacidolysis released monolignols from stems at different stages. Data are shown as percentages of the relative thioacidolysis yield. For A and B, all data represent means ± sd from biological replicates (n = 3). C, Histochemical staining of lignified tissues in internodes at different developmental stages. D, Wiesner staining at MS and labeling of tissues in B. distachyon stem. Chl, Chlorenchyma; CXE, connecting xylem elements; Ep, epidermis; IF, interfascicular fibers; lPa, lignified parenchyma; Me, mestome; MX, metaxylem; nlPa, nonlignified parenchyma; Ph, phloem; PX, protoxylem; VF, vascular fibers; Xpa, xylem parenchyma.
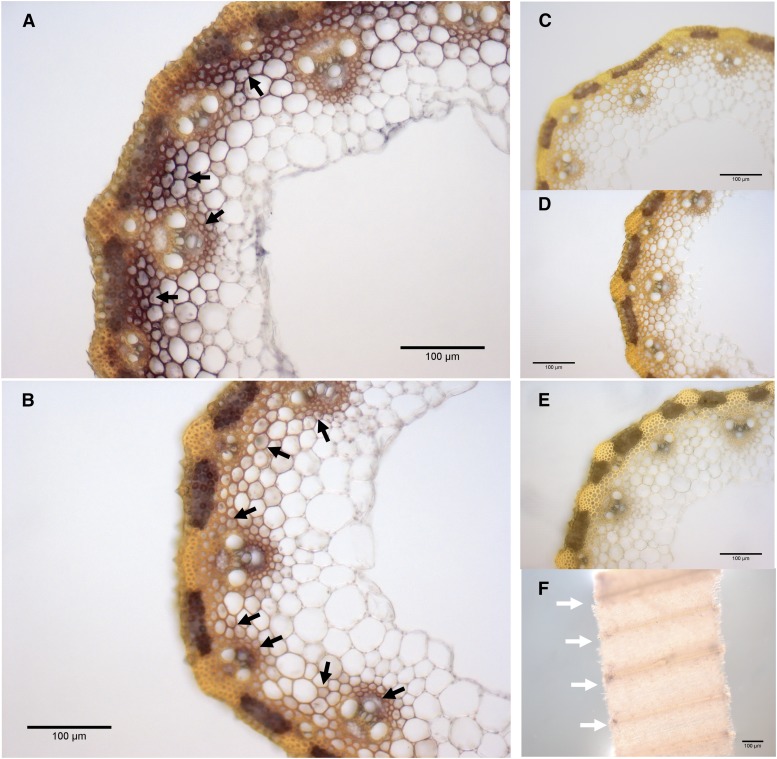
To more accurately localize BdLAC5 and BdLAC6 transcripts at the tissue level, in situ hybridization experiments were performed with appropriate RNA probes applied to internode sections collected at EFS. At this stage, the sclerenchyma (interfascicular fiber) tissue, a few parenchyma cells, and vascular bundles are lignified (Fig. 3C). The intense signal of the BdLAC5 antisense probe was detected in most of these tissues (Fig. 4A) when compared with the negative control performed with the sense probe (Fig. 4C). The metaxylem and protoxylem areas were poorly labeled, a phenomenon that could be accounted for by the fact that these cells are fully functional and cytosol free at EFS (Bollhöner et al., 2012; Schuetz et al., 2013). The most intense signal of BdLAC5 transcripts was observed in most sclerenchyma cells and some lignifying neighboring parenchyma cells of the interfascicular area, whereas the nonlignified phloem was not labeled (Fig. 4A). When the hybridization test was carried out on whole leaves (Fig. 4F), only foliar veins were labeled, a result that further confirms the location of BdLAC5 transcripts in lignifying areas. Most signals of BdLAC6 transcripts were observed to parallel those of BdLAC5, but with a lower intensity (Fig. 4B). In conclusion, the maximum levels of BdLAC5 and BdLAC6 transcripts were found in cells and stages undergoing the most active lignification, with BdLAC5 being expressed to a much higher extent than BdLAC6 in stems.
Figure 4.
Tissue localization of BdLAC5 and BdLAC6 transcripts. Transcripts of BdLAC5 and BdLAC6 were detected by in situ hybridization at the heading and flowering stages (EFS). A, Cross sections of the second internode were labeled with BdLAC5 antisense probes. B, BdLAC6 antisense probes. C, BdLAC5 sense probes. D, BdLAC6 sense probes. E, Cross section of internodes without labeled probes. F, Whole-mount leaves labeled by BdLAC5 antisense probes. Black arrows show immunolabeling signal. White arrows show veins. Bars = 100 μm.
BdLAC5 and BdLAC6 Proteins Are Localized in the Apoplasm
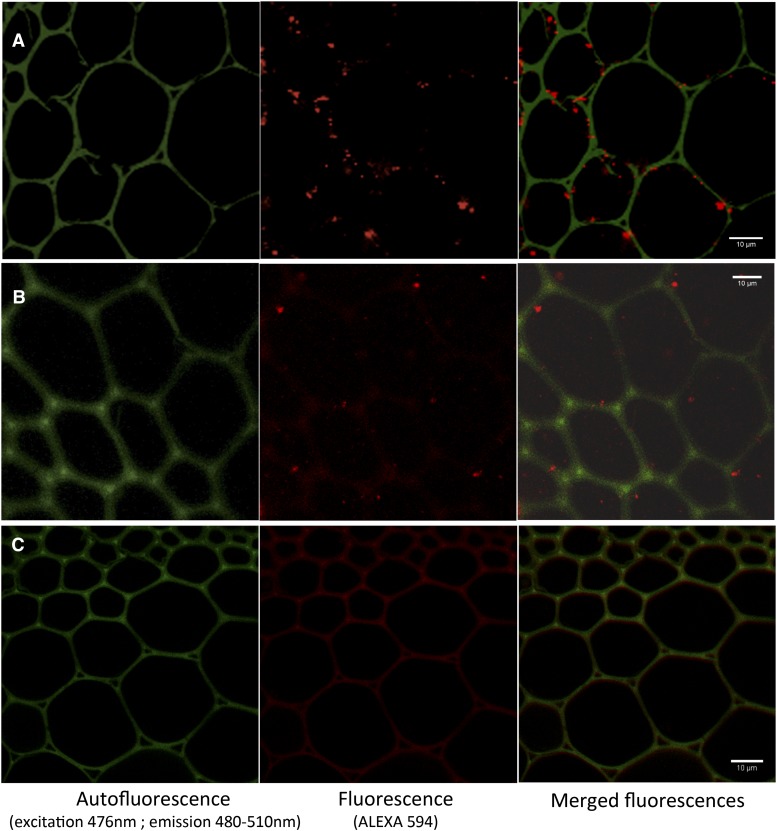
The BdLAC5 gene (1,719-bp coding sequence) encodes a predicted 572-amino acid polypeptide (theoretical molecular mass and pI, 62.7 kD and 8.96), while the BdLAC6 gene (1,689-bp coding sequence) encodes a predicted 561-amino acid polypeptide (theoretical molecular mass and pI, 62.1 kD and 9.34). According to the prediction software, both BdLAC5 and BdLAC6 are hydrophilic proteins that could be glycosylated and exported to the cell wall (Supplemental Fig. S2). Little experimental evidence is published regarding the subcellular location of laccases (Berthet et al., 2011; Cesarino et al., 2013). It is worth noting that Pang et al. (2013) observed the AtLAC15 protein fused to GFP in the vacuole instead of the cell wall, which would make sense if AtLAC15 is involved in the polymerization of proanthocyanidins (Pang et al., 2013). More recently, it was shown that AtLAC17 and AtLAC4 are located in secondary cell walls throughout protoxylem tracheary element differentiation (Schuetz et al., 2014). We attempted to detect BdLAC5 and BdLAC6 subcellular location using immunolabeling with two specific primary antibodies against these laccases. We focused our attention on the interfascicular fiber cells, because vascular bundles have high levels of autofluorescence. This immunolabeling assay revealed anti-BdLAC5-labeled particles in or next to the walls of the interfascicular fibers (Fig. 5). BdLAC6 showed a similar pattern, but fewer particles were detected, presumably due to the lower expression of BdLAC6. However, these data suggest that the two laccases are secreted to the apoplasm and accumulate in secondary cell walls of lignified cells, a result in accordance with the location of AtLAC4 and AtLAC17 in secondary cell walls of tracheary elements (Schuetz et al., 2014).
Figure 5.
Subcellular localization of BdLAC5 and BdLAC6 in lignifying interfascicular fibers. Immunolocalizations are imaged with confocal microscopy. A, Immunolocalization of Alexa Fluor 594 after the detection of BdLAC5 primary antibody. B, Immunolocalization of Alexa Fluor 594 after the detection of BdLAC6 primary antibody. C, Control immunofluorescence with Alexa Fluor 594 secondary antibody without primary antibody. Green indicates autofluorescence of cell wall (excitation, 476 nm; emission, 480–510 nm), and red indicates fluorescence of Alexa Fluor 594. Bars = 10 μm.
Identification of BdLAC5 and BdLAC6 Mutants Affected in the Highly Conserved C-Terminal Domain
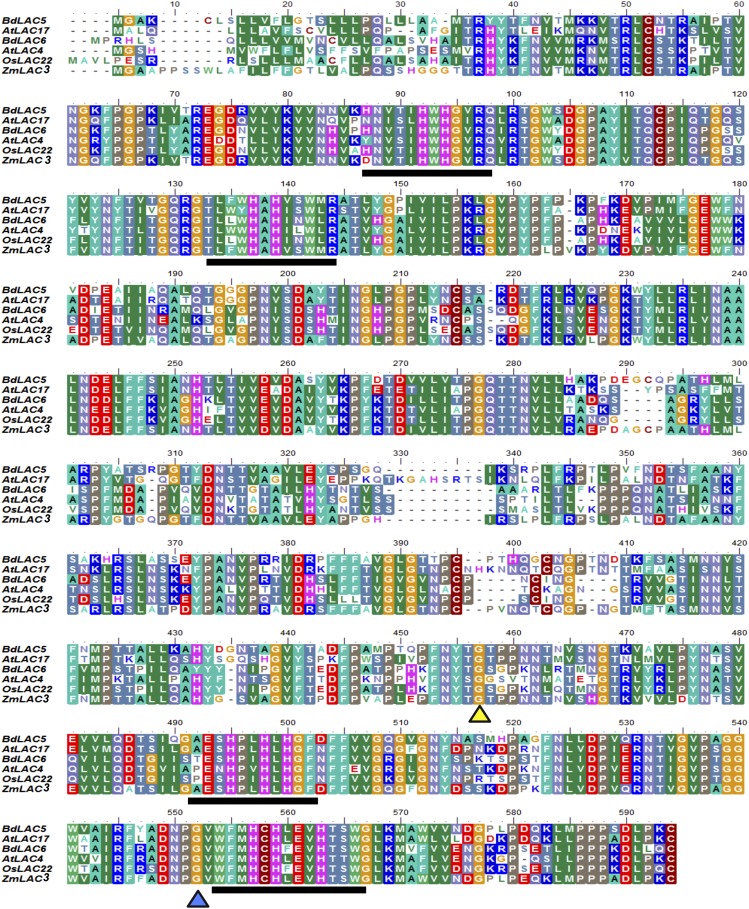
A targeting-induced local lesion in genome screen was carried out on the Versailles B. distachyon mutant collection in order to identify mutations in BdLAC5 and BdLAC6 genes by reverse genetics. This screen led to the identification of several lines with mutations in BdLAC5 and BdLAC6 (Dalmais et al., 2013). Among these mutants, one BdLAC5 mutant, Bd4442, and one BdLAC6 mutant, Bd5024, were selected on the rationale that mutations result in the loss of the highly conserved and essential laccase C-terminal domain. To be active, laccase needs to bind four copper ions (McCaig et al., 2005; Reiss et al., 2013). One copper ion site is in the C-terminal domain of the protein, and it is involved in the oxidation of the reducing substrate and, thus, necessary for oxidase activity (Durão et al., 2006). The mutant Bd4442 has a mutation in the 5′ splice site of the last intron of the BdLAC5 (Bradi1g66720) gene model (Dalmais et al., 2013). In order to verify the effect of this mutation on the transcript, we performed RT-PCR analysis to amplify the BdLAC5 mRNA in Bd4442. When compared with the wild-type sample, the BdLAC5 mRNA from Bd4442 displayed an insertion of 13 nucleotides from the last intron, which is predicted to cause a truncation of the C-terminal domain of the BdLAC5 protein (Supplemental Fig. S3). The mutant Bd5024 possesses a single G-to-T nucleotide substitution at position 1,270 of the BdLAC6 complementary DNA (cDNA), which introduces a premature stop codon at Gly-424. Consequently, the two selected lines share truncated laccase proteins, which likely lack the highly conserved C-terminal domain (Fig. 6).
Figure 6.
Multiple alignment of deduced amino acid sequences from B. distachyon and other plant laccases. Amino acid sequences of both B. distachyon laccases, maize (ZmLAC3), rice (Oryza sativa; OsLAC22), and Arabidopsis (AtLAC4 and AtLAC17) laccases were aligned with ClustalW software. The copper-binding domain motifs are underlined. Triangles represent the locations of amino acid changes in BdLAC5 of Bd4442 (blue) and the location of the premature stop codon in Bd5024 (yellow).
Impact of Mutations in BdLAC5 and BdLAC6 on Plant Phenotype and Lignification
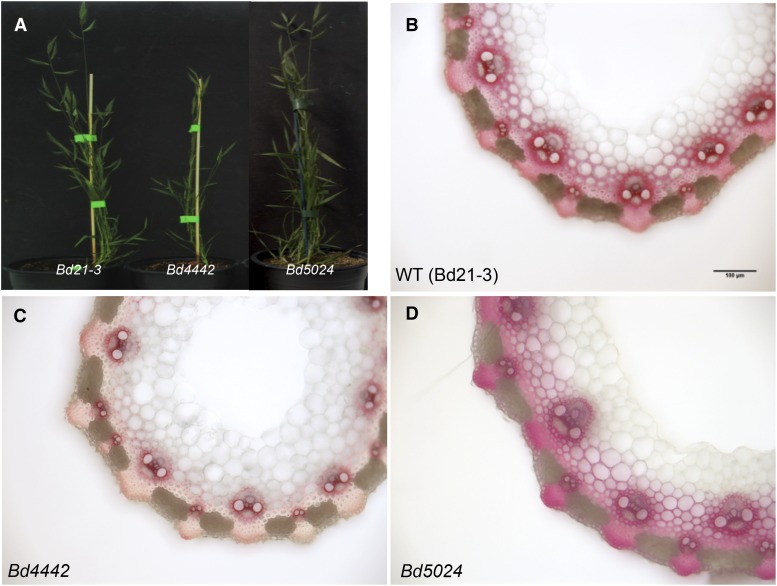
Compared with the wild-type sample, the Bd4442 and Bd5024 homozygous mutants did not display any drastic alteration of growth and development in our culture conditions (Fig. 7A). However, closer examination revealed that the stem height and internode diameter of the Bd4442 line were systematically smaller than those of the wild-type line (Supplemental Table S4). At the tissue level, the area of the vascular bundles from the homozygous Bd4442 line was also found to be reduced slightly as compared with the wild type.
Figure 7.
Phenotypes of mutants and wild-type plants. A, Developmental phenotypes of Bd4442, Bd5024, and the wild type. B to D, Transverse sections of second internodes stained with Wiesner reagent. Bar = 100 μm. For B to D, photographs were taken at the same magnification.
The lignin level of extractive-free mature stems of Bd4442 and Bd5024 mutants was measured by the Klason lignin method (Dence, 1992). Consistent with the histochemical results, the BdLAC6-deficient Bd5024 line did not display any alteration to the lignin content of mature stems. By contrast, disrupting the BdLAC5 gene in the Bd4442 mutant induced a modest but significant reduction of the culm lignin level (Table I). This reduction (by about 10% compared with the wild-type level) was systematically observed for independent cocultures of Bd4442 and wild-type lines carried out over a 2-year period and confirmed by the TGA lignin assays (Supplemental Table S5). The analysis of cell wall polysaccharides suggested that the decreased lignin content in Bd4442 stems is balanced by a slightly increased hemicellulose content (Supplemental Fig. S4). The lignin structure of the wild-type and mutant lines was also investigated by thioacidolysis (Lapierre, 1993). Compared with the corresponding wild-type sample, the frequency of the S thioacidolysis monomers released from Bd4442 mature stems was found to be systematically higher (Table I). This higher S frequency has been also reported in Atlac17 (Berthet et al., 2011), an Arabidopsis mutant affected in the expression of AtLAC17 (a presumed Arabidopsis ortholog of BdLAC5).
Table I. Lignin analyses for the wild type and BdLAC5 (Bd4442) and BdLAC6 (Bd5024) misregulated mutants.
Lignin content and structure for extractive-free mature stems were measured by Klason and thioacidolysis methods. Values are means ± sd from individually analyzed plants (n = 3).
| Culture and Genotype | Lignin Content | Relative Frequency of Lignin-Derived Thioacidolysis Monomers |
||
|---|---|---|---|---|
| H | G | S | ||
| % | ||||
| First culture | ||||
| Wild type | 17.63 ± 0.01 | 2.8 ± 0.1 | 30.9 ± 0.8 | 66.3 ± 0.9 |
| Bd4442 | 15.71 ± 0.04a | 2.6 ± 0.2 | 25.3 ± 0.4a | 72.2 ± 0.6a |
| Bd5024 | 17.53 ± 0.14 | 2.8 ± 0.1 | 33.3 ± 0.3 | 63.9 ± 0.3 |
| Second culture | ||||
| Wild type | 16.93 ± 0.57 | 3.0 ± 0.2 | 30.1 ± 2.5 | 66.9 ± 2.7 |
| Bd4442 | 15.60 ± 0.07a | 1.6 ± 0.2a | 22.2 ± 3.5a | 76.3 ± 3.5a |
| Third culture | ||||
| Wild type | 17.59 ± 0.49 | 3.3 ± 0.1 | 31.0 ± 1.0 | 65.7 ± 1.0 |
| Bd4442 | 16.19 ± 0.20a | 2.7 ± 0.1 | 26.5 ± 0.5a | 70.8 ± 0.5a |
Significantly different from the corresponding wild-type sample (one-way ANOVA, Tukey’s honestly significant difference) at P < 0.05.
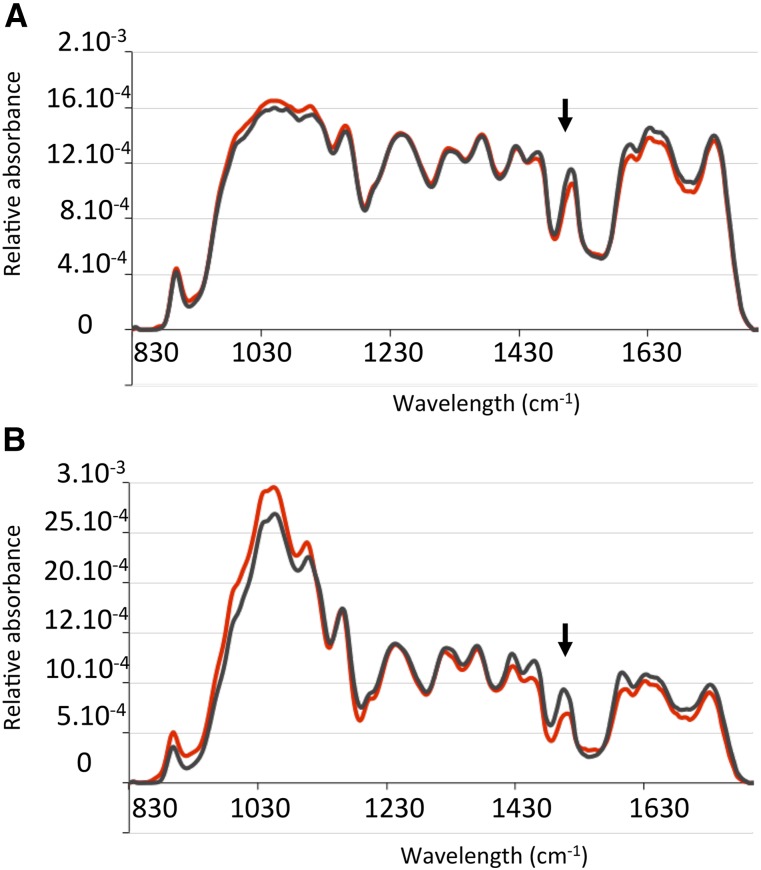
We further investigated the impact of the mutation in Bd4442 at the tissue level by histochemical staining and Fourier transform infrared (FTIR) microspectrophotometry. Compared with the wild type, Wiesner staining of Bd4442 stems at MS was substantially weaker, particularly in the sclerenchyma (Fig. 7). By contrast, this test did not show obvious differences between the Bd5024 and wild-type lines. To further investigate the topochemistry of lignification in Bd4442 culms as compared with control plants, FTIR absorbance spectra of vascular bundle or interfascicular fiber areas were recorded with a spectra-microscope system as described previously (Mouille et al., 2003; Sibout et al., 2005). Relative to the wild type, the most striking impact of the Bd4442 mutation on the FTIR spectra was a substantial reduction of the lignin-specific aromatic signal at 1,508 cm−1 and mainly in the interfascicular fiber area (Fig. 8; Supplemental Fig. S5). By contrast, this lignin peak was reduced to a much lower extent in the Bd4442 vascular bundle area. Taken together, the histochemical and FTIR results revealed that the lignification of B. distachyon culm was affected by the Bd4442 mutation, more specifically in interfascicular fibers.
Figure 8.
FTIR analysis of interfascicular fibers and vascular bundles. FTIR spectra were acquired from vascular bundles (A) and interfascicular fibers (B) in cross sections of internodes at MS. Black lines indicate wild-type plants, and red lines indicate Bd4442. Arrows shows absorbance at 1,508 cm−1. Each spectrum corresponds to the normalized means of biological replicates (n = 6).
From these analyses, we may conclude that BdLAC5 disruption results in a decreased lignification of mature stems, mainly in the interfascicular fibers, and that the biosynthesis of G lignin units is affected to a higher extent than that of S lignin units. The latter result might be accounted for either from some substrate specificity of the BdLAC5 enzyme and/or a spatiotemporally regulated expression of the BdLAC5 gene at the time when G lignin units are deposited.
It is well established that grass lignins are not only acylated by p-coumaric acid (CA) but also are covalently linked to feruloylated arabinoxylans, consisting of a xylan backbone with Ara substituents partially acylated by ferulic acid (FA; Ralph, 2010). At the onset of lignification, these FA esters act as lignin initiation sites by oxidatively driven coupling mainly to coniferyl alcohol (Jacquet et al., 1995). The impact of the BdLAC5 and BdLAC6 mutations on the level of CA and FA ester linked to stem cell walls was studied by mild alkaline hydrolysis according to previously published methods (Bouvier d’Yvoire et al., 2013; Petrik et al., 2014). While the level of CA esters was found to be slightly higher in Bd5024 and unchanged in Bd4442, mild alkaline hydrolysis released about 40% more FA from Bd4442 than from the wild type (Table II). This higher content of FA units only ester linked to Bd4442 cell walls was accompanied by a lower amount of FA units ether linked to lignins, as revealed by severe alkaline hydrolysis (Barrière et al., 2004a, 2004b). The amount of FA ethers in the Bd4442 and wild-type samples was 5.89 ± 0.36 and 6.93 ± 0.48 mg g−1, respectively. According to model studies carried out with artificially lignified maize cell walls, these changes could be linked to the reduced lignification of Bd4442 cell walls (Grabber et al., 2000). In addition and together with peroxidases, BdLAC5 might participate in the oxidatively driven coupling of FA esters and lignin units at the onset of lignification. Its misregulation together with the reduced lignification might further limit this cross-coupling mechanism.
Table II. Amounts of CA and FA released by mild alkaline hydrolysis of mature stems from the wild type and BdLAC5 (Bd4442) and BdLAC6 (Bd5024) misregulated mutants.
Values are means ± sd from individually analyzed plants (n = 3).
| Culture and Genotype | Compound |
|
|---|---|---|
| CA | FA | |
| mg g−1 | ||
| First culture | ||
| Wild type | 7.17 ± 0.04 | 4.69 ± 0.03 |
| Bd4442 | 7.50 ± 0.43 | 6.48 ± 0.41a |
| Bd5024 | 8.20 ± 0.18a | 4.42 ± 0.09 |
| Second culture | ||
| Wild type | 6.83 ± 0.52 | 5.31 ± 0.44 |
| Bd4442 | 6.52 ± 0.31 | 7.69 ± 0.15a |
| Third culture | ||
| Wild type | 9.42 ± 0.21 | 5.05 ± 0.07 |
| Bd4442 | 9.48 ± 0.23 | 7.42 ± 0.11a |
Significantly different from the corresponding wild-type sample (one-way ANOVA, Tukey’s honestly significant difference) at P < 0.05.
Complementation of the Bd4442 Mutant with the BdLAC5 Wild-Type Allele Restores Lignification and Saccharification Yield
The definitive demonstration that BdLAC5 is a lignin-specific laccase involved in the lignification of B. distachyon culms was provided by complementation experiments. We produced transgenic lines overexpressing a BdLAC5 wild-type gene copy driven by a maize ubiquitin promoter into Bd4442. We obtained few transgenic lines suggesting the recalcitrance of this line to embryogenesis. However, we selected one transformed line with detectable levels of the transgene (Supplemental Fig. S6). This line showed a complete restoration of lignin level, lignin structure, and FA esters (Table III). This line also restored phenotype (Supplemental Table S4) and lignin histochemical staining (Supplemental Fig. S7). Not unexpectedly, the saccharification yield of the extractive-free mature stems was found to be improved for the Bd4442 mutant and comparable to that of the wild-type sample for the complemented line (Table IV). The weight loss induced by a cellulase treatment without any pretreatment was found to be 30.5 ± 1.3% for Bd4442 samples versus 23.5% ± 1.4% for wild-type samples and 20.6% ± 0.4% for the complemented line (mean ± sd values for two independent experiments each performed on three different samples per line).
Table III. Lignin and FA analyses for the wild type and the BdLAC5 (Bd4442) mutant line as compared with the Bd4442 complemented line3 (CP3) mutant line complemented with the BdLAC5 wild-type allele (T2 transformant).
Lignin content and structure for extractive-free mature stems were measured by Klason and thioacidolysis methods; FA level ester linked to the cell walls was measured by mild alkaline hydrolysis. Values are means ± sd from individually analyzed plants (n = 3). All the plants were obtained from the same culture experiment in the greenhouse.
| Genotype | Lignin Content | Relative Frequency of Lignin-Derived Thioacidolysis Monomers |
FA Amount | ||
|---|---|---|---|---|---|
| H | G | S | |||
| % | mg g−1 | ||||
| Wild type | 18.40 ± 0.33 | 3.0 ± 0.1 | 27.1 ± 1.1 | 66.3 ± 0.9 | 5.05 ± 0.07 |
| Bd4442 | 16.43 ± 0.19a | 3.1 ± 0.1 | 23.7 ± 0.5a | 73.4 ± 0.6a | 7.42 ± 0.11 |
| Bd4442 CP3 | 18.15 ± 0.23 | 3.1 ± 0.1 | 32.6 ± 0.5 | 64.4 ± 0.5 | 5.88 ± 0.12 |
Significantly different from the corresponding wild-type sample (one-way ANOVA, Tukey’s honestly significant difference) at P < 0.05.
Table IV. Saccharification assays of extractive-free mature stem from the wild type and the BdLAC5 (Bd4442) mutant line as compared with the Bd4442 CP3 mutant line complemented with the BdLAC5 wild-type allele (T2 transformants).
Saccharification was evaluated both by the weight loss percentage and by the amount of Glc released from the samples. The data represent means ± sd from biological replicates (two independent experiments performed each on three different samples per line). Letters indicate significant differences analyzed by one-way ANOVA (Tukey’s honestly significant difference, P < 0.05).
| Genotype | Weight Loss Percentage | Glc |
|---|---|---|
| mg g−1 | ||
| Bd21-3 wild type | 23.5 ± 1.4 a | 68.5 ± 3.9 a |
| Bd4442 homozygous | 30.5 ± 1.3 b | 113.6 ± 4.4 d |
| Bd4442 CP3 | 20.6 ± 0.4 c | 75.3 ± 3.3 a,b |
CONCLUSION
To our knowledge, this study provides the first evidence that a laccase enzyme, BdLAC5, is involved in the lignification of B. distachyon, a model plant for important grass crops. Indeed, in the BdLAC5-misregulated Bd4442 mutant line identified in this work, the alteration of the C-terminal domain of BdLAC5 induces significant alterations to the lignification of mature culms, with a 10% lower lignin level, a slight increase of the frequency of S lignin units, and a substantial increase of measurable FA esters. Despite a similar expression pattern of BdLAC6 and BdLAC5, it is difficult to draw the same conclusion for the role of BdLAC6 in lignification. This laccase is expressed at a much lower level than BdLAC5, and redundancy might prevent any detectable impact on lignin content in the Bdlac6 mutant. Only a double mutant would reveal a role for both genes in lignin formation. B. distachyon possess 29 laccases in its genome, and nine of them belong to the same cluster as BdLAC5 and BdLAC6. More functional experiments are required to determine precisely which other enzymes participate in lignification; however, this study provides evidence that, as in Arabidopsis, laccases are involved in the lignification of B. distachyon, the model plant for grasses.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Mutants were identified in the collection of chemically induced Brachypodium distachyon mutants at the Research Center of the Institut National de la Recherche Agronomique-Versailles-Grignon (Dalmais et al., 2013). B. distachyon plants (accession Bd21-3) were grown in a greenhouse under long-day conditions (18 h of light, 400-W sodium lamps). Day and night temperatures were 23°C and 18°C, respectively. For sample harvesting, four stages (Supplemental Fig. S8) were chosen as in previous articles (Matos et al., 2013): TS, 17 d after germination, when the plants are tillering and the first true internode is elongating; EFS, approximately 35 d after germination, when an inflorescence appears with immature flowers; MS, approximately 60 d after germination, when seeds are filled and the oldest leaves are turning yellow; and SS, approximately 120 d after germination, when the whole plant is dead and dry.
Histochemical Staining
All the histochemical staining was performed on sections cut in the middle of the second internode from the top, except at TS, where the first internode above the crown was selected. Samples were fixed in 50% (v/v) ethanol, 10% (v/v) formalin, and 5% (v/v) acetic acid solution overnight and then embedded in 7% (w/v) agarose before being transversely sectioned at a thickness of 100 μm using a vibratome (VT1000S; Leica). Lignin deposition and composition were investigated histochemically by Maüle and Wiesner staining as published previously (Bouvier d’Yvoire et al., 2013). All sections were observed with a Zeiss AxioPlan 2 microscope system with automatic exposure times.
Phenotype Measurements and Image Analysis
All the measurements were performed on the tallest stems. Stem height was measured with a ruler, from the crown (the bottom of the stem) to the top of peduncle. The internode diameter was measured with a digital caliper on the second internode from the top or on the first internode above the crown at TS. ImageJ software was used for the quantification of cell wall thickness and vascular bundles. Significant differences were analyzed by one-way ANOVA (Tukey’s honestly significant difference, P < 0.05).
Lignin Content and Structure Determination
One or two main stems of each plant were collected and ground after removing spikelets and leaves. Ground samples were sequentially extracted at 60°C with 50 mL of ethanol, water, and ethanol. At each step, the samples were vortexed. These steps were repeated twice. Samples were then dried and used for the following analysis.
The protocol of the TGA lignin assay was optimized from Hatfield and Fukushima (2005) and Suzuki et al. (2009). Extractive-free cell wall residues of stem were treated with TGA. Briefly, 10 to 15 mg of plant cell wall residues was treated with TGA at 80°C for 8 h. After washing with water and extracting with NaOH (37°C overnight), the purified lignin complex was precipitated using concentrated HCl. The pellet was dissolved in 1 mL of 1 m NaOH. After 6-fold dilution with 1 m NaOH, the solution was measured by UV spectrophotometry at 280 nm. The calibration curves were made by lignin standards purified from B. distachyon. The relative lignin concentration was determined by dividing the amount of lignin calculated based on the calibration curve by the sample weight.
The Klason lignin content was measured according to Dence (1992). The lignin structure and composition were studied by thioacidolysis, as described previously (Lapierre et al., 1999). The lignin-derived thioacidolysis monomers were identified by gas chromatography-mass spectrometry as their trimethylsilylated derivatives. All the analyses were performed with at least three biological replicates. Significant differences were inferred by one-way ANOVA (Tukey’s honestly significant difference, P < 0.05).
Monosaccharide Composition and Linkage Analysis of Polysaccharides
Neutral monosaccharide composition was determined on 5 mg of dried alcohol-insoluble material after hydrolysis in 2.5 m trifluoroacetic acid for 1.5 h at 100°C as described by Harholt et al. (2006). To determine the cellulose content, the residual pellet obtained after the monosaccharide analysis was rinsed twice with 10 volumes of water and then hydrolyzed with H2SO4 as described by Updegraff (1969). The released Glc was diluted 500-fold and then quantified using high-performance anion-exchange chromatography-pulsed-amperometric detection as described by Harholt et al. (2006).
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted using the Qiagen Plant RNAeasy kit with an additional DNase step. RNA samples were quantified using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). One microgram of total RNA was used for RT using the SuperScript Reverse Transcriptase II kit (Invitrogen). The absence of genomic contamination was confirmed by PCR with primers targeting introns. Quantitative RT-PCR was performed on the Eppendorff Realplex2 Mastercycler using the SYBR Green kit (Bio-Rad) and the following conditions: 95°C for 3 min, followed by 40 cycles of 95°C for 20 s, 59°C for 20 s, and 72°C for 20 s. Information on primers is available in Supplemental Table S4. PCR products were verified by sequencing. The normalized expression levels of target genes were calculated by Q-Gene software (http://www.biotechniques.com/softlib/qgene.html) based on the formula for the amplification efficiency of PCR of Muller et al. (2002). The B. distachyon ubiquitin gene BdUBI4 (Hong et al., 2008) was used as the reference gene (Supplemental Table S6). The results were based on three independent biological replicates.
RNA in Situ Hybridization
Primer sequences used for the digoxygenin (DIG)-labeled probe amplification are shown in Supplemental Table S1. Antisense probes were synthesized by in vitro transcription with T7 RNA polymerase using a DIG RNA labeling kit (Roche Applied Science) according to the manufacturer’s protocol. A sense probe was used as a negative control.
Second internodes at the flowering stage were fixed, embedded, and cross sectioned as described above. In situ hybridization was carried out as described by Chapelle et al. (2012) with minor changes: after washing and dehydration of the tissues, sections were prehybridized for 1 h at 60°C with hybridization solution (50% [v/v] formamide, 5× SSC, 100 μg mL−1 tRNA, 50 μg mL−1 heparin, and 0.1% [v/v] Tween). Sections were then hybridized with DIG-labeled antisense or sense probes (1:1,000) overnight at 60°C and washed with a series of SSC solutions. Immunodetection of the DIG-labeled probe was performed with an anti-DIG antibody coupled with alkaline phosphatase using the nitroblue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate kit according to the manufacturer’s instructions (Roche Applied Science). All sections were observed with a Zeiss AxioPlan 2 microscope system with automatic exposure time.
Phylogeny Tree and Three-Dimensional Structure Prediction
An unrooted phylogenetic tree was constructed with PhyML in the Phylogeny.fr platform (http://www.phylogeny.fr) with Blosum62 as a substitution model, with 1,000 bootstraps after loading the protein ClustalW alignment (Dereeper et al., 2008). The final shape of the tree was produced after submitting the Newick format tree to TreeDyn. Multiple sequence alignment was performed with ClustalW software (http://www.ebi.ac.uk/Tools/clustalw2).
Genetic Complementation of the BdLAC5 Splicing Mutant Bd4442
The full-length cDNA including the 5′- and 3′-untranslated regions of BdLAC5 was cloned from B. distachyon stem cDNA using specific primers (Supplemental Table S5). The purified PCR product was ligated into the pDONR207 plasmid via the BP Gateway (Invitrogen) reaction and confirmed by sequencing. The resulting pDONR207-BdLAC5 plasmid was used to transfer BdLAC5 into pIPKb2 (Himmelbach et al., 2007) for B. distachyon transformation by the LR Gateway (Invitrogen) reaction. pIPKb2-BdLAC5 was electroporated into Agrobacterium tumefaciens (AGL1 strain). The BdLAC5 splicing mutant (Bd4442) was genetically transformed following the protocol of Vogel and Hill (2008). Transgenic plants were selected on Murashige and Skoog growth medium containing 40 mg mL−1 hygromycin.
FTIR Spectroscopy
Fixed internodes were cut into 50-µm-thick sections with a vibratome and then were rinsed abundantly with distilled water for 2 min and dried at 37°C for 20 min. FTIR spectra were collected from a 60- × 60-µm window targeting vascular bundles or interfascicular fibers. For each genotype, three sections from three different plants were analyzed. Normalization of the spectral data and statistical analyses were performed as described by Mouille et al. (2003). Student’s t test was performed to estimate statistical differences between mutants and the wild type.
Cell Wall Saccharification
Saccharification assays were performed as described by Berthet et al. (2011). For each sample, 30 mg of extractive-free cell wall residues was incubated with 4 mL of acetate buffer, pH 4.5, containing 4 mg mL−1 commercial cellulase (Onozuka-R10; Serva) and 0.5 mg mL−1 NaN3 for 3 d at 45°C on a carousel. After centrifugation, the recovery of the supernatant for Glc was determined via enzymatic assay with the bioMérieux Kit. The pellet was washed twice with water, then freeze dried and weighed to evaluate the weight loss.
Immunolabeling
Based on their amino acid sequences, the C-terminal domains were used to design the potentially antigenic peptide sequences. The peptide sequence 5′-AGGWVAIRFYADNPGVWFMH-3′ was used to produce anti-BdLAC5, and peptide sequence 5′-KMVFVVENGKRPSETLI-3′ was used to produce anti-BdLAC6. Antibodies were produced by Genescript.
Internodes at flowering and heading stages were collected, fixed, and embedded as described above. After embedding, the internodes were sectioned with a thickness of 30 μm. The immunolabeling was carried out as described by Verhertbruggen et al. (2009). Slides were incubated in Evans blue solution (0.001% [v/v] phosphate-buffered saline) for 10 min to decrease the autofluorescence of lignified cells. Sections were labeled with the primary anti-BdLAC5 or anti-BdLAC6 antibody from rabbit (1:1,000), and the secondary antibody was an anti-rabbit antibody conjugated to Alexa Fluor 594 (Invitrogen; 1:500). This dye was found to be the best marker because of its low interference with the autofluorescence of B. distachyon cell walls (data not shown). The labeled sections were viewed with a confocal laser-scanning microscope (TCS SP5 Confocal; Leica).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression profiles of BdLAC5 and BdLAC6 obtained from PlaNET database (http://aranet.mpimp-golm.mpg.de/bradinet/).
Supplemental Figure S2. Transmembrane segment prediction of BdLAC5 and BdLAC6.
Supplemental Figure S3. The aberrant transcript of BdLAC5 and its deduced amino acid sequence in Bd4442.
Supplemental Figure S4. Hemicellulose and cellulose contents in the extract-free cell wall of wild-type and Bd4442 mutants.
Supplemental Figure S5. FTIR absorbance of different tissues at 1508 cm−1.
Supplemental Figure S6. Genomic characterization of the transgenic line.
Supplemental Figure S7. Staining of lignified tissues in control plants and the complemented line.
Supplemental Figure S8. Phenotypes of B. distachyon (accession no. Bd21-3) at different development stages.
Supplemental Table S1. The LACCASE family genes in B. distachyon.
Supplemental Table S2. Amino acid sequences of laccases used in this study.
Supplemental Table S3. List of genes coexpressed with BdLAC5.
Supplemental Table S4. Anatomical comparison in different genotypes.
Supplemental Table S5. Quantification of thioglycolic acid lignin.
Supplemental Table S6. Primers.
Supplementary Material
Acknowledgments
We thank Jonathan Griffiths (Institut National de la Recherche Agronomique, Institut Jean-Pierre Bourgin) for reading the article, the reviewers for useful comments and corrections, and Olivier Grandjean (Institut National de la Recherche Agronomique, Institut Jean-Pierre Bourgin) for help with confocal microscopy.
Glossary
- RT
real-time
- TS
tillering stage
- EFS
early flowering stage
- MS
maturity stage
- TGA
thioglycolic acid
- SS
senescence stage
- cDNA
complementary DNA
- FTIR
Fourier transform infrared
- CA
p-coumaric acid
- FA
ferulic acid
- DIG
digoxygenin
- H
p-hydroxyphenyl
- G
guaiacyl
- S
syringyl
Footnotes
This work was supported by the Institut National de la Recherche Agronomique program Amelioration de la Ligno-Cellulose; by the European Commission’s Seventh Framework Programme project RENEWALL (grant no. 211982), the Knowledge-Based Bio Economy project CELLWALL, and BRAVO (grant no. ANR–14–CE19–0012–01); and by the China Scholarship Council (to Y.W.).
References
- Barrière Y, Emile JC, Traineau R, Surault F, Briand M, Gallais A (2004a) Genetic variation for organic matter and cell wall digestibility in silage maize: lessons from a 34-year long experiment with sheep in digestibility crates. Maydica 49: 115–126 [Google Scholar]
- Barrière Y, Ralph J, Méchin V, Guillaumie S, Grabber JH, Argillier O, Chabbert B, Lapierre C (2004b) Genetic and molecular basis of grass cell wall biosynthesis and degradability: II. Lessons from brown-midrib mutants. C R Biol 327: 847–860 [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollhöner B, Prestele J, Tuominen H (2012) Xylem cell death: emerging understanding of regulation and function. J Exp Bot 63: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M, Bouchabke-Coussa O, Voorend W, Antelme S, Cézard L, Legée F, Lebris P, Legay S, Whitehead C, McQueen-Mason SJ, et al. (2013) Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J 73: 496–508 [DOI] [PubMed] [Google Scholar]
- Bragg JN, Wu J, Gordon SP, Guttman ME, Thilmony R, Lazo GR, Gu YQ, Vogel JP (2012) Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS ONE 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparrós-Ruiz D, Fornalé S, Civardi L, Puigdomènech P, Rigau J (2006) Isolation and characterisation of a family of laccases in maize. Plant Sci 171: 217–225 [Google Scholar]
- Cesarino I, Araújo P, Sampaio Mayer JL, Vicentini R, Berthet S, Demedts B, Vanholme B, Boerjan W, Mazzafera P (2013) Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J Exp Bot 64: 1769–1781 [DOI] [PubMed] [Google Scholar]
- Chapelle A, Morreel K, Vanholme R, Le-Bris P, Morin H, Lapierre C, Boerjan W, Jouanin L, Demont-Caulet N (2012) Impact of the absence of stem-specific β-glucosidases on lignin and monolignols. Plant Physiol 160: 1204–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, Wang Y, Darracq O, d’Yvoire MB, Cézard L, Légée F, Blondet E, Oria N, et al. (2013) A TILLING platform for functional genomics in Brachypodium distachyon. PLoS ONE 8: e65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dence CW. (1992) The determination of lignin. InLin S, Dence C, eds, Methods in Lignin Chemistry. Springer, Berlin, pp 33–61 [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durão P, Bento I, Fernandes AT, Melo EP, Lindley PF, Martins LO (2006) Perturbations of the T1 copper site in the CotA laccase from Bacillus subtilis: structural, biochemical, enzymatic and stability studies. J Biol Inorg Chem 11: 514–526 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem 48: 6106–6113 [DOI] [PubMed] [Google Scholar]
- Handakumbura PP, Matos DA, Osmont KS, Harrington MJ, Heo K, Kafle K, Kim SH, Baskin TI, Hazen SP (2013) Perturbation of Brachypodium distachyon CELLULOSE SYNTHASE A4 or 7 results in abnormal cell walls. BMC Plant Biol 13: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield R, Fukushima RS (2005) Can lignin be accurately measured? Crop Sci 45: 832–839 [Google Scholar]
- Himmelbach A, Zierold U, Hensel G, Riechen J, Douchkov D, Schweizer P, Kumlehn J (2007) A set of modular binary vectors for transformation of cereals. Plant Physiol 145: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet G, Pollet B, Lapierre C (1995) New ether-linked ferulic acid-coniferyl alcohol dimers identified in grass straws. J Agric Food Chem 43: 2746–2751 [Google Scholar]
- Lapierre C. (1993) Applications of new methods for the investigation of lignin structure. InJung HG, Buxton DR, Hatfield RD, Ralph J, eds, Forage Cell Wall Structure and Digestibility. American Society of Agronomy, Madison, WI, pp 133–163 [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leple JC, Boerjan W, Ferret V, De Nadai V, et al. (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos DA, Whitney IP, Harrington MJ, Hazen SP (2013) Cell walls and the developmental anatomy of the Brachypodium distachyon stem internode. PLoS ONE 8: e80640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig BC, Meagher RB, Dean JFD (2005) Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221: 619–636 [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1374, 1376, 1378–1379 [PubMed] [Google Scholar]
- Pang Y, Cheng X, Huhman DV, Ma J, Peel GJ, Yonekura-Sakakibara K, Saito K, Shen G, Sumner LW, Tang Y, et al. (2013) Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238: 139–154 [DOI] [PubMed] [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, et al. (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol 129: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss R, Ihssen J, Richter M, Eichhorn E, Schilling B, Thöny-Meyer L (2013) Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 8: e65633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL (2014) Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol 166: 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B (2013) Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 64: 11–31 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Suzuki Y, Yamamoto N, Hattori T, Sakamoto M, Umezawa T (2009) High-throughput determination of thioglycolic acid lignin from rice. Plant Biotechnol 26: 337–340 [Google Scholar]
- Terashima N, Fukushima K, He L, Takabe K (1993) Comprehensive model of the lignified plant cell wall. InJung HG, Buxton DR, Hatfield RD, Ralph J, eds, Forage Cell Wall Structure and Digestibility. American Society of Agronomy, Madison, WI, pp 247–270 [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Verhoef R, Schols HA, McCleary BV, McKee L, Gilbert HJ, Knox JP (2009) Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J 59: 413–425 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang C, Zhu M, Yu Y, Zhang Y, Wei Z (2008) Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tiss Organ Cult 93: 303–310 [Google Scholar]
- Zhang K, Lu K, Qu C, Liang Y, Wang R, Chai Y, Li J (2013) Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS ONE 8: e61247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.