Characterization of a rice mutant defective in autophagy highlights its importance in nitrogen remobilization from senescent leaves, biomass increase, and nitrogen use efficiency in the vegetative plant.
Abstract
Much of the nitrogen in leaves is distributed to chloroplasts, mainly in photosynthetic proteins. During leaf senescence, chloroplastic proteins, including Rubisco, are rapidly degraded, and the released nitrogen is remobilized and reused in newly developing tissues. Autophagy facilitates the degradation of intracellular components for nutrient recycling in all eukaryotes, and recent studies have revealed critical roles for autophagy in Rubisco degradation and nitrogen remobilization into seeds in Arabidopsis (Arabidopsis thaliana). Here, we examined the function of autophagy in vegetative growth and nitrogen usage in a cereal plant, rice (Oryza sativa). An autophagy-disrupted rice mutant, Osatg7-1, showed reduced biomass production and nitrogen use efficiency compared with the wild type. While Osatg7-1 showed early visible leaf senescence, the nitrogen concentration remained high in the senescent leaves. 15N pulse chase analysis revealed suppression of nitrogen remobilization during leaf senescence in Osatg7-1. Accordingly, the reduction of nitrogen available for newly developing tissues in Osatg7-1 likely led its reduced leaf area and tillers. The limited leaf growth in Osatg7-1 decreased the photosynthetic capacity of the plant. Much of the nitrogen remaining in senescent leaves of Osatg7-1 was in soluble proteins, and the Rubisco concentration in senescing leaves of Osatg7-1 was about 2.5 times higher than in the wild type. Transmission electron micrographs showed a cytosolic fraction rich with organelles in senescent leaves of Osatg7-1. Our results suggest that autophagy contributes to efficient nitrogen remobilization at the whole-plant level by facilitating protein degradation for nitrogen recycling in senescent leaves.
Nitrogen is an essential nutrient that strongly influences plant growth and productivity. A large input of nitrogen fertilizer is required to maintain high crop yields but comes with high costs to both the farmer and the environment (Good et al., 2004; Brauer and Shelp, 2010). Nitrogen use efficiency (NUE), which can be defined in several ways, is recognized as an important target for improvement to promote sustainable and productive agriculture (Good et al., 2004; Masclaux-Daubresse et al., 2010; Kant et al., 2011). Thus, many efforts have been made to study the steps involved in nitrogen usage by plants, namely uptake, translocation, assimilation, and recycling.
Remobilization of assimilated nitrogen from senescent leaves greatly affects NUE in cereal crops, such as wheat (Triticum aestivum), barley (Hordeum vulgare), and rice (Oryza sativa; (Mae, 1997; Gregersen et al., 2008). During the vegetative growth stage, a large portion of the plant’s nitrogen is allocated to leaves (Mae and Ohira, 1981). In C3 plants, 75% to 80% of leaf nitrogen is found in chloroplasts, mainly as photosynthetic proteins (Makino and Osmond, 1991; Makino et al., 2003). The photosynthetic CO2-fixing enzyme Rubisco accounts for 12% to 35% of the leaf nitrogen (Evans and Seemann, 1989). Active influx of nitrogen occurs during leaf expansion and maturation, when chloroplasts are increasing in number and size and when the majority of Rubisco is biosynthesized (Mae et al., 1983). During leaf senescence, chloroplasts stop dividing, decrease in size and number, and finally differentiate into gerontoplasts (Krupinska, 2006). Rubisco and other photosynthetic proteins are rapidly degraded at this stage; the released nitrogen is sequentially remobilized into new leaves and other organs and ultimately stored in grains in cereal crops (Wittenbach, 1978; Yoneyama and Sano, 1978; Friedrich and Huffaker, 1980).
The mechanisms of Rubisco degradation in senescent cereal leaves have long been studied as a hallmark of nitrogen remobilization (Feller et al., 2008) but are still not fully elucidated. In the model plant Arabidopsis (Arabidopsis thaliana), recent studies have revealed that autophagy plays an important role in the degradation of chloroplasts and Rubisco in senescent leaves (Ishida et al., 2014). Autophagy is an evolutionarily conserved degradation system in eukaryotic cells (Nakatogawa et al., 2009), and several types and features of autophagy have been characterized. The best-known case occurs under nutrient starvation conditions, when intracellular components such as proteins and organelles are nonselectively sequestered by the autophagosome, transported into the vacuole, and then degraded and recycled. Autophagy-related (ATG) genes, which are essential for autophagy progression, were originally identified in yeast (Saccharomyces cerevisiae; Tsukada and Ohsumi, 1993), opening the door to the study of autophagy in diverse eukaryotes, including plants (Li and Vierstra, 2012; Liu and Bassham, 2012). More than 30 ATG genes have been identified in yeast, 15 of which are involved in the core machinery for membrane formation, which is essential for any type of autophagy (Nakatogawa et al., 2009). Orthologs of yeast ATG genes have been identified in each eukaryotic genome reported to date, and orthologs of most core ATG genes are found in both Arabidopsis and rice (Avin-Wittenberg et al., 2012; Yoshimoto, 2012). Reverse genetic approaches with Arabidopsis ATG-defective mutant (Atatgs) demonstrated that plants contain an autophagy system with molecular machineries homologous to those of yeast, and recent studies have explored plant-specific factors and the physiological roles of autophagy (Floyd et al., 2012; Yoshimoto, 2012).
The plant-specific organelle, the chloroplast, is also degraded by ATG-dependent autophagy. There are two pathways of chloroplast-targeted autophagy, namely piecemeal-type autophagy via Rubisco-containing bodies (RCBs) and whole-organelle autophagy termed chlorophagy (Ishida et al., 2014). RCBs are a specific type of autophagic bodies that contain stromal material from chloroplasts but do not contain thylakoid membranes or their proteins (Chiba et al., 2003; Ishida et al., 2008). These two chloroplastic autophagy pathways are partially responsible for the decreases in size and number of chloroplasts during the induced senescence of individually darkened leaves (Wada et al., 2009).
Recent studies on whole plants have clearly demonstrated the importance of autophagy in vegetative growth, seed production, and nitrogen remobilization in Arabidopsis (Guiboileau et al., 2012, 2013). Autophagy controls carbon and nitrogen status and protein concentration in leaves of Arabidopsis (Guiboileau et al., 2013). It is of interest to elucidate the role and contribution of autophagy to nitrogen management in plants other than Arabidopsis.
Rice is one of the most important food crops in the world and has long been used for investigations of nitrogen remobilization during senescence. At the vegetative stage in rice, the proportion of remobilized nitrogen out of the total nitrogen in a newly expanded leaf ranges from 30% to 100%, depending on the amount of external nitrogen supplied to the plant (Mae and Ohira, 1983; Mae, 1997). Such high utilization of remobilized nitrogen allows for balanced growth, even in the face of changes in nitrogen availability during the growth cycle (Mae, 1997). We recently isolated a retrotransposon Tos17-insertional OsATG7-knockout rice mutant (Osatg7-1) that is defective in autophagy (Kurusu et al., 2014). It showed a male-sterile phenotype, different from autophagy-defective mutants in Arabidopsis (Hanamata et al., 2014; Kurusu et al., 2014). Here, we focused on the growth and nitrogen usage of Osatg7-1 at the vegetative stage under normal nutrient conditions. Together with our recent study that directly demonstrated the progression of ATG-dependent autophagy and its involvement in chloroplast and Rubisco degradation in rice plants (Izumi et al., 2015), our data suggest that autophagy contributes to efficient vegetative growth and nitrogen utilization through protein degradation and nitrogen remobilization in senescent leaves of rice.
RESULTS
The Reduction of Biomass Production and NUE in the Osatg7 Mutant
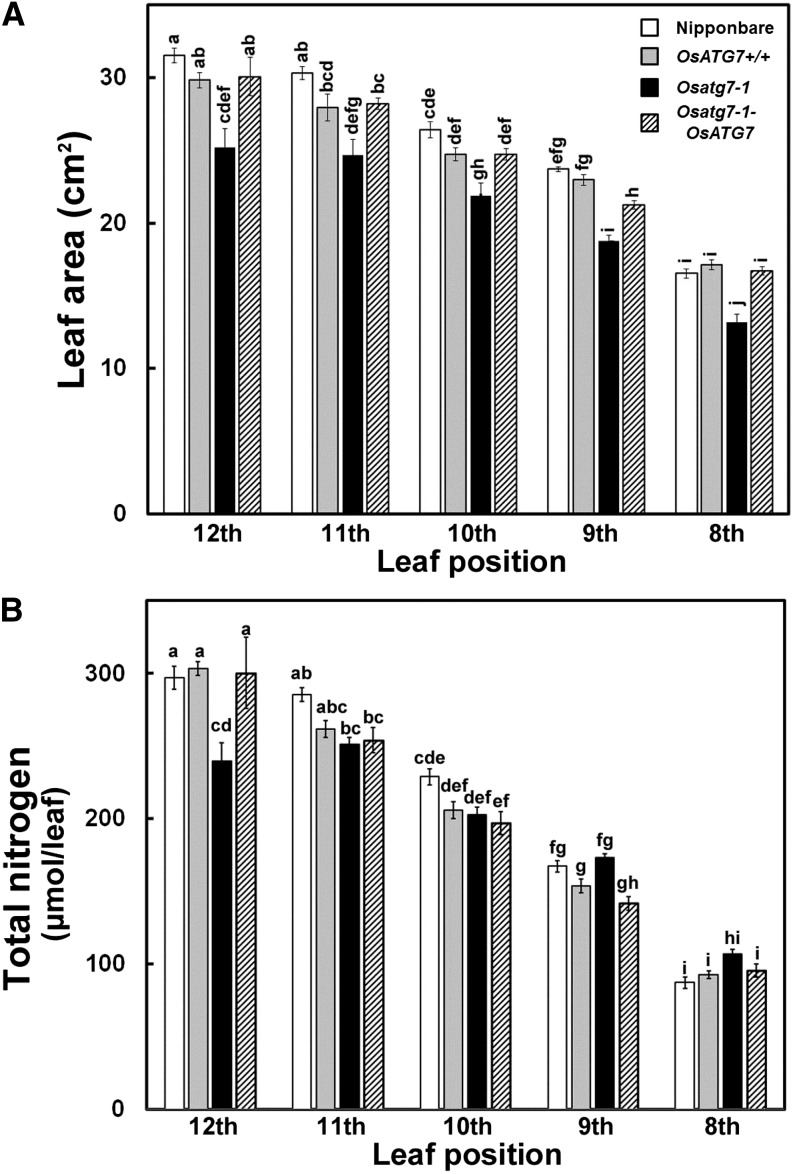
We first monitored the effects of autophagy disruption on vegetative growth in rice. Autophagy-disrupted mutants, Osatg7-1 (Kurusu et al., 2014), and control plants were grown hydroponically under ample-nutrient conditions (including 2 mm nitrogen) until the late vegetative stage (Fig. 1). Osatg7-1 was obviously smaller in both the shoots and roots throughout the growth stage compared with control plants, which included the original parental cultivar (Nipponbare), a wild-type line that was identified from the segregating mutant population (OsATG7+/+), and a complementation line (Osatg7-1-OsATG7; Fig. 1, A and B). Osatg7-1 was significantly smaller than the other genotypes not only in vertical size such as plant height and root length, but also in terms of tiller number and total leaf area (Fig. 2).
Figure 1.
The Osatg7-1 mutant shows reduced size in the vegetative stage. The mutant was grown hydroponically for 77 d after germination with the following control plants: parental cv Nipponbare, a wild-type line identified from the segregating mutant population (OsATG7+/+), and a complementation line (Osatg7-1-OsATG7). Shoots (A) and roots (B) are pictured; four plants are planted in each pot. Bars = 20 cm.
Figure 2.
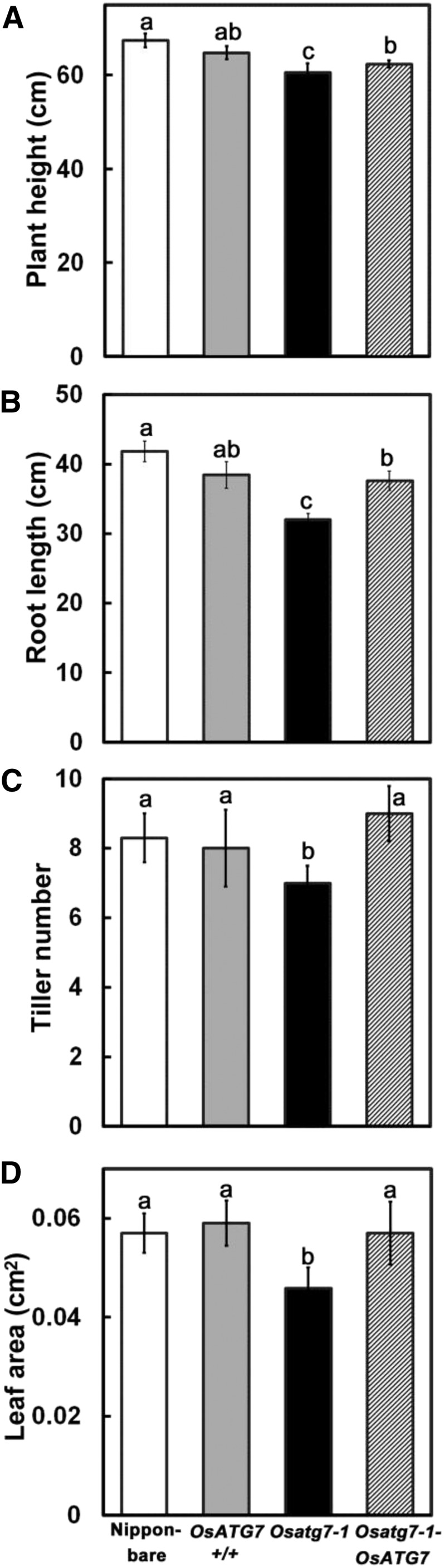
Comparison of plant height, root length, tiller number, and leaf area between Osatg7-1 mutants and control plants. The size of Osatg7-1 was reduced throughout the plant body. Plants were sampled at 77 d after germination, and plant height (A), root length (B), tiller number (C), and leaf area (D) were measured. Data are means ± sd (n = 8). Different letters denote significant differences in the parameter analyzed by Tukey’s honestly significant difference test (α = 0.05).
We next examined growth of Osatg7-1 under different nitrogen supply conditions (Supplemental Fig. S1). When the supplied nitrogen was even more abundant (6 mm) than in the ample condition (2 mm), Osatg7-1 grew to the same size as OsATG7+/+ (Supplemental Fig. S1). By contrast, under nitrogen-insufficient conditions (0.5 mm), the growth differences between Osatg7-1 and OsATG7+/+ were exacerbated. Therefore, the reduced size of Osatg7-1 under standard growth conditions appears to be largely related to nitrogen nutrition.
Consistent with the above data, the total biomass of Osatg7-1 was less than that of the control plants under ample-nutrient conditions (2 mm nitrogen; Fig. 3A). Whereas Osatg7-1 shows delayed heading in the reproductive stage (Kurusu et al., 2014), leaf aging in Osatg7-1 during the vegetative stage proceeded as in OsATG7+/+ (Supplemental Fig. S2). This trait suggested that the biomass reduction of Osatg7-1 in the vegetative stage was not associated with a delay of development. The whole-plant nitrogen content in Osatg7-1 was not significantly different from that in the control plants (Fig. 3B). The nitrogen concentration in whole plant (N%DW) was significantly higher in Osatg7-1 than in the control plants (Fig. 3C). As an indicator for NUE, nitrogen usage index (UI) was calculated by dividing shoot dry weight by shoot N%DW (Fig. 3D). UI is suitable for estimating the NUE at the vegetative stage (Good et al., 2004; Brauer and Shelp, 2010), and high UI indicates a plant with high biomass and low nitrogen concentration. Osatg7-1 showed a lower UI than the control plants. These plants were not significantly different in dry matter concentration on a whole-plant or per-leaf basis or in leaf thickness (Supplemental Fig. S3). These findings imply that the tissue structure and the major carbon content are similar in these lines. Together, these results suggest that autophagy contributes to nitrogen utilization within the plant rather than nitrogen uptake from the culture solution during the vegetative stage in rice.
Figure 3.
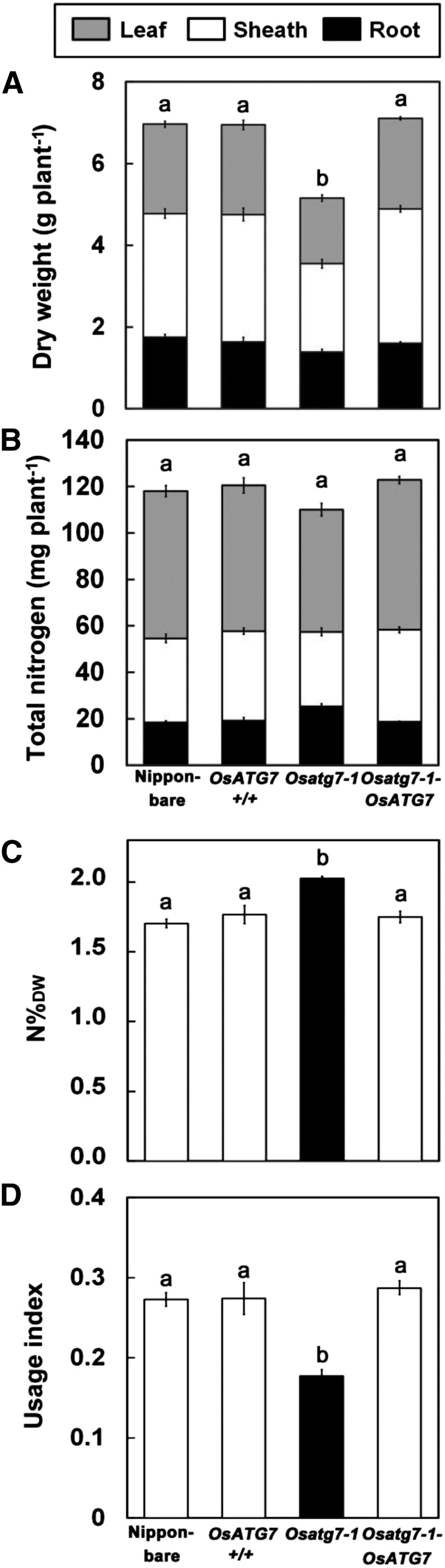
Reduced biomass and nitrogen UI of the Osatg7-1 mutant. All measurements were taken at 77 d after germination. Dry weight (A) and total nitrogen (B) were measured in leaf (gray), sheath (white), and root (black) separately. C, Nitrogen concentration in the whole plant is represented as N%DW (milligrams nitrogen per 100 mg dry weight). D, NUE was evaluated as UI, which is calculated by division of the shoot dry weight by the shoot N%DW. Data are means ± se (n = 4). Different letters denote significant differences (total value of the plant in the case of A and B) by Tukey’s honestly significant difference test (α = 0.05).
The Osatg7 Mutant Shows Early Visible Leaf Senescence, But the Nitrogen Concentration Remains High in Senescent Leaves
Nitrogen remobilization from senescing leaves is an important factor for high NUE at the level of the whole plant. As shown in Supplemental Figure S2, leaves on the main stem of Osatg7-1 emerged and expanded at essentially the same rate as those of OsATG7+/+. Therefore, we directly compared the progress of senescence and the decrease of components in Osatg7-1 and OsATG7+/+ using leaves at different positions on the main stem. Rice leaves begin senescence sequentially in the order of emergence at the vegetative stage. There is also an age gradient from the tip (old) to the base (young) within a single leaf. In both OsATG7+/+ and Osatg7-1, visible leaf senescence occurred in the same order, but it was clearly accelerated in the mutant (Fig. 4). In the lower seventh leaves, only the tip was withered and dried in OsATG7+/+, compared with up to half of the leaf in Osatg7-1 (Fig. 4). These phenotypes of early senescence and cell death in Osatg7-1 under favorable growth conditions are similar to those commonly observed in atg mutants in Arabidopsis (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2009).
Figure 4.
Visible leaf senescence appears early in the Osatg7-1 mutant. Leaves of main stems in 77-d-old plants are shown. The 12th leaves are the uppermost fully expanded leaves. Bars = 10 cm.
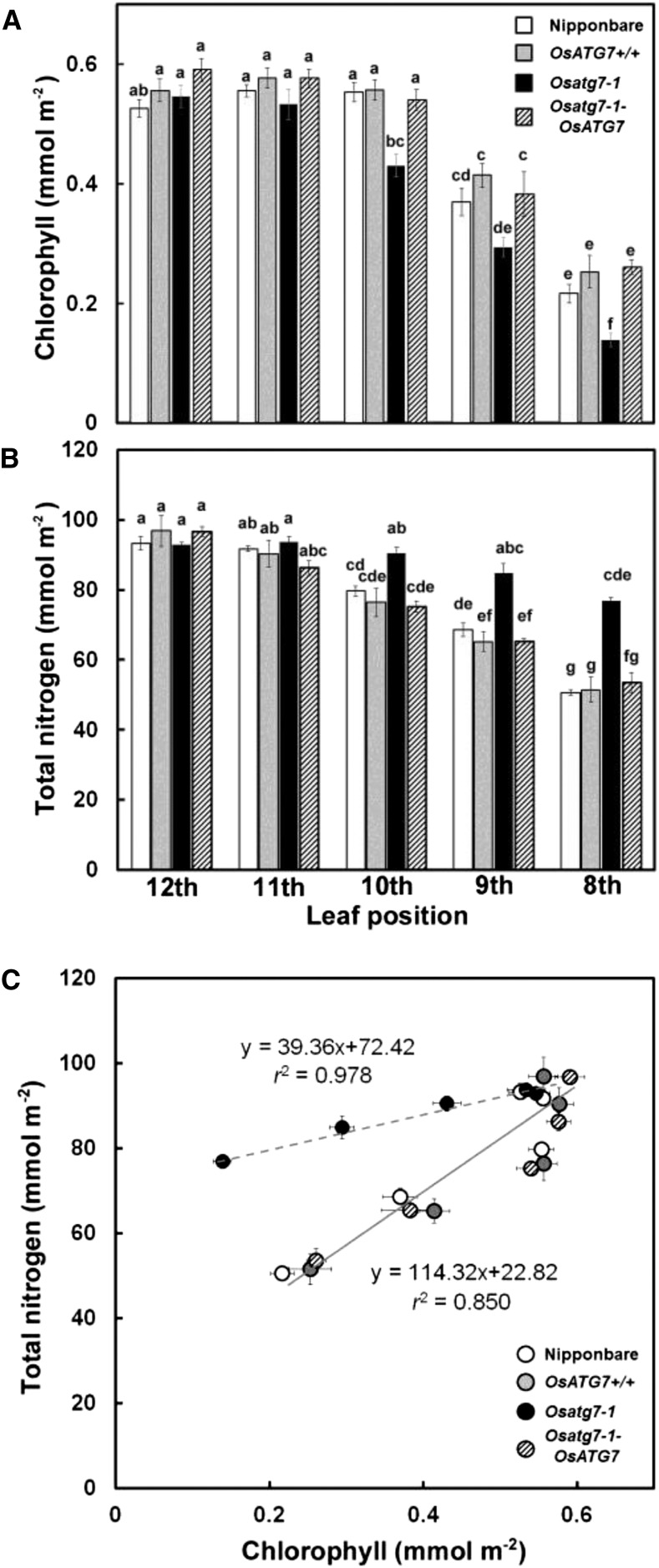
We measured chlorophyll and total nitrogen concentrations from the uppermost fully expanded leaves (12th leaves) to the lower senescent leaves (eighth leaves) of Osatg7-1 and the control plants (Fig. 5). There were no differences in chlorophyll and total nitrogen concentrations in the uppermost 12th leaves between Osatg7-1 and the other control plants (Fig. 5, A and B). Chlorophyll concentrations gradually decreased from the upper to lower leaves in each plant, and the decrease was especially accelerated in the lower leaves of Osatg7-1 (Fig. 5A). By contrast, the total nitrogen concentration of Osatg7-1 remained high in the lower leaves, with levels for each lower leaf significantly higher than those in control plants (Fig. 5B). Figure 5C shows the relationship between total nitrogen and chlorophyll in each plant, and only the gradient (senescence pattern) of Osatg7-1 was significantly different from that of the other plants. The soluble-protein nitrogen concentration in senescent leaves of Osatg7-1 was remarkably higher than that of other plants (Supplemental Fig. S4A). Only in the eighth leaves was the insoluble nitrogen concentration of Osatg7-1 significantly higher than that of the other plants (Supplemental Fig. S4B). There were no differences between Osatg7-1 and the control plants in the concentration of trichloroacetic acid (TCA)-soluble nitrogen (Supplemental Fig. S4C), which is mainly composed of low-molecular-weight nitrogen compounds such as free amino acids and inorganic nitrogen (Sudo et al., 2003).
Figure 5.
Defective autophagy affects processes of leaf senescence. Age-dependent changes in chlorophyll and total nitrogen were compared between Osatg7-1 and control plants. Chlorophyll (A) and total nitrogen (B) were determined in the whole leaf homogenates of the individual 12th to eighth leaves on the main stem of cv Nipponbare, OsATG7+/+, Osatg7-1, and Osatg7-1-ATG7. Data are means ± se (n = 4). Different letters in A and B denote significantly different values (Tukey’s honestly significant difference test, α = 0.05). In C, the data from A and B is used to show the relationship between total nitrogen and chlorophyll in each plant. The gradients shown by the regression lines (senescing pattern) of each plant were statistically analyzed by Student’s t test (α = 0.05), and only Osatg7-1 had a significantly different gradient line from the others. The dashed and black lines with equations are the regression lines for Osatg7-1 and the control plants, respectively.
Suppression of Nitrogen Remobilization during Leaf Senescence in the Osatg7 Mutant
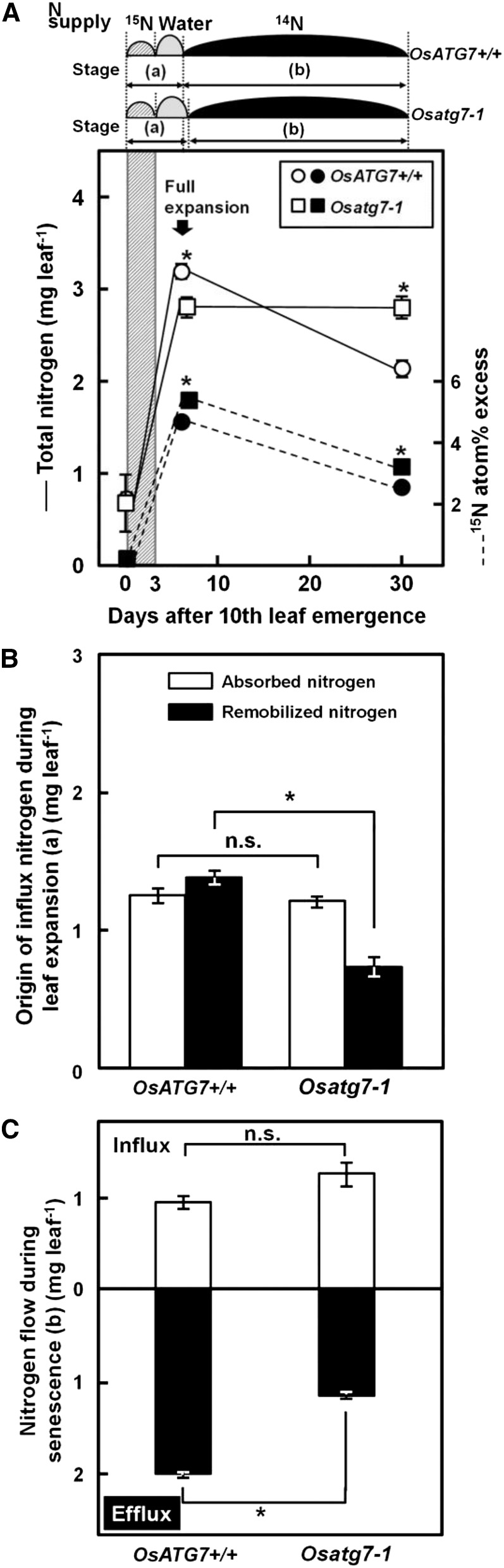
Next, we performed 15N pulse chase analysis to estimate the nitrogen flow in and out of the 10th leaves of OsATG7+/+ and Osatg7-1 from the leaf emergence to senescence (Fig. 6). The experiment was conducted similarly to that of Mae et al. (1983). 15N-labeled (15NH4)2SO4 was supplied at the time of 10th leaf emergence for 3 d, and then the culture solution was changed to pH-adjusted water until these leaves were fully expanded (expanding stage; Fig. 6A, a). After full expansion, plants were grown under ample-nutrient conditions with nonlabeled nitrogen (2 mm) until 30 d after leaf emergence (senescence stage; Fig. 6A, b). As shown in Supplemental Figure S2, leaf aging, i.e. the timing of leaf emergence and of full expansion, is almost identical between OsATG7+/+ and Osatg7-1. In this experiment, however, full expansion was delayed in Osatg7-1 by 1 d compared with OsATG7+/+ (Fig. 6A). Total nitrogen increased throughout the developing stage in both OsATG7+/+ and Osatg7-1, and the increase in nitrogen from leaf emergence was significantly greater in OsATG7+/+ than in Osatg7-1. However, the nitrogen concentration (per dry weight or per unit leaf area) was almost the same at full expansion (OsATG7+/+, 9.32 ± 0.21 mg nitrogen g–1 dry weight, and Osatg7-1, 9.10 ± 0.15 mg nitrogen g–1 dry weight; means ± se, n = 4). In the senescence stage, total nitrogen decreased by one-third in OsATG7+/+ but was stable in Osatg7-1 (Fig. 6A). These results were consistent with the nitrogen levels of upper and lower leaves (Fig. 5B). 15N atom% excess, the relative abundance of 15N among total nitrogen isotopes (15N + 14N), also greatly increased during the expansion stage, which indicates that there was influx of nitrogen absorbed in the roots (Fig. 6A). Based on this 15N atom% excess, the origin of nitrogen entering the leaves during expansion was separately estimated as absorbed nitrogen and remobilized nitrogen (Fig. 6B). The absorbed nitrogen was not significantly different between OsATG7+/+ and Osatg7-1; however, the remobilized nitrogen in Osatg7-1 was significantly (40%) less than in OsATG7+/+. Therefore, during leaf expansion, OsATG7+/+ depended upon remobilized nitrogen for 51% of its total leaf nitrogen, whereas remobilized nitrogen accounted for only 35% in Osatg7-1. During the leaf senescence stage, 15N atom% excess decreased in both OsATG7+/+ and Osatg7-1 (Fig. 6A), indicating replacement of nitrogen by a balance of influx and efflux. This nitrogen influx and efflux was separately estimated on the assumption that the nitrogen influx during this stage included a background level of 15N atom% excess (Fig. 6C). The nitrogen influx was not significantly different in the two lines, whereas the nitrogen efflux was significantly (42%) decreased in Osatg7-1.
Figure 6.
Leaf nitrogen remobilization is interrupted during vegetative stage in the Osatg7-1 mutant. Nitrogen flow in the 10th leaf of OsATG7+/+ and Osatg7-1 was estimated by 15N pulse chase analysis using elemental analyzer/isotope ratio mass spectrometry. Total nitrogen and 15N atom% excess of the 10th leaves were measured at three time points (just after leaf emergence, full expansion, and senescence; A). 15N-labeled (15NH4)2SO4 was supplied for 3 d (indicated as gray slashed column) when the 10th leaves were emerged. After 15N supply, culture medium was changed to pH-adjusted water until the 10th leaves were fully expanded (for 3 d in OsATG7+/+ and for 4 d in Osatg7-1). From full expansion onward, nutrient solution including nonlabeled nitrogen was supplied once a week until 30 d after 10th leaf emergence. White and black symbols, Total nitrogen amount and 15N atom% excess, on left and right axes, respectively; circles, OsATG7+/+; squares, Osatg7-1. The origin of nitrogen flow in the 10th leaves during leaf expansion (a) was estimated in B. Nitrogen influx and efflux during leaf senescence (b) were also estimated in C. In each graph, data are means ± se (n = 3–4). Asterisks indicate significant difference analyzed by Student’s t test (P < 0.05). Calculations in B and C are described in “Materials and Methods.” n.s., Not significant.
Remaining Proteins and Cell Components in Senescent Leaves of the Osatg7 Mutant
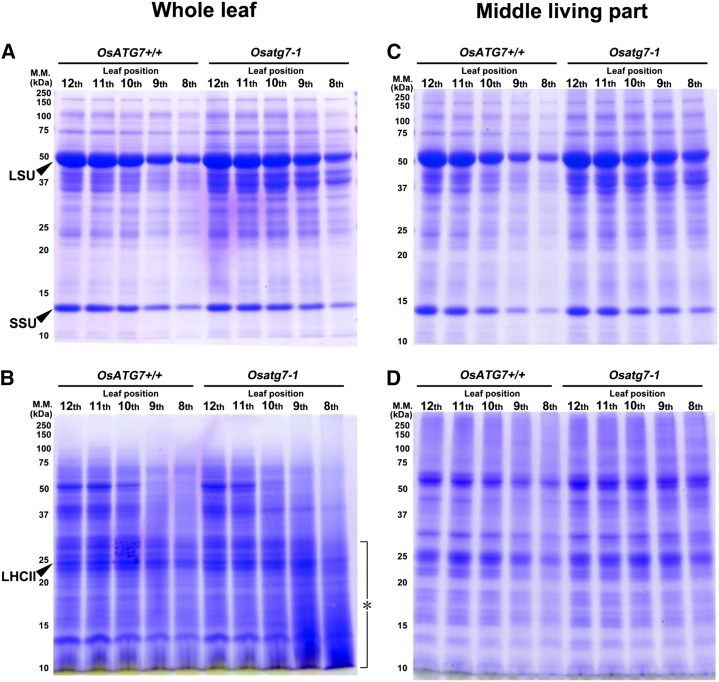
Age-dependent changes in polypeptide profiles and contents of OsATG7+/+ and Osatg7-1 were analyzed by SDS-PAGE (Fig. 7). Major polypeptides of soluble proteins gradually decreased from the upper to the lower leaf positions in OsATG7+/+, while relatively more remained in the lower leaves of Osatg7-1 (Fig. 7A), which corresponded well with the age-dependent changes of soluble protein nitrogen in those lines (Supplemental Fig. S4A). In the insoluble fraction, major polypeptides gradually decreased from the upper to the lower leaves in both lines (Fig. 7B). However, the senescent ninth and eighth leaves of Osatg7-1 accumulated smears of small (≤30 kD) polypeptides (Fig. 7B, asterisk), which were not found in OsATG7+/+. Based on their intensity in the gel, the accumulation of the smears seemed to correspond to the increase of insoluble nitrogen in the senescent leaves of Osatg7-1 (Supplemental Fig. S4B). To examine whether the smear was derived from the proteins that abnormally remained in the senescing leaves of Osatg7-1, we analyzed the polypeptide profile from the middle, living part of the leaf (Fig. 7, C and D). The quantifications of soluble protein nitrogen and insoluble nitrogen in the same samples are shown in Supplemental Figure S5. The smears found in the whole leaf extract were not detected in the extract from the living part of the leaves of Osatg7-1 (Fig. 7, B and D). In senescent leaves, the insoluble nitrogen level in the middle living part of Osatg7-1 was significantly higher than that of OsATG7+/+ (Supplemental Fig. S5B), but the difference between the two lines in the middle, living part was not greater than that in whole-leaf extracts (Supplemental Fig. S4B). Thus, the smear was most likely composed of protein derivatives precipitated mainly from the withered leaf tip. Moreover, the soluble proteins in the living part of the leaves of Osatg7-1 were more stable for each leaf position compared with the same analysis using whole-leaf extracts (Fig. 7, A and C; Supplemental Figs. S4A and S5A). It is possible that soluble proteins in the withered leaf tip were precipitated into the insoluble fraction in the whole-leaf extracts, and thus the level of soluble proteins was underestimated relative to the level of remaining protein that could be determined from living leaf tissue.
Figure 7.
Age-dependent degradation of bulk proteins is disrupted in the Osatg7-1 mutant. Soluble (A and C) and insoluble (B and D) proteins were extracted from whole leaves (A and B) and the middle, living parts (10 cm of the middle section of leaves; C and D) at different positions on the plant. Proteins from equal leaf area were applied to all lanes in the same gel for analysis by SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250. The asterisk indicates accumulation of smears of small polypeptides in lower leaves of Osatg7-1. Major leaf polypeptides in the soluble fraction (LSU and small subunit [SSU] of Rubisco) and insoluble fraction (LHCII) are indicated with black arrowheads. Sizes of molecular mass (M.M.; kilodaltons) markers are indicated on the left of the stained gel.
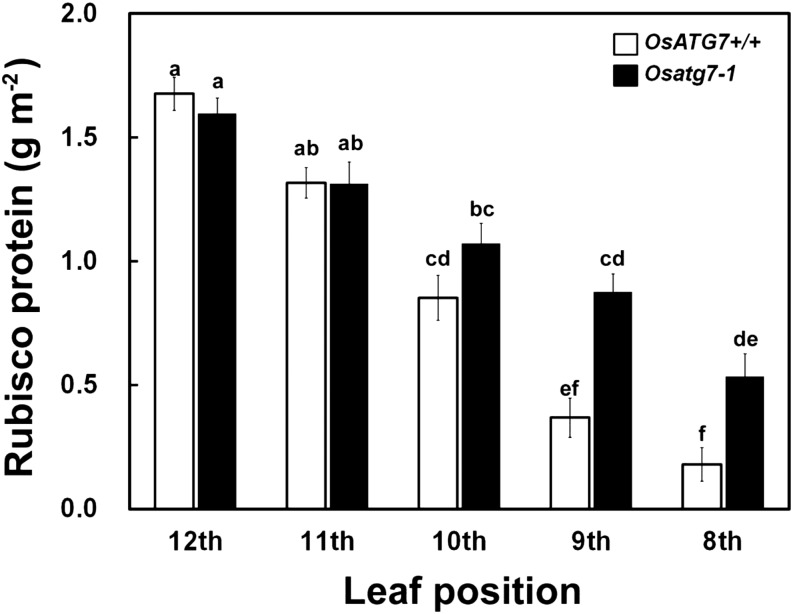
To address this, we determined the Rubisco protein concentrations in the middle, living parts of leaves (Fig. 8). In the OsATG7+/+ leaves, Rubisco markedly decreased toward the lower leaf positions. By contrast, in the leaves of Osatg7-1, Rubisco decreased to a lesser degree, so that in the senescent eighth leaf, the Rubisco level in Osatg7-1 was about 2.5 times higher than that in OsATG7+/+. This difference of Rubisco protein corresponded to the approximately 20% difference in whole soluble protein between the lines (Fig. 8; Supplemental Fig. S5A). Thus, it appears that the abnormally persisting Rubisco protein in Osatg7-1 is a large factor in the inhibition of nitrogen remobilization. Age-dependent changes of other chloroplastic proteins, including Gln synthetase (GS2) as a stromal protein and light-harvesting complex II (LHCII), coupling factor1 of ATPase (CF1), and cytochrome f (Cyt f) as thylakoid membrane proteins, were also semiquantitatively analyzed by immunoblotting (Supplemental Fig. S6). There were no significant differences in levels of LHCII, CF1, and Cyt f in any leaf positions, whereas the GS2 levels in senescent ninth and eighth leaves of Osatg7-1 were higher than those of OsATG7+/+ (Supplemental Fig. S6). These results indicated that, among chloroplast proteins, thylakoid membrane proteins decreased at a similar rate in Osatg7-1 as in OsATG7+/+. However, polypeptides other than Rubisco and GS2 were also stably detected in the senescent leaves of Osatg7-1 (Fig. 7C), which suggested that there was wide-ranging impairment of the degradation of soluble proteins in the mutant leaves.
Figure 8.
Age-dependent degradation of Rubisco protein is disrupted in the Osatg7-1 mutant. Rubisco protein concentration was determined in the middle, living parts (10 cm of the middle section) of leaves from different positions in OsATG7+/+ (white) and Osatg7-1 (black) plants. Data are means ± se (n = 4). Different letters in the graph denote significant difference compared with values for leaves analyzed by Tukey’s honestly significant difference test (α = 0.05).
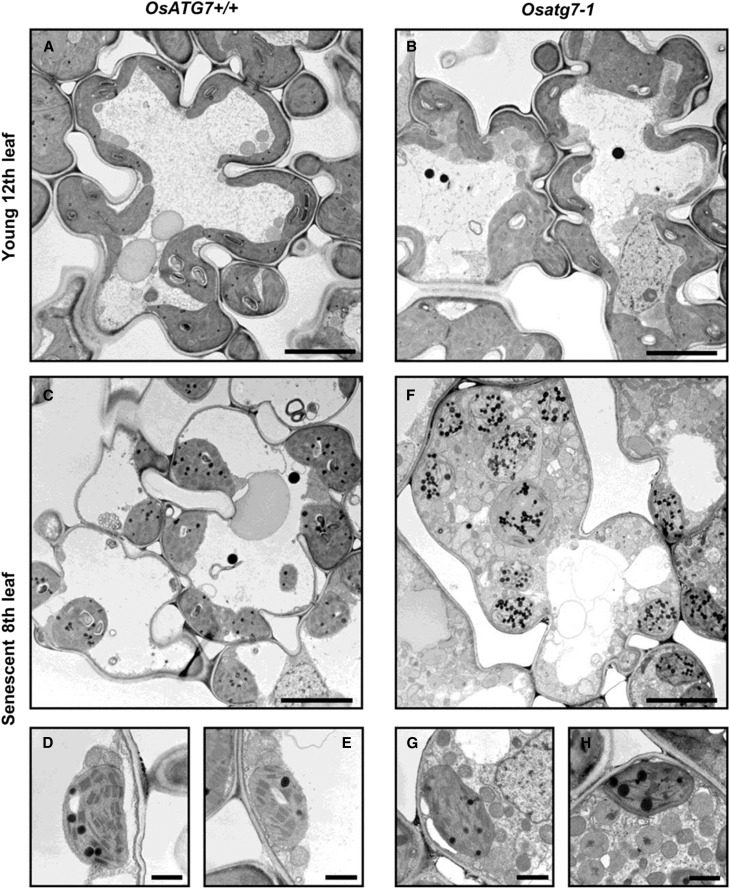
We next compared the intracellular components of mesophyll cells in leaves of Osatg7-1 with those of OsATG7+/+ by transmission electron microscopy (Fig. 9). In young 12th leaves, mesophyll cells of both OsATG7+/+ and Osatg7-1 exhibited no apparent differences (Fig. 9, A and B). In senescent eighth leaves, OsATG7+/+ had a smaller cytosolic fraction with fewer organelles and smaller chloroplasts than the young leaves (Fig. 9, C–E). By contrast, Osatg7-1 contained a rich cytosolic fraction filled with numerous and various organelles, including chloroplasts, mitochondria, peroxisomes, and Golgi apparatus (Fig. 9, F–H). These results suggested that the degradation of bulk leaf cell components was impaired in the senescent leaves of Osatg7-1. In addition, plastoglobules were more abundant in the senescent chloroplasts of Osatg7-1 than in those of OsATG7+/+ (Fig. 9, C–E and F–H), which might reflect enhanced catabolism of chlorophylls and proteins within chloroplasts in Osatg7-1 (Lundquist et al., 2012).
Figure 9.
Ultrastructure of mesophyll cells in young and senescent leaves of OsATG7+/+ and Osatg7-1. Mesophyll cells of senescent Osatg7-1 leaves have a cytosol with numerous organelles. Samples from the uppermost young 12th (A and B) and senescent eighth leaves (C–H) of OsATG7+/+ (A and C–E) and Osatg7-1 (B and F–H) at 77 d after germination were used for transmission electron microscope analysis. Images are focused on the cytosolic components in D, E, G, and H. Bars = 5 μm (A–C and E) and 1 μm (D, E, G, and H).
Leaf Photosynthesis in the Osatg7 Mutant
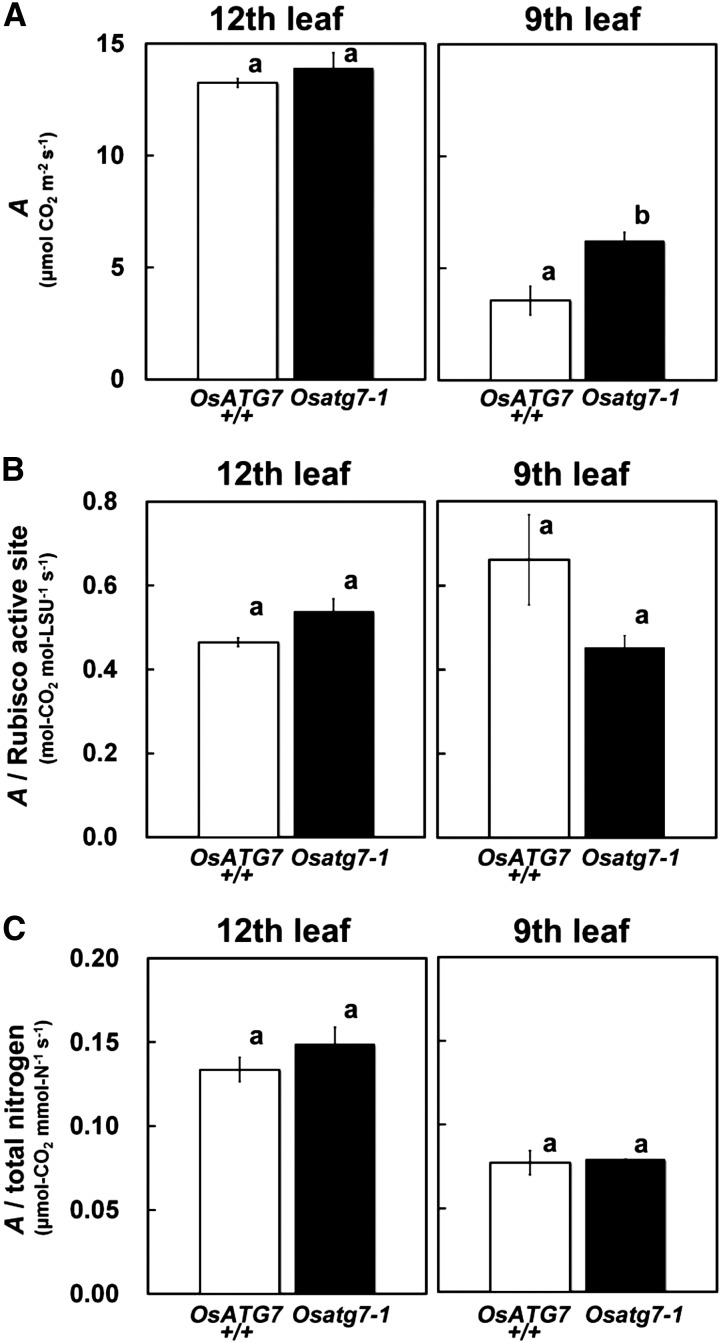
We measured the CO2 assimilation rate (A) under light-saturated conditions (photosynthetic photon flux density [PPFD] = 1,500 μmol m−2 s−1) at internal CO2 concentration of the leaf (pCi) = 20 Pa in the 12th and ninth leaves of OsATG7+/+ and Osatg7-1 (Fig. 10), because A at pCi = 20 Pa is limited by Rubisco (von Caemmerer and Farquhar, 1981). There were no differences in A between OsATG7+/+ and Osatg7-1 in the uppermost 12th leaves (Fig. 10A). However, A in the senescent ninth leaves of Osatg7-1 was significantly higher than that of OsATG7+/+ (Fig. 10A). There was no significant difference in A per Rubisco active site between the two lines, and the values in the ninth leaves were similar to those in the 12th leaves (Fig. 10B). There was no significant difference in A per total nitrogen between the two lines in either the 12th or ninth leaves (Fig. 10C). Recently, it was reported that Arabidopsis atg mutants accumulate degradation products of Rubisco-large subunit (LSU) and chloroplastic Gln synthetase (GS2) in senescent leaves (Guiboileau et al., 2013). We therefore examined age-dependent changes in immunoblot profiles of Rubisco-LSU and Gln synthetase polypeptides and found that there was no specific accumulation of degradation products of those polypeptides in senescent leaves of Osatg7-1 (Supplemental Fig. S7). Collectively, these results indicate that the abnormally persistent Rubisco protein exists mostly in an intact (potentially functional) form in the senescent leaves of Osatg7-1.
Figure 10.
The Rubisco and nitrogen that remain in senescent leaves of Osatg7-1 are functional for photosynthesis. The A (A) and its ratio to Rubisco active sites (B) and to total nitrogen (C) were measured at pCi = 20 Pa and PPFD = 1,500 μmol m–2 s–1 in the middle, living parts of the uppermost fully expanded 12th leaves and the lower senescent ninth leaves of 77-d-old plants. Data are means ± se (n = 4). Different letters in each graph denote significant difference based on Student’s t test (α = 0.05).
To estimate the photosynthetic capacity of a whole leaf, we compared leaf area and total nitrogen per leaf at different leaf positions between the control plants and Osatg7-1 (Fig. 11). The leaf area of Osatg7-1 was smaller than that of control plants at each leaf position (Fig. 11A). As a result, the total nitrogen per leaf of Osatg7-1 was significantly lower than that of the control plants at the uppermost 12th leaves, and it was similar between Osatg7-1 and control plants in the lower leaves (Fig. 11B). These data suggest that the photosynthetic capacity of the most active center leaves of Osatg7-1 is lower than that of the control plants and that this underlies the reduced biomass of the mutants compared with the control plants.
Figure 11.
Comparison of leaf area and total nitrogen in leaves of Osatg7-1 and control plants. Leaf area (A) and total nitrogen content per leaf (B) from 12th to eighth leaves on the main stem of cv Nipponbare, OsATG7+/+, Osatg7-1, and Osatg7-1-ATG7 are shown. Data are means ± se (n = 4). Different letters denote significant difference based on Tukey’s honestly significant difference test (α = 0.05).
DISCUSSION
The data presented here indicate that autophagy supports the vegetative growth of rice by facilitating efficient nitrogen usage within the plant, specifically nitrogen remobilization from senescent leaves. In spite of containing the same amount of nitrogen as the control plants, Osatg7-1 showed reduced biomass. This indicates that defective autophagy causes a decrease of the UI (NUE) during the vegetative stage in rice. Although leaves in Osatg7-1 were smaller than in OsATG7+/+, there were no significant differences in total nitrogen, chlorophyll, and Rubisco concentrations or in A per unit leaf area between the uppermost fully expanded leaves of OsATG7+/+ and Osatg7-1. Thus, photosynthetic NUE was calculated to be the same between the two lines in those young leaves. Therefore, autophagy appears to be relatively less important for the development of photosynthetic function during leaf expansion. By contrast, significant differences between OsATG7+/+ and Osatg7-1 plants were found in the lower senescent leaves. Consistent with accelerated visible leaf senescence, the gradual decrease of chlorophyll concentration in the lower leaves was accelerated in Osatg7-1. Although the decrease of chlorophyll concentration is generally taken as a visible symptom of nitrogen remobilization, the two phenomena are not always correlated. In actuality, the protein degradation for nitrogen remobilization occurs in advance of chlorophyll breakdown during leaf senescence in rice (Makino et al., 1983). Accordingly, the early yellowing leaves of Osatg7-1 would be expected to contain less nitrogen than the corresponding leaves of the control plants. The decrease in leaf nitrogen of the lower senescing leaves of the control plants reflects a typical pattern for age-dependent nitrogen remobilization. However, the nitrogen level was abnormally high in the senescent leaves of Osatg7-1. 15N pulse chase analysis showed that the abnormal persistence of nitrogen in the senescent leaves of Osatg7-1 resulted from decreased nitrogen remobilization during leaf senescence. This represents strong evidence implicating autophagy as playing a critical role in nitrogen remobilization during leaf senescence in rice.
The growth of cereal leaves strongly depends on nitrogen nutrition. It was reported that 45% of the structural nitrogen in the 12th leaf of rice was derived from remobilized nitrogen under conditions of hydroponic culture with 2 mm nitrogen (Mae and Ohira, 1981). Here, we cultivated the rice in basically the same conditions as in that previous report (Mae and Ohira, 1981) and similarly found that the 10th leaves of OsATG7+/+ contained 51% remobilized nitrogen (Fig. 6B). By contrast, remobilized nitrogen was reduced by 35% in Osatg7-1 compared with OsATG7+/+, resulting in a significantly decreased nitrogen supply for newly expanding leaves. Under nitrogen-limited culture conditions, rice plants exhibit decreases in both leaf area and nitrogen content per unit leaf area (Makino et al., 1997). However, leaf nitrogen concentration in the uppermost fully expanded leaf of Osatg7-1 was the same as in OsATG7+/+ (Fig. 5). Instead, Osatg7-1 grew fewer tillers and smaller leaves at each stage, which would help to maintain the nitrogen concentration (Figs. 2 and 11A). Therefore, the responses to remobilized nitrogen limitation do not seem to be the same as those for nitrogen supply limitation in rice.
During the lifespan of a leaf, the leaf nitrogen status, especially the Rubisco protein concentration, determines photosynthetic capacity (Makino et al., 1983). On a per unit leaf area basis, the potential photosynthetic capacity (reported as A) of the uppermost fully expanded leaves was the same in OsATG7+/+ and Osatg7-1, whereas A of the middle, living parts of lower senescent leaves was greater in Osatg7-1 than in OsATG7+/+, reflecting the higher concentrations of total nitrogen and Rubisco in the former (Fig. 10). The persistent high nitrogen in the senescent leaves of Osatg7-1 could have a positive effect on leaf photosynthesis. In actuality, however, Osatg7-1 had reduced leaf size at each leaf position (Fig. 11A). Therefore, compared with the control plants, the total nitrogen contents of the whole leaf of Osatg7-1 were lower in the top fully expanded leaf (12th leaf) and were not significantly different even in the lower senescent leaves (eighth and ninth leaves; Fig. 11B). Taking nitrogen content to represent photosynthetic capacity, this would indicate that, on the individual leaf basis, the most active center leaf of Osatg7-1 had reduced photosynthetic capacity compared with the control plants. Even in the lower leaves, there would be no significant difference in photosynthetic capacity between lines. Leaf senescence was accelerated in Osatg7-1, and a portion of the nitrogen was precipitated in the withered leaf tip (Fig. 7). This precipitated nitrogen did not seem to be active for photosynthesis. In addition, lower leaves are shaded by upper leaves of the same plant or leaves of neighboring plants even in the pot culture conditions used here (Fig. 1); thus, they usually cannot perform to the same potential as upper leaves. Considering these factors, the photosynthetic output of an individual leaf of Osatg7-1 is estimated not to be more than that of the control plants throughout the leaf lifespan. This could underlie the lower overall plant biomass of Osatg7-1 compared with that of the control plants.
Leaf soluble proteins are the predominant nitrogen source for remobilization and are rapidly degraded from the early stage of leaf senescence. The data presented here indicate that Osatg7-1 is impaired in the degradation of soluble proteins in senescing leaves and consequently experiences reduced nitrogen remobilization. The age-dependent Rubisco decrease is delayed in Osatg7-1 (Fig. 8). This is consistent with previous reports showing that Rubisco is mobilized to the vacuole via RCBs by autophagy for degradation in senescing leaves of Arabidopsis (Ono et al., 2013) and that this pathway also functions in rice (Izumi et al., 2015). However, the age-dependent decrease in soluble proteins and nitrogen did not seem to be completely blocked, which suggests that alternative ATG-independent pathways contribute to the protein recycling in senescent leaves of rice.
With regard to insoluble proteins, the level of insoluble nitrogen in the senescent leaves of Osatg7-1 was slightly higher than that in control plants (Supplemental Fig. S5B). In addition to RCBs, autophagy in Arabidopsis transports and degrades whole chloroplasts including thylakoid membranes in the vacuole (chlorophagy; Wada et al., 2009). However, the levels of some thylakoid membrane proteins (LHCII, CF1, and Cyt f) in Osatg7-1 were slightly but not significantly higher than in OsATG7+/+ (Supplemental Fig. S6, B–D). This suggests that the contribution of chlorophagy to the degradation of thylakoid proteins is not greater than the contribution of RCBs to the degradation of stromal proteins such as Rubisco and GS2. These results are also suggestive of other autophagy-independent proteolysis pathways, such as chloroplast-localized proteases, in Osatg7-1. In addition, the level of TCA-soluble nitrogen was not significantly different between control plants and Osatg7-1 at each leaf position (Supplemental Fig. S4C). Together with the nitrogen efflux analyzed during leaf senescence (Fig. 6C), these fractionated nitrogen data suggest that Osatg7-1 is able to accomplish amino acid transport for nitrogen remobilization but is partially impaired in the process of protein degradation to produce transportable nitrogen.
The 15N pulse chase analysis indicated that the nitrogen remaining in the senescent leaves of Osatg7-1 resulted from the suppression of nitrogen remobilization (Fig. 6). However, could any other factors be responsible? For example, Osatg7-1 showed early leaf senescence and cell death. In Arabidopsis, autophagy deficiency causes enhanced cell death in senescence and immunity responses due to excessive salicylic acid signaling (Yoshimoto et al., 2009). Double mutants of atg5 NahG or atg5 sid2, in which salicylic acid signaling is dampened, show normal leaf senescence and partially recovered vegetative and seed biomass, but those mutants do not recover grain NUE and NRE from vegetative tissue to seeds (Guiboileau et al., 2012). Therefore, it appears that autophagy itself, rather than early senescence, is the main factor affecting nitrogen remobilization in Arabidopsis (Guiboileau et al., 2012). At present, it is not known if early leaf senescence in Osatg7-1 is caused by the same mechanisms as in Atatg mutants, and that merits further investigation.
Recent studies have revealed that autophagy is the main factor affecting nitrogen remobilization from senescing leaves to seeds in Arabidopsis (Guiboileau et al., 2012). The contribution of autophagy to nitrogen remobilization from leaves to seeds needs to be analyzed and evaluated further in rice. Autophagy plays a critical role in pollen development, and the male sterility phenotype of Osatg mutants (Kurusu et al., 2014) prevented us from examining the contribution of autophagy-mediated nitrogen remobilization to seeds of rice in this study. In rice, remobilized nitrogen from vegetative organs accounts for 70% to 90% of total panicle nitrogen (Mae, 1997). Therefore, in addition to its critical role in anther development in rice, autophagy may also be important as a remobilized nitrogen supply system, which would influence seed quality and quantity. Anther-specific rescue of autophagy in Osatg genes or development of reduced-autophagy lines (OsATG antisense or RNA interference) may help to elucidate the contribution to rice seed maturation of autophagy in vegetative tissues.
In conclusion, our results establish autophagy as a key process for the maintenance of high NUE during vegetative growth in rice. Thus, proper and specific enhancement of autophagy in senescing leaves might improve NUE, especially in relatively lower-NUE crop species and varieties. To adopt this strategy for crop improvement, we will need to further elucidate the regulatory mechanisms for autophagy in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tos17-insertional rice (Oryza sativa) atg7 mutant (Osatg7-1; OsATG7–/–; Kurusu et al., 2014) and the wild-type (OsATG7+/+) plants were identified in the seed pool obtained from heterozygous plants (OsATG7+/–) by genomic PCR using the following primer mixture: OsATG7 forward primer 5′-AAAAAGAAGGGAGAGCTCGCAGT-3′, Tos-17 forward primer 5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′, and OsATG7 reverse primer 5′-TCACAAACCACCAACTCAGGC-3′. The original parental cultivar of Tos17-insertional mutants, cv Nipponbare, and the OsATG7 genome complementation line (Osatg7-1-OsATG7; Kurusu et al., 2014) were also used as control plants for Osatg7-1. Plants were grown hydroponically in an environmentally controlled growth chamber with a 14-h photoperiod from natural light and supplemental lighting and day/night temperature of 27°C /22°C. The nutrient composition and applied conditions were the same as previously described (Mae and Ohira, 1981). For nitrogen-abundant and -insufficient conditions, the nitrogen supply (normally 1 mm NH4NO3 for 2 mm nitrogen) was replaced by 1.5 mm NH4NO3 with 3.0 mm NaNO3 for 6 mm nitrogen and 0.25 mm NH4NO3 for 0.5 mm nitrogen, respectively. Plant biomass and NUE were measured repeatedly with four plants in three independent cultivations. In other experiments, two independent biological replicates were carried out with four plant samples. In each set of experiments, similar results were obtained, and representative results are shown in figures.
Biomass Measurement and Assessment of NUE
Plants were sampled by separation into leaf blades (represented as leaf), leaf sheaths (sheath), and roots (root) at 77 d after germination. After their sizes and leaf area were measured, all samples were kept in a dry oven at 80°C for a week, and dry mass was measured. The nitrogen contents were determined by the Kjeldahl method as previously described (Makino et al., 1997). Dry samples were first ground to powder, and the aliquots were used for nitrogen determination. NUE was calculated and represented as nitrogen UI [UI = DWShoot (Leaf + Sheath)/NShoot/DWShoot].
Analysis of Age-Dependent Change of Leaf Components
The 12th to eighth fresh leaves on the main stem were sampled 77 d after germination, frozen in liquid N2, and kept at –80°C until analysis. A set of age-gradient leaves was sampled from the same plant, and four sets of leaves (from four plants) were used in one series of experiments. To sample the middle, living part, leaf tips and bottoms were cut off, and middle parts (10 cm long) were used for experiments.
The nitrogen contents were determined by the Kjeldahl method as previously described (Makino et al., 1997). Samples were homogenized in 50 mm sodium phosphate buffer (pH 7.0) containing 0.8% (v/v) 2-mercaptoethanol, 2 mm iodoacetic acid, and 5% (v/v) glycerol (Makino et al., 1988), and the total nitrogen contents were determined from a portion of the homogenate. Chlorophyll contents were determined from another portion of the homogenate according to the method of Arnon (1949). The homogenates were separated into soluble fractions and insoluble fractions by centrifugation. The insoluble nitrogen was determined using the insoluble fraction after resuspension with homogenization buffer. After the nitrogen content in the soluble fraction was determined, an aliquot of the soluble fraction was added to an equal volume of 10% (w/v) TCA to precipitate the soluble proteins. After centrifugation at 2,000g for 30 min, the soluble protein nitrogen was determined from the pellet. The free nitrogen content was calculated by subtracting the soluble protein nitrogen from soluble fraction nitrogen. Protein samples were prepared from the same homogenates as used for total nitrogen and chlorophyll determinations. For the subsequent SDS-PAGE analysis, portions of the soluble and resuspended insoluble fractions were added to an equal volume of SDS sample buffer (200 mm Tris-HCl containing 2% [v/v] SDS, 20% [v/v] glycerol, and 5% [v/v] 2-mercaptoethanol) and boiled for 3 min. These samples were kept at –30°C until analysis.
15N Pulse Chase Analysis
The 10th leaves on the main stem were used for 15N pulse chase analysis. When most of the 10th leaves were just emerged from the sheath of the ninth leaves, the nutrient solution was replaced with one containing 15N-labeled 1 mm (15NH4)2SO4 (10.6 atom% excess) for 3 d. Then, plants were cultured in pH-adjusted water (pH 5.3) until full expansion of the 10th leaves. After full expansion, regular nutrient solution containing 1 mm NH4NO3 was resumed until 30 d after the emergence of the 10th leaves. During the experiment, the 10th leaves were sampled at three time points, leaf emergence (15N supply), full expansion, and 30 d after leaf emergence. All samples were kept in a dry oven at 80°C for a week, and dry mass was measured. Dried samples were ground to powder, and approximately 1-mg aliquots were wrapped in tin foil and applied to an elemental analyzer/isotope ratio mass spectrometry system (Thermo Scientific, Flash2000/Delta V Advantage).
Absorbed and remobilized nitrogen during the leaf expansion stage (Fig. 6A, a) were estimated using the following equations.
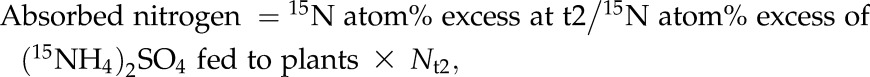 |
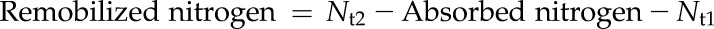 |
Influx and efflux nitrogen during the leaf senescence stage (Fig. 6A, b) was estimated as follows.
 |
Nt1, Nt2, and Nt3 are total nitrogen of the 10th leaves at sampling time point t1, t2, and t3. t1 is leaf emergence, t2 is leaf full expansion, and t3 is 30 d after leaf emergence. As the background 15N atom% excess in the samples, 15N atom% excess at t1 was analyzed by the same method and considered in the equations using the average value 0.36.
SDS-PAGE, Immunoblotting, and Rubisco Determination
SDS-PAGE was performed as previously described (Ishida et al., 1999). Immunoblotting was performed as previously described (Towbin et al., 1979). Antibodies used were affinity-purified anti-LSU of Rubisco (Ishida et al., 1997), anti-maize (Zea mays) chloroplastic Gln synthetase (gift from Dr. Hitoshi Sakakibara; Ishida et al., 2002), anti-rice LHCII (Hidema et al., 1992), anti-rice CF1, and anti-rice Cyt f (Hidema et al., 1991). Protein signals were detected by enhanced chemiluminescence, and semiquantitative analysis was performed using a LAS-4000 Mini/Multigauge system (Fujifilm). Rubisco content was determined spectrophotometrically by formamide extraction of the Coomassie Brilliant Blue R-250-stained subunit bands corresponding to the large and small subunits of Rubisco after SDS-PAGE using calibration curves made with Rubisco purified from rice leaves (Makino et al., 1994).
Transmission Electron Microscopy
Immediately after sampling, the middle, living part of the 12th and ninth leaves of the control and Osatg7-1 were fixed with 50 mm cacodylate buffer (pH 7.4) containing 2% (v/v) glutaraldehyde and 2% (v/v) paraformaldehyde overnight at 4°C, after which samples were rinsed with 50 mm cacodylate buffer and postfixed with 2% (v/v) osmium tetroxide for 3 h at 4°C. After dehydration by ethanol gradient treatment, samples were embedded and polymerized in resin at 60°C for 2 d. The blocks were ultra-thin sectioned at 80 nm with a diamond knife attached to an ultramicrotome (Ultracut UCT, Leica Microsystems). These sections were placed on copper grids and stained with 2% (w/v) uranyl acetate. The grids were observation using a transmission electron microscope (JEM-1200EX, JEOL), and images were captured by a CCD camera (VELETA, Olympus).
Gas Exchange Measurement
As were measured with a portable gas exchange system (LI-6400, Li-Cor) as previously described (Hirotsu et al., 2004). As the size of the chamber was 2 × 3 cm (width × length), the rates were measured at middle, living parts of the leaves. The relative humidity in the chamber was maintained at 60% to 70% with a dew point generator (LI-610, Li-Cor). Irradiance was provided by a light-emitting diode light source (6400-02B, Li-Cor) and adjusted to a PPFD of 1,500 μm mol quanta m–2 s–1 at the leaf surface in the chamber. All measurements were performed at a pCi of 20 Pa. Gas exchange parameters were calculated according to previously described equations (von Caemmerer and Farquhar, 1981).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of nitrogen nutrition on growth of Osatg7-1.
Supplemental Figure S2. Leaf aging of Osatg7-1 and OsATG7+/+.
Supplemental Figure S3. Evaluation of tissue dry matter concentrations between Osatg7-1 and control plants.
Supplemental Figure S4. Age-dependent changes in the fractionated nitrogen prepared from whole leaf extracts.
Supplemental Figure S5. Age-dependent changes in the fractionated nitrogen prepared from middle, living part of the leaves.
Supplemental Figure S6. Age-dependent changes in thylakoid proteins.
Supplemental Figure S7. Age-dependent changes in immunoblot profiles of Rubisco LSU and GS.
Supplementary Material
Acknowledgments
We thank Dr. Hitoshi Sakakibara for the gift of anti-GS2 antibodies and Dr. Louis Irving for advice about statistical analysis.
Glossary
- NUE
nitrogen use efficiency
- RCB
Rubisco-containing body
- N%DW
nitrogen concentration in whole plant
- UI
usage index
- TCA
trichloroacetic acid
- A
CO2 assimilation rate
- LSU
large subunit
Footnotes
This work was supported in part by KAKENHI (grant nos. 24380037, 25119703, and 26660286 to H.I.) and the GRENE/NC-CARP project (to A.M.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Articles can be viewed without a subscription.
References
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Honig A, Galili G (2012) Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249: 285–299 [DOI] [PubMed] [Google Scholar]
- Brauer EK, Shelp BJ (2010) Nitrogen use efficiency: re-consideration of the bioengineering approach. Botany 88: 103–109 [Google Scholar]
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T (2003) Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol 44: 914–921 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Evans JR, Seemann JR (1989) The allocation of protein nitrogen in the photosynthetic apparatus: costs, consequences, and control. In Briggs WR, ed, Photosynthesis. Alan R. Liss, Inc., New York, pp 183–205 [Google Scholar]
- Feller U, Anders I, Mae T (2008) Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot 59: 1615–1624 [DOI] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, Macintosh GC, Bassham DC (2012) What to eat: evidence for selective autophagy in plants. J Integr Plant Biol 54: 907–920 [DOI] [PubMed] [Google Scholar]
- Friedrich JW, Huffaker RC (1980) Photosynthesis, leaf resistances, and ribulose-1,5-bisphosphate carboxylase degradation in senescing barley leaves. Plant Physiol 65: 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9: 597–605 [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB, Krupinska K (2008) Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol (Stuttg) (Suppl 1) 10: 37–49 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux-Daubresse C (2013) Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol 199: 683–694 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanamata S, Kurusu T, Kuchitsu K (2014) Roles of autophagy in male reproductive development in plants. Front Plant Sci 5: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidema J, Makino A, Kurita Y, Mae T, Ojima K (1992) Changes in the levels of chlorophyll and light-harvesting chlorophyll a/b protein of PS II in rice leaves aged under different irradiances from full expansion through senescence. Plant Cell Physiol 33: 1209–1214 [Google Scholar]
- Hidema J, Makino A, Mae T, Ojima K (1991) Photosynthetic characteristics of rice leaves aged under different irradiances from full expansion through senescence. Plant Physiol 97: 1287–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu N, Makino A, Ushio A, Mae T (2004) Changes in the thermal dissipation and the electron flow in the water-water cycle in rice grown under conditions of physiologically low temperature. Plant Cell Physiol 45: 635–644 [DOI] [PubMed] [Google Scholar]
- Ishida H, Anzawa D, Kokubun N, Makino A, Mae T (2002) Direct evidence for non-enzymatic fragmentation of chloroplastic glutamine synthetase by a reactive oxygen species. Plant Cell Environ 25: 625–631 [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A (2014) Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 1837: 512–521 [DOI] [PubMed] [Google Scholar]
- Ishida H, Makino A, Mae T (1999) Fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species occurs near Gly-329. J Biol Chem 274: 5222–5226 [DOI] [PubMed] [Google Scholar]
- Ishida H, Nishimori Y, Sugisawa M, Makino A, Mae T (1997) The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol 38: 471–479 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Wada S, Kondo E, Kurusu T, Kuchitsu K, Makino A, Ishida H (2015) Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol 167: 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Rothstein SJ (2011) Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J Exp Bot 62: 1499–1509 [DOI] [PubMed] [Google Scholar]
- Krupinska K. (2006) Fate and activities of plastids during leaf senescence. InThe Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 433–449 [Google Scholar]
- Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al. (2014) OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10: 878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Liu YM, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae T. (1997) Physiological nitrogen efficiency in rice: Nitrogen utilization, photosynthesis, and yield potential. InPlant nutrition for sustainable food production and environment. Springer, Dordrecht, The Netherlands, pp 51–60 [Google Scholar]
- Mae T, Makino A, Ohira K (1983) Changes in the amounts of ribulose bisphosphate carboxylase synthesized and degraded during the life-span of rice leaf (Oryza sativa L). Plant Cell Physiol 24: 1079–1086 [Google Scholar]
- Mae T, Ohira K (1981) The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol 22: 1067–1074 [Google Scholar]
- Mae T, Ohira K (1983) Origin of the nitrogen a growing rice leaf and its relation to nitrogen nutrition. Jpn J Soil Sci Plant Nutr 54: 401–405 [Google Scholar]
- Makino A, Mae T, Ohira K (1983) Photosynthesis and ribulose 1,5-bisphosphate carboxylase in rice leaves: changes in photosynthesis and enzymes involved in carbon assimilation from leaf development through senescence. Plant Physiol 73: 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K (1988) Differences between wheat and rice in the enzymic properties of ribulose-1,5-bisphosphate carboxylase/oxygenase and the relationship to photosynthetic gas exchange. Planta 174: 30–38 [DOI] [PubMed] [Google Scholar]
- Makino A, Nakano H, Mae T (1994) Responses of ribulose-1,5-bisphosphate carboxylase, cytochrome f, and sucrose synthesis enzymes in rice leaves to leaf nitrogen and their relationships to photosynthesis. Plant Physiol 105: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Osmond B (1991) Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol 96: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Sakuma H, Sudo E, Mae T (2003) Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant Cell Physiol 44: 952–956 [DOI] [PubMed] [Google Scholar]
- Makino A, Sato T, Nakano H, Mae T (1997) Leaf photosynthesis, plant growth and nitrogen allocation in rice under different irradiances. Planta 203: 390–398 [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Ono Y, Wada S, Izumi M, Makino A, Ishida H (2013) Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ 36: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Sudo E, Makino A, Mae T (2003) Differences between rice and wheat in ribulose-1, 5-bisphosphate regeneration capacity per unit of leaf-N content. Plant Cell Environ 26: 255–263 [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387 [DOI] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA. (1978) Breakdown of ribulose bisphosphate carboxylase and change in proteolytic activity during dark-induced senescence of wheat seedlings. Plant Physiol 62: 604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T, Sano C (1978) Nitrogen nutrition and growth of the rice plant. II. Considerations concerning the dynamics of nitrogen in rice seedlings. Soil Sci Plant Nutr 24: 191–198 [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.