A candidate effector protein contributes to fungal aggressiveness by targeting and inhibiting a host small heat shock protein.
Abstract
Pathogens secrete effector proteins to establish a successful interaction with their host. Here, we describe two barley (Hordeum vulgare) powdery mildew candidate secreted effector proteins, CSEP0105 and CSEP0162, which contribute to pathogen success and appear to be required during or after haustorial formation. Silencing of either CSEP using host-induced gene silencing significantly reduced the fungal haustorial formation rate. Interestingly, both CSEPs interact with the barley small heat shock proteins, Hsp16.9 and Hsp17.5, in a yeast two-hybrid assay. Small heat shock proteins are known to stabilize several intracellular proteins, including defense-related signaling components, through their chaperone activity. CSEP0105 and CSEP0162 localized to the cytosol and the nucleus of barley epidermal cells, whereas Hsp16.9 and Hsp17.5 are cytosolic. Intriguingly, only those specific CSEPs changed localization and became restricted to the cytosol when coexpressed with Hsp16.9 and Hsp17.5, confirming the CSEP-small heat shock protein interaction. As predicted, Hsp16.9 showed chaperone activity, as it could prevent the aggregation of Escherichia coli proteins during thermal stress. Remarkably, CSEP0105 compromised this activity. These data suggest that CSEP0105 promotes virulence by interfering with the chaperone activity of a barley small heat shock protein essential for defense and stress responses.
Powdery mildew fungi are obligate biotrophs that cause significant economic loss in temperate areas by infecting more than 9,000 dicot and 650 monocot species, including grape (Vitis vinifera), tomato (Solanum lycopersicum), barley (Hordeum vulgare), and wheat (Triticum aestivum; Takamatsu, 2013). The barley powdery mildew fungus (Blumeria graminis f. sp. hordei [Bgh]) reproduces and survives on the epidermal cell layer of wild and cultivated Hordeum spp. Under favorable conditions, the conidia germinate and produce a primary and an appressorial germ tube. The appressorial germ tube swells at the end and forms an appressorium, from which a penetration peg develops. From here, a penetration hypha penetrates the plant cell wall and forms a specialized feeding structure, the haustorium, which is vital for the obligate biotrophic interaction. The fungus utilizes the haustorium to acquire nutrients from the plant cell, and it has been suggested as a site of effector biosynthesis and delivery to the plant cell (Panstruga, 2003; Stergiopoulos and de Wit, 2009). Pathogens deliver effector molecules to the host cell to manipulate cellular function to redirect nutrient transport and to interfere with the host defense mechanisms (Kamoun, 2006; Thomma et al., 2011; Ahmed et al., 2015).
A genomic study supported by proteomics and transcriptomics identified around 500 candidate secreted effector proteins (CSEPs) from Bgh. Some of them share a Y/F/WxC motif in the N-terminal part of the mature protein (Bindschedler et al., 2009, 2011; Godfrey et al., 2010; Spanu et al., 2010; Pedersen et al., 2012). Only a few CSEPs have been studied and shown to play a role in virulence. A method for the genetic transformation of powdery mildew fungi is not available. Therefore, these results were obtained using host-induced gene silencing (HIGS), an RNA interference (RNAi) method in which hairpin constructs targeting fungal transcripts are expressed in the attacked host cell (Nowara et al., 2010; Zhang et al., 2012a; Pliego et al., 2013). For example, CSEP0055 interacts with the barley pathogenesis-related protein PR17c and promotes fungal aggressiveness by suppressing defense (Zhang et al., 2012a). Similarly, an RNase-like effector candidate, CSEP0264, suppresses defense by obstructing pathogen-induced host cell death (Pliego et al., 2013). In addition, two avirulence (Avr) proteins, Avrk1 and Avra10, that lack conventional N-terminal signal sequences have been identified. These avirulence proteins trigger host cell death when recognized by their cognate Resistance (R) proteins, Mlk1 and Mla10 (Ridout et al., 2006).
Plants utilize a wide range of responses to defend themselves against invasion by pathogens and parasites. These include the expression of heat shock proteins (Hsps), which allow the cells to adapt to and survive severe environmental changes (Didelot et al., 2006; Gupta et al., 2010). Hsps are highly conserved molecular chaperones that stabilize intracellular proteins and facilitate the refolding of misfolded or denatured proteins during stress, such as that inflicted by microbial attack (Didelot et al., 2006). Two well-studied ATP-dependent chaperones, Hsp70 and Hsp90, play significant roles in hypersensitive response (HR) induction and nonhost resistance in Nicotiana benthamiana (Kanzaki et al., 2003; Jelenska et al., 2010). Moreover, Hsp90 stabilizes and is required for the function of numerous R proteins, such as I-2, RPM1, N, and Rx (Hubert et al., 2003; Lu et al., 2003; Liu et al., 2004; de la Fuente van Bentem et al., 2005; Kadota and Shirasu, 2012). ATP-independent small heat shock protein (sHsp) chaperones also protect the cell from stress-induced protein aggregation and misfolding (McHaourab et al., 2009; Basha et al., 2012). sHsps are characterized by a conserved domain of approximately 90 residues forming an α-crystallin domain flanked by a variable N-terminal sequence and a short C-terminal extension (Basha et al., 2012). They assist in the Hsp70/Hsp90-mediated refolding process by maintaining the misfolded or denatured proteins in a folding-competent state (Lee et al., 1997; Van Ooijen et al., 2010). The monomeric size of sHsps ranges between 12 and 42 kD. However, in most cases, they form large oligomers of eight to 32 subunits (Basha et al., 2012). The oligomers are activated to a higher substrate affinity state during stress, and they bind nonnative proteins of similar weight (Basha et al., 2006; Tyedmers et al., 2010). While the main function of sHsps is to provide thermotolerance, some are also involved in defense against pathogens. For instance, the tomato sHsp20, Required for Stability of I-2 (RSI2), is involved in Fusarium oxysporum resistance by stabilizing the tomato resistance protein, I-2 (Van Ooijen et al., 2010). The Nicotiana tabacum sHsp, Ntshsp17, and its N. benthamiana ortholog, Nbshsp17, are required for basal immunity against Ralstonia solanacearum (Maimbo et al., 2007).
In this study, we are now able to demonstrate sHsps being targeted by pathogen effectors. We found that two barley powdery mildew effector candidates, CSEP0105 and CSEP0162, contributed to Bgh infection success. Two barley sHsps, Hsp16.9 and Hsp17.5, were identified as host targets for these two CSEPs. Notably, Hsp16.9 showed in vitro chaperone activity, which was specifically suppressed by CSEP0105.
RESULTS
CSEP0105 and CSEP0162 Promote Bgh Virulence
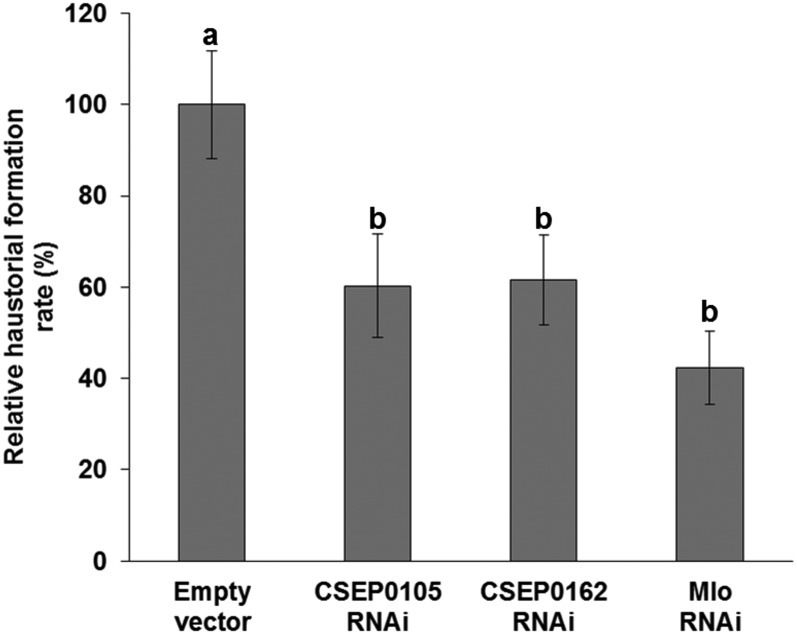
Effectors promote the virulence of pathogens, for instance by suppressing host defense and by enhancing nutrient uptake. Thus, we studied the contribution of Bgh CSEPs to fungal infection success using HIGS (Nowara et al., 2010). Among the 500 described CSEPs (Godfrey et al., 2010; Pedersen et al., 2012), a number were selected based on their level of expression, the presence or absence of nuclear localization signals, and to which CSEP family they belong. Our data revealed that independent silencing of approximately half of them reduced Bgh aggressiveness significantly. Of these, RNAi of CSEP0105 and CSEP0162 (Supplemental Fig. S1) reduced the Bgh haustorial formation rate by approximately 40% relative to empty vector controls (Fig. 1). The Mlo RNAi positive control reduced the haustorial formation rate by approximately 60% (Fig. 1). Using the SI-FI software tool (http://labtools.ipk-gatersleben.de/), the CSEP0162 RNAi construct was predicted to generate an off-target silencing effect on CSEP0163 and CSEP0164. These CSEPs are highly similar to CSEP0162 and belong to the same CSEP family 4, which has a total of 19 members (Pedersen et al., 2012). Thus, the reduced haustorial formation rate caused by the CSEP0162 RNAi construct may be due to the silencing of more members of this family. CSEP0105 belongs to CSEP family 31, consisting of four members (Pedersen et al., 2012). Despite this, no off target was predicted in the Bgh transcriptome for the CSEP0105 RNAi construct. In addition, neither of the RNAi constructs was predicted to have an off target in the barley transcriptome (version 12, released on March 19, 2011), as determined by the small interfering RNA scan software (http://bioinfo2.noble.org/RNAiScan.htm). Therefore, the reduced infection rates observed after silencing indicate that CSEP0105 and CSEP0162 contribute to Bgh infection success, most likely as effectors.
Figure 1.
Silencing of CSEP0105 and CSEP0162 by HIGS reduces the Bgh haustorial formation rate. One-week-old barley leaves were bombarded with an RNAi construct and a GUS reporter construct. Two days later, leaves were infected with Bgh, and at 3 dpi, they were stained with a 5-bromo-4-chloro-3-indoxyl-β-d-glucuronic acid, cyclohexylammonium salt solution and scored for fungal haustorial formation rate, which was calculated as the number of GUS-expressing cells with haustoria divided by the total number of GUS-expressing cells. The relative haustorial formation rate of each construct was obtained by comparing with the empty vector control of each experiment, which was set to 100%. Data represent means ± se of five independent experiments, except for Mlo RNAi (n = 3). A total of 1,821, 2,102, 1,779, and 617 cells were assessed for the empty vector, CSEP0105, CSEP0162, and Mlo RNAi, respectively. Bars marked with different letters are significantly different at P < 0.01.
CSEP0105 and CSEP0162 Are Highly Expressed during and after Haustorial Formation
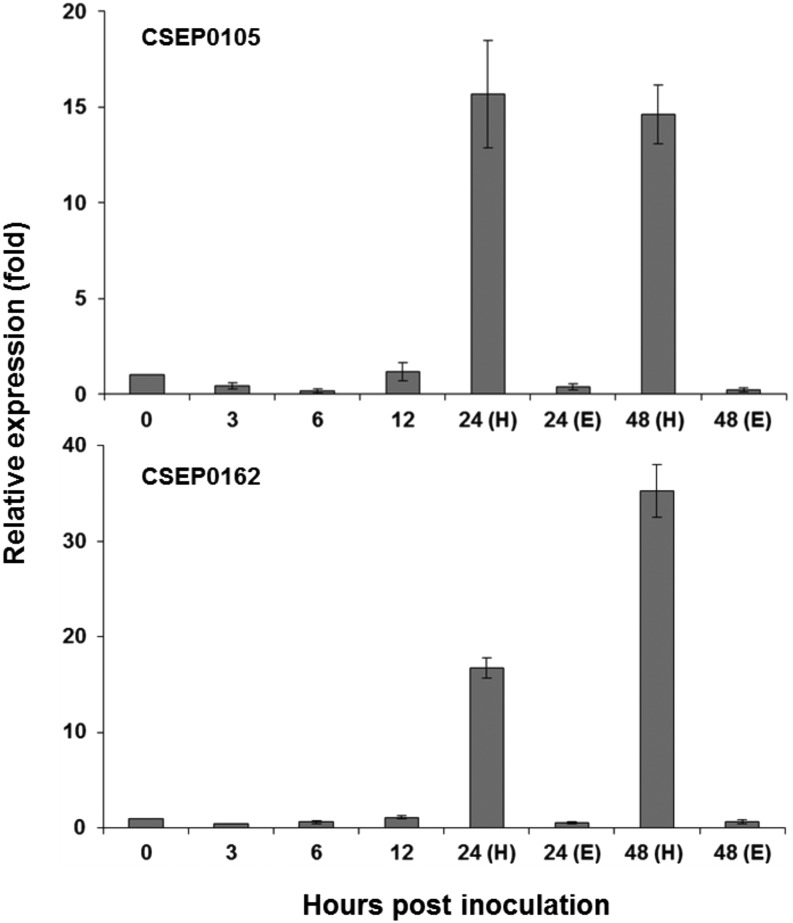
Effectors are differentially expressed and assumed to be required at various stages of infection. To determine the expression patterns of CSEP0105 and CSEP0162 at different fungal development stages, we performed a time-course experiment in a compatible Bgh/barley interaction. The time course corresponds to ungerminated conidia (0 h post inoculation [hpi]), primary germ tube formation (3 hpi), appressorial germ tube formation (6 hpi), penetration (12 hpi), haustorial formation (24 hpi), and secondary penetration (48 hpi). For the first four time points, total RNA from the entire infected leaves was analyzed. Since haustorial development is in progress at 24 hpi, haustorial and epiphytic expressions were analyzed separately at 24 and 48 hpi. A quantitative PCR expression analysis revealed that the CSEP0105 and CSEP0162 transcripts were predominantly expressed in haustoria and only poorly in the epiphytic tissue (Fig. 2). Although present at earlier time points, both CSEP transcripts were dramatically up-regulated during haustorial formation and secondary penetrations at 24 and 48 hpi (Fig. 2).
Figure 2.
Expression patterns of CSEP0105 and CSEP0162 at different stages of Bgh development. Total RNA was isolated from Bgh-infected (isolate DH14) barley leaves (cv Golden Promise) at 0, 3, 6, 12, 24, and 48 hpi. H and E denote haustorial and epiphytic expression. Expression of Bgh glyceraldehyde 3-phosphate dehydrogenase was used to normalize the CSEP expression in each sample. Relative expression was determined compared with time point 0 hpi, arbitrarily set to 1. Three biological and two technical repetitions were included for each time point. Data shown are means ± se of three independent biological repetitions.
CSEP0105 and CSEP0162 Interact with the Barley Hsp16.9 and Hsp17.5 Proteins
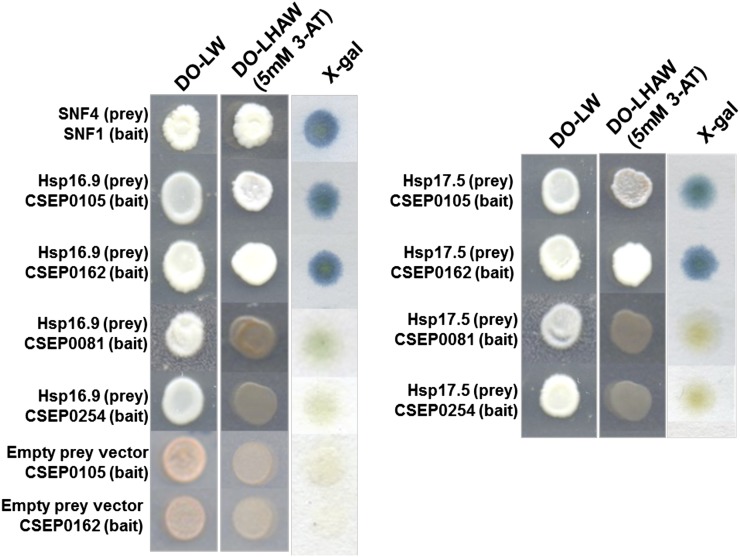
CSEP0105 and CSEP0162 have predicted N-terminal signal peptides, and they are expected to be secreted from the fungus. Thus, we anticipated that both CSEPs could have barley host targets and that interactions with these will promote disease. To identify such targets, we conducted yeast two-hybrid (Y2H) screens for both CSEPs. Approximately 1.75 × 106 clones were screened for each CSEP bait construct using a prey complementary DNA (cDNA) library generated from Bgh-infected barley leaves (Zhang et al., 2012a). This led to the identification of one prey clone, encountered twice, using CSEP0162 as bait (Fig. 3). The clone has a 456-bp cDNA insert, encoding a full-length protein that has 98.7% amino acid identity to a barley class I sHsp with a molecular mass of 16.9 kD (i.e. HvHsp16.9-CI; GenBank accession no. AK362925; Reddy et al., 2014). The identified cDNA has 14 nucleotide substitutions compared with HvHsp16.9-CI, where 12 are synonymous and two are nonsynonymous (Supplemental Fig. S2). With this number of variants, we consider the identified gene to be different from HvHsp16.9-CI; thus, we named it HvHsp16.9-CI-H (H for homolog; Fig. 4). Another prey clone was identified using CSEP0105 as bait. This prey clone carried a 477-bp cDNA insert, which encodes the full-length protein of another class I sHsp of 17.5 kD, HvHsp17.5-CI (AK250749). HvHsp16.9-CI-H and HvHsp17.5-CI are 71% identical at the amino acid level and are hereafter named Hsp16.9 and Hsp17.5. To assess the specificity of these interactions, a targeted Y2H assay was performed between CSEP0105, CSEP0162, and two negative control CSEPs (CSEP0081 and CSEP0254) as baits and Hsp16.9 and Hsp17.5 as preys. The Hsps were found to interact with both CSEP0105 and CSEP0162 but not with the negative controls, indicating that the interactions were specific (Fig. 3). 3-Amino-1,2,4-triazole (3-AT) is a competitive inhibitor of an enzyme catalyzing the sixth step of His biosynthesis. This inhibitor is normally added to the selective medium at a concentration of 1 mm to rule out false positives and weak interactors (James et al., 1996; Gietz et al., 1997). Yeast (Saccharomyces cerevisiae) cells with Hsp16.9 and Hsp17.5/CSEP0105 and CSEP0162 Y2H protein combinations grew at 5 mm 3-AT, indicating strong protein interactions.
Figure 3.
CSEP0105 and CSEP0162 interact with the barley Hsp16.9 and Hsp17.5 proteins in a Y2H assay. Yeast was transformed with bait and prey constructs. Interactions were selected on dropout (DO) medium lacking Leu (L), His (H), adenine (A), and Trp (W), supplemented with 5 mm 3-AT. The β-galactosidase assay (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-gal]) was performed on a filter paper print of DO-Leu-Trp-grown yeast. SNF4/SNF1 interaction was used as a positive control, and CSEP0081 and CSEP0254 were used as negative controls.
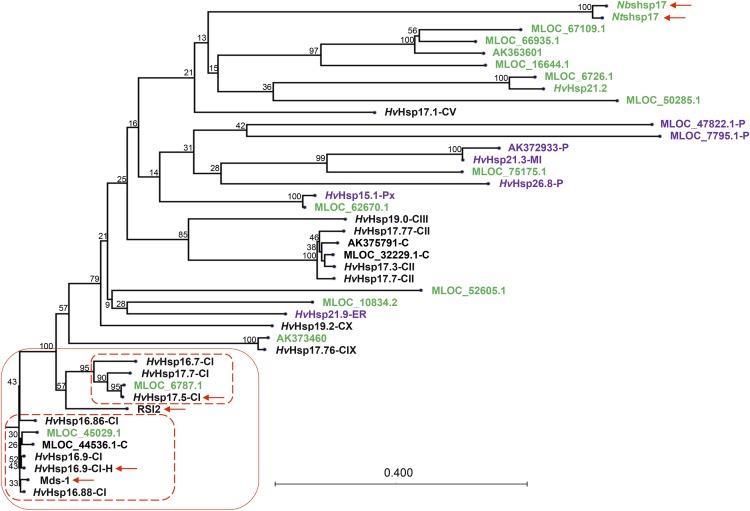
Figure 4.
Amino acid-based phylogenetic tree of all 37 identified full-length barley sHsps, tomato RSI2, Ntshsp17, and Nbshsp17. The lower left branches mainly contain cytosolic (black) sHsps with high similarity to each other. Those with noncytosolic and unknown localizations are shown in purple and green, respectively. Letters following the molecular weight indicate their localizations: C, cytosolic; ER, endoplasmic reticulum; MI, mitochondria; P, chloroplast; and Px, peroxisome. The two sHsps identified in the Y2H screen, RSI2, Nthsp17, Nbhsp17, and Mds1 are indicated by arrows. The naming follows Reddy et al. (2014) for those starting with HvHsp. The MLOC names are from the annotation of the barley genome. There are four sequences solely based on cDNA clones, with GenBank accession numbers given.
To confirm these results using another protein-protein interaction method, a bimolecular fluorescence complementation (BiFC) assay (Hu et al., 2002) was carried out in N. benthamiana and barley plants. These experiments supported that Hsp16.9 and Hsp17.5 interact with both CSEP0105 and CSEP0162 but not with CSEP0254 (Supplemental Fig. S3). The interactions were observed only for the N-terminal yellow fluorescent protein (nYFP)-CSEP fusions combined with the C-terminal cyan fluorescent protein (cCFP)-Hsp fusions, which appeared to form a stable CSEP-Hsp BiFC aggregate proximal to the nucleus. As a positive control, we used dimerization of 14-3-3 (Aitken, 2006). As a negative control, interactions of 14-3-3 with CSEP0105 and CSEP0162 were tested (Supplemental Fig. S3). Together, the Y2H and BiFC results provide strong evidence that CSEP0105 and CSEP0162 interact with both Hsp16.9 and Hsp17.5 proteins in yeast and in planta.
Using a genome-wide sequence survey, Reddy et al. (2014) discovered 18 full-length sHsps that are differentially regulated during drought stress and seed development in barley. We searched and identified 19 additional full-length sHsps (Supplemental Table S1) in the recently published barley genome (International Barley Genome Sequencing Consortium, 2012) and the full-length barley cDNAs at the National Center for Biotechnology Information (NCBI; Matsumoto et al., 2011). A phylogenetic tree generated based on the amino acid sequences of these 37 barley sHsps, as well as tomato RSI2, NtsHsp17, and NbsHsp17, revealed that Hsp16.9 and Hsp17.5 belong to a subgroup of 10 highly homologous cytosolic sHsps in barley (Fig. 4). Within this group, Hsp16.9 belongs to a clade of six sHsps, between which the identity is above 90% (Fig. 4), while Hsp17.5 belongs to another clade of four sHsps that have more than 78% identity.
sHsps function as pathogen defense components (Maimbo et al., 2007; Van Ooijen et al., 2010). Thus, to investigate the role of Hsp16.9 and Hsp17.5 proteins, we generated a single RNAi construct that can target the transcripts of both sHsps and performed transient-induced gene silencing in epidermal cells using particle bombardment. This experiment did not result in any susceptibility difference between the sHsp-silenced and empty vector control transformed cells.
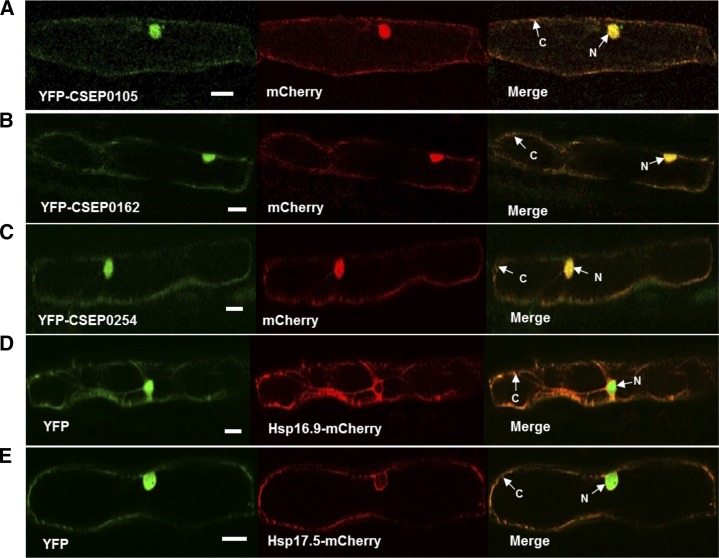
CSEP0105 and CSEP0162 Localize to the Cytosol and the Nucleus, While Hsp16.9 and Hsp17.5 Are Cytosolic
To investigate the subcellular localizations of CSEP0105, CSEP0162, and CSEP0254 in the host cell, N-terminal YFP fusions to the mature CSEPs (lacking the signal peptide) were constructed. The fusion constructs were expressed in barley epidermal cells using particle bombardment together with a free mCherry marker, which localizes to the cytosol and the nucleus. The YFP-CSEP0105, YFP-CSEP0162, and YFP-CSEP0254 fusion proteins accumulated both in the cytosol and the nucleus, as there was a complete overlap between the YFP and mCherry fluorescent signals (Fig. 5, A–C). Similar localizations were observed when these CSEPs were expressed as C-terminal YFP fusions. Usually, effectors localize together with their host targets in order to execute their virulence function. Therefore, to determine in which of these locations the CSEP targets reside, we expressed C-terminally mCherry-tagged Hsp fusions in barley epidermal cells. This revealed that both the Hsp16.9-mCherry and Hsp17.5-mCherry fusion proteins accumulated exclusively in the cytosol, in contrast to the coexpressed yellow fluorescent marker that localized both in the cytosol and the nucleus (Fig. 5, D and E). This indicated that Hsp16.9 and Hsp17.5 are cytosolic proteins, as predicted by Reddy et al. (2014), and that their interactions with CSEP0105 and CSEP0162 likely occur in the cytosol.
Figure 5.
CSEP0105, CSEP0162, and CSEP0254 have cytosolic and nuclear localization in barley epidermal cells, whereas Hsp16.9 and Hsp17.5 are cytosolic. Ubiquitin promoter-driven expression constructs were generated for all. A to C, Constructs encoding signal peptide-lacking CSEPs fused to the C terminus of YFP were cotransformed with a free mCherry construct into barley leaf epidermal cells using particle bombardment. Two to 4 d later, cells were analyzed using a Leica SP5 confocal laser scanning microscope. All the YFP-CSEP fusion signals are in the cytosol (C) and the nucleus (N). D and E, Constructs encoding full-length Hsps fused to the N terminus of mCherry were cotransformed with a free YFP construct. Both Hsp-mCherry signals are in the cytosol. The free mCherry and YFP markers localize to the cytosol and nucleus. Bars = 20 µm.
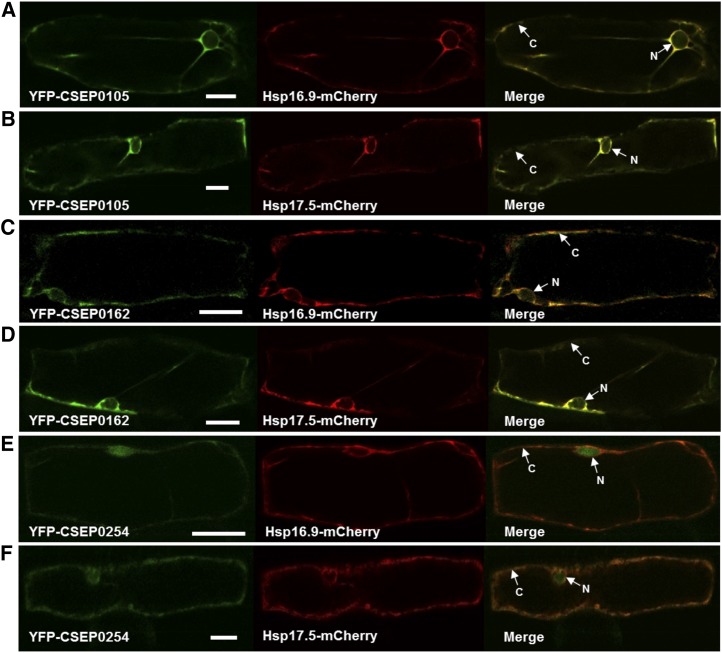
CSEP0105 and CSEP0162 Are Targeted Exclusively to the Cytosol When Coexpressed with Hsp16.9 and Hsp17.5
The protein-protein interaction and localization results shown in Figures 3 and 5 encouraged us to examine the effect of their interactions on the localizations of the CSEPs and Hsps. Therefore, the YFP-CSEP0105 and YFP-CSEP0162 fusion constructs were coexpressed with either the Hsp16.9-mCherry or Hsp17.5-mCherry fusion construct in barley leaf epidermal cells. Our data revealed that the accumulation of the two Hsps was unaltered by the presence of the interacting CSEPs (Fig. 6). Intriguingly, YFP-CSEP0105 and YFP-CSEP0162 fusion proteins remained in the cytosol and did not translocate to the nucleus in the presence of Hsp16.9-mCherry and Hsp17.5-mCherry fusion proteins (Fig. 6, A–D). Meanwhile, the noninteracting CSEP, YFP-CSEP0254, kept its nuclear and cytosolic localization when coexpressed with either Hsp16.9-mCherry or Hsp17.5-mCherry (Fig. 6, E and F). This suggests that CSEP0105 and CSEP0162 are exclusively directed to the compartment where their host targets localize. This observation also provides additional evidence for the physical interaction between the two partners, substantiating that the sHsp/effector candidate interactions are authentic.
Figure 6.
CSEP0105 and CSEP0162 accumulate exclusively in the cytosol when coexpressed with Hsp16.9 and Hsp17.5. The same YFP-CSEP and Hsp-mCherry fusion constructs were used as described in Figure 5. The YFP-CSEP constructs were cotransformed with each Hsp-mCherry construct into barley leaf epidermal cells using particle bombardment. Two to 4 d later, individual cells were visualized using a Leica SP5 confocal laser scanning microscope. CSEP0254 was included as a negative control. The YFP-CSEP0105, YFP-CSEP0162, Hsp16.9-mCherry, and Hsp17.5-mCherry signals are in the cytosol (C), whereas YFP-CSEP0254 is in the cytosol and the nucleus (N). Bars = 20 µm.
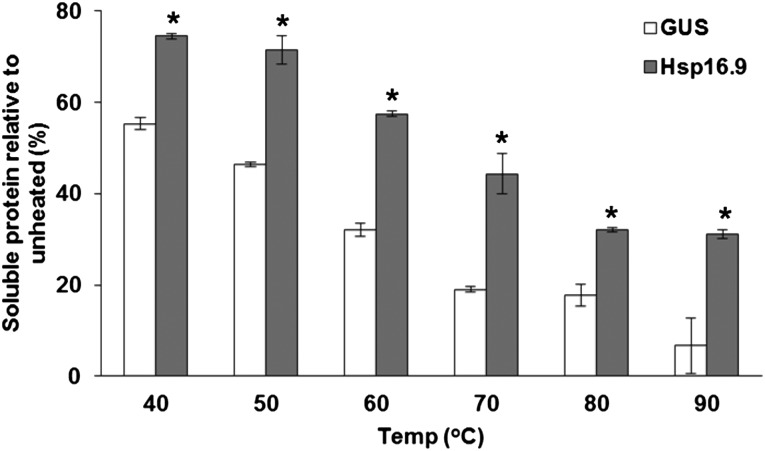
Hsp16.9 Prevents Heat-Induced Protein Aggregation in Vitro
sHsps have chaperone activity that prevents protein aggregation during heat stress. To investigate whether Hsp16.9 is a bona fide chaperone, we used a thermal stability assay of Escherichia coli cellular proteins. Recombinant Hsp16.9 protein fused with a His tag was expressed in E. coli. After heating the total cellular protein fraction in a cell-free extract from GUS (control) expressing E. coli at 40°C for 15 min, approximately 55% of the proteins remained soluble. After heating at 90°C, only 7% of the proteins remained soluble (Fig. 7). However, in lysates containing Hsp16.9, 75% of the proteins were soluble after the 40°C treatment, and remarkably, 31% of the proteins were soluble after the 90°C treatment (Fig. 7). This strongly suggests that Hsp16.9 is effective in preventing the aggregation of bacterial cellular proteins. Hsp17.5 was not tested for its chaperone activity due to insolubility of the His-tagged fusion protein in vitro.
Figure 7.
Hsp16.9 protects the heat denaturation of E. coli proteins in vitro. Thermostability is shown for total protein from E. coli expressing GUS or Hsp16.9. Cell-free total protein extracts (250 µg mL−1) were heated for 15 min. Heat-denatured proteins were removed by centrifugation, and the protein content of the supernatant fractions was determined. Values are relative to the unheated controls. Data shown are means of three replicates ± se. Asterisks show significant differences (P < 0.05) between the GUS control and Hsp16.9 at each heat treatment analyzed by Duncan’s multiple range test. The experiment was repeated once with a similar result.
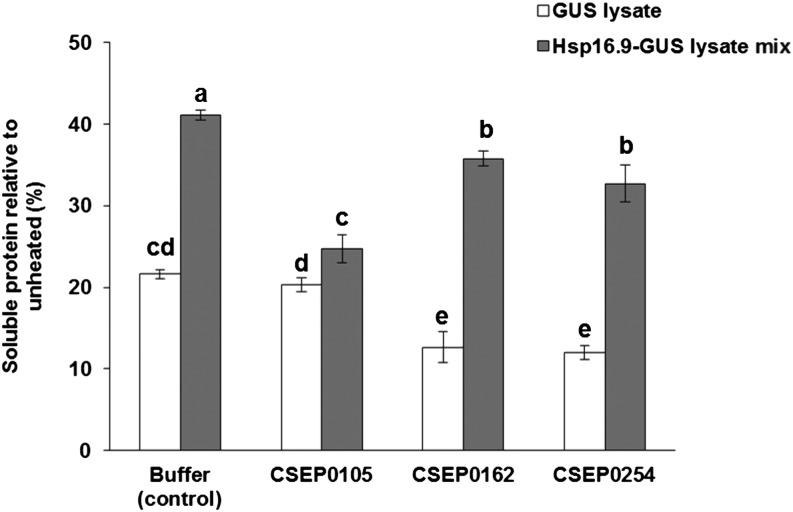
CSEP0105 Interferes with Hsp16.9 Chaperone Activity
Effectors interfere with the functions of their targets. As CSEP0105 and CSEP0162 were found to interact with Hsp16.9, we wanted to test their effect on its chaperone activity. For this purpose, we performed a thermal stability assay of E. coli cellular proteins similar to that described above, with and without the CSEPs. We chose to perform this experiment at 60°C. Based on the results in Figure 7, we concluded that sHsp-mediated protection from protein aggregation is sufficiently high to be able to determine an effect of the CSEPs at this temperature. Furthermore, we expect that heating at 60°C will not damage the CSEPs. In this experiment, 20% of the proteins were soluble after heating the control extract, whereas this value was 40% for the Hsp16.9-containing extract (Fig. 8). This corroborated the chaperone activity of Hsp16.9. When CSEP0105 was added to the control lysate and heated, no change in thermal stability was seen. Interestingly, CSEP0105 essentially prevented Hsp16.9 from improving the thermal stability (Fig. 8). This reveals that CSEP0105 has a detrimental effect on the chaperone activity of Hsp16.9. While CSEP0105 specifically interfered with Hsp16.9-mediated increased protein solubility, CSEP0162 and CSEP0254 also reduced the solubility of cellular proteins. However, the effects of these CSEPs were smaller than that of CSEP0105 and not associated with Hsp16.9 activity, as they occurred in both E. coli protein samples (Fig. 8). This can reflect that CSEP0162 and CSEP0254 interfere with endogenous E. coli chaperones or that they form insoluble complexes with E. coli proteins. Such effects were not seen for CSEP0105, confirming its specific interaction with Hsp16.9.
Figure 8.
CSEP0105 inhibits the chaperone activity of Hsp16.9. Thermostability is shown at 60°C for total protein from E. coli expressing GUS or Hsp16.9 with and without CSEPs. Purified CSEPs were added in a 1:2 protein molar ratio with Hsp16.9 in the cellular lysate (250 µg mL−1). CSEP0254 was added as a negative control. Values are relative to the unheated controls. Data shown are means of three replicates ± se. Means with different letters are significantly different (P < 0.05). Duncan’s multiple range test was used to compare all the means. The experiment was repeated once with a similar result.
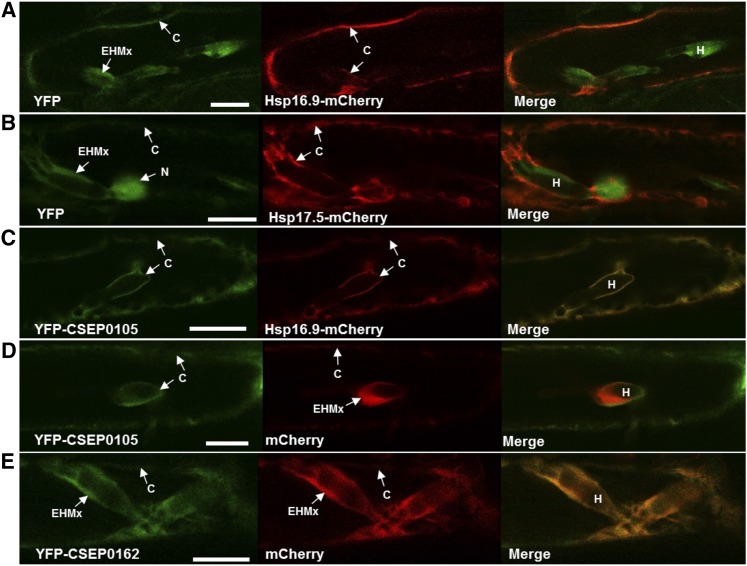
CSEP0105 Keeps Its Cytosolic Localization in Infected Barley Cells, Whereas CSEP0162 Accumulates in the Extrahaustorial Matrix
Since CSEP0105 reduced the chaperone activity of Hsp16.9, we further studied the localization of these physically and functionally interacting proteins in infected cells. For this purpose, the YFP-CSEP and Hsp-mCherry constructs were expressed in barley epidermal cells as described above. One day after the particle bombardment, the leaves were inoculated with Bgh; 2 to 3 d later, the protein localization was determined in cells containing fungal haustoria. To our surprise, we could detect free YFP and mCherry in the extrahaustorial matrix (EHMx) between the extrahaustorial membrane (EHM) and the fungal haustorium (Fig. 9, A, B, D, and E). Hsp16.9-mCherry and Hsp17.5-mCherry fusion proteins retained their cytosolic localization and did not enter the nucleus, as discussed above for uninfected cells (Fig. 5, D and E), and they did not enter the EHMx either (Fig. 9, A–C; Supplemental Fig. S4, A and B). However, an interesting difference was observed for the two CSEPs. YFP-CSEP0105 was cytosolic and nuclear, while YFP-CSEP0162 in addition was observed in the EHMx, where it overlapped with free mCherry (Fig. 9, C–E; Supplemental Fig. S4, B–D). Exclusion of YFP-CSEP0105 from the EHMx occurred independent of sHsp overexpression. Since YFP-CSEP0105 is smaller (38.8 kD) than YFP-CSEP0162 (42.7 kD), the localization difference cannot be a matter of protein size. Yet, the data suggest that CSEP0105 colocalized with the Hsp, which it may inhibit. This is parallel to the colocalization in the unstressed cells (Fig. 6).
Figure 9.
Localization of CSEP0105, CSEP0162, Hsp16.9, and Hsp17.5 proteins in Bgh-infected barley cells. Confocal images show cells transiently cotransformed by particle bombardment with the respective constructs as described in Figure 5. The Hsp16.9-mCherry and Hsp17.5-mCherry fluorescent signals are in the cytosol (C). YFP-CSEP0105 fluorescent signal is in the cytosol when coexpressed with Hsp16.9-mCherry and in the cytosol and nucleus (N) when coexpressed with the free mCherry construct. YFP-CSEP0162, as well as the free YFP and mCherry marker proteins, localized to the cytosol, nucleus, and EHMx. The fluorescent signals labeled EHMx also include the cytosol surrounding the haustoria (H). Since the nucleus and haustoria were not found in a single confocal plane, images showing nuclei of the same cells are presented in Supplemental Figure S3. Bars = 20 µm.
DISCUSSION
Previously, it has been suggested and supported by BiFC that Bgh CSEP0055 interacts with the barley pathogenesis-related protein, PR17 (Zhang et al., 2012a), and that the Bgh effector candidates, BEC3 and BEC4, interact with a thiopurine methyltransferase and an E2 ubiquitin-conjugating enzyme, respectively (Schmidt et al., 2014). These three effector candidates all contributed to Bgh virulence. Here, we provide evidence that CSEP0105 and CSEP0162 enhance Bgh virulence. Interestingly, these CSEPs are unrelated and they belong to CSEP families 31 and 4, respectively (Pedersen et al., 2012). The expression of both CSEPs occurred predominantly in the haustoria, and they were highly up-regulated during and after the formation of this fungal structure. This result is consistent with previous proteomic and transcriptomic analyses, where both CSEPs were identified exclusively in the haustorial proteome and not in sporulating hyphae (Bindschedler et al., 2011; Pedersen et al., 2012). This signifies that the two CSEPs could be required at the late infection stage, during haustorial development and secondary penetration.
Using the Y2H system, the barley Hsp16.9 and Hsp17.5 proteins were identified as interactors of both CSEP0105 and CSEP0162. The growth of the yeast cells at 5 mm 3-AT indicated strong interactions between these proteins. These results were verified by in planta expression using the BiFC method in N. benthamiana and barley plants, which has been proven as a reliable method to show protein interactions in living cells. In this assay, the CSEPs and sHsps were fused to complementing N- and C-terminal halves of a YFP molecule. The interaction between a CSEP and an Hsp brought the two halves of the YFP in close contact, allowing them to irreversibly reconstitute an intact, fluorescent YFP molecule. The observation of YFP fluorescence, therefore, showed that the CSEPs and sHsps formed a complex in planta. A drawback of the BiFC system is that YFP reconstitution is permanent, and for this reason, interaction dynamics cannot be studied (Hu et al., 2002; Kerppola, 2008). Therefore, we cannot draw conclusions on the timing or strength of CSEP-sHsp complex formation. The BiFC-CSEP-sHsp protein complexes localized to large aggregates proximal to the nucleus (Supplemental Fig. S3). This phenomenon is most likely caused by the irreversible formation of BiFC-sHsp-CSEP complexes, while the coexpression of the proteins fused to conventional fluorophores (Supplemental Fig. S4) does not result in the formation of these aggregates.
Meanwhile, more conclusive evidence for these interactions comes from a coexpression study showing that the localization of CSEPs is following the sHsps. When expressed alone, the CSEPs accumulate both in the cytosol and the nucleus, although they lack predicted nuclear localization signals (Pedersen et al., 2012). However, when coexpressed with the two cytosolic sHsps, the interacting CSEPs no longer localized to the nucleus, while a noninteracting, negative control CSEP was not affected. This strongly indicates that these interactions occur in the cytosol, where the host targets reside. Although the endogenous Hsp16.9 and Hsp17.5 proteins were also expected to interact with the overexpressed CSEPs, they appeared to occur in insufficient quantities to bind and trap all CSEP protein in the cytosol. Collectively, we demonstrate that sHsps are targets of pathogen effectors. Previously, the Pseudomonas syringae HOPI1 effector was demonstrated to bind and recruit cytosolic Hsp70 to the chloroplast, where they form large complexes. Such complex formation, combined with the HOPI1-stimulated Hsp70 ATP hydrolysis activity, enabled the pathogen to hijack the plant defense and stress machinery to promote virulence (Jelenska et al., 2010).
The EHMx is a separate compartment that exists between the host cell and haustoria-forming microbes. Effectors synthesized from such microbes are secreted into the EHMx, where some of them may execute their virulence function. Others will be transferred over the EHM to the host cytosol to execute their mission there. The localization of CSEP0162 to the EHMx suggests that this CSEP perhaps has a virulence role in the matrix, in addition to the cytosol, where we have shown it to target sHsps. If Bgh-expressed CSEP0105 is transferred over the EHM to the plant cell, our data would suggest that it only interacts with its target in the cytosol.
The phylogenetic tree illustrated that Hsp16.9 and Hsp17.5 belong to a subgroup of cytosolic sHsps in barley that consists of 10 members with high similarity (Fig. 4). Therefore, other members of this subgroup might interact with the two CSEPs. In contrast, CSEP0105 and CSEP00162 are distinct and belong to different CSEP families. It is well established that effectors from the same or different pathogens target a common cellular hub to gain efficient suppression of host defense and facilitate pathogen fitness (Mukhtar et al., 2011; Weßling et al., 2014). For example, six P. syringae effectors (AvrB, AvrRpm1, AvrRpt2, AvrPto, AvrPtoB, and HopF2) all target Arabidopsis (Arabidopsis thaliana) RIN4 (Deslandes and Rivas, 2012). Thus, our finding that two unrelated Bgh effectors target identical proteins of the stress machinery agrees with accumulating data that effectors converge on common host hubs that are important for defense.
Generally, misfolded proteins expose hydrophobic residues, which otherwise are buried in the protein structure. Such hydrophobic surfaces have a strong tendency to induce protein aggregation (Tyedmers et al., 2010). sHsps recognize and bind such hydrophobic surfaces to refold the protein, either alone or with the help of other ATP-dependent chaperones (Basha et al., 2012; Fu, 2014). Thus, in order to investigate whether the interactions we found resemble the binding of sHsps with their substrate, we predicted the hydrophobic surface areas for the two CSEPs that interact with sHsps and two noninteracting CSEPs (CSEP0081 and CSEP0254). Protein structure models were created for the four CSEPs using the IntFOLD server (Roche et al., 2011). These models were analyzed with the Swiss-PdbViewer 4.1.0 (Guex and Peitsch, 1997) to detect potential hydrophobic patches on the surfaces of the proteins. This predicted that CSEP0081, CSEP0105, CSEP0162, and CSEP0254 have more or less similar surface hydrophobicity of 22%, 23%, 19%, and 15%, respectively. As the exposed hydrophobic amino acids occur singly or in groups of a few and only make up small patches of hydrophobicity (less than 200 Å2), hydrophobic effects cannot explain the CSEP-Hsp interaction. Hence, we consider that the CSEPs bind the Hsps according to other principles.
Similar to the well-studied high-molecular-weight Hsps, sHsps are also involved in basal defense responses and HR-mediated immunity in plants (Maimbo et al., 2007; Van Ooijen et al., 2010). Silencing of Nbshsp17 significantly reduced the expression of defense-related genes, such as PR1, PR4, and the ethylene-responsive element-binding protein, which are marker genes for salicylic acid, methyl jasmonate, and ethylene signaling, respectively (Maimbo et al., 2007). In addition, silencing of Nbshsp17 increased the growth of both virulent and avirulent R. solanacearum isolates and accelerated wilt symptoms. However, the HR induced by avirulent R. solanacearum, Pseudomonas cichorii, and INF1 was not affected in Nbshsp17-silenced plants. Accordingly, Nbshsp17 is involved in HR-independent basal defense responses in N. benthamiana plants (Maimbo et al., 2007). In contrast, the tomato sHsp20 (RSI2) is required for HR-dependent defense responses (Van Ooijen et al., 2010). Silencing of RSI2 compromises the HR triggered by an autoactive version of the I-2 R gene. This phenotype is correlated with reduced I-2 protein accumulation, which led the authors to conclude that RSI2 is required for the stabilization of I-2 and/or other signaling components involved in the execution of the HR (Van Ooijen et al., 2010). The phylogenetic tree in Figure 4 shows that Nbshsp17 is distantly related to RSI2 and the two CSEP-interacting barley sHsps. Plants possess a large number of highly diverse sHsps with various subcellular localizations (Fig. 4; Jiang et al., 2009; Basha et al., 2010). Hsp16.9, Hsp17.5, and RSI2 are cytosolic class I proteins, whereas Nbshsp17 is clustered with potentially noncytosolic sHsps (Fig. 4). Therefore, RSI2 and Nbshsp17 could have a specialized role in R-gene mediated immunity and basal defense, respectively. However, the role of RSI2 as a component of basal defense has not been investigated and cannot be excluded.
Barley Hsp16.9 and Hsp17.5 have 68% amino acid identity to the full-length RSI2, and they have 81% and 83% identity to the α-crystallin domain of RSI2, respectively. In general, sHsps diverge more in the N-terminal region, upstream of the α-crystallin domain (Basha et al., 2012). Thus, as they share a high sequence similarity, localization, and, for Hsp16.9, experimentally verified chaperone activity, we expect that Hsp16.9 and Hsp17.5 proteins could have similar biochemical functions to RSI2 in immunity. However, there was no observable change in the susceptibility of barley after introduction of the Hsp16.9 and Hsp17.5 RNAi construct. This is likely due to sHsp redundancy caused by other sHsps (Fig. 4) not targeted by the RNAi construct, which would mask the effect. This notion is consistent with the observation that CSEP0105 and CSEP0162 each interacts with at least two sHsps. In fact, a recent study in wheat implicates Mds1, a close homolog of barley Hsp16.9 and Hsp17.5 (Fig. 4), in resistance to powdery mildew (Liu et al., 2013). Here, knockdown of Mds1 made wheat resistant to powdery mildew. This observation is not necessarily in conflict with sHsps playing a positive role in defense and being effector targets. Knocking out positive defense genes often activates defense (Nishimura et al., 2003; Stein et al., 2006; Zhang et al., 2008), which can be mediated by R proteins detecting the absence of effector targets (Mackey et al., 2002; Jones and Dangl, 2006; Zhang et al., 2012b).
We found that CSEP0105 hampered the chaperone activity of Hsp16.9 in vitro. By hydrophobic interactions, the melittin peptide binds and outcompetes the hydrophobic binding site of an eye lens α-crystallin protein, thereby diminishing its chaperone activity (Sharma et al., 1998, 2000; Santhoshkumar and Sharma, 2001). Our predictions indicate that CSEP0105 is not particularly rich in exposed hydrophobic amino acids; therefore, it remains to be seen how this protein interferes with the chaperone activity of Hsp16.9. Yet, we speculate that it can interfere with the function of multiple sHsps. On the other hand, CSEP0162 did not have a specific effect on the chaperone activity of Hsp16.9, potentially because its sHsp inhibition is not revealed in our assay.
To summarize, we found that CSEP0105 and CSEP0162 promoted Bgh virulence in barley, suggesting them to be effector proteins. In addition, both CSEPs were demonstrated to interact with the barley Hsp16.9 and Hsp17.5 proteins. CSEP0105 compromised the chaperone activity of Hsp16.9, which we assume to be the mechanism by which it suppresses defense.
MATERIALS AND METHODS
Plant and Fungal Materials
One-week-old barley (Hordeum vulgare ‘Golden Promise’) seedlings were used for the HIGS, BiFC, localization, and expression profiling studies, in combination with the virulent isolate, DH14, of Blumeria graminis f. sp. hordei. This fungal isolate was maintained on cv Golden Promise by weekly inoculum transfers. Barley seedlings were grown at 16 h of light (20°C, 150 µE m−2 s−1)/8 h of darkness (15°C). Two- to 4-week-old Nicotiana benthamiana plants were used for BiFC studies.
Construction of Gateway Entry Clones
Gateway entry clones were produced for the coding sequences of effector candidates CSEP0081 (BGHDH14_bgh03006), CSEP0105 (BGHDH14_bgh03459), CSEP0162 (BGHDH14_bgh03874), and CSEP0254 (BGHDH14_bgh05751), with and without signal peptides, as available in the EnsemblFungi database (http://fungi.ensembl.org/index.html), and for Hsp16.9-CI-H and Hsp17.5 (AK250749), all with and without stop codons. The sequences were PCR amplified on fungal and plant cDNAs using the primer pairs described in Supplemental Table S2. The PCR products were TOPO cloned into either the pCR8/GW/TOPO or the pENTR/D-TOPO entry vector. From there, the inserts were transferred to destination vectors using Gateway LR cloning reactions (Invitrogen). All entry and destination clones were confirmed by sequencing.
HIGS and Transient-Induced Gene Silencing
Gene silencing in Bgh and barley was conducted using the particle bombardment technique as described by Nowara et al. (2010) and Douchkov et al. (2005), respectively. The RNAi constructs were generated in the 35S promoter-driven hairpin destination vector, pIPKTA30N (Douchkov et al., 2005). They were cotransformed with a GUS reporter gene construct into barley epidermal cells. Seven leaves were transformed for each construct using particle bombardment. Two days later, the leaves were inoculated with Bgh, and 3 d after that, they were stained for GUS activity. Transformed (blue) cells were assessed microscopically for the presence of haustoria as an indication of fungal aggressiveness. Haustorial formation rates were calculated as the number of blue cells with haustoria divided by the total number of blue cells. The empty vector pIPKTA30N and the Mlo RNAi (pIPKTA36) construct were used as negative and positive controls, respectively. Relative haustorial formation rates were calculated as the haustorial formation rate for each construct divided by the haustorial formation rate for the empty vector control treatment. The data were analyzed using the software package SAS (version 9.4; SAS Institute) by logistic regression (PROC GENMOD, corrected for overdispersion), using a generalized linear model and assuming a binomial distribution. The probability of Bgh haustorial formation was modeled as a linear function of an intercept parameter, a CSEP effect, and an experiment effect. The significance of differences between each construct and the empty vector RNAi were analyzed using Pearson’s χ2 test.
Y2H Screen
A pAD-GAL4-2.1-based prey cDNA library, which was generated from Bgh-infected barley leaves (Zhang et al., 2012a), was used for the Y2H screens. The CSEP bait constructs were made in the pDEST-AS2-1 destination vector, producing fusions to the C terminus of the DNA-binding domain of the GAL4 transcription factor (Robertson, 2004).
The J69-4a yeast (Saccharomyces cerevisiae) strain, which contains HIS3 and ADE2 reporter genes, was used for the Y2H screen (James et al., 1996). The CSEP0105 and CSEP0162 bait constructs were transformed independently into J69-4a and selected on dropout medium lacking Trp. Subsequently, the clones with the bait plasmid were retransformed with the prey cDNA library. Positive clones were selected on dropout medium lacking Trp, Leu, His, and adenine and supplemented with 2 mm 3-AT. Positive clones were subjected to 2, 3, and 5 mm 3-AT to select strong interactors. Prey plasmids from the positive clones that grew at 5 mm 3-AT were extracted and retransformed to bait-containing clones to confirm their interactions. Expression of another reporter gene, LacZ, was examined using the Y190 strain as described by Bai and Elledge (1997). The yeast clones grown in dropout-Trp-Leu liquid medium were transferred to filter paper and incubated with a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside solution at 30°C for 10 to 12 h to test the synthesis of β-galactosidase. The prey plasmids from the double-confirmed clones were sequenced and analyzed by BLAST screens in the NCBI database. The interaction of SNF1 (bait) and SNF4 (prey) was used as a positive control (Durfee et al., 1993). CSEP0081 and CSEP0254 were used as negative bait controls. The dropout medium is composed of Glc (20 g L−1); yeast nitrogen base (6.7 g L−1); dropout mix minus Trp, Leu, His, and adenine (2 g L−1); supplemental amino acids; agar (15 g L−1); and water.
BiFC
Coding sequences for full-length barley Hsp and fungal CSEP, lacking signal peptides and with and without stop codons, were transferred to 35S promoter-driven BiFC binary Ti destination vectors (Liu et al., 2009) as N- and C-terminal fusions of nYFP and cCFP. nYFP denotes amino acids 1 to 172 of YFP, and cCFP denotes amino acids 154 to 239 of CFP. Constructs were transiently transformed to N. benthamiana using Agrobacterium tumefaciens-mediated transformation. All eight possible combinations were evaluated for each CSEP/Hsp combination. Fluorescent signal was evaluated using a Leica SP5 confocal laser scanning microscope at 488-nm excitation; the emission was detected between 518 and 540 nm, according to Hu and Kerppola (2003). Construct combinations that gave positive results from the N. benthamiana BiFC assay were cotransformed into barley leaf epidermal tissues together with an mCherry transformation marker using particle bombardment and evaluated for protein interaction.
Localization
The CSEP coding sequences with and without stop codons were transferred to a ubiquitin promoter-driven overexpression destination vector to mediate fusion to the N and C termini of YFP. The coding sequences for Hsp16.9 and Hsp17.5 were transferred to a ubiquitin promoter-driven overexpression destination vector to mediate fusion to the N terminus of mCherry. Particle bombardment was used to transform the constructs into barley epidermal cells. Fluorescent signal was monitored using a Leica SP5 confocal laser scanning microscope. Lasers of 514 and 543 nm were used to excite YFP and mCherry, respectively. The YFP and mCherry emissions were detected between 524 to 539 nm and 590 to 640 nm, respectively. Free YFP and mCherry markers, which localize to the cytosol and the nucleus, were used as controls.
Identification of sHsps in the Barley Genome and Clustering Analysis
To identify sHsps in barley, we used the 19 barley sHsps described by Reddy et al. (2014) and rice (Oryza sativa) sHsps obtained from the Phytozome server (http://www.phytozome.net/) as queries in BLASTp searches of the high-confidence barley genome sequence predicted by the International Barley Genome Sequencing Consortium (2012; http://mips.helmholtz-muenchen.de/plant/barley/download/index.jsp) and of barley ESTs at NCBI, HarvEST (Close et al., 2004), and PLEXdb (Dash et al., 2012). An amino acid-based alignment and a phylogenetic tree were made for the identified sHsps as well as the tomato (Solanum lycopersicum) RSI2 (Van Ooijen et al., 2010), Ntshsp17, and Nbshsp17 (Maimbo et al., 2007) using the CLC main workbench (CLC Bio). Protein localizations were predicted using TargetP (Emanuelsson et al., 2007), WoLF PSORT (Horton et al., 2006), and PlantLoc (http://cal.tongji.edu.cn/PlantLoc/index.jsp). Only sHsps that were predicted consistently by the three softwares are presented here.
RNA Isolation, Reverse Transcription-PCR, and Quantitative PCR
Total RNA was isolated from Bgh-infected barley leaves at 0, 3, 6, 12, 24, and 48 hpi using the polyvinylpolypyrrolidone method (Chen et al., 2000). RNA was isolated from total infected leaves for the first four time points and from the epiphytic fungal material and the separated leaves containing the haustoria for the 24- and 48-hpi time points. This separation was done by dipping the Bgh-infected leaves into 10% (w/v) cellulose acetate (in acetone). The leaves were dried for 10 min, and cellulose acetate strips containing the epiphytic fungal material were detached from the leaves. cDNA was synthesized using the SMART MMLV RT kit (Clontech) following the manufacturer’s instructions with the gene-specific primers described in Supplemental Table S2. Transcript quantification was performed on a Stratagene MX3000P real-time PCR detection system using the FIREPol EvaGreen qPCR-kit (Solis Biodyne). Each reaction consisted of 20 µL containing 4 µL of 5× HOT FIREPol EvaGreen qPCR Mix Plus, 2 µL of cDNA, 1 µL of 10 mm solution of each gene-specific primer (Supplemental Table S2), and 12 µL of water. Reactions for each CSEP and glyceraldehyde 3-phosphate dehydrogenase (reference gene) were combined on one 96-well plate. PCRs were carried out using the following thermocycle: 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 20 s at 58°C, and 20 s at 72°C. The dissociation curves of the PCR products were recorded between 55°C and 95°C. Amplification efficiencies of the reference gene and genes of interest were between 90% and 100%. The results were analyzed using Stratagene MX3000 qPCR analysis software. Relative expression was determined compared with time point 0 hpi (ungerminated conidia), arbitrarily set to 1. Three biological and two technical repetitions were included for each time point.
Chaperone Activity of Recombinant Barley Hsp16.9 on Escherichia coli Proteins
A thermal aggregation assay of E. coli cellular proteins was performed according to Yu et al. (2005). The coding sequence for the full-length Hsp16.9 was transferred to a pDest17 (His tag) expression destination vector. The construct was transformed into the E. coli strain Rosetta, and expression was induced by adding 1 mm isopropyl-β-d-thiogalactopyranoside. GUS expression was used as a negative control, where the entry clone was obtained from the Gateway LR clonase kit (Invitrogen). The cells were harvested and resuspended in buffer A (50 mm Tris-HCl, 100 mm NaCl, and 10% [v/v] glycerol) with 1× Protease inhibitor cocktail (Roche). A cell-free E. coli protein extract was generated from the cell suspension by sonication for 10 × 15 s at setting 5 of 10, followed by centrifugation at 12,500g for 10 min at 4°C to remove cell debris. The cell-free cellular extract (500 µL of 250 µg mL−1 total protein) was subjected to a chaperone activity assay using different heat treatments (40°C, 50°C, 60°C, 70°C, 80°C, and 90°C) for 15 min. Heat-treated proteins were centrifuged at 12,500g for 10 min to separate the denatured (pelleted) proteins from the nondenatured (supernatant) proteins. The protein content of the supernatant was measured using the Quick Start Bradford dye reagent (Bio-Rad). Three replicates were included for each heat treatment, and three measurements were taken for each replicate. The data were analyzed using a general linear model (PROC GLM) using the software package SAS (version 9.4; SAS Institute). Means of GUS- and Hsp16.9-containing extracts at each heat treatment were separated by Duncan’s multiple range test.
To study the effect of the CSEPs on Hsp16.9 chaperone activity, purified CSEPs were mixed with the cell-free cellular lysates (250 µg mL−1 total protein) containing Hsp16.9 or GUS. Since Hsp16.9 expression was high in E. coli, we diluted the Hsp16.9-containing extract with a GUS expression E. coli lysate in a 1:1 total protein ratio. Subsequently, 5 µL of purified CSEP in 40 mm glutathione was added to 50 µL of cellular lysate, making a 1:2 protein molar ratio with Hsp16.9. As a negative control, 5 µL of 40 mm glutathione was added. The mixture was incubated for 1 h at 4°C with gentle agitation and centrifuged at 12,500g for 10 min. Subsequently, a chaperone activity assay was performed as above using 60°C as the heating temperature. Three replicates were included for each treatment, and two measurements were taken for each replicate. The data were analyzed using the SAS software as described above. All means were separated using Duncan’s multiple range test.
Protein Purification
The coding sequences for CSEPs without signal peptides were transferred to the pDest15 overexpression destination vector to fuse a glutathione S-transferase tag to their N termini. The constructs were transformed into the Rosetta E. coli strain. Cultures of 500 mL were prepared from 5 mL of overnight cultures and grown for 4 to 5 h at 37°C at 150 rpm. Protein expression was induced by adding 0.5 mm isopropyl-β-d-thiogalactopyranoside, and cultures were grown for 18 h at 18°C at 150 rpm. The cells were harvested and suspended in 25 mL of buffer A containing 0.5% (v/v) Triton X-100 and 1× Protease inhibitor cocktail (Roche). Cells were sonicated for 10 × 15 s at setting 5 of 10. Cellular debris was pelleted by centrifugation at 13,000g for 10 min at 4°C. The supernatant was diluted by adding 25 mL of buffer A containing 1× protease inhibitor cocktail. Subsequently, 100 µL of glutathione S-transferase beads slurry (Protino Glutathione Agarose 4B; Macherey-Nagel) was added to the protein extract, which was incubated by rolling for 2 h at 4°C. The beads were pelleted and washed twice with buffer A containing 0.2% (v/v) Triton X-100. Protein bound to the beads was eluted in 100 µL of 40 mm glutathione, pH 8.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK250749, AK362925, AK363601, AK372933, AK375791, and AK373460.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nucleotide and amino acid sequences of CSEP0105 and CSEP0162.
Supplemental Figure S2. Nucleotide and amino acid alignments of HvHsp16.9-CI and HvHsp16.9-CI-H.
Supplemental Figure S3. Interaction of CSEP0105 and CSEP0162 with the barley Hsp16.9 and Hsp17.5 proteins in a BiFC assay.
Supplemental Figure S4. Localization of CSEP0105, CSEP0162, and Hsp16.9 proteins in Bgh-infected barley cells.
Supplemental Table S1. List of sHsps identified in barley.
Supplemental Table S2. Primer sequences for plasmid construction and gene expression assay.
Supplementary Material
Acknowledgments
We thank Dr. Patrick Schweizer (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for providing the RNAi vector and the Mlo RNAi construct, Dr. Masumi Robertson (Commonwealth Scientific and Industrial Research Organization Plant Industry) for the Gateway Y2H vectors (pDEST-ACT2 and pDEST-AS2-1), Dr. Anja Fuglsang (University of Copenhagen) for the BiFC binary Ti destination vectors, and Dr. Paul Schulze-Lefert (Max Planck Institute for Plant Breeding Research) for the YFP and mCherry fusion Gateway vectors. Imaging data were collected at the Center for Advanced Bioimaging, University of Copenhagen.
Glossary
- Bgh
Blumeria graminis f. sp. hordei
- HIGS
host-induced gene silencing
- HR
hypersensitive response
- RNAi
RNA interference
- hpi
hours post inoculation
- Y2H
yeast two-hybrid
- cDNA
complementary DNA
- 3-AT
3-amino-1,2,4-triazole
- BiFC
bimolecular fluorescence complementation
- NCBI
National Center for Biotechnology Information
- EHMx
extrahaustorial matrix
- EHM
extrahaustorial membrane
Footnotes
This work was supported by the Danish Strategic Research Council, the Danish Council for Independent Research/Technology and Production Sciences, and the Department of Plant and Environmental Sciences, University of Copenhagen.
Articles can be viewed without a subscription.
References
- Ahmed AA, McLellan H, Aguilar GB, Hein I, Thordal-Christensen H, Birch P (2015) Engineering barriers to infection by undermining pathogen effector function or by gaining effector recognition. InCollinge DB, ed, Biotechnology for Plant Disease Control. Wiley, New York (in press) [Google Scholar]
- Aitken A. (2006) 14-3-3 proteins: a historic overview. Semin Cancer Biol 16: 162–172 [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ (1997) Searching for interacting proteins with the two-hybrid system. I. The yeast two-hybrid system series. InAdvances in Molecular Biology. Oxford University Press, Oxford, pp 11–28 [Google Scholar]
- Basha E, Friedrich KL, Vierling E (2006) The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem 281: 39943–39952 [DOI] [PubMed] [Google Scholar]
- Basha E, Jones C, Wysocki V, Vierling E (2010) Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J Biol Chem 285: 11489–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E (2012) Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci 37: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Burgis TA, Mills DJ, Ho JT, Cramer R, Spanu PD (2009) In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol Cell Proteomics 8: 2368–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, McGuffin LJ, Burgis TA, Spanu PD, Cramer R (2011) Proteogenomics and in silico structural and functional annotation of the barley powdery mildew Blumeria graminis f. sp. hordei. Methods 54: 432–441 [DOI] [PubMed] [Google Scholar]
- Chen GYJ, Jin S, Goodwin PH (2000) An improved method for the isolation of total RNA from Malva pusilla tissues infected with Colletotrichum gloeosporioides. J Phytopathol 148: 57–60 [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res 40: D1194–D1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Vossen JH, de Vries KJ, van Wees S, Tameling WIL, Dekker HL, de Koster CG, Haring MA, Takken FLW, Cornelissen BJC (2005) Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J 43: 284–298 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655 [DOI] [PubMed] [Google Scholar]
- Didelot C, Schmitt E, Brunet M, Maingret L, Parcellier A, Garrido C (2006) Heat shock proteins: endogenous modulators of apoptotic cell death. InStarke K, Gaestel M, eds, Molecular Chaperones in Health and Disease, Vol 172 Springer, Berlin, pp 171–198 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7: 555–569 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fu X. (2014) Chaperone function and mechanism of small heat-shock proteins. Acta Biochim Biophys Sin (Shanghai) 46: 347–356 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA (1997) Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol Cell Biochem 172: 67–79 [PubMed] [Google Scholar]
- Godfrey D, Böhlenius H, Pedersen C, Zhang Z, Emmersen J, Thordal-Christensen H (2010) Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Gupta SC, Sharma A, Mishra M, Mishra RK, Chowdhuri DK (2010) Heat shock proteins in toxicology: how close and how far? Life Sci 86: 377–384 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Nkai K (2006) Protein subcellular localization prediction with WoLF PSOR. InProceedings of Asian Pacific Bioinformatics Conference APBC06. pp 39–48 [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT (2010) Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci USA 107: 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xu J, Zhang H, Zhang X, Shi J, Li M, Ming F (2009) A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ 32: 1046–1059 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K (2012) The HSP90 complex of plants. Biochim Biophys Acta 1823: 689–697 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44: 41–60 [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H, Terauchi R (2003) Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol Plant Pathol 4: 383–391 [DOI] [PubMed] [Google Scholar]
- Kerppola TK. (2008) Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37: 465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Khajuria C, Li J, Trick HN, Huang L, Gill BS, Reeck GR, Antony G, White FF, Chen MS (2013) Wheat Mds-1 encodes a heat-shock protein and governs susceptibility towards the Hessian fly gall midge. Nat Commun 4: 2070. [DOI] [PubMed] [Google Scholar]
- Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem 279: 2101–2108 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A (2007) Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol 145: 1588–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, Kikuta A, Kamiya K, Yamamoto M, Ikawa H, et al. (2011) Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol 156: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaourab HS, Godar JA, Stewart PL (2009) Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry 48: 3828–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22: 3130–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R. (2003) Establishing compatibility between plants and obligate biotrophic pathogens. Curr Opin Plant Biol 6: 320–326 [DOI] [PubMed] [Google Scholar]
- Pedersen C, Ver Loren van Themaat E, McGuffin LJ, Abbott JC, Burgis TA, Barton G, Bindschedler LV, Lu X, Maekawa T, Wessling R, et al. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego C, Nowara D, Bonciani G, Gheorghe DM, Xu R, Surana P, Whigham E, Nettleton D, Bogdanove AJ, Wise RP, et al. (2013) Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol Plant Microbe Interact 26: 633–642 [DOI] [PubMed] [Google Scholar]
- Reddy PS, Kavi Kishor PB, Seiler C, Kuhlmann M, Eschen-Lippold L, Lee J, Reddy MK, Sreenivasulu N (2014) Unraveling regulation of the small heat shock proteins by the heat shock factor HvHsfB2c in barley: its implications in drought stress response and seed development. PLoS ONE 9: e89125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18: 2402–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M. (2004) Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol 136: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche DB, Buenavista MT, Tetchner SJ, McGuffin LJ (2011) The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res 39: W171–W176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhoshkumar P, Sharma KK (2001) Phe71 is essential for chaperone-like function in α A-crystallin. J Biol Chem 276: 47094–47099 [DOI] [PubMed] [Google Scholar]
- Schmidt SM, Kuhn H, Micali C, Liller C, Kwaaitaal M, Panstruga R (2014) Interaction of a Blumeria graminis f. sp. hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target. Mol Plant Pathol 15: 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KK, Kumar GS, Murphy AS, Kester K (1998) Identification of 1,1′-bi(4-anilino)naphthalene-5,5′-disulfonic acid binding sequences in α-crystallin. J Biol Chem 273: 15474–15478 [DOI] [PubMed] [Google Scholar]
- Sharma KK, Kumar RS, Kumar GS, Quinn PT (2000) Synthesis and characterization of a peptide identified as a functional element in alphaA-crystallin. J Biol Chem 275: 3767–3771 [DOI] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ, et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I, de Wit PJ (2009) Fungal effector proteins. Annu Rev Phytopathol 47: 233–263 [DOI] [PubMed] [Google Scholar]
- Takamatsu S. (2013) Molecular phylogeny reveals phenotypic evolution of powdery mildews (Erysiphales, Ascomycota). J Gen Plant Pathol 79: 218–226 [Google Scholar]
- Thomma BPHJ, Nürnberger T, Joosten MHAJ (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B (2010) Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Van Ooijen G, Lukasik E, Van Den Burg HA, Vossen JH, Cornelissen BJC, Takken FLW (2010) The small heat shock protein 20 RSI2 interacts with and is required for stability and function of tomato resistance protein I-2. Plant J 63: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Kim KP, Park SM, Hong CB (2005) Biochemical analysis of a cytosolic small heat shock protein, NtHSP18.3, from Nicotiana tabacum. Mol Cells 19: 328–333 [PubMed] [Google Scholar]
- Zhang WJ, Pedersen C, Kwaaitaal M, Gregersen PL, Mørch SM, Hanisch S, Kristensen A, Fuglsang AT, Collinge DB, Thordal-Christensen H (2012a) Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol Plant Pathol 13: 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lenk A, Andersson MX, Gjetting T, Pedersen C, Nielsen ME, Newman MA, Hou BH, Somerville SC, Thordal-Christensen H (2008) A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol Plant 1: 510–527 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012b) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.