Abstract
Transcription through early-elongation checkpoints requires phosphorylation of negative transcription elongation factors (NTEFs) by the cyclin-dependent kinase (CDK)9. Using CDK9 inhibitors and global run-on sequencing (GRO-seq), we have mapped CDK9 inhibitor-sensitive checkpoints genome-wide in human (Homo sapiens) cells. Our data indicate that early-elongation checkpoints are a general feature of RNA polymerase (pol) II-transcribed human genes and occur independently of polymerase stalling. Pol II that has negotiated the early-elongation checkpoint can elongate in the presence of inhibitors but, remarkably, terminates transcription prematurely close to the terminal polyadenylation (poly(A)) site. Our analysis has revealed a hitherto-unsuspected poly(A)-associated elongation checkpoint, which has major implications for the regulation of gene expression. Interestingly, the pattern of modification of the carboxyl-terminal domain (CTD) of pol II terminated at this novel checkpoint largely mirrors the pattern normally found downstream of the poly(A) site, suggesting common mechanisms of termination.
RNA polymerase (pol) II elongation is blocked soon after initiation of transcription of many protein-coding genes due to the concerted action of the negative transcription elongation factors (NTEFs), DRB-sensitivity-inducing factor (DSIF) and negative elongation factor (NELF)1-3. The switch to productive elongation requires the activity of positive elongation factor b (P-TEFb), comprising cyclin-dependent kinase (CDK)9 and a Cyclin T. P-TEFb phosphorylates NELF, causing it to be released, and phosphorylates DSIF, converting it into a positive elongation factor1-3. P-TEFb also phosphorylates serine (Ser)2 of the Tyr1Ser2Pro3Thr4Ser5Pro6Ser7 repeat of the carboxyl-terminal domain (CTD) of the large subunit of pol II, which helps recruit RNA processing factors to emerging transcripts4,5. This potentially rate-limiting elongation “checkpoint” can, therefore, hold back pol II to create a kinetic “window of opportunity” for the recruitment of factors needed for RNA 5′ capping, productive elongation and RNA processing. In addition, this checkpoint facilitates the rapid and synchronous expression of genes during development6 and c-myc, NF-kB and HIV Tat recruit P-TEFb to activate transcription elongation1.
Pol II pausing also occurs at intron-exon junctions to facilitate recruitment of factors involved in splicing7-9 and a transcription checkpoint is associated with yeast pre-spliceosome formation8. In addition, pol II pausing at the end of the transcription unit is thought to couple 3′ processing to transcription termination10,11.
We set out to elucidate the contribution of CDK9 to the control of transcription elongation genome-wide in human HeLa cells. Our data indicate that productive elongation of the vast majority of pol II-transcribed human genes requires negotiation of a CDK9-dependent checkpoint early in the transcription cycle. Early elongation checkpoints are generally (but not exclusively) located within ~200 bp of the transcription start site (TSS), and occur whether or not pol II is obviously paused close to the TSS. These results are consistent with the effect of CDK9 inhibitors in HEK293 cells 12, mouse embryonic stem (mES) cells13 and Drosophila14 and emphasize that CDK9 is a critical kinase for transcription of metazoan pol II-transcribed genes.
Surprisingly, our analysis also revealed an additional major elongation control point close to the terminal poly(A) site, beyond which pol II cannot effectively proceed in the presence of CDK9 inhibitors. CTD phosphorylation associated with premature termination induced by the inhibitors is similar to the CTD phosphorylation pattern associated with normal termination downstream of the poly(A) site. The inhibitors also cause loss of association of CDK9, Spt5 and the poly(A) factors Ssu72 and CstF64, whereas association of the termination factor Pcf11increases.
Kinase-dependent elongation checkpoints at terminal poly(A) sites provide a point late in the transcription cycle to regulate production of poly(A) mRNA and could, for example, facilitate rapid control of mRNA production from long genes in response to environmental signals/developmental cues.
Results
KM and DRB inhibit transcription of GAPDH
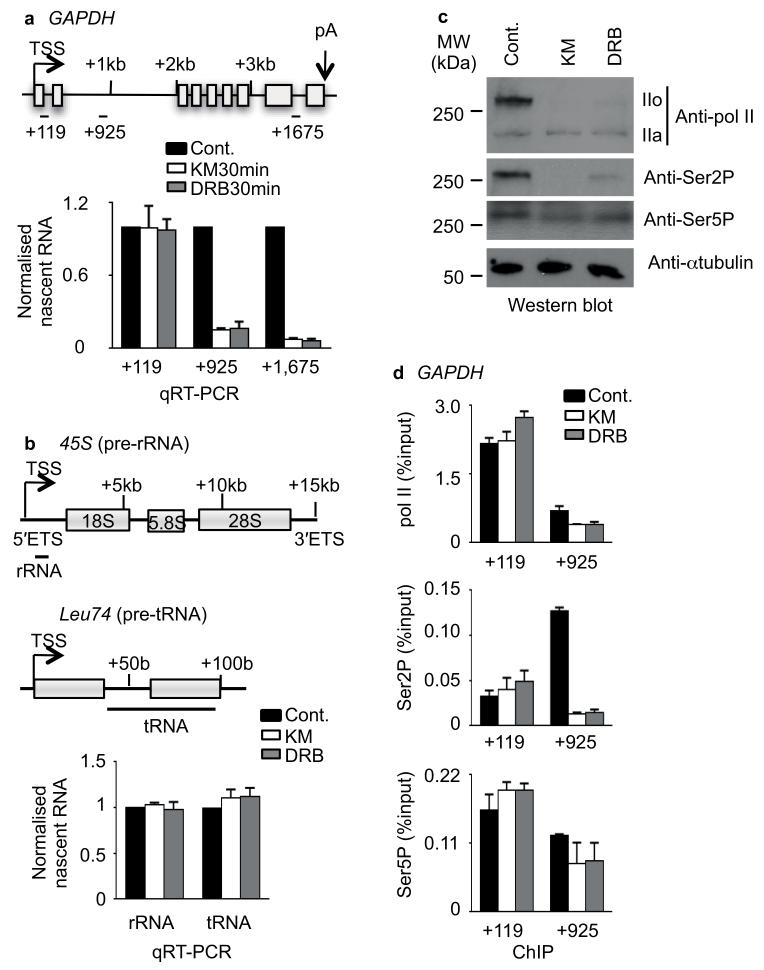
To investigate the contribution of CDK9 to transcription elongation, we treated HeLa cervical adenocarcinoma cells with two drugs known to inhibit CDK9; KM05382 (KM) and 5,6-dichlorobenzimidazone-1-β-D-ribofuranoside (DRB)15 for 15, 30, 45 and 60 min. In accordance with previous findings16,17, these inhibitors caused loss of nascent transcripts from GAPDH and inhibition was complete after 30 minutes treatment (Fig. 1a, Supplementary Fig. 1 and Supplementary Table). Nascent transcripts from the 5′ ETS of pol I–transcribed ribosomal (r)RNA genes and a pol III-transcribed transfer (t)RNA gene were not adversely affected by the treatment (Fig. 1b). This data is consistent with the inability of pol II to negotiate a transcription start site (TSS)-proximal checkpoint when CDK9 is inhibited. CTD hyperphosphorylation (pol IIo) and CTD Ser2P were greatly reduced in cell extract after 30 minutes of drug treatment, whereas Ser5P was less affected (Fig. 1c). Ser2P levels in the GAPDH gene body were also greatly reduced after 30 minutes of drug treatment while Ser5P levels were not (Fig. 1d and Supplementary Fig. 1c).
Figure 1. KM and DRB inhibit transcription of GAPDH.
(a) Quantitative reverse transcription (qRT)-PCR analysis of nascent RNA from GAPDH after KM05382 (KM) and DRB treatment. Top, gene schematic of GAPDH marks the transcription start site (TSS) and polyadenylation site (pA) by arrows, exons by boxes and introns by lines. The middle of amplicons used for chromatin immunoprecipitation (ChIP) experiments is also indicated in kilo base pairs (kb) here and in subsequent figures. Bottom, the value of untreated sample (Cont.) has been normalised to 1 to facilitate comparison of the level of nascent transcript along GAPDH. Error bars, s.e.m. (n = 3 biological replicates). (b) qRT-PCR analysis of nascent transcripts from 45S (pre-ribosomal-RNA) and Leu74 (pre-transfer RNA) after treatment of cells with KM or DRB. Error bars, s.e.m. (n = 3 biological replicates). (c) Western blot analysis of cell extract after KM and DRB treatment, using the indicated antibodies (a full image of a replicate experiment for the anti-pol II panel is shown in Supplementary Data Set 1). Hypophosphorylated (IIa) and hyperphosphorylated (IIo) pol II are noted. α-tubulin serves as a loading control. (d) ChIP analysis of pol II, Ser2P and Ser5P on GAPDH performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates).
In conclusion, 30 minutes of drug treatment is sufficient for effective CDK9 inhibition.
CDK9 inhibitors globally affect early elongation
The role of P-TEFb in transcription-coupled RNA processing4 precludes the use of steady state RNA analysis to study the role of this complex in transcription. Accordingly, we mapped CDK9-reversible elongation checkpoints on pol II-dependent genes in HeLa cells using global run-on sequencing (GRO-Seq)18 coupled with KM and DRB treatment. GRO-seq allows the position and the orientation of engaged RNA polymerases to be mapped across the genome, providing a direct measure of transcription18.
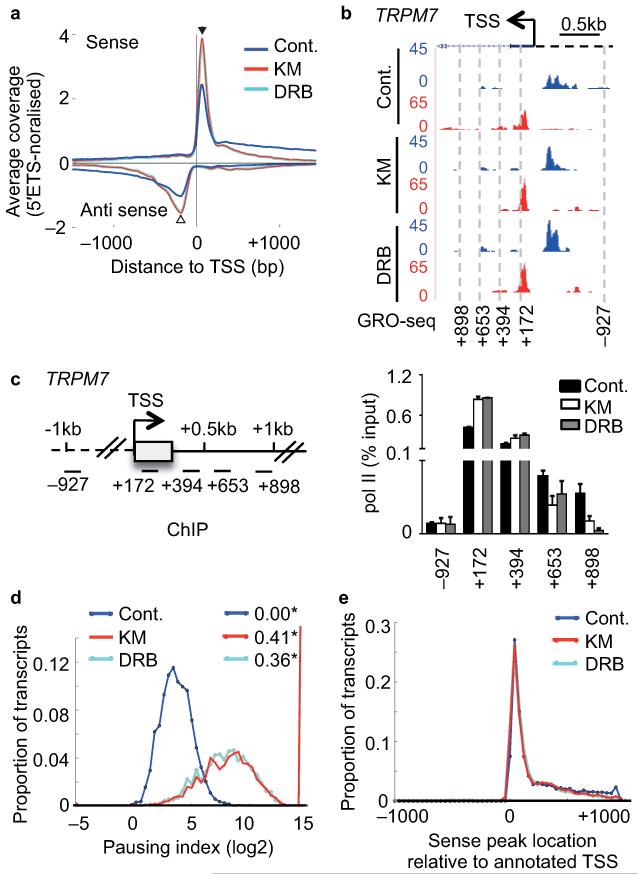
We carried out GRO-seq18 on nuclei isolated after 30 minutes of drug treatment, generated libraries of nascent RNAs and sequenced them using Illumina technology. We processed and normalised GRO-seq reads to reads in the 5′ ETS of the 45S rRNA gene (Supplementary Fig. 2a,b,c,d and Fig. 1b). The read profiles for two GRO-seq replicates using untreated (control) HeLa cells are very similar (Supplementary Fig. 2e,f,g) and the results are consistent with the results of a previous study using primary human lung fibroblast (IMR90) cells18. Also in line with this previous study18, the results of our metagene analysis indicate that most genes have a peak of GRO-seq reads, which corresponds to a peak of active paused pol II, close to the TSS in both sense and antisense directions. The sense peak is located 65bp downstream of the TSS and antisense peak 200bp upstream (Fig. 2a and Supplementary Fig. 2e,h), which is similar to the location of these peaks on genes in IMR90 cells18.
Figure 2. CDK9 inhibitors globally affect early elongation.
(a) Metagene profile of average global run-on sequencing (GRO-seq) coverage (normalised to 45S-5′ETS) of non-overlapping protein-coding genes (>5kb) centred at the TSS with and without KM or DRB treatment of cells. Antisense reads are depicted as negative values. (b) GRO-seq profile of TRPM7 with treatment indicated on the UCSC genome browser. Forward strand reads are noted in blue and reverse strand reads in red here and in subsequent figures. The direction of sense transcription is marked by an arrow. (c) Left, gene schematic of TRPM7. Right, qRT-PCR analysis of pol II ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates). (d) Distribution of log2 of pausing index for individual genes after treatment. Pausing index is calculated as ratio of read density in the TSS-proximal peak to that in the first 5kb of the gene body. * values indicate the proportion of genes with no reads in the 5kb body. (e) Distribution of positions of peaks of sense signal (in 100bp windows) relative to the annotated TSS for each gene after treatment.
Treatment of cells with KM and DRB caused a notable increase of GRO-seq reads close to the TSS in both directions and loss of transcription within 500 bp of the TSS for the majority of genes (Fig. 2a). This metagene profile is exemplified by the TRPM7 and DHX9 GRO-seq and pol II ChIP profiles, which show an increase both in the level of GRO-seq transcripts and pol II association close to the TSS (Fig. 2b,c and Supplementary Fig. 3a,b). Inhibition of transcription beyond a TSS-proximal checkpoint is reflected by a substantial increase in the metagene pausing index after drug treatment (Fig. 2d). Antisense transcription upstream of the TSS was also globally promoter-proximal restricted by drug treatment (Fig. 2a) indicating that antisense transcription is CDK9-dependent genome-wide, in line with previous findings on individual genes19. The position of the sense (Fig. 2e) and antisense (Supplementary Fig. 3c) peaks relative to the TSS was the same before and after treatment, indicating that CDK9 inhibition reinforces TSS-proximal pausing without affecting the location of the pause. These results indicate that the drugs have a profound effect on the ability of newly-initiated pol II to make the transition to productive elongation on protein-coding genes genome-wide, presumably due to inhibition of CDK9.
The promoter-proximal response to P-TEFb inhibitors varies
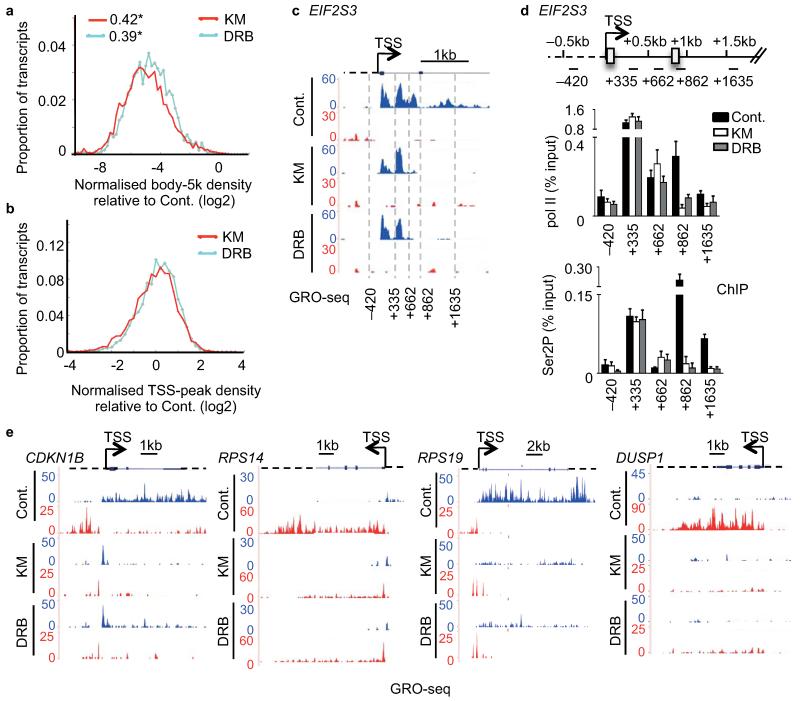
The metagene analysis (Fig. 2a) exemplifies the most common profiles induced by the inhibitors but masks the behaviour of genes with a different profile. Metagene analysis of the ratio of reads in untreated samples to those in treated samples in the first 5kb of the gene body gave negative values between −2 and −8 (Fig. 3a). Thus, the number of reads in the gene body dropped for all genes after treatment but the relative drop varied for different genes. In the TSS-proximal region, this ratio varied from −2 to +2 (Fig. 3b), indicating that the TSS-proximal peak diagnostic of pol II pausing is not always reinforced after treatment with KM and DRB. This is exemplified by the results of GRO-seq and pol II ChIP analysis of EIF2S3 (Fig. 3c,d) and PLK2 (Supplementary Fig. 4a,b).
Figure 3. The promoter-proximal response to P-TEFb inhibitors varies.
(a,b) Distribution of log2 of the ratio between treated and control samples for normalised read density in the first 5kb of the gene body (a) and in the TSS-proximal peak (b). * values in (a) indicate the proportion of genes with no reads in the 5kb body. (c) GRO-seq profile of EIF2S3. (d) Top, gene schematic of EIF2S3. Bottom, qRT-PCR analysis of pol II and Ser2P ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates). (e) GRO-seq profile of CDKN1B, RPS14, RPS19 and DUSP1.
In addition, not all genes had TSS-proximal pol II peaks before drug treatment (e.g. CDKN1B, RPS14, RPS19 and DUSP1) (Fig. 3e). However, the CDK9 inhibitors effectively abolished transcription in the body of these genes, indicating that TSS-proximal CDK9-dependent checkpoints also occur on genes without promoter-proximal pausing (Fig. 3e). Moreover, the response of pol II transcribing these genes was not uniform, with drug treatment causing either a marked increase in transcription close to the TSS (CDKN1B and RPS14), or a loss of reads close to the TSS and throughout the gene (RPS19 and DUSP1) (Fig. 3e). There is therefore more than one flavour of response to P-TEFb inhibitors early in the transcription cycle.
The first sense checkpoint was also encountered relatively far (~1 kb) from the TSS in some cases: EIF2S3 (Fig. 3c,d) and DHX9 (Supplementary Fig. 3a,b). The drop of GRO-seq reads at this “later than normal” promoter-proximal elongation checkpoint is consistent with the loss of pol II ChIP signals (Fig. 3c,d and Supplementary Fig. 3a,b). Moreover, a drop in Ser2P levels occurred close to this “late” promoter-proximal elongation checkpoint on EIF2S3 (Fig. 3d and Supplementary Fig. 4c), rather than close to the TSS as found for most genes, including PLK2 (Supplementary Fig. 4b). In all cases, Ser5P was much less affected than Ser2P (Supplementary Fig. 4b,c). Although two transcription start sites could account for the two peaks of pol II at the beginning of EIF2S3, there is no evidence of this from transcript analysis or TATA-binding protein (TBP) association studies (UCSC Genome Browser 2013).
Our data indicate that a P-TEFb-dependent checkpoint early in the transcription cycle is a general pol II control mechanism but that the position of the checkpoint and the behaviour of pol II in response to CDK9 inhibition is not uniform. In addition, our data indicate that pol II stalling in the absence of CDK9 inhibitors is neither necessary for nor diagnostic of a CDK9-dependent checkpoint early in transcription of human genes.
Uncovering a poly(A)-associated elongation checkpoint
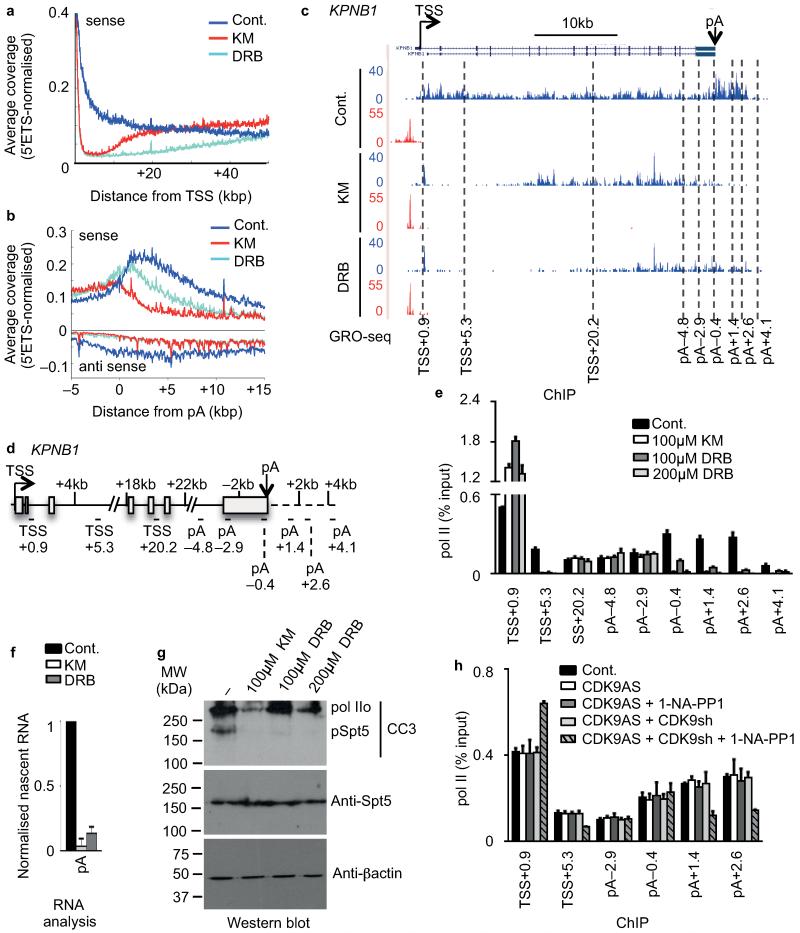
Metagene analysis indicates that in the body of long genes, GRO-seq signals returned to control levels beyond ~20 kbp after 30′ of KM and beyond ~50 kbp after 30′ of DRB treatment (Fig. 4a and Supplementary Fig. 5a). Pol II that has already negotiated the early elongation checkpoint at the start of drug treatment can therefore continue transcription in the presence of CDK9 inhibitors. Transcription came back later in DRB-treated cells than in KM-treated cells, due to differential rates of entry into the nucleus or differential effects of the two drugs on elongation rate. Considering that it may take DRB 5 to 10 minutes to enter the nucleus and fully inhibit CDK9, pol II had travelled approximately 50kb (Fig. 4a) in 20-25 minutes without active CDK9. The elongation rate of pol II under these conditions would correspond to ~2-3kb/min, which is within the range of measurements of elongation rate by other methods13,20,21.
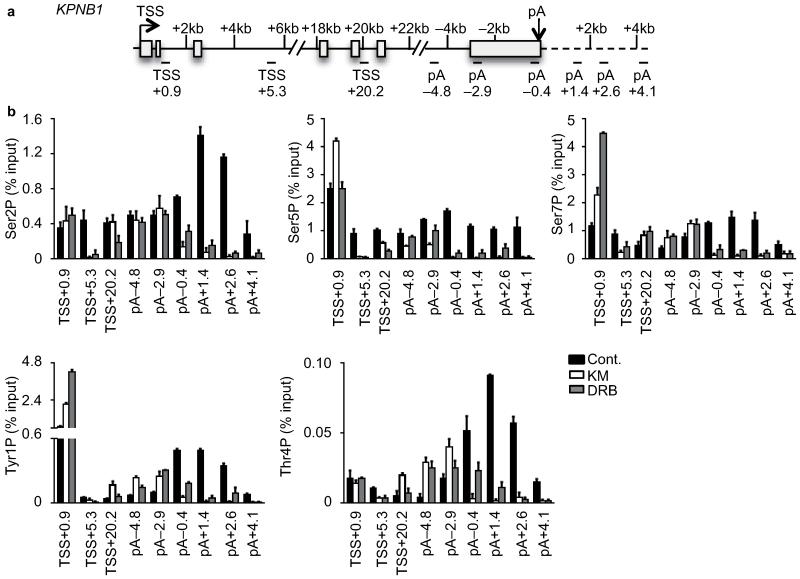
Figure 4. Uncovering a poly(A)-associated elongation checkpoint.
(a,b) Metagene profile of average GRO-seq coverage (normalised to 45S-5′ETS) of long protein-coding transcripts (>50kb) after treatment centred at TSS (0) (a) and on the terminal poly(A) cleavage site (pA) (b). (c) GRO-seq profile of KPNB1 with treatment indicated. The middle of amplicons used for subsequent analysis is indicated in kilo base pair (kb) related to the TSS or to the pA site. (d) Gene schematic of KPNB1. (e) qRT-PCR analysis of pol II ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM, 100 μM DRB or 200 μM DRB. Error bars, s.e.m. (n = 3 biological replicates). (f) qRT-PCR analysis of nascent RNA in HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates). (g) Western blot analysis of cell extract after treatment of cells with KM or DRB, using the indicated antibodies. The CC3 antibody has been shown to recognize hyperphosphorylated (IIo) pol II and phosphorylated Spt544. β-actin serves as a loading control. (h) qRT-PCR analysis of pol II ChIP in control (Cont.) or myc-tagged analog-sensitive CDK9 (CDK9AS)-expressing HEK 293 cell. Cells were also transfected with endogenous CDK9 mRNA-specific shRNA (shRNA) and treated with 1-NA-PP1 as indicated. Error bars, s.e.m. (n = 3 biological replicates).
In line with previous observations18, a broad sense peak is evident downstream of the poly(A) site in the absence of drug treatment (Fig. 4b and Supplementary Fig. 5a). This peak may reflect pol II pausing before termination22,23. Unexpectedly, elongation of transcription beyond the poly(A) site was markedly reduced by the inhibitors, particularly KM, as shown by the metagene analysis of genes longer than 50 kb, whereas transcription upstream of the poly(A) site was unaffected (Fig.4b). A KM and DRB-sensitive kinase is therefore required for elongation of transcription past the poly(A) site, in addition to the requirement for a KM/DRB-sensitive kinase early in the transcription cycle. These findings provide the first evidence of the existence of a poly(A)-associated checkpoint.
GRO-seq and pol II ChIP analysis of KPNB1 (Fig. 4c,d,e) and DHX9 (Supplementary Fig. 5b,c) indicate that drug treatment did not affect the pol II levels upstream of the poly(A) site on these genes, whereas pol II levels at the poly(A) site and downstream were drastically reduced. The level of nascent transcripts past the poly(A) site of KPNB1 and DHX9 was also drastically reduced (Fig. 4f and Supplementary Fig. 5c). These results indicate that pol II prematurely terminated close to the poly(A) site, possibly as a result of CDK9 inhibition.
The differential effect of KM and DRB on the levels of pol II after the poly(A) site both at the genome-wide level (Fig. 4b) and on KPNB1 and DHX9 (Fig. 4c,e and Supplementary Fig. 5b,c), may reflect differential inhibition of CDK9 by the two drugs. Increasing the concentration of DRB decreased the amount of pol II transcribing past the KPNB1 poly(A) site (Fig. 4e). In addition, 100 μM of KM and 200 μM DRB inhibited Spt5 phosphorylation more effectively than 100 μM DRB (Fig. 4g). The CDK9 inhibitor, Flavopiridol13 also effectively inhibited KPNB1 transcription past the poly(A) site (Supplementary Fig. 5d). However, it is also possible that these inhibitors affect kinases other than CDK9. For example, CDK12 is Ser2 kinase that is recruited to the 3′ end of protein-coding genes and is inhibited by Flavopiridol in vitro 24-27. To assess the effect of more specific inhibition of CDK9, we ectopically overexpressed, in human embryonic kidney (HEK) 293 cells, a form of CDK9 that has the gatekeeper amino acid Phe103 mutated to an alanine residue to make the kinase sensitive to the analog 1-NA-PP1 (analog-sensitive CDK9; CDK9AS)28. After siRNA-mediated knockdown (KD) of endogenous CDK9 (Fig. 4h and Supplementary Fig. 5e), CDK9AS represents the majority of cellular CDK9 (Supplementary Fig. 5f). KM has the same effect on pol II levels on KPNB1 in both HeLa and HEK 293 cells and 1-NA-PP1 has no effect on transcription of KPNB1 in HEK 293 cells when CDK9AS is not overexpressed (Fig. 4h and Supplementary Fig. 5g). However, when CDK9AS is overexpressed, 1-NA-PP1 reduces the level of pol II after the poly(A) site (Fig. 4h), suggesting that specific inhibition of CDK9 is at least partly responsible for the premature termination caused by KM, DRB and Flavopiridol. Inhibition of CDK9AS by 1-NA-PP1 causes a less severe defect in elongation at both the beginning and the end of KPNB1 than KM, DRB or Flavopiridol, which could be explained by some remaining endogenous CDK9 activity or incomplete inhibition of CDK9AS by 1-NA-PP1. Alternatively, additional kinase(s) regulate transcription beyond the poly(A) site. The differential effects of KM and DRB at the 3′ end of genes would therefore be due to differential inhibition of a kinase(s) other than CDK9 that also play(s) a role in the control of elongation at the end of genes.
These data support the notion that CDK9 activity is required for efficient transcription through a poly(A)-associated elongation checkpoint to the normal end of the transcription unit, although other KM, DRB and Flavopiridol-sensitive kinases may also be involved.
CTD phosphorylation and premature transcription termination
Termination of transcription induced by CDK9 inhibitors is likely caused by premature release of elongation factor(s) or premature recruitment of termination factor(s). The CTD of pol II acts as a platform to recruit various factors involved in transcriptional and cotranscriptional events and its properties are modulated by post-translational modifications5. Changes in the CTD modification pattern caused by drug treatment may therefore be associated with premature termination. In the absence of CDK9 inhibitors, Ser2P and Thr4P increased towards the 3′ end of KPNB1, Ser5P and Tyr1P were enriched at both the 5′ and 3′ ends, whereas Ser7P was quite constant along the gene (Fig. 5 and Supplementary Fig. 6a,b). These results are in line with genome-wide analyses for these modifications29,30. When ratioed to the pol II level, Tyr1P, Ser2P and Thr4P were higher after the poly(A) site than immediately before, Ser5P decreased, whereas Ser7P levels remained the same (Supplementary Fig. 6a,b).
Figure 5. CTD phosphorylation and premature transcription termination.
(a) Gene schematic of KPNB1. (b) qRT-PCR analysis of Ser2P, Ser5P, Ser7P, Tyr1P and Thr4P ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates).
Changes in the CTD phosphorylation pattern on KPNB1 after drug treatment can only be investigated where the level of pol II is detectable (Fig. 4e, Fig. 5 and Supplementary Fig. 6a). The cellular levels of Tyr1P, Thr4P and Ser7P were not drastically affected by the drugs (Supplementary Fig. 6c). The level of Ser2P before the poly(A) site was not markedly affected by the drugs, whereas Ser5P decreased before the poly(A) site with 100 μM KM causing a greater effect than 100 μM DRB. Ser7P increased with both drugs. The peaks of Tyr1P and Thr4P found downstream of the poly(A) site in the absence of CDK9 inhibition were shifted to upstream of the poly(A) site by drug treatment. The increase in the level of Tyr1P and Thr4P upstream of the poly(A) site was greater after 100μM KM treatment than 100μM DRB treatment, suggesting that the increase in these modifications is somehow associated with premature termination.
Thus, the CTD of pol II associated with premature termination is phosphorylated on Tyr1, Ser2, Thr4, Ser5 and Ser7, which closely mirrors the signature of CTD phosphorylation on pol II before termination downstream of a functional poly(A) site, suggesting that common mechanisms are involved in these termination processes.
Recruitment of poly(A) and elongation factors
Four major complexes are involved in recognition of the poly(A) site and RNA cleavage; the cleavage and polyadenylation specific factor (CPSF), the cleavage stimulatory factor (CSTF), the cleavage factor I (CFI) and the cleavage factor II (CFII)31.
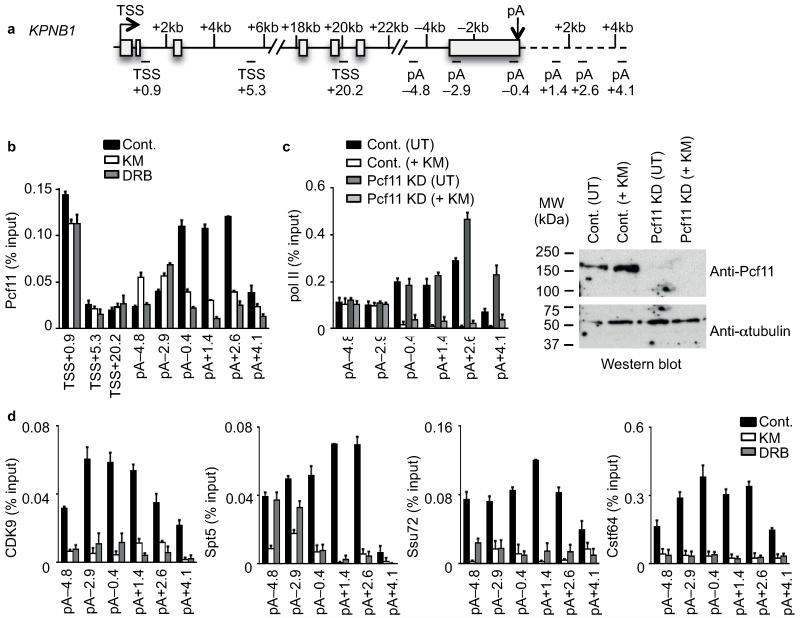
The Pcf11 component of cleavage factor II (CFII) is required for termination in vivo32 and promotes pol II release from template DNA in vitro33,34. The peak of Pcf11association shifted from downstream to upstream of the poly(A) site after drug treatment (Fig. 6a,b and Supplementary Fig. 7). Pcf11 could therefore be involved in premature termination caused by CDK9 inhibitors. However, Pcf11 KD induced a termination defect, as expected32, but did not prevent KM-mediated premature termination (Fig. 6c).
Figure 6. Recruitment of poly(A) and elongation factors.
(a) Gene schematic of KPNB1. (b) qRT-PCR analysis of Pcf11 ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates). (c) Left, qRT-PCR analysis of pol II in control (Cont.) or Pcf11 knockdown (KD) cells with or without KM treatment of cells. Error bars, s.e.m. (n = 3 biological replicates). Right, western blot analysis of extracts from control or Pcf11 knockdown cells with or without KM treatment of cells, with the indicated antibodies. α-tubulin serves as a loading control. (d) qRT-PCR analysis of CDK9, Spt5, Ssu72 and Cstf64 ChIP performed on HeLa cells untreated (Cont.) or treated with 100 μM KM or DRB. Error bars, s.e.m. (n = 3 biological replicates).
Ssu72 Ser5P phosphatase, which is a subunit of the CPSF complex, CstF64, which is a subunit of the CSTF complex, CDK9 and Spt5 were all lost from the KPNB1 poly(A) site region after treatment with KM or DRB (Fig. 6d, Supplementary Fig. 7), implicating these factors in transcription past the poly(A) site. The cellular level of these factors was not adversely affected (Supplementary Fig. 6c).
Loss of Ssu72 and Cstf64 suggests that polyadenylation of transcripts is affected by the inhibitors. Loss of CDK9 and Spt5 suggests that Spt5 and its phosphorylation by CDK9 may be critical to negotiate a transcription checkpoint close to the poly(A) site at the functional end of protein-coding genes.
Discussion
Our characterization of the contribution of CDK9-dependent step(s) to the control of transcription in human cells has defined kinase-dependent checkpoints close to the TSS of pol II-transcribed genes in both sense and antisense directions genome-wide. Our analysis has also revealed a hitherto-unsuspected kinase-dependent transcription elongation checkpoint near the end of genes.
The role of P-TEFb early in the transcription cycle
The development of genome-wide run-on analysis (GRO-seq) has allowed actively-engaged pol II to be distinguished from unengaged pol II at the 5′ end of genes18. This technique therefore facilitates a more accurate characterization of pol II behaviour immediately after initiation. Our results of GRO-seq combined with a short treatment of cells with CDK9 inhibitors indicate that the vast majority of human protein-coding-type genes analysed, including those for long non-coding RNAs (eg Malat), have a CDK9-dependent promoter-proximal elongation checkpoint. Our findings are in agreement with the large amount of in vitro and in vivo data indicating that CDK9 inhibitors have a drastic effect on pol II transcription1-3,12-14. In addition, most, but not all, protein-coding-type genes also have CDK9-sensitive antisense transcription initiating in the TSS region. Thus, pol II initiating in either direction comes under the control of NTEFs, suggesting that their recruitment is linked to initiation of transcription of this type of gene. Our results are consistent with the results of other genome-wide studies using CDK9 inhibitors in human, mouse and Drosophila cells12,13,14.
Productive elongation of genes with no obvious pol II pausing is CDK9-dependent. The lack of promoter-proximal pausing therefore does not mean that an early elongation checkpoint is absent but rather that initiation and the transition to productive elongation are balanced. Our data also indicate that promoter-proximal checkpoints can be located further than 200bp downstream of the TSS. The exact response of pol II to CDK9 inhibitors at the beginning of genes and the position of the checkpoint is likely to depend on the complex kinetic interplay of pol II, transcription factors, P-TEFb and NTEFs on each gene. For example, if NELF and/or DSIF are recruited slowly, relative to the speed of pol II, the checkpoint will occur further from the TSS, as has been shown for pol II pausing in Drosophila35. Our data is therefore consistent with the notion that P-TEFb activity is a general requirement for productive elongation of mammalian genes, whether or not pol II is obviously paused after initiation.
Transcription elongation in the absence of active CDK9
Our results and those of Jonkers13, indicate that pol II continues elongation of transcription in both HeLa and mES cells in the presence of CDK9 inhibitors. Thus, the continued activity of CDK9 appears to be dispensible for transcription once the early-elongation checkpoint has been passed. CDK9 phosphorylates the Nelf-E subunit of NELF, the Spt5 subunit of DSIF and Ser2 of the pol II CTD heptapeptide. NELF is found associated only with the beginning of genes as it is released from the elongation complex after phosphorylation36. We detect phosphorylation of Ser2 of the CTD of elongating pol II after treatment of cells with CDK9 inhibitors, arguing that this mark persists or is placed by a kinase unaffected by the drugs. It is likely that some Spt5 phosphorylation persists up to the point of premature termination in the presence of the inhibitors, as Spt5P is an important elongation factor for pol II after the early elongation checkpoint37,38.
After pol II transcribes a poly(A) site, it pauses to allow cleavage and polyadenylation of the nascent transcript, which is followed by termination of transcription11. The finding that treatment with CDK9 inhibitors causes pol II to prematurely terminate transcription close to the poly(A) site genome-wide was unexpected. The drop in the levels of both CDK9 and Spt5 associated with the region of the poly(A) site caused by CDK9 inhibitors is consistent with the notion that Spt5 and its phosphorylation by CDK9 is required for efficient transcription beyond the poly(A) site.
CTD phosphorylation and termination of transcription
The pattern of CTD phosphorylation associated with premature termination in response to CDK9 inhibition is similar to CTD phosphorylation of pol II terminating downstream of a poly(A) site, particularly with respect to the peak of Tyr1P and Thr4P and the decrease of Ser5P29,30. This pattern of phosphorylation could be a cause or consequence of termination. For example, a premature increase in phosphorylation of Tyr1 and, or Thr4 could cause release of necessary elongation factors or premature recruitment of termination factors.
We were surprised to find Tyr1P at both ends of KPNB1 as this modification is specifically associated with the body of yeast genes39. However, a recent paper has shown that phosphorylation of Tyr1 is associated with the 5′ and 3′ end of mammalian genes genome-wide30. In yeast, Tyr1P helps to recruit and, or stabilize binding of the elongation-associated factor SPT6 and inhibit binding of the termination factors Pcf11 and Rtt10339. We find that the increase in Tyr1P at the 3′end of KPNB1 correlates rather with an increase in Pcf11 recruitment and premature termination rather than a termination defect. These results emphasize a clear difference in the effect of Tyr1P on Pcf11 recruitment in yeast and human cells.
The increase in Ser7P caused by CDK9 inhibitors could be due to loss of Ssu72, which has been shown to dephosphorylate Ser7P40.
Effect of CDK9 inhibitors on poly(A) site-associated factors
Premature termination near the poly(A) site is accompanied by loss of the poly(A) factors Ssu72 and CstF64. CstF64 recognizes the less conserved U- or GU-rich sequence located 30 nucleotides downstream of the cleavage site of the poly(A) signal31. Ssu72 is a component of CPSF and the yeast homologue has CTD Ser5P and Ser7P phosphatase activity40,41. Loss of these factors suggests that no functional poly(A) site is available and consequently no functional polyadenylation complex is formed.
Pcf11 is required for termination in vivo and facilitates premature termination of transcription of HIV templates32,42. The increase in the level of Pcf11 upstream of the poly(A) site following CDK9 inhibition is, however, not necessary for premature termination.
The loss of CDK9 and Spt5 from the end of the KPNB1 gene in response to drug treatment provides a mechanistic explanation for the failure of pol II to efficiently elongate to the point of normal termination.
The poly(A)-associated elongation checkpoint
Our data supports the model that phosphorylation of one or more targets of CDK9 is required for pol II to efficiently elongate beyond the poly(A) site. Targets of CDK9 involved in regulation of transcription include Ser2 of the pol II CTD, the NelfE subunit of NELF and the Spt5 subunit of DSIF. The level of Ser2P upstream of the poly(A) site is not affected by drug treatment and NelfE is not present at the end of genes36. Spt5 instead is detected at the end of genes genome-wide36. This raises the possibility that phosphorylation of Spt5 is required for transcription through both the early-elongation checkpoint and the poly(A)-associated checkpoint. In support of this notion, 100 μM KM more effectively inhibits phosphorylation of Spt5 and the association of Spt5 upstream of the KPNB1 poly(A) site than 100 μM DRB, which could account for the differential effect of these inhibitors on transcription beyond the poly(A) site. Our analysis does not exclude the possibility that kinases other than CDK9 are inhibited by the drugs. Although inhibition of CDK9AS affects elongation of transcription at both the beginning and end of KPNB1, KM causes transcription to terminate further upstream from the poly(A) site, implicating additional KM-sensitive kinases in transcription through a poly(A)-associated checkpoint.
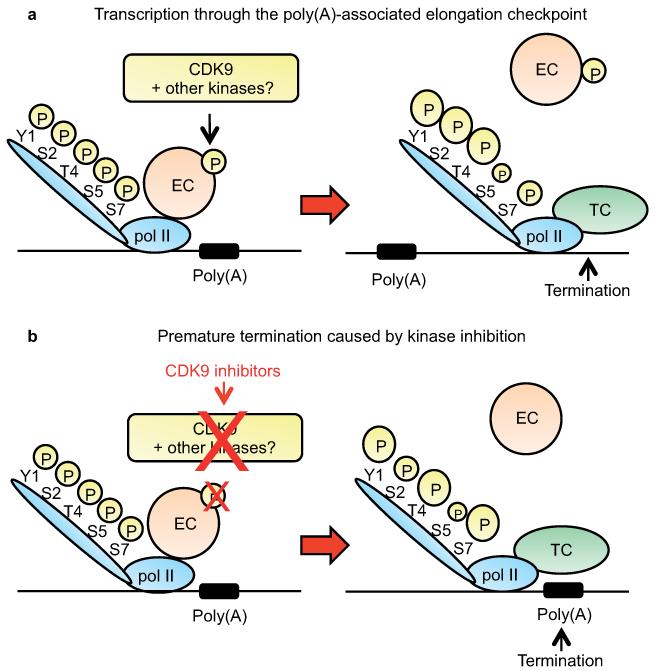
Our current working model (Fig. 7) is that inhibition of CDK9, and possibly additional kinases, causes loss of CDK9 and Spt5P near the poly(A) site, compromising the elongation efficiency of pol II. Additionally, specific loss of other elongation factors in this region of genes 43 and, or recruitment of termination factors10,11 through, for example, alterations in pol II CTD modification may cause pol II to terminate prematurely close to the poly(A) site.
Figure 7. Transcription through the poly(A)-associated checkpoint.
(a) At the 3′ end of the genes, a component(s) of the elongation complex (EC) (e.g. Spt5) must be phosphorylated to allow pol II to transcribe past the poly(A) site. Downstream of the poly(A) site, the elongation complex leaves pol II, phosphorylation of Tyr1 (Y1), Ser2 (S2) and Thr4 (T4) of the CTD is increased, phosphorylation of Ser5 (S5) is slightly decreased and phosphorylation of Ser7(S7) is unaffected, either causing or as a consequence of recruitment of the termination complex (TC). (b) In the presence of kinase inhibitors, the lack of phosphorylation of a component(s) of the EC (e.g. Spt5) prevents transcription past the poly(A) site. In this case, phosphorylation of Y1, T4 and S7 increases and S5 decreases upstream of the poly(A) site, largely mirroring the pattern of CTD phosphorylation found downstream of the poly(A) site in untreated cells. This change in CTD phosphorylation may play a role in loss of the EC and, or recruitment of the TC, inducing premature termination. The size of the circled P (phosphate) reflects the relative level of phosphorylation.
Soon after pol II transcribes a legitimate poly(A) signal, the commitment to make a viable mRNA becomes irreversible if all the necessary splicing and polyadenylation factors are available; the point of no return. A kinase-regulated poly(A)-associated transcription elongation checkpoint therefore constitutes a powerful final control mechanism at the level of transcription to halt production of a processed mRNA.
Online Methods
Cell culture and transient transfection
HeLa and HEK 293 cells were grown in DMEM medium supplemented with 10% fetal calf serum, 100U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine at 37°C and 5% CO2. HeLa cells were treated with 100 μM KM05283 (Maybridge), 100 μM or 200 μM DRB (Sigma) or 1 μM flavopiridol (Sanofi) as indicated.
For ectopic expression of CDK9AS, a pcDNA3 vector encoding CDK9AS with 3 Myc epitope tags at the C-terminus (Supplementary Fig. 6a) was co-transfected into HEK293 cells (~7×106) together with a pcDNA3 vector encoding cyclinT1, a pLKO.1 vector encoding CDK9 shRNA (Sigma) or a control plasmid carrying puromycin resistance cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were treated with neomycin (0.4 mg/ml) and puromycin (3 μg/ml) to select transfected cells. Selected cells were treated for 30 min with 10 μM 1-NA-PP1. As a negative control, HEK293 cells were co-transfected with empty neomycin-resistant pcDNA3.
siRNAs targeting Pcf11 (Dharmacon siGENOME SMART pool) were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions for 48 hours and cells were treated with 100 μM KM for 30 min.
RNA preparation
RNA was extracted from 6 × 106 control or KM/DRB-treated HeLa cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. Reverse-transcription (RT) was performed with 1μg of RNA using random hexamers with the SuperScriptIII kit (Invitrogen) according to the manufacturer’s instructions. cDNA was amplified by quantitative (q)PCR using QuantiTect SYBR Green PCR kit (Qiagen). The level of 7SK RNA was used to normalise the samples. Experiments were replicated 3 times to ensure reproducibility and each RNA sample was measured in triplicate by qPCR. The sequence of primers is given in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as described 45 using antibodies against IgG (sc-2027, Santa Cruz) as an IP negative control, pol II (N-20, Santa cruz), Ser2P (Ab5095, Abcam), Ser5P (ab5131, Abcam), Tyr1P (Active Motif 61384), Thr4P (Active motif 61362), Ser7P (Active motif 61088), Pcf11 (A303-706A, Bethyl Laboratories), Ssu72 (sc-69614, Santa Cruz), Cstf64 (A301-092A, Bethyl Laboratories), Spt5 (sc-28678X, Santa Cruz) and CDK9 (sc-8338, Santa Cruz). Validation for all antibodies is provided on the manufacturers’ websites. ChIP samples were analysed by real-time qPCR using QuantiTect SYBR Green PCR kit (Qiagen) and Rotor-Gene RG-3000 (Corbett Research). Signals are presented as percentage of total DNA after removing the background signal from the IP with the IgG antibody. The sequence of primers used for qRT-PCR is given in Supplementary Table 1. Experiments were replicated 3 times to ensure reproducibility and each ChIP sample was measured in triplicate by qPCR.
Western blot
Western blot analysis was performed as previously described 46 using approximately 30 μg of proteins from cells boiled in Laemmli buffer (50mM Tris pH6.8, 2% sodium dodecyl sulphate, 5% β-mercaptoethanol, 10% glycerol, 0.1% Bromophenol Blue) and antibodies against pol II (sc-9001, Santa Cruz), pSer2 (Ab5095, Abcam), pSer5 (ab5131, Abcam), mitotic phosphoproteins/CC3 (ab78272, Abcam), which recognizes Spt5P 44, Spt5 (sc-28678, Santa Cruz), α-tubulin (200-301-880, Rockland), actin (sc-1615, Santa Cruz), Pcf11 (A303-706A, Bethyl Laboratories), Tyr1P (Active Motif 61384), Thr4P (Active motif 61362), Ser7P (Active motif 61088), Cstf64 (A301-092A, Bethyl Laboratories) and CDK9 (sc-484, Santa Cruz). Validation for all antibodies is provided on the manufacturers’ websites.
Global Run-On Sequencing (GRO-seq)
1.5 × 107 HeLa cells treated or not with 100 μM KM or 100 μM DRB for 30 minutes (min) were harvested by centrifugation, washed with PBS, and resuspended in HLB (10 mM Tris pH 7.5, 10 mM NaCl, 2.5 mM MgCl2) + 0.5% NP-40 + 2 units (U)/ml SUPERase In (Ambion). After 5 min incubation on ice, nuclei were pelleted through a cushion of HLB + 0.5% NP-40 + 10% sucrose + 2U/ml SUPERase In. Nuclei were resuspended in an equal volume of transcription buffer (10 mM Tris pH 8, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 20U SUPERase In, 1% Sarkosyl, 500 uM ATP, GTP and Br-UTP, 2 uM CTP). The transcription reaction was carried out at 30°C for 5 min and terminated by centrifugation for 30 seconds (sec). Nuclei were resuspended in HSB (10 mM Tris pH 7.5, 0.5 M NaCl, 10 mM MgCl2) containing 10U of RNase-free DNase and incubated at 30°C for 20 min. Protein was digested by the addition of 200 μg/ml of proteinase K in buffer (20 mM Tris-HCl pH 7.4, 2% SDS, 10 mM EDTA) at 55°C for 1 hour (h). RNA were extracted twice with acid phenol:chloroform, once with chloroform and precipitated with 3 volumes of −20°C ethanol containing 30 mM NaCl and glycogen. The pellet was washed with 75% ethanol before resuspending in 20 μl of DEPC-treated water. Base hydrolysis, immunoprecipitation of Br-U RNA and end-repair were performed as described 18. The run-on library was prepared by the High-Throughput Genomics Group at The Wellcome Trust Centre for Human Genetics and sequenced using Illumina HiSeq.
GRO-seq read processing
Samples from four GRO-seq analyses-two untreated controls, KM-treated and DRB-treated-were multiplexed together and sequenced on a single lane of Illumina Hiseq using a standard 51 nucleotides (nt) paired-end reads protocol. The obtained GRO-seq reads were processed as described. First, the sequences of the adapters used for sequencing were removed from the 3′ end of the read sequence. The resulting reads which were shorter than 20 nt were then discarded (Supplementary Fig. 2a). The pre-processed reads were mapped to the hg19 assembly of the human genome using bowtie aligner, allowing for up to 2 mismatches and keeping only uniquely mapped reads (bowtie options: -m 1 -v 2 –best –strata). High numbers of read-pairs mapping to the same genomic location indicated potential PCR duplicates, originating from non-linear amplification during library preparation. Reads were therefore de-duplicated using Picard Mark-duplicates tool. Furthermore, a relatively high number of reads mapped to region corresponding to mature products of non-coding RNA genes (especially ribosomal (r)RNA), indicating potential contamination of nuclear run-on samples with these highly abundant RNA species. All reads overlapping annotated non-coding RNA genes (downloaded from the UCSC track) were therefore removed before metagene profiling and segment analysis (Supplementary Fig. 2b).
Normalisation between samples
The total number of mapped reads cannot be used for normalisation of coverage tracks between the samples as normalisation needs to be based on a quantity which is expected to remain unchanged by treatment conditions. As KM and DRB treated samples inhibits transcription of the majority of genes, using such normalisation would lead to an artificially enhanced signal at places where transcriptions remains. We have therefore normalised reads from pol II-transcribed genes to those from the 5′ ETS of the pol I-transcribed rRNA genes, where transcription is not affected by the inhibitors used (Figure 1b). Using the reads from the 5′ETS avoids reads due to contamination from stable and highly abundant 18S, 5.8S and 28S. We believe that an indirect validation of our approach is the finding that this leads to almost equal normalised read density upstream of pA sites of long (>50kb) genes in treated and untreated samples (Fig 4b), which is consistent with Pol II in this region transcribing efficiently in the presence of the drugs.
Reads were mapped independently to the 45S pre-rRNA sequence (downloaded from NCBI, Reference Sequence: NR_046235.1), using the same bowtie settings as above without deduplication (Supplementary Fig. 2c). All samples were then normalised as for 1 million reads mapped to the 5′ETS sequence (Supplementary Fig. 2d). Normalised bigwig coverage tracks were visualised using the UCSC genome browser.
Genome annotation
To construct metagene pileup profiles and to analyse the transcription status of individual genes, we used protein-coding transcripts from Ensembl 68 annotation of human genome (downloaded from http://www.ensembl.org/info/data/ftp/index.html). Only transcripts with a unique start and unique end and longer than 2 kb (unless indicated otherwise in the figure legend) were analysed. Analysis of individual genes was further restricted to transcripts with a normalised sense read-count in the first kilobase (kb) downstream of the TSS above the threshold of 10 and at least 10 times higher than the signal from 1 kb upstream of the TSS.
Metagene profiles, TSS-proximal peaks and transcript segment analysis
To create metagene profiles, 5′ETS normalised read coverage was integrated for each transcript at corresponding base-pair positions, relative to either annotated transcription start site (TSS) or annotated terminal polyadenylation (poly(A)) sites or normalised as if each transcript would be 10 kb in length. For calculation of the signal in different transcripts segments, only the 5′end of each read was counted and 5′ETS normalised. Considering the short time of drug treatment and the resulting recovery of signal on long genes, a transcription level for each transcript was scored between +1 kb and +6 kb downstream of the TSS (gene body). To locate and quantify TSS-proximal peaks, an interval of 1 kb upstream and downstream of the annotated TSS was scanned using a 100 bp sliding window. The location of the maximum signal and its 5′ETS normalised read density was identified. Pausing index was calculated as a ratio of signal in the TSS-proximal peak and average read density in the 5 kb gene body. The transcription termination profile was quantified using a ratio of normalised readcounts in the 2 kb intervals downstream and upstream of the annotated transcript end, which corresponds to the terminal poly(A).
Supplementary Material
Acknowledgements
We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust Grant 090532/Z/09/Z and UK Medical Research Council Hub grant G0900747 91070) for the generation of the sequencing data and A. Heger and CGAT group members for allowing use of their code repository for the analysis. We also thank P. Cook, C. Norbury and D. O’Reilly for critical reading of the manuscript. This work was supported by Wellcome Trust Grants 088542/Z/09/Z and 092483/Z/10/Z and Sir Edward Penley Abraham Trust Fund grants (S.M.) and a scholarship from the Malaysian Government (N.F.I.).
Footnotes
Accession codes
Sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-3360.
Normalised deduplicated bigwig coverage tracks are visualized in the UCSC genome browser session:http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=martind&hgS_otherUserSessionName=hg19%20Groseq%20paper
References
- 1.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: Promoter-proximal pausing and beyond. Biochim Biophys Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Guo J, Price DH. RNA polymerase II transcription elongation control. Chem Rev. 2013;113:8583–603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–41. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–90. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 6.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–11. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell. 2014;53:779–90. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–35. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–69. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–94. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saponaro M, et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–49. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriques T, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52:517–28. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancebo HS, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–44. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff S, Al-Rawaf H, O’Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29:4002–13. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng B, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A. 2011;108:10460–5. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol. 2009;16:1123–4. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- 21.Wada Y, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A. 2009;106:18357–61. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anamika K, Gyenis A, Poidevin L, Poch O, Tora L. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS One. 2012;7:e38769. doi: 10.1371/journal.pone.0038769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–8. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–16. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–72. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davidson L, Muniz L, West S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28:342–56. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosken CA, et al. The structure and substrate specificity of human Cdk12/Cyclin K. Nat Commun. 2014;5:3505. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez MS, Kliegman JI, Shokat KM. The logic and design of analog-sensitive kinases and their small molecule inhibitors. Methods Enzymol. 2014;548:189–213. doi: 10.1016/B978-0-12-397918-6.00008-2. [DOI] [PubMed] [Google Scholar]
- 29.Hintermair C, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012 doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Descostes N, et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife. 2014;3:e02105. doi: 10.7554/eLife.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2:321–35. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West S, Proudfoot NJ. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res. 2008;36:905–14. doi: 10.1093/nar/gkm1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–80. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. Kinetic Competition between Elongation Rate and Binding of NELF Controls Promoter-Proximal Pausing. Mol Cell. 2013;50:711–22. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–15. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W, et al. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007;35:4064–75. doi: 10.1093/nar/gkm430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–5. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 40.Zhang DW, et al. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem. 2012;287:8541–51. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–94. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Klatt A, Henderson AJ, Gilmour DS. Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev. 2007;21:1609–14. doi: 10.1101/gad.1542707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–68. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 44.Lavoie SB, Albert AL, Handa H, Vincent M, Bensaude O. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J Mol Biol. 2001;312:675–85. doi: 10.1006/jmbi.2001.4991. [DOI] [PubMed] [Google Scholar]
References for Online Methods
- 45.Medlin J, et al. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–65. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–34. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.