SUMMARY
In experimental science, organisms are usually studied in isolation, but in the wild they compete and cooperate in complex communities. We report a system for cross-kingdom communication by which bacteria heritably transform yeast metabolism. An ancient biological circuit blocks yeast from using other carbon sources in the presence of glucose. [GAR+], a protein-based epigenetic element, allows yeast to circumvent this glucose repression and use multiple carbon sources in the presence of glucose. Some bacteria secrete a chemical factor that induces [GAR+]. [GAR+] is advantageous to bacteria because yeast cells make less ethanol, and is advantageous to yeast because their growth and long-term viability is improved in complex carbon sources. This cross-kingdom communication is broadly conserved, providing a compelling argument for its adaptive value. By heritably transforming growth and survival strategies in response to the selective pressures of life in a biological community, [GAR+] presents a unique example of Lamarckian inheritance.
INTRODUCTION
When glucose is present, an ancient biological circuit in yeast cells turn off pathways for the utilization of other carbon sources (Johnston, 1999). Many organisms possess such catabolite repression mechanisms (Gorke and Stulke, 2008; Stulke and Hillen, 2000). However, in Saccharomyces cerevisiae this repression is especially stringent (Gancedo, 1998). Indeed, the extreme nature of this trait has led to man's exploitation of yeast for converting sugar into ethanol (Mortimer, 2000). We recently identified a prion, [GAR+], that bypasses this quintessential feature of yeast metabolism (Brown and Lindquist, 2009), allowing cells to utilize glycerol as a carbon source even in the presence of glucose.
Prions are proteins with the unusual ability to stably adopt multiple conformations, at least one of which is self-perpetuating (Shorter and Lindquist, 2005). This provides the basis for a paradigm-shifting mode of inheritance: biological traits that are based on self-templating changes in protein structures rather than on changes in nucleic acid sequence (Halfmann and Lindquist, 2010; Prusiner, 1982). The [GAR+] prion is named for its ability to bypass glucose-associated repression. The brackets denote [GAR+]'s characteristic cytoplasmic pattern of inheritance, the italics reference its function as a genetic element, and the capital letters signify its dominance in genetic crosses. In [GAR+] cells, a small fraction of the major plasma membrane proton pump, Pma1, adopts an altered conformation and forms a complex with Std1, a less abundant protein involved in glucose signaling (Brown and Lindquist, 2009). Because Pma1 is a ten-pass transmembrane protein, the large oligomeric complexes that it forms with Std1 have been difficult to precisely characterize physically, but unlike other well known prions they do not appear to be amyloid in character. However, [GAR+] exhibits the other defining features of a prion: the phenotype it confers is: 1) dominant in genetic crosses, 2) inherited in a non-Mendelian fashion through meiosis, 3) transferred to mating partners without the exchange of nuclei, 4) does not involve the mitochondrial genome and 5) a molecular chaperone protein (in this case Hsp70) is involved in its propagation from one generation to the next (Brown and Lindquist, 2009). Thus, [GAR+] behaves as a prion - a self-templating, protein-based element of epigenetic inheritance.
We have previously hypothesized (Halfmann and Lindquist, 2010; Shorter and Lindquist, 2005; True and Lindquist, 2000) that prions and prion-like epigenetic elements might serve sophisticated bet-hedging functions. In brief: microorganisms face the constant challenge of fluctuating conditions in their natural environments. The prion conformations of diverse proteins appear in yeast populations at low spontaneous rates. When they do, their self-perpetuating changes in protein function create new epigenetic traits. If such traits happen to be detrimental in a particular environment, only those few individuals in the population that have converted to the prion state would be lost. However, if the trait happens to be beneficial, it would allow cells to persist when they might otherwise perish. Cells can lose these epigenetic states spontaneously, providing a complementary survival advantage if the environment should change to disfavor them. Finally, the intrinsic link between environmental stress and protein homeostasis increases the rates at which these epigenetic states are gained and lost precisely when cells are ill suited to their environments (Tyedmers et al., 2008; Newnam et al. 2011; Chernova et al. 2011; Holmes et al. 2013; Cox et al. 1988). Thus, prions would afford a natural route for cells to diversify their phenotypes exactly when such diversity might be most beneficial.
The common presence of prions in wild strains of yeast (Halfmann et al., 2012) and their ability to create complex adaptive traits (Holmes et al. 2013; Halfmann et al., 2012; True and Lindquist, 2000) argue in favor of this notion. Mathematical models with realistic assumptions of prion switching rates and distributions of fitness effects provide additional support (Lancaster et al. 2010; Griswold & Masel, 2009; Jarosz et al., this issue). However, it must be admitted that our understanding of the risks and benefits that such heritable epigenetic traits confer is still primitive. Here we take advantage of genetic, biochemical, and ecological approaches to investigate such questions for [GAR+]. We find that [GAR+] is induced by cross-kingdom chemical communication between yeast and diverse bacteria. This prion-based social interaction confers strong adaptive advantages to both organisms and is evolutionarily conserved. Thus, chemical induction of [GAR+] provides an unprecedented mechanism to govern the appearance of new heritable traits in response to the dynamics of life in a biological community.
RESULTS
S. hominis induces a heritable change in yeast carbon utilization
When glucose is present, yeast cells turn off pathways required for the utilization of other carbon sources such as glycerol (Johnston, 1999). This is conveniently demonstrated in the laboratory by their inability to grow on glycerol medium when it contains trace quantities of non-metabolizable glucose mimetics such as glucosamine (GlcN) (Kunz and Ball, 1977). Even when present in very small quantities, GlcN completely prevents cells from growing on glycerol (Ball et al., 1976; Kunz and Ball, 1977). When cells harbor the epigenetic prion element known as [GAR+], they circumvent glucose repression of growth on glycerol. The prion therefore allows them to grow robustly on glycerol in the presence of glucosamine (GLY + GlcN; Brown and Lindquist, 2009; Kunz and Ball, 1977).
Through a serendipitous accident we discovered that bacteria have the capacity to induce this epigenetic element in yeast. When yeast cells were plated onto GLY + GlcN, we noticed a circle of growth surrounding a contaminating bacterial colony. The yeast cells could grown on GLY + GlcN when re-streaked to fresh medium (data not shown). We then directly tested the ability of this contaminant, Staphylococcus hominis, to induce yeast to grow on such media (Fig. 1A). We first spotted S. hominis cells in a row across the top of a GLY + GlcN plate. Beneath this row of bacterial cells we plated identical rows of naive yeast cells in five-fold serial dilutions. When S. hominis was present on the plate, many yeast cells grew into colonies (Fig. 1A) and their growth followed a strict spatial gradient in proportion to their distance from the bacteria. On control GLY + GlcN plates virtually no colonies grew.
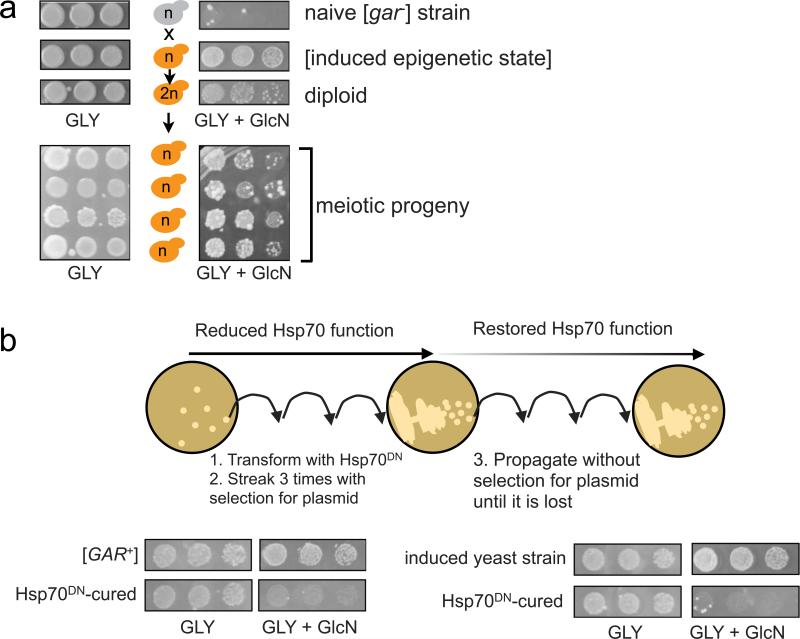
Figure 1. S. hominis induces a stable circumvention of glucose repression.
A) Growth of S. hominis adjacent to [gar−] S. cerevisiae on Gly + GlcN plates induces the yeast cells to acquire a glucosamine-resistant trait. Both organisms are plated in five-fold serial dilutions. B) This trait is stable after hundreds of generations of growth on non-selective media followed by re-testing on Gly + GlcN plates (top). See also Table S1.
This suggests that the bacteria produced a diffusible factor that allowed yeast cells to circumvent glucose repression. To determine whether this bacterial factor had simply influenced the metabolism of nearby yeast cells in a transient way or had induced a stable heritable [GAR+]-like trait, we isolated multiple colonies from such plates for further analysis. We propagated the cells for hundreds of generations on glycerol media without glucosamine and without bacteria. We then transferred them back to GLY + GlcN media. Through all these doublings, in the absence of any direct selective pressures from GlcN or bacteria, the yeast cells retained their ability to circumvent glucose repression (Fig. 1B).
Bacterial induction of the epigenetic trait occurs in diverse yeast strains
We tested whether the ability of yeast to respond to this bacterial signal and acquire the ability to grow on glycerol in the presence of GlcN is broadly distributed. We assembled a panel of 15 genetically diverse yeast strains from distinct wild origins: wine, beer, sake, soil, oak, and infected human patients (Table S1). Collectively, these strains harbor at least 100,000 polymorphisms (Schacherer et al, 2009; Liti et al, 2009). We grew each of these strains to mid-exponential phase and plated them in five-fold serial dilutions adjacent to rows of bacteria (S. hominis) on GLY + GlcN plates. In each strain, this exposure induced in some cells a heritable capacity to circumvent glucose repression on glycerol (Table S1). Thus, this cross-kingdom communication between bacteria and yeast has been broadly conserved over the evolutionary history of the species.
The heritable change in metabolism is due to induction of [GAR+]
The large number of yeast cells that heritably changed their growth properties in response to nearby bacteria made it highly unlikely that this trait arose from genetic mutations. Accordingly, we asked whether the bacteria might be inducing the de novo appearance of the spontaneous epigenetic element we have previously described as [GAR+]. To do so, we asked if the bacterially-induced trait had the same highly unusual inheritance features that are characteristic of [GAR+]. These features derive from the fact that [GAR+] and other prions are self-perpetuating changes in protein conformation. First, when a [PRION+] cell is mated to a [prion−] cell, the prion conformation propagates to proteins of the same type in mating partners and therefore manifests as a dominant trait. Second, because the trait is not based on mutations in DNA, it segregates in a non-Mendelian fashion to progeny in meiosis. Third, the maintenance of prions depends upon the continuous activity of chaperone proteins, which guide the folding of other proteins and thereby govern the self-perpetuation of prion conformations.
In genetic crosses, the trait that was induced by S. hominis to allow growth on GLY + GlcN was, indeed, dominant. That is, the diploids that were formed by mating such cells to naive [gar−] cells had the ability to grow on GLY + GlcN. Moreover, this trait was inherited in a non-Mendelian fashion and was transmitted to all the progeny of meiosis (Fig 2).
Figure 2. S. hominis induces the [GAR+] prion.
A) The GlcN-resistant trait induced by S. hominis has the same dominant, non-Mendelian pattern of inheritance as spontaneous [GAR+] in genetic crosses to [gar−] strains. B) Both the [GAR+] prion and the GlcN-resistant trait induced by S. hominis can be lost by transient reductions in Hsp70 activity caused by expression of a plasmid-borne dominant negative variant (K69M) of the Ssa1 protein. See also Fig. S1 and Fig.
Virtually all mammalian and fungal prions can propagate as distinct strains with ‘strong’ and ‘weak’ phenotypes (these are based on differences in the nature of the self-perpetuating protein template). In our experience, however, [GAR+] is unique in that both mating and meiosis destabilize the element and spark the diversification of ‘strong’ and ‘weak’ prion phenotypes (Brown and Lindquist, 2009). Indeed, with the bacterially-induced trait, freshly created diploids gave rise to mixed populations of ‘strong’ and ‘weak’ strains: some cells grew robustly and developed into large colonies and others grew more slowly and developed into small colonies. These strong and weak bacterially-induced phenotypes propagated faithfully during subsequent mitotic divisions. After meiosis however, while all progeny inherited the ability to grow on GLY + GlcN, they also displayed ‘strong’ and ‘weak’ phenotypes (Fig. 2A). Thus, S. hominis induced an epigenetic element with the same highly unusual patterns of inheritance as [GAR+].
Among the prions characterized to date, [GAR+] is also unique in its chaperone requirements. Instead of requiring the continuous maintenance of Hsp104 activities, it requires the continuous maintenance of high levels of Hsp70 function (Brown and Lindquist, 2009). To determine if the bacterially-induced trait had this same dependence on Hsp70, we made use of a dominant-negative point mutant that has been well characterized in many eukaryotes (Hsp70(K69M); Newmyer and Schmid, 2001). We transformed cells with a plasmid expressing this variant from a constitutive promoter, passaged the colonies three times on selective media and then relaxed this selection to obtain cells that had lost the plasmid (Fig. 2B). The transient inhibition of Hsp70 function cured cells of the epigenetic trait. Importantly, while this same regimen cures cells of [GAR+] (Fig. 2B), it does not affect the transmission of genetic mutations that circumvent glucose repression (Brown and Lindquist, 2009). Finally, we tested whether the bacterially-induced trait could be cured by transient inhibition of Hsp104. It was not (Fig. S1). We conclude that the heritable transformation of yeast metabolism induced by S. hominis is due to the induction of an element indistinguishable from [GAR+].
The prion-inducing activity of S. hominis is selective for [GAR+]
Does this interaction between bacteria and yeast lead to a general induction of prion switching? To test this, we grew S. hominis in co-culture with yeast cells harboring a well-studied reporter for the [PSI+] prion. This protein-based epigenetic element, formed by self-templating conformational changes in the Sup35 translation termination factor, leads to stop codon read-through. Many environmental stresses increase the rates of switching into and out of this prion state, owing to its reliance on protein homeostasis networks for propagation from one generation to the next (Tyedmers et al., 2008). However, co-culture with S. hominis did not induce [PSI+] above background levels in any of our experiments (Fig. S2). Co-culture with S. hominis also had no effect on the acquisition of [MOT3+] prion, which appeared at a frequency of ~10-4 in each case (Alberti et al., 2009; Holmes et al, 2013). Thus, S. hominis and other does not appear to simply cause a general increase in prion induction, but rather specifically induce [GAR+].
[GAR+] is induced by inter-kingdom chemical communication
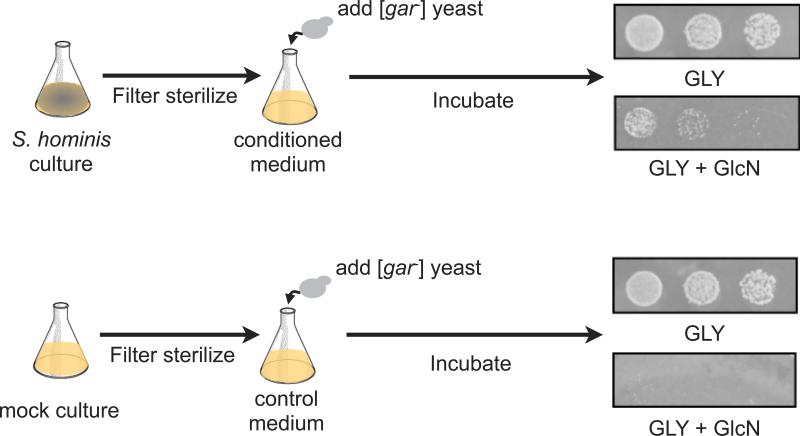
The steep spatial gradient of [GAR+] induction on agar plates suggested it was mediated by a diffusible factor. We investigated the nature of this inducing factor by exposing yeast cells that did not contain the prion, [gar−] cells, to filter-sterilized medium in which inducing bacteria had been grown (Fig. 3). Yeast cells were then pelleted, washed, and plated onto selective GLY + GlcN media. Transient exposure to conditioned medium greatly increased the number of [GAR+] colonies that appeared on such plates (Fig. 3). This was true even after very short exposures (1-4 h). Thus, conditioned medium did not simply enrich for the growth of pre-existing [GAR+] yeast cells, but induced its de novo appearance.
Figure 3. S. hominis induces [GAR+] via a diffusible factor.
Exposure to S. hominis-conditioned medium for 4 h leads to a large induction of the [GAR+] prion. Control ‘mock’ treated cells showed no such effect. The small number of colonies that do appear matches the frequency of spontaneous [GAR+] appearance in this W303 strain. 10-fold serial dilutions are plated on GLY + GlcN medium. See also Table S2.
We asked whether [GAR+] induction was due to a change in pH of the medium, or the presence of small molecules previously known to mediate cell-to-cell communication in microbes such as acyl-homoserine lactones (Ng and Bassler, 2009), farnesols (Hogan et al., 2004), or 2-phenylethanol (Chen and Fink, 2006). It was not (Table S2). Moreover, the inducing activity was stable to boiling, to extreme changes in pH, to repeated freeze/thaw cycles, and to exhaustive protease, RNAse, and DNAse digestion. Inducing activity could be recovered in the flow-through from 3 kDa filters. Further, it could be lyophilized and extracted into polar organic solvents such as methylene chloride (Table S2) and partially fractionated by reverse phase chromatography, although much of the activity was lost during fractionation. When we analyzed the partially active fractions that we did recover by mass spectrometry, they were chemically complex. Because biologically active natural products often involve multiple compounds and completely novel chemistries that are very difficult to unravel (Chen et al., 2002), identification of the inducing agent(s) will require a more extended analysis. Nonetheless, our data establish that bacteria can induce a heritable, epigenetic change in yeast metabolism via a highly robust, but previously unrecognized, form of chemical communication.
Evolutionarily diverse bacteria can induce [GAR+]
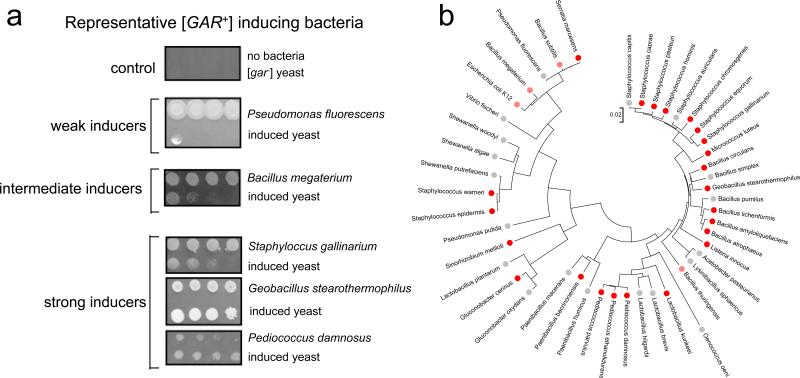
Next we asked if the secretion of a [GAR+]-inducing factor was an idiosyncratic property of S. hominis. We assembled wild isolates of evolutionarily and ecologically diverse bacterial species and asked if they also secreted a diffusible factor that allowed yeast cells to grow on GLY + GlcN media (Table S3). Remarkably, thirty percent of the bacteria we tested had this capacity (Fig. 4; Table S3).
Figure 4. [GAR+] inducing capacity is shared among many bacteria.
A) Yeast cells (as the second row in Fig. 1A) can be induced to acquire [GAR+] by many bacteria and with a wide range of efficiencies. B) Clustering of sequenced, inducing bacteria (and their closest non-inducing relatives) by 16S RNA sequence does not reveal conservation of inducing capacity by clade. Bright red dots represent strains that strongly induce [GAR+]; light red dots represent strains with intermediate inducing ability; grey dots represent strains with weak or no inducing ability. See also Fig. S3 and Table S3-S6.
To quantitatively compare the induction capacity of diverse bacteria, we first grew them to saturation. The bacterial cultures were then spotted next to 5-fold serial dilutions of yeast. Different bacterial species induced the trait with different efficiencies (Fig 4; Table S3). The bacteria did not simply reduce the strength of selection on GLY + GlcN. Cells that did not grow into a colony arrested after 1-3 divisions on GLY + GlcN both when grown alone and when grown near inducing bacteria (Fig. S3). Remarkably, growth in the vicinity of some bacterial species increased the frequency at which the trait appeared >10,000-fold over its spontaneous frequency (e.g. for ‘strong inducers’ in Fig. 4A). The strong inducers Listeria innocua, Sinorhizobium meliloti, and Bacillus megaterium did not alter the frequency of [PSI+] or [MOT3+] appearance (~10−6 for [PSI+] and ~10−4 for [MOT3+]). Inducing bacteria included both Gram-positive and Gram-negative organisms and, within these groups, did not cluster by clade (Fig. 4B).
Although there was no evident phylogenetic relationship among inducing bacteria, we did note a striking ecological relationship among some of the strongest prion-inducing bacteria. Vintners have characterized the bacterial species that are commonly found in arrested wine fermentations (Boulton et al., 1996) – fermentations that they classify as “unsuccessful.” Increased numbers of bacterial cells, and a much greater diversity of species, is a hallmark of these “unsuccessful” fermentations. We tested seven bacterial strains from such arrested fermentations for inducing activity. Four of the seven had an unusually strong induction capacity (Table S3). These included evolutionarily diverse bacterial species, such as Pediococcus damnosus and Lactobacillus kunkeei.
To determine if the induced ability to grow on GLY + GlcN media was due to the de novo appearance of [GAR+], we examined the heritability of this trait in colonies from twenty inductions employing diverse bacterial species (Table S3). We passaged the yeast colonies for hundreds of generations on glucose medium in the absence of bacteria (and without GlcN) and then returned them to selective GLY + GlcN plates. All cells we tested maintained the ability to circumvent glucose repression, establishing the long-term, stable inheritance of this trait. We further characterized the genetic behavior of five representative yeast strains, again choosing strains in which the trait had been induced by bacteria with diverse evolutionary origins (Staphylococcus gallinarium, Escherichia coli, Bacillus subtilis, Sinorhizobium meliloti, and Listeria innocua). In all of these strains, the ability to grow on GLY + GlcN medium showed the same highly unusual patterns of inheritance characteristic of [GAR+] (Table S3). Specifically, the trait was dominant and exhibited non-Mendelian inheritance to progeny of meiosis. Thus, the ability to induce [GAR+] is broadly distributed amongst bacteria.
Distinct pathways drive spontaneous and bacterial induction of [GAR+]
To identify yeast genes that are involved in perceiving the inducing signal and transmuting it into a new heritable trait, we performed a genome-wide screen. We employed a library of 4,848 isogenic yeast strains containing precise deletions of virtually all non-essential open reading frames (ORFs) in the genome (Winzeler et al., 1999). Using robotic pinning, we interspersed rows of such yeast mutants with rows of S. hominis and grew them on GLY + GlcN plates. We identified 60 yeast mutants that reduced [GAR+] induction (Table S4) and 30 that enhanced it (Table S5).
Most notably, proteins encoded by each of these gene sets were strongly enriched for interaction with Pma1 (P < 0.01; Chi-squared test), the major plasma membrane protein that we have previously reported to be a critical component in the spontaneous appearance of [GAR+] (Brown and Lindquist, 2009). However, there was otherwise little intersection among pathways that affected the bacterial induction of [GAR+] and those we previously found to influence its spontaneous appearance (Brown and Lindquist, 2009). Hence, while the systems for the spontaneous appearance of [GAR+] and its bacterial induction intersect at a common node, they are highly distinct.
The induction of [GAR+] in yeast confers selective advantages to bacteria
What change in yeast metabolism conferred by [GAR+] might drive the maintenance of this form of chemical communication by bacteria? The only substantial difference in gene expression we have detected between [GAR+] and [gar-] yeast cells is a ~40-fold reduction in the transcription of HXT3, a hexose transporter (Brown and Lindquist, 2009; Table S6 and Extended Experimental Procedures). We therefore used a fluorescent glucose analog to ask whether glucose uptake was reduced in [GAR+] cells. It was not (see Extended Experimental Procedures). This was not surprising, given that yeast have many hexose transporters (Ozcan and Johnston, 1999). However, HXT3 mutants have a unique influence on fermentation kinetics and ethanol production, separate and apart from their effects on glucose uptake (Karpel et al., 2008). Given that bacteria found in arrested wine fermentations had a particularly strong [GAR+] induction capacity, we wondered if [GAR+] might reduce the production of ethanol by yeast. If so, the reduced ethanol and residual sugar might provide more favorable conditions for bacterial growth.
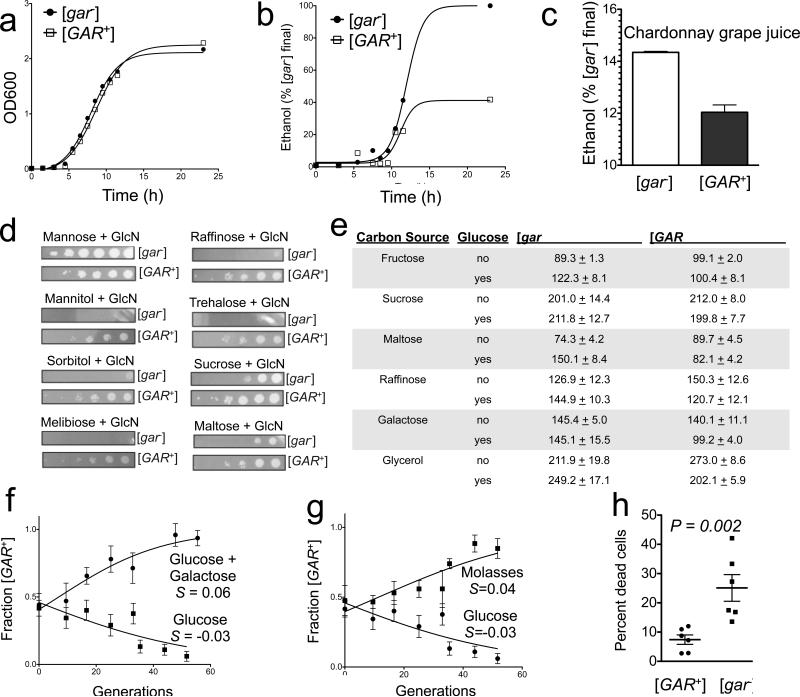
Indeed, although [GAR+] and [gar−] strains grew at similar rates in YPD (Fig. 5A) laboratory glucose medium, [GAR+] cells produced substantially less ethanol (Fig. 5B). This result also held true in Chardonnay grape juice (Fig. 5C). Next, we determined the concentration of ethanol that is toxic to the bacteria that have the capacity to induce [GAR+]. In exposures ranging from 1 to 72 h, with concentrations of ethanol that are typically reached in [GAR+] cultures, the prion-inducing bacteria we examined tended to grow well. With the concentrations of ethanol produced by [gar−] cultures they tended to grow poorly (Table S3).
Figure 5. [GAR+] reduces ethanol production yet confers benefit to yeast cells.
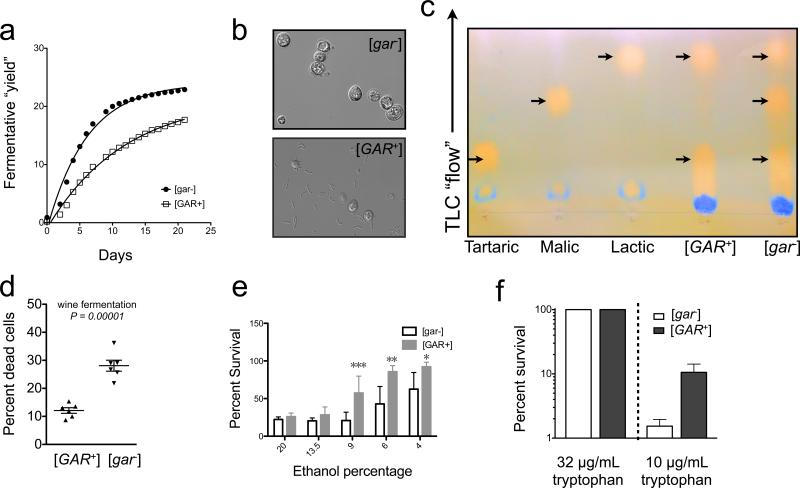
A) Representative growth curves of [GAR+] and [gar−] cells in rich medium (YPD) are indistinguishable. B) [GAR+] cells produce much less ethanol than isogenic [gar−] cells. Representative curves are shown. Similar results were obtained with both colorimetric and electrochemical detection. C) Similar effects occur in Chardonnay grape juice. Error bars represent the standard deviation obtained from three biological replicates. D) [GAR+] circumvents glucose repression of other carbon sources. E) [GAR+] confers advantage in mixtures of glucose and other carbon sources. F) Starting with equal numbers of cells, [GAR+] yeast are outcompeted by isogenic cells that do not harbor the prion in glucose alone. In contrast, [GAR+] strongly outcompetes [gar−] in a mixed carbon source environment (YP with 1.9% galactose and 0.1% glycerol in G); Molasses in D)). Calculated selection coefficients (S) are noted for [GAR+] in these conditions. Error bars are the standard deviation determined from three independent biological replicates. H) [GAR+] cells survive longer in aged cultures than isogenic [gar−]. Cultures were grown for 3 weeks in minimal grape must medium and viability was judged by the cells ability to export methylene blue and the formation of colony forming units. Error bars represent the standard deviation obtained from three biological replicates.
The induction of [GAR+] confers selective advantages to yeast
Why would yeast maintain a complex genetic network to respond to bacteria by switching to [GAR+], given that that the bacteria in their environment might exploit this switch to their advantage? To answer this question we investigated what advantages [GAR+] might confer to yeast cells. We first asked if [GAR+] would allow cells to circumvent glucose repression in carbon sources other than glycerol. To provide a stable non-metabolizable glucose signal, we again employed a low concentration of the glucose mimetic GlcN. Inclusion of GlcN prevented naïve [gar−] yeast cells from metabolizing all of these carbon sources. In every case, however, [GAR+] allowed yeast cells to circumvent glucose repression (Fig. 5D).
In nature, yeast cells generally do not grow on pure carbon sources (and, of course, they do not typically encounter glucose mimetics). They do frequently encounter mixtures of glucose and other carbon sources. Indeed, although [GAR+] provided no growth advantage in any pure carbon source we tested, it provided a substantial benefit in many mixed carbon sources (Fig. 5E). This advantage held true with commercially important substrates for fermentation (e.g. molasses) and was particularly evident when [GAR+] and [gar−] cells were grown in direct competition (Fig. 5 F-G; P=1.4 x 10−8). Therefore, as discussed at length in the accompanying paper (Jarosz et al., this issue), [GAR+] cells act as metabolic generalists, having high fitness across a much wider range of carbon source conditions than [gar−] cells, which are ‘specialists’ for the fermentation of glucose.
Finally, investigating additional benefits of [GAR+], we examined the long-term viability of cells in aged cultures (>2 weeks post saturation phase). We assessed cell density microscopically and measured viability by both plating for colony forming units and staining with methylene blue (Boulton et al., 1996). Strikingly, aged cultures of [GAR+] cells had much higher viabilities than those of [gar−] cells (Fig. 5H).
A few bacterial cells are sufficient to induce [GAR+] in individual yeast cells with high efficiency
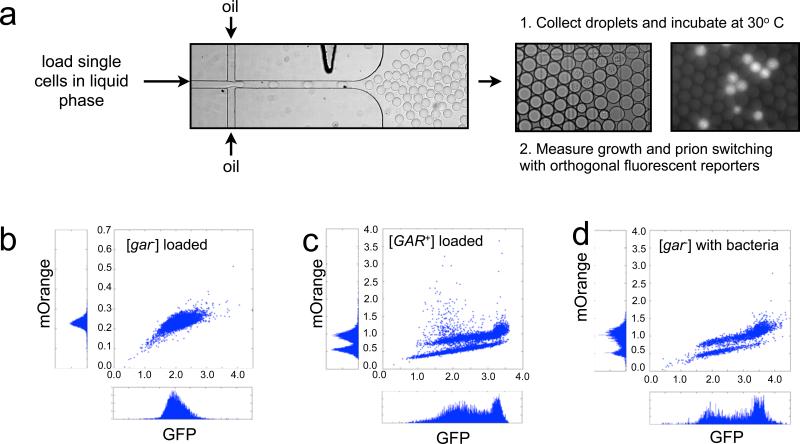
Next we asked whether the induction [GAR+] required the concerted action of millions of bacterial cells or could occur through interactions with just a few. To investigate we took advantage of droplet microfluidics to assess the dynamics of [GAR+] induction on a single-cell level. This technique enables rapid and massively parallel analysis of individual cells in millions of microscopic growth chambers. The microdroplets we used are formed by the encapsulation of growth medium suspended within a biocompatible, gas-permeable hydrocarbon phase (Koster et al., 2008). We loaded between one and three yeast cells in such droplets (with a volume of 65 picoliters), ultimately analyzing millions of cells (Fig. 6A). The cells harbored a constitutive fluorescent marker (mOrange) to report on cell number and an Hxt3-GFP fusion to report on prion status (Brown and Lindquist, 2009). Droplets were incubated for various times at 30 °C, and the growth of cells within them was periodically, and rapidly, analyzed by automated microscopic examination of the droplets as they passed through a micro-chamber in a continuous fluid stream.
Figure 6. Single cell dynamics of [GAR+] induction.
A) Schematic of microfluidic encapsulation of single yeast and bacterial cells in droplets and subsequent experiments. B-D) Scatterplots of mOrange vs. GFP fluorescence for ~106 droplets containing either [gar−] yeast, [GAR+] yeast, or [gar−] yeast and E. coli strain MG1655 (an inducing bacterium) after 48 h incubation. See also Figure S4.
When the droplets were composed of nonselective glycerol medium, both [gar−] and [GAR+] cells grew at similar rates, and to similar final culture densities, as they did in bulk culture (Fig. S4). We next encapsulated cells in droplets composed of [GAR+] selection medium (GLY + GlcN). In this medium, [GAR+] cells grew well, but most [gar−] cells underwent at most one cell doubling – vividly demonstrating, on a single-cell level, the extreme rapidity of glucose repression (Johnston et al., 1994). This finding also allowed us to directly measure the rate at which [gar−] cells switch to [GAR+] per-generation (see Extended Experimental Procedures). Most of the droplets showed no growth after 12, 24, or 48h in selective media, but ~one in 100,000 droplets contained cells that had grown to the same density as [GAR+] cells. Cells in such droplets had switched on the [GAR+] reporter (Fig. 6B vs. Fig. 6C; see also Extended Experimental Procedures). Next we co-encapsulated an average of three cells of an inducing bacterium (a wild strain of Escherichia coli) with each yeast cell. This translates to a ~50-fold excess of yeast cell mass (due to the much smaller size of bacterial cells). Even at early time points (12h post encapsulation), many of the droplets that contained both bacteria and yeast had the fluorescent signal characteristic of [GAR+] cells. By 48h, ~80% of such droplets did (Fig. 6D), representing an 80,000-fold increase over the spontaneous prion switching rate. Thus, the interaction between an individual yeast cell and small numbers of inducing bacterial cells is sufficient to elicit the prion with very high efficiency and this switch takes place rapidly, in the course of just a few cell doublings.
[GAR+] transforms community dynamics in natural fermentations
A natural condition in which bacteria and yeast inhabit the same community, in the fermentation of fruits, has been widely exploited by man to produce alcoholic beverages. Because of the economic incentives involved, the changing dynamics in these communities has been extensively studied. We took advantage of this knowledge base, examining the influence of [GAR+] on the stereotyped dynamics of wine fermentations. Such fermentations are populated by diverse microbes present on the grapes and in the winery (with additional yeast often being spiked in by vintners). The microbiological ecology and physical environment of wine fermentation is extremely dynamic and complex. In “successful” fermentations, from the perspective of man, S. cerevisiae quickly overtakes the bacteria and other fungi naturally present on the grapes, and efficiently converts sugars (glucose and fructose) into ethanol. Several stereotyped changes in growth conditions drive the outcome of such competitions including, most classically, ethanol exposure and nutrient depletion (Bisson et al., 2007). S. cerevisiae thrives in these conditions, which are non-permissive to most bacteria and non-Saccharomyces yeasts. Given that [GAR+] cells make less ethanol in laboratory conditions, we asked what effects [GAR+] might have in the context of winemaking.
We started with an Italian vineyard isolate that is used in winemaking (UCD932) and isolated isogenic [GAR+] variants on GLY + GlcN. We used these cells to perform fermentations with an unsulfured, unsterilized Chardonnay juice obtained from the vineyards at the University of California, Davis. We monitored multiple six-gallon fermentations inoculated with [GAR+] cells, and multiple six-gallon fermentations inoculated with [gar−] cells. We used an inoculum of 1 × 106 yeast cells per ml, a cell concentration standard for winemaking. The [gar−] cells vigorously fermented the Chardonnay juice, dominating other microbes and converting all of the glucose and fructose into ethanol (Fig. 7A; final glucose 0.6%; final ethanol 12.7%). In contrast, the [GAR+] fermentations displayed the characteristics typical of ‘arrested’ fermentations, leaving residual sugar and producing significantly less ethanol (Fig. 7A; final glucose 3.1%; final ethanol 8.4% ). Bacteria that had been present in the Chardonnay juice flourished in the [GAR+] fermentations (Figure 7B; estimated 108 per mL based on microscopic examination), and even completed malolactic fermentation (Figure 7C). They did not flourish in the [gar−] fermentations (Fig. 7B; estimated < 105 per mL based on microscopic examination).
Figure 7. The switch to [GAR+] alters population dynamics and confers adaptive advantages in natural fermentations.
A) Fermentations of natural grape juice seeded with [GAR+] yeast have a reduced fermentative yield (a composite measurement of ethanol production, CO2 loss, and residual sugar - see Boulton et al, 1996) compared to isogenic [gar−] yeast. B) Lees collected at the end of fermentations seeded with [GAR+] cells contains at least one hundred fold more bacteria than is normally obtained with [gar−] cells. C) Chromatography analysis of post-fermentation supernatant following inoculation with [gar−] and [GAR+] yeast cells (see Extended Experimental Procedures). [gar−] fermentations exhibit tartaric, malic, and lactic acids, indicating completion of fermentation, while [GAR+] fermentations exhibit little malic acid. D)Yeast that acquire [GAR+] have extended viability, judged by methylene blue staining after prolonged culture and confirmed by changes in colony forming capacity (see Extended Experimental Procedures). Each point represents the percent of dye-permeable yeast in a field of 100 cells. E). [GAR+] cells are more resistant to ethanol than [gar−] cells. 106 cells were incubated for 24 h at 30 °C in varying starting concentrations of ethanol in SD-CSM. Viability was judged by recovery of colony forming units. Error bars represent the standard deviation of six biological replicates. P-values by T-test: ***(P<0.0001), **(P<0.0005), *(P<0.005). F) [GAR+] cells have a strong growth advantage in limiting tryptophan relative to [gar−] cells. Approximately 106 cells were challenged with the indicated concentration of tryptophan in synthetic medium for 24h and then plated to rich medium. Colony forming units in limiting tryptophan were compared to a tryptophan-replete control (50 ug/mL). Error bars represent the standard deviation obtained from three biological replicates.
Pressed grapes naturally contain a wide diversity of fungi. Indeed, when we performed the same fermentations without inoculating them with additional yeast cells, the S. cerevisiae naturally present on the grapes took over the cultures and completed the conversion of sugars into ethanol. The fact that these same cells did not take over the fermentations inoculated with [GAR+] yeast suggests that [GAR+] yeast may produce factors that block the growth of these other fungi. This would ensure that such fungi do not participate in the dynamic sharing of “common goods” between inducing bacteria and [GAR+] yeast (Hardin, 1968; Kramer and Brewer, 1984; Rankin et al., 2007). That is, this finding suggests that [GAR+] yeast may have a mechanism for preventing “cheaters” from arising in the fermentations and stealing the profits of this prion-based mutualism.
Advantages of [GAR+] during wine fermentation
We periodically sampled these fermentations throughout their standard three-week course. The [GAR+] fermentations continued to be dominated by cells that retained the ability to grow on GLY + GlcN. That is, they remained [GAR+]. Cells from [gar−] fermentations remained [gar−]. At the end of these fermentations, yeast cells from the [GAR+] fermentations had much higher viability than those from the [gar−] fermentations (Fig. 7D), just as we had observed in aged laboratory cultures. Investigating possible explanations for this difference, we found that [GAR+] yeast cells survived the high ethanol concentrations characteristic of wine fermentations much better than isogenic [gar−] cells (Fig. 7E).
In addition to ethanol exposure, in long-term fermentations yeast cells also endure starvation for amino acids and other nutrients (Bisson et al., 2007). To directly assess whether [GAR+] provides an advantage against starvation we investigated a laboratory strain that was auxotrophic for tryptophan. The uptake of this amino acid is particularly dependent upon the proton gradient created by the critical [GAR+] determinant Pma1 (Vallejo and Serrano, 1989). Tryptophan prototrophs that also carry partial loss-of-function mutations in Pma1 cannot grow in media where tryptophan levels are low (Vallejo and Serrano, 1989). We found that [GAR+] cells had a strong advantage for growth in this condition relative to isogenic [gar−] cells (Fig. 7F). This finding raises the intriguing possibility that the changes in Pma1 function associated with [GAR+] constitute a gain-of-function rather than a loss of function. In any case, they establish that in the face of defining stresses of fermentation, the [GAR+] prion couples microbial dynamics to heritable changes in metabolic strategies. It does so in a manner that might act to the frustration of man, but acts to the benefit of bacteria and yeast alike.
DISCUSSION
We have discovered a system of chemical communication by which diverse bacteria can induce a stable, epigenetically inherited trait in a eukaryotic organism. This trait transforms one of the most basic metabolic decisions a cell makes: whether to send glucose through the respiratory chain and harvest its full ATP-generating potential, or to instead maximize the conversion of glucose to ethanol through fermentation. The ability of a chemical compound secreted into the environment to elicit this heritable epigenetic trait represents an extreme example of “Lamarckian evolution” in action.
This feat is accomplished via the de novo induction of a protein-based genetic element, the prion known as [GAR+]. The cross-kingdom chemical communication that induces [GAR+] involves multiple genes that are not required for the spontaneous induction of [GAR+] in the absence of bacteria. Given the role of Pma1 in the maintenance of this trait, and the central importance of this proton pump in diverse physiological processes of yeast, other features of [GAR+] biology likely remain to be uncovered. In any case, [GAR+] creates a mutualism that benefits both participants: bacteria gain a more hospitable growth environment, and yeast gain both increased longevity and the ability to metabolize a more diverse array of carbon sources.
The ability of bacteria to broadcast this prion-inducing signal has been conserved in evolutionarily diverse species. And, as demonstrated in the accompanying paper (Jarosz et al., submitted), the capacity of yeast to both receive this signal and transduce it into a new epigenetically determined metabolic state has also been broadly conserved. These observations provide a strong argument that the mutualism we have uncovered confers a benefit to both organisms in the complex natural communities they normally inhabit, ranging from woodlands, to rotting fruit, and even infected human patients.
It is also notable that the genetic network we found to control the chemical induction of this epigenetic switch in yeast includes several genes whose functions have remained elusive to date. The recent advent of genome-wide analyses in S. cerevisiae has ensured that the function of every gene in this organism has been extensively probed. Indeed, each viable yeast gene deletion strain has been tested under at least 400 diverse conditions (Hillenmeyer et al., 2008; Winzeler et al., 1999). Yet, despite the exhaustive nature of these analyses, several of the yeast genes that we find to be critical for the bacterial induction of [GAR+] had no previously reported function. This finding raises the intriguing possibility that portions of this genetic network exist expressly for the purpose of this (and perhaps others) form of chemical communication and social interaction.
We have previously suggested that prions provide sophisticated bet-hedging strategies to diversify heritable phenotypes and enhance survival in fluctuating environments (True and Lindquist, 2000). An opposing view is that they are simply ‘diseases’ of yeast, or even artifacts of laboratory culture (McGlinchey et al., 2011; Nakayashiki et al., 2005). We recently demonstrated that prions formed by diverse yeast proteins have adaptive value. They are widespread in ecologically diverse wild strains of yeast (Halfmann et al., 2012). Moreover, they change the capacity of cells to utilize a vast array of different nutrients and to survive many stressful environments (True and Lindquist, 2000; True et al., 2004; Halfmann et al., 2012; Suzuki et al., 2012). They also alter colony morphology, alter the ability of cells to invade growth substrates, and even affect physical associations in cellular communities (Halfmann et al., 2012; Holmes et al., 2013). But perhaps no argument for the adaptive value of these epigenetic elements is stronger than those provided here and in the accompanying paper: a prion-based mechanism is employed for a broadly conserved system of cross-kingdom chemical communication that heritably transforms growth and survival strategies in response to the selective pressures that are inherent to life in a biological community.
In natural environments organisms live in diverse communities, enduring a fluctuating metabolic landscape and competing with their neighbors for limited resources. In the lab they are generally studied in isolation and in simple, nutrient-replete conditions. This practice was initially driven by the laudable goal of eliminating confounding experimental variables, but it has left us blind to the rich biology arising from natural communities. From tumor biology (Arthur et al., 2012; Dapito et al., 2012) to metabolic disorders (Nicholson et al., 2012; Yatsunenko et al., 2012), social interactions between bacterial and human cells are now emerging as powerful effectors of human health. [GAR+] provides an unprecedented example of an epigenetic mechanism that heritably alters metabolic strategies in complex cellular communities, but we doubt it will prove unique.
EXPERIMENTAL PROCEDURES
Yeast and bacterial techniques
All fungi were propagated on standard laboratory media unless otherwise specified. GLY + GlcN plates were made as previously described (Ball et al., 1976; Brown and Lindquist, 2009; Kunz and Ball, 1977). Bacteria were grown in LB or MRS broth prior to plating. Bacterial induction of S. cerevisiae growth on GLY+ GlcN medium was measured by plating serial dilutions of each organism in adjacent rows on solid agar plates. Growth was measured after 5 days of incubation at 30 °C. Dominant negative Hsp70 was expressed from a GPD promoter on a plasmid marked with G418 resistance that was based on pAG42. Loss of this plasmid was accomplished by propagation on YPD for 75-100 generations and confirmed by loss of G418 resistance. Conditioned media are described in the Extended Experimental Procedures. Ethanol was measured using a colorimetric assay kit (R-Biopharm) and confirmed with a YSI biochemistry analyzer. Competition experiments were performed using cells that harbored LEU+ or URA+ markers and measuring the fraction of the total cells that retained either marker during the course of the competition. The [GAR+] status was confirmed at each sampling. Control competitions were also performed in which cells instead carried the other marker. This did not affect the outcome. Established reporter strains were used to measure frequencies of [PSI+] and [MOT3+] (Alberti et al, 2009).
Microfluidics experiments
Bacteria and yeast were loaded into microdroplets generated on devices made with soft lithography techniques. Further details are provided in the Extended Experimental Procedures.
Chardonnay fermentations
Chardonnay grape juice was obtained from crushing grapes at the UCD winery. 6-gallon fermentations were inoculated with [gar−] or [GAR+] cells at 1X106 cells/mL. Fermentation progress was tracked via density measurements with an Anton Parr DMA 35. Samples were taken periodically and analyzed using plating and staining methods described in the Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. A. Grossman, D. Newman, L. Joseph, B. Bassler, and G. C. Walker, who generously provided bacterial strains for this study. We also thank A. Regev, D. Thompson, B. Bevis, J. Valastyan, M. Taipale, L. Pepper, B. Vincent, K. Erbil, and members of the Lindquist and Bisson laboratories for materials, discussions, and/or critical reading of the manuscript. N. Azubuine and T. Nanchung provided a constant supply of plates and media. This work was funded by grants from the G. Harold and Leila Y. Mathers Foundation and HHMI. D.F.J. was supported as an HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG-1964-08) and by an NIH Pathway to independence award (K99 GM098600). S.L. is a Howard Hughes Medical Institute (HHMI) investigator.
REFERENCES
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal Inflammation Targets Cancer- Inducing Activity of the Microbiota. Science. 2012 doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball AJ, Wong DK, Elliott JJ. Glucosamine resistance in yeast. I. A preliminary genetic analysis. Genetics. 1976;84:311–317. doi: 10.1093/genetics/84.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson L, Karpel J, Ramakrishan V, Joseph L. Functional Genomics of Wine Yeast Saccharomyces cerevisiae. Advances in Food and Nutrition Research. 2007:65–121. doi: 10.1016/S1043-4526(07)53003-2. [DOI] [PubMed] [Google Scholar]
- Boulton R, Singleton V, Bisson L, Kunkee R. Principles and Practices of Winemaking. Chapman & Hall; New York: 1996. [Google Scholar]
- Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes & Development. 2009;23:2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes & Development. 2006;20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O’Dell A, McNally JG, Liebman SW, et al. Prion Induction by the Short-Lived, Stress- Induced Protein Lsb2 Is Regulated by Ubiquitination and Association with the Actin Cytoskeleton. Molecular Cell. 2011;43:242–252. doi: 10.1016/j.molcel.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiology and Molecular Biology Reviews : MMBR. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nature Reviews Microbiology. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI+], even with realistic rates of yeast sex. PLoS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Hardin G. The Tragedy of the Commons. Science. 1968;162:1243–1248. [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Molecular Microbiology. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–65. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends in Genetics: TIG. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel JE, Place WR, Bisson LF. Analysis of the Major Hexose Transporter Genes in Wine Strains of Saccharomyces cerevisiae. American Journal of Enology and Viticulture. 2008;59:265–275. [Google Scholar]
- Koster S, Angile FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten CA, Pisignano D, Griffiths AD, et al. Drop-based microfluidic devices for encapsulation of single cells. Lab on a Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- Kramer RM, Brewer MB. Effects of group identity on resource use in a simulated commons dilemma. Journal of Personality and Social Psychology. 1984;46:1044–1057. doi: 10.1037//0022-3514.46.5.1044. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Ball AJ. Glucosamine resistance in yeast. II. Cytoplasmic determinants conferring resistance. Molecular & General Genetics: MGG. 1977;153:169–177. doi: 10.1007/BF00264732. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Research. 2000;10:403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. Journal of Cell Biology. 2001;152:607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. Journal of Molecular Biology. 2011;408:432–448. doi: 10.1016/j.jmb.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annual Review of Genetics. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiology and Molecular Miology Reviews: MMBR. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Rankin DJ, Bargum K, Kokko H. The tragedy of the commons in evolutionary biology. Trends in Ecology & Evolution. 2007;22:643–651. doi: 10.1016/j.tree.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nature Reviews Genetics. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Stulke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annual Review of Microbiology. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biology. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo CG, Serrano R. Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast. 1989;5:307–319. doi: 10.1002/yea.320050411. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.