Abstract
The International Committee on Taxonomy of Viruses (ICTV) recognized the Polydnaviridae in 1991 as a virus family associated with insects called parasitoid wasps. Polydnaviruses (PDVs) have historically received limited attention but advances in recent years have elevated interest because their unusual biology sheds interesting light on the question of what viruses are and how they function. Here, we present a succinct history of the PDV literature. We begin with the findings that first led ICTV to recognize the Polydnaviridae. We then discuss what subsequent studies revealed and how these findings have shaped views of PDV evolution.
Keywords: insect, parasitoid, mutualist, genome, evolution
Introduction
In the recent inaugural issue of The Annual Review of Virology (ARV), Summers (2014) examined the question of what viruses are and the ever-changing ways virologists have defined them since their discovery in the nineteenth century. In the same issue, we reviewed the Polydnaviridae (Strand and Burke, 2014), which was recognized as a family of insect viruses by the International Committee on Taxonomy of Viruses (ICTV) in 1991 but has largely languished in obscurity in the broader virology literature. Even among insect virologists, polydnaviruses (PDVs) have historically received limited attention because their life cycle makes them difficult to work with and their unusual biology was a disincentive for labs vested in other, primarily model, species. Yet advances in recent years have elevated interest in PDVs, precisely because their unusual biology sheds interesting light on virus evolution and what the essential qualities of viruses are. These considerations also underlie why we were asked to provide a review on PDVs for the 60th anniversary issue of Virology. We cannot avoid overlap here with other recent summaries including the aforementioned ARV article (Beckage and Drezen, 2012; Burke and Strand, 2012a; Strand and Burke, 2012; Strand and Burke, 2013; Gundersen-Rindal et al., 2013; Herniou et al., 2013; Strand and Burke, 2014). However in keeping with an anniversary issue, we orient this paper a bit differently by discussing the PDV literature in largely historical order. We begin with the findings that first led ICTV to recognize PDVs as viruses. We then discuss what later studies found and how these results have progressively shaped views of PDV evolution.
Early years: formal recognition of PDVs as a virus family
The study of PDVs began in the late 1960s and 1970s when particles resembling viruses were observed by electron microscopy (EM) in the reproductive tracts of a few insect species called parasitoid wasps (Hymenoptera) (Rotheram, 1967; Vinson and Scott, 1975; Stoltz et al., 1976). These insects are well known to entomologists because of their widespread abundance, high species diversity, and importance as biocontrol agents for many pest species in agriculture and forestry. In contrast, they are generally not familiar to other life scientists including virologists because of their small size and specialized habits. In brief then, parasitoid wasps are defined as insects that are free-living during their adult stage, which reproduce by laying eggs on or in the bodies of other arthropods referred to as hosts (Godfray, 1994; Pennacchio and Strand, 2006). Wasp progeny develop into adults by feeding parasitically on a single host and the host usually dies as a consequence of being parasitized. Most parasitoid wasps are also specialists that parasitize only one or a few host species.
The Hymenoptera is one of the largest insect orders (>200,000 species) and is divided into several superfamilies and many families. Most of these taxa consist primarily or exclusively of parasitoids. Studies in the late 1970s and early 1980s, however, suggested that PDVs are only associated with wasps in one superfamily, the Ichneumonoidea, which is divided into two families named the Braconidae and Ichneumonidae (Krell and Stoltz, 1979; 1980; Stoltz and Vinson, 1979). Studies during this period also noted that PDV particles from braconid and ichneumonid wasps morphologically differ from one another with the former having cylindrical, often tailed nucleocapsids surrounded by a single envelope that resembled some non-occluded baculoviruses (see below), and the latter having fusiform nucleocapsids with two envelopes (Stoltz and Vinson, 1979).
Despite their dissimilar morphology, early studies also showed that PDVs from braconids and ichneumonids share several features including a common life cycle. Both persist in all cells of braconid or ichneumonid wasps as integrated proviruses (Stoltz, 1990; Fleming and Summers, 1991). Both also only replicate in pupal and adult stage female wasps in nuclei of cells located in the ovaries called calyx cells. Replication produces large numbers of virions that are released by lysis of calyx cells in the case of braconids or budding in the case of ichneumonids. Virions are then stored at high density in the lumen of the reproductive tract (Stoltz and Vinson, 1979). Nucleic acid analysis showed that virions from braconid and ichneumonid wasps contain multiple circular, double-stranded DNAs that are non-equimolar in abundance. The number and size of DNA segments was noted to vary between wasp species (Krell and Stoltz, 1979; Stoltz and Vinson, 1979; Krell et al., 1982) with subsequent studies estimating aggregate sizes for these DNAs to range from ~150 kb to more than 600 kb (see below).
Braconid and ichneumonid wasps use their ovipositors to inject eggs containing the proviral genome, PDV particles, and other secretions into the body cavity of the hosts they parasitize, which are primarily larval stage Lepidoptera (moths and butterflies). Experiments in the late 1970s and 1980s showed that PDVs rapidly infect host cells and discharge their DNAs into nuclei, which is followed by expression of viral genes (Stoltz and Vinson, 1979; Fleming et al., 1983; Blissard et al. 1986). Experiments further demonstrated that survival of wasp offspring depends on infection of the host by PDVs and associated viral gene expression because wasp offspring die in the absence of infection by its associated PDV. This is because most PDVs disable immune defenses, which prevent hosts from killing wasp offspring (Edson et al. 1981; Guzo and Stoltz, 1987; Davies et al., 1987). PDVs were noted to also alter the growth of hosts (Stoltz and Vinson, 1979; Beckage and Riddiford, 1982). Yet, parallel studies showed that PDVs do not replicate in the hosts of wasps (Theilmann and Summers, 1986). The molecular basis for altering host physiology in the absence of replication was not understood in these early studies. The biological significance of these traits, however, was interpreted to mean that PDVs are only transmitted vertically through the germline of wasps, and wasp survival depends on the genes replication-defective PDV virions deliver to hosts. The reliance of PDVs and wasps on one another for survival further suggested they form a mutualistic association (Stoltz and Vinson, 1979; Edson et al., 1981; Fleming, 1992).
In summary, the first studies of PDVs referred to them as nuclear secretions or particles, which implicitly suggested they could be either non-viral or viruses. Thereafter, the literature up to the early 1990s strongly concluded PDVs were viruses because they: 1) replicate in the calyx cells of wasps, 2) morphologically look like viruses, 3) package nucleic acid, 4) are infectious, and 5) contain genes that are transcribed after infection of hosts (Stoltz and Vinson, 1979; Fleming, 1992) (Fig. 1). That no other known viruses packaged segmented, circular dsDNA genomes or exhibited a mutualistic association with another organism (wasps) further suggested PDVs were a new family (Fig. 1). Thus, based on their “poly-DNA” genomes, the family Polydnaviridae was proposed in 1984 along with a description of its key characters (Stoltz et al., 1984). ICTV ultimately adopted this proposal while recognizing two genera: the Bracovirus (BV) associated with wasps in the family Braconidae and the Ichnovirus (IV) associated with wasps in the family Ichneumonidae (Francki, 1991).
Fig. 1.
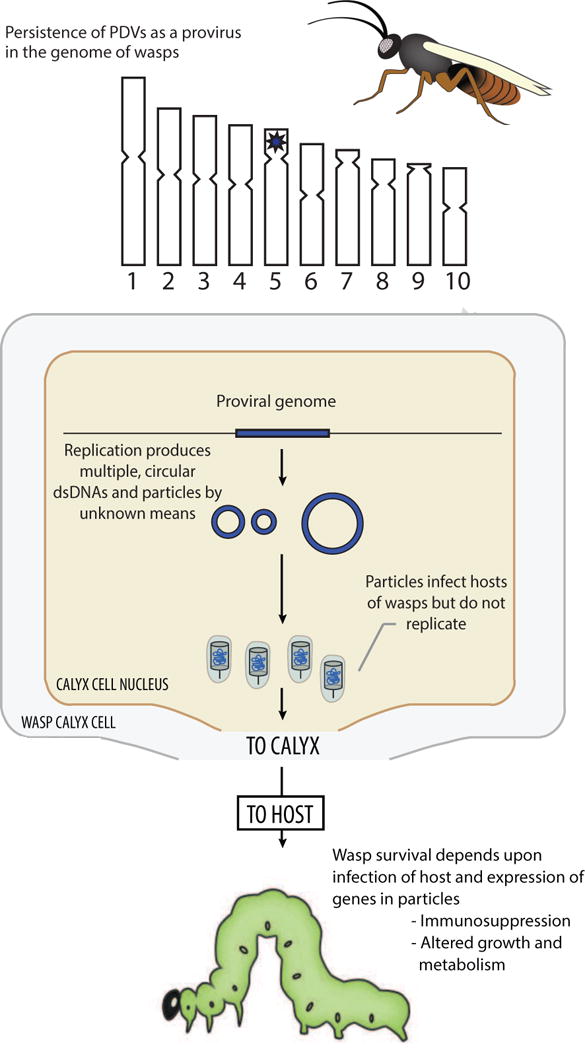
Key characteristics of PDVs in 1991 when the family Polydnaviridae was formally recognized by ICTV. The upper part of the figure shows an adult female wasp whose hypothetical genome consists of ten chromosomes. Data at this time indicated that each PDV associated with a given wasp species was genetically distinct and persisted as an integrated provirus. Based on other known dsDNA viruses with a proviral phase, PDV proviral genomes were implicitly assumed to persist as a large, linear dsDNA that was integrated in the wasp genome (*). The middle part of the figure shows the nucleus of a calyx cell. Data showed that particles packaging multiple circular dsDNAs were produced in calyx cells by unknown means followed by storage of particles in a domain referred to as the calyx. The lower part of the figure shows a larval stage lepidopteran host. Data generated prior to 1991 showed that wasps inject PDV particles into hosts, which infect different types of cells and express genes that cause physiological alterations wasp offspring depend upon for survival. Data generated prior to 1991 also showed that no replication of PDVs occurs in the hosts of wasps.
1990s-early 2000s: Experimental struggles and phylogenetic insights from wasps
Studies in the 1990s and early 2000s identified several PDV genes transcribed in hosts and provided new information on the physiological roles of PDVs in parasitism (Strand and Noda, 1991; Strand et al., 1992; Harwood et al., 1994; Li and Webb, 1994; Strand, 1994; Shelby and Webb, 1994; Doucet and Cusson, 1996; Asgari et al., 1996; Strand et al., 1997; Johner and Lanzrein, 2002; Chen et al., 2003; Glatz et al., 2003: Beck and Strand, 2003). Additional information was generated about the DNAs packaged into BV virions including their integration in the genome of wasps, their amplification, and packaging into virions (Albrecht et al., 1994; Gruber et al., 1996; Pasque-Barre et al., 2002; Belle et al., 2002; Beck et al., 2007). Studies of IVs also showed that some DNA segments packaged into virions undergo a recombination process that produces nested segments (Xu and Stoltz, 1993; Cui and Webb, 1997). Overall though, the experimental study of BVs and IVs struggled to make headway for several reasons. The most serious of these was that replication only occurs in the ovaries of very small wasps (most 5 mm or less in size) with also no permissive cell lines available for propagating PDVs. These factors greatly constrained the availability of material for study and the types of experiments that could be conducted. They also rendered many standard genetic and molecular virology methods available at the time useless for characterizing PDV genes.
In contrast, important insights were generated during this period by studying the phylogenetics of wasps that carry PDVs. In the case of braconids, all BV-carrying species were shown to reside in a small number of subfamilies that form a monophyletic assemblage called the microgastroid complex (Whitfied, 1992). This complex was also estimated to have diverged ~75 million years ago (MYA) from other subfamilies of braconids, which lack BVs (Whitfield, 2002). More recent data estimate the microgastroid complex diverged ~100 MYA and contains ~50,000 species, which makes it among the largest natural taxa of animals on Earth (Murphy et al. 2008; Theze et al., 2011; Rodriguez et al. 2013). Studies of ichneumonids estimated that 14,000 species in two subfamilies carry IVs (Quicke et al., 2009). It remains unclear whether these subfamilies form a monophyletic assemblage, but higher order data clearly indicated that IV-carrying ichneumonids and BV-carrying braconids are distantly related (Quicke et al., 2009; Heraty et al., 2011). These results were meaningful to the PDV literature for two reasons. First, they established that all BV-carrying species evolved from a common ancestor 100 MYA, which also suggested the BV-wasp association is ancient. Second, they suggested BVs and IVs evolved independently (i.e. the Polydnaviridae is not a natural taxon). This conclusion was consistent with the different morphologies of BV and IV particles, but if correct also meant the shared life cycle of BVs and IVs reflected convergent evolution driven by their similar roles in parasitism of hosts by wasps (Webb and Strand, 2005; Strand, 2010).
Mid-2000s: Sequencing the DNAs in particles further muddy the PDV waters
Molecular data from PDVs that could corroborate findings from the phylogenetic study of wasps were not available until the mid-2000s when technical advances finally made it possible to sequence, assemble, and analyze the complex population of DNAs present in BV and IV particles from different wasp species (Espagne et al., 2004; Webb et al., 2006; Lapointe et al., 2007; Tanaka et al., 2007; Desjardins et al., 2008; Chen et al., 2011). These sequencing results showed that the DNAs in BV and IV particles mirrored wasp phylogeny with gene content from closely related species being more similar to one another than to more distantly related species. Comparisons between BVs and IVs further showed they largely package different genes, which fully supported an independent origin. Yet consistent with their similar life cycle, sequencing also identified several architectural features besides segmentation that DNAs in BV and IV particles shared. These included low coding densities, a strong A:T bias, and the presence of many genes that have diversified through duplication events into multimember families.
The most important finding, however, was that almost no genes with homology to known viral genes were present on the DNAs in BV or IV particles including none with predicted functions in DNA replication, transcription, or virion formation (Espagne et al., 2004; Webb et al., 2006; Lapointe et al., 2007; Tanaka et al., 2007; Desjardins et al., 2008; Chen et al., 2011). Instead, most genes were either orphans or shared homology with genes from insects or other eukaryotes. Several of these genes were also related to factors in eukaryotic signaling pathways, which suggested they were virulence factors that caused some of the physiological alterations that occur in parasitized hosts. Subsequent studies supported this by demonstrating experimentally how certain BV and IV gene products interact with host immune molecules or cells (Beck and Strand, 2005; Thoetkiattikul et al., 2005; Beck and Strand, 2007; Ibrahim and Kim, 2008; Labropoulou et al., 2008; Kwon and Kim, 2008; Cooper et al., 2011; Magkrioti et al., 2011; Bitra et al., 2012; Gueguen et al., 2013), while implicating other genes in altering host growth, metabolism or endocrine physiology (Provost et al., 2004; Falabella et al., 2006; Kim et al., 2013; Presad et al., 2013).
That many of the genes present in BV and IV particles shared homology with genes present in insects was initially interpreted to mean they derived from wasps (see Stoltz and Krell, 2012). Comparative and phylogenetic data analyses, however, revealed a much more complex picture by showing that the genes in BV and IV particles have been acquired at different times in evolutionary history and from different sources (Huguet et al., 2012). Some are recent acquisitions from wasps (Desjardins et al., 2008; Burke and Strand, 2014), whereas others have been acquired by horizontal gene transfer from organisms outside of the Arthropoda or are ancient and of uncertain ancestry (Huguet et al., 2012; Burke and Strand, 2012a; Serbielle et al., 2013; Herniou et al., 2013). In keeping with this variable origin and history of gene acquisition, many genes have introns and other features associated with eukaryotic ancestry while several others are small and intronless as seen for genes of viral origin.
Overall then, the DNAs in BV and IV particles largely differ from one another in gene content although both contain primarily if not exclusively genes with demonstrated or hypothesized roles in altering the physiology of parasitized hosts (Fig. 2). This supported the independent origins of BV and IVs, explained why neither entity replicates outside of wasps, and further supported that BV and IV particles function as replication-defective gene delivery vectors that wasps use to parasitize hosts. The larger message, however, was that BV and IV particles showed no evidence of viral ancestry (Fig. 2). This supported the perspective that BVs and IVs are not viruses and should not be recognized by ICTV (Federici and Bigot, 2003; Whitfield and Asgari, 2003).
Fig. 2.
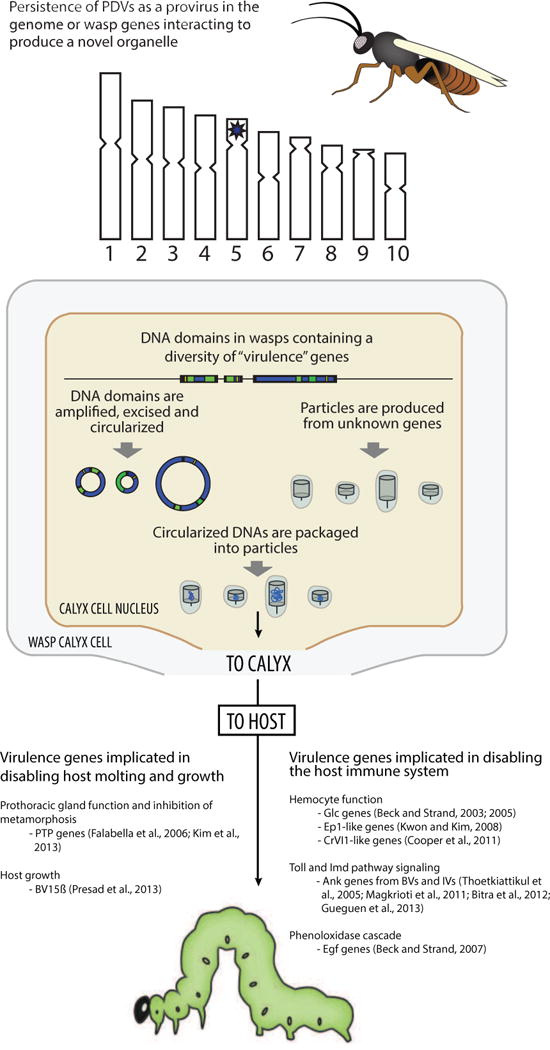
Key characteristics of PDVs as understood in the mid-2000s. Data generated during the 1990s and 2000s showed that the DNAs in BV and IV particles contained numerous virulence genes but almost no genes of recognizable viral origin. Little information was generated during this period regarding PDV proviral genomes but data did suggest the DNAs in BV particles were excised from the wasp genome and amplified in calyx cells before packaging into particles. The genes regulating these events or that were involved in particle formation remained unknown. In contrast, several studies provided insights about the identity and function of the virulence genes that are expressed in the hosts of wasps.
Late 2000s: transcriptome and functional data establish a viral origin for BVs
Entering the late 2000s, the aphorism “absence of evidence is not evidence of absence” very much applied to the PDV literature. That is, the absence of “viral” genes in particles did not alter that replication-like events occur in wasp calyx cells or that BVs and IVs look and, in several respects, function like viruses (Fig. 2). So the issue shifted to identifying the genes involved in particle formation in calyx cells. The first major breakthrough came from producing ovary cDNA libraries for two BV-carrying wasps, Cotesia congregata and Chelonus inanitus, Sanger sequencing a subset of clones, and then coupling these data to proteomic analyses of the particles (designated as CcBV and CiBV) produced by each species (Bezier et al., 2009; Wetterwald et al., 2010). A third, more comprehensive data set generated by Illumina sequencing and Orbitrap LC-MS technology was thereafter produced for the braconid Microplitis demolitor and its associated BV named MdBV (Burke and Strand, 2012b; Burke et al., 2013).
Data from these species identified 42 transcripts specifically expressed in ovaries (=calyx cells) during replication with weak (19–41%) but recognizable identity to genes from another family of large, enveloped DNA viruses that infect insects called nudiviruses (Wang and Jehle, 2009). Although poorly studied, the Nudiviridae is also the sister group to the Baculoviridae, which is among the best-studied families of insect viruses (Theze et al., 2011; Rohrmann, 2013). All baculoviruses examined to date share 37 core genes of which about half are required for replication (Herniou et al., 2003; Rohrmann, 2013). These include a DNA polymerase (DNApol) that replicates the viral genome, four subunits of a novel DNA dependent RNA polymerase (lef-4, lef-8, lef-9, p47), and several structural genes with promoter sequences that are specifically recognized by the viral RNA polymerase. These include vp39, vlf-1, p74 and pif genes that are capsid or envelope components. No functional data are available from nudiviruses but data from six sequenced nudivirus genomes indicate they share 20 baculovirus core genes including DNApol, the RNA polymerase subunits, and the aforementioned structural genes (Wang and Jehle, 2009; Rohrmann, 2013).
Nudivirus-like genes upregulated in calyx cells at the onset of replication include the four RNA polymerase subunits, which is then followed by expression of the nudivirus-like structural genes (Burke and Strand, 2012a). Three lines of evidence further indicated these nudivirus-like genes retain ancestral functions. First, proteins corresponding to all of the baculovirus-like capsid and envelope genes were detected in CcBV, CiBV, and MdBV particles (Bezier et al., 2009; Wetterwald et al., 2010; Burke et al., 2013). Second, loss of function studies in M. demolitor using RNA interference indicated the RNA polymerase subunit genes produced a functional enzyme that transcribes predicted structural genes but not wasp genes, while vp39, p74, and pif-1 were required to produce functionally normal virions (Burke et al., 2013). Third, BV and nudivirus virions have very similar morphology, which was first noted in early EM studies at a time when nudiviruses were not yet recognized but instead were considered as a type of ‘non-occluded’ baculovirus (Stoltz and Vinson, 1979).
Other results identified unique features of BV replication relative to baculoviruses and nudiviruses. These include the absence from transcriptome data sets of any homologs of baculovirus/nudivirus genes involved in viral DNA replication except for a helicase in M. demolitor. This suggested replication of the DNAs packaged into BV particles relies primarily on wasp DNA replication machinery (Burke and Strand, 2012b). While early studies suggested that amplification of the DNAs packaged into virus particles also involved excision of proviral segments from the genome of calyx cells (Gruber et al., 1996; Savary et al., 1999), more recent data indicate this is not the case (Louis et al., 2013). Other data showed that multiple variants of a tyrosine recombinase named integrase (int) known from nudiviruses but not baculoviruses is part of the BV core gene set. These int genes are structurally related to vlf-1, and both exhibit recombinase functions required for proper processing of the circularized DNAs that are packaged into virions (Burke et al., 2013). Finally, while all of the above nudivirus-like genes are transcribed in wasp calyx cells during replication, most of the virulence genes on the DNAs packaged into virions are not (Bitra et al., 2011; Burke and Strand, 2012b). Yet many or most of these genes are transcribed in the permissive hosts of wasps following infection (Provost et al., 2011; Bitra et al., 2011; Chevignon et al., 2014).
Similar approaches with three IV-carrying wasps identified no nudivirus or baculovirus-like genes but did identify genes corresponding to structural proteins in IV virions (Volkoff et al., 2010). These genes share no recognizable homology with any known viral structural gene but they do exhibit features suggestive of being of viral origin (Volkoff et al., 2010). Whether the ancestor of IVs is now extinct or undiscovered is unclear. Comprehensive expression studies also indicate that many virulence genes on DNAs in IV particles are transcribed in permissive hosts but with small exception such as rep genes (Theilmann and Summers, 1988; Rasoolizadeh et al., 2009) are not transcribed in wasps (Volkoff et al., 2010; Doremus et al., 2014).
Relationships between BVs, nudiviruses and baculoviruses
By 2012, indisputable connections had been established between BVs and nudiviruses. The monophyly of macrogastroid braconids combined with other data further supported that all BV-carrying wasps descend from a common nudivirus-wasp ancestor, and that nudiviruses diverged from baculoviruses ~300 MYA (Theze et al., 2011). Interest thus turned to asking more questions about how BVs evolved and what features underlie their unique biology relative to their ancestors. All baculoviruses and nudiviruses replicate and package a single large circular dsDNA (>100 kb) genome into virions (Wang and Jehle, 2009; Rohrmann, 2013). Like other large DNA viruses, baculoviruses and nudiviruses also exhibit high diversity in gene content outside of core genes due to repeated acquisition and loss of genes from exogenous sources that are selectively advantageous for specialization onto different hosts. In turn, different lineages of baculoviruses and nudiviruses have evolved in response to host speciation. BVs obviously differ from baculoviruses and nudiviruses in regard to the multiple, circular dsDNAs they replicate and package as well as the genes on these DNAs. Yet they are similar in the sense that all BVs share a set of core genes required for replication, while exhibiting overall high diversity in gene content due to virulence genes that have been acquired from diverse sources and at different times that have functions in parasitism of hosts.
Most baculoviruses and nudiviruses are virulent pathogens that establish systemic, fatal infections by introducing their circular genomes into the nuclei of infected cells (Rohrmann, 2013). In the case of baculoviruses, the viral genome persists as an episome in infected cells with early genes like DNApol and the RNA polymerase subunits being transcribed followed by expression of structural genes that result in replication of the genome and virion formation. Virions are then released by either lysis or budding to infect other cells. On the other hand, a few nudiviruses such as HzNV-1 and -2 are known that preferentially infect the reproductive systems of insects and in vitro establish persistent infections by integrating into the genome of infected cells (Wang and Jehle, 2009; Burand et al., 2012). HzNV-1 can also be reactivated to produce lytic infections (Wu et al., 2010; 2011). This finding suggested to labs studying BVs that a nudivirus ancestor may have initially established a persistent infection in a braconid wasp by integrating its circularized dsDNA genome into the germline as a linear proviral DNA (Drezen et al., 2012; Strand and Burke, 2012; 2013). Thereafter, rearrangements must have occurred that separated the nudivirus-like replication genes from virulence genes located on multiple DNAs. Changes further limited the expression and function of replication genes (and thus virus replication) to wasp calyx cells, while allowing delivery of a species-specific set of genes to the hosts of wasps via replication-defective virions. Understanding the nature of these alterations required finding and characterizing the different components of BV proviral genomes.
BAC clone sequencing identifies some features of BV proviral genomes
Most textbooks state that a provirus is a virus genome that is integrated into the DNA of a host cell. The biology of BVs obviously differs from most viruses but in keeping with the above definition BV proviral genomes consist of the core genes which have functions in formation of virions plus the DNAs and associated genes that are amplified, circularized, and packaged into virions (Strand and Burke, 2012). The DNAs in virions are also usually referred to in the PDV literature as proviral segments when integrated in the genome of wasps and as the encapsidated form of the genome when packaged in virions. The first insights into BV proviral genome architecture actually preceded the transcriptome studies that identified the nudivirus-like genes. It came from screening bacterial artificial chromosome (BAC) libraries made against two wasp species in the genus Glyptapanteles (Glyptapanteles indiensis, G. flavicoxis) that produce BVs named GiBV and GfBV (Dejardins et al., 2007; 2008). These data showed that the circularized DNA segments in particles reside as linear DNAs in the genomes of wasps, which are organized into 6 loci (Dejardins et al., 2007; 2008). Two of these loci contained multiple segments arranged tandemly while the other loci contained one or two segments. Subsequent BAC clone sequencing from two species in the genus Cotesia (Cotesia congregata, C. sesamiae) plus additional data from Microplitis demolitor show the same general organizational features although the total number of proviral loci and the number of proviral segments per locus differ between species (Bezier et al., 2013; Burke et al., 2014). Despite differences in gene content, the overall similarity in architecture of proviral segment loci between GfBV, GiBV CcBV, and MdBV also suggested shared ancestry.
Within species comparisons indicate adjoining proviral segments in a given locus are more similar to one another than to more distantly related segments. This suggested proviral DNAs have primarily evolved through tandem duplication events over evolutionary time followed by sequence divergence (Desjardins et al., 2008; Bezier et al., 2013; Burke et al., 2014). A second conserved feature gleaned from these studies is that direct repeats containing the tetramer AGCT flank each segment, which earlier studies identified as the site where each segment circularizes for packaging into nucleocapsids (Savary et al., 1997; Annaheim and Lanzrein, 2007). A study published in 2011 also showed that MdBV DNAs rapidly integrate into the genome of host cells following infection via an inverted repeat present in circularized segments (Beck et al., 2011). Thus, to distinguish between these two features, we named the conserved repeats flanking each proviral segment in the genome of M. demolitor wasp integration motifs (WIMs), and the inverted repeat that identifies the site of integration of circularized segments into the genome of a host cell as host integration motifs (HIM) (Beck et al., 2011). Analysis of MdBV proviral segments showed that most contain a HIM (Burke et al., 2014), while database analyses show that HIMs are also present in proviral segments of CcBV, GiBV, and GfBV.
Largely absent, however, from these proviral segment domains were the nudivirus-like genes identified in transcriptome studies with only one structural gene (odv-e66-like) found between locus 1 and 2 in C. congregata (Bezier et al., 2009). In contrast, BAC clone sequencing did identify an 18 kb domain in the genome of C. congregata that contained 10 nudivirus-like genes, including several that are structural components of CcBV virions, plus 5 other nudivirus-like genes outside of this cluster that were flanked by wasp DNA (Bezier et al., 2009). All of these nudivirus-like genes were also intronless.
The collective picture from these data is that BV proviral genomes consist of two functional components: proviral segments organized into multiple loci that are packaged into virions and nudivirus-like core genes that are required for virion formation (Fig. 3). The close proximity of several nudivirus-like genes to one another suggested this domain may be part of the original site of integration of a nudivirus into the genome of the ancestor of microgastroid braconids (Bezier et al., 2009). Finding a nudivirus-like gene in proximity to one of the proviral segments in C. congregata also suggested that proviral segment loci and nudivirus-like core genes are physically linked in the genomes of wasps (Belle et al., 2003; Bezier et al., 2009; Herniou et al., 2013) (Fig. 3).
Fig. 3.
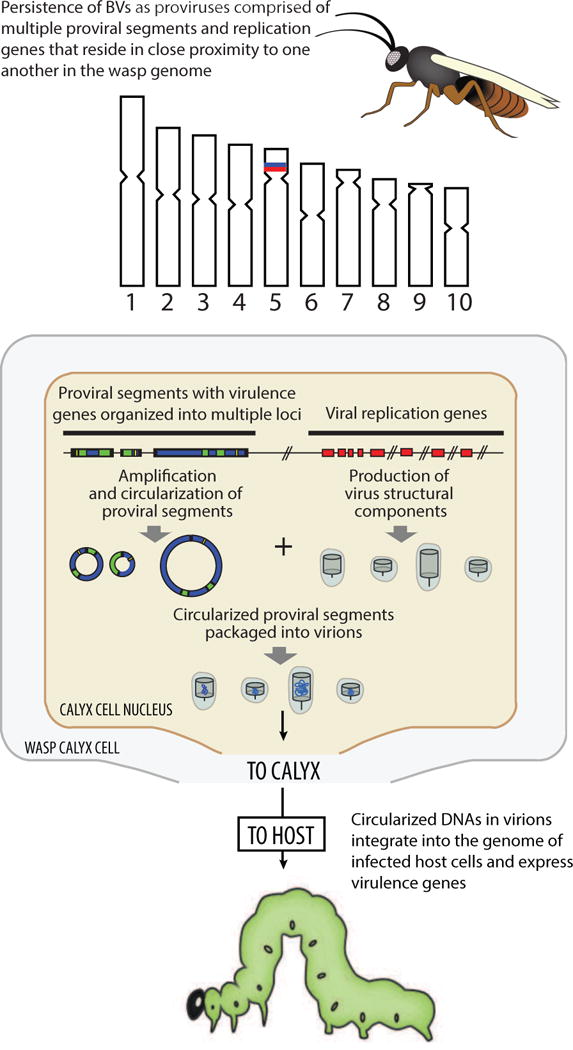
Key characteristics of BVs as understood in 2013. Sequence data established that BV proviral segment loci are organized into multiple loci, which are amplified without excision in calyx cells. Multiple nudivirus-like core genes had also been identified and shown to be required for virion formation. Proviral segment loci and nudivirus-like genes were hypothesized to reside in close physical proximity to one another in the genomes of wasps although precise locations remained unknown (hash marks). Experiments also established that circularized DNAs in BV virions integrate into the genome of infected host cells.
Whole genome sequencing of wasps indicates BV proviral genomes are dispersed
The strength of the preceding BAC data is they identified how proviral segments are organized while also showing that some nudivirus-like genes are clustered. Their weakness is they provided too little genomic context to ascertain how physically close or distant different proviral loci and nudivirus-like genes are to one another in the genomes of wasps. BAC clone sequencing also failed to identify many of the nudivirus-like genes identified in transcriptome studies, including several particularly important factors for replication like most of the RNA polymerase subunit genes. Thus, the only way to generate an overall picture of proviral genome architecture was whole genome sequencing of wasps.
This was recently done for Microplitis demolitor with assembly of its ~258 Mb genome revealing three key findings (Burke et al., 2014). First, these data showed that the eight loci containing MdBV proviral segments are flanked by large distances of wasp genomic DNA, which indicates each locus is physically distant from the other. Second, they identified all of the nudivirus-like genes previously described in transcriptome studies (Burke and Strand, 2012a). All of these genes are intronless. Twenty of these genes reside in a 75 kb cluster flanked by wasp genes with introns, which indicates the M. demolitor genome contains a nudivirus-like gene cluster. A subset of these genes are also present in the same order and orientation as the nudivirus cluster identified in C. congregata genome. However, most of the remaining nudivirus-like genes including helicase, all of the RNA polymerase subunits, and several structural genes required for virion formation are located singly on different scaffolds in the M. demolitor genome. Third, almost none of these nudivirus-like genes reside in proximity to any proviral segment locus, which indicates the MdBV proviral genome is overall highly dispersed (Fig. 4).
Fig. 4.
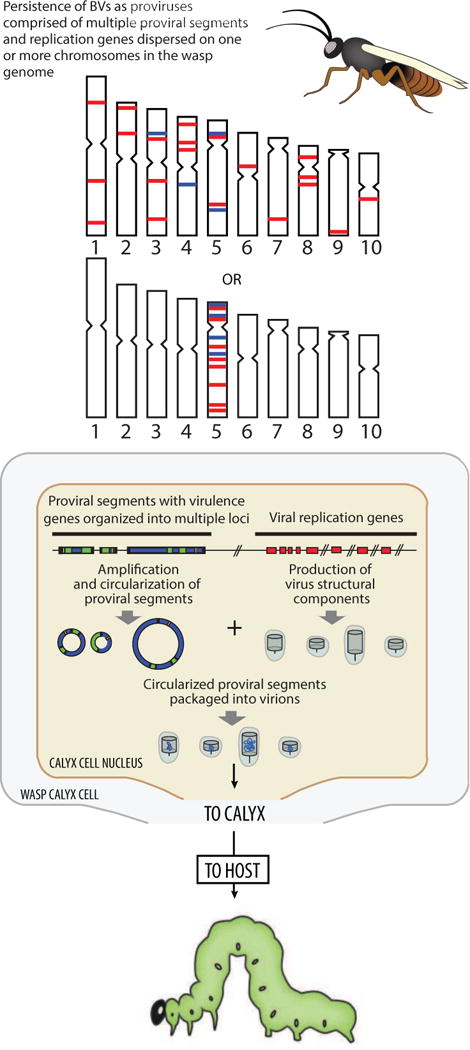
Key characteristics of BVs as understood in 2014. Whole genome sequencing of Microplitis demolitor established that proviral segment loci and nudivirus like genes are overall widely dispersed in the wasp genome on either one or multiple chromosomes.
The conserved synteny of predominantly structural genes in the nudivirus-like cluster of M. demolitor and C. congregata lends further support this domain represents the initial integration site for the nudivirus ancestor while suggesting that maintenance of these genes in a cluster is functionally important for virion formation. These data also indicate the MdBV and CcBV nudivirus-like clusters have remained stable since divergence 53 Mya, which suggests dispersal of the other nudivirus-like genes occurred relatively early in BV evolution. In contrast, whether the dispersed nudivirus-like genes identified in M. demolitor reside in similar locations in other species will remain unknown until additional comparative data are available. Also unknown from the current assembly of the M. demolitor genome is whether the different components of the MdBV genome reside on one or multiple wasp chromosomes (Fig. 4). Nonetheless, they indicate BV proviral genome components are organized in a manner dissimilar from their ancestors or other entities referred to as viruses that have a proviral phase.
What does genome dispersal mean for function?
At present, the literature indicates most nudivirus-like core genes retain their ancestral functions despite their dispersal in the genomes of wasps. Genome dispersal is also, obviously, not a barrier to high-level replication in calyx cells, which in the case of M. demolitor considerably exceeds replication levels of baculoviruses in permissive host cells (Burke and Strand, 2012b; Burke et al., 2013). Two features, likely inherited from the nudivirus ancestor, make this possible. First, the RNA polymerase subunits, once expressed and assembled to form a holoenzyme, specifically transcribe the nudivirus-like structural genes through promoter recognition regardless of their location in the wasp genome (Burke et al., 2014). Second, currently unknown DNA replication machinery plus the two nudivirus-like integrases (int-1, vlf-1) likely use the direct repeat boundaries (WIMs) to recognize all proviral segments for proper amplification, processing and packaging into virions. In contrast, the absence of these motifs from any of the DNA domains where nudivirus-like genes reside results in none being processed and packaged into particles.
The evolutionary events that produced this dispersed architecture are unclear. The absence of nudivirus-like genes in BV proviral segments suggests this feature must have arisen early in BV evolution (Drezen et al., 2012). Yet recognition of WIM domains by the nudivirus-like integrases suggests these flanking motifs derive from the ancestor. One possibility for dispersal is the ancestral nudivirus genome duplicated after integration. Another is several copies of the genome initially integrated, followed by elimination of conserved replication genes from some and elimination of WIMs from others (Strand and Burke, 2012; Gudersen-Rindal et al., 2013, Herniou et al., 2013; Strand and Burke, 2014). Subsequent duplications of proviral segments within loci together with acquisition of genes from diverse sources were thereafter selected for in different lineages, as wasps adapted to parasitism of particular hosts or complexes of hosts, and hosts reciprocally evolved to resist parasitism. Promoter mutation in association with selection may also underlie expression of virulence genes in the hosts of wasps and corresponding absence of expression in wasps themselves (Drezen et al., 2012).
Selection has clearly maintained the conserved roles of the nudivirus-like genes in particle formation although their dispersal and current locations in the M. demolitor genome may reflect random processes of genome dynamics (Burke et al., 2014). On the other hand, the separation of nudivirus-like genes required for virion formation from the proviral segments containing virulence genes has also potentially been favored because it is advantageous to wasps in their interactions with hosts. In effect, dispersal assures vertical transmission of all the genes and proviral segments required to produce virions, but prevents any replication machinery from being packaged into particles. Thus, no productive virus infection, which would likely be fatal to wasp offspring, can occur in the hosts wasps parasitize.
A number of important functional questions remain unanswered (Strand and Burke, 2014). Based on the baculovirus literature, we hypothesize wasp RNA polymerase II transcribes the BV integrase and RNA polymerase genes required for virion formation, but what restricts expression to only calyx cells is unknown. The absence of any baculovirus/nudivirus-like DNA polymerase in the M. demolitor genome further strengthens conclusions that a wasp DNA polymerase(s) amplifies proviral DNAs prior to their processing by integrases and packaging into particles. However, the wasp polymerase(s) responsible also remains unknown. A third functional issue not understood is what are the promoter features on proviral segments that prevent them from being transcribed in wasps yet allows them to be transcribed in the hosts wasps parasitize.
Concluding remarks
We began this summary by citing Summers (2014) who concludes that the answer to the question “What is a virus?” depends on the scientific discourse at a given time. Modern virologists still struggle with simple one sentence definitions for what viruses are, yet fully recognize that viruses exhibit a broad continuum of interactions with hosts. On one end of this continuum are entities with nucleic acid genomes (RNA or DNA) that replicate inside a living cell to produce particles that can horizontally transfer the genome to other cells. These characteristics are also the essential qualities most virologists today associate with being a virus (Cann, 1997; Summers, 2014). At the other end of this continuum are ancient endogenous virus elements (EVEs), which are DNA sequences derived from viruses that have integrated and become fixed in the host germline resulting in only vertical inheritance (Katzourkakis and Gifford, 2010). Most EVEs in animal genomes are fragments rendered non-functional by accumulation of neutral mutations, although a few individual viral genes or regulatory elements are known that have been coopted by hosts for new beneficial functions (Feschotte and Gilbert, 2012). No virologist though would say such fragments are viruses because they lack all of the essential qualities listed above. Entities like BVs, prokaryotic gene transfer agents (GTAs), and certain retroviruses and phages that persist for long periods as vertically transmitted proviruses fall somewhere between these extremes (Roosinck, 2011; Lang et al., 2012; Feschotte and Gilbert, 2012; Strand and Burke, 2013).
BVs are clearly ancient EVEs that benefit wasps, yet do so by continuing to function in many respects like a virus, which is what led ICTV to recognize the Polydnaviridae to begin with. Current knowledge regarding BVs and IVs supports the need to revise the Polydnaviridae, but the essential qualities of BVs (and IVs) remain unchanged from 1991. What has changed is that the components, which make up BVs are now known. These include definitive evidence that most genes required for replication are of viral origin, while genes on proviral segments have a mixture of origins including acquisition by horizontal transfer from wasps or other eukaryotes plus some genes and motifs like WIMs that suggest they too originated from the nudivirus ancestor. Nudiviruses and baculoviruses also consist of genes that are ‘viral’ in the sense that the proteins they encode are required for replication, while the majority of genes in their genomes are either of ancient origin and uncertain ancestry or are acquisitions from arthropods or other organisms (Rohrmann, 2013). Thus, what differs between BVs and their ancestors is not so much gene content as genome organization (Burke et al., 2014). Nudivirus and baculovirus genes, like those of most viruses, reside on a single stretch of DNA that can be replicated, packaged into particles and transmitted to another cell. BVs encode the same types of genes, which in many cases have the same functions, yet dispersal prevents BVs from existing independently of wasps. We previously noted that non-viral microbes like bacteria, which have evolved into vertically transmitted mutualists do not persist by integrating into the genome of their host, but they do exhibit alterations in genome organization and function that prevent them from existing independently (Burke et al., 2014). Long-standing discussions also exist in the literature whether such entities are organelles or bacteria with current sentiment supporting the latter (Andersson, 2000; McCutcheon and Moran, 2012). Can an entity exhibiting most essential qualities associated with viruses be such if derived from genes dispersed in the genome of another organism? It undoubtedly depends on the virologist answering the question.
Highlights.
Polydnaviruses were recognized as a virus family in 1991.
Here we review the polydnavirus literature in largely historical order.
The early literature established key features of polydnavirus biology.
Studies in 1990s and early 2000s generated functional and phylogenetic insights. y Studies in the mid-2000s raised issues about the origins of polydnaviruses.
Recent studies provide key data on origins, genome structure, and function.
Acknowledgments
We thank J. A. Johnson for her assistance in figure preparation. Some work discussed here was also supported by grants from the National Science Foundation (IOS 1145953), US Department of Agriculture (2009-35302-05250) and National Institutes of Health (F32 AI096552).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albrecht U, Wyler T, Pfister-Wilhelm R, Gruber A, Stettler P, Heiniger P, Schumperli D, Lanzrein B. PDV of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J Gen Virol. 1994;75:3353–3363. doi: 10.1099/0022-1317-75-12-3353. [DOI] [PubMed] [Google Scholar]
- Annaheim M, Lanzrein B. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers, and abundance of proviral and excised segments. J Gen Virol 2007. 2007;8:450–57. doi: 10.1099/vir.0.82396-0. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Evolutionary genomics: is Buchnera a bacterium or an organelle? Curr Biol. 2000;10:R866–R868. doi: 10.1016/s0960-9822(00)00816-2. [DOI] [PubMed] [Google Scholar]
- Asgari S, Hellers M, Schmidt O. Host haemocyte inactivation by an insect parasitoid: transient expression of a polydnavirus gene. J Gen Virol. 1996;77:2653–2662. doi: 10.1099/0022-1317-77-10-2653. [DOI] [PubMed] [Google Scholar]
- Beck M, Strand MR. RNA interference silences Microplitis demolitor bracovirus genes and implicates glc1.8 in disruption of adhesion in infected host cells. Virology. 2003;314:521–535. doi: 10.1016/s0042-6822(03)00463-x. [DOI] [PubMed] [Google Scholar]
- Beck M, Strand MR. Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect high five and S2 cells. J Virol. 2005;79:1861–70. doi: 10.1128/JVI.79.3.1861-1870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck MH, Strand MR. A novel protein from a polydnavirus inhibits the insect prophenoloxidase activation pathway. Proc Natl Acad Sci USA. 2007;104:19267–72. doi: 10.1073/pnas.0708056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck MH, Inman RB, Strand MR. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology. 2007;359:179–89. doi: 10.1016/j.virol.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Beck MH, Zhang S, Bitra K, Burke GR, Strand MR. The encapsidated genome of Microplitis demolitor bracovirus integrates into the host pseudoplusia includes. J Virol. 2011;85:11685–11696. doi: 10.1128/JVI.05726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckage NE, Drezen JM. Parasitoid Viruses. Academic Press; San Diego: 2012. [Google Scholar]
- Beckage NE, Riddiford LM. Effects of parasitism by Apanteles congregatus on the endocrine physiology of the tobacco hornworm, Manduca sexta. Gen Comp Endocrinol. 1982;47:308–322. doi: 10.1016/0016-6480(82)90238-6. [DOI] [PubMed] [Google Scholar]
- Belle E, Beckage NE, Rousselet J, Poirie M, Lemeunier F, Drezen JM. Visualization of polydnavirus sequences in a parasitoid wasp chromosome. J Virol. 2002;76:5793–5796. doi: 10.1128/JVI.76.11.5793-5796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, et al. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science. 2009;323:926–30. doi: 10.1126/science.1166788. [DOI] [PubMed] [Google Scholar]
- Bezier A, Louis F, Jancek S, Periquet G, Theze J, et al. Functional endogenous viral elements in the genome of the parasitoid wasp Cotesia congregata: insights into the evolutionary dynamics of bracoviruses. Philos T Roy Soc B. 2013;368:20130047. doi: 10.1098/rstb.2013.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitra K, Zhang S, Strand MR. Transcriptomic profiling of Microplitis demolitor bracovirus reveals host, tissue, and stage-specific patterns of activity. J Gen Virol. 2011;92:2060–2071. doi: 10.1099/vir.0.032680-0. [DOI] [PubMed] [Google Scholar]
- Bitra K, Suderman RJ, Strand MR. Polydnavirus Ank proteins function as IkB mimics that subvert the insect Imd signaling pathway. PLoS Pathogens. 2012;8:e1002722. doi: 10.1371/journal.ppat.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blissard GW, Vinson SB, Summers MD. Identification, mapping and in vitro translation of Campoletis sonorensis virus mRNAs from parasitized Heliothis virescens larvae. J Virol. 1986;57:318–327. doi: 10.1128/jvi.57.1.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burand JP, Kim W, Afonso CL, Tulman ER, Kutish GF, et al. Analysis of the genome of the sexually transmitted insect virus Helicoverpa zea nudivirus 2. Viruses. 2012;4:28–61. doi: 10.3390/v4010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Strand MR. Polydnaviruses of parasitic wasps: domestication of viruses to act as gene delivery vectors. Insects. 2012a;3:91–119. doi: 10.3390/insects3010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Strand MR. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor bracovirus. J Virol. 2012b;86:3293–3306. doi: 10.1128/JVI.06434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Strand MR. Systematic analysis of a wasp parasitism arsenal. Mol Ecol. 2014;23:890–901. doi: 10.1111/mec.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Thomas SA, Eum JH, Strand MR. Polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathogens. 2013;9:e1003348. doi: 10.1371/journal.ppat.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Walton K, Robertson H, Whitfield JB, Strand MR. Widespread genomic organization of an ancient viral integration event. PLoS Genetics. 2014;10:e1004660. doi: 10.1371/journal.pgen.1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann AJ. Principles of Molecular Virology. 2. Academic Press; New York: 1997. [Google Scholar]
- Chen YF, Gao F, Ye XQ, Wei SJ, Shi M, et al. Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology. 2011;414:42–50. doi: 10.1016/j.virol.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Chen YP, Taylor PB, Shapiro M, Gundersen-Rindal DE. Quantitative expression analysis of a Glyptapanteles indiensis polydnavirus protein tyrosine phosphatase gene in its natural lepidopteran host Lymantria dispar. Insect Mol Biol. 2003;12:271–280. doi: 10.1046/j.1365-2583.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- Chevignon G, Theze J, Cambier S, Poulain J, Da Silva C, Bezier A, Musset K, Moreau SJ, Drezen J-M, Huguet E. Functional annotation of Cotesia congregata bracovirus: identification of viral genes expressed in parasitized host immune tissues. J Virol. 2014;88:8795–8812. doi: 10.1128/JVI.00209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TH, Bailey-Hill K, Leifert WR, McMurchie EJ, Asgari S, et al. Identification of an in vitro interaction between an insect immune suppressor protein (CrV2) and G alpha proteins. J Biol Chem. 2011;286:10466–75. doi: 10.1074/jbc.M110.214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Webb BA. Homologous sequences in the Campolitis sonorensis polydnavirus genome are implicated in replication and nesting of the W segment family. J Virol. 1997;71:8504–8513. doi: 10.1128/jvi.71.11.8504-8513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DH, Strand MR, Vinson SB. Changes in differential haemocyte count and in vitro behaviour of plasmatocytes from host Heliothis virescens caused by Campoletis sonorensis PDV. J Insect Physiol. 1987;33:143–153. [Google Scholar]
- Dejardens CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fuester RW, et al. Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol. 2007;26:61. doi: 10.1186/1471-2180-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fadrosh DW, et al. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 2008;9:R183. doi: 10.1186/gb-2008-9-12-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremus T, Cousserans F, Gyapay G, Jouan V, Milano P, Wajnberg E, Darboux I, Consoli FL, Volkoff AN. Extensive transcription analysis of the Hyposoter didymator ichnovirus genome in permissive and non-permissive lepidopteran host species. PLoS One. 2014;12:e104072. doi: 10.1371/journal.pone.0104072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet D, Cusson M. Role of calyx fluid in alterations of immunity in Choristoneura fumiferana larvae parasitized by Tranosema rostrale. Comp Biochem Physiol. 1996;114:311–317. [Google Scholar]
- Drezen J-M, Herniou EA, Bezier A. Evolution and origin of polydnavirus virulence genes. In: Beckage NE, Drezen J-M, editors. Parasitoid Viruses Symbionts and Pathogens. Academic Press; San Diego: 2012. pp. 15–31. [Google Scholar]
- Edson KM, Vinson SB, Stoltz DB, Summers MD. Virus in a parasitoid wasp: suppression of the cellular immune response in the parasitoid’s host. Science. 1981;211:582–583. doi: 10.1126/science.7455695. [DOI] [PubMed] [Google Scholar]
- Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, et al. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science. 2004;306:286–89. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- Falabella P, Cacciaupi P, Varricchio P, Malva C, Pennacchio P. Protein tyrosine phosphatases of Toxoneuron nigriceps bracovirus as potential disrupters of host prothoracic gland function. Arch Insect Biochem Physiol. 2006;61:157–69. doi: 10.1002/arch.20120. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nature Rev Genetics. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- Federici BA, Bigot Y. Origin and evolution of polydnaviruses by symbiogenesis of insect DNA viruses in endoparasitic wasps. J Insect Physiol. 2003;49:419–32. doi: 10.1016/s0022-1910(03)00059-3. [DOI] [PubMed] [Google Scholar]
- Fleming JGW. Polydnaviruses: Mutualists and pathogens. Annu Rev Entomol. 1992;37:401–425. doi: 10.1146/annurev.en.37.010192.002153. [DOI] [PubMed] [Google Scholar]
- Fleming JAGW, Blissard GW, Summers MD, Vinson SB. Expression of Campoletis sonorensis virus in the parasitized host Heliothis virescens. J Virol. 1983;48:74–78. doi: 10.1128/jvi.48.1.74-78.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JG, Summers MD. PDV DNA is integrated in the DNA of its parasitoid wasp host. Proc Natl Acad Sci USA. 1991;88:9770–9774. doi: 10.1073/pnas.88.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki RIB. Classification and nomenclature of Viruses: Fifth Report of the International Committee on Taxonomy of Viruses for the Virology Division of the International Union of Microbiological Societies. Springer-Verlag; Berlin: 1991. [Google Scholar]
- Glatz R, Schmidt O, Asgari S. Characterization of a novel protein with homology to C-type lectins expressed by the Cotesia rubecula bracovirus in larvae of the lepidopteran host, Pieris rapae. J Biol Chem. 2003;278:19743–19750. doi: 10.1074/jbc.M301396200. [DOI] [PubMed] [Google Scholar]
- Godfray HCJ. Parasitoids. Princeton: Princeton Univ. Press, Princeton, USA; 1994. [Google Scholar]
- Gruber A, Stettler P, Heiniger P, Schumperli D, Lanzrein B. Polydnavirus DNA of the braconid wasp Chelonus inanitus is integrated in the wasp’s genome and excised only in later pupal and adult stages of the female. J Gen Virol. 1996;77:2873–79. doi: 10.1099/0022-1317-77-11-2873. [DOI] [PubMed] [Google Scholar]
- Gueguen G, Kalamarz ME, Ramroop J, Uribe J, Govind S. Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. PLoS Pathogens. 2013;9:e1003580. doi: 10.1371/journal.ppat.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen-Rindal D, Dupuy C, Huguet E, Drezen JM. Parasitoid polydnaviruses: evolution, pathology and applications. Biocontrol Sci Tech. 2013;23:1–61. [Google Scholar]
- Guzo D, Stoltz DB. Observations on cellular immunity and parasitism in the tussock moth. J Insect Physiol. 1987;33:19–31. [Google Scholar]
- Harwood SH, Grosovsky AJ, Cowles EA, Davis JW, Beckage NE. An abundantly expressed hemolymph glycoprotein isolated from newly parasitized Manduca sexta larvae is a PDV gene product. Virology. 1994;205:381–392. doi: 10.1006/viro.1994.1659. [DOI] [PubMed] [Google Scholar]
- Heraty J, Ronquist F, Carpenter JM, Hawks D, Schulmeister S, et al. Evolution of the hymenopteran megaradiation. Mol Phylogen Evol. 2011;60:73–88. doi: 10.1016/j.ympev.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Herniou EA, Olszewski JA, Cory JS, O’Reilly DR. The genome sequence and evolution of baculoviruses. Annu Rev Entomol. 2003;48:211–234. doi: 10.1146/annurev.ento.48.091801.112756. [DOI] [PubMed] [Google Scholar]
- Herniou EA, Huguet E, Theze J, Bezier A, Periquet G, et al. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos T Roy Soc B. 2013;368:20130051. doi: 10.1098/rstb.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet E, Serbielle C, Moreau JM. Evolution and origin of polydnavirus virulence genes. In: Beckage NE, Drezen J-M, editors. Parasitoid Viruses Symbionts and Pathogens. Academic Press; San Diego: 2012. pp. 63–78. [Google Scholar]
- Ibrahim AM, Kim Y. Transient expression of protein tyrosine phosphatases encoded by Cotesia plutellae bracovirus inhibits insect cellular immune responses. Naturwissenschaften. 2008;95:25–32. doi: 10.1007/s00114-007-0290-7. [DOI] [PubMed] [Google Scholar]
- Johner A, Lanzrein B. Characterization of two genes of the polydnavirus of Chelonus inanitus and their stage-specific expression in the host Spodoptera littoralis. J Gen Virol. 2002;83:1075–1085. doi: 10.1099/0022-1317-83-5-1075. [DOI] [PubMed] [Google Scholar]
- Katzourkakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genetics. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hepat R, Lee D, Kim Y. Protein tyrosine phosphatase encoded in Cotesia plutellae bracovirus suppresses larva-to-pupa metamorphosis of the diamondback moth Plutella xylostella. Comp Biochem Physiol A. 2013;166:60–69. doi: 10.1016/j.cbpa.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Krell PJ, Stoltz DB. Unusual baculovirus of the parasitoid wasp Apanteles melanoscelus: isolation and preliminary characterization. J Virol. 1979;29:1118–1130. doi: 10.1128/jvi.29.3.1118-1130.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell PJ, Stoltz DB. Virus-like particles in the ovary of an ichneumonid wasp: purification and preliminary characterization. Virology. 1980;101:408–418. doi: 10.1016/0042-6822(80)90454-7. [DOI] [PubMed] [Google Scholar]
- Krell PJ, Summers MD, Vinson SD. A virus with a multipartite superhelical DNA genome from the ichneumonid parasitoid Campoletis sonorensis. J Virol. 1982;43:859–870. doi: 10.1128/jvi.43.3.859-870.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B, Kim Y. Transient expression of an EP1-like gene encoded in Cotesia plutellae bracovirus suppresses the hemocyte population in the diamondback moth, Plutella xylostella. Dev Comp Immunol. 2008;32:932–42. doi: 10.1016/j.dci.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Labropoulou V, Douis V, Stefanou D, Magrioti C, Swevers L, et al. Endoparasitoid wasp bracovirus-mediated inhibition of hemolin function and lepidopteran host immunosuppression. Cell Microbiol. 2008;10:2118–28. doi: 10.1111/j.1462-5822.2008.01195.x. [DOI] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O, Beatty JT. Gene transfer agents: phage-like elements of genetic exchange. Nature Rev Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe R, Tanaka K, Barney WE, Whitfield JB, Banks JC, et al. Genomic and morphological features of a banchine polydnavirus: comparison with bracoviruses and ichnoviruses. J Virol. 2007;81:6491–6501. doi: 10.1128/JVI.02702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Webb BA. Apparent functional role for a cysteine-rich polydnavirus protein in suppression of the insect cellular immune response. J Virol. 1994;68:7482–7489. doi: 10.1128/jvi.68.11.7482-7489.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis F, Bezier A, Periquet G, Ferras C, Drezen JM, et al. The bracovirus genome of the parasitoid wasp Cotesia congregata is amplified within 13 replication units, including sequences not packaged into particles. J Virol. 2013;87:9649–60. doi: 10.1128/JVI.00886-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magkrioti C, Iatrou K, Labropoulou V. Differential inhibition of BmRelish1-dependent transcription in lepidopteran cells by bracovirus ankyrin-repeat proteins. Insect Bich Mol Biol. 2011;41:993–1002. doi: 10.1016/j.ibmb.2011.09.008. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Murphy N, Banks JC, Whitfield JB, Austin AD. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol Phylogenet Evol. 2008;47:378–95. doi: 10.1016/j.ympev.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Pasquier-Barre F, Dupuy C, Huguet E, Moneiro F, Moreau A, Poire M, Drezen J-M. Polydnavirus replication: The EP1 segment of the parasitoid wasp Cotesia congregata is amplified within a larger precursor molecule. J Gen Virol. 2002;83:2035–2045. doi: 10.1099/0022-1317-83-8-2035. [DOI] [PubMed] [Google Scholar]
- Pennacchio F, Strand MR. Evolution of developmental strategies in parasitic Hymenoptera. Annu Rev Entomol. 2006;51:233–58. doi: 10.1146/annurev.ento.51.110104.151029. [DOI] [PubMed] [Google Scholar]
- Presad SV, Hepat R, Kim Y. Selectivity of a translation inhibitory factor, CpBV15, in host mRNAs and subsequent alterations in host development and immunity. Dev Comp Immunol. 2013;18:152–162. doi: 10.1016/j.dci.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Provost B, Varricchio P, Arana E, Espagne E, Falabella P, et al. Bracoviruses contain a large multigene family coding for protein tyrosine phosphatases. J Virol. 2004;78:13090–103. doi: 10.1128/JVI.78.23.13090-13103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quicke DLJ, Laurenne NM, Fitton MG, Broad GR. A thousand and one wasps: a 28S rDNA and morphological phylogeny of the Ichneumonidae (Insecta: Hymenoptera) with an investigation inot alignment parameter space and elision. J Nat Hist. 2009;43:1305–21. [Google Scholar]
- Rasoolizadeh A, Beliveau C, Stewart D, Cloutier C, Couson M. Tranosema rostrale ichnovirus repeat element genes display distinct patterns in caterpillar and wasp hosts. J Gen Virol. 2009;90:1505–1514. doi: 10.1099/vir.0.008664-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez JJ, Fernandez-Triana JL, Smith AM, Janzen DH, Hallwachs W, et al. Extrapolations from field studies and known faunas converge on dramatically increased estimates of global microgastrine parasitoid wasp species richness (Hymenoptera: Braconidae) Insect Con Divers. 2013;6:530–536. [Google Scholar]
- Rohrmann GF. Baculovirus Molecular Biology. Bethesda: National Library of Medicine, National Center for Biotechnology Information; 2013. [PubMed] [Google Scholar]
- Roossinck MJ. The good viruses: viral mutualistic symbioses. Nature Rev Microbiol. 2011;9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- Rotheram SM. Immune surface of eggs of a parasitic insect. Nature. 1967;214:700. doi: 10.1038/214700a0. [DOI] [PubMed] [Google Scholar]
- Savary S, Beckage N, Tan F, Periquet G, Drezen JM. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J Gen Virol. 1997;78:3125–3134. doi: 10.1099/0022-1317-78-12-3125. [DOI] [PubMed] [Google Scholar]
- Serbielle C, Dupas S, Perdereau E, Hericourt F, Dupuy C, et al. Evolutionary mechanisms driving the evolution of a large polydnavirus gene family coding for protein tyrosine phosphatases. BMC Evol Biol. 2013;12:253. doi: 10.1186/1471-2148-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DB. Evidence for chromosomal transmission of polydnavirus DNA. J Gen Virol. 1990;71:1051–1056. doi: 10.1099/0022-1317-71-5-1051. [DOI] [PubMed] [Google Scholar]
- Stoltz DB, Vinson SB. Viruses and parasitism in insects. Adv Virus Res. 1979;24:125–71. doi: 10.1016/s0065-3527(08)60393-0. [DOI] [PubMed] [Google Scholar]
- Stoltz DB, Vinson SB, MacKinnon EA. Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. Can J Microbiol. 1976;22:1013–1023. doi: 10.1139/m76-148. [DOI] [PubMed] [Google Scholar]
- Stoltz D, Krell P. The origins and early history of polydnavirus research. In: Beckage NE, Drezen J-M, editors. Parasitoid Viruses Symbionts and Pathogens. Academic Press; San Diego: 2012. pp. 5–13. [Google Scholar]
- Stoltz DB, Krell P, Summers MD, Vinson SB. Polydnaviridae-a proposed family of insect viruses with segmented, double-stranded, circular DNA genomes. Intervirology. 1984;21:1–4. doi: 10.1159/000149497. [DOI] [PubMed] [Google Scholar]
- Strand MR. Microplitis demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia includens haemocytes. J Gen Virol. 1994;75:3007–3020. doi: 10.1099/0022-1317-75-11-3007. [DOI] [PubMed] [Google Scholar]
- Strand MR. Polydnaviruses. In: Asgari S, Johnson KN, editors. Insect Virology. Caister Academic Press; Norwich UK: 2010. pp. 171–197. [Google Scholar]
- Strand MR, Noda T. Alterations in the haemocytes of Pseudoplusia includens after parasitism by Microplitis demolitor. J Insect Physiol. 1991;37:839–850. [Google Scholar]
- Strand MR, Burke GR. Polydnaviruses as symbionts and gene delivery systems. PLoS Pathogens. 2012;8:e1002757. doi: 10.1371/journal.ppat.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand MR, Burke GR. Polydnavirus-wasp associations: evolution, genome organization, and function. Curr Opin Virol. 2013;3:587–94. doi: 10.1016/j.coviro.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Strand MR, Burke GR. Polydnaviruses: nature’s genetic engineers. Annu Rev Virol. 2014;1:333–354. doi: 10.1146/annurev-virology-031413-085451. [DOI] [PubMed] [Google Scholar]
- Strand MR, McKenzie DI, Grassl V, Dover BA, Aiken JM. Persistence and expression of Microplitis demolitor PDV in Pseudoplusia includens. J Gen Virol. 1992;73:1627–1635. doi: 10.1099/0022-1317-73-7-1627. [DOI] [PubMed] [Google Scholar]
- Strand MR, Witherell RA, Trudeau D. Two Microplitis demolitor polydnavirus mRNAs expressed in hemocytes of Pseudoplusia includens contain a common cysteine-rich domain. J Virol. 1997;71:2146–2156. doi: 10.1128/jvi.71.3.2146-2156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers WC. Inventing viruses. Annu Rev Virol. 2014;1:25–35. doi: 10.1146/annurev-virology-031413-085432. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Lapointe R, Barney WE, Makkay AM, Stoltz D, et al. Shared and species-specific features among ichnovirus genomes. Virology. 2007;363:26–35. doi: 10.1016/j.virol.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Theilmann DA, Summers MD. Molecular analysis of Campoletis sonorensis virus DNA in the lepidopteran host Heliothis virescens. J Gen Virol. 1986;67:1961–1969. doi: 10.1099/0022-1317-67-9-1961. [DOI] [PubMed] [Google Scholar]
- Theilmann DA, Summers MD. Identification and comparison of Campoletis sonorensis virus transcripts expressed from four genomic segments in the insect hosts Campoletis sonorensis and Heliothis virescens. Virology. 1988;167:329–341. [PubMed] [Google Scholar]
- Theze J, Bezier A, Periquet G, Drezen JM, Herniou EA. Paleozoic origin of insect large dsDNA viruses. Proc Natl Acad Sci U S A. 2011;108:15931–35. doi: 10.1073/pnas.1105580108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoetkiattikul H, Beck MH, Strand MR. Inhibitor κβ-like proteins from a polydnavirus inhibit NF-κβ activation and suppress the insect immune response. Proc Natl Acad Sci USA. 2005;102:11426–31. doi: 10.1073/pnas.0505240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson SB, Scott JR. Particles containing DNA associated with the oocyte of an insect parasitoid. J Invert Pathol. 1975;25:375–378. [Google Scholar]
- Volkoff AN, Jouan V, Urbach S, Samain S, Bergoin M, Wincker P, et al. Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog. 2010;6:e1000923. doi: 10.1371/journal.ppat.1000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jehle JA. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invert Pathol. 2009;101:187–93. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Webb BA, Strand MR. The biology and genomics of polydnaviruses. In: Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science. Vol. 6. Elsevier; Pergamon, Amsterdam: 2005. pp. 323–60. [Google Scholar]
- Webb BA, Strand MR, Dickey SE, Beck MH, Hilgarth RS, et al. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology. 2006;347:160–74. doi: 10.1016/j.virol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wetterwald C, Roth T, Kaeslin M, Annaheim M, Wespi G, et al. Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J Gen Virol. 2010;91:2610–19. doi: 10.1099/vir.0.022699-0. [DOI] [PubMed] [Google Scholar]
- Whitfield JB. Estimating the age of the polydnavirus-braconid wasp symbiosis. Proc Natl Acad Sci USA. 2002;99:7508–13. doi: 10.1073/pnas.112067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Asgari S. Virus or not? Phylogenetics of polydnaviruses and their wasp carriers. J Insect Physiol. 2003;49:397–405. doi: 10.1016/s0022-1910(03)00057-x. [DOI] [PubMed] [Google Scholar]
- Wu YL, Wu CP, Lee ST, Tang H, Chang CH, et al. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J Virol. 2010;84:1057–65. doi: 10.1128/JVI.01548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YL, Wu CP, Liu CYY, Hsu PW-C, Wu EC, et al. A non-coding RNA of insect Hz NV-1 virus establishes latent viral infection through microRNA. Scientific Rep. 2011;1:60. doi: 10.1038/srep00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Stoltz D. Polydnavirus genome segment families in the ichneumonid parasitoid Hyposoter fugitivus. J Virol. 1993;67:1340–1349. doi: 10.1128/jvi.67.3.1340-1349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


