Abstract
Poxviruses differ from most DNA viruses by replicating entirely within the cytoplasm. The first discernible viral structures are crescents and spherical immature virions containing a single lipoprotein membrane bilayer with an external honeycomb lattice. Because this viral membrane displays no obvious continuity with a cellular organelle, a de novo origin was suggested. Nevertheless, transient connections between viral and cellular membranes could be difficult to resolve. Despite the absence of direct evidence, the intermediate compartment (ERGIC) between the endoplasmic reticulum (ER) and Golgi apparatus and the ER itself were considered possible sources of crescent membranes. A break-through in understanding poxvirus membrane biogenesis has come from recent studies of the abortive replication of several vaccinia virus null mutants. Novel images showing continuity between viral crescents and the ER and the accumulation of immature virions in the expanded ER lumen provide the first direct evidence for a cellular origin of this poxvirus membrane.
Brief introduction to poxviruses
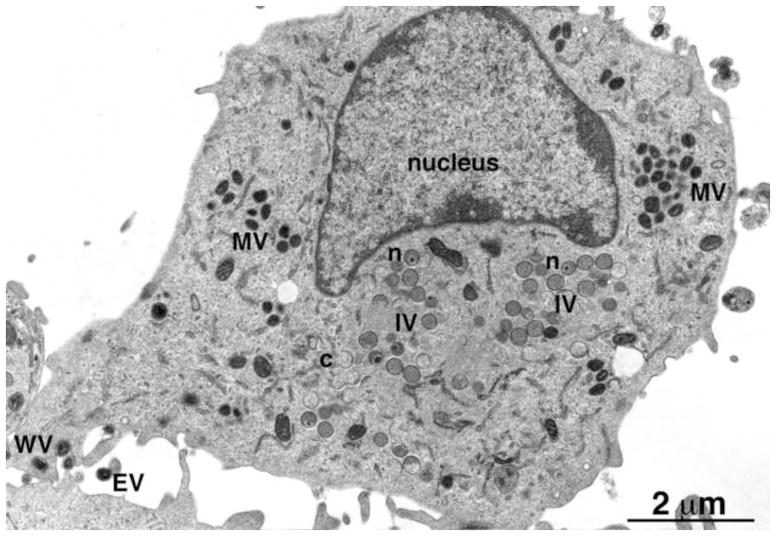
Poxviruses are large DNA viruses that infect vertebrates and invertebrates and include species that cause severe human disease (e.g. smallpox and monkeypox) and others that serve beneficial roles as vectors for vaccines against unrelated infectious agents (Damon, 2013; Moss, 2013a). The ability to reproduce entirely within the cytoplasm is a defining characteristic of the poxvirus family and depends on viral proteins for replication (Moss, 2013b) and transcription (Broyles and Knutson, 2010) of the large DNA genome. Approximately 100 genes, conserved in all chordopoxviruses, are required for reproduction in cultured cells (Upton et al., 2003; Xu et al., 2014); a similar number of less well conserved genes are important for optimal infection of animals (Haller et al., 2014; Smith et al., 2013). The cytoplasmic sites of viral DNA synthesis expand into factories where the intermediate and late stages of transcription and translation occur (Katsafanas and Moss, 2007). Crescent membranes appear within the factories and enlarge to form spherical immature virions (IVs) that condense into dense brick-shaped mature virions (MVs) (Gaylord and Melnick, 1953; Morgan et al., 1954; Morgan and Wyckoff, 1950). Depending on the poxvirus genus, MVs may be enclosed by an additional membrane derived from the trans-Golgi, endosomal cisternae or plasma membrane (Boulanger et al., 2000; Hiller and Weber, 1985; Schmelz et al., 1994; Tooze et al., 1993) to form wrapped virions (WVs) and transported to the cell periphery and exocytosed as extracellular enveloped virions (EVs) (Fig. 1).
Fig. 1.
Transmission electron microscopic image of a HeLa cell infected with VACV. Abbreviations: MV, mature virion; IV, immature virion; WV, wrapped virion; EV, extracellular enveloped virion; n, IV with nucleoid. Scale bar at bottom. Provided by A. Weisberg.
The structure and origin of the poxviral membrane delimiting crescents and IVs have intrigued virologists for more than half a century. The inability to discern connections between the viral membranes and cellular organelles (Fig. 2) led to the idea that viral membranes form de novo (Dales and Mosbach, 1968). However, the connections could be transient and an origin from cell membranes comprising the intermediate compartment (ERGIC) between the endoplasmic reticulum (ER) and Golgi apparatus (Sodeik and Krijnse-Locker, 2002) and from the ER itself (Husain et al., 2006) have been considered. This review focuses on recent studies that provide evidence for the formation of the crescent and IV membrane from the ER. Although the main features of morphogenesis are similar in all poxviruses, most research has been carried out with vaccinia virus (VACV). Several broad reviews of poxvirus structure and morphogenesis are available (Condit et al., 2006; Liu et al., 2014; Moss, 2013a; Roberts and Smith, 2008).
Fig. 2.
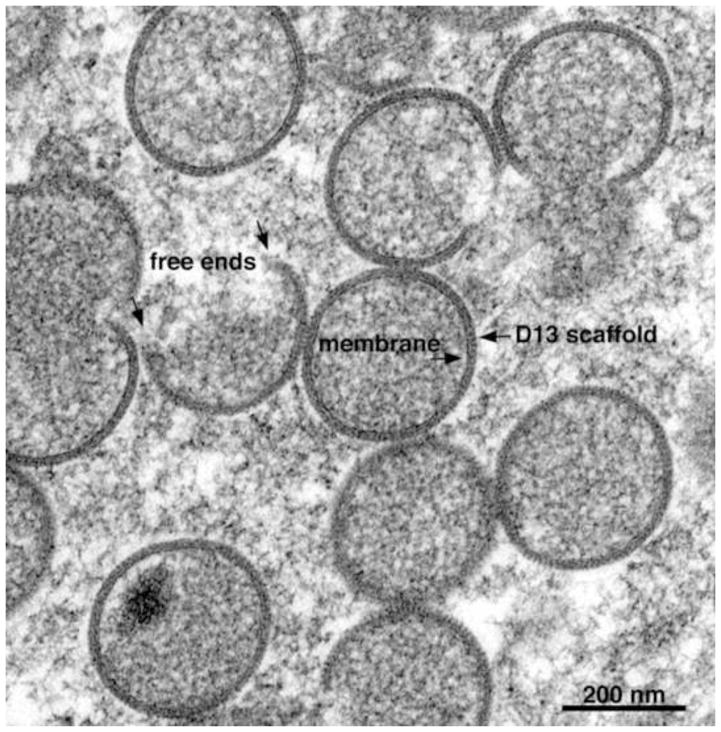
Transmission electron microscopic image of a cell infected with VACV showing IVs forming within a virus factory. Free ends, lipoprotein membrane and D13 scaffold are labeled with arrows. Scale bar at bottom. Provided by A. Weisberg.
One membrane or two?
Electron micrographs from the 1950s describe clusters of spherical IVs and dense brick-shaped MVs (Gaylord and Melnick, 1953; Morgan et al., 1954; Morgan and Wyckoff, 1950). In some images, the IVs appear to have a double membrane with an outer dense layer of 4 to 6 nm and an inner of about the same thickness (Higashi et al., 1960). In other images (Dales and Mosbach, 1968) there appears to be a single membrane sheet coated with dense spicules, rather than two lipid membranes. The latter interpretation is supported by studies with the drug rifampicin, which prevents the formation of the spicule layer and provides clear images of a single membrane bilayer (Grimley et al., 1970; Moss et al., 1969; Nagayama et al., 1970). Nevertheless, the concept of a double-membrane was revived in a series of subsequent publications that posit two membranes so tightly apposed as to give the illusion of a single membrane (Krijnse-Locker et al., 1996; Risco et al., 2002; Salmons et al., 1997; Sodeik et al., 1995; Sodeik et al., 1993; Sodeik et al., 1994). However, careful measurements of the thickness of the IV membrane (Hollinshead et al., 1999) and freeze-fracture (Heuser, 2005) fully support the prior single lipid membrane model and deep-etch and immunoelectron microscopy demonstrate that the “spicule layer” is a honeycomb lattice comprised of VACV D13 protein trimers external to the single membrane (Heuser, 2005; Szajner et al., 2005). The controversy ended when proponents of the double-membrane and others reported electron tomography images confirming a single membrane (Chichon et al., 2009; Chlanda et al., 2009).
De novo membrane biogenesis or acquisition from host membranes?
Failure to detect continuity between the crescent membrane and cellular organelles led to the conclusion that the open-ended sheets are formed de novo (Dales and Mosbach, 1968). Although seemingly a heretical notion, in view of the formation of all known membranes from pre-existing ones, a de novo viral origin could not be dismissed out of hand since poxviruses are complex and encode proteins for many other functions including genome replication, transcription and disulfide bond formation (Moss, 2013a). Moreover, no viral protein components of the IV or MV are known to have signal peptides or to be glycosylated, which are signatures of trafficking through the secretory pathway of the cell. However, immunogold electron microscopic studies localized some VACV membrane proteins to the rough ER and the ERGIC, suggesting that the latter might contribute to the formation of the viral membrane (Krijnse-Locker et al., 1996; Rodríguez et al., 1996; Salmons et al., 1997; Sodeik et al., 1995; Sodeik and Krijnse-Locker, 2002). Smooth ER membranes labeled with protein disulfide isomerase (PDI) and viral proteins have also been found in close proximity to crescents (Chlanda et al., 2009; Husain et al., 2006). In addition, the association of VACV membrane proteins with microsomes was demonstrated by in vitro translation (Betakova et al., 1999a; Krijnse-Locker et al., 1996). However, analysis of purified MVs failed to detect cellular membrane proteins (Chung et al., 2006; Krauss et al., 2002; Resch et al., 2007) and the above studies only provide circumstantial evidence for participation of the ERGIC or ER in viral membrane formation.
Further studies were intended to discriminate functionally between possible contributions of different cellular organelles to viral membrane assembly. The fungal metabolite brefeldin A did not prevent the formation of IVs and MVs, although wrapping of the latter with Golgi membranes was impaired (Ulaeto et al., 1995). The target of brefeldin A is the guanine nucleotide exchange factor (GBF1) (Donaldson et al., 1992; Helms and Rothman, 1992). Inhibition of GBF1 induces the retrograde transport of proteins from Golgi membranes to the ER and collapse of the Golgi apparatus (Lippincott-Schwartz et al., 1989). Therefore, the brefeldin A study does not discriminate between roles of the ER, ERGIC or Golgi network in VACV IV formation. In contrast, Sar1 GTPase is an essential component of coatomer protein II (COPII)-mediated cargo transport from the ER to the ERGIC and other post-ER compartments (Aridor et al., 2001; Kuge et al., 1994). Overexpression of a dominant negative Sar1 protein had no effect on formation of VACV IVs and MVs but like brefeldin A blocked wrapping of MVs by Golgi membranes (Husain and Moss, 2003). The Sar1 protein inhibitor result indicates that transport of viral proteins from the ER to the ERGIC or beyond is unnecessary for formation of IV and MV membranes, whereas such transport is necessary for the subsequent addition of the wrapping membrane. The drug H-89, a serine/threonine protein kinase inhibitor that prevents assembly of Sar1 protein into the ER membrane, and cerulenin, an inhibitor of lipid biogenesis, prevented infectious virus formation following attempted reversal of an early morphogenesis block (Punjabi and Traktman, 2005). However, the stage at which morphogenesis is affected by these drugs was not described and H-89 may have multiple protein kinase targets.
Although the dominant negative Sar1 protein experiment shows that transport from ER to post-ER compartments is unnecessary for IV formation (Husain and Moss, 2003), they do not prove that the ER itself is needed. An important next experiment would have been to determine the effect of blocking import of proteins to the ER. Unfortunately, neither a specific drug nor a potent dominant negative inhibitor of this step is available for animal cells. An alternative approach is to determine whether proteins could traffic to viral membranes through the ER. Since IV and MV proteins lack cleaved signal peptides, one strategy was to attach a signal peptide followed by a Flag epitope tag to the N-terminus of the viral A9 protein, which localizes in ER and IV membranes with the same topology (Husain et al., 2006). Cleavage of the signal peptide would put the Flag tag at the N-terminus and recognizable by a specific monoclonal antibody. Since signal peptidase is associated with the ER membrane, detection of the N-terminal Flag tag in IV membranes provided evidence for trafficking of the chimeric A9 protein through the ER to the viral membrane (Husain et al., 2006). Furthermore, addition of a COPII binding site to the A9 protein diverted it from viral to Golgi membranes also providing evidence of a functional ER connection (Husain et al., 2006). Additional experiments indicate that transit to the viral membrane might be a default pathway for proteins that are synthesized without a COPII signal in the virus factory (Husain et al., 2007).
The major membrane proteins of crescents and IVs
The two major transmembrane protein components of VACV crescents and IVs, A17 and A14, are expressed late in infection. Both proteins are co-translationally inserted into microsomal membranes in vitro and can be detected by immunoelectron microscopy on ER and ERGIC membranes in addition to crescents and IVs in vivo (Betakova and Moss, 2000; Krijnse-Locker et al., 1996; Rodriguez et al., 1997; Unger et al., 2013; Wolffe et al., 1996). Disulfide-bonded dimers of A14 interact with A17 directly or indirectly and both proteins are phosphorylated by the F10 kinase (Betakova et al., 1999b; Mercer and Traktman, 2003; Rodriguez et al., 1997; Szajner et al., 2004; Unger et al., 2013). In vitro studies suggest that the N- and C-termini of A17 face the cytoplasm (Betakova and Moss, 2000; Betakova et al., 1999a; Krijnse-Locker et al., 1996), whereas those of A14 may be luminal (Mercer and Traktman, 2003; Salmons et al., 1997) although the precise topologies of both proteins on IVs needs further investigation. In addition, the N- and C-termini of A17 are trimmed by the I7 proteinase (Ansarah-Sobrinho and Moss, 2004; Betakova et al., 1999b; Rodriguez et al., 1993; Takahashi et al., 1994). The N-terminus of A17 is required for IV formation and interacts with D13 trimers, which form the external honeycomb lattice (Bisht et al., 2009; Heuser, 2005; Szajner et al., 2005; Unger et al., 2013).
Conditional lethal mutants have been employed to investigate the stage at which A17 and A14 participate in viral membrane formation. When expression of A17 is repressed, morphogenesis is blocked at an early stage with small vesicles or tubules accumulating adjacent to large, dense bodies of viroplasm (Rodríguez et al., 1996; Wolffe et al., 1996). Small vesicles as well as empty crescents and incomplete IVs also accumulate when A14 is repressed (Rodriguez et al., 1998; Traktman et al., 2000). It is uncertain whether the empty crescents and partial IVs represent leaky repression or the true null phenotype. At the restrictive temperature, morphogenesis of temperature-sensitive mutants of the F10 kinase (Lin and Broyles, 1994), which phosphorylates A17 (Betakova et al., 1999b; Derrien et al., 1999) and A14 (Betakova et al., 1999b), is blocked prior to formation of small vesicles suggesting that F10 might have additional targets (Traktman et al., 1995; Wang and Shuman, 1995). A similar, but less stringent phenotype occurs when expression of F10 is repressed at 37°C; raising the temperature increased the stringency suggesting that some target of the kinase may be temperature sensitive in the absence of phosphorylation (Punjabi and Traktman, 2005; Szajner et al., 2004). Rescue of the F10 mutant in trans depends on an intact active kinase site (Punjabi and Traktman, 2005; Szajner et al., 2004).
Rifampicin and the D13 scaffold protein
The antibiotic rifampicin, an inhibitor of prokaryotic DNA-dependent RNA polymerase, has an unrelated anti-poxviral activity. In the presence of the drug, irregular sheets of membrane form adjacent to masses of viroplasm that contain the core proteins (Grimley et al., 1970; Moss et al., 1969; Nagayama et al., 1970). Washout of the drug leads to rapid coating of the membrane with the spicule layer even if protein synthesis is inhibited (Moss et al., 1969). The finding that rifampicin-resistant mutants map to the D13 protein suggested that the latter was a component of the spicule layer (Baldick and Moss, 1987; Charity et al., 2007; Tartaglia et al., 1986). Indeed, the effect of repression of D13 expression is identical to that of rifampicin (Zhang and Moss, 1992). In the presence of rifampicin, D13 accumulates in separate inclusions (Sodeik et al., 1994). Subsequent studies demonstrated that the so-called spicule layer is a honeycomb lattice comprised of trimers of D13 (Heuser, 2005; Szajner et al., 2005). D13 has no transmembrane domain and associates with the viral membrane through interaction with the N-terminus of A17 (Bisht et al., 2009; Unger et al., 2013). This interaction is supported by a recent finding that duplication or over expression of A17 confers resistance to rifampicin (Erlandson et al., 2014). The removal of D13 trimers during the transition from IV to MV is associated with processing of A17 (Bisht et al., 2009).
The structure of the D13 trimer was determined by cryoelectron tomography (Hyun et al., 2007) and X-ray crystallography (Bahar et al., 2011; Hyun et al., 2011). The analyses reveal a double β-barrel “jelly-roll” subunit arranged as pseudo-hexagonal trimers with similarity to the capsid proteins of icosahedral viruses. Most mutations in D13 that confer resistance to rifampicin map in contiguous membrane proximal regions. However, the binding site for rifampicin has not yet been determined.
Viral membrane assembly proteins (VMAPs [pronounced VeeMAPs])
A group of viral proteins that are involved in related steps in formation of the IV membrane have been termed VMAPs (Maruri-Avidal et al., 2013c). The VMAPs identified to date are the A6, A11, A30.5, H7 and L2 proteins (Table 1), which are conserved in all chordopoxviruses. Except for L2, the VMAPs are expressed exclusively following genome replication. Three VMAPs (L2, A30.5 and A11) have hydrophobic domains near their C-termini enabling association with ER and viral membranes. Properties of individual VMAPs are presented below and their roles, determined by construction of mutant viruses, are discussed in the subsequent section.
Table 1.
VMAPs
| Protein | kDa | Expression | Interaction | ERa | Reference |
|---|---|---|---|---|---|
| A6 | 43 | Post-rep | A11 | {Meng, 2007 #11530}{Meng, 2012 #14846} | |
| A11 | 36 | Post-rep | A6, A32 | + | {Resch, 2005 #10762}{Maruri-Avidal, 2013 #15396}{Wu, 2012 #15081} |
| A30.5 | 4.8 | Post-rep | L2 | + | {Maruri-Avidal, 2013 #15517} |
| H7 | 17 | Post-rep | {Satheshkumar, 2009 #12746}{Meng, 2013 #15392} | ||
| L2 | 10 | Pre-rep | A30.5 | + | {Maruri-Avidal, 2011 #14510; Maruri-Avidal, 2011 #14355}{Maruri-Avidal, 2013 #15207} |
Association with ER determined by immunofluorescence confocal microscopy
L2, a small 10-kDa protein with two near C-terminal hydrophobic domains, is first expressed early during infection (Maruri-Avidal et al., 2011a). L2 colocalizes with the ER throughout the cytoplasm of both infected and uninfected cells with the N-terminus exposed to the cytoplasm (Maruri-Avidal et al., 2011b)(S. Hyun, L. Maruri-Avidal and B. Moss, unpublished). Although small amounts are detected in purified MVs (Maruri-Avidal et al., 2011b), immunoelectron microscopy shows that the L2 within virus factories is predominantly at the edges of crescents and nearby tubular membranes containing ER markers (Maruri-Avidal et al., 2011a).
A30.5 is a 42-amino acid, largely hydrophobic protein that is expressed post-replicatively and immunopurifies with and cross-links to L2 (Maruri-Avidal et al., 2013c). An ORF encoding a similar size hydrophobic protein is conserved in the same genomic location in all chordopoxviruses, although the small size and predominance of hydrophobic amino acids preclude statistical confirmation of homology. A30.5 colocalizes with the ER in both infected and uninfected cells and in the former is also associated with L2 and the edges of crescents and nearby ER membranes.
A11 is a 40-kDa protein, with two hydrophobic domains near the C-terminus, conserved in all poxviruses. A11 was first described as interacting with itself and with the A32 DNA packaging protein in a VACV genome-wide yeast two-hybrid screen (McCraith et al., 2000). Although the interaction of A11 with A32 was confirmed by immunoprecipitation (Resch et al., 2005), the biological significance is unknown. Further studies indicate that A11 is expressed post-replicatively, localizes in viral factories, and is absent from purified virions (Resch et al., 2005). A11 weakly associates with A6, an interaction that is necessary for membrane and virus factory association of A11 (Wu et al., 2012). ER colocalization occurs only within virus factories, in contrast to L2 and A30.5, which associate with ER throughout the cell (Maruri-Avidal et al., 2013b). In uninfected cells A11 is broadly distributed in the cytoplasm and does not colocalize with ER even if A6 or H7 are co-transfected, although A11 post-translationally associates with microsomes in a cell-free expression system (Maruri-Avidal et al., 2013b).
A6 is a 43-kDa protein, with no predicted transmembrane domain, that is conserved in all chordopoxviruses, expressed post-replicatively and to a minor extent packaged in MV cores (Meng et al., 2007). As mentioned above, A6 has been shown to associate with the A11 protein (Wu et al., 2012).
H7 is a 40-kDa protein, with no predicted transmembrane domain, that is conserved in all chordopoxviruses and expressed post-replicatively (Satheshkumar et al., 2009). H7 is not detected in highly purified MVs and is not strongly retained in viral factories following synthesis (Satheshkumar et al., 2009). The crystal structure of H7 reveals a novel fold comprised of seven α-helices and a highly curved three-stranded antiparallel β-sheet (Kolli et al., 2014). A basic patch representing a phosphoinositide-binding site is essential for binding to phosphatidylinositol-3- and 4-phosphate in vitro (Kolli et al., 2014).
Analysis of conditional lethal VMAP mutants
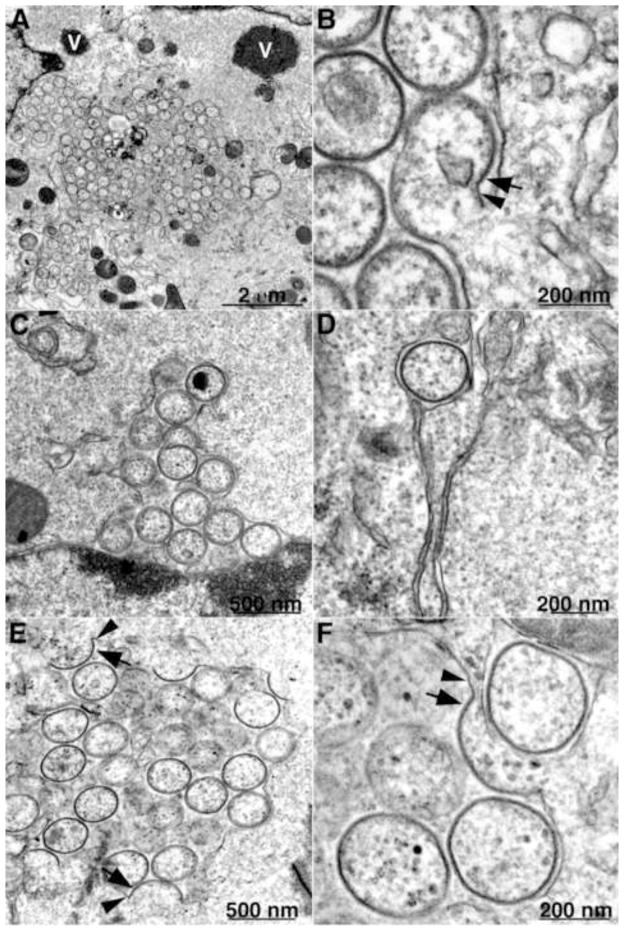
The roles of individual VMAPs have been investigated primarily by making conditional lethal inducible and/or deletion mutants. L2, A30.5 and A11 null mutants have similar if not identical phenotypes. Initial studies with an inducible L2 mutant demonstrated that expression of the protein is required for virion assembly (Maruri-Avidal et al., 2011a; Maruri-Avidal et al., 2011b). Large masses of dense viroplasm containing core proteins or their precursors, some of which have short, spicule-coated membrane arcs at their periphery that appear to represent the beginning of crescent formation, form in the absence of inducer. Although no typical IVs or MVs are seen, IV-like structures devoid of internal dense material appear in close association with ER membranes. In order to determine whether the aberrant structures represent incomplete repression of L2 synthesis, a L2 deletion mutant that could replicate in a stably transfected L2 cell line was constructed (Maruri-Avidal et al., 2013a). The phenotype of the deletion mutant is identical to that of the inducible mutant. Fig. 3 contains an image of a cell infected with the L2 deletion mutant showing a dense inclusion with short crescent-like arcs on the surface; Fig. 4A shows a cluster of IV-like structures surrounded by a smooth membrane, as well as separate inclusions of dense viroplasm. The external spicule layers of the IV-like structures as well as connections with the smooth membrane are resolved at high magnification (Fig. 4B). Similar strategies were used to construct and analyze an A11 inducible mutant (Maruri-Avidal et al., 2013b; Resch et al., 2005) and an A30.5 deletion mutant (Maruri-Avidal et al., 2013c), both of which have the same phenotype as the L2 deletion mutant. Immunoelectron microscopy demonstrated the presence of A17, A14 and D13 in the crescent- and IV-like membranes and calnexin, an ER marker in the associated smooth membranes (Maruri-Avidal et al., 2013a; Maruri-Avidal et al., 2013c). Images of cells infected with the A30.5 deletion mutant are shown in Fig. 4C–F. In Fig. 4C, the membrane surrounding the cluster of IV-like structures appears continuous with the external nuclear membrane, which connects with the ER. Fig. 4D and 4E show a single and multiple IV-like particles within the ER lumen, respectively. Clear connections between spicule-coated crescent membranes and the smooth membranes are revealed in Fig. 4E and 4F and have been confirmed by tomography (A. Weisberg, B.T. Hansen, F. H. Hoyt, L. Maruri-Avidal, B. Moss and E. R. Fischer, unpublished). The convex surfaces of the crescents face the lumen so that the IVs pinch off into the expanded luminal space. Discontinuities in the ER membrane apparently allow access of D13 but not dense viroplasm into the lumen. For reasons that are not yet understood, the IV-like structures are more abundant in RK-13 cells than in BS-C-1 and HeLa cells.
Fig. 3.
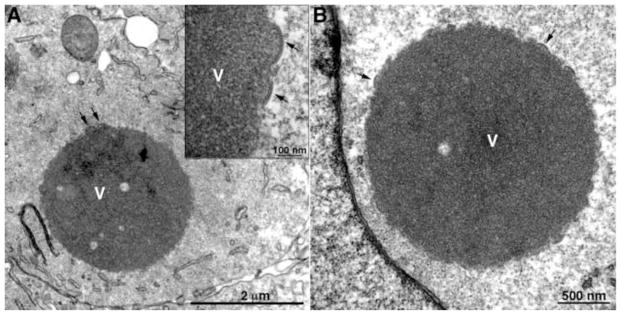
Transmission electron microscopic images of a BS-C-1 cell infected with a VACV L2 deletion mutant showing dense inclusions. Arrows point to short crescents. V, dense inclusion of viroplasm. Inset, high magnification of portion of inclusion. Scale bars shown. Adapted from (Maruri-Avidal et al., 2013a)
Fig. 4.
Transmission electron microscopic images of cells infected with a VACV L2 and A30.5 deletion mutants showing association of ER and IV-like structures. (A, B) VACV L2 deletion mutant infection of BS-C-1 cells. (C–F) VACV A30.5 deletion mutant infection of RK-13 cells. Arrows, membrane with spicule layer; Arrowheads, smooth membrane. Scale bars shown. Adapted from (Maruri-Avidal et al., 2013c).
Dense cytoplasmic inclusions some of which had short membrane arcs similar to that shown in Fig. 3 instead of IVs and MVs were found in cells infected with conditional lethal inducible A6 (Meng et al., 2012) and H7 (Satheshkumar et al., 2009) mutants. The phenotype of an H7 deletion mutant appeared to be more stringent as the dense inclusions lacked the short membrane arcs (Meng et al., 2013). The presence of IV-like structures similar to those in cells infected with L2, A30.5 and A11 have not been reported for either A6 or H7 mutants.
The VMAP mutants exhibit additional defects that seem to be secondary to the block in viral membrane formation and therefore will only be mentioned here. These include destabilization of certain membrane proteins including those that form the entry-fusion complex and mislocalization and glycosylation of others (Meng et al., 2007; Resch et al., 2005) (Maruri-Avidal et al., 2011a; Maruri-Avidal et al., 2013a; Maruri-Avidal et al., 2011b; Meng et al., 2012; Satheshkumar et al., 2009)}(Maruri-Avidal et al., 2013c).
A model for poxvirus membrane biogenesis
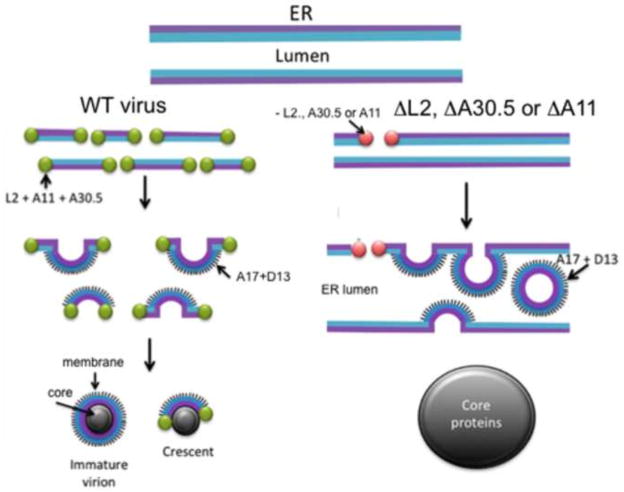
Electron micrographs from cells infected with VMAP null mutants are the first to demonstrate a direct connection between poxviral membranes and ER. The model for viral membrane formation depicted in Fig. 5 is based on the premise that a step in the normal viral membrane biogenesis pathway has been interrupted revealing ER connections. This hypothesis is supported by the identical phenotype of three different null mutants. The model posits that during a WT virus infection the L2, A30.5, A11, H7 and A6 VMAPs participate with cellular proteins to produce interruptions or stabilize naturally occurring transient breaks in a subsection of ER within the virus factory. As shown by immunoelectron microscopy, the L2, A30.5 and A11 VMAPs cap the free ends of membrane segments containing A17 and A14, which elongate by fusion with additional ER membrane segments to form crescents and spherical IVs. The model suggests that fragmentation of the ER membrane precedes crescent formation making it difficult or impossible to see ER connections during infection with WT virus. In contrast, the ER membranes containing A17 and A14 remain largely intact when L2, A30.5 or A11 is not synthesized. In cells infected with the VMAP null mutants, occasional breaks in the ER membrane allow the association of D13 with the N-terminus of A17, which is on the luminal side of the ER. The curvature imposed by D13 and A17, leads to the formation of IV-like spheres that pinch off and collect in the modified ER lumen. The sequestration of viral membranes in the lumen prevents their interaction with core proteins, which aggregate into masses of dense viroplasm mostly unadorned by viral membranes. A novel prediction of the model is that the outer surface of the IV and MV represents the luminal side of the ER. This topology can explain the exposure of some phosphatidylserine, which is more abundant on the luminal side of the ER (Dominski et al., 1983; Fairn et al., 2011) on the surface of the MV (Cluett and Machamer, 1996) (Mercer and Helenius, 2008).
Fig. 5.
Model for formation of viral membranes during infection with wild-type (WT) and mutant VACV. L2, A30.5, A11, A17, A14 and D13 refer to VACV proteins; ΔL2, ΔA30.5 and ΔA11 refer to VACV deletion mutants. Adapted from (Maruri-Avidal et al., 2013c).
The above model represents a starting point for further research. An important question is how breaks occur in the ER membrane. One possibility is that the ER is constantly being remodeled but that the breaks are rapidly sealed in uninfected cells. In this scenario, the main role of the VMAPs may be to cap the ends of the membrane fragments and prevent resealing. Alternatively, the VMAPs may actively induce breaks perhaps in association with cell proteins. Additional work is needed to define the roles of individual VMAPs, particularly those that do not exhibit direct membrane binding. The prediction that the surface of the IV represents the luminal side of the ER was unanticipated and needs to be confirmed for infection with wild-type virus. This “inside-outside” topology raises questions regarding the orientations of A17 and A14 and suggests that ER breakage may precede the insertion of the major viral proteins. Although the novel viral membrane-ER connections were demonstrated with null mutants of three different viral proteins, putative cell proteins involved need to be identified.
Highlights.
The origin of poxviral membranes has perplexed researchers for half a century
Viral membranes normally appear as open sheets unconnected to cellular organelles
A break-through has come from recent studies of several vaccinia virus null mutants
Connections of viral membranes to the ER persist in cells infected with mutants
Acknowledgments
The author thanks A. Weisberg and L. Maruri-Avidal for discussions during the course of the research that led to the model of poxvirus membrane biogenesis. A. Weisberg kindly provided the electron microscopy images. The work was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansarah-Sobrinho C, Moss B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J Virol. 2004;78:6335–6343. doi: 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott-Schwartz J, Balch WE. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001;152:213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick CJ, Moss B. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted NH2 terminus of a gene encoding an Mr 62,000 polypeptide. Virology. 1987;156:138–145. doi: 10.1016/0042-6822(87)90444-2. [DOI] [PubMed] [Google Scholar]
- Betakova T, Moss B. Disulfide bonds and membrane topology of the vaccinia virus A17L envelope protein. J Virol. 2000;74:2438–2442. doi: 10.1128/jvi.74.5.2438-2442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betakova T, Wolffe EJ, Moss B. Membrane topology of the vaccinia virus A17L envelope protein. Virology. 1999a;261:347–356. doi: 10.1006/viro.1999.9870. [DOI] [PubMed] [Google Scholar]
- Betakova T, Wolffe EJ, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L protein kinase. J Virol. 1999b;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H, Weisberg AS, Szajner P, Moss B. Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis. J Virol. 2009;83:9140–9150. doi: 10.1128/JVI.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger D, Smith T, Skinner MA. Morphogenesis and release of fowlpox virus. J Gen Virol. 2000;81:675–687. doi: 10.1099/0022-1317-81-3-675. [DOI] [PubMed] [Google Scholar]
- Broyles SS, Knutson BA. Poxvirus transcription. Future Virol. 2010;5:639–650. [Google Scholar]
- Charity JC, Katz E, Moss B. Amino acid substitutions at multiple sites within the vaccinia virus D13 scaffold protein confer resistance to rifampicin. Virology. 2007;359:227–232. doi: 10.1016/j.virol.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichon FJ, Rodriguez MJ, Risco C, Fraile-Ramos A, Fernandez JJ, Esteban M, Carrascosa JL. Membrane remodelling during vaccinia virus morphogenesis. Biol Cell. 2009;101:401–414. doi: 10.1042/BC20080176. [DOI] [PubMed] [Google Scholar]
- Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. Vaccinia virus proteome: Identification of proteins in vaccinia virus intracellular mature virion particles. J Virol. 2006;80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluett EB, Machamer CE. The envelope of vaccinia virus reveals an unusual phospholipid in Golgi complex membranes. J Cell Sci. 1996;109 (Pt 8):2121–2131. doi: 10.1242/jcs.109.8.2121. [DOI] [PubMed] [Google Scholar]
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- Dales S, Mosbach EH. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- Damon I. Poxviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2160–2184. [Google Scholar]
- Derrien M, Punjabi A, Khanna R, Grubisha O, Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: Involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski J, Binaglia L, Dreyfus H, Massarelli R, Mersel M, Freysz L. A study on the topological distribution of phospholipids in microsomal membranes of chick brain using phospholipase C and trinitrobenzenesulfonic acid. Biochimica et biophysica acta. 1983;734:257–266. doi: 10.1016/0005-2736(83)90123-2. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Finazzi D, Klausner RD. Brefeldin-A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Erlandson KJ, Cotter CA, Charity JC, Martens C, Fischer ER, Ricklefs SM, Porcella SF, Moss B. Duplication of the A17L locus of vaccinia virus provides an alternate route to rifampin resistance. J Virol. 2014;88:11576–11585. doi: 10.1128/JVI.00618-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylord WH, Jr, Melnick JL. Intracellular forms of pox viruses as shown by the electron microscope (Vaccinia, Ectromelia, Molluscum Contagiosum) J Exp Med. 1953;98:157–172. doi: 10.1084/jem.98.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley PM, Rosenblum EN, Mims SJ, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: Accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect Genet Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JB, Rothman JE. Inhibition by Brefeldin-A of a golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Heuser J. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J Cell Biol. 2005;169:269–283. doi: 10.1083/jcb.200412169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi N, Ozaki Y, Ichimiya M. Electron microscopy of pox virus-to-cell adsorption and the ultrastructure of developmental forms of pox virus. J Ultrastruct Res. 1960;3:270–281. doi: 10.1016/s0022-5320(60)80014-7. [DOI] [PubMed] [Google Scholar]
- Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinshead M, Vanderplasschen A, Smith GL, Vaux DJ. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M, Moss B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J Virol. 2003;77:11754–11766. doi: 10.1128/JVI.77.21.11754-11766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M, Weisberg AS, Moss B. Existence of an operative pathway from the endoplasmic reticulum to the immature poxvirus membrane. Proc Natl Acad Sci USA. 2006;103:19506–19511. doi: 10.1073/pnas.0609406103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M, Weisberg AS, Moss B. Sequence-independent targeting of transmembrane proteins synthesized within vaccinia virus factories to nascent viral membranes. J Virol. 2007;81:2646–2655. doi: 10.1128/JVI.02631-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun JK, Accurso C, Hijnen M, Schult P, Pettikiriarachchi A, Mitra AK, Coulibaly F. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Path. 2011;7:e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun JK, Coulibaly F, Turner AP, Baker EN, Mercer AA, Mitra AK. The structure of a putative scaffolding protein of immature poxvirus particles as determined by electron microscopy suggests similarity with capsid proteins of large icosahedral DNA viruses. J Virol. 2007;81:11075–11083. doi: 10.1128/JVI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli S, Meng X, Wu X, Shengjuler D, Cameron CE, Xiang Y, Deng J. Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication. J Virol. 2014 doi: 10.1128/JVI.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss O, Hollinshead R, Hollinshead M, Smith GL. An investigation of incorporation of cellular antigens into vaccinia virus particles. J Gen Virol. 2002;83:2347–2359. doi: 10.1099/0022-1317-83-10-2347. [DOI] [PubMed] [Google Scholar]
- Krijnse-Locker J, Schleich S, Rodriguez D, Goud B, Snijder EJ, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin s, Broyles SS. Vaccinia protein kinase 2: a second essential serine/threonine protein kinase encoded by vaccinia virus. Proc Natl Acad Sci USA. 1994;91:7653–7657. doi: 10.1073/pnas.91.16.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cooper T, Howley PM, Hayball JD. From crescent to mature virion: vaccinia virus assembly and maturation. Viruses. 2014;6:3787–3808. doi: 10.3390/v6103787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruri-Avidal L, Domi A, Weisberg AS, Moss B. Participation of vaccinia virus L2 protein in the formation of crescent membranes and immature virions. J Virol. 2011a;85:2504–2511. doi: 10.1128/JVI.02505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruri-Avidal L, Weisberg AS, Bisht H, Moss B. Analysis of viral membranes formed in cells infected by a vaccinia virus L2-deletion mutant suggests their origin from the endoplasmic reticulum. J Virol. 2013a;87:1861–1871. doi: 10.1128/JVI.02779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruri-Avidal L, Weisberg AS, Moss B. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J Virol. 2011b;85:12431–12441. doi: 10.1128/JVI.05573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruri-Avidal L, Weisberg AS, Moss B. Association of the vaccinia virus A11 protein with the endoplasmic reticulum and crescent precursors of immature virions. J Virol. 2013b;87:10195–11206. doi: 10.1128/JVI.01601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruri-Avidal L, Weisberg AS, Moss B. Direct formation of vaccinia viral membranes from the endoplasmic reticulum in the absence of the newly characterized L2-interacting A30.5 protein. J Virol. 2013c;87:12313–12326. doi: 10.1128/JVI.02137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Embry A, Rose L, Yan B, Xu C, Xiang Y. Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly. J Virol. 2012;86:5603–5613. doi: 10.1128/JVI.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wu X, Yan B, Deng J, Xiang Y. Analysis of the role of vaccinia virus H7 in virion membrane biogenesis with a H7-deletion mutant. J Virol. 2013;87:8247–8253. doi: 10.1128/JVI.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XZ, Embry A, Sochia D, Xiang Y. Vaccinia virus A6L encodes a virion core protein required for formation of mature virion. J Virol. 2007;81:1433–1443. doi: 10.1128/JVI.02206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- Mercer J, Traktman P. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J Virol. 2003;77:8857–8871. doi: 10.1128/JVI.77.16.8857-8871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Ellison S, Rose H, Moore D. Structure and development of viruses observed in the electron microscope. II Vaccinia and fowl pox viruses. J Exp Med. 1954;100:301–310. doi: 10.1084/jem.100.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Wyckoff RW. The electron microscopy of fowl pox virus within the chorioallantoic membrane. J Immunol. 1950;65:285–295. [PubMed] [Google Scholar]
- Moss B. Poxviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2013a. pp. 2129–2159. [Google Scholar]
- Moss B. Poxvirus DNA Replication. Cold Spring Harbor Perspect Biol. 2013b;5:a010199. doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Rosenblum EN, Katz E, Grimley PM. Rifampicin: A specific inhibitor of vaccinia virus assembly. Nature. 1969;224:1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- Nagayama A, Pogo BGT, Dales S. Biogenesis of vaccinia: separation of early stages from maturation by means of rifampicin. Virology. 1970;40:1039–1051. doi: 10.1016/0042-6822(70)90150-9. [DOI] [PubMed] [Google Scholar]
- Punjabi A, Traktman P. Cell biological and functional characterization of the vaccinia virus F10 kinase: Implications for the mechanism of virion morphogenesis. J Virol. 2005;79:2171–2190. doi: 10.1128/JVI.79.4.2171-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. Protein composition of the vaccinia virus mature virion. Virology. 2007;358:233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Resch W, Weisberg AS, Moss B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J Virol. 2005;79:6598–6609. doi: 10.1128/JVI.79.11.6598-6609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, Rodriguez JR, Lopez-Iglesias C, Carrascosa JL, Esteban M, Rodriguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008;16:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Rodríguez D, Risco C, Rodríguez JR, Carrascosa JL, Esteban M. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J Virol. 1996;70:7641–7653. doi: 10.1128/jvi.70.11.7641-7653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D, Rodriguez JR, Esteban M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JR, Risco C, Carrascosa JL, Esteban M, Rodriguez D. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J Virol. 1997;71:1821–1833. doi: 10.1128/jvi.71.3.1821-1833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JR, Risco C, Carrascosa JL, Esteban M, Rodriguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmons T, Kuhn A, Wylie F, Schleich S, Rodriguez JR, Rodriguez D, Estban M, Griffiths G, Locker JK. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satheshkumar PS, Weisberg A, Moss B. Vaccinia virus H7 protein contributes to the formation of crescent membrane precursors of immature virions. J Virol. 2009;83:8439–8450. doi: 10.1128/JVI.00877-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Benfield CTO, de Motes CM, Mazzon M, Ember SWJ, Ferguson BJ, Sumner RP. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles EG, Griffiths G. Assembly of vaccinia virus: Incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B, Doms RW, Ericsson M, Hiller G, Machamer CE, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B, Griffiths G, Ericsson M, Moss B, Doms RW. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J Virol. 1994;68:1103–1114. doi: 10.1128/jvi.68.2.1103-1114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B, Krijnse-Locker J. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 2002;10:15–24. doi: 10.1016/s0966-842x(01)02256-9. [DOI] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J Cell Biol. 2005;170:971–981. doi: 10.1083/jcb.200504026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Moss B. Evidence for an essential catalytic role of the F10 protein kinase in vaccinia virus morphogenesis. J Virol. 2004;78:257–265. doi: 10.1128/JVI.78.1.257-265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- Tartaglia J, Piccini A, Paoletti E. Vaccinia virus rifampicin-resistance locus specifies a late 63,000 Da gene product. Virology. 1986;150:45–54. doi: 10.1016/0042-6822(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- Traktman P, Caligiuri A, Jesty SA, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: Construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaeto D, Grosenbach D, Hruby DE. Brefeldin A inhibits vaccinia virus envelopment but does not prevent normal processing and localization of the putative envelopment receptor P37. J Gen Virol. 1995;76:103–111. doi: 10.1099/0022-1317-76-1-103. [DOI] [PubMed] [Google Scholar]
- Unger B, Mercer J, Boyle KA, Traktman P. Biogenesis of the vaccinia virus membrane: genetic and ultrastructural analysis of the contributions of the A14 and A17 proteins. J Virol. 2013;87:1083–1097. doi: 10.1128/JVI.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. 2003;77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Moore DM, Peters PJ, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Meng X, Yan B, Rose L, Deng J, Xiang Y. Vaccinia virus virion membrane biogenesis protein A11 associates with viral membranes in a manner that requires the expression of another membrane biogenesis protein, a6. J Virol. 2012;86:11276–11286. doi: 10.1128/JVI.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Zikos D, Osterrieder N, Tischer BK. Generation of a complete single-gene knockout bacterial artificial chromosome library of cowpoxv irus and identification of its essential genes. J Virol. 2014;88:490–502. doi: 10.1128/JVI.02385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]