Abstract
Membrane fusion is an essential step when enveloped viruses enter cells. Lipid bilayer fusion requires catalysis to overcome a high kinetic barrier; viral fusion proteins are the agents that fulfill this catalytic function. Despite a variety of molecular architectures, these proteins facilitate fusion by essentially the same generic mechanism. Stimulated by a signal associated with arrival at the cell to be infected (e.g., receptor or co-receptor binding, proton binding in an endosome), they undergo a series of conformational changes. A hydrophobic segment (a “fusion loop” or “fusion peptide”) engages the target-cell membrane and collapse of the bridging intermediate thus formed draws the two membranes (virus and cell) together. We know of three structural classes for viral fusion proteins. Structures for both pre- and postfusion conformations of illustrate the beginning and end points of a process that can be probed by single-virion measurements of fusion kinetics.
Keywords: virus entry, fusion mechanism, fusion protein
Introduction
Enveloped viruses require membrane fusion to enter a cell. They expose on their surface many copies of a fusion protein, held in a “prefusion conformation” by constraints that come either from another part of the same protein or from a different viral protein. Two events lead to a fusogenic conformational transition. One (“priming”) makes the transition possible, often by virtue of a proteolytic cleavage; the other (“triggering”) initiates the transition, usually as a result of ligand binding. The ligand can be a proton, in the case of low-pH induced conformational changes (example: influenza virus); it can be a co-receptor on the cell surface or in an internal compartment to which the entering virus traffics (example: HIV); or it can be a distinct protein on the virion surface, itself triggered to signal fusion, often by interaction with the cell-surface receptor for the virus in question (example: paramyxoviruses). Viral fusion proteins are “suicide enzymes”, because they undergo an irreversible priming step and act only once; intracellular fusion proteins, such as the well-known SNAREs, recycle, through the agency of an ATP-dependent protein such as NSF, and priming is therefore reversible.
Why is catalysis of fusion necessary? Although thermodynamically favorable, fusion of two membranes must overcome a kinetic barrier, due to a repulsive “hydration force”, which increases steeply as the distance between the surfaces of the two bilayers falls below 20 Å[1,2]. Because of this barrier, the two membranes require a source of free energy other than thermal fluctuation to bring them closer together than the 20 Å spacing at which the hydration force becomes very strong. Bilayer fusion proceeds through a so-called “hemifusion intermediate”, in which apposed leaflets have merged, but not yet distal ones[3–5]. Considerable evidence supports the picture shown in Fig. 1, for the hemifusion transition and for subsequent fusion pore formation. The productive, hemifused state is simply a narrow stalk, minimizing the area of close contact and hence minimizing the work done to overcome hydration force repulsion. Widening of the stalk into a “hemifusion diaphragm” is probably a kinetic dead end, at least if the diaphragm is more than a few lipid molecules wide[6].
Figure 1.
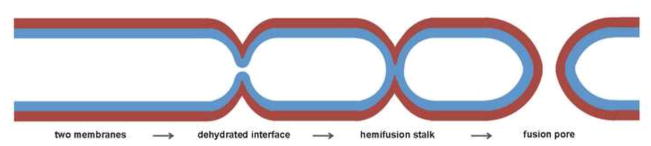
Steps in fusion of two lipid bilayers. Apposed leaflets in blue; distal leaflets in brown.
Viral fusion proteins fall into a small number of structural classes -- three reasonably well characterized ones at the time of this review[7]. The first of these three includes many of the best studied human pathogens, such as influenza virus[8] and HIV-1[9]. The proteins are trimers of a single-chain precursor, which requires a proteolytic cleavage to make it fusogenic. The cleavage, which may simply eliminate a single peptide bond, generates two fragments. The N-terminal fragment, in many cases a receptor-binding domain (e.g., the HA1 fragment of influenza virus hemagglutinin or the gp120 fragment of HIV-1 envelope protein), constrains the C-terminal, fusogenic fragment (e.g., HA2 or gp41), until triggered to release it. The latter bears a hydrophobic “fusion peptide” at or near its newly generated N-terminus and a transmembrane anchor, which holds it in the viral membrane, near its C-terminus.
Most members of the second structural class of fusion proteins -- those found on flaviviruses, alphaviruses, and bunyaviruses -- are in an icosahedrally symmetric array on the mature virion[10–14]. Although the virion of rubella virus is less regular that those of the closely related alphaviruses[15], its fusion protein has a characteristic class II structure[16]. For flaviviruses and alphaviruses, the priming event is cleavage of a second viral surface protein, which is, in effect, a “chaperone” that blocks any response to conditions of triggering[17,18]. When cleavage has inactivated the chaperone, triggering (exposure to reduced pH in case of both flavi- and alphaviruses) induces a reorgnization of the surface lattice and trimerization of the fusion protein. The hydrophobic segment that engages the target membrane during the fusogenic conformational change is an internal “fusion loop”.
Members of the third class of fusion proteins, found on rhabdoviruses (G protein), herpesviruses (gB), and group 1 alphabaculoviruses (gp64) combine certain features of the first two[19]. Herpesvirus gB is part of a larger fusion complex that includes several other proteins; the rhabdovirus G proteins are the sole surface proteins of those viruses. There is no obvious priming event, and most of the conformational transition in G that induces rhabdovirus fusion is reversible[20]. The proteins, which have two spatially adjacent, hydrophobic fusion loops on each subunit, are trimeric in both pre- and postfusion conformations, and they do not form a regular array on the virion surface.
Stuctures and fusogenic structural transitions
Class I: Priming by cleavage of trimeric, single-chain precursor
The classic, and still best characterized, example is influenza virus hemagglutinin (HA) [8,21]. Fig. 2a shows the pre- and post-fusion ectodomains of HA1:HA2, joined schematically to their transmembrane anchors; Fig. 3 shows the presumed sequence of events that links the two conformations. A crucial stage of the interpolated transition is an extended intermediate, in which the fusion peptide at the N-terminus of HA2 has engaged the target membrane, creating a bridge between the two bilayers destined to fuse. Evidence for this intermediate is strong, but indirect. Studies on other class I fusion proteins leave little doubt that a moderately long-lived, extended, so-called “prehairpin” intermediate is a general, on-pathway state.
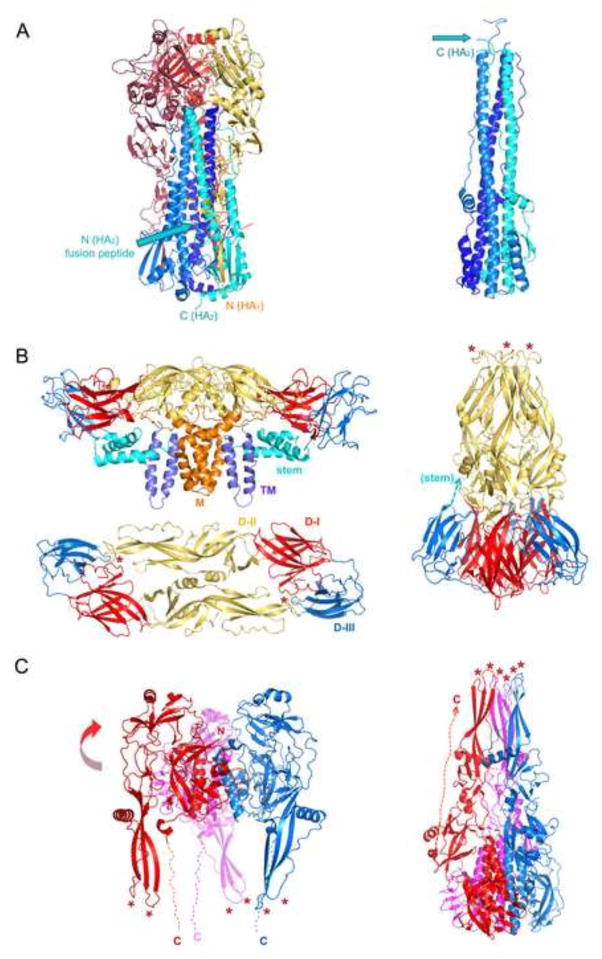
Figure 2.
Pre- and postfusion structures (left and right, respectively) of representative fusion-proteins from each of the three known classes, with approximate positions of membrane shown. A. Class I: influenza virus HA ectodomain (Protein Databank entries 2YPG and 1QU1 for pre- and postfusion forms of the ectodomain, respectively). HA1 chains in shades of red/gold and HA2 chains in shades of blue (paired as red-blue, gold-cyan, dark red-marine blue). The N terminus of HA1 and the C-terminus of the HA2 ectodomain are labeled. Blue arrow: position of fusion peptides inserted near threefold axis in prefusion form. Only HA2 is shown on the right. The N-terminus (blue arrow; note that the fusion peptide is not part of the structure shown) and C-terminus of the cyan-colored subunit are indicated. B. Class II: dengue virus type 2 E protein (3J27, 1OAN and 1OK8). The tangential (“side”) view shows a dimer of the complete E polypeptide chain and the M polypeptide chain from the cryoEM model (3J27); the radial (“top”) view shows just the “stem-less” ectodomain (1OAN). Colors: domain I: red; domain II: yellow; domain III: blue; stem: cyan; transmembrane anchor: slate; M: orange. Colors for domains I, II and III are the same in the postfusion representation. A dashed cyan arrow on the postfusion trimer shows where the stem emerges from domain III. Red asterisks: fusion loops. Class III: VSV G ectodomain (2J6J and 2CMZ). The three chains are in red, blue, and magenta. Dashed lines show the location of a disordered, C-terminal segment that connects the folded protein to the transmembrane anchor. Only the red-subunit C-terminal segment is shown on the right. The curve red arrow indicates that in the transition from the conformation on the left to the conformation one the right, the domain bearing the fusion loops flips over by about 180° to engage the host-cell membrane. Red asterisks: fusion loops. Figure made with Pymol (Schrödinger).
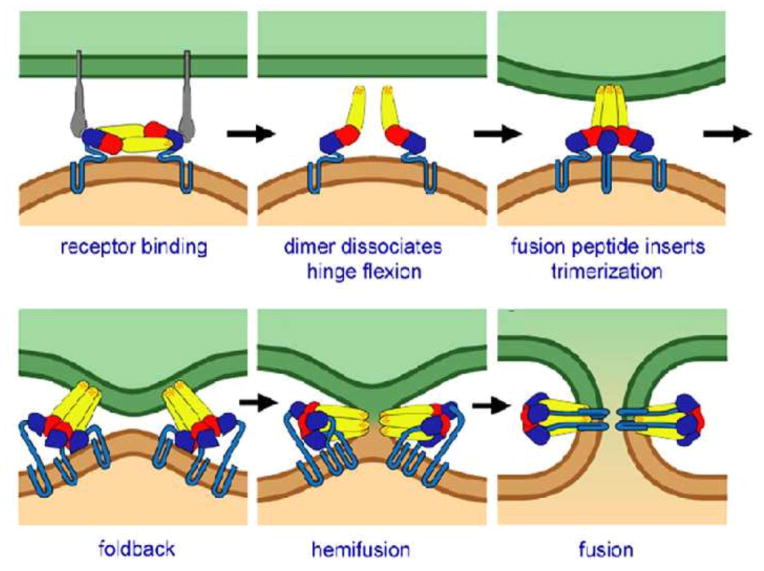
Figure 3.
Schematic diagram of stages of influenza virus fusion. (1) Receptor attachment. (2) Separation of HA1 heads triggered by reduced pH. (3) Extended intermediate. (Only HA2 shown, although HA1 remains tethered to it through a disulfide bond.) Fusion peptides engage the target membrane. Arrows show incipient folding back of the coiled-coil. (4) Hemifusion induced by refolding of HA2. (5) Final refolding steps stabilize nascent fusion pore.
To get from this intermediate to the observed postfusion state, the long central helix breaks, and the segment between the break and the membrane reconfigures so that it runs back along the central coiled-coil, ultimately drawing together the fusion peptide and the C-terminal transmembrane anchor -- along with the two membranes in which they reside[22,23]. An important characteristic, emphasized originally in a model for HIV gp41-mediated fusion, is that the zipping up of the three C-terminal “outer-layer” segments is asymmetric[24]. In none of the known postfusion structures do these segments interact with each other around the outside of the postfusion trimer-- consistent with an asymmetric collapse. Full threefold symmetry is regained at the end of the transition, when a fusion pore has opened and the membrane-proximal parts of the structure have clicked into place[23]. The full transition turns HA2 “inside out”, in the sense that most of the central coiled-coil in the postfusion structure comes from parts of the polypeptide chain that were on the outside of the trimer in the prefusion structure, while the outer part of the overall HA2 postfusion hairpin comes from parts of the polypeptide chain that were on the inside of the trimer before the transition.
Other class I fusion proteins appear to conform, with some variation, to the scheme shown in Fig. 3 for influenza virus HA. Recently determined structures for a prefusion conformation HIV-1 gp120:gp41 envelope protein confirm earlier proposals that its fusogenic conformational change would follow an HA-like sequence[25–28], but the triggering events are more complex. The relatively simple trigger for influenza virus HA is proton binding, leading to a transition when the endosomal pH falls below about 5.5; for the HIV-1 envelope protein, receptor (CD4) binding induces an initial conformation change that exposes a co-receptor binding surface, and only when co-receptor (CXCR4 or CCR5) has engaged the latter does the full transition proceed. The postfusion structure of the gp41 ectodomain trimer is quite simple: a central, three-chain coiled-coil (with the fusion peptide at its N-terminus), a relatively short loop with a conserved disulfide, and an outer layer of three helices (one from each chain), leading to the transmembrane segment[24,29]. This six-helix bundle, defined originally by biophysical experiments on the SIV protein[30] and subsequently by x-ray crystallography, has been a prototype for other six-helix bundle postfusion structures. The HA2 and Ebola GP2 postfusion structures[22,31] both show, however, that the essential characteristic of all these proteins is not six helices, although the central element is usually a coiled-coil, but rather three “hairpins”, to ensure proximity of the membrane-associated elements at each end of the ectodomain polypeptide chain.
The paramyxovirus fusion protein, F, requires cleavage to perform its fusogenic function, but unlike influenza HA0 or HIV gp160, activation at physiological temperature and pH depends on receptor binding by a second viral surface protein, variously known as HN,H or G[32]. Cleavage of F generates a smaller, N-terminal F2 and a larger, C-terminal F1. The latter has a fusion peptide followed by a segment at its N-terminus that becomes the long, central helix of the postfusion coiled-coil and a further, helix-forming segment just before the transmembrane anchor near its C-terminus[33]. The partner protein is a tetramer, with a four-chain coiled-coil stalk and a globular head that closely resemble influenza virus neuraminidase[34–37]. When the head binds receptor, changes in its orientation with respect to the stalk expose surfaces that in turn bind F1, inducing the fusogenic conformational transition[38–42]. Mix-and-match experiments, co-expressing the relevant proteins on the surface of cells and assaying for cell-cell fusion, show that activation is, in general, specific for the autologous receptor-binding protein[43,44].
The Ebola virus envelope glycoprotein, GP, which becomes GP(1) and GP(2) upon furin cleavage, adds further complexity to the sequence in Fig. 3[45]. Although the initial cleavage occurs in the producing cell, generating a trimer of disulfide-linked GP(1):GP(2) subunits[46], fusion activation requires cathepsin-mediated cleavage after uptake into the new host cell, stripping GP(1) of much of its mass[47], and subsequent interaction with a specific, endosome/lysosome-resident receptor, the Niemann-Pick C1 (NPC1) protein, a cholesterol transporter[48,49]. This last interaction may be the fusion trigger. The postfusion conformation of GP(2) is a trimer of hairpins, formally analogous to the structure of other class I fusion proteins, but the fusion peptide is not directly at the N-terminus of GP(2)[31].
During the fusion reaction for the various groups of viruses with class I fusion proteins, specific steps differ, both in intracellular location and in biochemical character, but current data appear to be consistent with the following general picture. A required priming step is specific cleavage of a trimeric precursor protein, often by a furin-like activity in the trans-Golgi network (TGN) of the producing cell or on its surface, but in some cases by a soluble extracellular protease (which may cleave subunits left intact during passage through the TGN) or potentially by an endosomal protease in the target cell. The triggering step may be as simple as proton binding or as complex as interaction with receptor-activated partner proteins or with an intracellular receptor, following further proteolytic processing. The trigger releases constraints imposed by the N-terminal fragment on the C-terminal, fusogenic fragment. The latter then undergoes a series of fusion-inducing conformational changes, passing through an extended trimeric intermediate that bridges the two fusing membranes and progressing to a final state in which the N-terminal (or N-proximal) fusion peptides and the C-terminal transmembrane anchors are all at one end of a trimer of hairpin-configured subunits.
Confusion sometimes attends the use of the word “metastable” in connection with the conformational changes just described. The uncleaved precursor is stable -- its conformation, achieved on folding in the endoplasmic reticulum (ER), is a free energy minimum. (This statement is not necessarily true of the ectodomain alone -- the evolved free-energy minimum includes anchoring in the membrane.) Cleavage renders the protein metastable, because now the minimum free energy state of the system is different -- it is the rearranged, trimeric, C-terminal fragment in its postfusion state, plus the three released N-terminal fragment (or effectively released if, as is the case with influenza virus HA, there remains a disulfide tether). In the absence of coupling to a partner membrane, the C-terminal fragment will simply invert, inserting its fusion peptides into the viral membrane.
Comparison of unprimed and primed influenza virus HA in the prefusion conformation shows that cleavage (priming) produces only a very local structural change[50]. The hydrophobic N-terminus of HA2 (the fusion peptide), exposed in a loop on HA0, tucks into a pocket along the threefold axis of the HA trimer, so that he three fusion peptides in the trimer contact each other[21]. The cleaved and uncleaved conformations of a paramyxovirus F protein are likewise essentially the same[51].
Class II: Priming by cleavage of heterodimeric partner protein (“chaperone”)
Flaviviruses and alphaviruses have icosahedrally symmetric outer coats, which conceal an underlying membrane[12–14,52–54]. The viral fusion protein, designated E for flaviviruses and E1 for alphaviruses, is in 1:1 association with a second protein (prM and pE2, respectively) -- a chaperone that prevents the fusogenic conformational change. The chaperone and fusion protein, sequential in the polyprotein precursors encoded by these plus-sense RNA viruses, are cleaved from each other during synthesis in the ER by signal protease. Further cleavage of the chaperone, by furin in the trans-Golgi network (TGN), allows the fusion protein to undergo a low-pH induced, fusogenic transition when the particle arrives in an acidified endosome of a new host cell[17,18].
E and E1 have essentially the same folded structure[12,55]. A central, beta-sandwich domain (“domain I”) organizes their common, three-domain fold (Fig. 2b). Domain II, formed by two long, disulfide-stabilized loops that emanate from the central domain I, bears at its tip a hydrophobic “fusion loop”. Domain III, an Ig-like structure that probably recognizes cellular receptors, terminates in a segment called the “stem”, which connects it with the transmembrane anchor. The stem of alphavirus E1 is relatively short; the longer stem of flavivirus E forms two amphipathic helices, submerged in the outer leaflet of the lipid bilayer[56].
Unlike the fusion proteins, the chaperone proteins in the two groups of viruses, prM and pE2, have unrelated overall structures[57–59]. Nonetheless, both protect the fusion peptide in the immature form of the particle and both extend along the surface of the fusion protein, so that the C-terminal, membrane anchors of the chaperone and fusion proteins are adjacent. In addition to covering the E1 fusion loop, alphavirus pE2 has a receptor-binding domain, which bears apparent homology with domain III of flavivirus E.
Flaviviruses assemble by budding into the ER and mature by passage through the Golgi and the trans-Golgi network (TGN). Immature flavivirus particles, recovered from cells by inhibiting furin cleavage, have a “spikey” appearance at neutral pH, with the 180 prM:E heterodimers clustered into 60 projecting trimers[60]. At the reduced pH of the TGN (about 5.7), the E subunits rearrange into a collapsed, herringbone-like packing indistinguishable from the packing of E on mature virions, but with prM associated[61]. The reorganization exposes the furin sensitive site on prM, allowing cleavage to pr+M. At pH<6, the pr fragment, which covers the fusion loop, remains in place. Only when the pH returns to neutral -- i.e., when the particle emerges from the cell -- does pr dissociate, allowing fusion to take place whenever the particle again experiences a low-pH environment.
Alphaviruses assemble by budding at the plasma membrane. Thus, the processing of the glycoproteins occurs before, rather than after, their incorporation into particles. Cleavage of pE2 to E3+E2 yields an E1:E2:E3 product, from which E3 dissociates in some cases and not in others. The budded particle has 240 E1:E2 or E1:E2:E3 subunits, organized into the (T=4) icosahedral lattice as trimeric “spikes”, in which E2 mediates the threefold contact, surrounded by E1[58,59].
In their postfusion conformation, class II fusion proteins are trimers [62,63] (Fig. 2b). To undergo a fusogenic conformational change, the flavivirus E proteins, packed as homodimers in the virion surface, must therefore dissociate into monomers and rearrange; the alphavirus E1 proteins, packed as heterodimers on the periphery of the E1:E2 trimeric spikes, must likewise dissociate and rearrange. Fig. 4 shows a proposed scheme for the fusogenic conformational change of a flavivirus E protein and for the relationship between the conformational transition and changes in the two fusing membranes. As in the scheme described above for influenza virus HA, there is an extended intermediate, with the fusion loop at the tip of domain II inserted into the target membrane. The refolding of each subunit in the trimer leads to a structure in which the fusion loops and the transmembrane anchors are adjacent -- essentially the equivalent of the hairpin-like final conformation of class I fusion proteins.
Figure 4.
Schematic diagram of stages of dengue virus fusion (modified from [62]).
Bunyaviruses have two surface glycoproteins, GN and GC, arrayed in T=12 icosahedral lattice. The structure of GC from Rift Valley fever virus (RVFV)[64] confirms earlier suggestions that it is a class II fusion protein[65]. When it crystallizes, RVFV GC forms dimers very similar to those of flavivirus E. The homology suggests that GN is a chaperone of some kind, but their heterodimeric relationship and their packing in the surface lattice of the virion are not yet clear.
The genome organization of pestiviruses and hepaciviruses relate them to flaviviruses and somewhat more closely to each other, and it had been assumed until structures appeared that the proteins designated E1 and E2 might resemble prM and E, respectively. The structure of bovine diarrhea virus E2 is not a class II fusion protein however, nor does it have an obvious fusion loop[66,67], and hepatitis C virus (HCV) E2 resembles neither class II fusion proteins nor pestivirus E2[68]. A potential commonality is that E1 and E2 may act together as a fusogen, with E1 providing the hydrophobic segment that engages the target membrane.
Class III: Triggering but no priming
The G-protein of VSV[69], gB of herpes simplex virus[70] and Epstein-Barr virus (EBV)[60], and gp64 of the insect-cell baculovirus[71] define a third class of viral fusion proteins. Like those of class II (and unlike those of class I), the folded structures of all known members are similar. Most of the ectodomain is a set of successively embedded domains, organized so that a subunit exposes the two fusion loops, each at the tip of a β-hairpin. Only for VSV G do we know the structures of both pre- and postfusion forms (Fig. 2f) [69,72]. The two interconvert as the pH is lowered or raised, with a transition around pH6.5. Reversibility is the thermodynamic counterpart of the absence of cleavage (or any other irreversible modification), either of the fusion protein itself or of a co-folded chaperone such as prM or pE2.
The relatively complex domain organization of class III proteins presents a “fusion module” (domain I) with two long β-hairpins bearing hydrophobic loops at their tips[19]. One can consider the most of the ectodomain as a β-sandwich core module (domain IV) with a succession of nested inserts (domains III, II and I) and a very long C-terminal extension (sometimes called domain V). The transition between pre- and post-fusion configurations is a rotation of domains I and II with respect to domain IV and some refolding of domain III. The transition resembles influenza-virus HA2 refolding in some respects: an extension of the central coiled-coil correlates with the rotation of domains I and II, and folding back of the C-terminal segment brings together the transmembrane segment and the domain-I hydrophobic fusion loops. Thus, the “trimer of hairpins” description applies to the postfusion form of these proteins, as it does to those of the other two classes.
The ectodomain of VSV-G is monomeric in solution at pH>7.5[73], but trimeric in the crystal structure -- probably because of the very high effective protein concentration at which the crystals grew. Anchored in the virion membrane at neutral pH, the full-length molecule may be in an equilibrium between monomer and trimer. A pH<6.5, a postfusion trimer is the stable conformation. A short segment (about two heptads) of the central coiled-coil is present in both the prefusion and postfusion trimers, but inspection of the structures, analysis of protein conformational equilibria in solution, and electron microscopy of virions all suggest the relevance of a monomeric, extended intermediate, in which domains I and II have rotated to present the fusion loops to the apposing target membrane and in which domain III has undergone some of its conformational reorganization. Association of soluble ectodomain with liposomes at low pH is reversible if the pH returns to neutral[73].
VSV-G is the only protein on the virion surface, and its fusion activity requires no cofactors in vitro other than protons (low pH). The herpesvirus fusion machinery is more complex: in addition to gB, fusion requires the heterodimeric gH/gL and receptor or co-receptor binding, usually by yet a fourth protein (gD of HSV, gp42 of EBV, etc.). Structures of these triggering factors have shown how they respond to receptor binding, but not yet how that signal transfers to gB[74–79].
Coupling protein conformational change with lipid-bilayer fusion
The pre- and postfusion structures of a viral fusion protein tell us about the configuration of a fusion catalyst before and after the event, but do not show directly how the catalyst facilitates the event itself. The one universal feature of all the postfusion structures, regardless of their other characteristics, is spatial proximity of the two membrane-associated elements -- the fusion loop(s) or peptide and the transmembrane anchor. This adjacency is probably the strongest evidence that a triggered conformational change in the protein overcomes the hydration-force barrier by coupling release of free energy from refolding to the spatial separation of the two fusing membranes. Together with related studies of SNARE-mediated fusion, the structures leave little doubt that this essentially mechanochemical description is correct.
Bilayer perturbations
Coupling refolding to close approach of two bilayers need not be the only contribution that fusion proteins make to facilitating membrane merger. Perturbations in the target bilayer, caused by insertion of a fusion loop or fusion peptide, could enhance the likelihood of a transition to hemifusion in the apposed bit of membrane surface (the “dehydration” to “stalk” step, Fig. 1), for example by stabilizing curvature. Experimental evidence for this frequently invoked possibility remains indirect. Moreover, we know from studies of non-protein fusogens that hemifusion stalk formation can proceed without such contributions, if the two membranes approach each other closely enough. The mechanism of fusion induced by polyethylene glycol is desolvation of the inter-membrane space -- effectively what fusion proteins do when they force two bilayers together -- without any apparent requirement for lipid-headgroup interaction or other disruption of local membrane organization[80–82].
We can have some idea of potential contributions to membrane curvature by considering what we know about fusion-peptide or fusion-loop insertion. The conformation of the 23-residue influenza virus HA fusion peptide bound on a dodecyl-phosphatidylcholine micelle is a hairpin of two tightly packed, antiparallel α-helices[83]. Hydrophobic residues project into the micelle and somewhat more polar residues, including two glutamic acids, project away from it. If the same orientation holds in the outer leaflet of a lipid bilayer, three HA trimers (see below), contributing a total of nine such hairpins to a nascent hemifusion stalk, would displace (laterally) about 3500 Å2 of lipid, less than 10% of the area of even a very sharp hemispherical cap and a small part of the difference between the areas of the inner and outer leaflets. The flavivirus E-protein fusion loops likewise insert only into the proximal membrane leaflet, occupying a similar total area. Thus, in both cases, the contribution of partial protein insertion to membrane curvature in the transition state is probably modest. At the hemifusion stalk stage, however, the HA fusion peptides from three trimers could occupy as much as 30% of the total lateral area (if they were to migrate into it from the target membrane) and hence affect either the transition to this intermediate or the transition out of it.
Many viral fusion proteins have hydrophobic “membrane proximal regions” at the junction between ectodomain and transmembrane anchor. The “MPER” of HIV gp41 and the membrane-proximal segment of the flavivirus stem are good examples. In both of those cases, experimental data suggest a change in conformation or exposure of the membrane proximal region during the fusogenic transition[56,62,84]. The effect of this change on the viral membrane could in principle accelerate fusion, but we do not yet have a structure for the postfusion membrane proximal region of a viral fusion protein, and only for the flaviviruses (dengue) do we know its prefusion structure[52,56].
Extended protein intermediates
Insertion of the fusion loop or peptide into the target membrane, demonstrated for several viral fusion proteins by photochemical crosslinking experiments, necessarily implies that some conformational intermediate bridge from one membrane to the other. Because of the “inside-out” character of the influenza virus HA2 conformational change (Fig. 3), the extended inner helical bundle must form before the outer layer can surround it. That extension in turn projects the fusion peptide, initially tucked into a cavity along the axis of the prefusion HA2, toward the target membrane and exposes it for insertion. A similar argument applies to HIV gp41. Moreover, inhibition of the gp41 mediated fusion by peptides representing the outer layer of the postfusion six-helix bundle (e.g., the T-20 peptide, developed as a drug, entfuvirtide or Fuzeon) or by reagents designed to represent the inner helical bundle, shows directly that the inner bundle directs the zipping up of the outer-layer helices and that “extended” is a reasonable description of the bridging intermediate[85]. An engineered trimer, designed to represent the gp41 extended intermediate based on the known final structure, has properties consistent with its intended mimicry[84].
Flavivirus E proteins must hinge outwards to present their fusion loops to the target membrane. Considerable evidence, both direct observation by electron microscopy and inference from properties of particles in solution, confirms this expectation[86,87].
Fusion dynamics
Hemifusion intermediate
The key intermediate for the bilayer comes when the protein has nearly completed its conformational rearrangement. The notion of a hemifusion stalk emerged from estimates of membrane distortability, considering the bilayer as a continuous elastic medium, with more direct experimental evidence subsequently coming from x-ray crystallographic studies of lipid phase transitions[3]. In kinetic studies, hemifusion can be detected by transfer of a marker such as a fluorescent dye from one membrane to the other, without concomitant transfer of a different marker from the interior of one fusing compartment (e.g., a liposome) to a common, fused interior.
Influenza virus HA-mediated fusion gets “stuck” at the hemifusion stage, if the protein is anchored by a lipid tail instead of a transmembrane helix[88]. Moreover, truncation of the transmembrane anchor (and deletion of the short interior tail) so that it no longer emerges from the inner bilayer leaflet likewise stalls the low-pH triggered reaction at the hemifusion step[89]. That is, the presence of some hydrophilic protein segment, even just one amino-acid residue, on the far side of the membrane appears greatly to accelerate fusion pore opening. Evidence that a fusion pore can flicker open and closed from the hemifusion stalk configuration[90] then suggests that influenza virus HA accelerates fusion not only by overcoming the hydration force barrier to hemifusion but also by trapping the fusion pore in an open state -- i.e., by reducing the kinetic barrier that separates hemifusion from full fusion. One plausible mechanism can be imagined by inspection of Fig. 3. Separation of the two interior compartments, even at the hemifusion stalk stage, prevents completion of ectodomain “zippering”, because the hydrophilic tail cannot move away from its side of the membrane. If that tail can move into an open pore, it will allow the zippering transition to finish, creating a barrier to any reversal and thus inhibiting reclosure of the pore, which will instead widen irreversibly and establish full continuity of the two previously separate, membrane-bound compartments. Interaction between the membrane proximal segment of the flavivirus E-protein stem and the fusion loops of the trimer at a late stage in the fusogenic transition might provide a similar driving force[91–93].
Catalytic mechanism
Structure-based analyses of fusion kinetics probe how fusion proteins accelerate membrane merger, just as experiments on enzyme kinetics probe how enzymes catalyze chemical reactions. Observations recorded in a single-virion format yield not just the average rate of a process for which one has a suitable reporter, but also how that rate varies from particle to particle. From the distribution of rates one can extract information about the number of independent events required for the process being studied. The effects on the rate distributions of mutations introduced into the fusion protein then connect the mechanism with fusion-protein structure.
Single-virion fusion studies of influenza-virus fusion with receptor-containing target membranes confirm that hemifusion (detected by dequenching of a fluorophore incorporated into the viral membrane) precedes fusion (detected by loss of a soluble fluorophore from the virion interior)[94]. Three independent, rate-limiting molecular events, interpreted as membrane-coupled conformational rearrangements in three HA trimers, contribute to the hemifusion step; one rate-limiting step governs the transition from hemifusion to fusion-pore opening. Effects of mutations that affect interactions of the fusion peptide show that (for the strains and mutational variants studied) expulsion of the fusion peptide from its pocket near the threefold axis determines the rate at which an extended intermediate forms[95]. The mutual contacts among the three fusion peptides should make this step a single, cooperative transition for each trimer.
A dense array of HA covers the surface of an influenza virus particle, and the contact zone between a virus particle and a target membrane will contain many more than three HA trimers[96]. When exposed to low pH (in laboratory experiments, by an abrupt pH drop; in a cell, by acidification of the endosome that contains the virion), individual HA trimers have a pH-dependent likelihood of exposing their fusion peptides, engaging the target membrane, and transitioning to the membrane-bridging, extended intermediate conformation. Experimental data and corresponding simulations show that hemifusion requires three adjacent trimers to extend in this way, so that their collapse to the stable postfusion conformation can drive membrane deformation[95]. Thus, within a contact zone of 100 HA trimers (about the expected number for a typical, slightly elongated influenza-virus particle produced by mammalian cells in culture), substantially more than three will have undergone the transition to extended intermediate before three neighbors have done so, but the presence of non-bridging trimers between them will prevent any concerted action. A single trimer, or two adjacent, extended trimers cannot collapse and bring about hemifusion, because the elastic properties of the two membranes resist their pull. Only when they are joined by a third trimer is the free energy recovered by the protein conformational change great enough to overcome the resistance of the membranes. At that point, the transition to hemifusion is rapid. Cooperativity is a consequence of the common insertion of the three neighboring HAs in the target membrane at one end and in the viral membrane at the other. It does not require specific trimer-trimer contacts.
Can three HA trimers generate a force sufficient to surmount the kinetic barrier to hemifusion, generally estimated to exceed 50 kcal/mol? Since the restoring force exerted by the two membranes resists collapse of a single HA from an extended intermediate to a trimer of hairpins, the pull from that collapse must not be so strong that it causes the fusion peptide to withdraw from the target membrane. The binding free energy for association of a single HA fusion peptide with a lipid bilayer is about 8 kcal/mol[97], so the pull on three fusion peptides from collapse of a trimer should not exceed 24 kcal/mol. Three HA trimers should therefore be just enough to overcome a 50–70 kcal/mol barrier.
Single-particle kinetic studies of flavivirus fusion yield a qualitatively similar picture[86]. E dimers dissociate, allowing the subunits to project outwards from the virion surface and to expose the fusion loop at the tip of domain II. This step is fast and reversible. The rate-limiting molecular step is trimerization, which depends in turn on the effective surface concentration of activated (i.e., outward projecting) monomers in the contact zone between the virus particle and the membrane with which it is fusing. The hemifusion rate then depends, as it does with influenza HA mediated fusion, on reaching a threshold number of adjacent trimers. The critical number of trimers for West Nile virus, as determined with virus-like particles, and for its close relative, Kunjin virus, is 2. The rate of hemifusion does not appear to depend on the overall curvature of the viral membrane, as it is the same for virus-like particles of two different diameters[86]..
The mechanisms just described include long-lived, extended intermediates, “waiting” for one or more extended neighbors. At 20–25°C, the waiting time for a typical extended intermediate is about a ~30–60 s for both influenza and West Nile viruses. Potentially much longer delays have been ascribed to the extended intermediate in HIV fusion, based on the time (ca. 15 min) between contact and acquisition of insensitivity to peptide inhibitors that target the prehairpin structure[98]. That interval is an upper limit, however, as it includes the time between receptor binding and target- membrane engagement of the extended (“prehairpin”) gp41. Moreover, the reported measurements involved cell-cell fusion, rather than virion-cell fusion, and hence a heterogeneous contact zone. If, like exposure of the HA fusion peptide, a slow step precedes fusion-peptide insertion into the target membrane, the extended-state lifetime may be considerably less than the time between binding and collapse (assumed to be the event that occludes the inhibitor binding surface). HIV particles have very few envelope trimers[99], and an alternative explanation for a very long-lived intermediate might be a requirement that two (or more) extended gp41 trimers come within some critical distance of each other to exert cooperative pull on the membranes they bridge. Then further steps, such as association of the neighboring envelope spikes with receptor and co-receptor, to trigger their activation and extension, would increase the delay between initial encounter and hemifusion. Finally, if two (or more) extended gp41 trimers are rarely close enough to cooperate and if a single trimer contributes 25–30 kcal/mol to overcoming a 50 kcal/mol barrier to hemifusion, the remaining barrier of 20–25 kcal/mol will imply delay times of a minute or longer. The infectivity of particles that display a mixture of wild-type and cleavage defective Env indeed suggests that HIV fusion may require only one fusogenic spike[100].
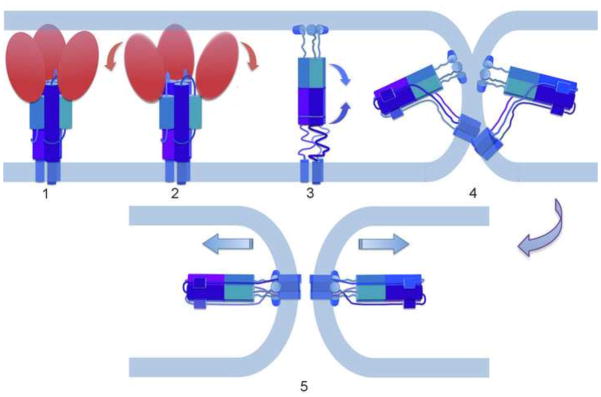
The SNARE protein complexes that catalyze synaptic-vesicle fusion pause in the “zippering” step when the two membranes are in contact, stalled by a specific component of the complex[101]. Binding of Ca2+ ions, the triggering signal, releases this inhibition, allowing rapid, synchronized fusion of docked vesicles. Although docking appears to bring the vesicle membrane into very close contact with the target membrane, it does not induce hemifusion[6]. The millisecond delay between triggering and release suggests, however, that the stalled zippering has already overcome most of the hydration-force barrier -- i.e., that the reaction has stalled close to the transition state. The diagrams in Fig. 5 compare fusion catalysis by influenza HA, flavivirus E and SNARE complexes, as a way to suggest their essential common features. Like serine proteases with unrelated three-dimensional structures (e.g., elastase and subtilisin), the essence of catalysis and the nature of the transition state is the same, despite the quite different large-scale molecular architectures of the catalysts.
Figure 5.
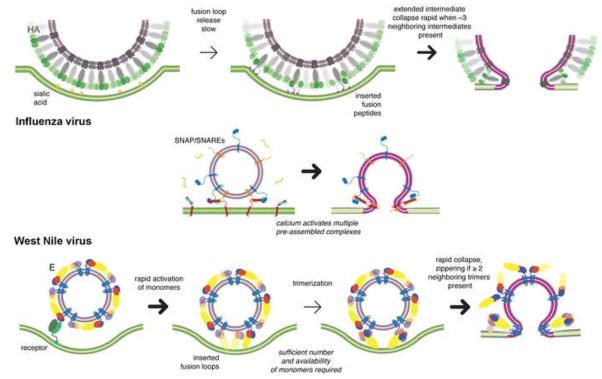
Comparison of fusion as catalyzed by influenza virus HA, SNAREs, flavivirus E protein (modified from [83]).
Highlights (Harrison review for 60th anniversary issue).
Viral fusion proteins overcome the high energy barrier to lipid bilayer merger
Different molecular structures but the same catalytic mechanism.
Review describes properties of three known fusion-protein structural classes
Single-virion fusion experiments elucidate mechanism
Acknowledgments
The author thanks members of his research group, in particular Luke Chao and Tijana Ivanovic, whose work shaped the point of view described in this review, and acknowledges support of NIH grants CA13202, AI057159, and AI109740. The author is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parsegian VA, Fuller N, Rand RP. Measured work of deformation and repulsion of lecithin bilayers. Proc Natl Acad Sci U S A. 1979;76:2750–2754. doi: 10.1073/pnas.76.6.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rand RP, Parsegian VA. Physical force considerations in model and biological membranes. Can J Biochem Cell Biol. 1984;62:752–759. doi: 10.1139/o84-097. [DOI] [PubMed] [Google Scholar]
- 3.Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci U S A. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Huang HW. Observation of a membrane fusion intermediate structure. Science. 2002;297:1877–1879. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 5.Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diao J, Grob P, Cipriano DJ, Kyoung M, Zhang Y, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. Elife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 9.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 10.von Bonsdorff CH, Harrison SC. Sindbis virus glycoproteins form a regular icosahedral surface lattice. J Virol. 1975;16:141–145. doi: 10.1128/jvi.16.1.141-145.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Bonsdorff CH, Pettersson R. Surface structure of Uukuniemi virus. J Virol. 1975;16:1296–1307. doi: 10.1128/jvi.16.5.1296-1307.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, et al. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, et al. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battisti AJ, Yoder JD, Plevka P, Winkler DC, Prasad VM, et al. Cryo-electron tomography of rubella virus. J Virol. 2012;86:11078–11085. doi: 10.1128/JVI.01390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuBois RM, Vaney MC, Tortorici MA, Kurdi RA, Barba-Spaeth G, et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- 17.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72 ( Pt 6):1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- 19.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roche S, Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 21.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 22.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci U S A. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 25.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 30.Blacklow SC, Lu M, Kim PS. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 31.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 32.Jardetzky TS, Lamb RA. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol. 2014;5:24–33. doi: 10.1016/j.coviro.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 35.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 37.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 38.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, et al. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, et al. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A. 2012;109:E2625–2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, et al. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol. 2013;87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, et al. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 2013;9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu XL, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White JM, Schornberg KL. A new player in the puzzle of filovirus entry. Nat Rev Microbiol. 2012;10:317–322. doi: 10.1038/nrmicro2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cote M, Misasi J, Ren T, Bruchez A, Lee K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 51.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, et al. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure. 2006;14:75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Ge P, Yu X, Brannan JM, Bi G, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 58.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 59.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, et al. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 62.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 63.Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 64.Dessau M, Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc Natl Acad Sci U S A. 2013;110:1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garry CE, Garry RF. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes) Theor Biol Med Model. 2004;1:10. doi: 10.1186/1742-4682-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Wang J, Kanai R, Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc Natl Acad Sci U S A. 2013;110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 70.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 71.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 72.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 73.Albertini AA, Merigoux C, Libersou S, Madiona K, Bressanelli S, et al. Characterization of monomeric intermediates during VSV glycoprotein structural transition. PLoS Pathog. 2012;8:e1002556. doi: 10.1371/journal.ppat.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 75.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, et al. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure. 2009;17:223–233. doi: 10.1016/j.str.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci U S A. 2010;107:22641–22646. doi: 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sathiyamoorthy K, Jiang J, Hu YX, Rowe CL, Mohl BS, et al. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog. 2014;10:e1004309. doi: 10.1371/journal.ppat.1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evans E, Metcalfe M. Free energy potential for aggregation of mixed phosphatidylcholine/phosphatidylserine lipid vesicles in glucose polymer (dextran) solutions. Biophys J. 1984;45:715–720. doi: 10.1016/S0006-3495(84)84213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evans E, Needham D. Attraction between Lipid Bilayer Membranes in Concentrated Solutions of Nonadsorbing Polymers: Comparison of Mean-Field Theory with Measurements of Adhesion Energy. Macromolecules. 1988;21:1822–1831. [Google Scholar]
- 82.Kuhl T, Guo Y, Alderfer JL, Berman AD, Leckband D, et al. Direct Measurement of Polyethylene Glycol Induced Depletion Attraction between Lipid Bilayers. Lagmuir. 1996;12:3003–3014. [Google Scholar]
- 83.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci U S A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weinstock MT, Francis JN, Redman JS, Kay MS. Protease-resistant peptide design-empowering nature’s fragile warriors against HIV. Biopolymers. 2012;98:431–442. doi: 10.1002/bip.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chao LH, Klein DE, Schmidt AG, Pena JM, Harrison SC. Sequential conformational rearrangements in flavivirus membrane fusion. Elife. 2014:4. doi: 10.7554/eLife.04389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Sheng J, Austin SK, Hoornweg TE, Smit JM, et al. Structure of Acidic pH Dengue Virus Showing the Fusogenic Glycoprotein Trimers. J Virol. 2015;89:743–750. doi: 10.1128/JVI.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 89.Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chanturiya A, Chernomordik LV, Zimmerberg J. Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc Natl Acad Sci U S A. 1997;94:14423–14428. doi: 10.1073/pnas.94.26.14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of flavivirus entry derived from the E protein stem. J Virol. 2010;84:12549–12554. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6:e1000851. doi: 10.1371/journal.ppat.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klein DE, Choi JL, Harrison SC. Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol. 2013;87:2287–2293. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci U S A. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. Elife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Han X, Lai AL, Bushweller JH, Cafiso DS, et al. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 100.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J Virol. 2006;80:4388–4395. doi: 10.1128/JVI.80.9.4388-4395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]