Abstract
DNA packaging into empty viral procapsids by ATP-driven motor proteins applies widely among viruses. Recent fluorescence studies of phage T4 reveal: 1) the small terminase subunit (TerS) synapses pac homologs by a two ring mechanism to gauge DNA maturation and allow packaging by the large terminase subunit (TerL); 2) translocation of linear DNA is efficient by TerL acting alone; expansion of the procapsid is controlled by the portal-terminase assembly; 3) both ends of the packaged DNA are held at the portal, showing a loop of DNA is packaged; 4) transient spring-like compression of B form to A form-like DNA accompanies translocation; 5) the C-terminal domain of TerL is docked to the portal and moves toward it when stalled; 6) a portal bound resolvase can release stalled Y-DNA compression and allow translocation in vitro; and 7) ATP powered translocation on A form dsDNA is supported by recent hexameric helicase studies.
Following the early demonstration (using DNA packaging essential gene 49:EndoVII (Holliday junction resolvase) mutants) that prohead filling could be completed in bacteriophage T4 infected bacteria (Luftig et al., 1971), diverse mechanisms were proposed to explain prohead filling to high DNA density. Among others, DNA replication, prohead expansion, DNA binding to prohead internal proteins, and scaffold-core protein proteolysis and exit were proposed to drive packaging energetically. Despite attractive features of coupling DNA packaging to these aspects of phage development in vivo (Fig. 1), these proposed mechanisms were shown not to apply. For example, the phage T4 internal proteins found with the DNA in the filled bacteriophage capsid are dispensable for packaging, and, as generally found to be the case where tested, these packaged proteins enter the prohead before the DNA. Most are then ejected along with the DNA into the host for early roles in infection; surprisingly, large and multimeric proteins packaged into the early precursor prohead scaffold or core can pass through a narrow diameter portal and tail tube channel into the host (Black and Thomas, 2012).
Fig. 1.
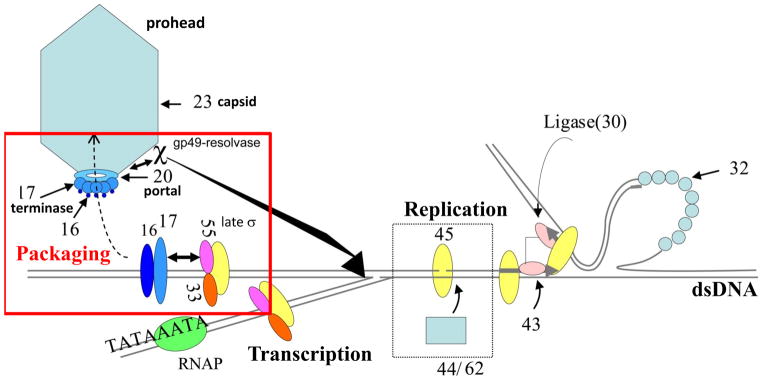
Regulatory integration in vivo of packaging, late transcription, and replication among T4-type phages. The phage T4 terminase large subunit ATPase motor protein gp17 (TerL) and the small terminase DNA recognition protein gp16 (TerS) initiate packaging. A gp20 portal bound packaging essential gp49:Endonuclease VII resolvase removes X and Y structures blocking packaging on a branched DNA concatemer in vivo. The gp55 late sigma factor interacts with TerL and appears to be essential for initiating concatemer packaging.
Bacteriophage T4 was the first phage shown to package its DNA by a terminase motor docked to a procapsid portal. Early structural studies revealed a unique vertex ring dodecamer structure that could be extracted from the prohead (Muller-Salamin et al., 1977). Order of function studies (Jarvik and Botstein, 1973) showed that the T4 portal had an early essential function in assembling an empty prohead and then a late essential function in packaging coupled to the terminase-ATPase motor (Hsiao and Black, 1977). A torsional DNA compression terminase-portal motor was proposed to use gyrase-like DNA supercoiling to drive packaging by energizing the DNA (Black and Silverman, 1978). An alternative early hypothetical portal-centric mechanism based on a symmetry mismatch at the portal vertex proposed a rotary portal motor; this model was long favored, possibly in part as rotary motor mechanisms were in fact demonstrated for ATP synthase and the bacterial flagella motor (Hendrix, 1978; Simpson et al., 2000). Only comparatively recently has rotation of the portal been shown not to be a feature of the motor mechanism in T4 (Baumann et al., 2006) (Fig. 2) or in φ29 (Hugel et al., 2007). Instead two linear terminase motor mechanisms for the T4 packaging motor were proposed simultaneously (Oram et al., 2008; Sun et al., 2008).
Fig. 2.
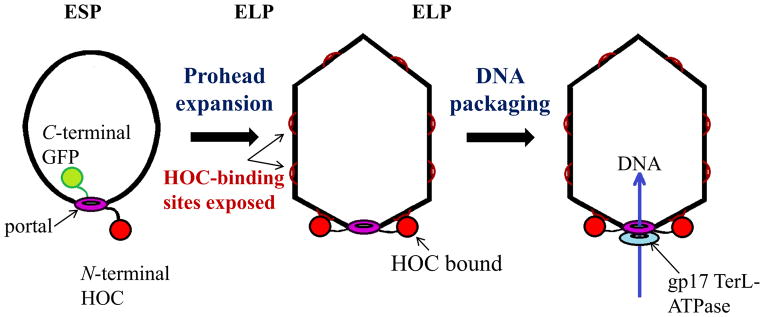
Evidence against a proposed prohead portal rotory motor mechanism. No rotation of six C-terminal portal-GFPs found as part of the portal dodecamer inside filled 500 mg/ml DNA active phages; no rotation of N-terminal HOC-gp20 portal protein tethered to HOC binding sites on expanded mature in vitro packaging active proheads. ESPs are immature empty small proheads, ELPs are mature and stable empty large proheads with HOC decoration protein binding sites exposed (Baumann et al., 2006).
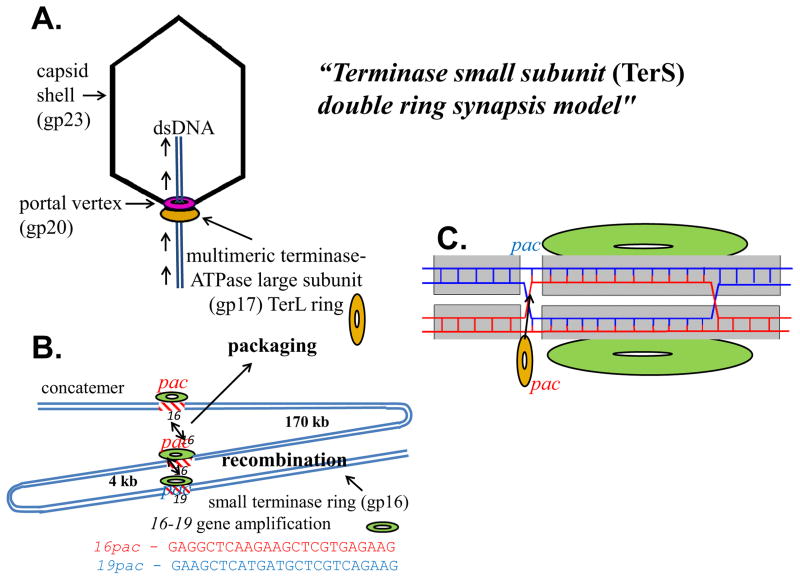
Many years of experimental study have supported the basic similarity of phage and viral DNA packaging motor mechanisms. The similar high resolution structures of proheads, portals, and large terminase subunits (TerL) support this view. Packaging is regulated differently in different phages; e.g. bacteriophage T4 packaging in vivo is connected to the DNA sliding clamp-late sigma gp55 complex (Black and Peng, 2006; Malys et al., 2002), tying it to both late transcription (Geiduschek et al., 1997) and replication. This interaction likely promotes TerL binding to the concatemeric DNA or to DNA repair, and this role may be similar to the essential packaging role of T7 and T3 RNAPs (Fig. 1). However T4 terminase small subunit gp16 (TerS) is essential in vitro and in vivo for packaging of circular or concatemeric DNA (i.e., DNA with no or few ends), the latter being the in vivo substrate that is subject to complex regulation of packaging initiation among pac site phages (Fig. 3) (Black and Peng, 2006). In pac site phages the TerS is required to initiate packaging at specific pac sites on concatemeric DNA by “handing off” the DNA to the large nuclease-containing terminase subunit (TerL) for packaging initiation cutting. In contrast, T4 TerL acting alone can package in vitro linear DNA with high efficiency.
Fig. 3.
Conserved bacteriophage T4 two subunit terminase pac site prohead packaging; (A) large terminase-ATPase subunit TerL alone packages linear DNAs in vitro; (B) small terminase subunit TerS (gp16) is required in vivo for concatemer (or circular DNA) packaging; a “synapsis model” proposes DNA concatemer maturation is assessed by TerS apposing two homologous pac site DNAs. Sequence specific terminase gene amplifications requiring TerS result from apposition of two homologous pac sites (16 and 19) under genetic selection for increased TerL synthesis; normally TerS gene 16–16 pac apposition would initiate packaging; and (C) a two ring two dsDNA TerS Holliday junction strand swap signal is proposed to initiate handoff to TerL for DNA cutting and packaging.
Substantial evidence supports a double terS protein ring double DNA pac site “synapsis model” as a mechanism to assess DNA concatemer maturation (Black, 1995)(Fig. 3). There is both genetic and biochemical evidence for this proposed mechanism for initiation of phage T4 packaging: i) purified TerS displays enhanced binding to a GC-rich sequence at the 3′ end of its gene that was identified as a pac site (Lin et al., 1997): ii) this sequence confers enhanced transduction of phage and plasmid DNAs containing it in vivo by a transducing derivative of T4 (Lin and Black, 1998); iii) under selection for increased synthesis of TerL the TerS protein is shown to be required for gene amplifications requiring recombination between homologous gene 16 and 19 pac sequences (See Fig. 3) (Wu et al., 1995). Such sequence specific amplifications require a phage internal alt protein knock-out to allow substantially more DNA to be packaged into the phage particle, apparently by increasing head volume, thus allowing multiple copies of the 16–19 region to be viable within the chromosome (Wu and Black, 1987; Wu et al., 1991); iv) a mature DNA restriction fragment containing a gene 16 pac site can be found in phage particles (Lin and Black, 1998); however site directed mutagenesis of the gene 16 amplification pac site that eliminates gene 16–19 amplifications is not lethal to the phage, suggesting backup pac sites (such as the homologous 19 pac site (See Fig. 3B) or other mechanisms (Wu and Black, 1995); and finally v) mass spectrometry and scanning transmission electron microscopy mass determination show single 11mer and double 22mer rings. These are proposed to be aplanar, lock washer like single- and double- ring structures that unstack to yield side-by-side rings (Lin et al., 1997). The single and double rings purified from an untagged protein expression vector are stable by mass spectrometry and have been shown to contain only protein (van Duijn, 2010). The two rings thus are not held together with DNA as hypothesized for another proposed two-ring TerS structure, although without direct experimental support (Sun et al., 2012). Unlike crystal structures of His-tagged TerS that by mass form 11mers and 12mers, the purified untagged rings are by mass only 11mers (not 12mers) and 22mers. Of course in vivo the TerS protein is expected to interact with DNA, possibly as shown in Fig. 3C.
An unexpected difference between TerL and TerS is that while the large terminase has long been known to be a signature homology feature among diverse phages (Black, 1989) [For informatics see Fig. 2 in (Serwer and Jiang, 2012)], the small terminase genes and gene product crystal structures display marked differences. Moreover major controversies abound as to how these crystal structures relate to TerS DNA binding in various pac site phages. Thus whether the pac DNA binding-site is at the N-terminus of terS (where there is generally a DNA binding motif) or at its C-terminus, and how this relates to number of monomers per crystal ring structure (8, 9, 10, 11, and 12 per single ring have been reported among different phages), as well as the very different TerS monomer structures [for a gallery of the single ring crystal structures see Fig. 2 in (Roy et al., 2012), and for the side-by-side double T4 TerS protein only rings, see Fig. 7 in (Lin et al., 1997)], and whether DNA moves through a central ring channel and/or wraps around the ring is debated (Buttner et al., 2012; Roy et al., 2012; Roy and Cingolani, 2012; Sun et al., 2012; Teschke, 2012; Zhao et al., 2010). Even more importantly, the relevance of the rings to function remains problematic since the single planar rings of variable numbers of monomers from different pac site phages have been produced at high protein expression levels or observed under crystallization conditions.
The T4 terminase small subunit TerS protein remains active when modified at both the N-terminal and C-terminal ends. In fact TerS-mCherry and TerS-GFP fusion proteins are highly fluorescent and readily purified as N-terminal His-tag proteins. Moreover both these His-tag expression vector synthesized proteins are active in vivo as judged by their complementation with gene 16 amber mutants. Full activity is also observed when TerS-mCherry and TerS-GFP fusion protein encoding genes replace the normal TerS gene by recombination into the T4 genome. Activity is retained despite the marked increase in molecular weight (18 to 45 kDa). The existence of fully functional N- and C-terminal TerS-fusions is contrary to proposals that the DNA moves through the small ring central channel (Zhao et al., 2010) and that DNA-binding is to the C-terminal end of the ring monomer (Roy et al., 2012). At least in phage T4 bulky C-terminal fusion additions to TerS would be expected to affect DNA–binding and DNA translocation through a narrow diameter TerS channel. We have also determined by FRET that a ring-like form of TerS is produced in vivo at low expression levels by co-infection using the two recombinant fluorescent TerS fusion synthesizing phage genomes; moreover, in contrast to wild type, when purified, a temperature sensitive (ts) mutant form of TerS forms rings at 20 °C (permissive) but not not 42 °C (non-permissive), strongly suggesting that TerS rings are produced and required for function in vivo (Dixit et al, ms in preparation). In fact, if it is assumed that a DNA-wrapped single ring or double ring (Fig. 3C) is the active TerS structure, the great variability in numbers of monomers and structures in different phage TerS rings may obscure quite similar functions and DNA structures. Our terS gene fusion results support the consensus view that DNA initially binds the N-terminal portion of the small subunit, where the DNA-binding motif is located (Lin et al., 1997), and then wraps around the small subunit ring without passing through the central channel.
Taken among all the pac site phages, the evidence favors a unitary nucleosome-like TerS ring DNA wrapping structure in which DNA bending may play a role in addition to sequence specific binding. In phage T4 the evidence discussed above for a double protein ring - double DNA pac structure is strong. It remains to be established whether the proposed critical signal for packaging initiation is the Holliday junction structure formed between swapped strands in the four-stranded pac site paired structure as shown in Fig. 3C. This proposed mechanism is favored by the pac sequence-specific, TerS-specific gene amplifications that can arise because of recombination between apposed homologous gene 16 and gene 19 pac sequences under selection, as shown in figure 3B. Alternatively the formation of the double DNA double protein ring may be sufficient, without strand swapping, to act as the signal for the TerL initiation cutting and subsequent packaging.
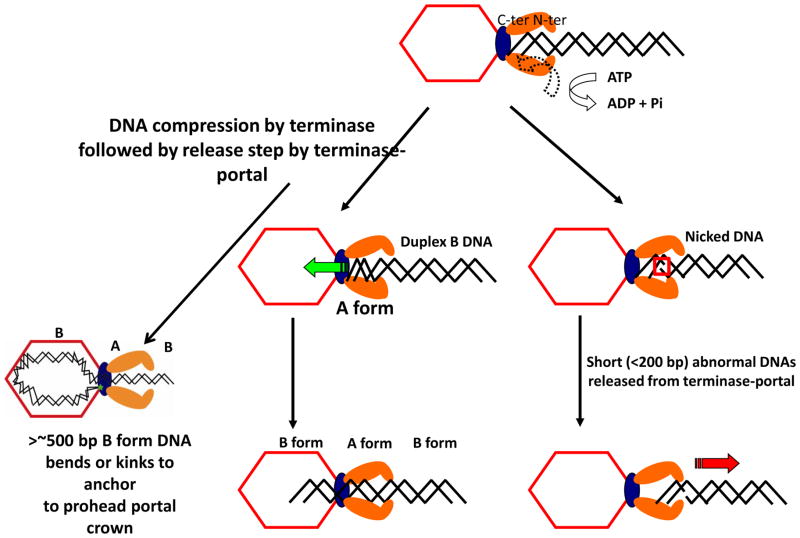
The DNA translocation mechanism of the prohead portal large terminase-ATPase motor protein TerL is under active investigation in several phages. The phage ϕ29, λ, and T4 motors all appear to produce comparable high (~60 pN) force and rates sufficient to fill their different size proheads in comparable times (Fuller et al., 2007; Smith et al., 2001), suggesting comparable mechanisms. Crystal structures of all the packaging proteins (portal, large and small terminase subunits) are now known in a number of phages (Lebedev et al., 2007; Simpson et al., 2000; Sun et al., 2008; Zhao et al., 2010; Kanamaru et al., 2004; Sun et al., 2007, Zhao et al, 2013). Phage T4 DNA translocation is driven by a two-component motor consisting of the prohead portal (gp20) docked with a packaging terminase-ATPase TerL. Our proposed linear DNA compression translocation motor is shown in Fig. 4 (Oram et al., 2008). A single ~170 kb DNA can be translocated into the empty prohead by the ATP powered TerL motor, or one or multiple short pieces of dsDNA can be translocated and retained in the prohead (Sabanayagam et al., 2007). The conserved bacteriophage T4 ATP based DNA translocation motor consists of a multimeric packaging terminase TerL (gp17) docked onto a unique prohead vertex containing a dodecameric portal ring. The portal ring can incorporate ring protein (gp20) fused to GFP at either the N- or C-terminal end into active proheads. Our work shows that the T4 portal is central in controlling the ESP (empty small prohead) to ELP (empty large prohead) maturation of the T4 major capsid protein lattice. Maturation is an essentially irreversible all-or-none concerted transformation of the major capsid protein to high stability (See Fig. 2). As compared to wild type, portal gp20-GFP (C-terminal fusion) protein containing proheads are stuck in the unexpanded ESP form until packaging expands them into the ELP form; GFP-gp20 (N-terminal fusion) proheads are however comparable to wild type in being mainly expanded. We proposed an “umbrella-runner” model for the altered portal-GFP structure that controls the whole procapsid lattice structure expansion. Comparing fusion and wild type proheads by nuclease assay and by FCS in this work, packaging was shown to be of comparable efficiency into the portal-fusion ESPs and into ELPs. The ELP structure of the prohead lattice is essential to protect the DNA against nuclease digestion but not to package it. These studies emphasize the central importance of the portal in virtually every aspect of prohead assembly and packaging, and show once more that there is no obligatory mechanistic coupling of prohead expansion to DNA packaging itself (Dixit et al., 2011; Ray et al., 2009).
Fig. 4.
Torsional compression model for DNA packaging involving transient B to A form spring-like DNA compression as the C-terminal domain of TerL is docked on the portal of the prohead. Although short abnormal DNAs are released by the motor, such DNAs can be anchored by a leader sequence by the terminase-portal motor complex. FRET measurements show once persistence length DNA (~500 bp or more) has been encapsidated the two DNA ends are held at the portal at a fixed distance regardless of DNA length as shown, thus that a loop of DNA is packaged, and thereby preventing knotting in the capsid.
Among the tailed dsDNA phages with comparable packaging mechanisms, phage T4 has several advantages as a model system. Phage T4 packaging can be carried out with up to 100% efficiency in vitro with small linear DNAs of any sequence and two highly purified components (TerL and portal containing prohead). Diffusible internal proteins within the active prohead can be fused to fluorescent GFP and allow measurement of packaging in real time. Fluorescence technologies (Fluorescence Correlation Spectroscopy (FCS), Förster Resonance Energy Transfer (FRET), and single molecule (sm) FRET) applied to small linear DNA substrates with precisely located fluorophore adducts have been shown to be applicable to understanding the T4 packaging complex in a novel application of FCS and single molecule technologies to assay packaging in real time (Sabanayagam et al., 2007). Fluorescence approaches combined with biochemical assays appear to have the most potential to establish actual motor dynamics, since they are capable of demonstrating structural changes to the DNA substrate as well as the motor proteins that accompany translocation. The combination of techniques provides more direct information of a dynamic process than can static crystal structures docked to rigid B form DNA.
It is known in other phages that packaging related headful non-sequence specific cutting of the concatemeric DNA substrate is determined by the portal TerL-nuclease interaction (Casjens et al., 1992; Isidro et al., 2004). How does this relate to the location of mature DNA ends in the full head? Our FRET measurements show that both ends of a single packaged DNA are co-localized to the portal and are separated by ~90 Å regardless of length (same distance between two dyes packaged on the ends of 5 kb or 50 kb DNA). Thus necessarily a loop of DNA is packaged, with one end tethered to the top of the portal the other to the bottom (Fig. 4), preventing knotting of ~170 kb DNA in the capsid. Multiple segments (five or more) of 100 bp DNAs can also be packed into a single prohead (Ray et al., 2010a; Sabanayagam et al., 2007). In such short DNAs abnormal DNA structure, even single nicks, causes release of the substrate, as shown in Fig. 4 (Oram et al., 2008). A longer leader structure in the DNA is required to anchor an abnormal (Y-junction) DNA substrate in the portal. Fluorescence resonance energy transfer (FRET) and fluorescence correlation spectroscopic (FRET-FCS) studies of a branched (Y-junction) DNA substrate with a prohead-anchoring leader segment and a single dye molecule situated at the junction point reveal that the “Y-DNA” stalls in proximity to the prohead portal fused to GFP at the C-terminal end (Ray et al., 2010b).
Y-DNA substrates containing energy transfer dye pairs separated in the Y-stem by 10 or 14 base pairs reveal that B-form DNA is locally compressed 22 or 24% by the linear force of the packaging motor, with comparable FRET increase (i.e. between stem double dye Ax and Cy shown in Fig. 5A) whether the FRET dye pairs are located same side or opposite sides of the helix. DNA B to A form linear compression is calculated to be 23% from their structures. These results support compression or “crunching” to A form rather than DNA kinking, melting, or “scrunching”–the latter term was first coined for a longer range transcription associated DNA structural change involving DNA melting (Revyakin et al., 2006) - for the decrease in distance between the two dyes that is shown by the FRET increase. Torsional compression of duplex DNA is thus implicated in the mechanism of DNA translocation even of the short DNAs studied that are expected to offer little resistance to the packaging motor. DNA compression, although less studied and understood than DNA melting or stretching, has been detected and the force measured as we referenced. To the extent found by us, transient B to A form compression is found to be well within the force generation of the T4 packaging motor (Ray et al., 2010b). Also, as we noted, DNA compression is a possible explanation of the non-integral (2.5 bp) step size of the ϕ29 packaging motor if the DNA is entering in A form steps (Liu et al., 2014b; Moffitt et al., 2009).
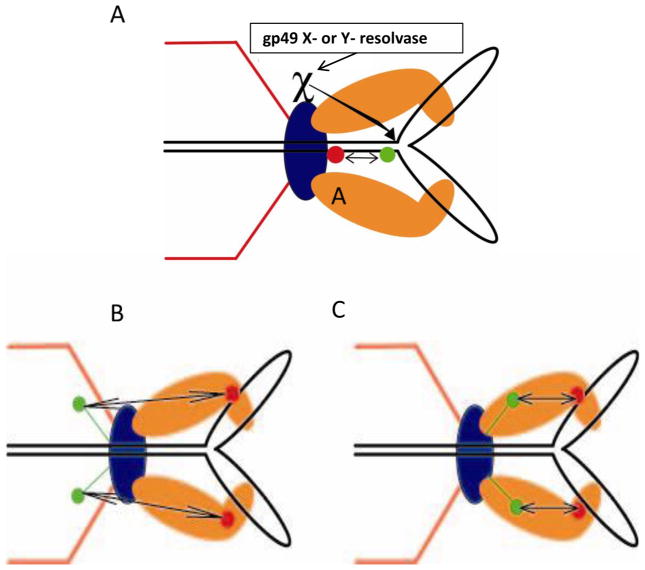
Fig. 5.
Substrate and motor dimensions and dynamics by FRET: (A) Increased FRET between stalled Y-stem DNA dye pairs supports B to A form compression; the gp49:Endonuclease VII X- or Y-resolvase decreases the Y-stem DNA FRET and allows completion of packaging. In the stalled anchored Y DNA the terminase moves toward the portal as judged by FRET increase between dye labeled terminase and (B) C-terminal GFP labeled gp20 portal containing proheads, and (C) N-terminal GFP labeled gp20 portal containing proheads that allow determination of the docking orientation in the two component prohead packaging motor.
To analyze further the dynamics of the T4 bacteriophage DNA packasome motor, gp49:Endo VII resolvase, which plays a central role in T4 packaging in vivo (see Fig. 1), has been used to release arrested Y-DNA substrates into the prohead (Dixit et al., 2011) (Fig. 5A). In vivo the T4 phage packaging motor deals with Y- or X- structures in the replicating concatemer substrate by employing the portal bound Holliday junction resolvase (the essential packaging gene 49:Endo VII resolvase) (Golz and Kemper, 1999) to trim and release these DNA roadblocks to packaging. Using double dye labeled packaging anchored 3.7 kb Y-DNAs or linear DNAs, we demonstrated FRET between the dye-labeled substrates and N-terminal GFP-portal containing proheads, and between GFP-portal and single dye labeled terminases (Fig. 5B&C). We showed using FRET-FCS that purified T4 gp49:EndoVII resolvase can release DNA compression in vitro. The enzyme can nick the Y-DNA roadblock so as to release it and to complete packaging into prohead portal packaging motor anchored and arrested double dye Y-DNA substrates. In addition, using active terminases labeled at the amino (N) - and carboxy (C) - terminal ends with a single ReAsH dye molecule, we showed by FRET the distance of the N-terminal GFP-labeled portal protein containing prohead at 6.9 nm from the N-terminus, and at 5.7 nm from the C-terminus of the terminase. Packaging with a C-terminal fluorescent terminase on a GFP-portal prohead, FRET showed a reduction in distance to the GFP-portal of 0.6 nm in the arrested Y-DNA as compared to linear DNA; the reduction was reversed by resolvase treatment. Conformational changes in both the motor proteins and the DNA substrate itself that are associated with the power stroke of the motor are consistent with our proposed linear motor mechanism employing a terminase to portal DNA grip-and-release. The FRET derived distances between terminase and portal are consistent with biochemical, immunological, and genetic evidence for these interactions (Lin et al., 1999).
A conformational change in the DNA is found not only in stalled Y-DNA, but accompanies active translocation of linear DNA into the prohead. T4 TerL packaging is inhibited in vivo and in vitro by a number of intercalating dyes (IDs) such as 9-aminoacridine, ethidium bromide, and YOYO1 (a DNA high affinity dimeric ethidium bromide-like derivative). By use of FCS, nuclease, and Typhoon image assays, we find packaging strips all detectable YOYO1 from the DNA when it enters the prohead in vitro, whereas YOYO1 is still bound to the fast diffusing unpackaged substrate as measured by FCS. Comparably bulky covalently base attached dyes on duplex DNA (densely attached using dye-modifiable 5′ aminohexylacrylamido-dCTP) (Liu et al., 2014a), and other dyes attached at the 5′-ends are successfully translocated by the terminase into the prohead (Ray et al., 2010b) Thus removal of all IDs from the substrate upon packaging strongly suggests that a transient conformational change in base pair stacking of the duplex accompanies linear DNA translocation, a novel finding for a DNA translocase that, unlike a helicase, is not expected to separate the two strands and thereby necessarily displace IDs. In support of B to A form compression as this change is the observation that whereas YOYO1 binds to B it does not bind to A form DNA as judged by NMR. Moreover YOYO1 binds to DNA more than 1000 fold more strongly than ethidium bromide and remains on DNA during electrophoresis (Glazer et al., 1990). However little difference is observed in packaging inhibition by the two IDs, supporting a qualitative structural change in the translocating DNA molecule to explain removal of all the YOYO1 from packaged DNA (Dixit et al., 2012). TerL mutations at only three sites in the N-terminal domain of the enzyme can lead to an enzyme much more resistant to packaging inhibition (plating differences of ~107 relative to wild type) by the above IDs. amber mutations have been introduced at each of these sites (res. 96, 249, and 267) to determine amino acid changes that confer resistance. Despite substantial sequence and three dimensional separation in the TerL crystal structure, these three sites show marked coordination through genetic studies; e.g. F249L is required to allow W267F (or Y (Dixit et al., 2012)). ID inhibition appears also to be under coordinated control at the three sites. The simplest interpretation is that these residues are located near to points of contact between terminase and the translocating DNA in the N-terminal ATPase domain where force is applied to the DNA, probably by insertion of a binding lobe into the major groove of the dsDNA or by contact of this lobe with the 5′ strand. Although force generation by contact with N-terminal ATPase domain residues fits better to the overall terminase and helicase studies as discussed in Dixit et al 2013, there is no direct evidence in any terminase for specific DNA-amino acid contact.
A crystal stucture of T4 TerL showed a two domain C-terminal nuclease and N-terminal ATPase protein (Sun et al., 2008), consistent with a biochemical characterization of the two domain activities (Kondabagil et al., 2006). However the structure is of an inactive and truncated protein; the Sf6 TerL crystal structures of active and ATP bound terminases may be more biologically relevant since they show important structural and possibly functional differences from the T4 TerL (Zhao et al., 2013). Another area of controversy is that our FRET distance measurements strongly support that the C-terminal nuclease domain of the TerL subunit docks to the prohead portal clip region (Dixit et al., 2011; Dixit et al., 2012; Dixit et al., 2013), an orientation that is opposite to a proposed N-terminal terminase domain docked mechanism (Sun et al., 2008). However this N-terminal domain cryo-EM docking reconstruction could well be incorrect because of low (<30 Å) resolution; in view of all other data, this orientation requires confirmation (Sun et al., 2008). The FRET-based orientation, where the C-terminal nuclease domain is docked to portal in the active translocation complex (Dixit et al., 2013) is consistent with the orientations determined in several other phages and with substantial biochemical work (Lin et al., 1999). For elucidation of the terminase mechanism, determination of the N- versus C- terminal domain docking orientation, relative to the portal, is absolutely critical. The published proposed N-terminal orientation appears to be inconsistent with a concurrent “electrostatic model” of DNA translocation proposing a piston-like up and down motion of the C-terminal domain to pump DNA through the portal, if instead the C-terminal nuclease domain is docked on the portal. In addition, a C-terminal terL docking does not appear consistent with proposed specific terS C-terminal to TerL N-terminal contacts proposed and generally found among terminase subunits (Sun et al., 2012).
Our proposed DNA compression or “DNA crunching” linear motor with transient compression of B form to A form-like DNA during packaging (Fig. 4) is supported by direct experimental measurements. These include FRET increases between closely positioned dye pairs in a Y-stem DNA in a stalled translocation complex that closely fit to the amount of B to A compression expected from the B and A structures. Also a qualitative structural change to A form in actively translocating DNA is supported by expulsion of strongly binding YOYO-1 and other intercalating dyes from the packaged DNA (Dixit et al., 2011; Dixit et al., 2012; Ray et al., 2010b). Interestingly, “the scrunchworm hypothesis” (Harvey, 2015) proposes that cycling transitions between B and A form dsDNA by a terminase dehydration-hydration motor provides the force to drive packaging. In support of B to A form terminase motors, it should be noted that the T4 motor does not package dsRNA or DNA:RNA heteroduplexes of 20 bps (both of these are expected to be A form), whereas 20 bp DNAs are packaged efficiently when ATP is supplied (Oram et al., 2008). The latter observation strongly suggests that the motor pushes multiple segments by a “helper mechanism” rather than pulls single ~68 (or ~52) Å 20 bp dsDNAs segments through the ~100 Å long terminase translocation channel. Translocation by a pulling mechanism would be expected to require a longer dsDNA segment. A DNA spring which transiently compresses B to A form requires that two segments of the DNA be gripped, whether by the terminase and portal, as proposed (Oram et al, 2008), or by two parts of the terminase is unknown. However the portal is highly likely to have a catch that prevents DNA leakage out of the prohead, whether or not this is also the second grip and release spring hold.
Bulk measurements in phages ϕ29 and T3 show that, on average, one ATP is consumed per ~2 bps packaged (Guo et al, 1987, Morita et al, 1993). A more recent optical tweezers-derived mechanism coupling four non-integral 2.5 bp steps per burst of 10 bps DNA packaged is entirely hypothetical as relates to ATP utilization per bp step packaged. ATP consumption per 10 bp packaged with one extra non-hydrolyzed ATP per packaging ATPase DNA register-making pentamer is speculative because not directly addressed by experimental ATP consumption versus force measurements at the single molecule level (Moffitt et al., 2009). (Liu et al., 2014b). Indeed there is no information about the DNA step size per ATP consumed for any viral packaging enzyme or helicase (Schlierf and Ha, 2012) as pointed out in a commentary on a novel hexameric helicase ssDNA structure (Itsathitphaisarn et al., 2012). As discussed above the non-integral sub-step size could be explained by transient B to A form DNA “crunching” (Ray et al., 2010b).
Unanticipated structural and mechanistic support for our B to A form translocation proposal has come from recent work on the DnaB hexameric helicase proposing ATP-driven translocation on A form dsDNA and for a non-planar multimer, both similar to our experimental evidence and proposal (Itsathitphaisarn et al., 2012). As referenced for the DnaB enzyme, and also for T7 gp4 hexameric helicase (Jeong et al., 2013), several studies show that hexameric helicases are capable of energetic translocation along dsDNA, speculatively along A form dsDNA for DnaB, but definitely by crystal structure along A form ssDNA for DnaB. As relates to phage DNA packaging, transient compression of DNA to A form could help to explain expulsion of DNA binding proteins, as well as of intercalating dyes, from the DNA as it enters the capsid, as is generally found to be the case. In contrast, large proteins packaged before the DNA (e.g. β-galactosidase, alt) leave the T4 capsid without difficulty through the portal channel into an infected host cell with DNA (reviewed in Black and Thomas, 2012). The transient A form “crunching” and slipping motor mechanism could well also be advantageous by allowing DNA repair during packaging, as shown for the gp49:EndoVII (Holliday junction resolvase) Y-DNA repair (Oram et al., 2008) (Dixit et al., 2011). Indeed both these properties have potential evolutionary advantages that could account for the surprising finding that the phage DNA packaging motor apparently employs a B to A spring-like mechanism by translocating on high energy A form DNA.
Research Highlights.
small terminase subunit (TerS) synapses two pac homologs by a two ring mechanism
both ends of TerL packaged DNA are held at portal, showing loop of DNA is packaged
transient spring-like compression of B form to A form DNA accompanies translocation
the C-terminal domain of TerL is docked to the portal, moves toward it when stalled
portal bound resolvase releases stalled Y-DNA A compression to allow translocation
Acknowledgments
This review is dedicated to the memory of Lindsay M. Black, one of the three founding editors of Virology in 1955. The author’s research was supported by NIH, United States, grant R01 AI11676.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays. 1995;17:1025–1030. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem. 2006;281:25635–25643. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28:643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Thomas JA. Condensed genome structure. Adv Exp Med Biol. 2012;726:469–487. doi: 10.1007/978-1-4614-0980-9_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA. Structural basis for DNA recognition and loading into a viral packaging motor. Proc Natl Acad Sci U S A. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: endonuclease VII resolvase release of arrested Y-DNA substrates. J Biol Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci U S A. 2012;109:20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Thomas JA, Black LW. The C-terminal domain of the bacteriophage T4 terminase docks on the prohead portal clip region during DNA packaging. Virology. 2013;446:293–302. doi: 10.1016/j.virol.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Fu TJKGASGM, Tinker-Kulberg RL, Eckstein FaLDMJ. Transcriptional Activation by a Topologically Linkable Protein: Forging a Connection Between Replication and Gene Activity, Mechanisms of Trasnscription. Springer-Verlag; Berlin Heidelberg: 1997. [Google Scholar]
- Glazer AN, Peck K, Mathies RA. A stable double-stranded DNA-ethidium homodimer complex: application to picogram fluorescence detection of DNA in agarose gels. Proc Natl Acad Sci U S A. 1990;87:3851–3855. doi: 10.1073/pnas.87.10.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz S, Kemper B. Association of holliday-structure resolving endonuclease VII with gp20 from the packaging machine of phage T4. J Mol Biol. 1999;285:1131–1144. doi: 10.1006/jmbi.1998.2399. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Initiation events in in-vitro packaging of bacteriophage phi 29 DNA-gp3. J Mol Biol. 1987;197:219–28. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- Harvey SC. The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages. J Struct Biol. 2015;189:1–8. doi: 10.1016/j.jsb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CL, Black LW. DNA packaging and the pathway of bacteriophage T4 head assembly. Proc Natl Acad Sci USA. 1977;74:3652–3656. doi: 10.1073/pnas.74.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro A, Santos MA, Henriques AO, Tavares P. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol Microbiol. 2004;51:949–962. doi: 10.1046/j.1365-2958.2003.03880.x. [DOI] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik J, Botstein D. A genetic method for determining the order of events in a biological pathway. Proc Natl Acad Sci U S A. 1973;70:2046–2050. doi: 10.1073/pnas.70.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Rajagopal V, Patel SS. Switching from single-stranded to double-stranded DNA limits the unwinding processivity of ring-shaped T7 DNA helicase. Nucleic Acids Res. 2013;41:4219–4229. doi: 10.1093/nar/gkt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. Embo J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–127. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- Liu JL, Dixit AB, Robertson KL, Qiao E, Black LW. Viral nanoparticle-encapsidated enzyme and restructured DNA for cell delivery and gene expression. Proc Natl Acad Sci U S A. 2014a;111:13319–13324. doi: 10.1073/pnas.1321940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, Bustamante C. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014b;157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luftig RB, Wood WB, Okinaka R. Bacteriophage T4 head morphogenesis. On the nature of gene 49-defective heads and their role as intermediates. J Mol Biol. 1971;57:555–573. doi: 10.1016/0022-2836(71)90109-4. [DOI] [PubMed] [Google Scholar]
- Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–52. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Salamin L, Onorato L, Showe MK. Localization of minor protein components of the head of bacteriophage T4. J Virol. 1977;24:121–134. doi: 10.1128/jvi.24.1.121-134.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J Mol Biol. 2010a;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Oram M, Ma J, Black LW. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010b;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G. Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure. 2012;20:1403–1413. doi: 10.1016/j.str.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Cingolani G. Structure of p22 headful packaging nuclease. J Biol Chem. 2012;287:28196–28205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlierf M, Ha T. A helicase with an extra spring in its step. Cell. 2012;151:244–6. doi: 10.1016/j.cell.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Serwer P, Jiang W. Dualities in the analysis of phage DNA packaging motors. Bacteriophage. 2012;2:239–255. doi: 10.4161/bact.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Teschke CM. Themes and variations of viral small terminase proteins. Structure. 2012;20:1291–1292. doi: 10.1016/j.str.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn E. Current limitations in native mass spectrometry based structural biology. Journal of the American Society for Mass Spectrometry. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Wu CH, Black LW. Mutational analysis of the sequence-specific recombination box for amplification of gene 17 of bacteriophage T4. J Mol Biol. 1995;247:604–617. [PubMed] [Google Scholar]
- Wu CH, Lin H, Black LW. Bacteriophage T4 gene 17 amplification mutants: evidence for initiation by the T4 terminase subunit gp16. J Mol Biol. 1995;247:523–528. [PubMed] [Google Scholar]
- Wu DG, Black LW. Gene amplification mechanism for the hyperproduction of T4 bacteriophage gene 17 and 18 proteins. J Mol Biol. 1987;195:769–783. doi: 10.1016/0022-2836(87)90483-9. [DOI] [PubMed] [Google Scholar]
- Wu DG, Wu CH, Black LW. Reiterated gene amplifications at specific short homology sequences in phage T4 produce Hp17 mutants. J Mol Biol. 1991;218:705–721. doi: 10.1016/0022-2836(91)90260-d. [DOI] [PubMed] [Google Scholar]
- Zhao H, Christensen TE, Kamau YN, Tang L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc Natl Acad Sci U S A. 2013;110:8075–8080. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A. 2010;107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]