Abstract
In Drosophila, myoblast fusion is a conserved process in which founder cells (FCs) and fusion competent myoblasts (FCMs) fuse to form a syncytial muscle fiber. Mutants for the myogenic regulator Myocyte enhancer factor-2 (MEF2) show a failure of myoblast fusion, indicating that MEF2 regulates the fusion process. Indeed, chromatin immunoprecipitation studies show that several genes involved in myoblast fusion are bound by MEF2 during embryogenesis. Of these, the MARVEL domain gene singles bar (sing), is down-regulated in MEF2 knockdown pupae, and has five consensus MEF2 binding sites within a 9000-bp region. To determine if MEF2 is an essential and direct regulator of sing during pupal muscle development, we identified a 315-bp myoblast enhancer of sing. This enhancer was active during myoblast fusion, and mutation of two MEF2 sites significantly decreased enhancer activity. We show that lack of sing expression resulted in adult lethality and muscle loss, due to a failure of fusion during the pupal stage. Additionally, we sought to determine if sing was required in either FCs or FCMs to support fusion. Interestingly, knockdown of sing in either population did not significantly affect fusion, however, knockdown in both FCs and FCMs resulted in muscles with significantly reduced nuclei numbers, provisionally indicating that sing function is required in either cell type, but not both. Finally, we found that MEF2 regulated sing expression at the embryonic stage through the same 315-bp enhancer, indicating that sing is a MEF2 target at both critical stages of myoblast fusion. Our studies define for the first time how MEF2 directly controls fusion at multiple stages of the life cycle, and provide further evidence that the mechanisms of fusion characterized in Drosophila embryos is also used in the formation of the more complex adult muscles.
Keywords: Drosophila, myoblast fusion, MEF2, transcriptional regulation, MARVEL domain, singles bar
INTRODUCTION
Myoblast fusion is a conserved and critical process in the formation of mature, functional muscle fibers. Mammals and invertebrates share several of the key steps and components of the fusion process, from coalescence of myoblasts at sites of fusion, to membrane breakdown to generate the muscle syncytium (Richardson et al 2008). In Drosophila, myoblast fusion begins by the designation of a founder cell (FC) and fusion competent myoblasts (FCMs). FCs differentially express a subset of genes, that function to attract FCMs, and fusion of the FC and the initial FCMs to form an early multi-nucleated muscle cell constitutes the initial round of fusion (Chen and Olson 2004). Subsequent fusion of further FCMs to the nascent myotube complete myoblast fusion (Schroter et al 2004). Several of the genes involved in each step of the fusion process are conserved between Drosophila and vertebrates: for example, myoblast adhesion can be partially attributed to the Drosophila protein Sticks and stones, for which Nephrin is the vertebrate ortholog (Rochlin et al 2010); and the Drosophila protein Myoblast city is required during cytoskeletal rearrangement within fusing myoblasts (Erickson et al 1997), as are the vertebrate orthologs, Dock1/Dock2 (Rochlin et al 2010). Clearly, understanding the molecular mechanisms that regulate myoblast fusion in Drosophila can provide insight into the fusion process in vertebrates.
While numerous studies have identified genes required for embryonic myoblast fusion in Drosophila (Paululat et al 1999; Chen and Olson 2004; Abmayr and Pavlath 2012), less is known about the genes involved in the phase of fusion that occurs in the development of the adult muscles. For the adult thoracic muscles, the fusion process begins with the migration of adepithelial cells originating from the imaginal discs into the developing thorax. While most adult muscles arise from de novo fusion of pupal FCs and FCMs (Dutta et al 2004), the dorsal longitudinal muscles (DLMs) develop upon larval muscle templates (Fernandes et al 1991), where the larval muscles function as FCs (Dutta et al 2004). Of the few published studies on adult myoblast fusion, WASp, an actin nucleator required for embryonic myoblast fusion (Massarwa et al 2007; Schafer et al 2007), is required at the time of adult myoblast fusion prior to pre-fusion complex formation (Mukherjee et al 2011). The lack of WASp results in a complete hindrance of fusion in adult muscles (Mukherjee et al 2011). More recently Gildor et al (2012) showed that sticks and stones/hibris and dumbfounded/roughest have redundant functions in fusion of adult myoblasts. Thus, there are at least some commonalities in the mechanisms of myoblast fusion between embryos and pupae.
The transcriptional regulation of factors participating in adult myoblast fusion has not been investigated in detail. One candidate regulator is Myocyte enhancer factor-2 (MEF2). MEF2 is a conserved myogenic transcription factor that is critical for muscle differentiation in both skeletal and cardiac muscles (Potthoff and Olson 2007). There are four orthologs of MEF2 in mammals while Drosophila has a single MEF2 gene, but for which the encoded protein shares the conserved A/T rich binding domain and function as a regulator of muscle differentiation (Lilly et al 1995; Bour et al 1995). However, the genetic redundancy of MEF2 genes in vertebrates makes it difficult to study the context of MEF2 solely in relation to myoblast fusion events. In Drosophila, studies have indicated that MEF2 has an essential role in embryonic myoblast fusion, since mutation of Mef2 resulted in unfused myoblasts in β3-Tubulin-stained embryos (Bour et al 1995). Expression in Drosophila of Mef2 RNAi lines results in a lack of adult muscle formation and the accumulation of unfused myoblasts in Mef2 knockdown pupae, also indicating a requirement for MEF2 in the fusion of adult myoblasts (Bryantsev et al 2012; Soler et al 2012).
Embryonic chromatin immunoprecipitation-microarray (ChIP-chip) studies in Drosophila support the hypothesis that MEF2 is a direct regulator of fusion gene transcription (Sandmann et al 2006). The fusion genes blown fuse (blow) and lameduck (lmd) are bound by MEF2 during embryonic muscle development, and loss of MEF2 results in loss of their expression (Chen and Olson 2004; Sandmann et al 2006). Similarly, roughest (rst) is required for myoblast fusion (Strunkelnberg et al 2001) and responds to MEF2 activity in the embryo (Apitz et al 2005). Nevertheless, although ChIP-chip data suggests a critical role for MEF2 in the regulation of many fusion genes, binding data is not sufficient to determine if MEF2 is essential for fusion gene expression: the fusion gene sticks and stones (sns), an immunoglobulin family gene expressed in FCMs, has MEF2 binding sites both upstream and downstream of the gene, as determined by ChIP-chip analysis (Sandmann et al. 2006); however, sns expression in embryos is not MEF2 dependent (Bour et al 2000), suggesting that although MEF2 binds to the region, it is not necessary for sns gene expression. Instead other factors, or factors functioning redundantly with MEF2, must control sns transcription. In addition to sns, blown fuse expression is not affected in MEF2 mutants, indicating that MEF2 may not directly regulate fusion gene transcription despite the presence of MEF2 binding sites (Schroter et al 2006).
There is some evidence that fusion genes may also be regulated by MEF2 in the pupal stages of myoblast fusion. We recently demonstrated that knockdown of Mef2 function during pupal development resulted in a failure of adult myogenesis, including a complete lack of myoblast fusion. By using RT-PCR of RNA collected from control and Mef2 knockdown pupal myoblasts, the embryonic fusion gene singles bar (sing) was down-regulated in MEF2 knockdown samples (Bryantsev et al 2012). Estrada et al (2006) previously identified sing as encoding a protein with a conserved transmembrane protein known as a MARVEL domain. This domain is believed to function in junction formation between cells and vesicle trafficking in vertebrates (Sanchez-Pulido et al 2002) suggesting that sing may be involved in the formation of the pre-fusion complex. The findings from Bryantsev et al (2012) suggested firstly that MEF2 may be a direct and essential regulator of sing during myogenesis, and secondly that sing functions in myoblast fusion at both embryonic and pupal stages.
To test these hypotheses, we identify in this manuscript a 315-bp enhancer for sing expression that functions at both adult and embryonic stages of myoblast fusion. We show that sing expression is directly regulated by MEF2 via two conserved binding sites in the enhancer, and that the knockdown of sing during adult myoblast fusion results in lethality and drastically reduced muscle formation arising from a failure of myoblast fusion. We also demonstrate that, whereas sing expression is observed in FCs and FCMs in embryos, sing knockdown in both cell types is necessary for defects in fusion to be observed. Overall, our results identify a regulatory role for MEF2 in myoblast fusion at multiple stages of development, and identify sing as a fusion gene that functions during both the embryonic and adult stages.
MATERIALS AND METHODS
Drosophila Stocks and Crosses
Stocks were maintained on Jazz-Mix Drosophila Fly Food (Fisher Scientific). rp298-gal4 driver has been previously described (Nose et al., 1998; Gomez et al., 2000). Mef2-gal4 was from Dr. Aaron Johnson (University of Colorado at Denver), sns-gal4 was from Dr. Elizabeth Chen (Johns Hopkins University Medical School), and 1151-gal4 was from Dr L.S. Shashidara (Anant et al 1998). The UAS-sing RNAi lines, P{GD3396}v12203 and P{GD3396}v12202/TM3 were obtained from Vienna Drosophila RNAi Center. The Mef2 knockdown line, UAS-dcr; UAS-Mef2 RNAi(15550) was described in Bryantsev et al (2012). The Mef2 null allele, P544, was balanced over a CyO, wg-lacZ balancer chromosome to enable visualization of homozygous mutant embryos.
Transgenic Lines and Mutagenesis
The following PCR primers were used to generate the sing enhancer using genomic DNA as a template:
| Sing315-attB1: 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCTTCCGCATAGACA-3’ |
| Sing315-attB2: 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTACAGAACGAACCCGAAATTG-3’ |
Gateway technology was used to clone the construct into pDONOR-lacZ-attB vector. Mutagenesis of the MEF2 sites in the sing315-lacZ construct was made by Gene SOE-ing site directed mutagenesis (Horton, 1993). The following primers were used to mutate the MEF2 sites within the enhancer:
| Sing315-1_Mef2_mutation_forward: | 5’-AATTGCTGTTATGGTACCTACTGGAGATTG-3’ |
| Sing315-1_Mef2_mutation_reverse: | 5’-CAATCTCCAGTAGGTACCATAACAGCAATT-3’ |
| Sing315-2_Mef2_mutation_forward: | 5’-AATTGCTGTTATGGTACCTACTGGAGATTG-3’ |
| Sing315-2_Mef2_mutation_reverse: | 5’-ACCAGGTTTAGGTACCATCTGCCGATAC-3’ |
Constructs for generating transgenic lines were injected into Drosophila embryos according to the protocol published by Rubin and Spradling (1982).
In situ Hybridization
Embryos were collected on agar-grape juice plates at 25°C and fixed according to standard protocols (Patel, 1994). In situ hybridization experiments were modified from a previously described method by the Berkley Drosophila Genome Project (Weiszmann et al., 2009). RNA probes were made by amplification of sing from embryonic RNA using the following primers:
| Sing_forward_with_HindIII: | 5’-AAGCTTATCAGTTGCAATCAGACC-3’ |
| Sing_reverse_with_XhoI: | 5’-CTCGAGTGCTTTTGTCTGGCCG-3’ |
The resulting PCR product was cloned into pGEM-T Easy Vector systems (Promega) and linearized using restriction enzymes HindIII (New England BioLabs) and XhoI (New England BioLabs) for generation of sense and antisense probes, respectively.
Cryosectioning and Immunostaining
Frozen sections of pupal samples were stained as described by Morriss et al (2011). Briefly, pupae collected at 16, 18, 24, 30, and 48 hours after puparium formation (APF), and those collected just prior to eclosion, had pupal casings removed prior to being submerged in Tissue-Tek OCT Compound (Sakura). Samples were frozen in liquid nitrogen and stored at −80°C until ready for sectioning. Samples were horizontally sectioned at a thickness of 10–12µm, and sections collected on a slide. Sections were fixed for eight minutes on a rotator in a 1:10 solution of 37% (v/v) formaldehyde and PBS. Slides were washed in PBTx [0.2%(w/v) BSA, 0.05% (v/v) Triton X-100, PBS] before incubation in Triton-X/PBTx solution for 30 minutes. Slides were incubated in primary antibody (anti-MEF2 diluted 1:1000, anti-Beta-galactosidase (Promega) diluted 1:1000, anti-Phospho-histone H3 (Thermo Scientific) diluted 1:400, and anti-Lamin (University of Iowa Development Studies Hybridoma Bank) diluted 1:10) in a humid chamber overnight before PBTx washing. Alexa Fluor secondary antibodies (Life Technologies) were diluted 1:300 in PBTx and incubated with sections in the dark at room temperature for 2 hours. Rabbit anti-MEF2 was from Dr Bruce Paterson.
Fluorescence and Confocal Microscopy
Stained sections of pharate adults were imaged using an Olympus BX51 fluorescent microscope. High resolution images for nuclei counts were taken using a 20×, 0.8 NA objective lens on a Zeiss LSM710 confocal microscope, and images were captured using Zen software.
Nuclei Counts
Nuclei counts from confocal images of stained adult muscle sections were recorded using the ITCN plugin for ImageJ (Rasband, 2014). All images were taken at 200× magnification on the confocal microscope. The threshold for detection was set to 0.8, nuclei width was set at 16 pixels, and nuclei distance was set to 8 pixels. Criteria for region specification for counting were based upon the largest continuous area of myoblasts or indirect flight muscle. Counts were normalized by determining the area of the region observed, and converting the nuclei counts from counts per square pixel to counts per 10,000µm2. A Dunnett-Tukey-Kramer pairwise multiple comparison test was used to determine significance between genotypic groups at p=0.05 level. Statistics and graphs were generated and programmed in R using the DTK package (Lau, 2013; R Core Team, 2013).
Electrophoretic mobility shift assay
MEF2 protein was generated using the TNT Coupled Reticulocyte Lysate System (Promega) using the pSK-MEF2 plasmid (Lilly et al 1994). Details of binding conditions were as described in Gossett et al (1989). The MEF2 site from Act57B was used as a positive control (Kelly et al 2002). Wild-type and mutant probe sequences were as follows (top strand shown only):
| Sing315-1 | 5’-GGAATTGCTGTTCTAAATTTAGCTGGAGATTG-3’ |
| Sing315-2 | 5’-GGGTATCGGCAGCTATTTATAGAACCTGGTTG-3’ |
| Sing315-1 mut | 5’-GGAATTGCTGTTATGGTACCTACTGGAGATTG-3’ |
| Sing315-2 mut | 5’-GGGTATCGGCAGATGGTACCTAAACCTGGTTG-3’ |
RESULTS
A 315bp enhancer upstream of sing containing two conserved MEF2 binding sites is active in adult myoblasts
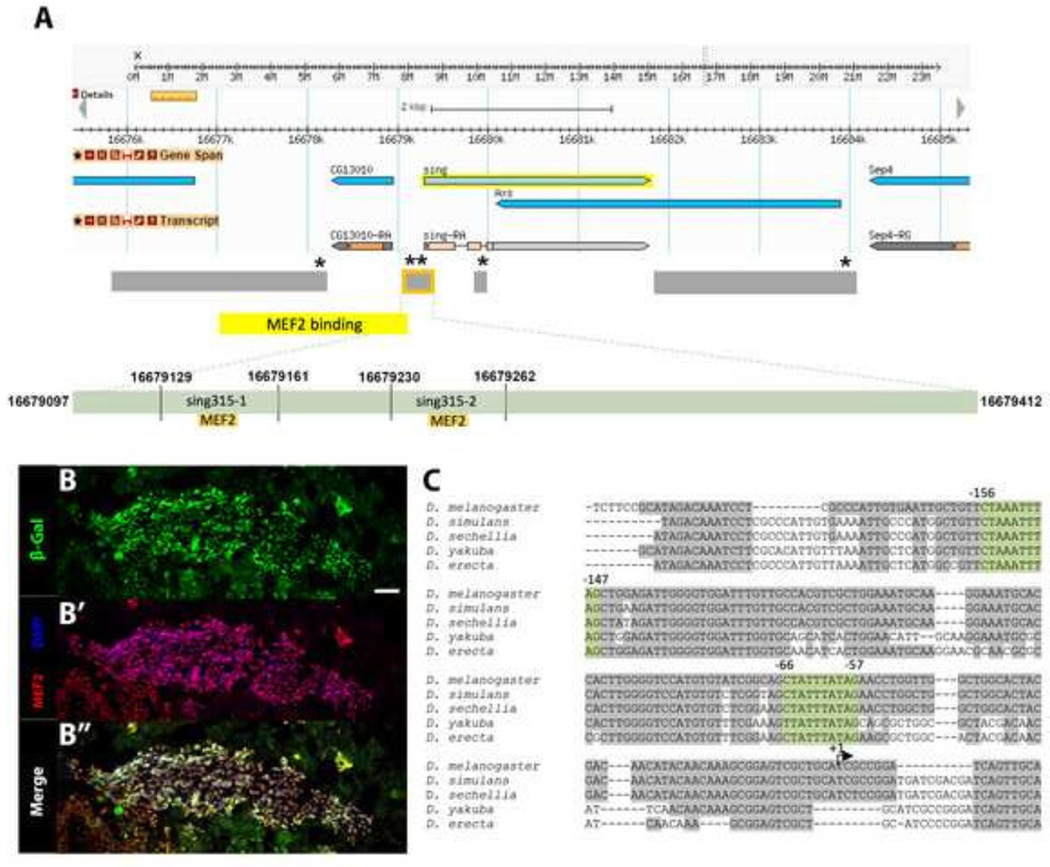
To test the hypothesis that sing is a direct transcriptional target of MEF2 during pupal muscle development, we first sought to identify sequences that control sing expression. Sandmann et al (2006) demonstrated, using ChIP-chip, that MEF2 bound to a ~4-kb region upstream of the sing transcription start site during embryogenesis (Figure 1A). Additionally, there are five consensus MEF2 binding sites in the region of the sing gene (asterisks on Figure 1A). We used these data as a starting point and amplified several fragments of genomic DNA to test for enhancer activity (Figure 1A).
Figure 1. A 315-bp enhancer of sing containing two conserved MEF2 binding sites is active in adult myoblasts.
(A) Diagram from Flybase.org of the genomic region surrounding sing, based upon Release 6 of the Drosophila genome. The regions tested for enhancer activity are shown in gray, and the genome region shown to bind MEF2 in ChIP-chip assays (Sandmann et al 2006) is shown in yellow. The 315-bp enhancer is outlined in orange. Asterisks indicate the approximate locations of consensus MEF2 binding sites. A more detailed view of the enhancer region is shown below, with the two MEF2 binding sites highlighted in orange. Coordinates above the putative MEF2 sites indicate the sizes of probes used in DNA binding assays. (B–B“) Horizontal section of 24h APF transgenic pupae carrying the sing315-lacZ reporter. A large area of cells was positive for βGal (green), which corresponded to swarming myoblasts positive for MEF2 (red). Scale bar, 20µm. (C) The D. melanogaster sing315 enhancer has two conserved MEF2 binding sites (highlighted in green) when compared to four other species of Drosophila.
To determine if the DNA fragments had enhancer activity in pupal myoblasts, we fused them to lacZ reporter genes and generated transgenic animals carrying the sing-lacZ constructs. Homozygotes for the transgenic constructs were aged to 24h after puparium formation (APF), and then frozen for cryosectioning and immunofluorescence. We chose 24h APF as the time point, since this is the period during pupal development when myoblast fusion is occurring (Atreya and Fernandes 2008). Moreover, high-throughout RNA sequencing of Drosophila at different stages of development indicates that 24h APF is the time at which peak pupal expression of sing is observed (St. Pierre et al 2014).
In order to visualize the location of sing-lacZ activity relative to the swarming myoblasts, cryosections of transgenic pupae were stained with DAPI, and with antibodies against β-Galactosidase (βGal) and MEF2. We found that there was strong reporter expression in the myoblasts for only one construct, a 315-bp region that we termed sing315 (outlined in orange in Figure 1A, B), demonstrating that the fragment of sing used in our assays had myoblast enhancer activity. Together with the observations from RNA sequencing analyses showing sing expression at this pupal time point (St. Pierre et al 2014), plus the detection of sing transcripts in pupal myoblasts (Bryantsev et al 2012), our data support the hypothesis that the 315-bp DNA fragment being tested is an enhancer for pupal myoblast expression of sing. Since none of the other fragments tested showed enhancer activity at adult nor embryonic stages (not shown), we conclude that sing315 is the predominant cis-regulatory region for sing.
To guide us in identifying important regulatory sequences within sing315, we next compared its sequence in D. melanogaster with the equivalent sequences in four other Drosophila species. We observed strong sequence similarity close to the transcriptional start site, as well as several areas of conservation elsewhere in the enhancer. Notably, the two consensus MEF2 binding sites, YTA(A/T)4TAR (Andres et al 1995), were 100% conserved across the five species tested in our alignments (Figure 1C), supporting the hypothesis that theMEF2 sites are important to sing expression. In more distantly-related Drosophila species, the enhancer is less well conserved, however the most promoter-proximal MEF2 site is always conserved (not shown).
Mutation of MEF2 sites in vitro and in vivo results in lack of MEF2 binding and diminished sing315 activity in adult myoblasts
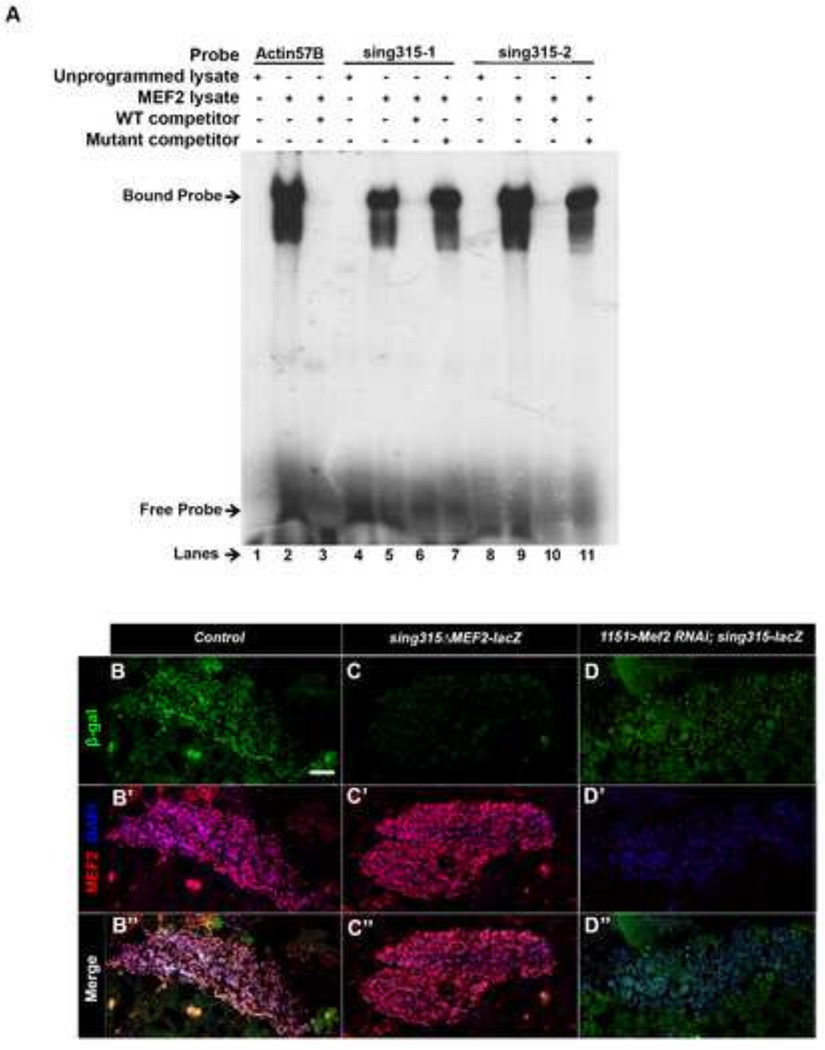
To determine if MEF2 is capable of binding to either of the MEF2 sites, MEF2 protein was generated in vitro and used for electrophoretic mobility shift assays (EMSA). Double-stranded DNA corresponding to a known MEF2 site from Drosophila Act57B (Kelly et al 2002), and to the two sites sing315-1 and sing315-2, were radioactively labeled with 32P and then used in EMSA (Figure 1A). Actin57B exhibited strong binding to MEF2 (Figure 2A, lane 2). The addition of unlabeled Actin57B at a 100 fold greater concentration resulted in a decrease in the intensity of the shifted band (Figure 2A, lane 3). When MEF2 was added to labeled sing315-1, strong binding to MEF2 was observed (Figure 2A, lane 5). MEF2 also showed robust binding with labeled sing315-2 probe (Figure 2A, lane 9). This confirmed that MEF2 is able to bind to both of the conserved MEF2 binding sites within the sing enhancer region.
Figure 2. MEF2 binds to the sing enhancer, and the MEF2 sites are required for enhancer activity.
(A) Electrophoretic mobility shift assay of MEF2 interacting with three different probes: Actin57B control (lanes 1–3), sing315-1 (lanes 4–7), and sing 315-2 (lanes 8–11). Wild type competitor was used in lanes 3, 6, and 10, and mutant competitor was used in lanes 7 and 11. MEF2 bound to the two sites in the sing enhancer and this interaction was sequence-specific, since wild-type sequences competed the interaction, whereas mutant sequences did not compete the interaction. The smear below the shifted band probably represents a minor modified or breakdown isoform of MEF2 interacting with the DNA. (B–B“) Horizontal section of 24h APF sing-lacZ animals stained to visualize MEF2, DAPI, and βGal in adult myoblasts. Note the accumulation of the βGal reporter in myoblasts. (C–C“) Horizontal section of 24h APF transgenic animals carrying sing-lacZ with both MEF2 binding sites mutated. Sections were stained as in B. Note the absence of βGal staining. (D–D“) Horizontal section of 24h APF 1151>dcr + Mef2-RNAi animals carrying sing315-lacZ. βGal staining was diminished in the absence of MEF2. Scale bar, 20µm.
To confirm that this binding was sequence-specific, we competed the MEF2-sing binding reactions with unlabeled wild type and mutant competitors, each at 100-fold greater concentration than the labeled probe. Both sing315-1 and sing315-2 showed almost a complete loss of MEF2 binding with the addition of the wild type competitor probe (Figure 2A, lanes 6 and 10). When the MEF2 binding sites were mutated in the mutant competitor, nearly all binding expression was recovered in both sing315-1 and sing315-2 (Figure 2A, lanes 7 and 11 respectively). This confirmed that MEF2 binding to both sites in sing315 was specific, and therefore supported our hypothesis that MEF2 is a regulator of sing expression.
Next, we wanted to determine if MEF2 was a regulator of sing315 expression in vivo. A construct of sing315 was generated in which both MEF2 binding sites were mutated. This construct was fused with a lacZ reporter, and inserted into the genome. Transgenic animals carrying the wild type sing315-lacZ construct, as well as those carrying the mutated sing-lacZ construct, were collected at 24h APF. Samples were sectioned and stained in parallel, to assess the relative lacZ expression levels controlled by the wild-type and mutant enhancers. In both sections, myoblasts could be observed based upon co-localization of MEF2 and DAPI (Figure 2B’, C’). However, when accumulation of βGal was visualized, there was a significant reduction in reporter activity in the MEF2 mutated version of sing315-lacZ compared to the non-mutated sing315-lacZ (Figure 2B“, C“).
We also generated animals carrying the sing315-lacZ reporter and in which Mef2 expression had been reduced using RNAi. We found that when MEF2 levels were strongly reduced, β-gal expression was diminished (Figure 2D–D“). These results paralleled our prior observations that expression of endogenous sing was dependent upon MEF2 (Bryantsev et al 2012), and therefore provided further support that MEF2 is a direct transcriptional regulator of sing expression during adult myogenesis.
Knockdown of sing during adult myoblast fusion results in reduced muscle formation and lethality at the pharate adult stage
Although MEF2 may be regulating other genes involved in the fusion process, the requirement of MEF2 for sing expression in pupal myoblasts provided one potential mechanism for the failure of myoblast fusion in Mef2 knockdown pupae. In this model, MEF2 activates sing expression, which in turn is required for adult myoblast fusion.
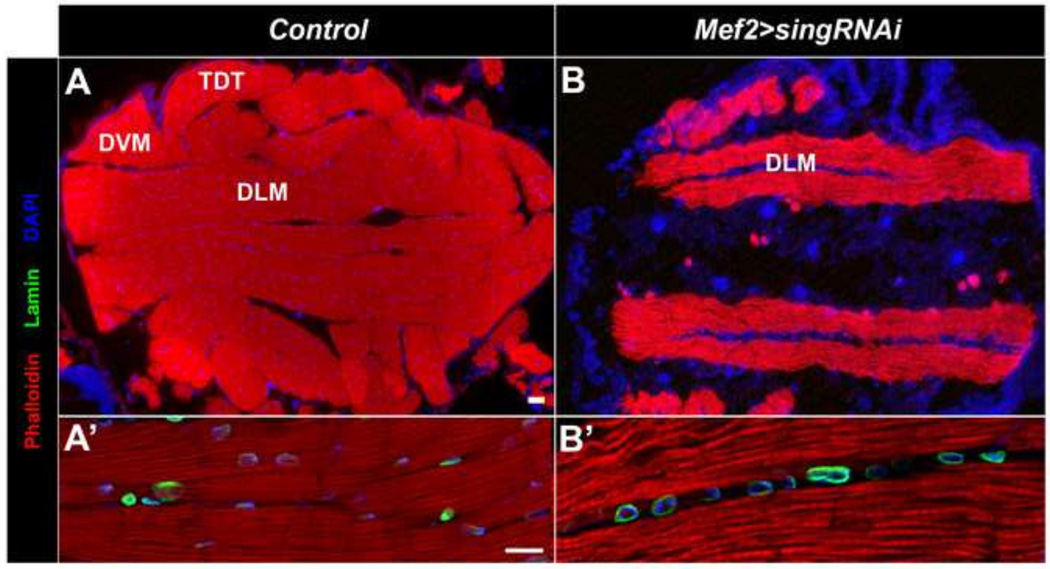
To determine if sing is critical to adult myoblast fusion, we knocked down sing expression using a Mef-gal4 driver crossed to UAS-sing RNAi. In an initial experiment, we allowed control and sing knockdown pupae to develop to the pharate adult stage, after which control animals eclosed from the pupal case, but sing knockdowns were lethal. We used a sing RNAi line for this experiment in which the RNAi is not predicted to have any off-target effects, providing evidence that the phenotypes we observed were due to loss of sing expression, and not due to effects upon other genes. When knockdown adults were sectioned and stained with Phalloidin, anti-Lamin, and DAPI, we observed a considerable reduction in muscle mass in the sing knockdowns compared to wild type (Figure 3A, B). Interestingly, the sing knockdowns still partially developed DLMs, although these muscles were smaller than normal. In the absence of significant fusion of myoblasts to the muscle templates, we propose that the muscles nevertheless grow and attempt to fulfill a role as DLMs. No other skeletal muscles were consistently observed in the sing knockdowns, indicating that sing function is essential for adult muscle development. In addition, the nuclei in the sing knockdown muscles were often clustered together, and always fewer in number compared to the homogenously dispersed nuclei in the wild type muscles (Figure 3A’, B’).
Figure 3. Knockdown of sing results in a failure of adult muscle formation.
(A) Horizontal section of wild type flies at the pharate adult stage. The muscles, stained for accumlation of F-actin, are large and contain numerous nuclei. (B) Horizontal sections of sing knockdown flies at the pharate adult stage show there is a significant failure of muscle formation in the knockdowns. (A’) Higher magnification of control sample at the pharate adult stage showed robust muscle formation with numerous nuclei per muscle fiber. (B’) Higher magnification in sing knockdown animals. The residual muscles that do form are the DLMs, which are smaller than their control counterparts, and only have sparse nuclei. In all panels Phalloidin (red) was used to visualize F-actin, and DAPI (blue) was used to visualize nuclei. Lamin (green) was detected to outline nuclei in A’ and B’ panels. DLM, Dorsal longitudinal muscle; DVM, Dorsoventral muscle; TDT, tergal depressor of the trochanter (jump muscle). Scale bar, 20 µm for A, B; 10 µm for A’, B’.
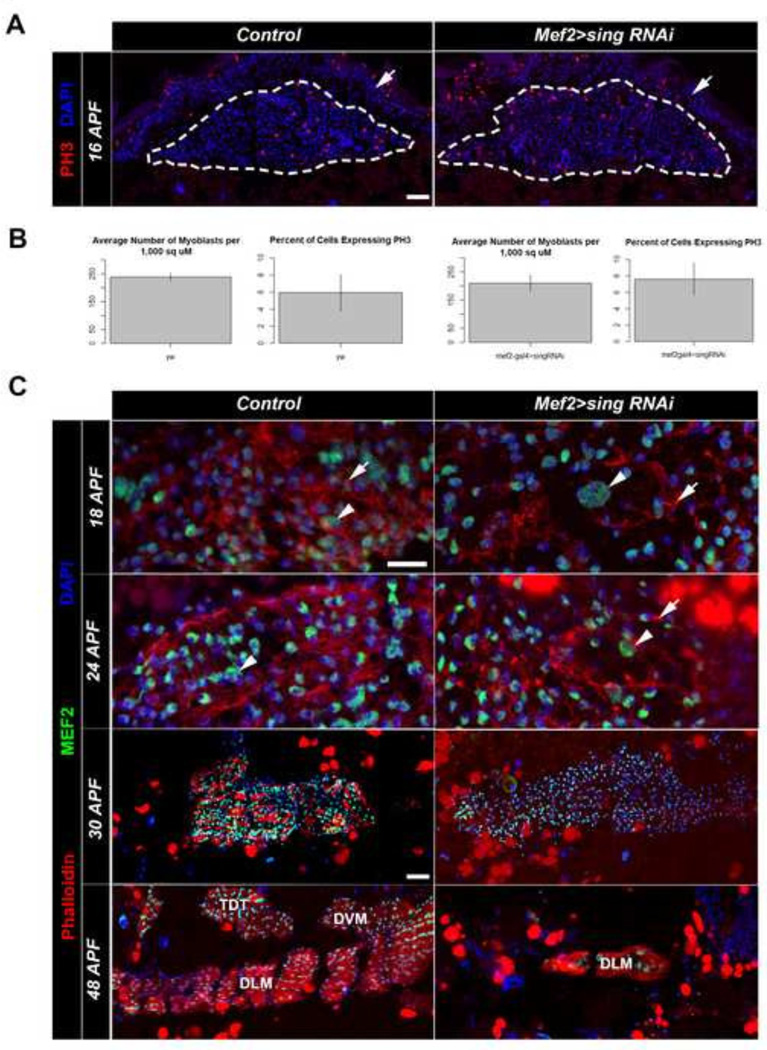
To determine if the phenotype observed in knockdown adults was a result of a fusion defect, we performed a time course analysis of muscle formation in control and sing knockdown pupae. At 16h APF, we sectioned and stained samples with DAPI and anti-PH3 to determine myoblast number and myoblast proliferation rates. We found that myoblast number was slightly reduced in the sing knockdowns, but this difference was minimal and was not significant. In addition, myoblast proliferation rates were not significantly different between control and knockdown (Figure 4A, B). These results indicated that the lack of muscle seen in the sing knockdowns could not be attributed to a smaller starting pool of myoblasts, nor was it due to a slower myoblast proliferation rate.
Figure 4. The adult sing knockdown phenotype results from a failure of myoblast fusion.
(A) Horizontal sections of control and sing knockdown animals at 16 APF respectively, stained for location of nuclei with DAPI (Blue), and for proliferating cells using anti-phospho-Histone3 (PH3, Red). Dotted lines indicate the pool of myoblasts, and arrowheads indicate the cuticle. Scale bar, 20µm. (B) Quantification of myoblast density and proliferation in control and sing knockdown animals. There is no statistical significance between the control and sing knockdown, p>0.05. (C) Time course of developing adult thoracic muscles through adult myoblast fusion, comparing Control and sing knockdown samples. Larger FC nuclei are often apparent (arrowheads). In the sing knockdown animals the FCs have few closely-apposed nuclei, indicating that myoblast fusion is not occurring. Note that F-actin foci (arrows) are apparent in both wild type and sing knockdown samples. DLM, dorsal longitudinal muscle; DVM, dorsoventral muscle; TDT, tergal depressor of the trochanter, or jump muscle. Scale bar, 10 µm for 18 APF, 24 APF; 20 µm for 30 APF, 48 APF.
We next assessed the formation of F-actin foci, a hallmark of fusing myoblasts. Knockdowns of sing at 18h APF compared to control had normal formation of actin foci on the developing templates (arrows, Figure 4C). Thus, the sing knockdown phenotype was not due to a failure of the FCMs to migrate to founder templates, nor due to a failure to initiate the process of fusion. Although actin foci formation appeared normal in the knockdown samples, a failure of fusion was evident at this time point because the developing templates contained founder cell nuclei (arrowheads, Figure 4C) that were surrounded by few myoblasts within the templates. This indicated that FCMs had not fused to the templates. To determine if the lack of fusion at 18h APF was due to a failure of fusion, or simply due to a delay in fusion, we also studies samples at 24h APF. At this later stage, the control templates had increased in size due to extensive fusion of FCMs with the templates, and by this stage F-actin foci were less evident in controls. In the sing knockdown, the templates were smaller, the F-actin foci were still apparent, and there was still little evidence fusion (Figure 4C). This result indicated that lack of sing expression caused a failure of myoblast fusion at the stage following the formation of F-actin foci. In addition, it suggested that when foci formed they remained stable when not resolved into a fusion event. Examination of stained sections staged to 30h APF and 48h APF revealed that the sing knockdown animals failed to form robust muscle compared to controls. In controls, the samples showed muscle forming at 30h APF due to the accumulation of dense F-actin, and at later stages the formation of the adult jump muscle (TDT) and indirect flight muscles (DLM, DVM) could be observed. In the sing knockdowns, it was difficult to discern any muscle formation based upon F-actin accumulation, other than a rudimentary DLM that must have arisen from the persistent larval templates. The defects in the knockdown animals arise presumably due to the lack of fusion in these samples. These results collectively suggested that the sing knockdown phenotype we characterized is indeed attributed to a fusion defect.
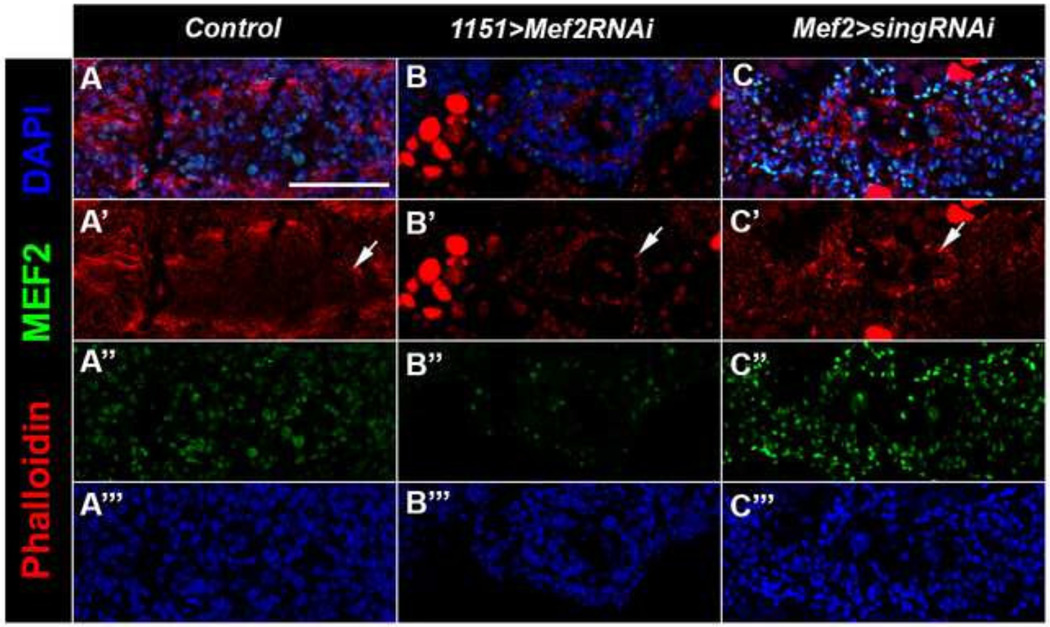
Taking all of this together, our data indicate that at least a part of the failure of fusion in Mef2 knockdowns arises from a failure of MEF2 to activate sing expression. In this model, it would be predicted that Mef2 knockdown myoblasts should not proceed past the formation of F-actin foci. To investigate this model, we sectioned and stained control, sing knockdown, and Mef2 knockdown animals at 24h APF, and determined if the Mef2 knockdown myoblasts were capable of forming F-actin foci. We observed foci outlining the template in the MEF2 knockdown samples compared to the controls (arrows, Figure 5A and B). This Mef2 knockdown phenotype was similar to that for sing knockdown (Figures 4C and 5C), consistent with the model described above.
Figure 5. F-actin foci are detected in sing and Mef2 knockdowns.
(A–C) Horizontal sections of samples aged to 24h APF stained with Phalloidin (red) to visualize F-actin, anti-MEF2 (green), and DAPI (blue) to visualize nuclei. Arrows mark F-actin foci. (A) Wild type control shows normal fusion of myoblasts to the larval templates. (B) Mef2 RNAi show smaller templates with fewer nuclei and pronounced actin foci. (C) sing RNAi shows actin foci at the periphery of the template. Scale bar, 50 µm.
Knockdown of sing in FCs and FCMs results in lethality and reduction in nuclei numbers
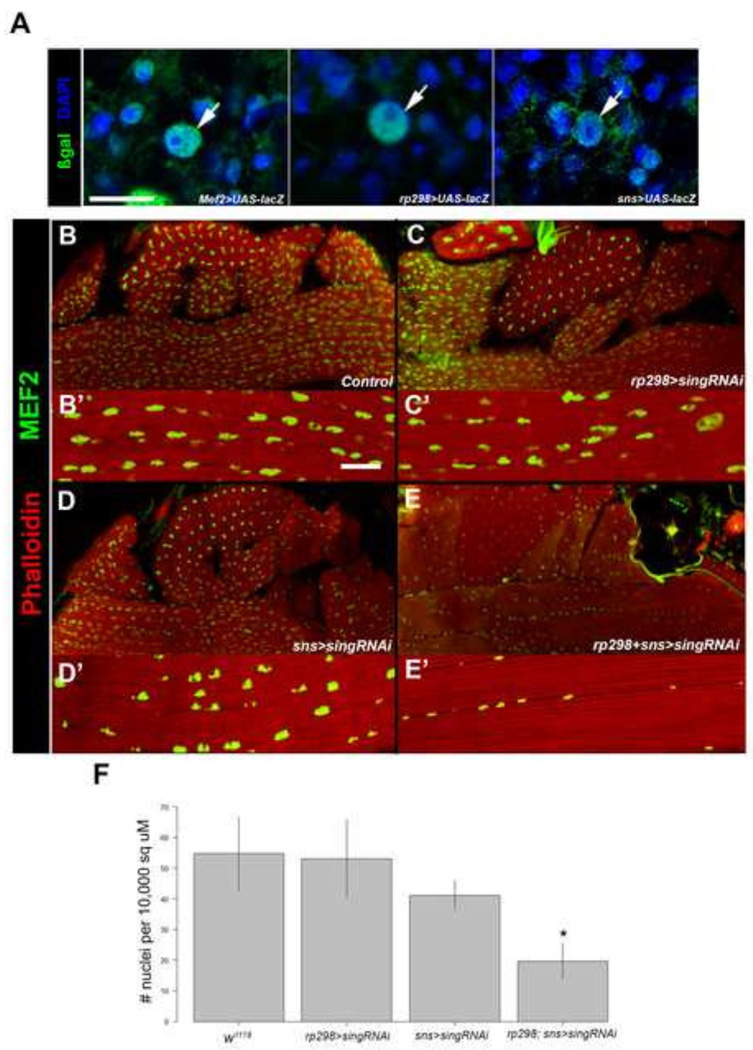
To test whether sing function is required in both the FCs and FCMs, or in just one cell type, we sought to knockdown sing expression individually in the FCs or the FCMs. To achieve this we used cell-specific Gal4 drivers for the FCs and FCMs. To assess the activities of the drivers, we first crossed each to UAS-lacZ and assessed reporter activity in pupae. As previously described, Mef2-gal4 was active in both the FCs and the FCMs (Figure 6A, left panel; Ranganayakulu et al 1998), and rp298-gal4 was active in the FC but not the FCMs (Figure 6A, center panel; Nose et al, 1998). An FCM driver, sns-gal4, directed lacZ expression in FCMs immediately surrounding the FCs, but more distantly-located FCMs did not show reporter activity (Figure 6A, right panel; Stute et al, 2006). We interpret this result to indicate that sns-gal4 becomes active in FCMs shortly prior to fusion. This activity mirrors expression of the endogenous sns gene in adult muscle development (Gildor et al 2012). We also observed reporter activity in the FCs of sns>lacZ samples, that we propose to arise from fusion of βGal-positive FCMs to the FC templates.
Figure 6. Knockdown of sing in both FCs and FCMs results in lethality and reduction in muscle nuclei numbers.
(A) Horizontal sections of 13h APF animal to show activities of gal4 drivers used in FCs (arrows) and FCCs. UAS-lacZ was crossed to each driver, and samples were stained with anti-β-galactosidase (green) and DAPI (blue) to visualize nuclei. Mef2-gal4 is active in all myoblasts; rp298-gal4 is active in FCs; and sns-gal4 is active in FCMs close to the template. βGal accumulation in founder cell nuclei of sns>lacZ samples probably arises from fusion of βGal-positive FCMs to the template. (B–E) Horizontal sections of pharate adults stained for accumulation of F-actin (Phalloidin) and MEF2. (B’–E’) Higher magnification views of muscle fibers and MEF2-positive nuclei (B–B’) Wild type; (C–C’) sing knockdown in founder cells; (D–D’) sing knockdown in fusion competent myoblasts. (E–E’) sing knockdown in founder cells plus fusion competent myoblasts. Note that muscle formation appears normal in all genotypes, but the size and number of nuclei is reduced in E and E’. (F) Quantification of average nuclei counts per unit area. Samples from the double driver are the only group that shows a significant reduction in nuclei number (*p>0.05). Scale bar, 20 µm.
We next used the cell-specific drivers to determine if we could uncover a role for sing in either the FCs of the FCMs. Using rp298-gal4, we expressed sing RNAi in just the FCs. The resulting progeny were 100% viable. When pharate adults were sectioned and stained for F-actin and MEF2, muscle formation was similar to that seen in wild type animals from the same stage (Figure 6B, C). This result suggested that sing knockdown in the FCs was not enough to halt adult myoblast fusion. Similarly, when sing expression was knocked down in only FCMs, using sns-gal4, the progeny were 100% viable and muscles formed normally (Figure 6D). These data suggested that sing might be required in either cell type, but that its presence is not essential in both FCs and FCMs.
To test this model, we also crossed flies in order to knock down sing simultaneously in FCs and FCMs. These progeny were lethal and died as pharate adults. Upon cryosectioning, whilst the muscles appeared robust, there was a clear reduction in the number of nuclei per muscle, and in many cases these nuclei appeared smaller than in other crosses (Figure 6E). To determine whether the number of nuclei present in the double-driver knockdown was significantly different from the other samples, the number of nuclei per 10,000 square microns was calculated from confocal images of control and knockdown muscles. ImageJ was used to count nuclei, and the results of each group were plotted on a bar graph (Figure 6F). A pairwise analysis of each group showed that the numbers of nuclei were significantly different in the double-driver group compared to each of the other samples; there was no significant difference seen between each of the other groups (Figure 6F). Since we previously showed myoblast proliferation rate and myoblast numbers remained unaffected in sing knockdowns, we hypothesize the lowered nuclei counts in the rp298+sns>sing RNAi samples resulted from reduced myoblast fusion occurring. This suggests that sing expression is required in either the FCMs or FCs, but not both. Additionally, the lack of sing in both cell types results in lethality and lowered nuclei counts.
sing315 is active during embryonic myoblast fusion and is regulated by MEF2
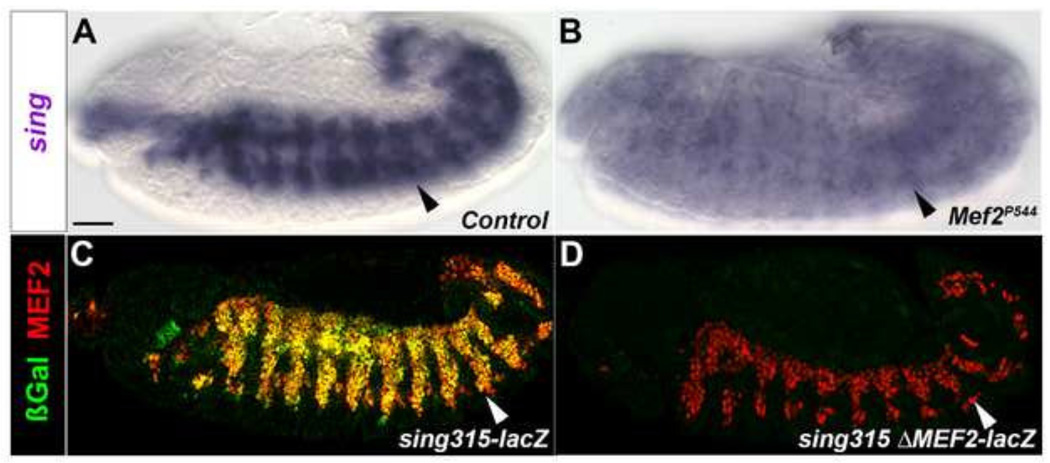
Given that sing function was first characterized in the embryo (Estrada et al., 2007), we determined if the enhancer for adult myoblasts also functioned at the embryonic stage. Using in situ hybridization, we first confirmed that sing was expressed in myoblasts at stage 13, as previously demonstrated by Estrada et al. (2007) (Figure 7A). Under the same conditions, we performed sing in situ hybridization in Mef2 mutants, to determine if sing expression depended upon Mef2 function. We saw diminished sing transcript levels in homozygous Mef2 mutant embryos (Figure 7B), consistent with our data in adults demonstrating that sing is genetically downstream of Mef2.
Figure 7. sing is directly regulated by MEF2 at the embryonic stage.
(A–D) Stage 13 embryos. (A) In situ hybridization to detect sing transcripts in control embryo, with sing transcripts observed in myoblasts (arrowhead). (B) sing expression in Mef2 null embryo is strongly diminished. (C) Immunofluorescent stain of sing-lacZ embryos displays co-localization of MEF2 and β-gal. (D) Immunofluorescent stain of sing-lacZ with mutated MEF2 sites lacks βGal accumulation in embryos. Scale bar, 50 µm.
To determine if the embryonic expression of sing arises from the sing315 enhancer, we carried out immunofluorescent staining of the sing-lacZ embryos. We observed mesoderm-specific expression of the lacZ reporter at stage 13, based upon co-localization of β-galactosidase and MEF2 (Figure 7C). In transgenic embryos carrying the sing-lacZ with both MEF2 sites mutated, there was no expression of the lacZ reporter at any stage of embryonic development (Figure 7D), indicating a direct role for MEF2 in activating sing at the embryonic stage as well as the pupal stage.
DISCUSSION
In this paper, we demonstrate that MEF2 is a transcriptional regulator of adult myoblast fusion, through direct activation of the fusion gene singles bar. We identify a 315-bp enhancer for sing, and show that mutation of conserved MEF2 sites in the enhancer results in a lack of enhancer activity during adult myoblast fusion. We also show that the knockdown of sing during adult muscle development results in pupal lethality and a strong reduction in muscle formation, and that this arises from a failure of fusion. Additionally we demonstrate that the 315bp sing-lacZ enhancer is functional during embryonic myoblast fusion and directly regulated by MEF2. Together our results show a direct role for MEF2 in myoblast fusion through the activation of sing.
Transcriptional control of myoblast fusion
The transcriptional regulation of myoblast fusion genes has received relatively little attention. MEF2 is thought to be a major activator of fusion gene expression, based upon both its requirement for fusion at embryonic and pupal stages (Bour et al 1995; Bryantsev et al 2012), and its direct interaction with a number of fusion genes during embryogenesis (Sandmann et al 2006). Here, we support these observations by demonstrating a direct and essential role for MEF2 in controlling sing expression and by indicating a requirement for sing in adult myoblast fusion. Together, our data and that previously published, provide a direct mechanistic link between MEF2 and myoblast fusion. While there are likely to be a number of additional MEF2 target genes that function in adult myoblast fusion, sing is the first such gene that has been demonstrated to be both required for adult myoblast fusion and that is directly regulated by MEF2.
Nevertheless, there are clearly a number of fusion genes whose expression is not absolutely dependent upon MEF2, either because their expression persists in Mef2 null embryos such as sns (Bour et al 2000), or because the fusion genes are not bound by MEF2 in embryonic ChIP-chip assays such as rost and mbc (Sandmann et al 2006). Moreover, adult myoblasts can at least proceed to the F-actin foci stage of myoblast fusion in the absence of MEF2 function, indicating that genes controlling earlier steps might be expressed independently of MEF2. Identification of additional transcription factors that regulate fusion, and their target genes, will provide a more detailed mechanistic insight into this process, and will also determine if a transcriptional network for fusion differs between FCs and FCMs.
We note that additional regulators of sing expression might still remain to be characterized. In addition to the MEF2 sites, other regions of the sing315 enhancer are evolutionarily conserved, including an E-box located between the two MEF2 sites. The E-box might be a target of activation by Twist, particularly since Sandmann et al (2007) identified sing as a target of Twist using ChIP-chip assays. On the other hand this E-box is not as well conserved in more divergent species (not shown), suggesting either that the E-box is not critical to sing activation, or that differing mechanisms for sing transcriptional activation might be used in more divergent species.
Sing function is required for adult myoblast fusion
Our studies also show a requirement for sing in adult myoblast fusion, with the sing knockdown showing a failure of fusion, muscle loss, and pupal lethality. Close examination of the persistent DLM muscles reveals that a limited amount of fusion has occurred. This may indicate that our sing knockdown is not a fully effective knockdown, and that a small quantity of sing transcript is enough for cells to pass the pre-fusion complex. Nevertheless, it is clear that there is a major requirement for Sing in the formation of the adult muscles.
The persistence of the DLMs can be accounted for by the observation that DLMs form from larval muscle templates, rather than from de novo fusion of myoblasts to newly-specified FCs (Fernandes et al 1991). It is interesting to note that large muscles can still be formed from the larval templates when there is little fusion, suggesting that relatively small numbers of nuclei can support the formation of a larger muscle fiber. Interestingly, in WASp pupal knockdowns where there was a failure of fusion, there was no overt formation of the DLM (Mukherjee et al 2011), which differs from our observations for the DLM. The differences in our observations may either result from some residual fusion taking place in the sing knockdowns; or from an additional requirement for WASp function at subsequent stages of muscle formation.
The function of sing in FCs and FCMs
sing is expressed in both the FCs and FCMs of the developing embryonic myoblasts (Estrada et al 2007). Our studies show that the knockdown of sing in both the FCs and FCMs, using either Mef2-Gal4 or a combination of rp298-Gal4 and sns-Gal4, resulted in adult lethality and lowered number of nuclei in the muscles. Nevertheless, the phenotype was much stronger using Mef2-Gal4, suggesting that this driver more effectively silenced sing expression, probably by the Mef2-Gal4 driver being active at a higher transcriptional level.
This conclusion impacts our interpretation of cell-specific knockdown studies, where we showed that knockdown of sing using drivers for FCs or FCMs did not significantly affect fusion, but that knockdown using the combined drivers affected fusion and muscle function. We interpret these results to mean that sing must be present in only one cell type for fusion to occur. Nevertheless we note that an alternative interpretation is that, only when the drivers were combined, was there sufficient RNAi produced to down-regulate sing expression. A resolution to these alternative explanations must await cell-specific drivers that are active at higher levels, or a more detailed molecular understanding of how Sing impacts myoblast fusion.
Highlights.
The MARVEL domain protein Singles Bar is required for adult muscle formation
sing knockdown myoblasts form actin foci but do not undergo fusion
sing transcription is regulated by Myocyte enhancer-factor-2 at embryonic and adult stages
MEF2 directly regulates a conserved 315-bp enhancer of sing
ACKNOWLEDGEMENTS
We thank Drs Susan Abmayr, Elizabeth Chen, Aaron Johnson, L.S. Shashidhara, and Bruce Paterson for reagents. This work was supported by GM061738 awarded by the NIH/NIGMS to RMC, and a March of Dimes Birth Defects Foundation grant awarded to RMC. BF was supported by the Initiatives to Maximize Student Diversity program, NIGMS grant R25 GM060201. We acknowledge technical support from the Molecular Biology Facility at the Department of Biology, UNM, supported by NIH grant P20 GM103452 from the Institute Development Award (IDeA) Program of NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Abmayr S, Pavlath G. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anant S, Roy S, VijayRaghavan K. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Developmental Biology. 1998;125(8):1361–1369. doi: 10.1242/dev.125.8.1361. [DOI] [PubMed] [Google Scholar]
- Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J. Biol. Chem. 1995;270(40):23246–23249. doi: 10.1074/jbc.270.40.23246. [DOI] [PubMed] [Google Scholar]
- Apitz H, Strunkelnberg M, de Couet HG, Fischbach K-F. Single-minded, Dmef2, Pointed, and Su(H) act on identified regulatory sequences of the roughest gene in Drosophila melanogaster. Dev Genes Evol. 2005;215:460–469. doi: 10.1007/s00427-005-0005-z. [DOI] [PubMed] [Google Scholar]
- Atreya K, Fernandes J. Founder cells regulate fiber number but not fiber formation during adult myogenesis in Drosophila. Developmental Biology. 2008;321(1):123–140. doi: 10.1016/j.ydbio.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Bour B, Chakravarti M, West J, Abmayr S. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes and Development. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes & Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Bryantsev A, Baker P, Lovato T, Jaramillo M, Cripps RM. Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila. Developmental Biology. 2012;362(2):191–207. doi: 10.1016/j.ydbio.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Olson E. Towards a molecular pathway for myoblast fusion is Drosophila. Trends in Cell Biology. 2004;14(8):452–460. doi: 10.1016/j.tcb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Dutta D, Anant S, Ruiz-Gomez M, Bate M. Founder myoblasts and fibre number during adult myogenesis in Drosophila. Development. 2004;131(15):3761–3772. doi: 10.1242/dev.01249. [DOI] [PubMed] [Google Scholar]
- Erickson M, Galletta B, Abmayr S. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. The Journal of Cell Biology. 1997;138(3):589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Maeland A, Gisselbrecht S, Bloor J, Brown N, Michelson A. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Developmental Biology. 2007;307(2):328–339. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Bate M, VijayRaghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Gildor B, Schejter ED, Shilo BZ. Bidirectional Notch represses fusion competence in swarming adult Drosophila myoblasts. Development. 2012;139(21):4040–4050. doi: 10.1242/dev.077495. [DOI] [PubMed] [Google Scholar]
- Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM. In vitro recombination and mutagenesis of DNA. In: White BA, editor. PCR Protocols: Current methods and applications. Vol. 15. Totowa, NJ: Humana Press; 1993. [Google Scholar]
- Kelly K, Meadows S, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mechanisms of Development. 2002;110(1–2):39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- Lau MK. DTK: Dunnett-Tukey-Kramer Pairwise Multiple Comparison Test Adjusted for Unequal Variances and Unequal Sample Sizes. R package version 3.5. 2013 http://CRAN.R-project.org/package=DTK. [Google Scholar]
- Lilly B, Galewsky S, Firulli A, Schulz R, Olson E. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc. Natl. Acad. Sci. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson B, Schulz R, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Developmental Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- McKinsey T, Zhang C, Olson E. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deactetylase 5. PNAS. 2000;97(26):14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J, Black B, Martin J, Olson E. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83(7):1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Morriss GR, Bryantsev AL, Chechenova M, LaBeau EM, Lovato TL, Ryan KM, Cripps RM. Analysis of skeletal muscle development in Drosophila. In: DiMario J, editor. Myogenesis: methods and protocols. New York: Springer; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Gildor B, Shilo BZ, VijayRaghavan K, Schejter E. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 2011;138(11):2347–2357. doi: 10.1242/dev.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A, Isshiki Takako, Takeichi Masatoshi. Regional Specification of Muscle Progenitors in Drosophila: The Role of the msh Homeobox Gene. Development. 1998;125:215–223. doi: 10.1242/dev.125.2.215. [DOI] [PubMed] [Google Scholar]
- Paululat A, Holz A, Renkawitz-Pohl R. Essential genes for myoblast fusion in Drosophila embryogenesis. Mechanisms of Development. 1999;83(1–2):17–26. doi: 10.1016/s0925-4773(99)00029-5. [DOI] [PubMed] [Google Scholar]
- Potthoff M, Olson E. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. URL http://www.R-project.org/ [Google Scholar]
- Ranganayakulu G, Elliott DA, Harvey RP, Olson EN. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development. 1998;125:3037–3048. doi: 10.1242/dev.125.16.3037. [DOI] [PubMed] [Google Scholar]
- Rasband WS. Image J U. S. National Institutes of Health. Bethesda, Maryland, USA: 1997–2014. http://imagej.nih.gov/ij/ [Google Scholar]
- Richardson B, Nowak S, Baylies M. Myoblast Fusion in Fly and Vertebrates: New Genes, New Processes and New Perspectives. Traffic. 2008;9(7):1050–1059. doi: 10.1111/j.1600-0854.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Shannon Y, Roy S, Baylies M. Myoblast Fusion: Where it takes more to make one. Developmental Biology. 2010;341(1):66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Price A, Taylor M, Bate M. Drosophila Dumbfounded: A Myoblast Attractant Essential for Fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends in Biochemical Science. 2002;27(12):599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jensen L, Jakobsen J, Karzynski M, Eichenlaub M, Bork P, Furlong E. A temporal map of transcription factor activity: Mef2 directly regulates target genes at all stages of muscle development. Developmental Cell. 2006;10(6):797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Developmental Biology. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schroter R, Buttgereit D, Beck L, Holz A, Renkawitz-Pohl R. Blown fuse regulates stretching and outgrowth but not myoblast fusion of the circular visceral muscles in Drosophila. Differentiation. 2006;74(9–10):608–621. doi: 10.1111/j.1432-0436.2006.00080.x. [DOI] [PubMed] [Google Scholar]
- Schroter R, Lier S, Holz A, Bogdan S, Klambt C, Beck L, Renkawitz-Pohl R. Kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131:4501–4509. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- Soler C, Han J, Taylor M. The conserved transcription factor Mef2 has multiple roles in adult Drosophila musculature formation. Development. 2012;139(7):1270–1275. doi: 10.1242/dev.077875. [DOI] [PubMed] [Google Scholar]
- St. Pierre SE, Ponting L, Stefancsik R, McQuilton P the FlyBase Consortium. FlyBase 102 advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42(D1):D780–D788. doi: 10.1093/nar/gkt1092. [FBrf0223749] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strünkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Stute C, Kesper D, Holz A, Buttgerei D, Renkawitz-Pohl R. Establishment of cell type specific Gal-4 driver lines for the mesoderm of Drosophila. D.I.S. 2006;89:111–115. [Google Scholar]
- Weiszmann R, Hammonds A, Celniker S. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nature Protocols. 2009;4:605–618. doi: 10.1038/nprot.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]