Summary
For many (if not all) bacterial and archaeal tailed viruses and eukaryotic Herpesvirdae the HK97-fold serves as the major architectural element in icosahedral capsid formation while still enabling the conformational flexibility required during assembly and maturation. Auxiliary proteins or Δ-domains strictly control assembly of multiple, identical, HK97-like subunits into procapsids with specific icosahedral symmetries, rather than aberrant non-icosahedral structures. Procapsids are precursor structures that mature into capsids in a process involving release of auxiliary proteins (or cleavage of Δ-domains), dsDNA packaging, and conformational rearrangement of the HK97-like subunits. Some coat proteins built on the ubiquitous HK97-fold also have accessory domains or loops that impart specific functions, such as increased monomer, procapsid, or capsid stability. In this review, we analyze the numerous HK97-like coat protein structures that are emerging in the literature (over 40 at time of writing) by comparing their topology, additional domains, and their assembly and misassembly reactions.
Keywords: HK-97 fold, capsid protein structure, bacteriophage, coat protein, accessory domain
The amazing and versatile HK97-fold
Viruses, in particular the tailed bacteriophages (Caudovirales), are by far the most abundant biological entities in the biosphere. Phages outnumber bacteria by a factor of ten with an estimated 1031 virus particles on Earth (Suttle, 2007; Wommack and Colwell, 2000). In addition, many phage genomes are integrated into bacterial chromosomes (prophages) with some species harboring as many as 20 prophages (Casjens, 2003). Structural comparison of the major subunits of viral capsids suggests common origins for viruses that infect hosts from different domains of life (Bamford et al., 2002; Bamford et al., 2005). One such viral structure-based lineage is the HK97-like group. Viruses belonging to this group share a common coat (or capsid) protein fold termed after the first coat protein structure solved by X-ray crystallography in this group, bacteriophage HK97 (Helgstrand et al., 2003; Wikoff et al., 2000) (Figure 1A, F). This lineage is usually only evident with structural studies, as proteins with this fold often share only 10 – 15 % or less sequence identity. Advances in electron cryomicroscopy (cryo-EM) and three-dimensional (3D) image reconstruction have accelerated the means by which structures of large, macromolecular complexes can be determined at subnanometer resolutions (Zhou, 2008), which has resulted in a plethora of structures of capsids having the HK97-fold (over 40 at time of writing, Table 1). The majority of capsids using the HK97-fold are of viruses infecting bacteria (bacteriophage or phage). However, this fold is also found in the coat protein floor domain of eukaryotic Herpesvirdae (shaded in Table 1), including Herpes simplex virus type 1 (HSV-1) (Baker et al., 2005; Zhou et al., 2000), murine cytomegalovirus (MCMV) (Hui et al., 2013), pseudorabies virus (PRV) (Homa et al., 2013), and rhesus monkey rhadinovirus (RRV) (Zhou et al., 2014), as well as the archaeal Haloarcula sinaiiensis tailed virus 1 (HSTV-1) (Pietila et al., 2013).
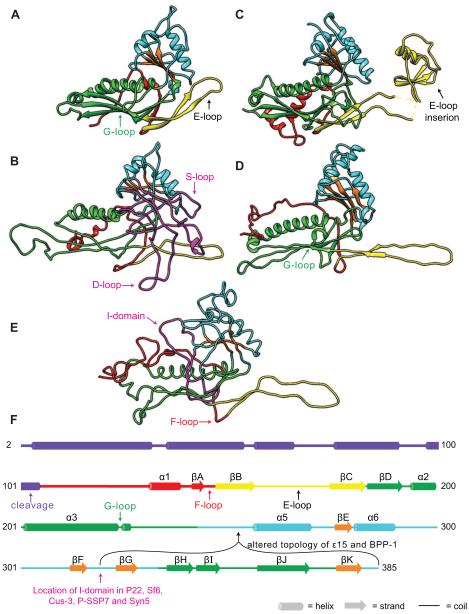
Figure 1. HK97 and related coat proteins.
HK97 coat protein in the immature state “Prohead II” (A, PDB entry 3E8K) is compared to P22 (B, from (Rizzo et al., 2014)), T4 (C, 1YUE), BPP-1 (D, 3J4U), and P-SSP7 (E, 2XD8) coat proteins. The common structural elements are colored correspondingly; N-arm (red), E-loop (yellow), P-domain (green), A-domain (cyan), β–hinge (orange). P22 and P-SSP7 coat proteins have insertions between βF and βG of the β-hinge (magenta in B and E). (F) Linear diagram of the secondary structure of HK97 coat protein based on PDB 3E8K with predicted secondary structure (PSIPRED) for the Δ-domain. β-strands E, F, G and K make up the β-hinge. The “backbone” is color-coded as in A – E.
Table 1.
Representative viruses with different T number classes having coat proteins with the HK97 fold.
| Capsid | T- number |
Maximum diameter (nm) |
Family | Unique properties | References |
|---|---|---|---|---|---|
|
T. maritima
encapsulin |
1 | 24 | – | 60° rotation of E-loop | (Sutter et al., 2008) |
| SsP2 | 3 | 32 | – | Crenarchaeota provirus | (Heinemann et al., 2011) |
|
M. xanthus
encapsulin |
3 | 32 | – | (McHugh et al., 2014) | |
| PfV encapsulin | 3 | 36 | – | Euryarchaeota | (Akita et al., 2007) |
| 029 | 3 Q=5 |
45 wide 54 long |
P | C-terminal addition of Big2 domain | (Morais et al., 2005) |
| C1 | 4 | 50 | P | Insertion domain and additional density across twofold axes |
(Aksyuk et al., 2012) |
| P2 | 7d | 60 | M | Also makes T=4 capsids (40 nm diameter) with P4 Sid | (Dearborn et al., 2012) |
| Sf6 | 7/ | 60 | P | I-domain | (Parent et al., 2012b) |
| A | 7/ | 60 | S | (Lander et al., 2008) | |
| T7 | 7* | 60 | P | Frameshift produces C-terminal extension in ~ 10% of proteins |
(Agirrezabala et al., 2007; Guo et al., 2014; lonel et al., 2011) |
| Syn5 | 7* | 60 | P | Small I-domain | (Gipson et al., 2014; Pope et al., 2007) |
| SPP1 | 7/ | 61 | S | Extended P-domain | (White et al., 2012) |
| HSTV-1 | 7/ | 62 | P | Infects Archaea | (Pietila et al., 2013) |
| K1E/K1-5 | 7* | 63 | P | C-terminal extension | (Leiman et al., 2007) |
| Gifsy-2 | 7/ | 63 | – | Prophage | (Effantin et al., 2010) |
| 80a | 7/ | 63 | S | P-loop in P-domain involved in threefold interactions |
(Spilman et al., 2011) |
| P-SSP7 | 7* | 65 | P | Long F-loop makes contact across twofold axes, extended N-arm |
(Liu et al., 2010) |
| TP901-1 | 7* | 66 | S | (Bebeacua et al., 2013) | |
| HK97 | 7/ | 66 | S | Covalent crosslinks in mature capsid |
(Gertsman et al., 2009; Helgstrand et al., 2003; Wikoff et al., 2000) |
| BPP-1 | 7/ | 67 | P | Altered topology, extended N-arm | (Zhang et al., 2013) |
| CUS-3 | 7/ | 69 | P | I-domain | (Parent et al., 2014) |
| £15 | 7/ | 70 | P | Altered topology | (Baker et al., 2013; Jiang et al., 2008) |
| CW02 | 7/ | 70 | P | (Shen et al., 2012) | |
| P22 | 7/ | 71 | P | I-domain | (Chen et al., 2011; Parent et al., 2010a) |
| Basilisk | 9 | 72 | S | (Grose et al., 2014) | |
| SIO-2 | 12 | 80 | S | (Lander et al., 2012) | |
| T5 | 13l | 90 | S | (Effantin et al., 2006) | |
| T4 | 13/ Q = 20 |
90 wide 200 long |
M | Insertion in E-loop and separate protein for penton position |
(Fokine et al., 2005b) |
| Bellamy | 13 Q = 24 |
95 wide 135 long |
M | Personal communication Roger Hendrix |
|
| SPO1 | 16 | 108 | M | (Duda et al., 2006) | |
| HSV-1 | 16 | 125 | H | Additional middle and upper domains |
(Baker et al., 2005; Zhou et al., 2000) |
| PRV | 16 | 125 | H | Additional middle and upper domains |
(Homa et al., 2013) |
| RRV | 16 | 130 | H | Additional middle and upper domains |
(Zhou et al., 2014) |
| MCMV | 16 | 131 | H | Additional middle and upper domains |
(Hui et al., 2013) |
| ϕM12 | 191 | 100 | M | (Stroupe et al., 2014) | |
| N3 | 19* | 119 | M | Personal communication Roger Hendrix |
|
| PAU | 25 | 130 | M | Personal communication Roger Hendrix |
|
| ϕRSL1 | 27 | 123 | M | (Effantin et al., 2013) | |
| PBS1 | 27 | 140 | M | Personal communication Roger Hendrix |
|
| ϕKZ | 27 | 145 | M | (Fokine et al., 2005a) | |
| 121Q | 28* | 140 | M | Personal communication Roger Hendrix |
|
| Phage G | 52* | 185 | M | Personal communication Roger Hendrix |
P – Podovirdae; M – Myovirdae; S – Siphovirdae; H – Herspesvirdae; ‘-‘ Unclassified family
Skew class, but handedness was not determined
l – laevo
d – dextro
In addition to viruses, the HK97-fold has also been observed in encapsulins, a large, yet mostly uncharacterized family of conserved protein nanocompartments (Akita et al., 2007; McHugh et al., 2014; Sutter et al., 2008). Structural studies of encapsulins from Thermotoga maritima (Sutter et al., 2008), Myxococcus xanthus (McHugh et al., 2014), and the archaeon Pyrococcus furiosus (Akita et al., 2007), revealed spherical particles with icosahedral symmetry composed of subunits with significant structural similarity to the coat protein of HK97. The T. maritima and M. xanthus (EncA) encapsulins were both shown to contain enzymes involved in defense against oxidative stress (McHugh et al., 2014; Sutter et al., 2008). Ferritin-like proteins (Flp) or dye-decolorizing peroxidases (DyP) are targeted to the interior of encapsulins via unique C-terminal extensions. Likewise, the P. furiosus virus-like (PfV) particles have a disordered N-terminal extension that has substantial sequence similarity with the Flp packaged in the encapsulins from T. maritima. In this case, the enzyme is linked with the capsid-forming domain so that assembly and packaging occur simultaneously. The encapsulin shell serves a protective role for the cell by sequestering toxic molecules (McHugh et al., 2014), and selectively controls movement of substrates into the nanocompartment. The encapsulin shell could also serve a protective role for the enzymes it carries, similar to the genome protective role of viral capsids (Sutter et al., 2008). The use of the HK97-fold by both viruses and prokaryotic cells points to the versatility and pliability of this building block.
Viruses using the HK97-fold share unexpected similarities beyond the fold of their coat proteins including the architecture of their virions, an internal scaffolding (or Δ-domain, purple in Figure 1F) protein-mediated assembly (Huang et al., 2011), active dsDNA packaging, and capsid maturation events. Proteins with the HK97-fold exhibit several common features including: an N-arm (N-terminal region, red in Figure 1) that can have α-helical content; an E-loop (extended loop, varying lengths, yellow in Figure 1) that is a two-stranded anti-parallel β-sheet (β2 and β3); a P-domain (peripheral domain, green in Figure 1) with a long (~ 20 – 30 amino acids) “spine helix” and an unusually long β-sheet; and an A-domain (axial domain, cyan in Figure 1) with a central β-sheet (referred to as the β-hinge, orange in Figure 1) surrounded by short helices and loops. Recently, two examples of HK97-like coat proteins with altered topologies were published (Baker et al., 2013; Zhang et al., 2013) (Figure 1D, F). In epsilon15 and BPP-1 coat proteins, the section corresponding to βG through βK (see numbering scheme in Figure 1F) in HK97 coat protein is inserted before the A-domain α-helices 5 and 6, with βF as the new C-terminus. Possibly, this altered topology will be more common as additional cryo-EM reconstructions of sufficiently high resolution to distinguish between the topologies are produced. Indeed, a lower resolution cryo-EM reconstruction of epsilon15 was previously misthreaded (Jiang et al., 2008) before a higher resolution cryo-EM structure from the same group revealed the altered topology (Baker et al., 2013). Despite the altered topology, the coat proteins of phages epsilon15, BPP-1, and HK97 have remarkably similar tertiary structures and quaternary arrangement of their subunits.
In capsids, HK97-like coat protein subunits are organized into pentons and hexons. (An exception would be for capsids with a triangulation (T) number of 1, which would be composed entirely of pentons. However, the only example of a T = 1 structure with the HK97-fold is the encapsulin nanocompartment (Sutter et al., 2008). A T = 1 viral capsid formed with subunits having the HK97-fold is unlikely to have enough internal capacity to contain a genome large enough for all the essential genes.) HK97-like coat proteins are roughly triangular in shape, with the A-domains clustered at the center of the capsomers (collective term for pentons and hexons) and the P-domains along the perimeter of the capsomers. The versatility of the HK97-fold is also evident when comparing the diameters of capsids with confirmed HK97-like coat proteins, which range from ~ 24 nm to ~ 185 nm (Table 1). This fold evolved to accommodate these dramatic differences in capsid diameter and the varying degrees of capsid curvature accompanying the spectrum of diameters. This range of capsid diameters is due to the different T numbers (ranging from 1 – 52 for capsids with confirmed HK97-like coat proteins) and thus the number of hexamers (0 – 510, respectively). Particles formed from subunits having the HK97-fold with certain T numbers have a handedness (T = 7, 13, 19, etc.). These are nearly always organized in laevo capsid lattices, but phage P2 uses the same fold to form T = 7 dextro lattices, again demonstrating the pliability of the fold as the chiral coat protein can accommodate even mirror image capsid configurations (Dearborn et al., 2012). While most tailed phages characterized by electron microscopy have isometric capsids, approximately 15% have prolate capsids (Ackermann and Prangishvili, 2012). Prolate heads, including those of T4 (Fokine et al., 2004) and 029 (Tao et al., 1998), are icosahedra elongated along a fivefold axis. It is not entirely clear how the length of these prolate capsids is determined, adding an additional conundrum to the assembly mechanism of these viruses that will not be discussed here.
Morphogenesis of bacteriophage P22
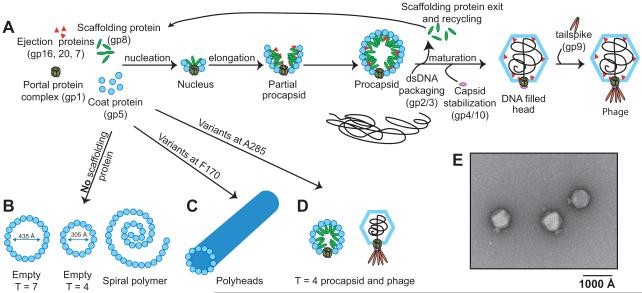
Bacteriophage P22 provides a well-studied example for assembly of capsids formed from subunits with the HK97-fold. Assembly of bacteriophage P22 in vivo (Figure 2A) proceeds via a nucleation-limited reaction in which 415 copies of coat protein (gp5, the product of gene 5) co-polymerize with 60-300 scaffolding proteins (gp8), the dodecameric portal protein complex (gp1), and ejection proteins (gp7, gp16 and gp20) (King et al., 1976; Prevelige and King, 1993). Coat protein monomers are added to the growing edge, eventually producing a closed, spherical, precursor structure (“procapsid”) in which the coat proteins are organized with T = 7 laevo quasi-symmetry. During assembly scaffolding protein catalyzes, stabilizes, and directs geometry of the procapsid. Without scaffolding protein, P22 coat protein polymerizes in vivo into aberrant forms such as T = 7 or T = 4 empty procapsids (shells) and aberrant spiral polymers (Earnshaw and King, 1978; Lenk et al., 1975; Thuman-Commike et al., 1998) (Figure 2B). While empty T = 7 procapsids have the correct icosahedral symmetry, they lack a portal protein complex and are therefore dead-end products. Scaffolding protein is not found in mature phage (Casjens and King, 1974), but exits during maturation and is therefore referred to as an assembly chaperone. The maturation event also involves packaging of dsDNA through the unique portal protein complex (Bazinet and King, 1988), capsid expansion, and a change in capsid morphology from one that is nearly spherical to one that is faceted (Earnshaw et al., 1976; Prasad et al., 1993). Considerable conformational rearrangements of coat protein N-arm, E-loop, and A-domain occur upon maturation. In P22 coat protein, the β-hinge has been shown to be the most dynamic region of the subunit during capsid maturation (Kang et al., 2006; Kang and Prevelige, 2005; Parent et al., 2010a; Teschke and Parent, 2010). This hinge appears to direct motion of the A-domain during hexon symmetrization that occurs simultaneously with expansion. Finally, plug proteins (gp4, gp10 and gp26) close the portal protein channel and tailspike proteins (gp9) are added to complete the mature infectious phage (Strauss and King, 1984) (Figure 2E).
Figure 2. Assembly and misassembly of bacteriophage P22.
In a normal infection, P22 coat protein assembles into mature, infectious phage (A, E) through a morphogenic pathway that includes an internal scaffolding protein-containing T = 7 procapsid. Aberrant assembly products are formed in the absence of scaffolding protein (B) or can be caused by amino acid substitutions in coat protein at F170 (C) or A285 (D).
Phage P22 has well characterized genetics, can be easily manipulated in vivo, and procapsid-like particles (procapsids without portal protein or ejection proteins) can be assembled in vitro from purified coat and scaffolding proteins. Therefore, P22 has long been used as a model system for coat protein and capsid structural studies, as well as for understanding assembly mechanisms of dsDNA viruses. This review will use phage P22 to describe assembly of capsids using the HK97-fold, pointing out variations along the way.
Assembly chaperoned by a scaffolding protein
For capsids with T numbers greater than 1, capsid subunits occupy non-identical, or quasi-equivalent, positions within the lattice (Caspar and Klug, 1962). The capsid sizes of bacterial and archaeal tailed viruses and Herpesvirdae have likely evolved over time to accommodate larger genomes (Hendrix, 2009). Increasing the capsid size can be accomplished by increasing the T number and thus the number of subunits. This results in a larger number of quasi-equivalent sites in the icosahedron, requiring polymorphism of the coat protein subunits. Tailed viruses and Herpesvirdae have evolved to use auxiliary proteins (scaffolding protein or core protein) or Δ-domains to restrict the conformational possibilities to achieve correctly sized capsids (Dokland, 1999).
The association between P22 coat and scaffolding proteins is regulated by electrostatic interactions (Parent et al., 2005; Parker and Prevelige, 1998; Zlotnick et al., 2012). About 30% of residues in the C-terminal helix-turn-helix domain of scaffolding protein (residues 283 – 303) are positively charged, and this region has been shown to interact with coat protein. Specifically, a basic residue (R293) of scaffolding protein is essential for coat protein binding and procapsid assembly (Cortines et al., 2011). Likewise, a single acidic amino acid in coat protein (D14) is indispensable for interaction with scaffolding protein (Cortines et al., 2014). Changing the essential coat protein D14 leads to a lethal phenotype for phage production and results in assembly of aberrant structures, similar to the spiral polymers seen in infections without scaffolding protein present (Earnshaw and King, 1978). A second scaffolding protein residue, K296, likely interacts with the coat protein N-arm residue E15. This additional interaction modulates the affinity between coat protein and scaffolding protein (Cortines et al., 2014). Coat protein D14 and E15 are modeled to be in a loop between two α-helices of the N-arm (Parent et al., 2010a). In the assembled structures, coat protein N-arms are positioned at icosahedral and quasi-threefold axes of symmetry on the inner surface of the procapsid. Identification of coat protein D14 as a primary interaction site for scaffolding protein confirmed previous cryo-EM data suggesting that the scaffolding protein-binding site of coat protein was at the so-called trimer tips (Thuman-Commike et al., 2000; Thuman-Commike et al., 1999). Therefore, coat protein and scaffolding protein have very specific interaction sites that direct robust assembly mainly through two amino acids, providing support for the hypothesis that virus capsid assembly is governed by seemingly simple interactions. The coat protein:scaffolding protein interaction may be simple, but it is also extremely sophisticated, as it ensures morphogenetic fidelity and efficiency of procapsid assembly. While an understanding of the specific interactions has been achieved for P22, the molecular mechanism by which coat protein assembly goes astray and how scaffolding protein prevents this is still not understood for any dsDNA virus.
Most tailed viruses including P22 as well as Herpesvirdae have a scaffolding protein that is separately expressed from the coat protein. However, some exceptions exist such as phages HK97 (Duda et al., 1995b) and T5 (Effantin et al., 2006; Huet et al., 2010) that have Δ-domains within their coat protein sequence that function like a scaffolding protein. The coat protein of phage HK97, which exists in equilibrium between pre-assembled hexons and pentons, co-polymerizes with the portal protein complex and a viral encoded protease to form a precursor particle (Prohead I) (Duda et al., 1995a; Duda et al., 1995b). The N-terminal 102 residues of HK97 coat protein (Δ-domain, purple in Figure 1F) assist in assembly and are subsequently removed by the protease (Conway et al., 1995). Secondary structure prediction of the Δ-domain (Figure 1F) suggests that it has high α-helical content, similar to many separately expressed scaffolding proteins (Dokland, 1999). N-terminal deletions of the Δ-domain revealed that it is absolutely essential for procapsid assembly (Oh et al., 2014). Deletion of the entire Δ-domain completely abolished assembly of all products, including capsomers, suggesting that it is important for penton and hexon formation and not just intercapsomer interactions. A crystal structure of Prohead I showed density on the inner surface of the shell that was attributed to the Δ-domain (Huang et al., 2011). This density is more disordered than the main part of the coat protein subunit, but clearly showed that interactions between Δ-domains occur within the same pentamer or hexamer, and not across capsomers. Thus, biochemical and structural studies agree that the Δ-domain is important for capsomer formation.
The HK97 Δ-domain also possesses some intramolecular chaperone-like activity, as removing it resulted in aggregation of the majority of the coat protein (Oh et al., 2014). HK97 coat protein interacts with the host’ s GroEL/ES chaperone complex to properly fold (Ding et al., 1995; Xie and Hendrix, 1995) and then likely requires its Δ-domain for capsomer formation. However, without the Δ-domain the monomeric coat protein is unstable and unable to assemble into capsomers, resulting in aggregation. Likewise, P22 scaffolding protein has been shown to exhibit chaperone-like activity for coat protein variants that have folding defects. Two global suppressors (su, D163G and T166I) of temperature-sensitive-folding (tsf) mutations in P22 coat protein rescue folding via enhanced interaction with scaffolding protein (Parent et al., 2004; Parent and Teschke, 2007). Thus, both HK97’s Δ-domain and P22’s scaffolding protein act as folding chaperones of their coat proteins by promoting assembly to a more stable state by partitioning the precursors to assembled particles before aggregation can occur.
Accessory domains and loops
In addition to the canonical HK97-like core, many coat proteins contain additional accessory domains or extended loops, likely imparting specific functions. These often stabilize intra and/or intercapsomer interactions (Fokine et al., 2005a; Fokine et al., 2005b). For example, the coat protein of phage T4 (Figure 1C) has a chitin-binding-like insertion domain in its E-loop (Fokine et al., 2004). This additional domain bridges neighboring molecules within one capsomer. Similarly, phage 029 coat protein has a group 2 bacterial immunoglobulin-like (BIG2) domain at its C-terminus (Morais et al., 2005) that links neighboring capsomers. While no biochemical analysis has been performed, the location of these domains strongly suggests they are involved in capsomer stabilization (T4) or capsid stabilization (029). Herpesvirdae coat proteins have middle and upper domains protruding from the floor domain to form towers, which are involved in interactions with several outer capsid proteins, including the triplex proteins (VP19C and VP23), a capsid decorator protein (VP26), and tegument proteins (Homa et al., 2013; Hui et al., 2013; Zhou et al., 1999; Zhou et al., 2000). The crystal structure of the 65 kDa “upper domain’ of HSV-1 coat protein has been determined (Bowman et al., 2003). The upper domains of hexons make electrostatic and polar contacts between adjacent subunits, while the interactions between the upper domains in pentons is weaker. The upper domain forms a stable structure and has been proposed to be the folding nucleus for this large protein, while allowing for the subunit flexibility required to assemble into both pentons and hexons (Bowman et al., 2003)
The coat proteins of phages P22 (Chen et al., 2011; Parent et al., 2010a), Sf6 (Parent et al., 2012b), and CUS-3 (Parent et al., 2014) each have an accessory domain inserted (Insertion domain, I-domain) between βF and βG of the β-hinge (Magenta in Figure 1B, F). Cryo-EM reconstructions revealed that the I-domains are positioned as surface-exposed protrusions from P22, Sf6, and CUS-3 capsids. The reconstructions of P22 and CUS-3 showed that their I-domains make intercapsomer contacts with a neighboring subunit across the twofold axes of symmetry (Chen et al., 2011; Parent et al., 2010a; Parent et al., 2014), while the Sf6 I-domain appears to make extensive intrahexon contacts with neighboring I-domains (Parent et al., 2012b). In P22, this density across the twofold axes was assigned to a loop (D-loop) in the I-domain (Chen et al., 2011); however, the I-domain was the least well-resolved region of the model. A recent NMR structure of the isolated P22 I-domain (coat protein residues 223 – 345) was used to improve the full-length coat protein model from Chen et al. 2011 (Rizzo et al., 2014) (Figure 1B). P22 I-domain adopts a six-stranded, antiparallel β-barrel with an accessory β-sheet, a short α-helix, and two long, disordered loops (Rizzo et al., 2012; Rizzo et al., 2014). One of these loops corresponds to the previously identified D-loop (Chen et al., 2011), although the residues are different from those previously published (residues 234 – 254 rather than 257 – 277). The D-loop, which is dynamic in isolated I-domain and intact monomeric coat protein, forms intermolecular salt-bridges across twofold axes of symmetry in the procapsid (Rizzo et al., 2014). The other loop (residues 281 – 291), which was not visualized in the previous coat protein models, contacts the A-domain and E-loop of the same subunit. Coat protein variants in this newly visualized loop produce petite capsids (discussed below), implicating it in capsid size determination. Thus, we named it the size determination loop, or S-loop.
With 40% sequence similarity between the I-domains of P22 and CUS-3 (Parent et al., 2014), it is possible that the CUS-3 I-domain has a similar overall fold to the I-domain of P22, including the presence of the D- and S-loops. The cryo-EM reconstruction of CUS-3 that shows density across the twofold axes, in a similar fashion as P22, also strengthens this hypothesis. However, the I-domain of Sf6 has significantly less sequence similarity to the I-domains of P22 (27%) and CUS-3 (22%). This may account for the differences in intracapsomer (Sf6) versus intercapsomer (P22 and CUS-3) interactions seen in the cryo-EM reconstructions. Higher resolution structures of CUS-3 and Sf6 I-domains are required to determine if these modules are structurally and functionally similar to each other and to P22 I-domain. Phage P-SSP7 and related phage Syn5 have a small insertion between βF and βG of the β-hinge (Figure 1E, F), which is the same position as P22’s I-domain (Gipson et al., 2014; Liu et al., 2010), suggesting an insertion at this position may be favored in evolution. The I-domain of these phages does not appear large enough to make contacts across the twofold axis of symmetry, indicating it has a different function for these phages.
Cryo-EM reconstructions of phage C1 (Aksyuk et al., 2012) and phage P-SSP7 (Liu et al., 2010) capsids revealed additional density not attributed to the HK97-fold, and distinct from the I-domain, at the twofold axes that results in interactions across capsomers. The additional density in P-SSP7 is due to an extended F-loop that contacts the P-domain of a subunit in an adjoining capsomer (Figure 1E). Similarly, the coat proteins of HK97, and some related phages, have a glycine rich (GDGTGDNLEG, first and last glycine residues are strictly conserved) G-loop that also reaches across twofold axes of symmetry to interact with the E-loop of an adjacent subunit (Tso et al., 2014) (Figure 1A). BPP-1 coat protein and the T. maritima encapsulin coat protein also have loops at a similar position, with the strictly conserved first and last glycine residues (GDSSIDAEKFMG and GCEKSGVKG, respectively). The HK97 G-loop appears to be rigid and is proposed to limit subunit conformational flexibility during capsomer assembly (Tso et al., 2014). Indeed, the interaction between the G- and E-loops was shown to be crucial for proper assembly, as deleting the G-loop or altering the residues involved in the interaction (D231 in the G-loop or K178 in the E-loop) inhibited Prohead I formation. Most of the variant coat proteins remained as soluble capsomers; however, some of the variants also produced tubes of coat protein (Tso et al., 2014). Interestingly, preliminary mutagenesis studies of the P22 D-loop, which also extends across twofold axes, have also identified a variant that results in coat protein tubes (D’Lima and Teschke, unpublished). Therefore, both the G-loop of HK97 and the D-loop of P22 may serve to control proper twofold interactions during assembly. The intercapsomer contacts made by the G-loop of HK97 are lost during maturation, suggesting they may be important for procapsid assembly but not stability of the mature capsid. The G-loop interaction may not be necessary in mature capsids as maturation produces a network of covalent crosslinks, unique to HK97.
In other viruses the N-arm of the HK97-fold is altered to increase capsid stability. The N-arm of most HK97-like proteins usually resides in the interior of the assembled capsids (red in Figure 1A, B, C, lies below the green P-domain); however in P-SSP7, BPP-1 and T7, the N-arms are elongated and positioned on the exterior of the mature capsids. In phage P-SSP7, the N-arm of the coat protein (red in Figure 1E) reaches up and over the top of the subunit to contact the E-loop of an adjacent subunit (Liu et al., 2010). In phage BPP-1, the N-arm (red in Figure 1D) extends up to contact one the phage’s cement proteins, which are added to the surface to stabilize the capsid (Zhang et al., 2013). In phage T7, the N-terminal helix unfolds and moves from the interior of the capsid to the exterior where it refolds into a hairpin structure. The hairpin, which tucks into a newly exposed pocket, is used to stabilize the capsid via intercapsomer interactions (Gou et al., 2014). As can be seen from the many examples presented above, accessory domains and extended loops provide many different ways to accomplish the same goals of stabilizing monomers, procapsids, or mature capsids.
When assembly goes awry: using aberrant products to understand proper assembly
For most capsids with HK97-like coat proteins, chemically identical subunits occupy non-identical sites in the icosahedron. This relies on coat protein plasticity as well as a mechanism by which identical subunits can be “switched” into slightly different conformations required to occupy quasi-equivalent sites in the icosahedron (Figure 3A), a process known as conformational switching. Conformational flexibility is also necessary for the rearrangements during the capsid maturation process. Thus, a fine balance is necessitated between this conformational polymorphism and stability of monomeric coat proteins.
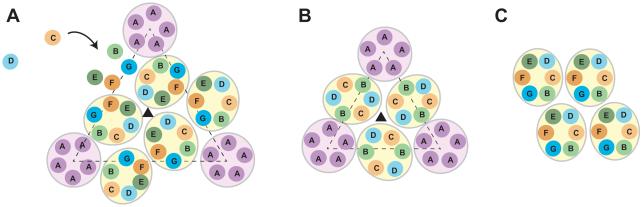
Figure 3. Capsid symmetry.
Icosahedral capsids are composed of twenty equilateral triangles (dashed triangles in A and B), whose size is proportional to the T number of the capsid. (A) The T = 7 procapsid contains 7 quasi-equivalent subunits (labeled A – G). Assembly of P22 occurs through addition of monomeric coat proteins to the growing edge of the capsid, forming hexons with a pronounced skew. (B) The T = 4 shell contains 4 quasi-equivalent subunits (labeled A – D). In the T = 4 structure, each of the twenty equilateral triangles is centered on a threefold axis. (C) An alternative arrangement of hexons identified in P22 tubes demonstrating the effect on capsomer organization driven by scaffolding protein.
Viral morphogenesis sometimes yields aberrant, off-pathway products. These non-infectious particles commonly arise from mutations in genes encoding structural proteins (e.g., an amber mutation in scaffolding protein or point mutations in coat protein). As mentioned above, in the absence of scaffolding protein, P22 coat protein will form empty, aberrant assemblies, consisting of both hexons and pentons, such as T = 4 and T = 7 icosahedral capsids, or spiral structures (Figure 2B). Similarly, mutations resulting in amino acid substitutions in coat protein that affect its flexibility or structure can influence the morphology of resulting products. Two residues of interest in P22 coat protein are F170 (Figure 2C) and A285 (Figure 2D), as substitutions at these positions result in polyheads and petite capsids, respectively.
Misplaced pentons: Evidence that scaffolding protein directs intercapsomer interactions
Analysis of wild-type (WT) P22 coat protein assembled in the absence of scaffolding protein suggests the assembly chaperone is inherently necessary for proper intercapsomer interactions. Empty T = 4 particles have both pentons and hexons; however, the pentons are placed too closely (Figure 3B). Structural comparisons of the T = 7 and T = 4 shells assembled in the absence of scaffolding protein revealed that each has similar penton and hexon clusters, but with slightly increased curvature of the T = 4 hexon (Thuman-Commike et al., 1998). Similarly, spiral polymers consist of pentamers and hexamers with seemingly random arrangement (Earnshaw and King, 1978). This indicates that WT coat protein is able to adopt both capsomers without assistance from scaffolding protein. However, scaffolding protein enhances proper arrangement of these capsomers to achieve the correct curvature to produce a T = 7 structure.
In P22, scaffolding protein also contributes to the stability of properly assembled procapsids. Recently, we completed a thermodynamic examination of procapsids and spiral polymers resulting from concentrating coat protein in vitro in the absence of scaffolding protein (Zlotnick et al., 2012). The association energy per coat protein in the procapsid was extrapolated to [scaffolding protein] = 0, indicating a pseudo-critical concentration (or Kd apparent) for assembly of coat protein into procapsids in the absence of scaffolding protein of 16 μM. The extrapolated value for procapsid formation in the absence of scaffolding protein was compared to the experimentally measured value for spontaneous assembly of coat protein (into spiral polymers) of ~8.5 μM. The difference between the 16 μM Kd apparent estimated for a coat protein:coat protein interaction in a procapsid and the 8.5 μM critical concentration observed for coat protein self-assembly suggests that interactions between subunits must be strained to obtain a procapsid. While the coat protein:coat protein interaction is stronger for the non-procapsid polymer than calculated for procapsid without scaffolding protein (shell), the overall stability of scaffolding protein-filled procapsids is much greater. Therefore, during procapsid assembly the energetic difference between coat protein:coat protein interactions is overcome by the contribution of each scaffolding protein to procapsid stability (~1.3 kcal/mol), with binding of approximately 100 scaffolding protein molecules required to produce a procapsid with similar overall stability to the non-procapsid polymer. Recently, ~112 scaffolding proteins were shown to be present in procapsids via charge detection mass spectrometry (Keifer et al., 2014). This is very close to the number required to achieve greater stability than the non-procapsid polymer. Therefore, scaffolding protein interactions with coat protein during assembly overcome the less than ideal coat protein interactions. The higher energy state of coat protein:coat protein interactions in an empty coat protein shell may be functionally important for the lattice to undergo the transition to produce the stable, mature head (Galisteo and King, 1993). Scaffolding protein exit from procapsids produces a structure with a higher energy that may be poised for subsequent conformational rearrangements occurring during maturation (Johnson, 2010; Zlotnick et al., 2012). In vivo, scaffolding protein exit is induced by DNA packaging, which could be responsible for the input of energy required to move the particle into a higher energy state. Metastable intermediates have been characterized in the maturation reaction of HK97 as well (Cardone et al., 2014).
The β-hinge and proper A-domain flexibility control intracapsomer interactions
The β-hinge is a conserved, four- or five-stranded β-sheet (up, up, down, up, up topology) that links the A- and P-domains of HK97-like proteins. In P22, three global su substitutions (D163G, T166I, and F170L) of coat protein folding defects highlight the β-hinge as important for coat protein folding (Aramli and Teschke, 1999). In the absence of their tsf parents, the su substitutions result in the assembly of coat protein into aberrant forms, indicating this region is also important for allowing the coat protein monomer to assemble into a functional product. D163G and T166I variants result in the production of spiral products and giant capsids, whereas the F170L coat protein was shown to promote formation of tubes of coat protein hexons (Figure 2C) (Parent et al., 2007; Suhanovsky et al., 2010), similar to those seen in other phages and viruses (Baker and Caspar, 1983; Bancroft et al., 1976; Georgopoulos et al., 1973; Parker et al., 1983; Steven et al., 1976). More drastic substitutions of alanine and lysine at residue F170 resulted in a stronger phenotype, compared to the F170L variant coat protein, as these variants made more numerous and longer tubes. Affinity chromatography and scaffolding protein reentry experiments revealed the coat protein variants had decreased ability to interact with scaffolding protein. Cryo-EM and the Iterative Helical Real Space Reconstruction (IHRSR) method (Egelman, 2000) were used to examine the structures of P22 polyheads formed from F170L and F170A coat protein variants (F170K polyheads were not stable enough for this analysis) (Parent et al., 2012a; Parent et al., 2010b). The polyheads consist of coat protein hexamers organized as helical arrays with rotational (Cn) symmetry. The polyhead hexamers clearly resemble hexons either in procapsids for the F170L coat protein variant, or in mature virions for the F170A coat protein variant. While the intrahexamer interactions do not appear to be drastically altered, the hexon:hexon interactions in the polyheads are quite distinct from those in the coat protein shells of procapsids and virions. In icosahedral T = 7 particles the hexons are related by threefold symmetry (Figure 3A), but in the polyheads the hexons are all identically oriented relative to the helix axis (Figure 3C). These differences in hexon orientation result from different intercapsomer interactions, specifically at the threefold axes, which are at least in part directed by scaffolding protein as the scaffolding protein-binding site on coat protein maps to the inner surface of the procapsid shell at the icosahedral and quasi-threefold axes of symmetry (Cortines et al., 2014; Thuman-Commike et al., 2000; Thuman-Commike et al., 1999). Assembly of polyheads by F170L coat protein, which shows the least pronounced defect in scaffolding protein binding, was inhibited with increasing concentration of scaffolding protein, indicating scaffolding protein is capable of correcting the improper threefold interactions. Therefore, as seen with spiral polymers, scaffolding protein is important to direct threefold interactions. Interaction with scaffolding protein thus affects the geometry and imposes the correct radius of curvature, resulting in assembly of proper products with exquisite fidelity. The nucleation complex for P22 procapsids has been proposed to be a dimer of scaffolding protein and a pentamer of coat protein, with both monomer and dimer scaffolding protein important for completion of procapsids (Prevelige et al., 1993; Tuma et al, 2007). Alternatively, we propose scaffolding protein drives the formation of specific, trimeric coat protein interactions based on the structural analysis of coat protein tubes (Parent et al., 2010b). These in vitro assembly experiments were done in the absence of the portal protein complex. In other phages and Herpesvirdae, the portal protein complex forms the nucleation complex (Black et al., 1994; Fu and Prevelige, 2009; Newcomb et al., 2005), but in P22 the role of portal protein as the nucleator is unclear and requires further investigation.
A defect in scaffolding protein binding is not enough to cause polyheads since WT coat protein does not form them in the absence of scaffolding protein. In addition to polyheads, the F170 variants produce some correctly sized procapsids in vivo and structural studies of these procapsids showed that F170A and F170K pentons differ from the pentons in WT procapsids, due to increased coat protein rigidity of the A-domain (Suhanovsky et al., 2010). We suggest that altering the amino acid at F170 disrupts formation of the β-hinge, thereby affecting A-domain flexibility, resulting in steric crowding at the center of the pentons, which have a smaller axial hole compared to hexons. Therefore, while scaffolding protein is important for organization of the capsomers (as seen with spiral polymers, petite shells, and polyheads), it appears that A-domain conformation regulates capsomer formation.
In phage T4, hexons and pentons are comprised of different proteins (gp23 and gp24, respectively), while in P22 both types of capsomer are made of the same protein (gp5). The T4 coat proteins are 21% identical in sequence and both have the HK97-fold (Fokine et al., 2005b). Consistent with the idea that the A-domain flexibility is critical for formation of pentons, amino acid substitutions in the A-domain of the T4 phage hexon protein (gp23) allow it to replace T4’s penton protein (gp24) (Fokine et al., 2004). These variants of gp23 may act by increasing the flexibility of its A-domain, thereby allowing both hexon and penton formation from a single protein. In the phage P4/P2 system, the same coat protein (gpN) is able to generate either T = 7 (phage P2) or T = 4 (phage P4) sized capsids (Dearborn et al., 2012; Dokland et al., 1992). Phage P4 expresses an external scaffolding protein called Sid, which imposes the greater curvature required for the smaller capsid through interaction with the coat protein A-domain. An internal scaffolding protein gpO directs the coat protein to form T = 7 capsids, but this interaction is transient and can be overridden by the more stable interaction of gpN with Sid (Dearborn et al., 2012). Therefore, A-domain conformation determines whether a subunit will assemble into a penton or hexon for phages P22, T4, and the P2/P4 system.
I-domain is involved in capsid size determination
F170L is a global su substitution that compensates for the decreased stability of many tsf coat proteins by restricting the flexibility of the A-domain. However, the more extreme alanine and lysine substitutions at position F170 resulted in a tsf phenotype, indicating that this flexibility is finely balanced. We exploited the tsf phenotype of F170A and F170K to identify other regions of coat protein critical for its folding and assembly. Suppressor searches of the polyhead forming coat protein variants identified position A285 in the I-domain. This domain is important for folding and stability of coat protein (Suhanovsky and Teschke, 2013; Teschke and Parent, 2010). However, in addition to fixing the folding and polyhead defects of the F170K substitution, the A285T suppressor substitution in an otherwise WT background resulted in petite capsids, suggesting an additional role for this domain and specifically for the S-loop. Electron micrographs of negatively stained particles from the A285T variant revealed the variant produced DNA filled T = 4 icosahedral particles along with smaller petite particles without DNA (Suhanovsky and Teschke, 2011). The improved coat protein model generated with the NMR structure of the I-domain revealed a possible intramolecular salt bridge between residue K286 of the S-loop and E159 of the A-domain though the low resolution of the cryo-EM maps precludes a definite assignment (Rizzo et al., 2014). Nevertheless, it is clear that increasing the volume at position 285 would result in a steric clash with the A-domain. Indeed, as the size of the side chain at 285 was increased (in the absence of the original F170K substitution) so did the prevalence of petite products. Bulkier substitutions (A285Y) lead to production of mostly incomplete particles with diameters similar to that expected for T = 1 particles. Steric clash with the A-domain may result in a slight twist in the orientation of the I-domain with respect to the coat protein core (HK97-like portion) in the petite particles compared to T = 7 procapsids. This torque of the I-domain caused by displacement by the A-domain could alter the D-loop position and therefore the twofold interactions between capsomers. The altered twofold interactions are accompanied by an increase in curvature and therefore a smaller diameter. A change in the interaction across the twofold axis of symmetry could also explain why the most particles formed with coat protein with the bulkiest substitutions at position 285 generated a great deal of incomplete particles. Therefore, bulkier substitutions at position 285 result in direct contact of the I-domain with the A-domain that must fix the conformation of the A-domain to allow both proper folding and penton formation. This comes at its own cost, which is manifested by improper orientation of the I-domain altering the bonding contacts between capsomers and producing petite capsids.
Concluding remarks
The HK97-fold, despite sequence, structural, and functional divergences, is a basic template for formation of a proteinacious shell utilized by viruses and prokaryotic cells alike. The protein shell, composed of HK97-like subunits, serves a protective role for the genomes of viruses infecting all three domains of life. Prokaryotic cells also utilize the HK97-fold to form encapsulin nanocompartments, and possibly gene transfer agent particles that contribute to high frequencies of horizontal gene transfer (Lang et al., 2012). The size of the particles assembled from proteins with the HK97-fold is highly variable, but precisely determined during assembly usually with the help of a scaffolding protein or Δ-domain. The capsid size has likely evolved to accommodate the size of the material to be packaged as T. maritima encapsulins package small ferritin-like proteins inside a particle with an inner volume of 4.18 × 106 Å3 while Phage G requires a much larger capsid (inner volume = 1.87 × 109 Å3) to package its 670 kb chromosome (Neitzey et al., 1993). In addition to varied sizes, an amazing diversity of loop and domain embellishments to the basic HK97-fold increase its functionality and often provide alternate ways to stabilize the assembled particles. Particles containing subunits with the HK97-fold will undoubtedly continue to be discovered with other variations for accomplishing folding, assembly, and capsid stabilization. As more structures are characterized, we will achieve a better understanding of how this versatile structural motif has evolved to support folding, procapsid assembly, and maturation in a variety of viruses.
*Tailed viruses and Herpesvirdae have coat proteins with the HK97-fold
*The HK97-fold has evolved to accommodate drastic differences in capsid diameter
*Embellishments to the HK97-fold provide alternative mechanisms to accomplish similar goals
Acknowledgements
This work was supported by NIH grant R01 GM076661. We thank Dr. Nadia D’Lima for helpful comments on this manuscript. We also thank Drs. Roger Hendrix and Nadia D’Lima for permission to describe some of their preliminary data.
Abbreviations
- cryo-EM
electron cryomicroscopy
- 3D
three-dimensional
- T
triangulation
- gp
gene product
- su
suppressor substitution
- tsf
temperature-sensitive-folding
- am
amber mutation
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann HW, Prangishvili D. Prokaryote viruses studied by electron microscopy. Archives of virology. 2012;157:1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, Nakaishi Y, Suzuki M, Namba K, Ono Y, Tsukihara T, Nakagawa A. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol. 2007;368:1469–1483. doi: 10.1016/j.jmb.2007.02.075. [DOI] [PubMed] [Google Scholar]
- Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG. Structural investigations of a Podoviridae streptococcus phage C1, implications for the mechanism of viral entry. Proc Natl Acad Sci U S A. 2012;109:14001–14006. doi: 10.1073/pnas.1207730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J. Biol. Chem. 1999;274:22217–22224. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc Natl Acad Sci U S A. 2013;110:12301–12306. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Caspar DLD. Polyoma virus 'hexamer' tubes consist of paired pentamers. Nature. 1983;302:446–448. doi: 10.1038/303446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Burnett RM, Stuart DI. Evolution of viral structure. Theor. Popul. Biol. 2002;61:461–470. doi: 10.1006/tpbi.2002.1591. [DOI] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Bancroft JB, McDonald JG, Rees MW. A mutant of cowpea chlorotic mottle virus with a perturbed assembly mechanism. Virology. 1976;75:293–305. doi: 10.1016/0042-6822(76)90029-5. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 1988;202:77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- Bebeacua C, Lai L, Vegge CS, Brondsted L, van Heel M, Veesler D, Cambillau C. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol. 2013;87:1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, editor. Molecular biology of bacteriophage T4. American Society for Microbiology; Washington D.C.: 1994. pp. 218–258. [Google Scholar]
- Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22:757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Duda RL, Cheng N, You L, Conway JF, Hendrix RW, Steven AC. Metastable intermediates as stepping stones on the maturation pathways of viral capsids. mBio. 2014;5:e02067. doi: 10.1128/mBio.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chen D-H, Baker ML, Hyrc CF, DiMaio F, Jakana J, Weimin W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King J, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. PNAS. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of a virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Cortines JR, Motwani T, Vyas AA, Teschke CM. Highly specific salt bridges govern bacteriophage P22 icosahedral capsid assembly: identification of the site in coat protein responsible for interaction with scaffolding protein. J Virol. 2014;88:5287–5297. doi: 10.1128/JVI.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortines JR, Weigele PR, Gilcrease EB, Casjens SR, Teschke CM. Decoding bacteriophage P22 assembly: identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology. 2011;421:1–11. doi: 10.1016/j.virol.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn AD, Laurinmaki P, Chandramouli P, Rodenburg CM, Wang S, Butcher SJ, Dokland T. Structure and size determination of bacteriophage P2 and P4 procapsids: function of size responsiveness mutations. J Struct Biol. 2012;178:215–224. doi: 10.1016/j.jsb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Duda RL, Hendrix RW, Rosenberg JM. Complexes between chaperonin GroEL and the capsid protein of bacteriophage HK97. Biochemistry. 1995;34:14918–14931. doi: 10.1021/bi00045a037. [DOI] [PubMed] [Google Scholar]
- Dokland T. Scaffolding proteins and their role in viral assembly. Cellular & Molecular Life Sciences. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO Journal. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 1995a;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr Biol. 2006;16:R11–13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995b;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison SC. Assembly of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J Mol Biol. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- Effantin G, Figueroa-Bossi N, Schoehn G, Bossi L, Conway JF. The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology. 2010;402:355–365. doi: 10.1016/j.virol.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Effantin G, Hamasaki R, Kawasaki T, Bacia M, Moriscot C, Weissenhorn W, Yamada T, Schoehn G. Cryo-Electron Microscopy Three-Dimensional Structure of the Jumbo Phage PhiRSL1 Infecting the Phytopathogen Ralstonia solanacearum. Structure. 2013;21:298–305. doi: 10.1016/j.str.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Kostyuchenko VA, Efimov AV, Kurochkina LP, Sykilinda NN, Robben J, Volckaert G, Hoenger A, Chipman PR, Battisti AJ, Rossmann MG, Mesyanzhinov VV. A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head. J Mol Biol. 2005a;352:117–124. doi: 10.1016/j.jmb.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA. 2005b;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CY, Prevelige PE., Jr. In vitro incorporation of the phage Phi29 connector complex. Virology. 2009;394:149–153. doi: 10.1016/j.virol.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisteo ML, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys. J. 1993;65:227–235. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos CP, Hendrix RW, Casjens S, Kaiser AD. Host participation in bacteriophage lambda head assembly. J. .Mol. Biol. 1973;76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson P, Baker ML, Raytcheva D, Haase-Pettingell C, Piret J, King JA, Chiu W. Protruding knob-like proteins violate local symmetries in an icosahedral marine virus. Nature communications. 2014;5:4278. doi: 10.1038/ncomms5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose JH, Belnap DM, Jensen JD, Mathis AD, Prince JT, Merrill BD, Burnett SH, Breakwell DP. The genomes, proteomes, and structures of three novel phages that infect the Bacillus cereus group and carry putative virulence factors. J Virol. 2014;88:11846–11860. doi: 10.1128/JVI.01364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakana J, Chiu W, Serwer P, Jiang W. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc Natl Acad Sci U S A. 2014;111:E4606–4614. doi: 10.1073/pnas.1407020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann J, Maaty WS, Gauss GH, Akkaladevi N, Brumfield SK, Rayaprolu V, Young MJ, Lawrence CM, Bothner B. Fossil record of an archaeal HK97-like provirus. Virology. 2011;417:362–368. doi: 10.1016/j.virol.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Van Etten JL, editor. Jumbo Bacteriophages. Lesser Known Large dsDNA Viruses. 2009 [PubMed] [Google Scholar]
- Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, Johnson JE. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011;408:541–554. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet A, Conway JF, Letellier L, Boulanger P. In vitro assembly of the T=13 procapsid of bacteriophage T5 with its scaffolding domain. J Virol. 2010;84:9350–9358. doi: 10.1128/JVI.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui WH, Tang Q, Liu H, Atanasov I, Liu F, Zhu H, Zhou ZH. Protein interactions in the murine cytomegalovirus capsid revealed by cryoEM. Protein & cell. 2013;4:833–845. doi: 10.1007/s13238-013-3060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionel A, Velazquez-Muriel JA, Luque D, Cuervo A, Caston JR, Valpuesta JM, Martin-Benito J, Carrascosa JL. Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J Biol Chem. 2011;286:234–242. doi: 10.1074/jbc.M110.187211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Johnson JE. Virus particle maturation: insights into elegantly programmed nanomachines. Curr Opin Struct Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Hawkridge AM, Johnson KL, Muddiman DC, Prevelige PJ. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass-spectrometry. J. Proteome Res. 2006;5:370–377. doi: 10.1021/pr050356f. [DOI] [PubMed] [Google Scholar]
- Kang S, Prevelige PJ. Domain Study of Bacteriophage P22 Coat Protein and Characterization of the Capsid Lattice Transformation by Hydrogen/Deuterium Exchange. J. Mol. Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Keifer DZ, Pierson EE, Hogan JA, Bedwell GJ, Prevelige PE, Jarrold MF. Charge detection mass spectrometry of bacteriophage P22 procapsid distributions above 20 MDa. Rapid communications in mass spectrometry : RCM. 2014;28:483–488. doi: 10.1002/rcm.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos. Trans. R. Soc. London B. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- Lander GC, Baudoux AC, Azam F, Potter CS, Carragher B, Johnson JE. Capsomer dynamics and stabilization in the T = 12 marine bacteriophage SIO-2 and its procapsid studied by CryoEM. Structure. 2012;20:498–503. doi: 10.1016/j.str.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O, Beatty JT. Gene transfer agents: phage-like elements of genetic exchange. Nature reviews. Microbiology. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 2007;371:836–849. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- Lenk E, Casjens S, Weeks J, King J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology. 1975;68:182–199. doi: 10.1016/0042-6822(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, Heymann JB, Winkler DC, Lam AS, Wall JS, Steven AC, Hoiczyk E. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014 doi: 10.15252/embj.201488566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in the tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Neitzey LM, Hutson MS, Holzwarth G. Two-dimensional motion of DNA bands during 120 degrees pulsed-field gel electrophoresis. Electrophoresis. 1993;14:296–303. doi: 10.1002/elps.1150140152. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Brown JC. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J Virol. 2005;79:10540–10546. doi: 10.1128/JVI.79.16.10540-10546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B, Moyer CL, Hendrix RW, Duda RL. The delta domain of the HK97 major capsid protein is essential for assembly. Virology. 2014;456-457:171–178. doi: 10.1016/j.virol.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Deedas CT, Egelman EH, Casjens SR, Baker TS, Teschke CM. Stepwise molecular display utilizing icosahedral and helical complexes of phage coat and decoration proteins in the development of robust nanoscale display vehicles. Biomaterials. 2012a;33:5628–5637. doi: 10.1016/j.biomaterials.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012b;427:177–188. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010a;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Ranaghan MJ, Teschke CM. A second-site suppressor of a folding defect functions via interactions with a chaperone network to improve folding and assembly in vivo. Mol Microbiol. 2004;54:1036–1050. doi: 10.1111/j.1365-2958.2004.04326.x. [DOI] [PubMed] [Google Scholar]
- Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS. Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. Phys Biol. 2010b:7. doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Polyhead formation in phage P22 pinpoints a region in coat protein required for conformational switching. Mol Microbiol. 2007;65:1300–1310. doi: 10.1111/j.1365-2958.2007.05868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Tang J, Cardone G, Gilcrease EB, Janssen ME, Olson NH, Casjens SR, Baker TS. Three-dimensional reconstructions of the bacteriophage CUS-3 virion reveal a conserved coat protein I-domain but a distinct tailspike receptor-binding domain. Virology. 2014;464-465C:55–66. doi: 10.1016/j.virol.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Teschke CM. GroEL/S substrate specificity based on substrate unfolding propensity. Cell Stress Chaperones. 2007;12:20–32. doi: 10.1379/CSC-219R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MH, Prevelige PE., Jr. Electrostatic interactions drive scaffolding/coat protein binding and procapsid maturation in bacteriophage P22. Virology. 1998;250:337–349. doi: 10.1006/viro.1998.9386. [DOI] [PubMed] [Google Scholar]
- Parker ML, Ralston EJ, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. II. Head structure and DNA size. J Virol. 1983;46:250–259. doi: 10.1128/jvi.46.1.250-259.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietila MK, Laurinmaki P, Russell DA, Ko CC, Jacobs-Sera D, Hendrix RW, Bamford DH, Butcher SJ. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc Natl Acad Sci U S A. 2013;110:10604–10609. doi: 10.1073/pnas.1303047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, Jiang W, Chiu W, Hatfull GF, Hendrix RW, King J. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a "horned" bacteriophage of marine synechococcus. J Mol Biol. 2007;368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Jr., Marieta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr., King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- Rizzo AA, Fraser LC, Sheftic SR, Suhanovsky MM, Teschke CM, Alexandrescu AT. NMR assignments for the telokin-like domain of bacteriophage P22 coat protein. Biomol NMR Assign. 2012 doi: 10.1007/s12104-012-9422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo AA, Suhanovsky MM, Baker ML, Fraser LC, Jones LM, Rempel DL, Gross ML, Chiu W, Alexandrescu AT, Teschke CM. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and CryoEM modeling. Structure. 2014;22:830–841. doi: 10.1016/j.str.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Domek MJ, Sanz-Garcia E, Makaju A, Taylor RM, Hoggan R, Culumber MD, Oberg CJ, Breakwell DP, Prince JT, Belnap DM. Sequence and structural characterization of great salt lake bacteriophage CW02, a member of the T7-like supergroup. J Virol. 2012;86:7907–7917. doi: 10.1128/JVI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman MS, Dearborn AD, Chang JR, Damle PK, Christie GE, Dokland T. A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80alpha capsids. J Mol Biol. 2011;405:863–876. doi: 10.1016/j.jmb.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Couture E, Aebi U, Showe MK. Structure of T4 polyheads. II. A pathway of polyhead transformation as a model for T4 capsid maturation. J. Mol. Biol. 1976;106:187–221. doi: 10.1016/0022-2836(76)90307-7. [DOI] [PubMed] [Google Scholar]
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- Stroupe ME, Brewer TE, Sousa DR, Jones KM. The structure of Sinorhizobium meliloti phage PhiM12, which has a novel T=19l triangulation number and is the founder of a new group of T4-superfamily phages. Virology. 2014;450-451:205–212. doi: 10.1016/j.virol.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Suhanovsky MM, Parent KN, Dunn DE, Baker TS, Teschke CM. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibility in conformational switching. Mol Microbiol. 2010;77:1568–1582. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhanovsky MM, Teschke CM. Bacteriophage P22 capsid size determination: roles for the coat protein telokin-like domain and the scaffolding protein amino-terminus. Virology. 2011;417:418–429. doi: 10.1016/j.virol.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhanovsky MM, Teschke CM. An intramolecular chaperone inserted in bacteriophage P22 coat protein mediates its chaperonin-independent folding. J Biol Chem. 2013;288:33772–33783. doi: 10.1074/jbc.M113.515312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses--major players in the global ecosystem. Nature reviews. Microbiology. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM, Parent KN. 'Let the phage do the work': using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–130. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jakana J, McGough A, Prevelige PE, Chiu W. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J. Virol. 2000;74:3871–3873. doi: 10.1128/jvi.74.8.3871-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PEJ. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys. J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys. J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso DJ, Hendrix RW, Duda RL. Transient contacts on the exterior of the HK97 procapsid that are essential for capsid assembly. J Mol Biol. 2014;426:2112–2129. doi: 10.1016/j.jmb.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HE, Sherman MB, Brasiles S, Jacquet E, Seavers P, Tavares P, Orlova EV. Capsid structure and its stability at the late stages of bacteriophage SPP1 assembly. J Virol. 2012;86:6768–6777. doi: 10.1128/JVI.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. Journal of Molecular Biology. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. eLife. 2013;2:e01299. doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Hui WH, Shah S, Jih J, O'Connor CM, Sherman MB, Kedes DH, Schein S. Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization. Structure. 2014;22:1385–1398. doi: 10.1016/j.str.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Suhanovsky MM, Teschke CM. The energetic contributions of scaffolding and coat proteins to the assembly of bacteriophage procapsids. Virology. 2012;428:64–69. doi: 10.1016/j.virol.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]