Abstract
Baculovirus infection of a host insect involves several steps, beginning with initiation of virus infection in the midgut, followed by dissemination of infection from the midgut to other tissues in the insect, and finally culminating in “melting” or liquefaction of the host, which allows for horizontal spread of infection to other insects. While all of the viral gene products are involved in ultimately reaching this dramatic infection endpoint, this review focuses on two particular types of baculovirus-encoded proteins: degradative enzymes and protease inhibitors. Neither of these types of proteins is commonly found in other virus families, but they both play important roles in baculovirus infection. The types of degradative enzymes and protease inhibitors encoded by baculoviruses are discussed, as are the roles of these proteins in the infection process.
Keywords: baculovirus, cathepsin, chitinase, matrix metalloprotease, P35, P49, ODV-E66, serpin
INTRODUCTION
Members of the virus family Baculoviridae contain large double-stranded DNA genomes (80-180 kbp) and infect invertebrates in the class Insecta. Like other DNA viruses, baculovirus genomes contain many genes that have easily recognizable homology with host genes, indicating gene transfer from the genomes of their hosts. The function of these host-derived genes benefits baculovirus infectivity, virulence, and their ability to disperse to other susceptible hosts. It has been hypothesized that at least for some genes, once the insect-derived gene is part of the virus genome, its function evolves to optimize virus infection or expand the virus host range (Lung and Blissard, 2005).
There are many unusual and interesting features associated with the baculoviruses (Clem and Passarelli, 2013). One of these is the existence of several types of baculovirus genes that encode degradative enzymes, which is uncommon amongst viruses. These enzymes facilitate baculovirus infection of insects through several processes, including penetration of the peritrophic matrix, a layer that protects epithelial cells in the insect gut, to establish primary infection; melanization, or darkening of tissue; and liquefaction of the infected cadaver at late stages of baculovirus infection. In addition, baculoviruses are one of only two virus families, the other being the poxviruses, which commonly encode protease inhibitors. We discuss the functions of two types of baculovirus-encoded protease inhibitors in curtailing innate defense mechanisms. Each of the more than seventy baculovirus genomes sequenced to date contains various combinations of these degradative enzyme and protease inhibitor genes. We review the functions of these proteins in virus infection and how viral pathogenesis may differ in viruses carrying different sets of degradative enzymes or protease inhibitors. Finally, we discuss the interplay amongst these gene products and how they ultimately cooperate to allow efficient virus replication and spread. The review does not cover viral-encoded nucleases.
BACULOVIRUS REPLICATION AND PATHOGENESIS
Only about two-thirds of the approximately 150 genes encoded by the prototype baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) have been characterized enough to know their specific function (Rohrmann, 2013). Approximately 15% encode proteins dedicated to gene transcription and nucleic acid replication and processing. Around 33% of the virally-encoded proteins have been identified as virion-associated proteins using proteomic methods (Hou et al., 2013). When 141 open reading frames (ORFs) were systematically deleted in Bombyx mori NPV (BmNPV), 86 of the mutant viruses were still able to replicate normally in cell culture (Ono et al., 2012), indicating that over half of baculovirus genes encode proteins that are either necessary for in vivo infection or serve auxiliary functions such as enhancing virulence or virus transfer between hosts. These numbers provide a glimpse into the diversity of functions and potential strategies that allow these viruses to outmaneuver their hosts.
The family Baculoviridae contains four recognized genera, Alphabaculovirus, nucleopolyhedroviruses (NPVs) that replicate in lepidopteran hosts; Betabaculovirus, granuloviruses (GVs), which also have lepidopteran hosts; Gammabaculovirus, NPVs that replicate in hymenopteran hosts, and Deltabaculovirus, which contains a single member that replicates in a dipteran host (Jehle et al., 2006). The host range of each baculovirus can vary from one to several dozen insect species (Harrison and Hoover, 2012), and gene acquisition has undoubtedly been instrumental in allowing infections in new hosts.
Baculovirus infection commences when an insect consumes vegetation contaminated with viral occlusion bodies (OBs), which consist of occlusion-derived virions (ODV) encased in an environmentally stable matrix. The proteinaceous component of the OB is mainly composed of a single protein, polyhedrin (called granulin in betabaculoviruses), that is abundantly synthesized at very late stages of virus replication. Depending on the virus, an OB contains between one and around 100 enveloped ODV. Once the OB reaches the midgut, the protective matrix is dissolved in the alkaline pH of the midgut lumen, releasing the ODV. To access host cells, the ODV must cross a barrier lining the midgut called the peritrophic matrix, a layer consisting of chitin and proteins that separates the midgut epithelium from the gut lumen. As discussed below, some viruses produce enhancins, metalloproteases that degrade the peritrophic matrix. ODV enter midgut epithelial cells by membrane fusion and this event requires a number of structural factors called per os infectivity factors (PIFs) to mediate successful virion attachment and entry. ODV replicate in the nucleus of midgut cells, and in most baculoviruses, a second type of enveloped virion is produced called budded virus (BV). The name BV reflects the fact that these virions acquire their envelopes and associated glycoproteins as they bud from the cellular plasma membrane. Infection can be restricted to midgut cells (in the case of deltabaculoviruses, gammabaculoviruses and a betabaculovirus), but for most baculoviruses, BV escape the midgut and extensively infect other tissues in the insect, where both BV and ODV are produced. Thus, the two virion forms have distinct functions during infection: BV spread infection between cells within an infected insect, while ODV are formed in the nucleus of infected cells, embedded in OBs, and released into the environment upon death of the host, spreading infection horizontally between insect hosts.
In the case of most baculoviruses, at the end of the replication cycle the insect cuticle becomes melanized (darkened) and the insect cadaver liquefies (Slack et al., 1995) (Hawtin et al., 1997; Kang et al., 1998; Wang et al., 2005). The complete dissolution of the infected insect allows the stable OBs to be efficiently released so they can be ingested by other feeding insects, initiating another infection cycle. Insect melanization and liquefaction requires the viral degradative enzymes chitinase and cathepsin, which work together in liberating OBs from the dead larvae. In viruses that do not establish a systemic infection, the mechanism for OB release is not clear but, as addressed below, the presence of other degradative enzymes may be involved in alternative mechanisms of release.
BACULOVIRUS-ENCODED DEGRADATIVE ENZYMES
Chitinase
Chitinases (EC 3.2.1.14) degrade chitin, a β-1,4-linked N-acetylglucosamine (GlcNAc) homopolymer, which constitutes an integral component of the exoskeleton, peritrophic matrix, certain organs (salivary glands and trachea), muscle joints and eggshells of arthropods. Different types of chitin may form either hard or soft barriers, and these are used to isolate organs or the organism from invading pathogens or a harsh environment (Muthukrishnan et al., 2012). Organisms produce specialized chitinases, each with different substrate preferences (Koga et al., 1999), and some that are enzymatically inactive but which bind chitin, perhaps to assist binding and catalysis of insoluble chitin fibers by active chitinases (Jollès and Muzzarelli, 1999; Koga et al., 1999; Vaaje-Kolstad et al., 2005). Insects utilize chitinases for growth and remodeling of exoskeleton and peritrophic matrix, which are essential structural and physiological invertebrate layers, so their activity must be stringently regulated (Kramer et al., 1985). In addition, some organisms that lack chitin produce chitinases as a defense against feeding and parasitism by insects and chitinous pathogens (Jollès and Muzzarelli, 1999).
Viral chitinases are family 18 glycohydrolases that belong to a family of multi-modular proteins found in diverse organisms including mammals, bacteria, plants, and fungi (Jollès and Muzzarelli, 1999). Most baculoviruses, specifically those that infect lepidopteran larvae, contain a chitinase gene (Table). The only other virus family known to encode chitinases are chlorella viruses that infect algae (Yamada et al., 1993). Chlorella virus chitinase is expressed as a late gene during virus replication and is likely involved in cell wall degradation needed for cell lysis and progeny virus release from these single-celled organisms (Hiramatsu et al., 1999). Baculovirus chitinases are also late genes (Hawtin et al., 1995; Hodgson et al., 2007), and are required for liquefaction of the host insect (Hawtin et al., 1997). Chlorella virus chitinases are not known to be virion-associated (Hiramatsu et al., 1999), but chitinases from the baculovirus AcMNPV have been detected in both OBs (Hawtin et al., 1995; Hawtin et al., 1997) and in BV (Wang et al., 2010).
Table.
Distribution of proteases and protease inhibitors in baculovirus genomesa
| Gene | Alphabaculoviruses | Betabaculoviruses | Deltabaculovirus | Gammabaculoviruses |
|---|---|---|---|---|
| chitinase | All Group I except: AgMNPV All Group II except: AdhoNPV |
All except: AdorGV ChocGV CrleGV PhopGV PxGV SpliGV |
None | None |
| cathepsin | All except: AgMNPV |
All except: AdorGV ChocGV HearGV PhopGV PxGV SpliGV |
None | None |
| mmp | None | All | None | None |
|
bacterial
collagenase- like |
Group I: None All Group II except: AdhoNPV OrerNPV |
None | None | None |
| enhancin | Group I: CfMNPV Group II: AgipMNPV AgseNPV EupsNPV HearMNPV LdMNPV LyxyMNPV MacoNPV-A MacoNPV-B |
AgseGV CfGV HearGV PsunG TnGV XcGV |
None | None |
| trypsin-like | None | None | None | All |
| odv-e66 | All Group I except: MaviMNPV PenuNPV All Group II except: ClbiNPV OrerNPV |
All except: ClanGV SpliGV CaLGV |
None | None |
| P35/P49 | Group I: AcMNPV PxNPVb RoNPVb BmNPV HycuNPV MaviNPV ThorNPV LsNPV Group II: SpltNPV SpliNPV |
CaLGV ChocGV ClanGV |
Nonec | None |
| serpin | Group I: None Group II: HespNPV |
None | None | None |
Virus name abbreviations: AcMNPV, Autographa californica MNPV; AdhoNPV, Adoxophyes honmai NPV; AdorGV, Adoxophyes orana granulovirus (GV); AgMPNV, Anticarsia gemmatalis MNPV; AgipMNPV, Agrotis ipsilon NPV; AgseGV, Agrotis segetum GV; AgseNPV, Agrotis segetum NPV; BmNPV, Bombyx mori NPV; CaLGV, Clostera anastomosis (L.) GV; CfGV, Choristoneura fumiferana GV; CfMNPV, Choristoneura fumiferana MNPV; ChocGV, Choristoneura occidentalis GV; ClanGV, Clostera anachoreta GV; ClbiNPV, Clanis bilineata NPV; CrleGV, Cyrptophlebia leucoreta GV; EupsNPV, Euproctis pseudoconspersa NPV; HearGV, Helicoverpa armigera GV; HearMNPV, Helicoverpa armigera NPV; HycuNPV, Hyphantria cunea NPV; LdMNPV, Lymantria dispar NPV; LsNPV, Leucania separata NPV; LyxyMNPV, Lymantria xylina NPV; Mamestra configurata NPV-A, -B; MaviNPV, Maruca vitrata NPV; OrerNPV, Orgyia ericae NPV; PenuNPV, Perina nuda NPV; PhopGV, Phthorimaea operculella GV; PsunGV, Pseudaletia unipuncta GV; PxGV, Plutella xylostella GV; SpliGV, Spodoptera litura GV; SpltNPV, Spodoptera litura NPV; SpliNPV, Spodoptera littoralis NPV; ThorMNPV, Thysanoplusia orichalcea MNPV; TnGV, Trichoplusia ni GV; XcGV, Xestia c-nigrum GV
PxNPV and RoNPV P35 homologs are 100% identical to AcMPNV P35. It is unclear whether these viruses are independent species, or only variants of AcMNPV.
An ORF in CuniNPV, ORF075, that was reported to be a P35 homolog does not have significant homology to P35
The domain structure of baculovirus chitinases is most similar to that of S. marcescens chitinase A (Henrissat, 1999), which is composed of an N-terminal chitin-binding immunoglobulin-like fold (~500 residues), and a C-terminal domain (~1100 residues) that contains the catalytic site (Hodgson et al., 2013; Young et al., 2005). Baculovirus chitinase N-terminal domains have several conserved aromatic surface residues important for insoluble chitin binding and substrate feeding of chitin chains into the catalytic pocket of the C-terminal α/β barrel (Young et al., 2005). If short (2-4 GlcNAc residues) chitin oligomer substrates are used, AcMNPV chitinase exhibits both endochitinase and exochitinase activities (Daimon et al., 2006; Daimon et al., 2007a; Hawtin et al., 1995; Thomas et al., 2000). However, Epiphyas postvittana NPV chitinase revealed only exochtinase-derived enzyme products (i.e., GlcNAc disaccharides) when colloidal chitin substrate hydrolysis products were resolved by thin-layer chromatography (Young et al., 2005).
The ancestral baculovirus chitinase may have been acquired from a lepidopteran intestinal bacterium (e.g., Serratia marcescens) (Hawtin et al., 1995; Kang et al., 1998) but has evolved to better suit baculovirus biology. Recent BLAST searches show higher similarity to a chitinase from some lepidopteran species. It has been proposed that this intron-less lepidopteran chitinase was horizontally acquired from bacteria or baculoviruses (Daimon et al., 2003).
Baculovirus chitinase retains high activity across a broader pH range (4 to 11) compared to its S. marcescens homolog, which is largely inhibited at pH values above 8 (Daimon et al., 2006; Hawtin et al., 1995). Baculovirus chitinases have acquired eukaryotic N-terminal secretory signal peptides, and those of alphabaculoviruses also have a C-terminal KDEL (or equivalent) endoplasmic reticulum (ER) retention motif (Saville et al., 2002; Thomas et al., 1998). The ER retention motif ensures that the viral chitinase accumulates in the infected cells (Thomas et al., 1998) until it is released due to cell lysis in the terminal stages of virus replication (Hodgson et al., 2011; Saville et al., 2002), when release of chitinase assists in liquefaction of the host insect. Earlier chitinase release could lead to premature insect host death and impair efficient virus multiplication. Most betabaculovirus chitinases lack ER retention signals and, as verified for Cydia pomonella granulovirus chitinase (Daimon et al., 2007a; Kang et al., 1998), these chitinases are likely to be continuously secreted during virus replication. Since most, if not all, alphabaculovirus chitinases contain predicted KDEL-like sequences while betabaculovirus chitinases typically lack these (Kang et al., 1998), this indicates that viruses in each genus utilize different mechanisms for regulating chitinase-mediated tissue dissolution. The advantages of regulating the timing of chitinase activity in infected hosts are further discussed below.
There is a clear role for baculovirus chitinases in the terminal stage of infection of host insects, (D'Amico et al., 2013; Hawtin et al., 1997; Vieira et al., 2012), but it has also been proposed that the packaging of chitinase in OBs may expedite virus transit through the chitinous peritrophic matrix and provide access to the midgut epithelial cells. However, deletion of the AcMNPV chitinase did not show any significant changes in infectivity or mortality of infected Trichoplusis ni larvae (Hawtin et al., 1997) .This indicates that chitinase is not required for the initial stages of infection in vivo and stresses its role in host liquefaction (Daimon et al., 2007b; Hawtin et al., 1997; Thomas et al., 2000; Wang et al., 2005). However, even if the viral chitinase is deleted, it is possible that low levels of host chitinase are incorporated into OBs and are able to assist in permeabilizing the peritrophic matrix (Hawtin et al., 1995). It is also possible that viral chitinase is only needed to help permeabilize the peritrophic matrix in certain hosts or host developmental stages. Several studies have shown that exogenous chitinases have the ability to affect the structure of the peritrophic matrix (Rao et al., 2004; Villalon et al., 2003; Wang and Granados, 2001). Feeding chitinase to insects prior to virus infection can reduce the integrity of the peritrophic matrix (Rao et al., 2004), reduce viral LD50 (Shapiro et al., 1987) and expression of either insect (Ding et al., 1998) or baculovirus (Corrado et al., 2008) chitinases in plants yielded protective activity against fungal and feeding insect pests. However, a role for the viral-encoded chitinases in peritrophic matrix degradation during virus infection has not been shown.
Cathepsin
Many viruses from diverse families encode proteases, but most of these are involved in viral polyprotein processing and often have discrete cellular and/or viral protein targets as substrates. It is less common for viruses to encode proteases that are involved in general degradation of host tissues. Baculovirus-encoded cathepsins (Table) are homologous to conserved eukaryotic endosomal cysteine proteases and analogous to other papain-like proteases that are expressed as zymogen precursors (Rawlings et al., 1992; Slack et al., 1995). The presumed ancestor of these baculovirus proteases was an endosomal cathepsin (Choe et al., 2006), a type of cysteine protease mostly known for their function within lysosomes but that more recently have been found to have other discrete roles in cells (Turk et al., 2012). Cathepsins can be further distinguished by their substrate-binding preferences (Choe et al., 2006). The baculovirus cathepsin protein sequence is most similar to cathepsin L (Slack et al., 1995), a cysteine endopeptidase that functions in protein turnover in the lysosome. BLAST protein searches indicate that the AcMNPV cathepsin is most closely related to cathepsins from insects in the orders Hymenoptera and Lepidoptera, indicating horizontal acquisition of an ancestral gene from an insect host.
The structure of baculovirus cathepsin has a less sterically-restricted substrate binding pocket than any of its cellular homologues (Hom et al., 2002; Ohkawa et al., 1994; Slack et al., 1995), expanding its potential versatility in substrate specificities, which presumably aids in efficient degradation of proteinacious host tissues. Baculovirus cathepsin can degrade diverse protein substrates (i.e., casein, collagen, gelatin) in vitro (Ohkawa et al., 1994; Slack et al., 1995). Although its pH optimum (5.0 to 5.5) resembles that of endosomal proteases, the viral cathepsin, unlike cellular cathepsins, still retains considerable activity at neutral pH (Hom and Volkman, 2000). Baculovirus cathepsin is transcribed at late times post infection, starting from about 9 to 12 hours post infection (Hill et al., 1995; Hodgson et al., 2007), and together with the viral chitinase, is critical for promoting terminal host cadaver liquefaction (Hawtin et al., 1997) (Ohkawa et al., 1994; Slack et al., 1995). In addition, cathepsin is also required for melanization of the host, which occurs at or near the time of death (Ohkawa et al., 1994; Slack et al., 1995). It is not clear whether melanization provides an advantage to the virus, or simply is a consequence of virus infection. It is possible that melanization of the host tissues helps protect the OBs from UV damage once the insect has liquefied, since OBs are rapidly inactivated when exposed to sunlight.
In addition to an expanded pH activity and broadened catalytic specificity of the baculovirus cathepsin, its subcellular trafficking, regulation of its activation, and release from infected cells are important properties that control the process of host liquefaction. Like cellular cathepsins, the viral protein has a signal peptide and localizes to the ER/Golgi where it is N-glycosylated at one or more conserved residues (Hom and Volkman, 2000; Slack et al., 1995). Cellular cathepsin zymogen precursors are often transported through the ER and trans-Golgi network to late endosomes. Proteolytic cleavage and removal of an approximately 100 residue prodomain is required to produce a catalytically active enzyme and similar processing occurs with the viral cathepsin (Hom and Volkman, 2000; Katunuma, 2010; Slack et al., 1995); this is an important regulation step that limits protease activity. It is only upon delivery to endosomal vesicles that cellular cathepsins mature into active proteases due to either low pH in the endosome or cleavage by active endosomal proteases. If retained in the ER artificially by modification of the KDEL ER retention motif, mammalian cathepsin D does not mature (Liaudet et al., 1994). The baculovirus cathepsin precursor can be activated in vitro by low pH or by treatment with chaotropic agents (i.e., SDS) (Hom and Volkman, 2000).
However, in spite of sequence similarities between mammalian and viral cathepsins, the proenzyme form of baculovirus cathepsin localizes to the ER of infected cells at late times post infection (Hodgson et al., 2009). Although it has been proposed to function in acidic endosomes (Ohkawa et al., 1994; Slack et al., 1995), the viral cathepsin has only been documented to localize to the ER. Therefore, the viral cathepsin, which was probably acquired from an invertebrate host, has been repurposed to fulfill a critical role in the baculovirus replication cycle by differentially regulating its trafficking and catalytic maturation mechanisms. Taken together, the viral cathepsin differs from its cellular counterparts in its localization, processing, and cellular release (discussed below).
Coordinated regulation of chitinase and cathepsin
Baculovirus chitinase and cathepsin need to function in a concerted and timely fashion to effectively degrade host tissues. If the timing of their enzymatic activity is altered, host tissues may be degraded too early during virus infection, compromising host survival and thereby limiting virus replication. During AcMNPV infection of late instar T. ni, 1 to 2 days after host death the insect exoskeleton becomes brittle, and the liquefied internal tissues ooze out until the entire cadaver is liquefied. In the process, OBs are released into the surroundings and are available to start a new infection cycle. In the absence of chitinase or cathepsin activity, host tissues are resilient to degradation long after the caterpillar succumbs to the infection (Daimon et al., 2007b; Hawtin et al., 1997; Katsuma et al., 2009; Ohkawa et al., 1994; Slack et al., 1995; Thomas et al., 2000). Notably, Anticarsia gemmatalis MNPV (AgMNPV), an alphabaculovirus closely related to AcMNPV, lacks both chitinase and cathepsin (Table). As expected, AgMNPV-infected Anticarsia gemmatalis caterpillars remain intact after death; however, if AgMNPV is engineered to express the chitinase and cathepsin genes from the baculovirus Choristeneura fumirerana MNPV (CfMNPV), liquefaction occurs (Lima et al., 2013). Carefully choreographed chitinase and cathepsin expression and activity during virus replication are crucial for the collaborative work of these enzymes.
Although most functionally-related baculovirus genes are not located adjacent to each other in the viral genome, chitinase and cathepsin genes are usually contiguous, often antiparallel to one another and sharing a small intergenic region. This genomic organization is exemplified in AcMNPV and other Group I alphabaculoviruses (Slack et al., 2004). Transcripts from each gene initiate only six nucleotides apart from each other, from back-to-back late viral TAAG promoter sequences (Hawtin et al., 1995; Hill et al., 1995). In AcMNPV, there are 22 examples of bidirectional ORFs that have their translation initiation codons separated by 14 to 550 bases (Norris, 2011). The function of many of the proteins encoded by these 22 AcMNPV bidirectional gene pairs is undetermined, and they may have different temporal transcription profiles. Whether transcription of any of these bidirectional genes, including chitinase and cathepsin, occurs simultaneously in opposing directions is uncertain. However, unpublished preliminary data show that if the entire chitinase ORF is truncated leaving its late promoter element, TAAG, adjacent to that of cathepsin, higher concentrations of cathepsin transcripts are observed. Reciprocal results are observed if the cathepsin late promoter element is altered. This implies that there is promoter occlusion between closely juxtaposed TAAG sequences, and may be a mechanism to prevent overexpression of either protein, which could be deleterious to the virus.
Alignment of Group I alphabaculovirus chitinase and cathepsin intergenic promoters reveals a well conserved sequence (Norris, 2011). As noted previously for AcMNPV and CfMNPV (Hill et al., 1995) the sequence between the cathepsin late TAAG RNA initiation sequence and predicted translational start sequence (ATG) in these viruses contains a highly conserved ATG sequence that is out-of-frame with the cathepsin ORF ATG. Although this upstream out-of-frame ATG is not conserved in Group II alphabaculoviruses or in betabaculoviruses, these other viruses instead often contain two in-frame ATGs within the first 5 to 10 codons. Lacking any further analysis of the chitinase and cathepsin transcription and translation profiles of individual viruses with 5' sequence differences, these observations nonetheless further suggest that the upstream chitinase and cathepsin sequence elements modulate expression kinetics of these adjacent, oppositely oriented ORFs.
The conserved adjacent genomic loci of chitinase and cathepsin and their harmonized expression suggest that their gene products may function together. It is intriguing that pro-cathepsin accumulates in the ER of infected cells instead of being secreted (Hodgson et al., 2009). Baculoviral pro-cathepsin lacks any known motifs or mechanisms that would function to block secretion of the polypeptide as it folds and accumulates in the ER. Reports exist documenting aggregated and inactive pro-cathepsin in cells infected with viruses that have chitinase inactivated by insertion of lacZ (Hawtin et al., 1997; Hom and Volkman, 2000; Katsuma et al., 2009), suggesting that the chitinase is needed as a pro-cathepsin folding chaperone (Hom and Volkman, 2000). These results are consistent amongst different research groups and in different virus-host systems and imply that chitinase binds pro-cathepsin as it folds in the ER. Results from bimolecular fluorescence complementation and pull-down experiments documented that direct association of AcMNPV chitinase and pro-cathepsin does occur in the ER of infected cells (Hodgson et al., 2011, 2013). However, the association of chitinase and pro-cathepsin in cells does not address whether chitinase is a bona fide folding chaperone. Cathepsin undergoes maturation and is active in cells infected with a dual chitinase/cathepsin negative AcMNPV bacmid (Kaba et al., 2004) repaired with cathepsin only (Hodgson et al., 2013). This challenges the hypothesis that chitinase is imperative for pro-cathepsin folding and maturation. A recombinant virus with a lacZ-inactivated chitinase may still transcribe and translate truncated peptides (Hawtin et al., 1997; Hodgson et al., 2013), which could potentially associate with pro-cathepsin in the ER, affecting pro-cathepsin folding or function.
Catalytic maturation of cathepsin is associated with cell death and lysis (Hom and Volkman, 2000). In this way, ER retention of cathepsin and chitinase could dictate programmed spatiotemporal regulation of chitinase and cathepsin-directed host tissue dissolution (Hodgson et al., 2011). Recent work indicates that maturation and activity of cathepsin plays an important role in inducing cell lysis and, thereby, its cellular release along with the ER-retained chitinase and nuclear OBs. This hypothesis is based on in vitro and in vivo data from different cathepsin-negative baculovirus constructs, from which a direct correlation between lack of viral cathepsin expression and reduced release of chitinase and OBs was observed (Hodgson, Ishimwe, Krell, and Passarelli, unpublished results). Although the precise events triggering maturation of cathepsin in cells are not known, these data indicate that activity of viral cathepsin is at least in part responsible for cellular disintegration and subsequent release of chitinase and cathepsin, which are prerequisites for host liquefaction and OB dissemination.
Betabaculovirus-produced chitinase is not retained in the ER (Daimon et al., 2007a; Salvador et al., 2014); thus, these viruses must employ a different molecular program that culminates in host tissue degradation. It is not known if betabaculovirus chitinases associate with the homologous pro-cathepsin in the ER, as it occurs in AcMNPV-infected cells, or if betabaculovirus pro-cathepsin has a mechanism for retention in infected cells. If chitinase and pro-cathepsin associate in betabaculovirus-infected cells, then chitinase/pro-cathepsin complexes lacking a KDEL-mediated ER retention sequence would co-secrete, as observed with AcMNPV expressing a ΔKDEL chitinase mutant (Hodgson et al., 2011).
A recent study described that AcMNPV cathepsin and chitinase are needed to degrade the basal lamina components surrounding silkworm silkglands prior to infection (Woltje et al., 2014). However, the infectivity or lethality of the AcMPNV viruses in the silkworm strains was not described, and it is possible that delays in infection in some tissues were interpreted as lack of infection. We have observed that Trichoplusia ni larvae infected with AcMNPV lacking cathepsin and chitinase have a significant delay in lethal time compared to control virus (Ishimwe, Hodgson, Passarelli, submitted for publication). It is unclear why the various basal lamina-lined organs would be differentially infected by cathepsin/chitinase-lacking viruses. The possibility still remains that since all organs are likely surrounded by extracellular matrix, baculoviral cathepsin and/or MMPs might universally aid in virus infection of the diverse tissues encountered by BVs spreading systemically through the insect hemolymph. However, this is not supported by studies assessing heterologous baculovirus expression of Sarcophaga cathepsin L (ScathL), which is constitutively secreted from infected cells. Although the ScathL expressing virus was more virulent than control AcMNPV, more rapid host mortality was not due to enhanced basal lamina degradation, but rather to extensive connective tissue degradation (Li et al., 2007). Given that the viral fibroblast growth factor (vFGF) homolog in AcMNPV is also involved in triggering host MMP activation and promoting disseminated infection (Means and Passarelli, 2010), it would be interesting to test if dual deletion of vfgf and either viral mmp or v-cath has additive effects on BV systemic dissemination and/or the rate of host mortality.
Matrix metalloprotease
Cellular MMP structure and function
MMPs are a family of zinc-dependent proteolytic enzymes that degrade proteinaceous components of the extracellular matrix. By degrading extracellular matrix components, MMPs play a pivotal role in a variety of physiological processes during normal development and pathological situations. The role of MMPs during healthy processes includes embryonic development, metamorphosis, bone and tissue remodeling, angiogenesis, and wound healing (Nagase and Woessner, 1999; Page-McCaw et al., 2007). Accelerated and uncontrolled MMP activity has been implicated with pathological conditions including chronic inflammation, fibrosis, cancer, cardiovascular and neurological diseases, bacterial and viral infections, and arthritis (Elkington et al., 2005; Frantz et al., 2010; Muschler and Streuli, 2010).
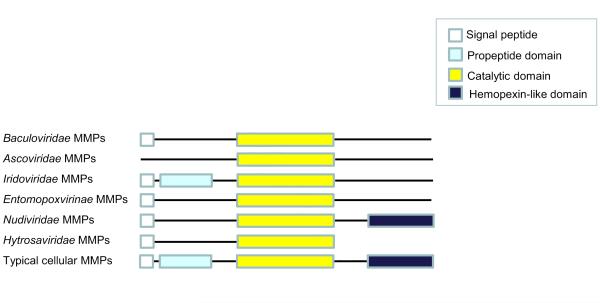
MMP proteins are structurally and functionally composed of three major domains (Figure). The propeptide domain located at the N terminus contains a conserved cysteine in the cysteine switch motif (PRCGV/NPD) that is involved in maintaining MMP latency (Van Wart and Birkedal-Hansen, 1990). The cysteine interacts with the zinc ion at the catalytic domain to preclude catalysis. The cysteine-Zn2+ interaction is disrupted when a protease cleaves the MMP, resulting in its activation. The MMP catalytic domain is characterized by the presence of a conserved zinc-binding motif (HEXGHXXGXXHS/T) that tethers a Zn2+ ion to the three conserved histidines in this motif (Dhanaraj et al., 1996). Lastly, a hemopexin-like domain that contributes to substrate specificity and binding of tissue inhibitors of MMPs (TIMPs) is found near the C terminus (Murphy et al., 1992). In addition, MMPs are usually secreted or membrane-associated enzymes and thus contain an N-terminal signal peptide sequence.
Figure. Comparison of the domain organization of viral and cellular MMPs.
The three major MMP domains are represented by rectangles of different colors and the presence of a signal peptide is shown as a white square. For simplification, the presence of a signal peptide was indicated when at least half of the species encoding mmps within a given virus family predicted a signal peptide. The putative MMP sequences in the following viruses do not predict a signal peptide: Iridoviridae: Invertebrate iridescent virus 22 (YP_008357315.1); Hytrosaviridae: Musca domestica salivary gland hypertrophy virus (YP_001883364.1); Entomopoxvirinae: MsEPV179 (NP_048249.1), Mythimna separata entomopoxvirus 'L' (YP_008003705.1); Baculoviridae: Xestia c-nigrum granulovirus (NP_059188.1), Pieris rapae granulovirus (YP_003429361.1), Epinotia aporema granulovirus (YP_006908552.1)
Baculoviruses encode several different kinds of degradative enzymes and protease inhibitors
Both types of genes are uncommon in viruses
Baculovirus degradative enzymes digest host tissues or enhance midgut infection
Baculovirus protease inhibitors block caspase activity and may suppress immune responses
MMPs have been studied extensively in mammals because of their link to human diseases, especially cancer. In humans, at least 24 MMP homologs with overlapping functions have been described (Ra and Parks, 2007). However, MMPs have also been identified in other vertebrate and invertebrate animals, as well as in plants. Drosophila melanogaster encodes two MMP homologs, which are involved in processes such as morphogenesis and metamorphosis (Page-McCaw et al., 2003). Other insects sometimes encode additional MMP homologs; for example, the genome of the red flour beetle, Tribolium castaneum, contains three MMPs (Knorr et al., 2009) and BLAST searches indicate that the mosquito Aedes aegypti contains eight putative MMP homologs.
Viral MMPs
Little is known about viral-encoded MMPs, because only a few viruses encode MMPs and these have not been extensively characterized. A search of the GenBank® protein database for putative viral MMPs revealed that only viruses from six families encode MMPs: Baculoviridae, Ascoviridae, Iridoviridae, Hytrosaviridae, Poxviridae (only those in the subfamily Entomopoxvirinae) and Nudiviridae (Ishimwe, Hodgson, Passarelli, submitted for publication). Interestingly, all these families consist of large double-stranded DNA viruses that infect insects, mainly in the orders Lepidoptera and Diptera. The presence of mmps in only insect viruses suggests a conserved function during insect virus pathogenesis. Two genes with sequences homologous to MMP catalytic domains were reported in the genome of Melanoplus sanguinipes entomopoxvirus (MsEPV) (Afonso et al., 1999). In contrast, all of the known baculoviruses that encode an MMP carry a single mmp gene. All species in the genus Betabaculovirus sequenced to date encode an mmp, but mmp homologs are absent in species from the other three baculovirus genera (Table) (Escasa et al., 2006; Ferrelli et al., 2012; Harrison and Popham, 2008; Hashimoto et al., 2000; Hayakawa et al., 1999; Lange and Jehle, 2003; Liang et al., 2011; Liang et al., 2013; Luque et al., 2001; Wang et al., 2008; Wormleaton et al., 2003; Zhang et al., 2012). Baculovirus MMPs have highest similarity to a single spider MMP and vertebrate MMPs, so it is not clear how the ancestral baculovirus mmp was acquired.
Viral MMPs do not completely share the conventional MMP domain organization found in typical cellular MMPs (Figure). While they contain the conserved MMP zinc binding motif at the catalytic site, MMPs in all virus families except Iridoviridae lack the conserved cysteine switch motif. Amongst cellular MMPs characterized to date, only human MMP-23 lacks this domain (Velasco et al., 1999). In addition, the hemopexin-like domain is absent in viral MMPs except for those in the Nudiviridae. This domain is found in all cellular MMPs except in human MMP-23 (Velasco et al., 1999), −7 and −26 (de Coignac et al., 2000) and Arabidopsis and soybean MMPs (Maidment et al., 1999; Pak et al., 1997). The hemopexin-like domain is known to be involved in the binding of TIMPs to MMPs in order to form an inhibitory complex (Gomez et al., 1997). Instead of a hemopexin-like domain, most viral MMPs have a long stretch of amino acids downstream of their catalytic site that does not contain any known functional domains. The lack of the cysteine switch and hemopexin-like domain suggests that most viral MMPs have evolved to overcome MMP regulatory controls, enabling them to be active immediately after synthesis. Hytrosaviridae MMPs have a truncated C-terminal domain similar to that of human MMP-7 and −26 (de Coignac et al., 2000). A predicted signal peptide which targets proteins to the secretory pathway is present at the N terminus of all baculovirus MMP sequences except those in Xestia c-nigrum granulovirus (XcGV), Pieris rapae granulovirus (PrGV) and Epinotia aporema granulovirus (EpapGV). Signal peptides are also present in all cellular MMPs except human MMP-23, which is associated with the cell plasma membrane by a signal anchor sequence (Ohnishi et al., 2001). Signal anchor sequences are not found in the predicted MMP sequences of XcGV, PrGV and EpapGV which suggest that, unlike human MMP-23, these enzymes are not membrane associated.
Based on substrate specificity and/or domain composition, MMPs have been classified into five major subfamilies: gelatinases, collagenases, stromelysins, matrilysins, and membrane-type MMPs. Baculovirus MMPs do not precisely fit into any of these subfamilies because of their nonconventional domain organization (Ishimwe, Hodgson, Passarelli, submitted for publication). However; numerous other MMPs including some human MMPs (e.g., MMP-12, −19, −20, −21, −23, −27, and −28) and insect MMPs also cannot be easily classified into any of these groups.
Role of baculovirus MMPs during virus infection
The function of baculovirus MMPs during virus infection has not been studied extensively. Gaining a complete understanding of the roles of MMPs in baculovirus infection will require inactivating MMPs in betabaculovirus genomes and testing the resulting phenotype in their natural hosts, something that is currently difficult, since there are not well-established cell culture systems to replicate betabaculoviruses. However, experiments in which betabaculovirus MMPs were expressed in more experimentally accessible alphabaculoviruses have shed some light on potential functions. To our knowledge, only two studies on baculovirus MMPs have been carried out. In both studies, the mmp was expressed in a heterologous baculovirus; Xestia c-nigrum granulovirus (XcGV)-MMP in BmNPV (Ko et al., 2000), and CpGV-MMP in Autographa californica M nucleopolyhedrovirus (AcMNPV) (Ishimwe, Hodgson, Passarelli, submitted for publication). Both recombinant MMP proteins were enzymatically active in vitro when assayed using synthetic substrate peptides (Ishimwe, Hodgson, Passarelli, submitted for publication) or Azo dye-impregnated collagen protease assay (azocoll) (Ko et al., 2000). XcGV-MMP could not be detected in the media of infected cells, consistent with the lack of a signal peptide (Ko et al., 2000). On the other hand, CpGV-MMP has a predicted signal peptide and was detected in the extracellular media of insect cells infected with recombinant AcMNPV expressing CpGV-MMP (Ishimwe, Hodgson, Passarelli, submitted for publication) . The heterologous expression of CpGV-MMP in AcMNPV did not appear to affect BV production or other processes in cultured cells, suggesting a possible function in the insect host (Ishimwe, Hodgson, Passarelli, submitted for publication).
Results from experiments using these engineered viruses suggest that baculovirus MMPs appear to share at least some redundant functions with the viral cathepsins. As discussed in the previous section of this review, deletion of cathepsin from either BmNPV or AcMNPV results in a complete lack of both larval melanization and liquefaction. Interestingly, insertion of either XcGV-MMP into BmNPV or CpGV-MMP into AcMNPV was able to rescue the melanization phenotype, but not liquefaction, in the absence of cathepsin (Ishimwe, Hodgson, Passarelli, submitted for publication) (Ko et al., 2000). It is not known exactly what triggers melanization in baculovirus-infected larvae. In insects, melanization normally occurs in response to tissue damage or pathogen infection, and is due to activation of phenoloxidase enzymes in hemolymph (Eleftherianos and Revenis, 2011). Since the melanization observed in baculovirus-infected larvae does not typically occur until several days after infection, around the time of larval death, it may simply be a response to tissue damage mediated by the enzymatic action of the viral cathepsin. Interestingly however, although MMP expression was not normally able to substitute for cathepsin in allowing larval liquefaction, expression of CpGV-MMP was able to partially rescue the liquefaction phenotype of an AcMNPV cathepsin mutant when the viral chitinase was modified by deletion of the KDEL motif, to allow its secretion from cells (Ishimwe, Hodgson, Passarelli, submitted for publication). This result suggests that expression of the viral cathepsin normally causes chitinase release, probably by promoting lysis of infected cells. Thus, it appears that CpGV-MMP was not able to substitute for cathepsin in promoting the release of chitinase from infected cells, but when chitinase was released by a different mechanism, CpGV-MMP was able to at least partially accomplish the functions of cathepsin that are required for liquefaction. Presumably this involves digestion of host tissues that can be carried out by either type of protease.
In alphabaculoviruses, which lack MMPs, chitinase and cathepsin work together to accomplish larval liquefaction. However, most of the betabaculoviruses express both a cathepsin and an MMP (Table). It seems unlikely that these viruses would carry two genes that have completely redundant functions, so this suggests that the MMPs probably play other roles in infection (although they may also contribute to liquefaction). It has been suggested that baculovirus MMPs may be involved in basement membrane degradation, which is not surprising since this is the major function of cellular MMPs (Ishimwe, Hodgson, Passarelli, submitted for publication) (Ko et al., 2000). Basement membranes underlie epithelia and surround muscle fibers and internal organs, and thus degradation of basement membranes could aid in systemic infection by breaking down these barriers. Betabaculovirus-encoded MMPs and/or cathepsins, if constitutively secreted from cells, might aid in BV systemic infection of tissues beyond the initial site of virus replication in the epithelia lining the gut lumen. This may work in a manner analogous to vFGF, which is encoded by all alphabaculoviruses and betabaculoviruses (except Maruca vitrata NPV), and triggers activation of host MMPs and tracheal cell basal lamina degradation to provide a conduit for BV systemic infection (Means and Passarelli, 2010). When vfgf is deleted from AcMNPV, there is a delay in host mortality due to less efficient BV escape from the primary site of infection (Detvisitsakun et al., 2007; Means and Passarelli, 2010).
In addition to defects in melanization and liquefaction, deletion of cathepsin from AcMNPV also caused a delay in host mortality, and expression of CpGV-MMP was able to rescue this delay (Ishimwe, Hodgson, Passarelli, submitted for publication). Furthermore, the simultaneous expression of cathepsin, v-chitinase and CpGV-MMP accelerated host mortality over that of wild type AcMNPV (Ishimwe, Hodgson, Passarelli, submitted for publication). XcGV-MMP expression did not significantly affect larval time of death when expressed in BmNPV, but this was apparently only tested in the absence of cathepsin (Ko et al., 2000). In addition, XcGV-MMP is not secreted from infected cells, which might affect its ability to substitute for cathepsin. Ko et al. speculated that XcGV-MMP might accumulate in a specific organelle and be released after lysis of infected cells. If true, this would imply that XcGV-MMP functions at later stages of infection. CpGV-MMP, on the other hand, is secreted from cells, presumably as it is synthesized. This suggests that CpGV-MMP is present in the extracellular environment at earlier stages of infection where it can degrade extracellular matrix components (Ishimwe, Hodgson, Passarelli, submitted for publication) The extracellular presence of CpGV-MMP may help the virus spread between tissues or organs, and thus accelerate host mortality.
A baculovirus protein with homology to bacterial collagenases
Analysis using HHpred (Soding et al., 2005) and BLAST (Altschul et al., 1990) reveals that all group II alphabaculoviruses sequenced to date, except for Adoxophyes honmai NPV and Orgyia ericae NPV, encode large proteins (>800 amino acids) with homology to collagenases from Clostridium. The homology extends over an approximately 400-amino acid region containing two putative zinc-binding histidine residues in the motif HEXXH and a conserved glutamic acid located 20 residues further towards the C-terminus, that acts as the third zinc ligand in this type of metalloprotease (Hooper, 1994). Clostridial collagenases are extracellular enzymes that act as virulence factors to facilitate host colonization (Matsushita and Okabe, 2001), but these enzymes contain additional domains involved in the degradation of collagen (Watanabe, 2004) that are absent in the viral homologs. Although some cellular MMPs are also called collagenases, the bacterial collagenases are a distinct type of enzyme, and there is no significant sequence similarity between the baculovirus collagenase-like proteins and baculovirus MMPs. This suggests that baculovirus collagenase-like proteins and MMPs may have non-overlapping functions. If these proteins are functional collagenases, they may have a role in tissue degradation. However, the role of baculovirus putative collagenases during virus infection has not been investigated, nor have they been tested for protease activity.
Enhancin
Baculovirus enhancins were first identified in betabaculoviruses, where they were found to enhance infection by alphabaculoviruses through degrading the peritrophic matrix (Derksen and Granados, 1988; Gallo et al., 1991; Tanada, 1959) (Corsaro et al., 1993). Enhancins are classified as metalloproteases based on the presence of the characteristic short consensus zinc-binding motif (HEXXH) (Hooper, 1994). Enhancin genes have been identified in the genomes of 15 baculoviruses; six of these are in the Betabaculovirus genus and the rest are from Alphabaculovirus. Most baculoviruses that encode enhancins contain a single enhancin gene; however, some genomes have more than one enhancin homolog. The largest number of enhancin homologs are found in Helicoverpa armigera GV (HearGV) and XcGV, which each harbor four enhancin genes (reviewed in (Slavicek, 2012)). Baculovirus enhancins show highest similarity to bacterial metalloproteases.
Enhancins are components of OBs. Protein localization studies determined that enhancin was a structural component of the OBs of Pseudaletia unipuncta GV (PsunGV), and enhancins E1 and E2 of Lymantria dispar MNPV (LdMNPV) were localized to the ODV envelope (Slavicek and Popham, 2005; Tanada et al., 1973; Yamamoto and Tanada, 1980). This fits with the observation that enhancins function during ODV infection of the midgut cells. Using antibodies and electron microscopy it was also shown that PsunGV enhancin associates with midgut cell membranes (Tanada et al., 1980; Uchima et al., 1988).
When an insect consumes OBs, enhancins are released and disrupt the insect peritrophic matrix which lines the midgut and acts as a barrier against pathogens (Adang and Spence, 1983). Trichoplusia ni GV (TnGV) enhancin was shown to degrade an intestinal mucin, a major component of the peritrophic matrix (Wang and Granados, 1997). In addition, the peritrophic matrix of larvae treated with TnGV enhancin showed significant alterations and permeability (Derksen and Granados, 1988; Gallo et al., 1991; Peng et al., 1999). Recombinant TnGV enhancin expressed in AcMNPV was able to improve AcMNPV infectivity (Gallo et al., 1991; Lepore et al., 1996). Similarly, expression of XcGV enhancin was able to increase the infectivity Xestia c-nigrum NPV (Derksen and Granados, 1988). Deletion of the enhancin genes in LdMNPV resulted in decreased virus infectivity (Bischoff and Slavicek, 1997; Popham et al., 2001). However, the findings in a recent study suggested that the ability of enhancins to improve virus infectivity is not limited to their ability to degrade the peritrophic matrix. An LdMNPV enhancin deletion virus was less infectious compared to wild-type LdMNPV when it was used to infect larvae lacking a peritrophic matrix (Hoover et al., 2010). This suggests that in addition to degrading the peritrophic matrix, enhancins can improve viral potency in other ways. Additional work is needed to determine additional functions of enhancins.
ODV-E66, a chondroitinase
The baculovirus protein ODV-E66 was originally identified as an ODV envelope protein (Hong et al., 1994). ODV-E66 homologs are present in most alphabaculoviruses and betabaculoviruses, with two copies present in some viruses (Rohrmann, 2013). Its N terminus was shown to be sufficient to traffic proteins to the inner nuclear membrane (Hong et al., 1997). The ODV-E66 protein has homology to hyaluronidases, and it was identified as a contaminating protein with hyaluronidase activity during purification of a human hyaluronidase from baculovirus-infected cells (Vigdorovich et al., 2007). In 2011, it was discovered that AcMPNV-infected cells contained chondroitinase activity in the cell supernatant, which was identified as an N-terminally truncated form of ODV-E66 (Sugiura et al., 2011). N-terminal processing occurs naturally during normal trafficking of the protein, so presumably the truncated form was present in the cell supernatant due to cell lysis. ODV-E66 has a novel specificity for 0S and 6S units of chondroitin sulfate chains (Sugiura et al., 2011). It is probably more properly considered a chondroitinase, since it has 600-fold higher activity on chondroitin than on hyaluronic acid, and insects do not contain hyaluronic acid (Sugiura et al., 2011).
ODV-E66 is the only known viral chondroitinase (Sugiura et al., 2011). It has been proposed that the presence of ODV-E66 in the ODV envelope may aid in degrading the peritrophic matrix, which contains low levels of chondroitin sulfate, during oral infection (Sugiura et al., 2011). Indeed, deletion of ODV-E66 from AcMNPV resulted in a mutant virus that replicated normally in cultured cells and killed Plutella xylostella larvae efficiently by injection of BV but required a 1000-fold higher dose for oral infection (Xiang et al., 2011). In addition, peptides with similarity to ODV-E66 reduced AcMNPV midgut infection in Heliothis virescens (Sparks et al., 2011). The crystal structure of ODV-E66 has been solved and it was shown to have structural similarity to polysaccharide lyase 8 proteins, with conserved residues involved in catalysis. It is most similar to bacterial chondroitin lyases, suggesting horizontal gene transfer from a bacterium during insect infection.
A trypsin-like protein encoded by gammabaculoviruses
Gammabaculoviruses encode a predicted trypsin-like protein (Duffy et al., 2006; Garcia-Maruniak et al., 2004; Lauzon et al., 2004), homologous to the hymenopteran Nasonia vitripennis (jewel wasp) trypsin-3-like protein. To our knowledge this trypsin-like protein has not been functionally characterized. Since gammabaculoviruses have a midgut-restricted infection pattern, it has been hypothesized that this enzyme performs a function in lieu of cathepsin and chitinase which are not encoded by gammabaculoviruses, allowing OB release from midgut-infected cells and their availability for the next replication cycle (Rohrmann, 2013). It is also possible that the trypsin-like protein serves to cleave a viral protein; for example, P74, which must be cleaved by trypsin in order to function in per os infectivity (Slack et al., 2008). However, the lone deltabaculovirus sequenced to date, Culex nigrapalpus NPV (CuniNPV), does not encode any of the proteases discussed above, yet it appears to effectively propagate between hosts. Details on the multiplication cycle and pathogenesis of CuniNPV are scarce, making it difficult to speculate on how its OBs are disseminated.
BACULOVIRUS-ENCODED PROTEASE INHIBITORS
Given the importance of proteases in many aspects of cellular and organismal biology, it is perhaps surprising that the existence of protease inhibitor genes is rare in virus genomes. With only a few isolated exceptions, to our knowledge the baculoviruses are one of only two families of viruses that encode protease inhibitors, the other being the poxviruses. The only exceptions that we are aware of are the adenovirus 100K assembly protein, which is able to inhibit human granzyme B (Andrade et al., 2003), and the presence of uncharacterized, but apparently intact, serpin-homologous genes in a few mimivirus genomes. There are a few other examples of viral genes with limited homology to certain serpin domains, but these do not appear to encode intact serpins.
Two types of protease inhibitors are found in both baculoviruses and poxviruses: 1) the P35/P49 family of caspase inhibitors, which are found in some baculoviruses and entomopoxviruses (Means et al., 2007), and 2) serpins, which are encoded by most members of the vertebrate-infecting chordopox viruses and in a single known baculovirus (Ardisson-Araujo et al., 2014; Rohrmann et al., 2013). It should be noted that while most baculoviruses encode homologs of the inhibitor of apoptosis (IAP) family of proteins, and some cellular IAPs can directly bind and inhibit caspases, the baculoviral IAPs do not appear to interact directly with caspases (Clem, 2007), and so are not included as protease inhibitors. Another type of apoptosis inhibitor encoded by some baculoviruses, called apsup, inhibits activation of caspases and immunoprecipitates with a caspase, but it has not been shown to be a direct protease inhibitor (Yamada et al., 2013).
Most protease inhibitors work by a reversible, “lock-and-key” mechanism of blocking access to the active site of the protease. However, both serpins and P35/P49 are irreversible mechanism-based inhibitors. It seems that when, on the rare occasions that viruses have acquired protease inhibitors, they have favored the retention of irreversible inhibitors. Perhaps this makes sense in light of the strategies of replication of these two groups of cytopathic viruses, which seek to gain ultimate control of host processes.
The caspase inhibitors P35 and P49
The P35 protein was first identified as an apoptosis suppressor protein in AcMNPV in 1991 (Clem et al., 1991). AcMNPV P35 is a 299 amino acid protein that is expressed both early and late during infection (Friesen and Miller, 1987) and is predominantly localized in the cytoplasm of infected cells (Hershberger et al., 1994). AcMNPV mutants that do not express P35 have a severely reduced ability to replicate in Spodoptera frugiperda, both in cultured cells and in insect larvae (Clem et al., 1991; Clem and Miller, 1993; Clem et al., 1994; Hershberger et al., 1992). However, in T. ni, whose cells do not undergo apoptosis easily, p35 mutant AcMNPV replicates normally both in vitro and in vivo (Clem and Miller, 1993; Hershberger et al., 1992). BmNPV p35 mutant-infected Bombyx mori-derived cells also exhibit apoptosis, but not as extensively as with AcMNPV infection of S. frugiperda cells, and BmNPV replication is not affected by mutation of p35 (Kamita et al., 1993). In addition to promoting viral replication by blocking apoptosis, P35 has also been shown to protect virions from caspase-mediated damage. Viruses lacking p35 produce unstable virions that have a delayed entry phenotype, but these phenotypes can be rescued by treating the cells used to produce p35 mutant virions with chemical caspase inhibitors (Bryant and Clem, 2009).
When P35 was first discovered, the mechanism by which it inhibited apoptosis was not known, but in 1995, investigators attempting to express a human caspase using AcMNPV as an expression vector found that cleaved P35 was covalently bound to the purified caspase, and further showed that P35 could directly inhibit caspases (Bump et al., 1995). Subsequent experiments demonstrated that P35 utilizes a mechanism-based strategy (sometimes referred to as a suicide inhibitor strategy) to inhibit caspases (Eddins et al., 2002) (Xu et al., 2001).
The first atomic structure of P35 was solved in 1999 and revealed a reactive site loop containing the caspase cleavage site extended from the rest of the structure (Fisher et al., 1999). Co-crystallization of P35 with initiator and effector caspases as well as biochemical evidence indicated that cleavage of the reactive site loop leads to conformational changes in P35 and the formation of a covalent thioester bond between P35 and the caspase (dela Cruz et al., 2001; Eddins et al., 2002; Riedl et al., 2001; Xu et al., 2001). In this way the mechanism is analogous to that used by serpins, although the two protein families have no detectable sequence or structural homology. However, the conformational change in P35 is not as drastic as that seen during serpin inhibition, with only two small loops shifting into place to trap the caspase (Stennicke et al., 2002).
The requirement for cleavage of P35 by the caspase explains why P35 is not able to prevent caspase activation, but rather can only inhibit the activity of caspases that have already been activated by cleavage (LaCount et al., 2000). The structure of P35 has evolved to have broad specificity for caspases, but an inability to inhibit other types of proteases. Thus P35 and P49 are exquisitely specific for caspases. For example, the serine protease granzyme B has the ability to cleave a similar set of substrates as caspases, but uses a completely different mechanism of proteolysis, and is resistant to inhibition by P35 (Stennicke et al., 2002). P35 appears to have evolved to take advantage of the unusual structure of the active site found in caspases, where there is an unusually long distance between the catalytic histidine and cysteine residues (Stennicke et al., 2002). P35 is able to inhibit many caspases from phylogenetically diverse organisms including nematodes, insects and mammals, which has made it a useful tool to study apoptosis and other processes in a variety of experimental systems. However, P35 is more efficient at inhibiting effector caspases than initiator caspases (Stennicke et al., 2002). For example, in AcMNPV-infected cells, activation of the effector caspase Sf-caspase-1 by the initiator caspase SfDronc is not prevented by P35, since P35 is unable to inhibit SfDronc (Huang et al., 2013; LaCount et al., 2000). In 1999, a homolog of P35 was reported in Spodoptera littoralis NPV and was named P49 (Du et al., 1999). AcMNPV P35 and SpliNPV P49 are 49% identical at the amino acid level, but P49 is 446 amino acids in length. Most of the additional sequence compared to P35 is in the central part of the P49 ORF. A homolog of P49 is also present in Spodoptera litura NPV (Yu et al., 2005). P49 is also a caspase inhibitor and is predicted to have a structure similar to that of P35 based on computer-assisted modeling and data obtained by site-directed mutagenesis (Pei et al., 2002). However, P49 is able to inhibit both initiator and effector caspases (Jabbour et al., 2002; Zoog et al., 2002), and can prevent Sf-caspase-1 activation by inhibiting Sf-Dronc (Huang et al., 2013).
The baculovirus serpin Hesp018
Serpins are a family of serine protease inhibitors that are encoded by animals, plants, parasites, bacteria, and certain viruses, which play important roles in processes such as immunity, blood coagulation, and tissue remodeling (Gettins, 2002). Like P35/P49, serpins utilize mechanism-based inhibition, which involves cleavage of a reactive center loop in the serpin, a major conformational shift in both the serpin and the protease, and formation of a covalent linkage between the serpin and the protease. Until recently, the only viruses known to encode serpin homologs were members of the vertebrate-infecting poxvirus subfamily, Chordopoxvirinae, and a few of the giant mimiviruses. In chordopoxviruses, serpins are involved in suppressing immunity of the vertebrate host (Lucas et al., 2009). Some poxvirus serpins such as CrmA are also able to inhibit caspases, even though caspases are cysteine proteases. However, a baculovirus serpin was recently reported in the genome of the Group II alphabaculovirus Hemileuca species NPV (HespNPV) (Rohrmann et al., 2013). This serpin homolog, Hesp018, has the conserved features of serpins and has been shown to be a functional serpin, being able to inhibit the serine proteases trypsin, chymotrypsin, and plasmin (Ardisson-Araujo et al., 2014). Recombinant Hesp018 protein is also able to inhibit phenoloxidase activation in Manduca sexta hemolymph (Ardisson-Araujo et al., 2014), suggesting that it might inhibit a humoral immune response in the natural host of HespNPV. Since the HespNPV genome was sequenced from an archived sample of OBs, and the virus has not been biologically characterized, it was necessary to express Hesp018 in AcMNPV in order to study its function during infection. Expression of Hesp018 in AcMNPV resulted in accelerated production of BV in Sf9 insect cells, and also a 4-fold decrease in the dose required for 50% lethality in T. ni larvae (Ardisson-Araujo et al., 2014), suggesting a possible role in pathogenesis.
CONCLUDING REMARKS
In some ways, baculoviruses can be thought of as the Ebola viruses of the insect world. Like Ebola virus, baculoviruses cause massive tissue damage in their hosts and use this as a way to spread from host to host. Unlike Ebola virus, however, baculoviruses encode enzymes that actively degrade host tissues, and in baculovirus infection the degradation occurs to the point where the host cadaver liquefies. This is an unusual strategy for virus propagation, but it obviously works well for these abundant and widespread viruses.
It has been proposed that as large DNA viruses continuously evolve, they are in a constant state of flux in regards to acquiring and discarding genes from their hosts (Roth and Andersson, 2012). If the expression of these genes serves a useful purpose, there is selective pressure for them to be maintained. Sometimes they may only provide an advantage in certain hosts or in certain situations, and sometimes their acquisition may allow viruses to expand their host range. The baculoviruses provide a fascinating example of the role of gene acquisition in viral pathogenesis and infection strategies. During their evolution, baculoviruses have acquired and/or lost various combinations of genes encoding degradative enzymes from their hosts, which have enhanced their abilities to efficiently infect and disseminate within and between hosts by disrupting chitin-containing structures and extracellular matrix, as well as degrading proteinaceous host tissues. Meanwhile, only a few viruses encode inhibitors of proteases, but the ones that are carried (serpins and P35/P49) appear to mainly function to inhibit host immune responses. Viral serpins were clearly obtained from host genomes, but it is not clear where the P35/P49 family was acquired from, since there are no recognizable cellular homologs. Regardless, it appears that the vast majority of viruses have devised alternative methods to avoid host immunity that does not require expression of protease inhibitors. It may be a disadvantage for most viruses to carry protease inhibitors that inhibit the host immune response, for example, if it leads to increased competition from other types of pathogens. Why these have evolved in baculoviruses and poxviruses, but not other viruses, remains a mystery.
There is variation in the types of degradative enzymes that are carried by different baculoviruses (Table), potentially indicating acquisition and/or loss of these genes many times throughout the evolution of these viruses. Most, but not all, alphabaculoviruses and betabaculoviruses encode chitinase and cathepsin (Table), and these have been shown to be necessary to liquefy an insect cadaver prior to virion dispersal in the environment. In addition to chitinase and cathepsin, betabaculoviruses also encode mmp genes. The role of viral MMPs during betabaculovirus pathogenesis is not clear but they may be involved in basal lamina component degradation to help provide access to various organs, or to aid in virus dispersal in the late stages of infection. Betabaculovirus infection patterns range from midgut-restricted to establishing systemic infections, but regardless of infection pattern, all sequenced betabaculoviruses carry mmp genes. Lack of mmp genes and/or acquisition of an alternative BV envelope protein may have contributed to altered pathogenicity and host range in the more recently evolved alphabaculoviruses.
On the other hand, gammabaculoviruses and deltabaculoviruses do not encode cathepsin, chitinase or mmp genes. These baculoviruses are midgut-restricted and are thought to spread from insect to insect by dispersal of the OBs following infectious diarrheal excretions. Deltabaculovirus-infected mosquito larvae live in an aqueous environment, which may facilitate easier transmission of OBs to other larvae via the oral-fecal route, thus making more superfluous the need for larvae liquefaction and tissue disintegration enzymes. Gammabaculoviruses all encode a trypsin-like protein gene (Table) with similarity to cellular trypsins (Duffy et al., 2006; Garcia-Maruniak et al., 2004; Lauzon et al., 2006). The function of these trypsin-like genes is not known and their conservation in gammabaculoviruses is less clear, since only a limited number of gammabaculovirus genomes have been sequenced.
Acknowledgments
We are grateful to the anonymous reviewers who provided helpful suggestions regarding this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adang MJ, Spence KD. Permeability of the peritrophic membrane of the Douglas fir tussock moth (Orgyia pseudotsugata) Comp. Biochem. Physiol. 1983;75A:233–238. [Google Scholar]
- Afonso CL, Tulman ER, Lu Z, Oma E, Kutish GF, Rock DL. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrade F, Casciola-Rosen LA, Rosen A. A novel domain in adenovirus L4-100K is required for stable binding and efficient inhibition of human granzyme B: possible interaction with a species-specific exosite. Mol Cell Biol. 2003;23:6315–6326. doi: 10.1128/MCB.23.17.6315-6326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardisson-Araujo DMP, Rohrmann GF, Ribeiro BM, Clem RJ. Functional characterization of hesp018, a baculovirus-encoded serpin gene. J. Gen. Virol. 2014 doi: 10.1099/vir.0.000041. In press. [DOI] [PubMed] [Google Scholar]
- Bischoff DS, Slavicek JM. Molecular analysis of an enhancin gene in the Lymantria dispar nuclear polyhedrosis virus. J Virol. 1997;71:8133–8140. doi: 10.1128/jvi.71.11.8133-8140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant B, Clem RJ. Caspase inhibitor P35 is required for the production of robust baculovirus virions in Trichoplusia ni TN-368 cells. J Gen Virol. 2009;90:654–661. doi: 10.1099/vir.0.007419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg AH, Miller LK, Wong WW. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- Clem RJ. Baculoviruses and apoptosis: a diversity of genes and responses. Current Drug Targets. 2007;8:1069–1074. doi: 10.2174/138945007782151405. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Miller LK. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 1993;67:3730–3738. doi: 10.1128/jvi.67.7.3730-3738.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ, Passarelli AL. Baculoviruses: sophisticated pathogens of insects. PLoS Pathog. 2013;9:14. doi: 10.1371/journal.ppat.1003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ, Robson M, Miller LK. Influence of infection route on the infectivity of baculovirus mutants lacking the apoptosis-inhibiting gene p35 and the adjacent gene p94. J. Virol. 1994;68:6759–6762. doi: 10.1128/jvi.68.10.6759-6762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado G, Arciello S, Fanti P, Fiandra L, Garonna A, Digilio MC, Lorito M, Giordana B, Pennacchio F, Rao R. The Chitinase A from the baculovirus AcMNPV enhances resistance to both fungi and herbivorous pests in tobacco. Transgenic Res. 2008;17:557–571. doi: 10.1007/s11248-007-9129-4. [DOI] [PubMed] [Google Scholar]
- Corsaro BG, Gijzen M, Wang P, Granados RR. Academic Press. New York, NY: 1993. Baculovirus enhancing proteins as determinants of viral pathogenesis, Parasites and Pathogens of Insects; pp. 127–145. [Google Scholar]
- D’mico V, Slavicek J, Podgwaite JD, Webb R, Fuester R, Peiffer RA. Deletion of v-chiA from a baculovirus reduces horizontal transmission in the field. Appl Environ Microbiol. 2013;79:4056–4064. doi: 10.1128/AEM.00152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon T, Hamada K, Mita K, Okano K, Suzuki MG, Kobayashi M, Shimada T. A Bombyx mori gene, BmChi-h, encodes a protein homologous to bacterial and baculovirus chitinases. Insect Biochem Mol Biol. 2003;33:749–759. doi: 10.1016/s0965-1748(03)00084-5. [DOI] [PubMed] [Google Scholar]
- Daimon T, Katsuma S, Kang W, Shimada T. Comparative studies of Bombyx mori nucleopolyhedrovirus chitinase and its host ortholog, BmChi-h. Biochem Biophys Res Commun. 2006;345:825–833. doi: 10.1016/j.bbrc.2006.04.112. [DOI] [PubMed] [Google Scholar]
- Daimon T, Katsuma S, Kang WK, Shimada T. Functional characterization of chitinase from Cydia pomonella granulovirus. Arch Virol. 2007a;152:1655–1664. doi: 10.1007/s00705-007-1000-7. [DOI] [PubMed] [Google Scholar]
- Daimon T, Katsuma S, Shimada T. Mutational analysis of active site residues of chitinase from Bombyx mori nucleopolyhedrovirus. Virus Res. 2007b;124:168–175. doi: 10.1016/j.virusres.2006.11.001. [DOI] [PubMed] [Google Scholar]
- de Coignac AB, Elson G, Delneste Y, Magistrelli G, Jeannin P, Aubry JP, Berthier O, Schmitt D, Bonnefoy JY, Gauchat JF. Cloning of MMP-26. A novel matrilysin-like proteinase. Eur J Biochem. 2000;267:3323–3329. doi: 10.1046/j.1432-1327.2000.01363.x. [DOI] [PubMed] [Google Scholar]
- dela Cruz WP, Friesen PD, Fisher AJ. Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage. J Biol Chem. 2001;276:32933–32939. doi: 10.1074/jbc.M103930200. [DOI] [PubMed] [Google Scholar]
- Derksen ACG, Granados RR. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virology. 1988;167:242–250. doi: 10.1016/0042-6822(88)90074-8. [DOI] [PubMed] [Google Scholar]
- Detvisitsakun C, Cain EL, Passarelli AL. The Autographa californica M nucleopolyhedrovirus fibroblast growth factor accelerates host mortality. Virology. 2007;365:70–78. doi: 10.1016/j.virol.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Dhanaraj V, Ye QZ, Johnson LL, Hupe DJ, Ortwine DF, Dunbar JB, Jr., Rubin JR, Pavlovsky A, Humblet C, Blundell TL. X-ray structure of a hydroxamate inhibitor complex of stromelysin catalytic domain and its comparison with members of the zinc metalloproteinase superfamily. Structure. 1996;4:375–386. doi: 10.1016/s0969-2126(96)00043-3. [DOI] [PubMed] [Google Scholar]
- Ding X, Gopalakrishnan B, Johnson LB, White FF, Wang X, Morgan TD, Kramer KJ, Muthukrishnan S. Insect resistance of transgenic tobacco expressing an insect chitinase gene. Transgenic Res. 1998;7:77–84. doi: 10.1023/a:1008820507262. [DOI] [PubMed] [Google Scholar]
- Du Q, Lehavi D, Faktor O, Qi Y, Chejanovsky N. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J Virol. 1999;73:1278–1285. doi: 10.1128/jvi.73.2.1278-1285.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy SP, Young AM, Morin B, Lucarotti CJ, Koop BF, Levin DB. Sequence analysis and organization of the Neodiprion abietis nucleopolyhedrovirus genome. J Virol. 2006;80:6952–6963. doi: 10.1128/JVI.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Lemongello D, Friesen PD, Fisher AJ. Crystallization and low-resolution structure of an effector-caspase/P35 complex: similarities and differences to an initiator-caspase/P35 complex. Acta Crystallogr D Biol Crystallogr. 2002;58:299–302. doi: 10.1107/s0907444901018753. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, Revenis C. Role and importance of phenoloxidase in insect hemostasis. J Innate Immun. 2011;3:28–33. doi: 10.1159/000321931. [DOI] [PubMed] [Google Scholar]
- Elkington PT, O’Kane CM, Friedland JS. The paradox of matrix metalloproteinases in infectious disease. Clin Exp Immunol. 2005;142:12–20. doi: 10.1111/j.1365-2249.2005.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escasa SR, Lauzon HA, Mathur AC, Krell PJ, Arif BM. Sequence analysis of the Choristoneura occidentalis granulovirus genome. J Gen Virol. 2006;87:1917–1933. doi: 10.1099/vir.0.81792-0. [DOI] [PubMed] [Google Scholar]
- Ferrelli ML, Salvador R, Biedma ME, Berretta MF, Haase S, Sciocco-Cap A, Ghiringhelli PD, Romanowski V. Genome of Epinotia aporema granulovirus (EpapGV), a polyorganotropic fast killing betabaculovirus with a novel thymidylate kinase gene. BMC Genomics. 2012;13:1471–2164. doi: 10.1186/1471-2164-13-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AJ, Cruz W, Zoog SJ, Schneider CL, Friesen PD. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. Embo J. 1999;18:2031–2039. doi: 10.1093/emboj/18.8.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen PD, Miller LK. Divergent transcription of early 3. and 94-kilodalton protein genes encoded by the Hind-III K genome fragment of the baculovirus Autographa californica nuclear polyhedrosis virus. 1987;J. Virol.61:2264–2272. doi: 10.1128/jvi.61.7.2264-2272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo LG, Corsaro BG, Hughes PR, Granados RR. In vivo enhancement of baculovirus infection by the viral enhancing factor of a granulosis virus of the cabbage looper, Trichoplusia-ni (Lepidoptera, Noctuidae) J Invertebr Pathol. 1991;58:203–210. [Google Scholar]
- Garcia-Maruniak A, Maruniak JE, Zanotto PMA, Doumbouya AE, Liu J-C, Merritt TM, Lanoie JS. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 2004;78:7036–7051. doi: 10.1128/JVI.78.13.7036-7051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- Harrison R, Hoover K. Baculoviruses and other occluded insect viruses. In: Vega FE, Kaya HK, editors. Insect Pathology. Academic Press; Waltham, MA: 2012. pp. 73–132. [Google Scholar]
- Harrison RL, Popham HJ. Genomic sequence analysis of a granulovirus isolated from the Old World bollworm, Helicoverpa armigera. Virus Genes. 2008;36:565–581. doi: 10.1007/s11262-008-0218-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Hayakawa T, Ueno Y, Fujita T, Sano Y, Matsumoto T. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275:358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- Hawtin RE, Arnold K, Ayres MD, Zanotto PM, Howard SC, Gooday GW, Chappell LH, Kitts PA, King LA, Possee RD. Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology. 1995;212:673–685. doi: 10.1006/viro.1995.1525. [DOI] [PubMed] [Google Scholar]
- Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, Kuzio JA, Possee RD. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology. 1997;238:243–253. doi: 10.1006/viro.1997.8816. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kazuhiro O, Seong S-I, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- Henrissat B. Classification of chitinases modules. Exs. 1999;87:137–156. doi: 10.1007/978-3-0348-8757-1_10. [DOI] [PubMed] [Google Scholar]
- Hershberger PA, Dickson JA, Friesen PD. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 1992;66:5525–5533. doi: 10.1128/jvi.66.9.5525-5533.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger PA, LaCount DJ, Friesen PD. The apoptotic suppressor P35 is required early during baculovirus replication and is targeted to the cytosol of infected cells. J. Virol. 1994;68:3467–3477. doi: 10.1128/jvi.68.6.3467-3477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Kuzio J, Faulkner P. Identification and characterization of the v-cath gene of the baculovirus, CfMNPV. Biochim Biophys Acta. 1995;27:275–278. doi: 10.1016/0167-4781(95)00195-6. [DOI] [PubMed] [Google Scholar]
- Hiramatsu S, Ishihara M, Fujie M, Usami S, Yamada T. Expression of a chitinase gene and lysis of the host cell wall during Chlorella virus CVK2 infection. Virology. 1999;260:308–315. doi: 10.1006/viro.1999.9824. [DOI] [PubMed] [Google Scholar]
- Hodgson JJ, Arif BM, Krell PJ. Reprogramming the chiA expression profile of Autographa californica multiple nucleopolyhedrovirus. J Gen Virol. 2007;88:2479–2487. doi: 10.1099/vir.0.82863-0. [DOI] [PubMed] [Google Scholar]
- Hodgson JJ, Arif BM, Krell PJ. Autographa californica multiple nucleopolyhedrovirus and Choristoneura fumiferana multiple nucleopolyhedrovirus v-cath genes are expressed as pre-proenzymes. J Gen Virol. 2009;90:995–1000. doi: 10.1099/vir.0.007740-0. [DOI] [PubMed] [Google Scholar]
- Hodgson JJ, Arif BM, Krell PJ. Interaction of Autographa californica multiple nucleopolyhedrovirus cathepsin protease progenitor (proV-CATH) with insect baculovirus chitinase as a mechanism for proV-CATH cellular retention. J Virol. 2011;85:3918–3929. doi: 10.1128/JVI.02165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JJ, Arif BM, Krell PJ. Role of interactions between Autographa californica multiple nucleopolyhedrovirus procathepsin and chitinase chitin-binding or active-site domains in viral cathepsin processing. J Virol. 2013;87:3471–3483. doi: 10.1128/JVI.01937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom LG, Ohkawa T, Trudeau D, Volkman LE. Autographa californica M nucleopolyhedrovirus ProV-CATH is activated during infected cell death. Virology. 2002;296:212–218. doi: 10.1006/viro.2002.1378. [DOI] [PubMed] [Google Scholar]
- Hom LG, Volkman LE. Autographa californica M nucleopolyhedrovirus chiA is required for processing of V-CATH. Virology. 2000;277:178–183. doi: 10.1006/viro.2000.0586. [DOI] [PubMed] [Google Scholar]
- Hong T, Braunagel SC, Summers MD. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology. 1994;204:210–222. doi: 10.1006/viro.1994.1525. [DOI] [PubMed] [Google Scholar]
- Hong T, Summers MD, Braunagel SC. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of occlusion derived virus. Proc. Natl. Acad. Sci. U S A. 1997;94:4050–4055. doi: 10.1073/pnas.94.8.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Hoover K, Humphries MA, Gendron AR, Slavicek JM. Impact of viral enhancin genes on potency of Lymantria dispar multiple nucleopolyhedrovirus in L. dispar following disruption of the peritrophic matrix. J Invertebr Pathol. 2010;104:150–152. doi: 10.1016/j.jip.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hou D, Zhang L, Deng F, Fang W, Wang R, Liu X, Guo L, Rayner S, Chen X, Wang H, Hu Z. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J. Virol. 2013;87:829–839. doi: 10.1128/JVI.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Civciristov S, Hawkins CJ, Clem RJ. SfDronc, an initiator caspase involved in apoptosis in the fall armyworm Spodoptera frugiperda. Insect Biochem Mol Biol. 2013;43:444–454. doi: 10.1016/j.ibmb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour AM, Ekert PG, Coulson EJ, Knight MJ, Ashley DM, Hawkins CJ. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ. 2002;9:1311–1320. doi: 10.1038/sj.cdd.4401135. [DOI] [PubMed] [Google Scholar]
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006 doi: 10.1007/s00705-006-0763-6. DOI 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- Jollès P, Muzzarelli RA. Chitin and chitinases. Springer; 1999. [Google Scholar]
- Kaba SA, Salcedo AM, Wafula PO, Vlak JM, van Oers MM. Development of a chitinase and v-cathepsin negative bacmid for improved integrity of secreted recombinant proteins. J Virol Methods. 2004;122:113–118. doi: 10.1016/j.jviromet.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kamita SG, Majima K, Maeda S. Identification and characterization of the p35 gene of Bombyx mori nuclear polyhedrosis virus that prevents virus-induced apoptosis. J. Virol. 1993;67:455–463. doi: 10.1128/jvi.67.1.455-463.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Tristem M, Maeda S, Crook NE, O'Reilly DR. Identification and characterization of the Cydia pomonella granulovirus cathepsin and chitinase genes. J Gen Virol. 1998;79:2283–2292. doi: 10.1099/0022-1317-79-9-2283. [DOI] [PubMed] [Google Scholar]
- Katsuma S, Nakanishi T, Daimon T, Shimada T. N-linked glycans located in the pro-region of Bombyx mori nucleopolyhedrovirus V-CATH are essential for the proper folding of V-CATH and V-CHIA. J Gen Virol. 2009;90:170–176. doi: 10.1099/vir.0.005835-0. [DOI] [PubMed] [Google Scholar]
- Katunuma N. Posttranslational processing and modification of cathepsins and cystatins. J Signal Transduct. 2010;375345:16. doi: 10.1155/2010/375345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr E, Schmidtberg H, Vilcinskas A, Altincicek B. MMPs regulate both development and immunity in the tribolium model insect. PLoS ONE. 2009;4:9. doi: 10.1371/journal.pone.0004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko R, Okano K, Maeda S. Structural and functional analysis of the Xestia c-nigrum granulovirus matrix metalloproteinase. J. Virol. 2000;74:11240–11246. doi: 10.1128/jvi.74.23.11240-11246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga D, Mitsutomi M, Kono M, Matsumiya M. Biochemistry of chitinases. In: Henrisset B, editor. Chitin and Chitinases. Birkhauser Verlag; Basel, Switzerland: 1999. pp. 111–123. [DOI] [PubMed] [Google Scholar]
- Kramer KJ, Dzidik-Tuber C, Koga D. Chitin metabolism in insects. In: Kerbut GA, Gilbert L, editors. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Pergamon, Oxford; 1985. pp. 75–115. [Google Scholar]
- LaCount DJ, Hanson SF, Schneider CL, Friesen PD. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J Biol Chem. 2000;275:15657–15664. doi: 10.1074/jbc.M000791200. [DOI] [PubMed] [Google Scholar]
- Lange M, Jehle JA. The genome of the Cryptophlebia leucotreta granulovirus. Virology. 2003;317:220–236. doi: 10.1016/s0042-6822(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Lauzon HA, Garcia-Maruniak A, Zanotto PM, Clemente JC, Herniou EA, Lucarotti CJ, Arif BM, Maruniak JE. Genomic comparison of Neodiprion sertifer and Neodiprion lecontei nucleopolyhedroviruses and identification of potential hymenopteran baculovirus-specific open reading frames. J Gen Virol. 2006;87:1477–1489. doi: 10.1099/vir.0.81727-0. [DOI] [PubMed] [Google Scholar]
- Lauzon HAM, Lucarotti CJ, Krell PJ, Feng Q, Retnakaran A, Arif BM. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 2004;78:7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore LS, Roelvink PR, Granados RR. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J Invertebr Pathol. 1996;68:131–140. doi: 10.1006/jipa.1996.0070. [DOI] [PubMed] [Google Scholar]
- Li H, Tang H, Harrison RL, Bonning BC. Impact of a basement membrane-degrading proteases on dissemination and secondary infection of Autographa californica multiple nucleopolyhedrovirus in Heliothis virescens (Fabricus) J. Gen. Virol. 2007;88:1109–1119. doi: 10.1099/vir.0.82691-0. [DOI] [PubMed] [Google Scholar]
- Liang Z, Zhang X, Yin X, Cao S, Xu F. Genomic sequencing and analysis of Clostera anachoreta granulovirus. Arch Virol. 2011;156:1185–1198. doi: 10.1007/s00705-011-0977-0. [DOI] [PubMed] [Google Scholar]
- Liang Z, Zhang X, Yin X, Song X, Shao X, Wang L. Comparative analysis of the genomes of Clostera anastomosis (L.) granulovirus and Clostera anachoreta granulovirus. Arch Virol. 2013;158:2109–2114. doi: 10.1007/s00705-013-1710-y. [DOI] [PubMed] [Google Scholar]
- Liaudet E, Garcia M, Rochefort H. Cathepsin D maturation and its stimulatory effect on metastasis are prevented by addition of KDEL retention signal. Oncogene. 1994;9:1145–1154. [PubMed] [Google Scholar]
- Lima AA, Aragao CW, de Castro ME, Oliveira JV, Sosa Gomez DR, Ribeiro BM. A recombinant Anticarsia gemmatalis MNPV harboring chiA and v-cath genes from Choristoneura fumiferana defective NPV induce host liquefaction and increased insecticidal activity. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0074592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A, Liu L, Dai E, Bot I, Viswanathan K, Munuswamy-Ramunujam G, Davids JA, Bartee MY, Richardson J, Christov A, Wang H, Macaulay C, Poznansky M, Zhong R, Miller L, Biessen E, Richardson M, Sullivan C, Moyer R, Hatton M, Lomas DA, McFadden G. The serpin saga; development of a new class of virus derived anti-inflammatory protein immunotherapeutics. Adv Exp Med Biol. 2009;666:132–156. doi: 10.1007/978-1-4419-1601-3_11. [DOI] [PubMed] [Google Scholar]
- Lung O, Blissard GW. A cellular Drosophila melanogaster protein with similarity to baculovirus F envelope fusion proteins. J. Virol. 2005;79:7979–7989. doi: 10.1128/JVI.79.13.7979-7989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque T, Finch R, Crook N, O'Reilly DR, Winstanley D. The complete sequence of the Cydia pomonella granulovirus genome. J Gen Virol. 2001;82:2531–2547. doi: 10.1099/0022-1317-82-10-2531. [DOI] [PubMed] [Google Scholar]
- Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM. Matrix metalloproteinase homologues from Arabidopsis thaliana. Expression and activity. J Biol Chem. 1999;274:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- Matsushita O, Okabe A. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon. 2001;39:1769–1780. doi: 10.1016/s0041-0101(01)00163-5. [DOI] [PubMed] [Google Scholar]
- Means JC, Passarelli AL. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing infection by baculoviruses. Proc. Natl. Acad. Sci. U S A. 2010;107:9825–9830. doi: 10.1073/pnas.0913582107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means JC, Penabaz T, Clem RJ. Identification and functional characterization of AMVp33, a novel homolog of the baculovirus caspase inhibitor p35 found in Amsacta moorei entomopoxvirus. Virology. 2007;358:436–447. doi: 10.1016/j.virol.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G, Allan JA, Willenbrock F, Cockett MI, O'Connell JP, Docherty AJ. The role of the C-terminal domain in collagenase and stromelysin specificity. J Biol Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- Muschler J, Streuli CH. Cell-matrix interactions in mammary gland development and breast cancer. Cold Spring Harb Perspect Biol. 2010;2:11. doi: 10.1101/cshperspect.a003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S, Merzendorfer H, Arakane Y, Kramer KJ. Chitin metabolism in insects. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press; Waltllham, MA: 2012. [Google Scholar]
- Nagase H, Woessner JF., Jr. Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Norris M. Characterization of the conserved chiA and v-cath bidirectional promoter of Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Molecular and Cellular Biology. The University of Guelph; 2011. p. 137. [Google Scholar]
- Ohkawa T, Majima K, Maeda S. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J Virol. 1994;68:6619–6625. doi: 10.1128/jvi.68.10.6619-6625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi J, Ohnishi E, Jin M, Hirano W, Nakane D, Matsui H, Kimura A, Sawa H, Nakayama K, Shibuya H, Nagashima K, Takahashi T. Cloning and characterization of a rat ortholog of MMP-23 (matrix metalloproteinase-23), a unique type of membrane-anchored matrix metalloproteinase and conditioned switching of its expression during the ovarian follicular development. Mol Endocrinol. 2001;15:747–764. doi: 10.1210/mend.15.5.0638. [DOI] [PubMed] [Google Scholar]
- Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. Phenotypic grouping of 14. BmNPVs lacking viral gene sequences. Virus Res. 2012;165:197–206. doi: 10.1016/j.virusres.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, Serano J, Sante JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003;4:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- Pak JH, Liu CY, Huangpu J, Graham JS. Construction and characterization of the soybean leaf metalloproteinase cDNA. FEBS Lett. 1997;404:283–288. doi: 10.1016/s0014-5793(97)00141-5. [DOI] [PubMed] [Google Scholar]
- Pei Z, Reske G, Huang Q, Hammock BD, Qi Y, Chejanovsky N. Characterization of the apoptosis suppressor protein P49 from the Spodoptera littoralis nucleopolyhedrovirus. J. Biol. Chem. 2002;277:48677–48684. doi: 10.1074/jbc.M208810200. [DOI] [PubMed] [Google Scholar]
- Peng J, Zhong J, R RG. A baculovirus enhancin alters the permeability of a mucosal midgut peritrophic matrix from lepidopteran larvae. J Insect Physiol. 1999;45:159–166. doi: 10.1016/s0022-1910(98)00110-3. [DOI] [PubMed] [Google Scholar]
- Popham HJ, Bischoff DS, Slavicek JM. Both Lymantria dispar nucleopolyhedrovirus enhancin genes contribute to viral potency. J Virol. 2001;75:8639–8648. doi: 10.1128/JVI.75.18.8639-8648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra HJ, Parks WC. Control of matrix metalloproteinase catalytic activity. Matrix Biol. 2007;26:587–596. doi: 10.1016/j.matbio.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Fiandra L, Giordana B, de Eguileor M, Congiu T, Burlini N, Arciello S, Corrado G, Pennacchio F. AcMNPV ChiA protein disrupts the peritrophic membrane and alters midgut physiology of Bombyx mori larvae. Insect Biochem Mol Biol. 2004;34:1205–1213. doi: 10.1016/j.ibmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Pearl LH, Buttle DJ. The baculovirus Autographa californica nuclear polyhedrosis virus genome includes a papain-like sequence. Biol Chem Hoppe Seyler. 1992;373:1211–1215. doi: 10.1515/bchm3.1992.373.2.1211. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Snipas SJ, Salvesen GS. Mechanism-based inactivation of caspases by the apoptotic suppressor p35. Biochemistry. 2001;40:13274–13280. doi: 10.1021/bi010574w. [DOI] [PubMed] [Google Scholar]
- Rohrmann GF. Baculovirus molecular biology. Third. National Center for Biotechnology Information; Bethesda, MD: 2013. [PubMed] [Google Scholar]
- Rohrmann GF, Erlandson MA, Theilmann DA. The genome of a baculovirus isolated from Hemileuca sp. encodes a serpin ortholog. Virus Genes. 2013;47:357–364. doi: 10.1007/s11262-013-0951-x. [DOI] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Poxvirus use a "gene accordion" to tune out host defenses. Cell. 2012;150:671–672. doi: 10.1016/j.cell.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Salvador R, Ferrelli ML, Sciocco-Cap A, Romanowski V. Analysis of a chitinase from EpapGV, a fast killing betabaculovirus. Virus Genes. 2014;48:406–409. doi: 10.1007/s11262-013-1019-7. [DOI] [PubMed] [Google Scholar]
- Saville GP, Thomas CJ, Possee RD, King LA. Partial redistribution of the Autographa californica nucleopolyhedrovirus chitinase in virus-infected cells accompanies mutation of the carboxy-terminal KDEL ER-retention motif. J Gen Virol. 2002;83:685–694. doi: 10.1099/0022-1317-83-3-685. [DOI] [PubMed] [Google Scholar]
- Shapiro M, Priesler HK, Robertson JL. Enhancement of baculovirus activity on gypsy moth (Lepidoptera: Lymantridae) by chitinase. J. Econ. Entomol. 1987;85:1113–1116. [Google Scholar]
- Slack JM, Kuzio J, Faulkner P. Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. J. Gen. Virol. 1995;76:1091–1098. doi: 10.1099/0022-1317-76-5-1091. [DOI] [PubMed] [Google Scholar]
- Slack JM, Lawrence SD, Krell PJ, Arif BM. Trypsin cleavage of the baculovirus occlusion-derived virus attachment protein P74 is prerequisite in per os infection. J Gen Virol. 2008;89:2388–2397. doi: 10.1099/vir.0.2008/002543-0. [DOI] [PubMed] [Google Scholar]
- Slack JM, Ribeiro BM, de Souza ML. The gp64 locus of Anticarsia gemmatalis multicapsid nucleopolyhedrovirus contains a 3. repair exonuclease homologue and lacks v-cath and ChiA genes. J Gen Virol. 2004;85:211–219. doi: 10.1099/vir.0.19617-0. [DOI] [PubMed] [Google Scholar]
- Slavicek JM. Baculovirus enhancins and their role in viral pathogenicity. In: Adoga MP, editor. Molecular Virology. InTech; 2012. pp. 147–168. [Google Scholar]
- Slavicek JM, Popham HJ. The Lymantria dispar nucleopolyhedrovirus enhancins are components of occlusion-derived virus. J Virol. 2005;79:10578–10588. doi: 10.1128/JVI.79.16.10578-10588.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks WO, Rohlfing A, Bonning BC. A peptide with similarity to baculovirus ODV-E66 binds the gut epithelium of Heliothis virescens and impedes infection with Autographa californica multiple nucleopolyhedrovirus. J Gen Virol. 2011;92:1051–1060. doi: 10.1099/vir.0.028118-0. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Ryan CA, Salvesen GS. Reprieval from execution: the molecular basis of caspase inhibition. Trends Biochem Sci. 2002;27:94–101. doi: 10.1016/s0968-0004(01)02045-x. [DOI] [PubMed] [Google Scholar]
- Sugiura N, Setoyama Y, Chiba M, Kimata K, Watanabe H. Baculovirus envelope protein ODV-E66 is a novel chondroitinase with distinct substrate specificity. J Biol Chem. 2011;286:29026–29034. doi: 10.1074/jbc.M111.251157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanada Y. Synergism between two viruses of the armyworm, Pseudaletia unipuncta. Haworth; Lepidoptera, Noctuidae: 1959. [Place of publication not identified] [Google Scholar]
- Tanada Y, Himeno M, Omi EM. Isolation of a factor, from the capsule of a granulosis virus, synergistic for a nuclear-polyhedrosis virus of the armyworm. J Invertebr Pathol. 1973;21:31–40. doi: 10.1016/0022-2011(73)90110-9. [DOI] [PubMed] [Google Scholar]
- Tanada Y, Inoue H, Hess RT, Omi EM. Site of action of a synergistic factor of a granulosis virus of the armyworm, Pseudaletia unipuncta. Journal of Invertebrate Pathology. 1980;35:249–255. [Google Scholar]
- Thomas CJ, Brown HL, Hawes CR, Lee BY, Min MK, King LA, Possee RD. Localization of a baculovirus-induced chitinase in the insect cell endoplasmic reticulum. J Virol. 1998;72:10207–10212. doi: 10.1128/jvi.72.12.10207-10212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CJ, Gooday GW, King LA, Possee RD. Mutagenesis of the active site coding region of the Autographa californica nucleopolyhedrovirus chiA gene. J Gen Virol. 2000;81:1403–1411. doi: 10.1099/0022-1317-81-5-1403. [DOI] [PubMed] [Google Scholar]
- Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchima K, Harvey JP, Omi EM, Tanada Y. Binding sites on the midgut cell membrane for the synergistic factor of a granulosis virus of the armyworm (Pseudaletia unipuncta) Insect Biochem. 1988;18:645–650. [Google Scholar]
- Vaaje-Kolstad G, Horn SJ, van Aalten DM, Synstad B, Eijsink VG. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem. 2005;280:28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci U S A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Pendas AM, Fueyo A, Knauper V, Murphy G, Lopez-Otin C. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J Biol Chem. 1999;274:4570–4576. doi: 10.1074/jbc.274.8.4570. [DOI] [PubMed] [Google Scholar]
- Vieira CM, Tuelher ES, Valicente FH, Wolff JL. Characterization of a Spodoptera frugiperda multiple nucleopolyhedrovirus isolate that does not liquefy the integument of infected larvae. J Invertebr Pathol. 2012;111:189–192. doi: 10.1016/j.jip.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Vigdorovich V, Miller AD, Strong RK. Ability of hyaluronidase 2 to degrade extracellular hyaluronan is not required for its function as a receptor for jaagsiekte sheep retrovirus. J Virol. 2007;81:3124–3129. doi: 10.1128/JVI.02177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalon JM, Ghosh A, Jacobs-Lorena M. The peritrophic matrix limits the rate of digestion in adult Anopheles stephensi and Aedes aegypti mosquitoes. Journal of Insect Physiology. 2003;49:891–895. doi: 10.1016/s0022-1910(03)00135-5. [DOI] [PubMed] [Google Scholar]
- Wang F, Zhang CX, Shyam Kumar V, Wu XF. Influences of chitinase gene deletion from BmNPV on the cell lysis and host liquefaction. Arch Virol. 2005;150:981–990. doi: 10.1007/s00705-004-0458-9. [DOI] [PubMed] [Google Scholar]
- Wang P, Granados RR. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci U S A. 1997;94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Granados RR. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch Insect Biochem Physiol. 2001;47:110–118. doi: 10.1002/arch.1041. [DOI] [PubMed] [Google Scholar]
- Wang R, Deng F, Hou D, Zhao Y, Guo L, Wang H, Hu Z. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 2010;84:7233–7242. doi: 10.1128/JVI.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Choi JY, Roh JY, Woo SD, Jin BR, Je YH. Molecular and phylogenetic characterization of Spodoptera litura granulovirus. J Microbiol. 2008;46:704–708. doi: 10.1007/s12275-008-0133-z. [DOI] [PubMed] [Google Scholar]
- Watanabe K. Collagenolytic proteases from bacteria. Appl Microbiol Biotechnol. 2004;63:520–526. doi: 10.1007/s00253-003-1442-0. [DOI] [PubMed] [Google Scholar]
- Woltje M, Bobel M, Rheinnecker M, Tettamanti G, Franzetti E, Saviane A, Cappellozza S. Transgenic protein production in silkworm silk glands requires cathepsin and chitinase of Autographa californica multicapsid nucleopolyhedrovirus. Appl Microbiol Biotechnol. 2014;98:4571–4580. doi: 10.1007/s00253-014-5543-8. [DOI] [PubMed] [Google Scholar]
- Wormleaton S, Kuzio J, Winstanley D. The complete sequence of the Adoxophyes orana granulovirus. Virology. 2003;311:350–365. doi: 10.1016/s0042-6822(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Xiang X, Chen L, Hu X, Yu S, Yang R, Wu X. Autographa californica multiple nucleopolyhedrovirus odv-e66 is an essential gene required for oral infectivity. Virus Res. 2011;158:72–78. doi: 10.1016/j.virusres.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Xu G, Cirilli M, Huang Y, Rich RL, Myszka DG, Wu H. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- Yamada H, Kitaguchi K, Hamajima R, Kobayashi M, Ikeda M. Novel apoptosis suppressor Apsup from the baculovirus Lymantria dispar multiple nucleopolyhedrovirus precludes apoptosis by preventing proteolytic processing of initiator caspase Dronc. J Virol. 2013;87:12925–12934. doi: 10.1128/JVI.02065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Shimomae A, Furukawa S, Takehara J. Widespread distribution of Chlorella viruses in Japan. Biosci Biotechnol Biochem. 1993;57:733–739. doi: 10.1271/bbb.57.733. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Tanada Y. Physicochemical properties and location of capsule components, in particular the synergistic factor, in the occlusion body of a granulosis virus of the armyworm, Pseudaletia unipuncta. Virology. 1980;107:434–440. doi: 10.1016/0042-6822(80)90310-4. [DOI] [PubMed] [Google Scholar]
- Young VL, Simpson RM, Ward VK. Characterization of an exochitinase from Epiphyas postvittana nucleopolyhedrovirus (family Baculoviridae) J Gen Virol. 2005;86:3253–3261. doi: 10.1099/vir.0.81262-0. [DOI] [PubMed] [Google Scholar]
- Yu M, Li Z, Yang K, Lin T, Gong Y, Pan L, Pang Y. Identification of the apoptosis inhibitor gene p49 of Spodoptera litura multicapsid nucleopolyhedrovirus. Virus Genes. 2005;31:145–151. doi: 10.1007/s11262-005-1786-x. [DOI] [PubMed] [Google Scholar]
- Zhang BQ, Cheng RL, Wang XF, Zhang CX. The Genome of Pieris rapae Granulovirus. J Virol. 2012;86:01431–01412. doi: 10.1128/JVI.01431-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoog SJ, Schiller JJ, Wetter JA, Chejanovsky N, Friesen PD. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. Embo J. 2002;21:5130–5140. doi: 10.1093/emboj/cdf520. [DOI] [PMC free article] [PubMed] [Google Scholar]