Background: Nanobodies represent a specific means to modulate enzyme function.
Results: HypE-specific nanobodies modulate AMPylation activity and identify HypE as localized to the nuclear envelope.
Conclusion: HypE function can be interfered with and probed for with new tools.
Significance: HypE nanobodies are the first known HypE activators and inhibitors.
Keywords: Histone Modification, Nuclear Membrane, Phage Display, Post-translational Modification (PTM), Recombinant Protein Expression, AMPylation, Fic Proteins, HypE, VHHs
Abstract
The covalent addition of mono-AMP to target proteins (AMPylation) by Fic domain-containing proteins is a poorly understood, yet highly conserved post-translational modification. Here, we describe the generation, evaluation, and application of four HypE-specific nanobodies: three that inhibit HypE-mediated target AMPylation in vitro and one that acts as an activator. All heavy chain-only antibody variable domains bind HypE when expressed as GFP fusions in intact cells. We observed localization of HypE at the nuclear envelope and further identified histones H2–H4, but not H1, as novel in vitro targets of the human Fic protein. Its role in histone modification provides a possible link between AMPylation and regulation of gene expression.
Introduction
Post-translational protein modifications represent a fundamental regulatory principle of controlling protein activities. Accordingly, modifications by proteases, kinases, methylases, or acetylases have been explored extensively, and their misregulation is often associated with severe pathology, including autoimmune diseases or cancer (1, 2). In contrast, AMPylases, which transfer an AMP moiety to a protein, are poorly understood. AMPylation, also referred to as adenylylation, was first described in the 1960s as a regulatory mechanism for controlling glutamine synthetase activity in Escherichia coli (1, 3). The transfer of AMP to a tyrosine residue in glutamine synthetase resulted in reversible enzyme inhibition. However, AMPylation is now known to have a role in the pathogenicity of several bacteria by translocating effector AMPylases into their host cells, where these effectors target host proteins (4–7). The bacterial effectors VopS of Vibrio parahaemolyticus and IbpA of Histophilus somni AMPylate host cell GTPases Rac1, Cdc42, and RhoA on Thr-35 and Tyr-32, respectively. The addition of a bulky AMP moiety to Thr-35 or Tyr-32 impairs binding of downstream effectors, resulting in inhibition of GTPase activity (5, 8).
Sequence comparisons and structural analysis of secreted bacterial AMPylators reveal the presence of a Fic (filamentation-induced by cAMP) domain that is essential for AMP transfer (9). All Fic domain-containing proteins (Fic proteins) share a conserved sequence motif (HXFX(D/E)GN(G/K)R). Mutation of the invariant histidine residue in VopS or IbpA abolishes their AMPylation activity (10). To regulate AMPylation, a conserved flexible α-helix (αinh) containing the sequence motif (S/T)XXXE(G/N) is positioned to obstruct the ATP-binding site. Although, in most Fic proteins, αinh is fused to the Fic domain as either an N-terminal (class II) or A C-terminal (class III) helix, αinh can also exist as an independent inhibitory antitoxin (class I), as shown for the VbhT-VbhA toxin-antitoxin system (10). Substitution of the glutamic acid present in αinh with glycine relieves intramolecular autoinhibition, resulting in Fic enzymes with significantly enhanced auto-AMPylation as well as enhanced target AMPylation.
Fic protein activity is not limited to the transfer of AMP onto small GTPases: the Legionella pneumophila Fic protein AnkX is also translocated into host cells, where it anchors a phosphorylcholine unit onto Ser-79 of Rab1 GTPases, thereby modulating host vesicle trafficking (11, 12). AvrAC from the plant pathogen Xanthomonas campestris covalently links UMP to RIPK and BIK1 kinases, thereby interfering with host immune defense (13). Finally, the bacterial Fic protein Doc, the toxin unit of the Doc-PhD toxin-antitoxin system present in E. coli, phosphorylates Thr-382 of the translation elongation factor EF-Tu, inhibiting translation (14, 15). Thus, Fic protein activity is not linked exclusively to the transfer of AMP onto target proteins. Rather, all Fic proteins share a common substrate preference (nucleoside triphosphates) for their enzymatic activities.
To date, some ∼2700 Fic proteins have been identified, mostly in bacteria. Eukaryotes, including Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens, encode at least one Fic protein each, with the exception of most fungi, which contain no known AMPylases (16, 17). The Drosophila Fic protein, referred to as CG9523 or dFic, possesses auto-AMPylation activity in vitro; this activity is diminished upon mutation of the conserved histidine residue within the Fic motif (18). Mutant flies that lack dFic are blind, but otherwise viable. Photoreceptor cells in dFic-deficient animals depolarize normally following light stimulation, but fail to activate post-synaptic neurons, indicative of a defect in neurotransmission. Expression of enzymatically active dFic on capitate projections of glia cells, but not neurons, rescues the null phenotype. The catalytic site of dFic resides in the endoplasmic reticulum (ER)4 lumen, where it AMPylates Thr-366 of BiP/GRP78, a conserved HSP70 family member important for quality control and regulation of the unfolded protein response in the ER (19). ER stress increases intracellular dFic and BiP levels. Conversely, AMPylation of BiP is reduced upon induction of the unfolded protein response, suggesting an inhibitory role for AMPylation in BiP activity.
The human genome encodes a single Fic protein named HypE (Huntingtin-associated yeast-interacting protein E; also referred to as FICD); it exhibits AMPylation activity against Cdc42, Rac1, and BiP in vitro (8). RNA expression data suggest that HypE is present in most cell types and tissues, albeit at low levels. Overexpression of wild-type HypE in cultured cells has few obvious consequences, suggesting a tight regulation of HypE activity by its inhibitory α-helix or by other, yet unidentified regulatory proteins (10). The physiological role of HypE remains elusive; however, HypE is assumed to interact with Huntingtin, a protein that, when mutated, causes Huntington disease (20), a neurodegenerative disorder.
The study of protein AMPylation in eukaryotes is limited in part by a dearth of appropriate tools to selectively track Fic proteins or modulate their activity. A threonine/AMP-specific mouse serum was used to confirm VopS-mediated AMPylation of Cdc42 and Rac1 and to study HypE-mediated BiP AMPylation (21). In addition, a set of small molecules inhibiting VopS in in vitro assays has been characterized (22). However, such tools are few in number. It thus remains a challenge to study the regulation and consequences of AMPylation in intact cells.
Camelid VHHs, derived from the variable domain of heavy chain-only antibodies, present a unique tool (23, 24). VHHs, also referred to as nanobodies, are smaller and more stable than their traditional antibody counterparts and are less dependent on disulfide bond formation for proper folding. They can therefore be expressed in the cytoplasm of bacteria (e.g. E. coli) or eukaryotic cells without loss of their antigen recognition properties (25). VHHs are easily modified and can be equipped with fluorescent or affinity tags to track their target antigens within the cell (26, 27). VHHs can serve as specific and efficient protein inhibitors or modulators and can induce changes in protein conformation within cells. Furthermore, VHHs can stabilize transient protein conformations, facilitating protein crystallization (28).
Here, we report the generation of HypE-specific VHHs that either inhibit or stimulate AMPylation activity in vitro. Three of the VHHs inhibit histone H3 AMPylation, whereas one VHH increases target AMPylation levels. Employing a HypE-specific mouse serum in combination with our VHHs, we show that HypE localizes to the nuclear envelope. Finally, we identify histones H2–H4, but not H1, as new targets for HypE-promoted protein AMPylation, suggesting a possible role for HypE in regulation of gene expression or DNA repair.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa, HEK293T, and A549 cells were cultured in DMEM supplemented with 10% FBS. Transduced HeLa and A549 cell lines were cultured in DMEM supplemented with 10% FBS and G418 (Difco) at a concentration of 100 μg/ml.
Sortase-based VHH Modification
Heptamutant Ca2+-independent sortase A from Staphylococcus aureus (SrtAstaph7M) was purified as described (29). Sortase reaction nucleophiles GGG-TAMRA, GGG-biotin, and GGG-Alexa Fluor 647 were synthesized as described (30, 31). In-solution sortase labeling reactions with SrtAstaph7M were performed overnight at 4 °C in 50 mm Tris-HCl (pH 7.5) and 150 mm NaCl; upon reaction completion, His6-tagged sortase was removed from the reaction mixture using nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen). Residual GGG nucleophiles were separated from dye-coupled VHHs by size exclusion chromatography on PD10 columns (GE Healthcare).
Immunoblotting
Cells were seeded in 6-well plates or 10-cm2 tissue culture dishes and transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. After incubation for 24–48 h, cells were harvested and lysed in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, and 0.1% SDS supplemented with protease inhibitor mixture (Roche Applied Science). Protein samples were subjected to SDS-PAGE, transferred to a PVDF membrane, and probed with appropriate antibodies or the respective VHH (1 μg/ml). Table 1 lists all antibodies used in this study. Chemiluminescent signals were detected using a Western Lightning ECL detection kit (PerkinElmer Life Sciences) and exposure to XAR-5 films (Kodak).
TABLE 1.
Antibodies used in this study
IF, immunofluorescence; WB, Western blotting.
| Provider | Catalog no. | Specificity | Dilution used |
|---|---|---|---|
| Primary antibodies | |||
| Sigma-Aldrich | AV46826-50UG | Human FICD | 1:3000 |
| Abcam | ab18579 | GST | 1:6000 |
| Santa Cruz Biotechnology | sc-805 | HA (clone Y-11) | 1:500 (IF), 1:5000 (WB) |
| Abcam | ab16048 | Lamin B1 | 1:100 |
| Secondary antibodies | |||
| Qiagen | 34460 | Penta-His | 1:10,000 |
| Fisher | OB4050-05 | Anti-rabbit IgG (H + L) | 1:25,000 |
| SouthernBiotech | 1038-05 | Anti-mouse IgG (H + L) | 1:25,000 |
Immunofluorescence Staining
HeLa or A549 cells grown on coverslips were either transfected with respective plasmids using Lipofectamine 2000 or grown to 70–90% confluence. Cells were fixed in 4% formaldehyde and PBS for 20 min, permeabilized for 15 min in PBS and 0.3% Triton X-100, and blocked with 1% BSA and PBS. Cells were incubated for 1 h at room temperature in 1% BSA and PBS containing primary antibodies, washed three times with PBS, and incubated for another hour at room temperature in 1% BSA and PBS supplemented with Alexa Fluor 488- or Alexa Fluor 647-coupled secondary antibodies. Alternatively, cells were stained with Alexa Fluor 647-coupled VHHs (10 μg/ml) for 1 h at room temperature. All images were collected on a PerkinElmer Ultraview multispectral spinning disk confocal microscope equipped with a Zeiss 1.4 numerical aperture oil immersion ×63 objective lens and a Prior piezoelectric objective focusing device for maintaining focus. Images were acquired with a Hamamatsu ORCA ER cooled CCD camera controlled with Volocity software. Post-acquisition image manipulations were made using Fiji software. For analysis of nuclear morphology, at least 10 randomly chosen frames from three independent samples were imaged, and nuclear morphology was assessed by eye.
Coupling of VHHs to Cyanogen Bromide-activated Beads
Beads were rinsed with 1 mm HCl for 15 min, mixed with VHHs diluted in coupling buffer (0.1 m NaHCO3 (pH 8.3) and 0.5 m NaCl), and incubated overnight at 4 °C with agitation. Beads were then washed five times with coupling buffer, blocked for 2 h at room temperature with 0.1 m Tris-HCl (pH 8.0), and subjected to at least five wash cycles with two alternating wash buffers (0.1 m acetic acid/sodium acetate (pH 4.0) and 0.5 m NaCl; and 0.1 m Tris-HCl (pH 8.0) and 0.5 m NaCl).
Competition ELISA
96-Well ELISA plates were coated with HypE or HypE E234G (0.5 μg/ml) overnight at 4 °C. Following washing and blocking with PBS, 0.1% Tween 20, and 4% milk for 2 h at 37 °C, test wells were supplemented with His-tagged unmodified VHHs (25 μg/ml) and incubated at room temperature for 1 h. Wells were then washed and supplemented with biotinylated VHHs (25 μg/ml), incubated at room temperature for 1 h, and analyzed using streptavidin-HRP to trigger a colorimetric readout.
Generation of Stable Cell Lines
Cell lines were engineered to allow doxycycline-inducible expression of introduced transgenes using the pINDUCER toolkit (32). Following lentiviral transduction of target cells (HeLa, A549) and subsequent selection for chromosomal integration with G418, polyclonal cell lines were assayed for GFP expression upon doxycycline addition.
Generation of HypE-specific Mouse Serum
BL52/B6 mice were subcutaneously primed with 100 μg of HypE in 100 μl of PBS supplemented with complete Freund's adjuvant, boosted by intraperitoneal injection 4 weeks later with 20 μg in PBS, and terminally bled 5 days later.
Generation of VHHs
VHH phage display library preparation and phage library panning were performed as described (33). Enriched VHH sequences were cloned into a pHEN expression vector downstream of an N-terminal pelB sequence and equipped with a C-terminal LPTEGG sortase motif.
VHH-HypE Interaction Tests
∼50–100 μg of recombinant HypE(187–437) or HypE(187–437) E234G was incubated with 50 μg of VHH-TAMRA at 4 °C for 2 h and analyzed on a Superdex S75 10/300 GL size exclusion column. Absorbance at 280 nm (proteins) and 545 nm (TAMRA) was recorded.
Protein Purification
HypE(187–437), HypE(187–437) E234G, and HypE(187–437) H363A
E. coli BL21(DE3) cells containing overexpression plasmids (pDuet) were grown in Terrific broth to A600 = 0.6, induced with 0.2 mm isopropyl β-d-thiogalactopyranoside, and incubated at 23 °C for 48 h with shaking at 200 rpm. Cells were pelleted, resuspended in lysis buffer A (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, protease inhibitor mixture, and DNase I (Roche Applied Science)), and lysed with a French press. Lysate was cleared by centrifugation, and the soluble fraction was run over pre-equilibrated Ni-NTA beads. Beads were washed with 150 ml of wash buffer A (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, and 10 mm imidazole) and eluted with 3 ml of elution buffer A (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, and 300 mm imidazole). Ni-NTA elution fractions were further purified using a Superdex S75 16/600 column and stored in 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 10% glycerol at 4 or −80 °C. GST-VopS was purified as described (5).
GST-HypEΔ83
E. coli BL21(DE3) cells containing overexpression plasmids were grown in LB medium to A600 = 0.6, induced with 0.2 mm isopropyl β-d-thiogalactopyranoside, incubated overnight at 30 °C, and purified as described (4).
His6-Histone H3-LPTEGG
E. coli BL21(DE3) cells containing overexpression plasmids were grown in LB medium to A600 = 0.6, induced with 1.0 mm isopropyl β-d-thiogalactopyranoside, and incubated at 37 °C for 3 h. Cells were pelleted, resuspended in lysis buffer B (50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 5% glycerol, protease inhibitor mixture, and DNase I), and lysed by sonication. Following centrifugation, the pellet was resuspended in 50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 5% glycerol, and 6 m urea. The resuspended pellet was centrifuged again, and the supernatant was incubated with pre-equilibrated Ni-NTA beads for 2 h at 4 °C. Beads were washed with 150 ml of wash buffer B (50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 5% glycerol, 6 m urea, and 10 mm imidazole) and eluted with 5 ml of elution buffer B (50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 5% glycerol, 6 m urea, and 250 mm imidazole). Eluted histones were dialyzed overnight against 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 10% glycerol and stored at 4 or −80 °C.
VHHs
1 liter of LB medium was inoculated overnight with a 10-ml culture of E. coli WK6 containing VHH-encoding pHEN6 plasmids. Protein expression was induced at A600 = 0.6 with 1.0 mm isopropyl β-d-thiogalactopyranoside, and bacteria were grown overnight at 30 °C. The following day, bacteria were pelleted, resuspended in 20 ml of 1× TES (0.2 m Tris-HCl (pH 8.0), 0.65 mm EDTA, and 0.5 m sucrose), and incubated at 4 °C for 1 h. Bacteria were then repelleted, resuspended in 60 ml of 0.25× TES, and incubated overnight at 4 °C. Following centrifugation for 30 min at 10,000 × g, the supernatant was incubated with pre-equilibrated Ni-NTA at 4 °C for 1 h. Beads were transferred into a gravity flow column, washed with 150 ml of wash buffer A, and eluted with 3 ml of elution buffer A. Elution fractions were further purified on a Superdex S75 16/600 column and stored at −80 °C.
In Vitro AMPylation Assays
5–10 μg of HypE(187–437), HypE(187–437) E234G, or HypE(187–437) H363A in 10 μl were first mixed with 25 μl of 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 7.5 mm MgCl2, and 0.1 mm DTT supplemented with 0.5 μCi of [α-33P]ATP. The reaction was allowed to proceed for 1 h at room temperature, followed by the addition of 1–5 μg of target protein (histones) in a 10-μl volume. The reaction mixture was incubated for another hour at room temperature, supplemented with 10 μl of reducing SDS sample buffer, boiled for 5 min, and analyzed on 10–15% SDS-polyacrylamide gels. For VHH inhibition assays, HypE(187–437) proteins were incubated with the respective VHHs for 1 h at room temperature prior to the addition of [α-33P]ATP-containing reaction buffer. Human histones H1, H2A, H2B, H3.1, H3.2, H3.3, and H4 were purchased from Sigma. Autoradiograph signal intensities were analyzed using the gel analysis macro included in the Fiji software package (34).
Cell Viability Assays and FACS Analysis
Cells were grown in 12-well plates, transfected with the indicated plasmids, and incubated for 36 h. Cells were then trypsinized and treated with LIVE/DEAD fixable violet dead cell stain (Life Technologies) according to the manufacturer's instructions. Data acquisition was performed with a BD LSR II system (BD Biosciences) using CellQuest Pro software (BD Biosciences). Data were analyzed with FlowJo (Tree Star Inc.).
RESULTS
Characterization of HypE-specific VHHs
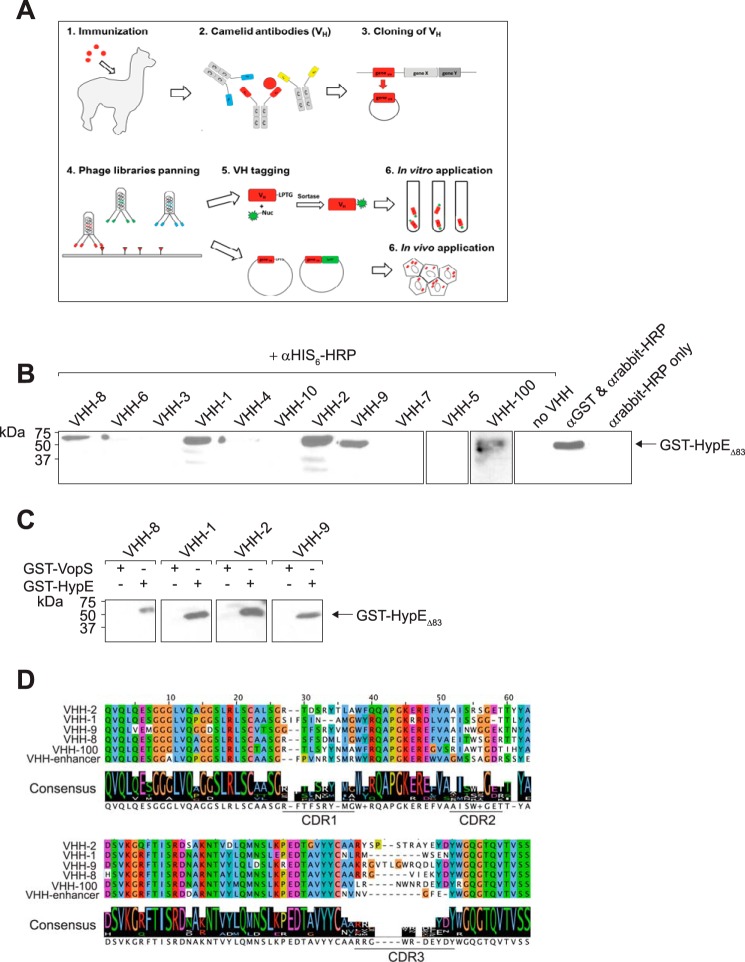
Camelid-derived VHHs are potent and specific tools for studying protein function (35). To generate HypE-specific VHHs, we immunized an alpaca with HypE(187–437) E234G, harvested its peripheral blood lymphocytes, and used VHHs amplified from this sample to generate a VHH phagemid library. Fig. 1A provides a schematic overview of the process. Phage display was then performed using immobilized HypE(187–437) or HypE(187–437) E234G as the bait protein. Sequencing of 192 individual VHH clones isolated after two subsequent rounds of panning revealed 17 clones present at least twice in the collection, with the most abundant clone recovered from six independent colonies. We subcloned 10 unique VHH sequences into a pHEN6 periplasmic expression vector and installed a C-terminal LPETGG-His6 tag to facilitate protein purification and render the VHH a substrate for a sortase-based transpeptidation reaction. All candidates were expressed with yields varying from 2 to 40 mg/liter of E. coli culture. To assay the specificity of the VHHs for HypE, we first tested the VHHs in immunoblots using the VHH as a primary antibody at a concentration of 1 μg/ml (Fig. 1B). Four of 10 VHHs detected GST-HypEΔ83 and showed no reactivity toward GST-VopS, indicating their specificity (Fig. 1C). Sequence alignment of the VHHs showed that, as expected, most sequence heterogeneity is found in the complementarity-determining regions (CDRs), particularly in CDR3 (Fig. 1D). Although VHH-8 and VHH-1 represent the most similar VHH pair, VHH-9 is the most distant representative based on sequence homology analysis (26).
FIGURE 1.
Initial characterization of HypE-specific VHHs by immunoblotting and overview of the generation of VHHs. A, schematic representation of VHH generation. B, an initial test of 10 putative HypE-specific VHHs in immunoblot assays. His6-VHHs were probed against GST-HypEΔ83 and detected using an anti-His6-HRP antibody. C, evaluation of cross-reactivity of VHHs binding to GST-HypEΔ83. His6-tagged VHHs were probed against GST-HypEΔ83 and GST-VopS and detected using an anti-His6-HRP antibody. D, sequence alignment of characterized anti-HypE VHHs, including an anti-GFP VHH (enhancer) as reference. CDR1–CDR3 are underlined.
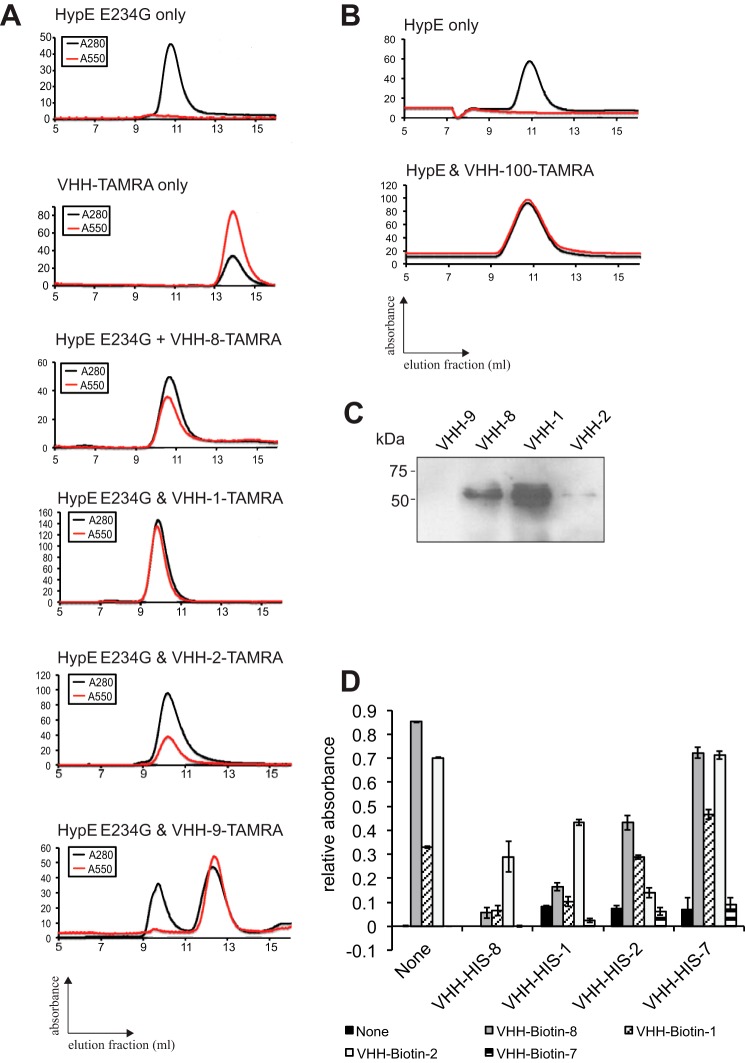
In the next step, we covalently linked a TAMRA moiety to the C terminus of our HypE-specific VHHs by sortagging, resulting in site-specifically, fluorescently labeled VHHs. These modified VHHs were then incubated with recombinant HypE(187–437) E234G at 1:1 molar ratio, and complex formation was analyzed by size exclusion chromatography (Fig. 2, A and B). We observed stable complex formation of HypE(187–437) E234G with VHH-8, VHH-1, and VHH-2 and of HypE(187–437) with VHH-100, as inferred from 1) a slight left shift in the HypE(187–437) E234G-corresponding elution peak (∼10.5 ml), 2) the disappearance of the VHH-TAMRA elution peak (∼14 ml), and 3) the shift in the TAMRA signal from the VHH elution peak to the HypE elution peak. In contrast, VHH-9 failed to form a complex with native HypE(187–437) E234G. To corroborate these results, we coupled four VHHs to CNBr-activated beads, immunoprecipitated HypE from HeLa cell lysates, and probed the precipitated fractions with a commercial anti-HypE polyclonal antibody. As indicated in Fig. 2C, beads coupled with VHH-1, VHH-2, and VHH-8, but not VHH-9, recovered HypE from cell lysates, albeit with different efficiencies. Furthermore, competition ELISAs showed that VHH-8 and VHH-1 competed for binding on HypE(187–437) E234G, suggesting that they recognize overlapping epitopes (Fig. 2D).
FIGURE 2.
Validation and characterization of HypE-specific VHHs. Recombinant HypE(187–437) E234G (A) and HypE(187–437) (B) were incubated with TAMRA-labeled VHHs. Complex formation was assessed by size exclusion chromatography. Black lines (A280) represent total protein, and red lines (A545) represent the TAMRA signal. C, endogenous HypE was immunoprecipitated from HeLa cell lysates using VHH-coupled beads and analyzed by Western blotting with a commercial anti-HypE antibody. D, competition ELISAs were performed using His6-VHHs for initial epitope masking and biotin-VHH for subsequent probing.
VHHs Inhibit or Stimulate HypE-mediated Target AMPylation in Vitro
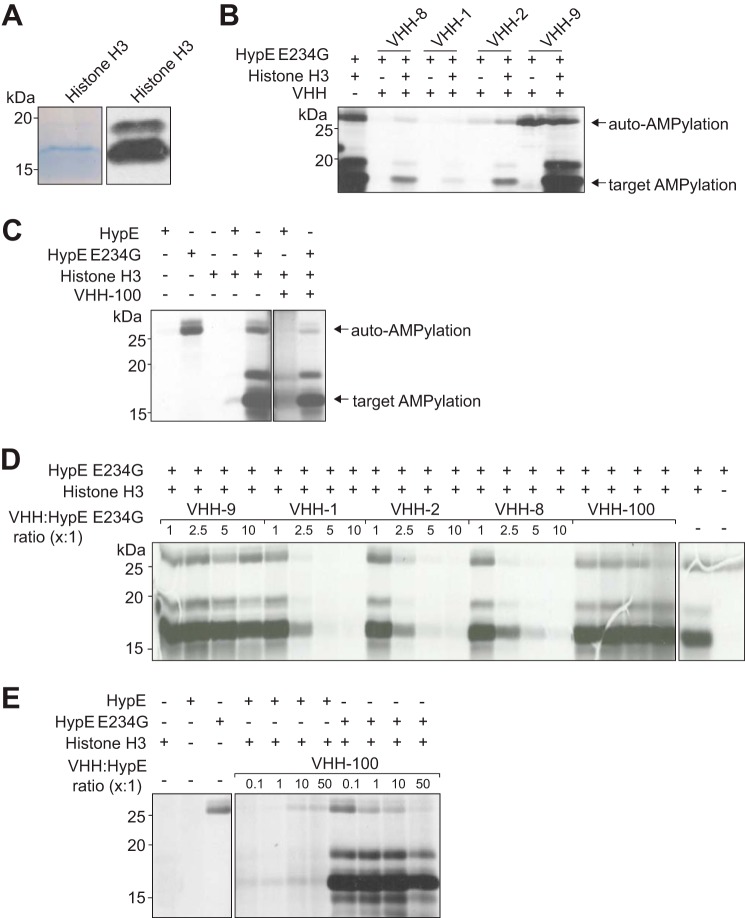
The Fic domain of HypE has been associated with AMPylation activity both in vitro and in vivo (19). To test if our VHHs interfere with auto-AMPylation and target AMPylation, we incubated recombinant HypE(187–437) E234G with excess VHH for 1 h before supplementing the reaction with [α-33P]ATP and recombinant human histone H3 as a substrate. The purity of the recombinant histone H3 preparation used in these assays is depicted in Fig. 3A, which shows the presence of some remaining contaminating polypeptides. VHH-8, VHH-1, and VHH-2 interfered with auto-AMPylation and target AMPylation to different extents (Fig. 3B). In contrast, VHH-9 and VHH-100 had no detectable effect on HypE(187–437) E234G target AMPylation, although VHH-100 decreased the auto-AMPylation activity of HypE(187–437) E234G (Fig. 3, B and C). Titration experiments showed a dose dependence of HypE(187–437) E234G inhibition, with VHH-1 emerging as the most potent, with almost complete inhibition at a 5:1 molar ratio of VHH to HypE(187–437) E234G (Fig. 3D). Notably, complex formation between HypE(187–437) and VHH-100 enhanced auto-AMPylation and target-AMPylation by HypE(187–437) (Fig. 3E), whereas VHH-1, VHH-2, and VHH-8 did not (data not shown). The VHHs described here represent the first examples of HypE-specific AMPylation inhibitors or enhancers.
FIGURE 3.
VHHs modulate HypE E234G-mediated target AMPylation. A, recombinant histone H3 was separated by SDS-PAGE and either visualized by Coomassie Brilliant Blue staining (left panel) or transferred to a PVDF membrane and probed with an anti-His6-HRP antibody (right panel). Recombinant HypE(187–437) E234G (B) or HypE(187–437) (C) was incubated with an excess of the respective VHHs for 1 h at room temperature, at which point [α-33P]ATP and histone H3 substrate were added, and the mixture was incubated for an additional hour. Incorporation of label was assessed by SDS-PAGE and autoradiography. D, dose dependence of VHH-mediated HypE inhibition. Recombinant HypE(187–437) E234G was incubated with the respective VHHs at the indicated ratios for 1 h at room temperature, combined with [α-33P]ATP and histone H3 substrate, and incubated for another hour. Sample autoradiography was assessed. E, dose dependence of VHH-mediated HypE activation. Recombinant HypE(187–437) or HypE(187–437) E234G was incubated with the respective VHHs at the indicated ratios for 1 h at room temperature, combined with [α-33P]ATP and histone H3 substrate, and incubated for another hour. Sample autoradiography was assessed.
Endogenous HypE Is Localized to the Nuclear Envelope
To complement our VHH-based approach with another HypE-specific tool, we generated a HypE-specific mouse serum, as commercially available sera had proved inadequate. This serum efficiently detected HA-tagged HypE constructs overexpressed in HeLa cells (Fig. 4A). We co-stained HeLa cells overexpressing HA-HypE with anti-HypE serum and an anti-HA antibody. We observed co-localization of both labels, demonstrating the serum's specificity for HypE and its utility in immunofluorescence (Fig. 4B). In cells that overexpress HypE, the protein was present in the nuclear envelope and associated structures. The latter likely represent the endoplasmic reticulum. To confirm the intracellular localization of HypE, we stained untransfected HeLa cells with anti-HypE serum and co-labeled the cells with an anti-lamin AC antibody. The majority of endogenous HypE resides in the nuclear lamina, as indicated by the extensive co-localization of HypE and the lamina-specific signal (Fig. 4C).
FIGURE 4.
Characterization of HypE-specific mouse serum. A, cell lysates of pHypE-, pHypE E234G-, or pHypE H363A-transfected cells were probed with HypE-specific serum. B, co-staining of fixed HeLa or A549 cells with HypE-specific serum, anti-lamin antibody, and DAPI (DNA). C, co-staining of pHypE-, pHypE E234G-, or pHypE H363A-transfected HeLa cells with HypE-specific serum, anti-HA antibody, and DAPI (DNA).
VHHs Recognize HypE in Live and Fixed Cells
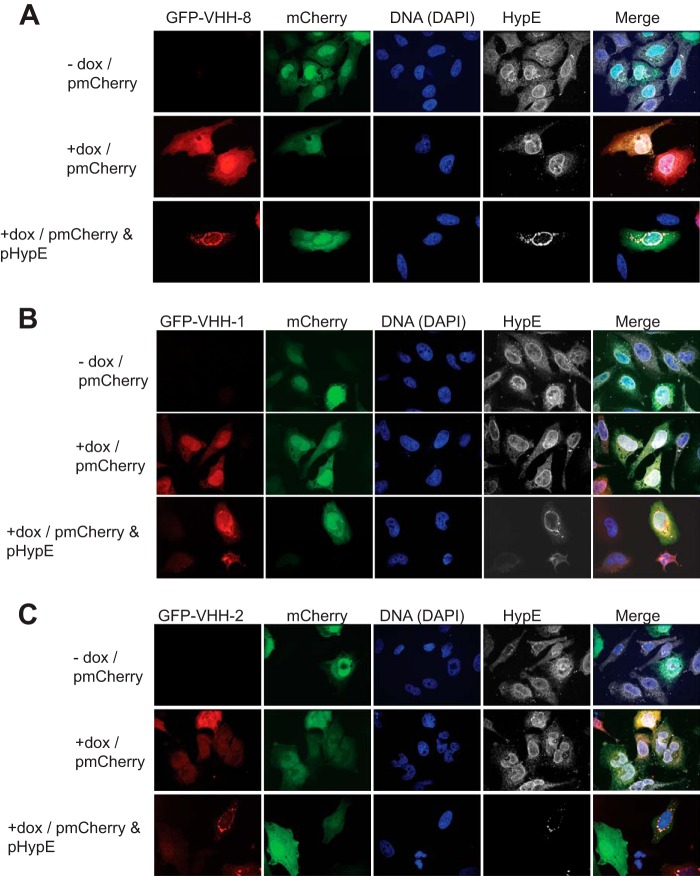
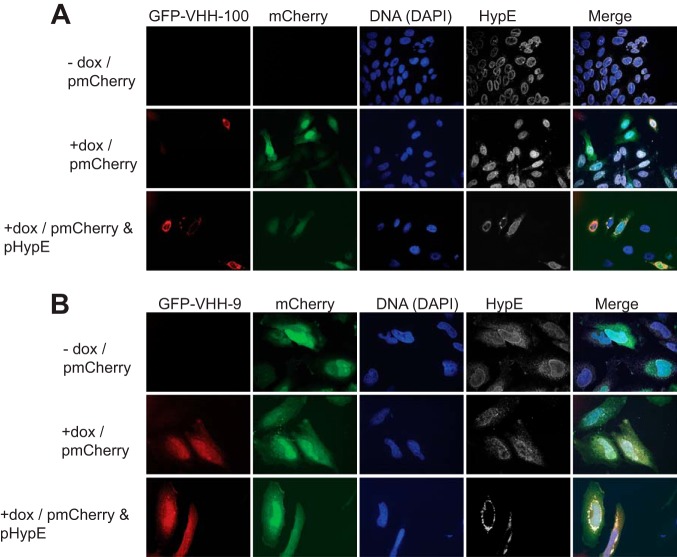
To determine whether our VHHs interact with HypE in an intact cellular environment, we generated HeLa cell lines that contain an inducible GFP-VHH expression cassette for the anti-HypE VHHs. Upon induction with doxycycline, GFP-VHHs were distributed throughout the cell, including the nucleus (Fig. 5, A–C, and Fig. 6, A and B). Ectopic expression of HA-tagged HypE constructs in cells that express GFP-VHH-8, GFP-VHH-1, GFP-VHH-2, and GFP-VHH-100 resulted in robust co-localization of the VHHs with HA-HypE, whereas the GFP signal remained diffuse in cells expressing GFP-VHH-9. To further test our VHHs, we engineered VHH-Alexa Fluor 647 fusions by sortagging (29) and used these to stain fixed HypE-overexpressing cells (Fig. 7, A and B). As with the GFP-tagged VHHs, we observed robust co-localization of our VHHs with HA-HypE, establishing that directly labeled VHHs are useful tools for microscopy.
FIGURE 5.
GFP-tagged VHHs bind to HypE in intact cells. pmCherry- or pmCherry/pHypE-transfected HeLa cells inducibly expressing GFP-tagged VHH-8 (A), VHH-1 (B), or VHH-2 (C) were stained with HypE-specific mouse serum. dox, doxycycline.
FIGURE 6.
GFP-tagged VHHs bind to HypE in intact cells. pmCherry- or pmCherry/pHypE-transfected HeLa cells inducibly expressing GFP-tagged VHH-100 (A) or VHH-9 (B) were stained with HypE-specific mouse serum. dox, doxycycline.
FIGURE 7.
Alexa Fluor 647-linked VHHs recognize HypE in fixed cells. pmCherry-transfected (A) or pmCherry/pHypE-transfected (B) cells were stained with HypE-specific mouse serum and Alexa Fluor 647-coupled VHHs.
HypE AMPylates Histones H2–H4, but Not H1
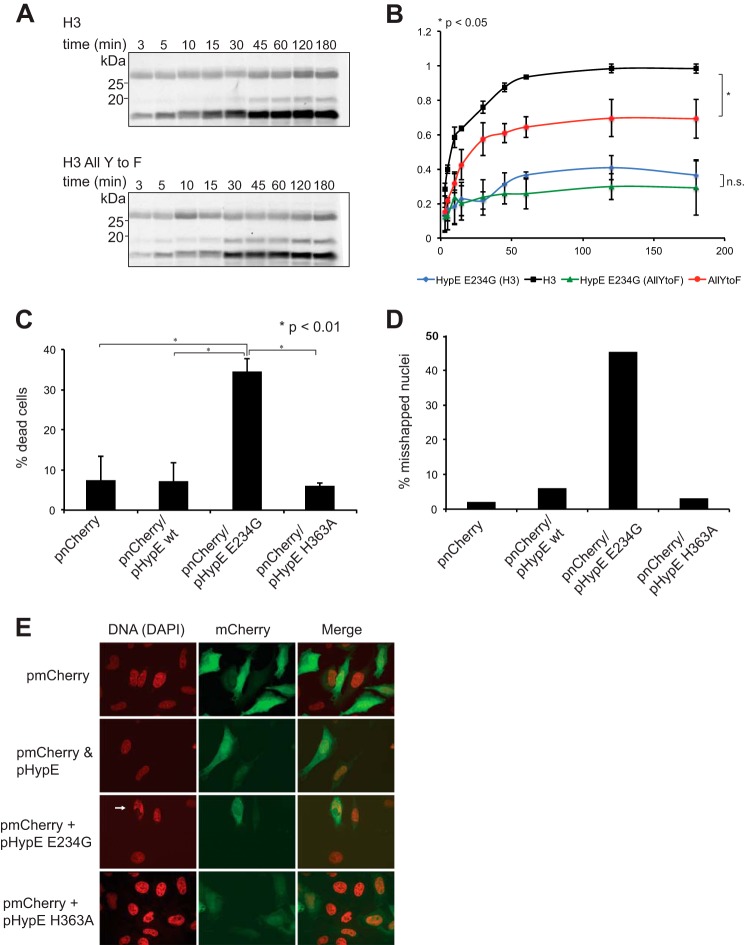
To explore the role of HypE in intact cells, we set out to identify novel HypE target proteins. As endogenous HypE localized to the nuclear envelope, we focused on nucleus-associated proteins, including histones. Histone H3 has been reported to be a substrate for HypE (22). We systematically tested representatives of all major histone classes (H1, H2A, H2B, H3.1, H3.2, H3.3, and H4) in in vitro AMPylation assays and observed that HypE(187–437) and HypE(187–437) E234G AMPylated histones H2A, H2B, H3.1, H3.2, H3.3, and H4, but not H1 (Fig. 8, A and B). Comparing the signal intensities observed for HypE(187–437)- and HypE(187–437) E234G-mediated histone H3 AMPylation, we calculated that HypE E234G showed an ∼30-fold increase in AMPylation activity compared with the wild-type enzyme. Unbiased motif identification using MEME with histones H2–H4 as positive input and histone H1 as negative input did not suggest the obvious presence of a conserved motif for AMPylation (36). However, as HypE has been associated with tyrosine AMPylation of small GTPases, we constructed and purified a set of mutant histone H3 constructs in which we exchanged all tyrosines for phenylalanines or retained a single tyrosine. In vitro AMPylation tests using these histone H3 mutants demonstrated that even the tyrosine-free version was still AMPylated by HypE E234G (Fig. 8C). However, we observed significantly decreased AMPylation levels for tyrosine-free histone H3, whereas the reduction in AMPylation observed for single tyrosine-containing histones was less pronounced (Fig. 8D). The fact that even the tyrosine-free histone version was AMPylated indicates at least one non-tyrosine site of AMPylation, likely Ser, Thr, or His. Time course experiments in which we compared the AMPylation of wild-type versus tyrosine-free histone H3 showed similar kinetics, although wild-type histone H3 was AMPylated at a significantly higher level (Fig. 9, A and B) compared with the tyrosine-free variant.
FIGURE 8.
HypE AMPylates histones H2A–H4, but not H1. Recombinant HypE(187–437) (A) and HypE(187–437) E234G (B) were incubated with [α-33P]ATP for 1 h and mixed with the respective histone substrates for another hour. Incorporation of label was assessed by SDS-PAGE and autoradiography. C, recombinant HypE(187–437), HypE(187–437) E234G, and HypE(187–437) H363A were incubated with [α-33P]ATP for 1 h and mixed with the respective histone mutant substrates for another hour. Incorporation of label was assessed by SDS-PAGE and autoradiography. D, quantification of histone mutant AMPylation by HypE(187–437) E234G. The average of three independent experiments is shown. *, p < 0.05. AllYtoF, all tyrosines substituted with phenylalanines; Y1only, Y2only, and Y3only, only a single tyrosine retained; n.s., not significant.
FIGURE 9.
HypE E234G AMPylates tyrosine-free histone H3 and induces cell death. Shown are the kinetics of HypE(187–437) E234G-mediated histone H3 and tyrosine-free histone H3 mutant AMPylation. A representative autoradiogram is depicted in A, and the quantification of histone AMPylation by HypE(187–437) E234G is shown in B (average of three independent experiments). *, p < 0.05. All Y to F, all tyrosines substituted with phenylalanines; n.s., not significant. C, A549 cells were transfected with the indicated plasmids, incubated for 48 h, stained with a vital dye, and analyzed by FACS. The average of three independent experiments is shown. D, distribution of misshaped nuclei in transfected HeLa cells. Collective data obtained from analysis of at least 10 frames from three independent samples are shown. E, staining of HeLa cells transfected with indicated plasmids for 48 h. The arrow indicates a typical abnormal nucleus in a HypE E234G-expressing cell.
Intracellular Hyper-AMPylation by HypE E234G Intoxicates Cells
In the course of our experiments, we noticed that cells ectopically expressing the hyperactive mutant HypE E234G showed obvious signs of cellular stress and increased cell death rates. To quantify these findings, we co-transfected HeLa cells with mCherry and pHypE constructs and assayed for cell death. Transfected cells were selected for analysis based on the presence of the mCherry signal. Overexpression of HypE E234G was toxic and significantly increased cell death (Fig. 9C). Furthermore, populations of cells that expressed HypE E234G displayed an increased number of cells with misshapen nuclei (Fig. 9D). The abnormal nuclei were of approximately the same size as regular nuclei, but were severely misshapen and often showed signs of a ruptured envelope or nuclear bleb-like structures (Fig. 9E), in contrast to apoptotic nuclei, which are characterized by DNA condensation and an increase in DNA staining intensity. Cells overexpressing either HypE or the active-site mutant HypE H363A showed neither increased death rates nor any signs of aberrant nuclear morphology, indicating that the observed changes are linked to increased AMPylation by HypE E234G. The HypE substrate(s) responsible for imposition of altered nuclear morphology remain to be identified.
DISCUSSION
Target AMPylation by Fic domain-containing enzymes represents a conserved protein modification mechanism. Although translocated bacterial Fic proteins perturb host cell signaling during infections, the D. melanogaster Fic protein, dFic, resides within the ER lumen, where it AMPylates BiP, an essential ER chaperone involved in regulation of the unfolded protein response (5, 19). However, despite the fact that the human Fic protein HypE was shown to AMPylate Cdc42, Rac1, and BiP in in vitro assays, the localization of HypE, its physiologically most relevant targets, and its role in regulating cell signaling mechanisms remain elusive.
This work describes camelid-derived VHHs specific for HypE that allow an examination of HypE localization and function. We established the specificity of the VHHs for HypE and demonstrated their versatility in the several applications reported here. The anti-HypE VHHs allowed us to specifically manipulate HypE function in vitro. We used a sortase-based approach to generate Alexa Fluor 647-, TAMRA-, and biotin-modified anti-HypE VHHs, demonstrating that their C-terminal modification does not impair function. The requirements for proper disulfide bond formation and glycosylation confound intracellular expression of full-sized antibodies and most antibody fragments in the reducing environment of the cytoplasm (37). VHHs can function without these modifications (25). The HypE-specific VHHs are efficiently expressed in HeLa cells fused at their C terminus to GFP. Co-localization of GFP-tagged VHHs in HeLa cells with ectopically expressed HypE constructs indicates that the GFP tag does not interfere with the ability of a VHH to bind to its designated target protein. Although our anti-HypE polyclonal serum readily detected endogenous HypE levels, our VHH-Alexa Fluor 647 fusions performed less well because the signal intensity of sortase-labeled VHHs is limited to a single fluorophore.
From the set of GFP-tagged VHHs, only VHH-8-GFP showed occasional co-localization with the nuclear envelope in untransfected cells. Endogenous HypE levels were low, and therefore, the more highly expressed VHH-GFP fusions remained free and dispersed throughout the cell, masking those few VHH-GFP molecules that interacted with HypE at the nuclear envelope.
The VHHs identified in this study represent the first HypE-specific inhibitors or enhancers with demonstrated in vitro activity. A recent study characterized small molecule inhibitors of VopS; however, these molecules failed to rescue VopS-induced cell rounding and subsequent cell death in tissue culture assays (22). There are other examples of inhibiting or neutralizing VHHs (35), but only a few VHHs have shown stimulatory activity, such as a pair of VHHs capable of increasing the zymogen activity of the carboxypeptidase TAFI (38). The HypE-specific VHHs remain functional when expressed in HeLa cells, as indicated by co-localization with HypE. As we still do not understand how endogenous HypE regulates cellular processes and the consequences of its inhibition, we cannot at present provide functional data that confirm the inhibitory or stimulatory activity of a VHH when expressed inside cells. Because the HypE-binding sites of the individual VHHs remain to be mapped, we can only speculate how these VHHs inhibit or stimulate AMPylation. Enzyme inhibition most likely arises from steric interference with or occlusion of the active site, avoiding proper nucleotide turnover and target binding. Stimulation of auto-AMPylation and AMP transfer may result from VHHs stabilizing an enzyme confirmation that resembles the mutant E234G state, with presumably more direct access to the active site. Recent structural work on HypE and HypE E234G suggested that HypE dimerizes in a Fic domain-dependent fashion. Mutation of Leu-258 abolishes HypE dimerization and greatly reduces auto-AMPylation (39). Other mutations in the dimerization interface inhibit HypE auto-AMPylation without fully blocking dimerization. Thus, our inhibitory HypE VHHs might occlude the dimerization interface and phenocopy the Leu-258 mutation. Solving the structure of a VHH-HypE co-crystal is likely to clarify the mechanism of action of the individual VHHs. The ability of VHHs to serve as crystallization chaperones will be a useful attribute in this regard (33).
We found that human HypE localizes to the nuclear envelope, in contrast to the ER lumenal localization reported for the Drosophila Fic protein dFic. Furthermore, recent work by Sanyal et al. (40) suggested that HypE may not only localize to the ER, but may also be present in the nuclear envelope, as evident from immunofluorescence images. The inner and outer nuclear membranes are contiguous with the endoplasmic reticulum, and certain proteins have been shown to freely diffuse between the two cellular compartments, as HypE may (41). We found that especially in cells overexpressing HypE, the protein not only localizes to the nuclear membrane, but also is seen in other compartments in the perinuclear space that are likely to represent the ER, possibly as a consequence of overexpression.
The mechanism that underlies the aberrant nuclear morphology observed in response to overexpression of HypE E234G remains to be identified. Neither HypE nor HypE H363A overexpression affects nuclear morphology. Rather, aberrant nuclear shape appears to be linked to the AMPylation activity of HypE. Similar nuclear blebbing is seen in fibroblasts derived from major lamin A-only knock-out mice (42). However, these aberrations are not linked to any obvious decrease in fitness. Thus, we assume that the changes in nuclear morphology do not account for increased cell death seen in cells that overexpress HypE E234G.
The localization of endogenous HypE suggests a possible role for this enzyme in genome-associated processes. Interestingly, most bacterial Fic proteins contain a DNA-binding domain such as a helix-turn-helix motif, suggesting a possible link between Fic protein activity and DNA biology.
Histone modifications by methylases, acetylases, and other enzymes regulate gene expression in eukaryotes. We identified histones H2A, H2B, H3.1, H3.2, H3.3, and H4, but not H1, as in vitro substrates for the human Fic protein HypE. The N-terminal histone tail extensions protrude from the nucleosome and are targeted by methylases and additional modifying enzymes. Histone H1 is not part of the nucleosome; instead, it stabilizes chromatin structure by binding DNA between individual nucleosome core particles. Histone H1 is the most variable histone across species, and N-terminal modifications are less prevalent. Histone modifications are not limited to small covalent additions such as methylation or phosphorylation: all core histones (H2–H4) as well as the linker histone H1 (43) can undergo poly(ADP-ribosylation). Histone poly(ADP-ribosylation) is most pronounced following genotoxic stress inducing DNA strand breaks (44). However, detection of histone poly(ADP-ribosylation) is complicated by the fact that only a small fraction of histones are modified, as may be the case for AMPylation as well.
Our in vitro AMPylation results demonstrate that histone AMPylation is not restricted to tyrosines, as a tyrosine-free mutant was still readily modified by HypE E234G. The use of [α-33P]ATP ensured that we were monitoring AMPylation, as distinct from phosphorylation. Furthermore, a mutant version of histone H3 containing phenylalanines instead of tyrosines was AMPylated less well than wild-type histone H3. Whether this reduction in AMPylation is indicative of tyrosine AMPylation or rather the consequence of altered folding remains to be tested.
The list of HypE substrates has been extended recently by the addition of BiP, an ER-located HSP70 family member (19, 40). BiP is modified on Ser-365 and Thr-366, suggesting that HypE is capable of AMPylation of serine, tyrosine, and threonine residues. Threonines are far more prevalent in histones and are obvious kinase targets such as Chk1 (45). How histone AMPylation relates to other modifications such as methylation, acetylation, ubiquitylation, and phosphorylation remains to be investigated. Despite considerable efforts, we have not been able to localize the precise site(s) of histone AMPylation by MS. Proteolytic digests of histone H3 with commonly used proteases result in very small peptides difficult to retain, ionize, and detect by MS (46), reflected also by the poor sequence coverage we observed in our MS experiments (data not shown). In addition, MS analysis of intact in vitro AMPylated histone H3 indicated that only a small fraction (<1%) of the input protein was modified, as reported for histone ADP-ribosylation, thus further complicating the detection of AMPylated histone H3-derived peptides by MS (data not shown).
We have described HypE-specific VHHs that inhibit or enhance HypE-promoted auto-AMPylation and target AMPylation. In addition, we have localized HypE to the nuclear envelope and established that histones H2–H4, but not H1, are novel HypE targets. Consequently, HypE might be involved in the regulation of gene expression, DNA repair, or other histone-associated processes in human cells. Whether histone AMPylation may prove to be an epigenetic mark will require more extensive genetic and biochemical analysis.
Acknowledgments
We are grateful to the members of the Ploegh laboratory for stimulating discussions and suggestions. Denise Alwing and Marie-Lyne Tatlock are acknowledged for technical assistance with protein purification. We thank Wendy Salmon (Keck imaging facility) for confocal imaging assistance and the Whitehead FACS facility for help with FACS-based assays. Finally, we acknowledge Christoph Dehio for providing HypE-encoding plasmids.
Footnotes
- ER
- endoplasmic reticulum
- VHH
- heavy chain-only antibody variable domain
- Ni-NTA
- nickel-nitrilotriacetic acid
- CDR
- complementarity-determining region.
REFERENCES
- 1. Brown M. S., Segal A., Stadtman E. R. (1971) Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the PII regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 68, 2949–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Shea J. J., Holland S. M., Staudt L. M. (2013) JAKs and STATs in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 368, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chock P. B., Rhee S. G., Stadtman E. R. (1980) Interconvertible enzyme cascades in cellular regulation. Annu. Rev. Biochem. 49, 813–843 [DOI] [PubMed] [Google Scholar]
- 4. Mattoo S., Durrant E., Chen M. J., Xiao J., Lazar C. S., Manning G., Dixon J. E., Worby C. A. (2011) Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other Fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J. Biol. Chem. 286, 32834–32842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 [DOI] [PubMed] [Google Scholar]
- 6. Palanivelu D. V., Goepfert A., Meury M., Guye P., Dehio C., Schirmer T. (2011) Fic domain-catalyzed adenylylation: insight provided by the structural analysis of the type IV secretion system effector BepA. Protein Sci. 20, 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao J., Worby C. A., Mattoo S., Sankaran B., Dixon J. E. (2010) Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., Dixon J. E. (2009) The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luong P., Kinch L. N., Brautigam C. A., Grishin N. V., Tomchick D. R., Orth K. (2010) Kinetic and structural insights into the mechanism of AMPylation by VopS Fic domain. J. Biol. Chem. 285, 20155–20163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engel P., Goepfert A., Stanger F. V., Harms A., Schmidt A., Schirmer T., Dehio C. (2012) Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482, 107–110 [DOI] [PubMed] [Google Scholar]
- 11. Mukherjee S., Liu X., Arasaki K., McDonough J., Galán J. E., Roy C. R. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campanacci V., Mukherjee S., Roy C. R., Cherfils J. (2013) Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J. 32, 1469–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J. M. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118 [DOI] [PubMed] [Google Scholar]
- 14. Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N. A., Loris R., Zenkin N. (2013) The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cruz J. W., Rothenbacher F. P., Maehigashi T., Lane W. S., Dunham C. M., Woychik N. A. (2014) Doc toxin is a kinase that inactivates elongation factor Tu. J. Biol. Chem. 289, 7788–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woolery A. R., Luong P., Broberg C. A., Orth K. (2010) AMPylation: something old is new again. Front. Microbiol. 1, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Itzen A., Blankenfeldt W., Goody R. S. (2011) Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem. Sci. 36, 221–228 [DOI] [PubMed] [Google Scholar]
- 18. Rahman M., Ham H., Liu X., Sugiura Y., Orth K., Krämer H. (2012) Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 15, 871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ham H., Woolery A. R., Tracy C., Stenesen D., Krämer H., Orth K. (2014) Unfolded protein response-regulated Drosophila Fic (dFic) reversibly AMPylates BiP during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059–36069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K. S., Knoblich M., Haenig C., Herbst M., Suopanki J., Scherzinger E., Abraham C., Bauer B., Hasenbank R., Fritzsche A., Ludewig A. H., Büssow K., Buessow K., Coleman S. H., Gutekunst C. A., Landwehrmeyer B. G., Lehrach H., Wanker E. E. (2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15, 853–865 [DOI] [PubMed] [Google Scholar]
- 21. Hao Y. H., Chuang T., Ball H. L., Luong P., Li Y., Flores-Saaib R. D., Orth K. (2011) Characterization of a rabbit polyclonal antibody against threonine-AMPylation. J. Biotechnol. 151, 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewallen D. M., Sreelatha A., Dharmarajan V., Madoux F., Chase P., Griffin P. R., Orth K., Hodder P., Thompson P. R. (2014) Inhibiting AMPylation: a novel screen to identify the first small molecule inhibitors of protein AMPylation. ACS Chem. Biol. 9, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leavy O. (2010) Therapeutic antibodies: past, present and future. Nat. Rev. Immunol. 10, 297. [DOI] [PubMed] [Google Scholar]
- 24. Dougan S. K., Ashour J., Karssemeijer R. A., Popp M. W., Avalos A. M., Barisa M., Altenburg A. F., Ingram J. R., Cragnolini J. J., Guo C., Alt F. W., Jaenisch R., Ploegh H. L. (2013) Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature 503, 406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harmsen M. M., van Solt C. B., van Zijderveld-van Bemmel A. M., Niewold T. A., van Zijderveld F. G. (2006) Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl. Microbiol. Biotechnol. 72, 544–551 [DOI] [PubMed] [Google Scholar]
- 26. Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C. S., Cardoso M. C., Leonhardt H., Hopfner K. P., Rothbauer U. (2010) Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 [DOI] [PubMed] [Google Scholar]
- 27. Caussinus E., Kanca O., Affolter M. (2012) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 [DOI] [PubMed] [Google Scholar]
- 28. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wuethrich I., Peeters J. G., Blom A. E., Theile C. S., Li Z., Spooner E., Ploegh H. L., Guimaraes C. P. (2014) Site-specific chemoenzymatic labeling of aerolysin enables the identification of new aerolysin receptors. PLoS ONE 9, e109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theile C. S., Witte M. D., Blom A. E., Kundrat L., Ploegh H. L., Guimaraes C. P. (2013) Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1800–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Swee L. K., Guimaraes C. P., Sehrawat S., Spooner E., Barrasa M. I., Ploegh H. L. (2013) Sortase-mediated modification of αDEC205 affords optimization of antigen presentation and immunization against a set of viral epitopes. Proc. Natl. Acad. Sci. U.S.A. 110, 1428–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A., Schiff R., Rosen J. M., Westbrook T. F., Elledge S. J. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sosa B. A., Demircioglu F. E., Chen J. Z., Ingram J., Ploegh H. L., Schwartz T. U. (2014) How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife 3, e03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mukherjee J., Tremblay J. M., Leysath C. E., Ofori K., Baldwin K., Feng X., Bedenice D., Webb R. P., Wright P. M., Smith L. A., Tzipori S., Shoemaker C. B. (2012) A novel strategy for development of recombinant antitoxin therapeutics tested in a mouse botulism model. PLoS ONE 7, e29941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sundberg E. J., Mariuzza R. A. (2002) Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 61, 119–160 [DOI] [PubMed] [Google Scholar]
- 38. Mishra N., Buelens K., Theyskens S., Compernolle G., Gils A., Declerck P. J. (2012) Increased zymogen activity of thrombin-activatable fibrinolysis inhibitor prolongs clot lysis. J. Thromb. Haemost. 10, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 39. Bunney T. D., Cole A. R., Broncel M., Esposito D., Tate E. W., Katan M. (2014) Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 10.1016/j.str.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanyal A., Chen A. J., Nakayasu E. S., Lazar C. S., Zbornik E. A., Worby C. A., Koller A., Mattoo S. (2015) A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J. Biol. Chem. 10.1074/jbc.M114.618348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. González J. M., Andrés V. (2011) Synthesis, transport and incorporation into the nuclear envelope of A-type lamins and inner nuclear membrane proteins. Biochem. Soc. Trans. 39, 1758–1763 [DOI] [PubMed] [Google Scholar]
- 42. Coffinier C., Jung H. J., Li Z., Nobumori C., Yun U. J., Farber E. A., Davies B. S., Weinstein M. M., Yang S. H., Lammerding J., Farahani J. N., Bentolila L. A., Fong L. G., Young S. G. (2010) Direct synthesis of lamin A, bypassing prelamin a processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J. Biol. Chem. 285, 20818–20826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burzio L. O., Riquelme P. T., Koide S. S. (1979) ADP-ribosylation of rat liver nucleosomal core histones. J. Biol. Chem. 254, 3029–3037 [PubMed] [Google Scholar]
- 44. Boulikas T. (1989) DNA strand breaks alter histone ADP-ribosylation. Proc. Natl. Acad. Sci. U.S.A. 86, 3499–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shimada M., Niida H., Zineldeen D. H., Tagami H., Tanaka M., Saito H., Nakanishi M. (2008) Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 [DOI] [PubMed] [Google Scholar]
- 46. Lin S., Garcia B. A. (2012) Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 512, 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]